Abstract
The discovery of histone demethylases has revealed the dynamic nature of the regulation of histone methylation. KDM2B is an important histone lysine demethylase that removes methyl from H3K36me2 and H3K4me3. It participates in many aspects of normal cellular processes such as cell senescence, cell differentiation and stem cell self-renewal. Recent studies also showed that KDM2B was overexpressed in various types of cancers. This review focuses primarily on the current knowledge of KDM2B and its function in cancer development.
Keywords: KDM2B, histone demethylase, epigenetic regulation, cancer
Introduction
Epigenetics is defined as mitotically heritable changes in gene expression without altering primary DNA sequence [1,2]. In general, DNA methylation, histone post-translational modifications, nucleosome positioning, and post-transcriptional gene regulation by non-coding RNAs are the main components of epigenetic regulation [3,4]. Among these epigenetic modifications, histone post-translational modifications largely control chromatin structure, recruit effector proteins, and play essential roles in cancer [5-9]. NH2-terminal histone tails from the nucleosome are the major dynamic and reversible sites for histone modifications, which mainly include methylation, acetylation phosphorylation, ADP-ribosylation, ubiquitination, and biotinylation [10,11].
Histone methylation, the most common histone modification, is highly dynamic in regulating gene transcription [12,13]. Moreover, histone methylation can either activate gene expression or silence gene expression that depends on its methylation site [14,15]. It has been demonstrated that methylation at H3K4, H3K36, and H3K79 was usually associated with gene activation, whereas methylation at H3K9, H3K27, and H4K20 was associated with gene silencing [16]. In general, histone methylation is performed by methyltransferases, thus resulting in mono-, di-, or trimethyl. However, it was thought to be irreversible for a long time until the first histone demethylase, LSD1 (Lysine-specific demethylase-1) was identified and characterized in 2004 [17-19]. Up to now, more than 20 demethylases have been found, and they are categorized into two families: LSD family and JmjC family [20]. Furthermore, the two families oxidatively remove methyl groups from histones with distinct mechanisms. The LSD family is flaying adenine dinucleotide (FAD) dependent monoamine oxidases [20], whereas, the JmjC family is Fe(II) and α-ketoglutarate (α-KG) dependent dioxygenases [21]. The lysine (K)-specific demethylase 2B (KDM2B) is one of the members of JmjC family. In this review, we summarize the structure and biological function of KDM2B, and highlight its key findings in cancer.
Structure of KDM2B
KDM2B, also known as JHDM1B/FBXL10/NDY1, is a conserved and ubiquitously expressed nuclear protein. It targets H3K36me2 and H3K4me3 for demethylation [22,23]. KDM2B contains multiple functional domains: an N-terminal JmjC domain, a CxxC domain, a PHD domain, an F-box domain, and seven leucine-rich repeats. The JmjC domain is necessary for the demethylation of H3K36me2. The CxxC zinc-finger domain is a DNA binding domain, which specifically recognizes CpG islands and recruits polycomb repressive complex 1 (PRC1) to target genes [24]. The PHD domain can act as an E3 ligase or a histone modification reader domain [25]. The F-box domain functions as a linker protein between a target protein and an E3 ubiquitin ligase (Figure 1) [26].
Figure 1.
