Abstract
Growing evidence has revealed that the initiation of various malignancies is closely associated with alternative splicing (AS) events in certain key oncogenes. However, in diffuse large B-cell lymphoma (DLBCL), there is still a great deal to learn about AS variants. In this study, 33,724 AS variant profiles were obtained from 16,278 genes in 48 DLBCL cases. A total of 10 AS variants were identified as overall survival (OS)- related events via multivariate Cox regression analysis. Notably, alternative donor (AD) sites in AS events in the low-risk group showed a significantly better outcome in DLBCL patients than in the high-risk group (P=0.0002). The area under the curve (AUC) of the receiver-operator characteristic curve (ROC) for ADs in DLBCL was 0.746. Furthermore, 66 related splicing factors were obtained to investigate their potential correlations with AS events. Factors SF1, HNRNPC, HNRNPD, and HNRNPH3 were significantly involved in different OS-related AS variants. Collectively, we constructed valuable prognostic predictors for DLBCL patients and mapped novel splicing networks for further investigation of the underlying mechanisms related to AS variants in DLBCLs.
Keywords: Alternative splicing, diffuse large B-cell lymphoma, prognosis, correlation network
Introduction
Alternative splicing (AS) is a spatiotemporal and pivotal post-transcriptional process. AS determinatively affects over 90% of multi-exon human genes and is important to such mechanisms as cell functions, cell differentiation, organogenesis [1-3]. A single gene can generate diverse mRNA isoforms through exon skipping, the use of alternative splicing sites, and intron retention [4-6]. The AS process can also enhance the diversity of both proteomes and transcriptomes [7,8].
In 2018, the estimated new cases and deaths from non-Hodgkin lymphoma ranked 7th and 9th, respectively, in both sexes [9]. As the most common subtype, diffuse large B-cell lymphoma (DLBCL) accounts for 30%-40% of non-Hodgkin lymphomas in adults [10-14]. Currently, prognoses for DLBCL patients can be slightly improved through the use of rituximab and a regimen consisting of a combination of cyclophosphamide, doxorubicin (hydroxydaunomycin), vincristine (Oncovin®), and prednisolone (a steroid) (CHOP) [15,16]. However, approximately 30% of advanced stage DLBCL patients are still refractory to standard chemotherapy. Almost 50% of relapsed cases fail to respond to high-dose chemotherapy and a great majority of relapsed patients die of lymphoma [17-19]. Thus, to improve prognoses, there is an urgent need for novel treatments that go beyond simple chemotherapy.
The International Prognostic Index developed a prognostic model of DLBCL based on several clinical parameters without molecular insights. However, in DLBCL patients, a growing body of evidence has identified various biological molecular markers and gene signatures that are correlated with prognostic significance. Studies have demonstrated that ICT1, Mad2, and decreased LMR could be unfavorable prognosis biomarkers for DLBCL [20-22]. Inversely, NLR and ERβ1 have been recommended as valuable prognostic markers for DLBCL [20,23].
Meanwhile, the underlying mechanisms of perturbed AS could influence DLBCL. Previous studies reported a smaller mRNA isoform, FOXP1, that was found to be primarily expressed in DLBCL and that represented an adverse factor in B-cell lymphomas [24]. Genes with AS PTPROt could significantly increase G0/G1 arrest via sense cloning of itself [25]. In addition, Leivonen et al. revealed that AS events in DLBCL have a direct impact on prognosis and could help in the discrimination of DLBCL subtypes [26].
In the current study, we systematically evaluated survival profiles of AS in DLBCL. DLBCL splicing networks were mapped for further research on potential mechanisms. All AS events and clinicopathological features of DLBCL were obtained from The Cancer Genome Atlas (TCGA).
Materials and methods
Alternative splicing data
RNA-Seq data for the DLBCL cohort were obtained from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). Splicing events for each DLBCL sample from TCGA were analyzed utilizing SpliceSeq, a Java application that visualizes and quantitates RNA-Seq reads and transcriptional splicing graphs [27]. The percent spliced in (PSI) value was applied to quantify various AS events from zero to one.
UpSet view and network construction
To analyze and visualize the intersected AS events, an UpSet plot was created using ImageGP (http://www.ehbio.com/ImageGP/). An interaction network of survival-related AS genes was constructed using Cytoscape (v 3.5.1). Meanwhile, potential pathway annotations were obtained from the Database for Annotation, Visualization and Integrated Discovery (DAVID, v 6.8) and the enrichment plot was mapped using ImageGP (http://www.ehbio.com/ImageGP/).
Survival analysis
The clinical parameters of 48 DLBCL patients were obtained from TCGA database. A total of 44 DLBCL patients with an overall survival (OS) time of more than 90 days were finally included in the study. To assess the association between AS events and OS, univariate Cox regression was carried out. Multivariate Cox regression was performed to select the independent prognostic factors (SPSS, v 22.0). The survival ROC package in R (v 3.2.4) was applied to estimate the significance of the prognostic predictors in DLBCL via generating receiver-operator characteristic curves (ROC) with censored data.
Correlations between splicing factor-related events and prognosis-related AS events
We obtained splicing factors from SpliceAid 2 (http://193.206.120.249/splicing_tissue.html). Expression data (level 3) for the splicing factors associated with AS were downloaded from TCGA. Correlations between splicing factor-related events and prognosis-related AS events were evaluated using Spearman’s rank-order correlation. The correlation diagrams and survival curves were generated with GraphPad (v 5.01). P-values of ≤ 0.05 were considered significant.