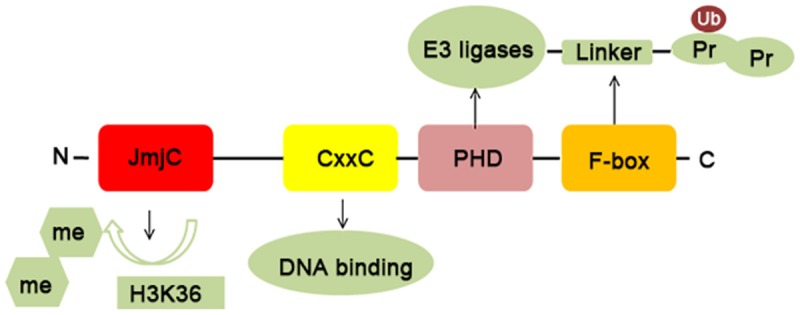
Basic protein structure and biochemical characteristics of KDM2B. KDM2B consists of four distinct domains: a JmjC domain, a CxxC domain, a PHD domain, and an F-box domain. The JmjC domain exerts a function in binding H3K36me2; The CxxC domain is a DNA binding domain and specifically recognizes CpG islands; The PHD domain has E3 ubiquitin ligase activity and acts as a histone modification reader domain; The F-box domain acts as a linker protein between a target protein and an E3 ubiquitin ligase.
Biological function of KDM2B
KDM2B inhibits cell senescence, promotes cell proliferation and migration
KDM2B was initially identified as a novel regulator of lifespan in mouse embryonic fibroblasts (MEFs) [27]. Knockdown of KDM2B in MEFs delayed cell proliferation and induced senescence. ChIP assays further confirmed that KDM2B directly inhibited p15Ink4b and this function was mediated by H3K36me2 demethylation. Subsequent studies demonstrated that KDM2B protected MEFs from senescence by repressing the expression of p16Ink4a and p19Arf [28]. In details, KDM2B overexpression upregulated EZH2 and histone H3K27me3, the latter complex facilitated the binding of Bmi1 and Ink4a/Arf locus, then silenced the expression of p16Ink4a and p19Arf. Besides, KDM2B demethylated the locus-associated H3K36me2 and H3K4me3, which inhibited the binding of RNA Pol II to Ink4a locus. In addition to these findings, other studies demonstrated that KDM2B could repress the expression of EZH2-associated miRNAs, let-7b and miR-101, then further inhibited cell senescence and promoted cell proliferation [29]. Moreover, the deletion of JmjC domain of KDM2B in germline stem cells also led to a reduction of cell proliferation [30]. Correlations analysis of the gene expression from the TCGA database revealed that KDM2B has significant correlations with many cell cycle related genes [31].
Another study indicated that KDM2B promoted cell migration via directly targeting migration-associated genes, such as Areg, Mdk, Lmnb1, Thbs1, Mgp and Cxcl12 [32]. To sum up, these findings provide a novel epigenetic mechanism of KDM2B in cell senescence, proliferation, and migration (Figure 2). As is well-known, cancer cells can evade senescence and activate proliferation signaling. We think KDM2B overexpression might contribute to tumor’s continuous proliferative signaling.
Figure 2.
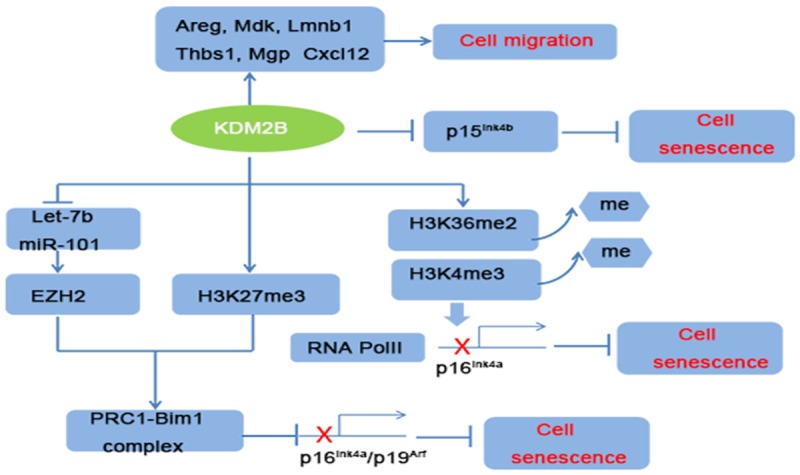
Mechanisms of KDM2B in regulating cell migration and senescence. KDM2B can bind directly to migration-associated genes to promote cell migration. Moreover, KDM2B promotes cell proliferation and inhibited cell senescence by repressing senescence-associated Ink4a/Arf/Ink4b locus and this function was via upregulation EZH2/H3K27me3 and H3K36me2/H3K4me3 demethylation.