Results
Number of AS events in the DLBCL cohort from TCGA
In the present study, out of 48 DLBCL cases, 7 AS events were classified, including alternate acceptor (AA) sites, alternate donor (AD) sites, alternate promoter (AP), alternate terminator (AT), exon skip (ES), mutually exclusive exon (ME) and retained intron (RI). As shown in Figure 1A, a total of 33,724 AS events from 16,098 genes were curated. Out of 5,086 genes, the maximum number of ES events was 12,101, and out of 150 genes, the minimum number of ME events was 156. Furthermore, via univariate Cox regression analysis, 1,262 AS events in 971 genes were considered significantly associated with OS in cases of DLBCL (P<0.05) (Figure 1B). We calculated 116 AAs in 110 genes, 84 ADs in 81 genes, 225 APs in 136 genes, 288 ATs in 165 genes, 475 ESs in 411 genes, 9 MEs in 9 genes, and 65 RIs in 59 genes. The intersecting sets in various AS events were performed by UpSet plot. As shown in Figure 1C, most genes that related to OS were collected from ES events. In addition, one single survival-related gene could appear in up to three types of AS events simultaneously. For example, gene SF1 significantly correlated to OS in AP, ES, and RI.
Figure 1.
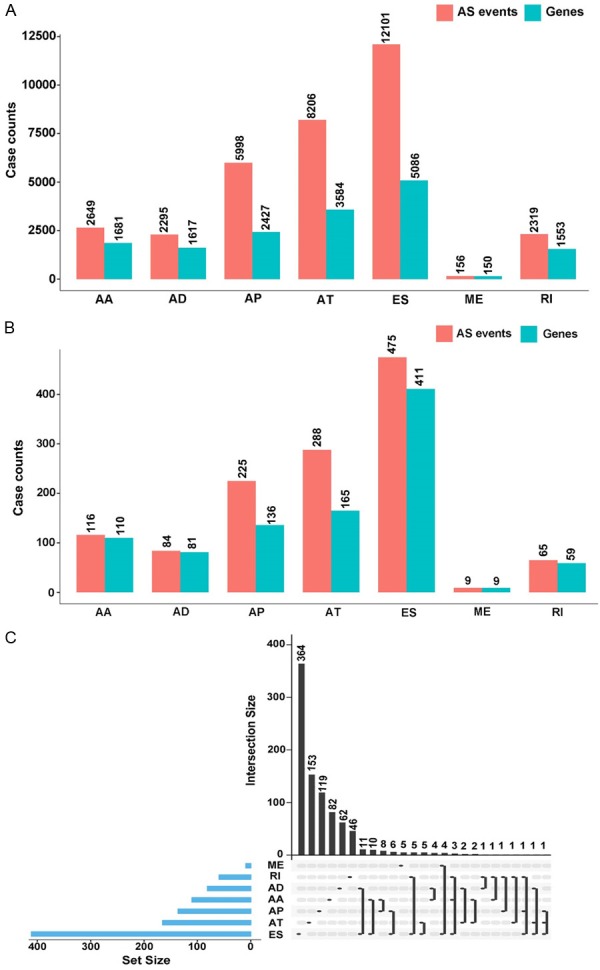
Number of AS variants and genes involved in DLBCL. A. A total of 33,724 AS events from 16,278 genes in AA, AD, AP, AT, ES, ME, and RI. B. 1,262 AS events from 971 OS-related genes in AA, AD, AP, AT, ES, ME, and RI. C. UpSet plot of different types of OS-related AS variants.
Underlying OS-related gene pathways
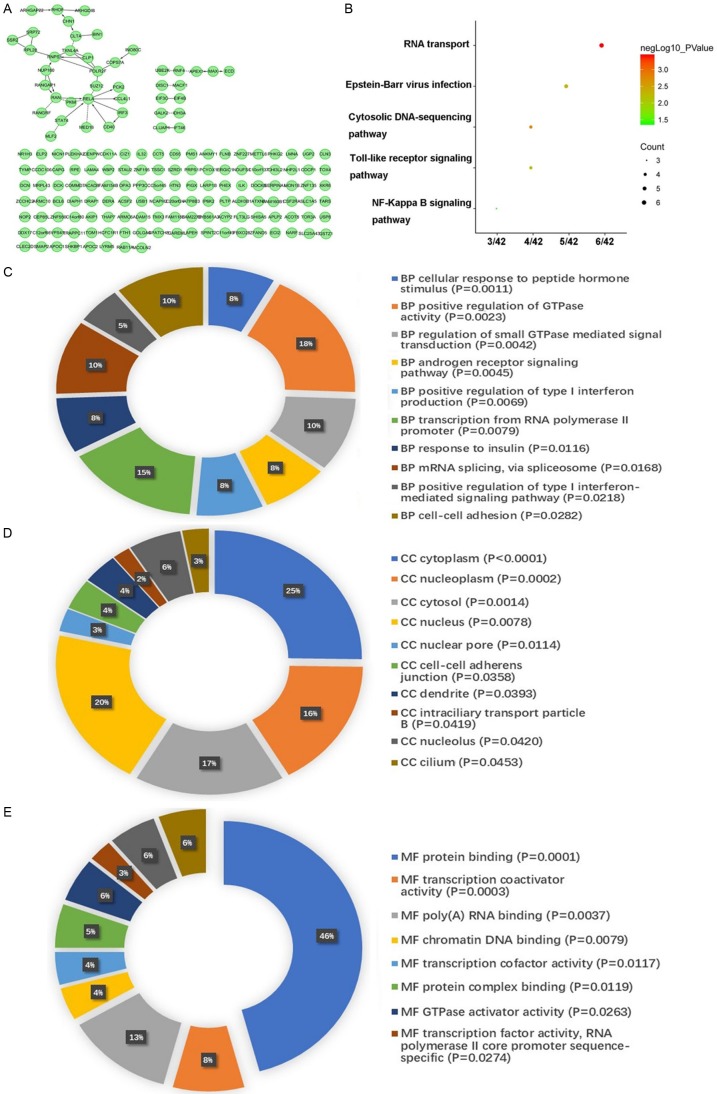
Two hundred and four (204) genes that significantly relate to OS were certified utilizing univariate Cox regression (P<0.01). As shown in Figure 2A, 42 of these selected genes interacted with different genes. For additional study, the Kyoto Encyclopedia of Genes and Genomes (KEGG) was used and gene ontology (GO) enrichment analysis was performed (Figure 2B-E). KEGG pathways indicated that 6 genes including EIF3C, EIF4B, NUP160, RAN, RANGAP1, and RNPS1 co-functioned in the most significant pathway, “RNA transport”. Meanwhile, the GO enrichment annotations showed that these 42 interacting genes were enriched in various crucial pathways, such as mRNA splicing, poly (A) RNA binding, and protein complex binding.