KDM2B prevents cell differentiation
It was confirmed that KDM2B could recruit polycomb repressive complex (PRC) to CpG islands (CGIs) [24,33,34]. CGIs were usually enriched in cell development and differentiation genes [35-37]. It was substantiated that KDM2B was highly expressed in mouse embryonic stem cells (mESCs) [38]. Conditional deletion of KDM2B in mESCs induced cell early differentiation. The mechanism was that KDM2B recruited PRC1 to the CGIs of early lineage-specific genes [38]. Interestingly, the role of KDM2B in maintaining the undifferentiated state of mESCs depended on its CxxC domain. Furthermore, KDM2B also inhibited differentiation signaling pathways to maintain the self-renewal of hematopoietic stem cells (HSCs) [39]. In addition, it has been reported that KDM2B overexpression inhibited the expression of the chondrogenic differentiation markers, COL1, COL2 and SOX9 [40]. Consistent with these findings, it was explored and found that KDM2B attracted a noncanonical PRC1 containing RING1B, SKP1 to the key adipogenic genes, such as Cdk1, Uhrf1, Pparg1, Pparg2, and prevented 3T3-L1 cells differentiate into adipose cells [41]. The role of KDM2B in adipogenesis was mediated by the F-box domain. Taken together, these results highlight the importance of KDM2B in cell differentiation and epigenetic regulation field (Figure 3). As is known to all, cancer stem cells are the root causes of tumorigrnicity, metastasis and chemoresistance [42,43]. Other studies have found that KDM2B could form a different PRC1 containing BCoR (Bcl-6-interacting co-repressor), SKP1, PCGF1 (polycomb group RING finger1) and RING1B in cancer cells [44-47]. Thus, KDM2B might be an important molecular mechanism in maintaining cancer stem cell self-renewal.
Figure 3.
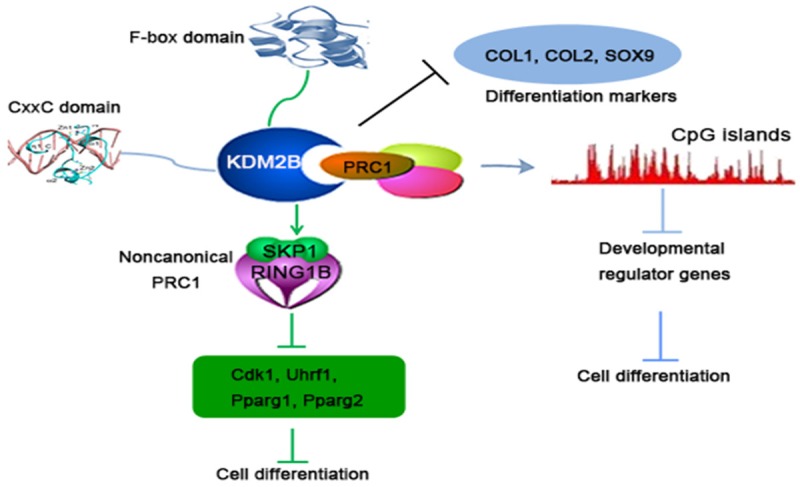
Different mechanisms of KDM2B-induced cell differentiation suppression. KDM2B could recruit PRC1 to CpG islands of developmental genes via its CxxC-ZF domain. In addition, KDM2B also attracts a noncanonical PRC1 to the key cell differentiation-associated genes through the F-box domain. Becides, KDM2B can also directly inhibit the expression of differentiation markers.
KDM2B is a critical regulator for the somatic cell reprogramming
As well as regulating stem cell self-renewal, KDM2B also promoted somatic cell reprogramming into pluripotent stem cells (iPSCs). Somatic cell fate can be reprogrammed into a pluripotent embryonic stem cell (ESC)-like state by the use of key transcription factors including Oct4, Sox2, Klf4 and c-Myc [48,49]. But the underlying mechanisms of somatic cell reprogramming remain poorly understood. Tao and his colleagues [50] reported that KDM2B can enable efficient generation of iPSCs, and enhanced Oct4 reprogramming by overcoming Ink4/Arf-triggered cell senescence. Moreover, during the process of reprogramming, KDM2B could cooperate with Oct4 to increase the expression of cell cycle-related miRNA cluster 302/367. Subsequent studies indicated that KDM2B enhanced Oct4-induced somatic reprogramming primarily through recruitment of a variant PRC1 to the CGIs of development genes [51]. KDM2B overexpression also enhanced the expression of Oct4 and Sox2. It has been reported that Oct4/Sox2 was frequently overexpressed in lung cancer, breast cancer, esophageal carcinoma, and lymphoma [52-55]. Thus, KDM2B might take part in Oct4/Sox2- associated cancers.
The multifaceted role of KDM2B in human tumors
Dysregulation of KDM2B has been found in ALL, AML, breast cancer, pancreatic cancer, gastric cancer, lung cancer, and bladder cancer [31,56-59]. It is worth mentioning that the regulatory mechanisms of KDM2B in these tumors are very complex and various.
KDM2B in malignant hematopoiesis
Previous study have revealed that KDM2B overexpression promoted the progression of MoMuLV-induced T cell lymphomas, acute lymphoblastic leukemia (ALL), and acute myelocytic leukemia (AML) [60-62]. Knockdown of KDM2B in ALL, CML, and AML cell lines not only decreased cell growth but also reduced lung metastasis in mice. In addition, KDM2B presented high levels in patients of AML, T-cell acute lymphoid leukemia, and B-cell acute lymphoid leukemia according to the Oncomine database. Other study revealed that KDM2B was upregulated in Hoxa9/Meis1-induced leukemic stem cells (LSCs) and was required for leukemic transformation and tumor progression [22]. The oncogenic function of KDM2B in leukemia was via silencing of p15Ink4b. Besides, KDM2B also combined with other members to form a non-canonical Polycomb complex (PRC1.1), and PRC1.1 was critically important for LSCs [63]. Consist with these findings, other research demonstrated that KDM2B transgenic mice could develop myeloid and B-lymphocytic leukemias [64]. Molecular mechanism revealed that KDM2B induced the expression of Nsg2, which led to failure cell differentiation. Meanwhile, KDM2B increased oxidative phosphorylation related genes to promote cell cycle progression. Considering that KDM2B is often overexpressed in human malignant hematopoiesis, it might be a strong driver of epigenetic regulator that guides the way to leukemogenesis.
KDM2B in gynecological cancers
Previous research showed that KDM2B was overexpressed in gynecological tumors [57,65]. Knockdown of KDM2B in adenocarcinoma cell lines led to the upregulation of miR-101, miR-200a/b/c, miR-181a/b/b and miR-203, and these miRNAs downregulated PRC1 and PRC2 subunits, then further induced G1 arrest, decreased cell size and cancer stem cell markers in breast cancer [57]. KDM2B was also highly expressed in basal-like triple-negative (ER-, PR- and Her2-) breast cancers, and was a poor prognostic factor [57]. Besides, functional genetic screens revealed that KDM2B was also a breast cancer anti-estrogen resistance (BCAR) gene [66]. However, in contrast, another research demonstrated that KDM2B acted as a tumor suppressor by controlling the ribosome biogenesis in breast cancer [67]. KDM2B knockdown in MDA-MB-231 cell triggered a more invasive and proliferative phenotype, suggesting the dual role of KDM2B in breast cancer.