Figure 2.
Correlation network and enrichment analysis of OS-correlated genes. A. Interacting network of OS-related genes. B. KEGG enrichment analysis of OS-related genes. C. Biological process enrichment analysis of OS-related genes. D. Cellular component enrichment analysis of OS-related genes. E. Molecular function enrichment analysis of OS-related genes.
Potential prognostic roles in DLBCL
In order to achieve some reliable prognostic predictors in DLBCL patients, we chose the five most significant univariate AS events among all seven types. Through the use of multivariate Cox regression analysis,10 AS events, including EIF3C and TYMP in AAs, TOX4 and USB1 in ADs, NHP2L1 and MLF2 in APs, STAT4 and POLR2F in ATs, and COX16 and SS18 in MEs (Table 1), were finally selected from AS events in 35 candidates. Unfortunately, no significant results were calculated in ES or RI events. Meanwhile, a prognostic index (PI) model was built to estimate the outcomes for DLBCL patients. Median PI values divided the patients into high and low-risk groups. As shown in Figure 3A-E, there was a trend toward longer OS for five different AS events in the low-risk group. The median OS days for AAs in the low-risk group were 1,972, and in the high-risk group, it was 651. In the AD low-risk and high-risk groups, the median OS was 4,989 and 352 days, respectively. For ATs, 1,252 days and 391 days represented the median OS in the low-risk and high-risk groups, respectively. Meanwhile, the median OS for both APs and MEs were calculated as 3,553 days and 391 days in the low-risk and high-risk groups, respectively. Moreover, ADs of AS events in the low-risk group indicated a significantly better outcome than in the high-risk group among DLBCL patients (P=0.0002). A combination of 10 events also verified considerable power in predicting good prognostic conditions in the low-risk group (P=0.002) (Figure 3F). The median survival days for the low-risk and high-risk groups were 6,425 and 595 days, respectively. Furthermore, ROC curves were used to compare the efficiencies of these 10 events. Areas under the curve (AUCs) of AAs, ADs, APs, ATs, MEs, and all five types of AS events were estimated (Figure 3G-L); APs showed high prognosis predicting efficiency with AUCs over 0.9.
Table 1.
The ten AS events in the final prognostic model
| Splice type | Gene symbol | AS ID | Exons | From exon | To exon | Hazard ratio | P-value |
|---|---|---|---|---|---|---|---|
| AA | EIF3C | 35828 | 5.1 | 4 | 5.2 | 1.08E-13 | 0.0014 |
| TYMP | 62854 | 2.2 | 1 | 2.3 | 1.2554 | 0.0031 | |
| AD | TOX4 | 26588 | 3.2 | 3.1 | 5.1 | 15.1828 | 0.0044 |
| USB1 | 36628 | 3.2 | 3.1 | 4 | 0.0093 | 0.0043 | |
| AP | NHP2L1 | 62447 | 1 | Null | Null | 7.2020 | 0.0213 |
| MLF2 | 19962 | 2 | Null | Null | 0.5920 | 0.0016 | |
| AT | STAT4 | 56602 | 26 | Null | Null | 0.7468 | 0.0032 |
| POLR2F | 62182 | 11 | Null | Null | 15.5894 | 0.0004 | |
| ME | COX16 | 28177 | 2|3 | 1 | 4 | 1.51E-09 | 0.0410 |
| SS18 | 94799 | 5|6 | 3 | 9 | 1.0407 | 0.0235 |
Figure 3.
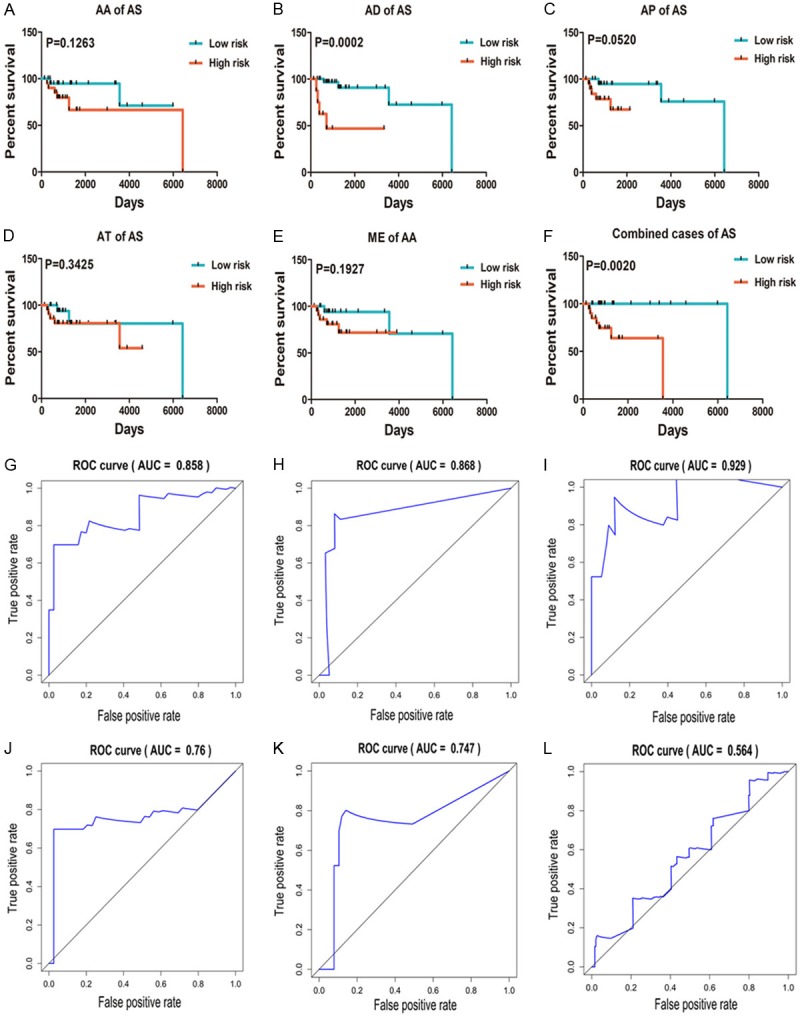
Kaplan-Meier and ROC curves for prognostic predictors of DLBCLs. A-E. Kaplan-Meier curves for AA, AD, AP, AT, and ME for DLBCL. The red line represents the high-risk group, and the green line represents the low-risk group. F. Kaplan-Meier curves for all five types of AS variants for DLBCL. The red line represents the high-risk group, and the green line represents the low-risk group. G-K. ROC curves with AUC for AA, AD, AP, AT, and ME in DLBCL. L. ROC curves for all five types of AS events.