In cervical cancer cell line Hela cells, KDM2B overexpression repressed ribosomal RNA genes, decreased cell size and cell proliferation [68]. And this function was mediated by H3K4me3 demethylation and c-Fos ubiquitylation [69]. However, it has been reported that KDM2B promoted proliferation and glycolysis by activating RIP3 and Crabp2 metabolic enzymes in HeLa cells [70]. These paradoxically phenomenon suggests the function of KDM2B in regulating cell proliferation might have different molecular mechanisms.
High levels of KDM2B was also found in ovarian cancer, and positively correlated with pathological grades [65]. Knockdown of KDM2B in ovarian cancer cells inhibited cell growth and migration.
KDM2B in pancreatic cancer and gastric cancer
Alexandros et al. [58] reported that KDM2B was markedly increased in pancreatic cancer (PDAC), and positively correlated with tumor grade. It is worth mentioning that, KDM2B was typically enriched in the poorly differentiated and invasive cancer cells. KDM2B knockdown suppressed cell proliferation and blocked xenograft tumor formation. Mechanistically, KDM2B interacted with Polycomb group proteins to silence cellular differentiation programs through H3K36 demethylation and H3K27 methylation; on the other hand, KDM2B activated the expression of MYC and KDM5A genes to promote PDAC progression.
As in PDAC, KDM2B was also commonly expressed in human gastric cancer. It was a poor prognostic factor both in intestinal and disuse Lauren types gastric cancer [71]. However, ChIP analysis demonstrated that KDM2B directly acted on the MYC promoter, further then inhibited glycolysis in gastric cancer [59]. To be concluded, these above reports imply the significance of KDM2B in tumors (Table 1).
Table 1.
Summary of the role of KDM2B in cancers so far
| Cancer type | Function of KDM2B | Related gene | References |
|---|---|---|---|
| Leukemia (ALL, AML) | Tumor cell growth and proliferation | p15Ink4b, Nsg2, PRC | [22,60-64] |
| Breast cancer | Oncogenic (Proliferation, Cancer stem cell, anti-estrogen resistance)/Tumor suppressive (Down regulation of ribosome biogenesis) | Oncogenic (miR-101, PRC1, PRC2, CD44, ALDH)/Tumor suppressive (45S pre-rRNA) | [57,66,67] |
| Cervical cancer | Oncogenic (Cell glycolysis)/Tumor suppressive (Cell proliferation) | Oncogenic (RIP3 and Crabp2)/Tumor suppressive (Ribosomal RNA, c-Fos) | [68-70] |
| Ovarian cancer | Oncogenic (Tumor growth, proliferation and metastasis) | EZH2 | [65] |
| Pancreatic cancer | Oncogenic (Tumor growth, proliferation and metastasis, differentiation) | PRC, EZH2, MYC | [58] |
| Gastric cancer | Oncogenic (Poor prognostic factor)/Tumor suppressive (Decreased cell growth and proliferation) | Oncogenic (mTOR and p70S6K)/Tumor suppressive (miR-448, MYC) | [59,71] |
| Lung, Bladder cancer | Oncogenic (Tumor growth, proliferation and metastasis) | miR-101, EZH2 | [79] |
| Nasopharyngeal carcinoma | Oncogenic | Oncogenic (mTOR and P70s6k) | [80] |
| Prostate cancer, Glioblastoma multiforme | Oncogenic (TRAIL-resistant) | Oncogenic (c-Fos/c-FLIP) | [77,78] |
Regulation of signaling pathways by KDM2B
As discussed above, KDM2B regulated the cell proliferation, invasion, and cancer stem cells of different cancers. Moreover, KDM2B has been actively involved in tumor-associated cell signaling pathways. AP-1 pathway was considered as an important regulator in tumor growth and metastasis [72-75]. AP-1 pathway consists of two proteins: c-Jun and c-Fos. C-Fos was an important regulator of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis [76]. KDM2B was reduced in TRAIL-sensitive cancer cell lines after treated with TRAIL, and KDM2B knockdown could change TRAIL-resistant cells into TRAIL -sensitive cells [77]. Further study confirmed that KDM2B restrained TRAIL-mediated apoptosis by repressing c-Fos/c-FLIP pathway. Besides, this anti-apoptotic of KDM2B was NF-kB pathway dependent. Recent research has shown that KDM2B decreased TRAIL response in glioblastoma multiforme (GBM) cells [78]. KDM2B silencing significantly enhanced the sensitivity of GBM cells to TRAIL, and promoted the activation of caspase-8, -3, -7 and PARP cleavage.