Network of splicing factor-related events
Splicing factors control different genes by affecting the transcriptome. Meanwhile, mRNA stability is regulated by interactions between trans-splicing and basal splicing factors [28]. In this study, we obtained 66 splicing factors, and 65 of these factors functioned in different types of the 193 AS events. Eventually, eight splicing factor related AS events were selected after univariate regression analysis. For deeper insight into correlations between splicing factor-related AS events and significant univariate AS events, PSI values of 71 significant univariate AS events (P<0.005) and eight splicing factor-related AS events were calculated. As shown in Figure 4A, eight splicing factor-related AS events (blue dots) positively correlated to 63 univariate events (red dots) and negatively correlated to 65 univariate events (green dots) (Table 2). Furthermore, survival analysis of four splicing factors involved in eight AS events were estimated (Figure 4B-E); however, no significant results were obtained.
Figure 4.
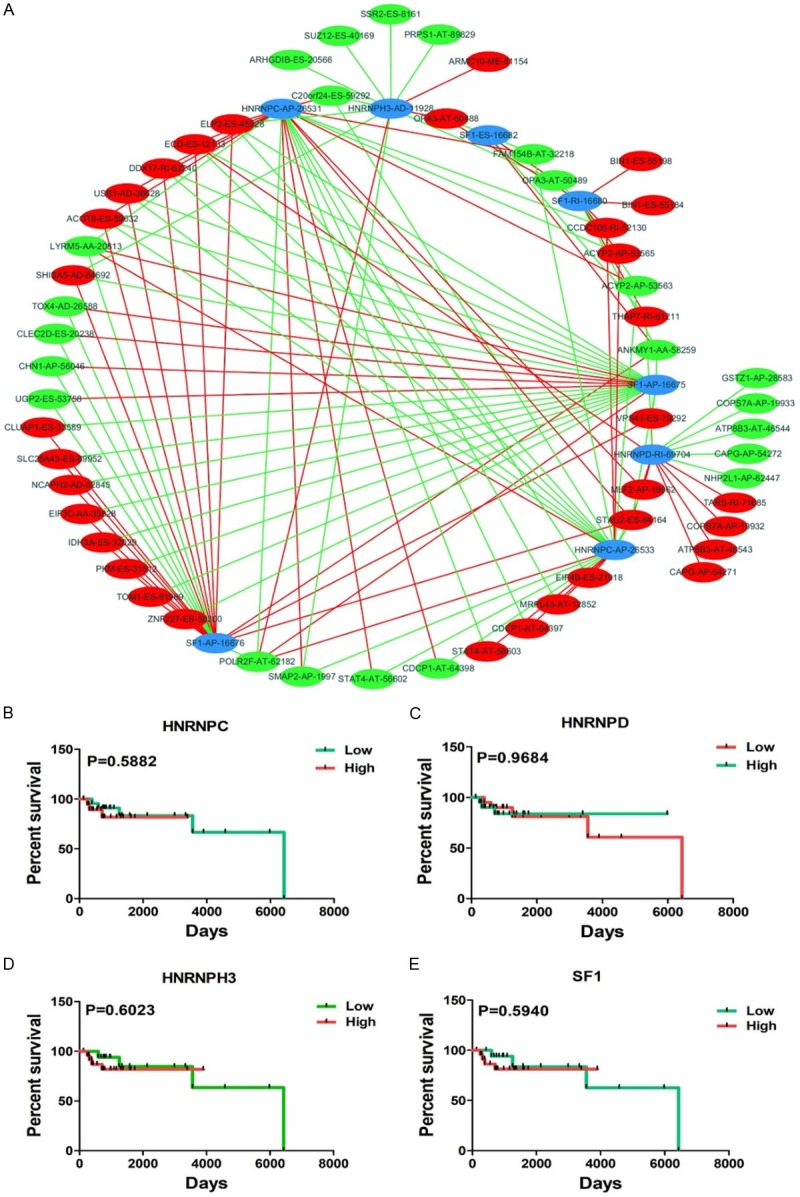
Correlation splicing network and Kaplan-Meier curves for OS splicing factors. A. Splicing network of OS-related AS variants. The blue dots represent the AS events from OS-related splicing factors. The red/green dots represent the AS events that are positively (red lines) or negatively (green lines) associated with AS events from splicing factors. B-E. Kaplan-Meier curves for four splicing factors in DLBCL. The red line represents the high level group and the green line represents the low level group.
Table 2.
Involvement of four splicing factors in various AS events
| Splicing factor-related AS events | Negatively correlated events | Correlation value | P-value | Positively correlated events | Correlation value | P-value |
|---|---|---|---|---|---|---|
| HNRNPH3-AD-11928 | C20orf24-ES-59292 | -0.513 | 0.0004 | OPA3-AT-50488 | 0.344 | 0.0222 |
| ARHGDIB-ES-20566 | -0.454 | 0.0020 | ARMC10-ME-81154 | 0.344 | 0.0223 | |
| ELP2-ES-45228 | -0.403 | 0.0110 | POLR2F-AT-62182 | 0.302 | 0.0463 | |
| SHISA5-AD-64692 | -0.375 | 0.0170 | ||||
| OPA3-AT-50489 | -0.344 | 0.0222 | ||||
| SMAP2-AP-1997 | -0.339 | 0.0245 | ||||
| SUZ12-ES-40169 | -0.331 | 0.0300 | ||||
| SSR2-ES-8161 | -0.318 | 0.0356 | ||||
| PRPS1-AT-89829 | -0.297 | 0.0499 | ||||
| HNRNPC-AP-26531 | POLR2F-AT-62182 | -0.452 | 0.0021 | SMAP2-AP-1997 | 0.519 | 0.0003 |
| EIF4B-ES-21918 | -0.395 | 0.0080 | MLF2-AP-19962 | 0.488 | 0.0008 | |
| CCDC106-RI-52130 | -0.395 | 0.0087 | ACOT8-ES-59632 | 0.429 | 0.0041 | |
| MRPL43-AT-12852 | -0.363 | 0.0153 | ELP2-ES-45228 | 0.416 | 0.0084 | |
| CDCP1-AT-64397 | -0.358 | 0.0169 | CDCP1-AT-64398 | 0.358 | 0.0169 | |
| ACYP2-AP-53565 | -0.336 | 0.0259 | USB1-AD-36628 | 0.357 | 0.0187 | |
| STAT4-AT-56603 | -0.324 | 0.0338 | ACYP2-AP-53563 | 0.346 | 0.0216 | |
| LYRM5-AA-20813 | -0.363 | 0.0351 | FAM154B-AT-32218 | 0.335 | 0.0279 | |
| STAT4-AT-56602 | 0.324 | 0.0338 | ||||
| DDX17-RI-62240 | 0.315 | 0.0375 | ||||
| C20orf24-ES-59292 | 0.309 | 0.0416 | ||||
| ECD-ES-12133 | 0.316 | 0.0442 | ||||
| STAU2-ES-84164 | 0.316 | 0.0471 | ||||
| HNRNPC-AP-26533 | SMAP2-AP-1997 | -0.519 | 0.0003 | POLR2F-AT-62182 | 0.452 | 0.0021 |
| MLF2-AP-19962 | -0.488 | 0.0008 | EIF4B-ES-21918 | 0.395 | 0.0080 | |
| ACOT8-ES-59632 | -0.429 | 0.0041 | CCDC106-RI-52130 | 0.395 | 0.0087 | |
| ELP2-ES-45228 | -0.416 | 0.0084 | MRPL43-AT-12852 | 0.363 | 0.0153 | |
| CDCP1-AT-64398 | -0.358 | 0.0169 | CDCP1-AT-64397 | 0.358 | 0.0169 | |
| USB1-AD-36628 | -0.357 | 0.0187 | ACYP2-AP-53565 | 0.336 | 0.0259 | |
| ACYP2-AP-53563 | -0.346 | 0.0216 | STAT4-AT-56603 | 0.324 | 0.0338 | |
| FAM154B-AT-32218 | -0.335 | 0.0279 | LYRM5-AA-20813 | 0.363 | 0.0351 | |
| STAT4-AT-56602 | -0.324 | 0.0338 | ||||
| DDX17-RI-62240 | -0.315 | 0.0375 | ||||
| C20orf24-ES-59292 | -0.309 | 0.0416 | ||||
| ECD-ES-12133 | -0.316 | 0.0442 | ||||
| STAU2-ES-84164 | -0.316 | 0.0471 | ||||
| SF1-AP-16675 | ELP2-ES-45228 | -0.546 | 0.0003 | UGP2-ES-53758 | 0.416 | 0.0062 |
| ZNF227-ES-50300 | -0.508 | 0.0013 | CHN1-AP-56046 | 0.384 | 0.0131 | |
| USB1-AD-36628 | -0.442 | 0.0030 | POLR2F-AT-62182 | 0.338 | 0.0268 | |
| TOM1-ES-61969 | -0.431 | 0.0039 | CLEC2D-ES-20238 | 0.337 | 0.0270 | |
| PKM-ES-31512 | -0.429 | 0.0041 | LYRM5-AA-20813 | 0.349 | 0.0430 | |
| ECD-ES-12133 | -0.435 | 0.0045 | TOX4-AD-26588 | 0.337 | 0.0445 | |
| ANKMY1-AA-58259 | -0.433 | 0.0047 | ||||
| IDH3A-ES-32029 | -0.374 | 0.0135 | ||||
| EIF3C-AA-35828 | -0.372 | 0.0141 | ||||
| NCAPH2-AD-62845 | -0.375 | 0.0144 | ||||
| SLC25A43-ES-89952 | -0.395 | 0.0154 | ||||
| DDX17-RI-62240 | -0.342 | 0.0247 | ||||
| SHISA5-AD-64692 | -0.338 | 0.0329 | ||||
| ACOT8-ES-59632 | -0.321 | 0.0381 | ||||
| CLUAP1-ES-33589 | -0.318 | 0.0404 | ||||
| VPS41-ES-79292 | -0.338 | 0.0469 | ||||
| STAU2-ES-84164 | -0.314 | 0.0483 | ||||
| SF1-AP-16676 | UGP2-ES-53758 | -0.416 | 0.0062 | ELP2-ES-45228 | 0.546 | 0.0003 |
| CHN1-AP-56046 | -0.384 | 0.0131 | ZNF227-ES-50300 | 0.508 | 0.0013 | |
| POLR2F-AT-62182 | -0.338 | 0.0268 | USB1-AD-36628 | 0.442 | 0.0030 | |
| CLEC2D-ES-20238 | -0.337 | 0.0270 | TOM1-ES-61969 | 0.431 | 0.0039 | |
| LYRM5-AA-20813 | -0.349 | 0.0430 | PKM-ES-31512 | 0.429 | 0.0041 | |
| TOX4-AD-26588 | -0.337 | 0.0445 | ECD-ES-12133 | 0.435 | 0.0045 | |
| ANKMY1-AA-58259 | 0.433 | 0.0047 | ||||
| IDH3A-ES-32029 | 0.374 | 0.0135 | ||||
| EIF3C-AA-35828 | 0.372 | 0.0141 | ||||
| NCAPH2-AD-62845 | 0.375 | 0.0144 | ||||
| SLC25A43-ES-89952 | 0.395 | 0.0154 | ||||
| DDX17-RI-62240 | 0.342 | 0.0247 | ||||
| SHISA5-AD-64692 | 0.338 | 0.0329 | ||||
| ACOT8-ES-59632 | 0.321 | 0.0381 | ||||
| CLUAP1-ES-33589 | 0.318 | 0.0404 | ||||
| VPS41-ES-79292 | 0.338 | 0.0469 | ||||
| STAU2-ES-84164 | 0.314 | 0.0483 | ||||
| SF1-ES-16682 | OPA3-AT-50488 | -0.454 | 0.0020 | OPA3-AT-50489 | 0.454 | 0.0020 |
| FAM154B-AT-32218 | -0.311 | 0.0424 | ANKMY1-AA-58259 | 0.356 | 0.0225 | |
| THAP7-RI-61211 | -0.299 | 0.0490 | ||||
| HNRNPD-RI-69704 | GSTZ1-AP-28583 | -0.407 | 0.0067 | TARS-RI-71685 | 0.528 | 0.0008 |
| COPS7A-AP-19933 | -0.344 | 0.0240 | COPS7A-AP-19932 | 0.344 | 0.0240 | |
| ATP8B3-AT-46544 | -0.332 | 0.0296 | ATP8B3-AT-46543 | 0.332 | 0.0296 | |
| THAP7-RI-61211 | -0.330 | 0.0306 | MLF2-AP-19962 | 0.323 | 0.0346 | |