In addition, gene-expression profiling in T-ALL cell lines revealed that KDM2B knockdown decreased MYC and EZH2-dependent pathway, while tumor-suppressor pathways including p53, TGFB1, and SMARCA4 were upregulated [61]. Previous studies have also shown that KDM2B -miR-101-EZH2 pathway was active in at least 25 human cancer cell lines, including bladder cancer and lung carcinomas [79]. And this pathway contributed to cell proliferation, migration, angiogenesis and cancer stem cell self-renewal in bladder cancer by increasing the expression of EMT-promoting transcription factors [31]. Moreover, KDM2B could promoted nasopharyngeal carcinoma progression by activating PI3K/mTOR pathway [80]. In gastric cancer cells, KDM2B knockdown also induced autophagy via PI3K/Akt/mTOR inhibition and ERK1/2 activation [71]. In addition, a lot of evidence verified that Wnt/β-catenin pathway was essential in cancers [81,82]. Another study confirmed that KDM2B could inhibit Wnt/β-catenin pathway by inducing degradation of non-phosphorylated β-catenin in the nucleus [83]. Whether KDM2B-Wnt/β-catenin pathway works in cancer still needs more research. Together, these findings indicated that KDM2B has both positive and negative effect in different oncogenic pathways (Figure 4).
Figure 4.
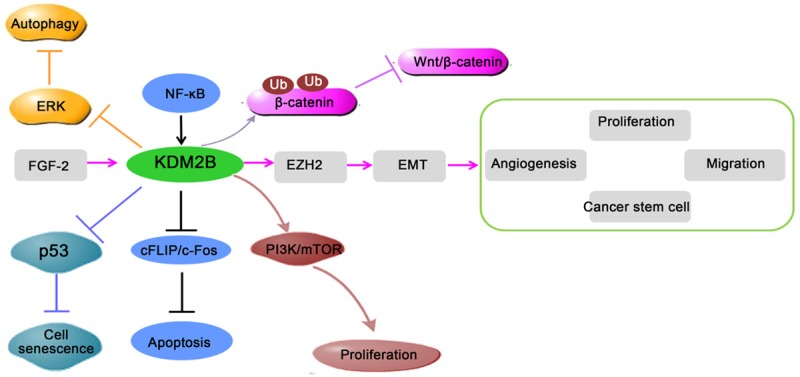
Regulation of cell signaling pathways involved in cancers by KDM2B. First, the activation of FGF-2-KDM2B-EZH2 pathway contributes to cell proliferation, migration, angiogenesis and self-renewal of cancer stem cells. Second, KDM2B inhibits cell apoptosis by repressing c-Fos/c-FLIP pathway. Finally, KDM2B also activates PI3K/Akt/mTOR pathway and inhibits P53 pathway in cancers. In contrast, KDM2B also inhibits Wnt/β-catenin signaling pathway by inducing degradation of β-catenin.