| CAPG-AP-54272 | -0.322 | 0.0355 | CAPG-AP-54271 | 0.322 | 0.0355 | |
| NHP2L1-AP-62447 | -0.303 | 0.0480 | STAU2-ES-84164 | 0.325 | 0.0410 | |
| ECD-ES-12133 | 0.319 | 0.0419 | ||||
| VPS41-ES-79292 | 0.34 | 0.0455 | ||||
| SF1-RI-16680 | ANKMY1-AA-58259 | -0.448 | 0.0033 | CCDC106-RI-52130 | 0.442 | 0.0030 |
| ACYP2-AP-53563 | -0.342 | 0.0229 | BIN1-ES-55198 | 0.372 | 0.0129 | |
| OPA3-AT-50489 | -0.330 | 0.0289 | THAP7-RI-61211 | 0.358 | 0.0169 | |
| ACYP2-AP-53565 | 0.348 | 0.0207 | ||||
| BIN1-ES-55184 | 0.366 | 0.0240 | ||||
| OPA3-AT-50488 | 0.33 | 0.0289 |
Discussion
Profound modulatory points in gene expression have significant effects on the development of cancers. As one of the regulatory factors in gene control, AS variants are involved in the reorganization of proteomes and can modulate the levels of a number of oncogenes and tumor inhibitor isoforms [29]. As an identified splicing factor oncoprotein, SF2/ASF amplificates in various cancers and generates the activation of proximal 5’ splicing sites. Up-regulated SF2/ASF also blocks aberrant exon-skipping and manages AS events in the tumor-suppressor BIN1 [30,31]. Meanwhile, accurate FOX2 binding sites have been correlated to alternative exons at silencer or enhancer proteins [32]. Furthermore, the level of splicing factors could be influenced by various cancer pathways; for example, hnRNPA1 and hnRNPA2 are up-regulated by c-Myc [29].
In DLBCL patients, a relatively small number of studies have reported pre-mRNA AS variants to be predictive biomarkers and AS events have also been associated with diagnosis and prognosis. Dysregulated CD44 levels can alter lymph node draining [33]. As an isoform of CD44, CD44H is strongly related to poor survival of DLBCL cohorts, and rituximab could decrease the significance of its prognosis [34]. Meanwhile, four likely splice variants of the STAT3 gene have all been represented in DLBCL, including the ΔS/S and α/β splicing variants [35]. Co-worked terms for S and ΔS could sustain activated B-cell-like DLBCL cell survival and modulate STAT3 targets, such as NFKBIA and NFKBIZ [36]. In addition, p63 directly impact cell apoptosis and differentiation. Isoform TAp63, which is a product of an upstream intronic promoter, has been found to be highly expressed in DLBCL cell lines [37]. Collectively, these results outline the critical role of AS variants in DLBCL. A wide variety of isoforms might be valuable candidates for further prospective research on DLBCL.
In the current study, the functional categories of interacting AS genes were analyzed using KEGG and GO. The KEGG enrichment results revealed that “RNA transport” was the most significant term at the P=0.0004 level. We found several consistent pathways, such as “transcription factor activity, RNA polymerase II core promoter sequence-specific”, “mRNA splicing, via spliceosome”, and “protein complex binding” related to AS events from GO annotations. Hence, in the future, gaining insight into these significant interacting AS-related genes in terms of DLBCL mechanisms will be of interest.
Out of 66 splicing factors, genes SF1, HNRNPC, HNRNPD, and HNRNPH3 were identified as the factors involved in OS correlated AS events. To date, it had been well established that protein SF1 was able to recognize the 3’ splice site by binding the branchpoints of certain introns and repressing transcription. Meanwhile, SF1 played a critical role in the retention of nuclear pre-mRNA and SF1 silencing closely related to AS variants of endogenous transcription [38-40]. In recent years, there has been an increase in the literature on SF1 in various cancers, such as in epithelial ovarian cancer [41] and testicular germ cell tumors [42]. In this study, we discovered that protein SF1 was simultaneously engaged in AP, ES, and RI events in DLBCL. However, several studies have indicated that SF1 was identified in just one type of malignancy and rarely seen in lymphoid malignancies [43,44]. Thus, the role of SF1 requires further investigation; our study offers a novel aspect of SF1 in DLBCL. Furthermore, the remaining factors, HNRNPC, HNRNPD, and HNRNPH3, are members of the heterogeneous nuclear ribonucleoproteins (hnRNPs). AS transcript variants of these three factors were previously revealed. Fawal et al. reported that AUF1/hnRNPD is the partner of anaplastic lymphoma kinase (ALK) [45]. Compared to conventional DLBCL patients, ALK positive DLBCL patients displayed unique clinical features and histological types [46]. Collectively, the aforementioned results imply that these four splicing factors might perform multiple functions and offer an orientation from which we might investigate the underlying mechanisms of AS variants in DLBCL.
Conclusion
Taken together, the results of this study revealed that OS-related AD of AS variants is a potential valuable predictor of DLBCL prognosis. In addition, correlation networks and enrichment analysis were performed to uncover the potential regulatory roles of key splicing factors. Considering this evidence, in DLBCL, oncoproteins spliced by certain AS events or the aberrant transcription of splicing factors would be provided. However, the mechanisms of OS-related AS variants and splicing factors are still not clearly understood. Further investigation needs to be conducted with regard to AS in DLBCL patients.
Acknowledgements
The present study was supported by the National Natural Science Foundation of Guangxi (2017GXNSFAA198107) and the Innovation Project of Graduate Education (YCSW2018103). The authors also wish to thank the public databases, such as The Cancer Genome Atlas, for the use of their data.
Disclosure of conflict of interest
None.
References
- 1.Zhu J, Chen Z, Yong L. Systematic profiling of alternative splicing signature reveals prognostic predictor for ovarian cancer. Gynecol Oncol. 2018;148:368–374. doi: 10.1016/j.ygyno.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Yao Y, Sun L, Zhou J, Miao M, Luo S, Deng G, Li J, Wang J, Tang J. Snail driving alternative splicing of CD44 by ESRP1 enhances invasion and migration in epithelial ovarian cancer. Cell Physiol Biochem. 2017;43:2489–2504. doi: 10.1159/000484458. [DOI] [PubMed] [Google Scholar]
- 3.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu CT, Chiou CY, Chiu HC, Yang UC. Fine-tuning of microRNA-mediated repression of mRNA by splicing-regulated and highly repressive microRNA recognition element. BMC Genomics. 2013;14:438. doi: 10.1186/1471-2164-14-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Q, Gao J, Li Z. Identification of a novel alternative splicing transcript variant of the suppressor of fused: relationship with lymph node metastasis in pancreatic ductal adenocarcinoma. Int J Oncol. 2016;49:2611–2619. doi: 10.3892/ijo.2016.3753. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi QY, Deng G, Chen N, Bai ZS, Yuan JS, Wu GH, Wang YW, Wu SJ. Metformin inhibits development of diabetic retinopathy through inducing alternative splicing of VEGF-A. Am J Transl Res. 2016;8:3947–3954. [PMC free article] [PubMed] [Google Scholar]
- 8.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 9.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 10.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Guo P, Ma D, Lin X, Fang Q, Wang J. Overexpression of heme oxygenase-1 induced by constitutively activated NF-kappaB as a potential therapeutic target for activated B-cell-like diffuse large B-cell lymphoma. Int J Oncol. 2016;49:253–264. doi: 10.3892/ijo.2016.3529. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Z, Li X, Zhu Y, Gu W, Xie X, Jiang J. Prognostic significance of MiRNA in patients with diffuse large B-cell lymphoma: a meta-analysis. Cell Physiol Biochem. 2016;39:1891–1904. doi: 10.1159/000447887. [DOI] [PubMed] [Google Scholar]
- 13.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Li Z. New insights into microRNAs involves in drug resistance in diffuse large B cell lymphoma. Am J Transl Res. 2015;7:2536–2542. [PMC free article] [PubMed] [Google Scholar]
- 15.Sun F, Zhu J, Lu S, Zhen Z, Wang J, Huang J, Ding Z, Zeng M, Sun X. An inflammation-based cumulative prognostic score system in patients with diffuse large B cell lymphoma in rituximab era. BMC Cancer. 2018;18:5. doi: 10.1186/s12885-017-3931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Wu X, Liu X, Yang P, Xu J, Chai Y, Guo Q, Wang Z, Zhang L. Prognostic significance of monocytes and monocytic myeloid-derived suppressor cells in diffuse large B-cell lymphoma treated with R-CHOP. Cell Physiol Biochem. 2016;39:521–530. doi: 10.1159/000445644. [DOI] [PubMed] [Google Scholar]
- 17.Mularek O. Cytometrical investigations of neurocytes nuclei in the brains of neonatal rats exposed to x-ray irradiation during various periods of fetal life. Radiobiol Radiother (Berl) 1968;9:495–499. [PubMed] [Google Scholar]
- 18.Friedberg JW. Relapsed/refractory diffuse large B-cell lymphoma. Hematology Am Soc Hematol Educ Program. 2011;2011:498–505. doi: 10.1182/asheducation-2011.1.498. [DOI] [PubMed] [Google Scholar]
- 19.Cheng C, Wang T, Song Z, Peng L, Gao M, Hermine O, Rousseaux S, Khochbin S, Mi JQ, Wang J. Induction of autophagy and autophagy-dependent apoptosis in diffuse large B-cell lymphoma by a new antimalarial artemisinin derivative, SM1044. Cancer Med. 2018;7:380–396. doi: 10.1002/cam4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie W, Wu M, Fu T, Li X, Wang Z, Hu Y, Zhu L, Zhang G. ICT1 predicts a poor survival and correlated with cell proliferation in diffuse large B-cell lymphoma. Gene. 2017;627:255–262. doi: 10.1016/j.gene.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Gao K, Lei W, Dong L, Xuan Q, Feng M, Wang J, Ye X, Jin T, Zhang Z, Zhang Q. Lymphocyte-to-monocyte ratio is associated with prognosis of diffuse large B-cell lymphoma: correlation with CD163 positive M2 type tumor-associated macrophages, not PD-1 positive tumor-infiltrating lymphocytes. Oncotarget. 2017;8:5414–5425. doi: 10.18632/oncotarget.14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F, Liu S, Zhou Y, Shen H, Zuo X. Mad2 overexpression is associated with high cell proliferation and reduced disease-free survival in primary gastrointestinal diffuse large B-cell lymphoma. Hematology. 2016;21:399–403. doi: 10.1080/10245332.2015.1101970. [DOI] [PubMed] [Google Scholar]
- 23.Hasni MS, Berglund M, Yakimchuk K, Guan J, Linderoth J, Amini RM, Enblad G, Okret S. Estrogen receptor beta1 in diffuse large B-cell lymphoma growth and as a prognostic biomarker. Leuk Lymphoma. 2017;58:418–427. doi: 10.1080/10428194.2016.1193853. [DOI] [PubMed] [Google Scholar]
- 24.Brown PJ, Ashe SL, Leich E, Burek C, Barrans S, Fenton JA, Jack AS, Pulford K, Rosenwald A, Banham AH. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood. 2008;111:2816–2824. doi: 10.1182/blood-2007-09-115113. [DOI] [PubMed] [Google Scholar]
- 25.Aguiar RC, Yakushijin Y, Kharbanda S, Tiwari S, Freeman GJ, Shipp MA. PTPROt: an alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood. 1999;94:2403–2413. [PubMed] [Google Scholar]
- 26.Leivonen SK, Taskinen M, Cervera A, Karjalainen-Lindsberg ML, Delabie J, Holte H, Lehtonen R, Hautaniemi S, Leppa S. Alternative splicing discriminates molecular subtypes and has prognostic impact in diffuse large B-cell lymphoma. Blood Cancer J. 2017;7:e596. doi: 10.1038/bcj.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan MC, Cleland J, Kim R, Wong WC, Weinstein JN. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics. 2012;28:2385–2387. doi: 10.1093/bioinformatics/bts452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta SK, Carmi S, Waldman Ben-Asher H, Tkacz ID, Naboishchikov I, Michaeli S. Basal splicing factors regulate the stability of mature mRNAs in trypanosomes. J Biol Chem. 2013;288:4991–5006. doi: 10.1074/jbc.M112.416578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 32.Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, Lucier JF, Thibault P, Rancourt C, Tremblay K, Prinos P, Chabot B, Elela SA. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16:670–676. doi: 10.1038/nsmb.1608. [DOI] [PubMed] [Google Scholar]
- 33.Salles G, Zain M, Jiang WM, Boussiotis VA, Shipp MA. Alternatively spliced CD44 transcripts in diffuse large-cell lymphomas: characterization and comparison with normal activated B cells and epithelial malignancies. Blood. 1993;82:3539–3547. [PubMed] [Google Scholar]
- 34.Wei X, Xu M, Wei Y, Huang F, Zhao T, Li X, Feng R, Ye BH. The addition of rituximab to CHOP therapy alters the prognostic significance of CD44 expression. J Hematol Oncol. 2014;7:34. doi: 10.1186/1756-8722-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turton KB, Annis DS, Rui L, Esnault S, Mosher DF. Ratios of four STAT3 splice variants in human eosinophils and diffuse large B cell lymphoma cells. PLoS One. 2015;10:e0127243. doi: 10.1371/journal.pone.0127243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng M, Turton KB, Zhu F, Li Y, Grindle KM, Annis DS, Lu L, Drennan AC, Tweardy DJ, Bharadwaj U, Mosher DF, Rui L. A mix of S and DeltaS variants of STAT3 enable survival of activated B-cell-like diffuse large B-cell lymphoma cells in culture. Oncogenesis. 2016;4:e184. doi: 10.1038/oncsis.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sethi I, Romano RA, Gluck C, Smalley K, Vojtesek B, Buck MJ, Sinha S. A global analysis of the complex landscape of isoforms and regulatory networks of p63 in human cells and tissues. BMC Genomics. 2015;16:584. doi: 10.1186/s12864-015-1793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crisci A, Raleff F, Bagdiul I, Raabe M, Urlaub H, Rain JC, Kramer A. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 2015;43:10456–10473. doi: 10.1093/nar/gkv952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corioni M, Antih N, Tanackovic G, Zavolan M, Kramer A. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucleic Acids Res. 2011;39:1868–1879. doi: 10.1093/nar/gkq1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Dong W, Wang J, Cai J, Wang Z. Human omental adipose-derived mesenchymal stem cell-conditioned medium alters the proteomic profile of epithelial ovarian cancer cell lines in vitro. Onco Targets Ther. 2017;10:1655–1663. [Google Scholar]
- 42.Zhu R, Heaney J, Nadeau JH, Ali S, Matin A. Deficiency of splicing factor 1 suppresses the occurrence of testicular germ cell tumors. Cancer Res. 2010;70:7264–7272. doi: 10.1158/0008-5472.CAN-10-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogusz AM. EBV-negative monomorphic B-cell posttransplant lymphoproliferative disorder with marked morphologic pleomorphism and pathogenic mutations in ASXL1, BCOR, CDKN2A, NF1, and TP53. Case Rep Hematol. 2017;2017:5083463. doi: 10.1155/2017/5083463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimoto A, Okada Y, Boroevich KA, Tsunoda T, Taniguchi H, Nakagawa H. Systematic analysis of mutation distribution in three dimensional protein structures identifies cancer driver genes. Sci Rep. 2016;6:26483. doi: 10.1038/srep26483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 46.Salat H, Din NU, Moatter T, Kayani N, Ahmed A. Anaplastic lymphoma kinase protein positive diffuse large B cell lymphoma; a developing world experience. Pathol Res Pract. 2017;213:649–653. doi: 10.1016/j.prp.2017.02.017. [DOI] [PubMed] [Google Scholar]