KDM2B as a potential therapeutic target
Since histone methylation is reversible, a lot of histone demethylase inhibitors had been used for anti-cancer treatment [84-86]. For instance, N-oxalylglycine 2 (NOG) and its derivatives were identified to inhibit KDM4A, KDM2A, KDM2C and KDM2D [87]. 2,4-pyridinedicarboxylic acid (2,4-PDCA) suppressed KDM5B [88]. Moreover, CPI-455 and GSK-J1/GSK-J4 were reported to be selective inhibitors of KDM5 and KDM6 subfamilies [89,90]. However, the selective and specific inhibitors for KDM2B have not yet been found. It was due to the histone demethylases have high structural similarity. Therefore, more research is necessary to develop novel KDM2B inhibitors. Nevertheless, Quanticel Pharmaceuticals have patented a series of pyridine derivatives as KDM2B inhibitors in 2016 [91]. 1,7-naphthyridones has exhibited high selectivity of KDM4C and KDM2B isoforms [92]. We think KDM2B inhibitors might be promising agents for anti-cancer therapy in the future.
Conclusions
A plethora of evidences have demonstrated the significant role of KDM2B in regulating cell cycle progression, cell differentiation, stem cell self-renewal, and cell apoptosis. It is also clear that elevated expression of KDM2B promotes cancer cell proliferation, metastasis, cancer stem cells self-renewal, and drug resistance (Figure 5). However, KDM2B could also decrease cancer cell proliferation by inhibiting the expression of oncogenes. KDM2B appears to function as a double-edged sword in the regulation of cancer development (Figure 6).
Figure 5.
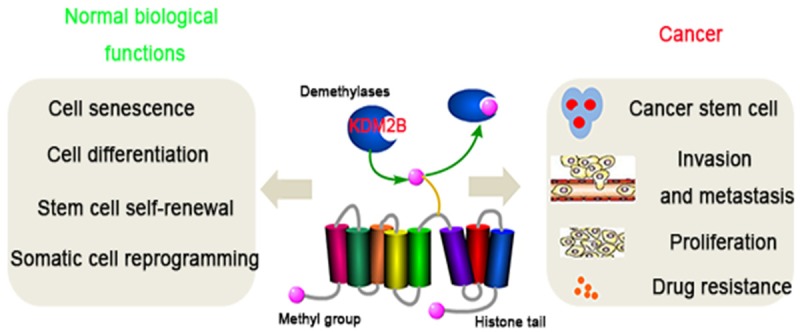
Pivotal role of KDM2B in normal biological functions and cancer. As an important histone lysine demethylase, KDM2B is very important in regulating cell senescence, cell differentiation, stem cell self-renewal and somatic cell reprogramming. The dysregulation of KDM2B might contribute to cancer progression including cell proliferation, metastasis and drug resistance.
Figure 6.
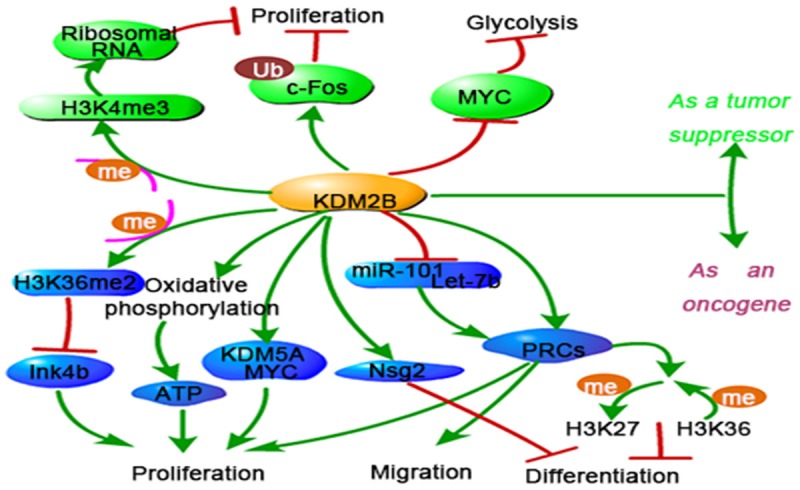
Overview of KDM2B mediated molecular mechanisms in cancer development. KDM2B acts as a double-edged sword in cancer development. On one hand, KDM2B inhibits p15Ink4b pathway, increases oxidative phosphorylation and enhances KDM5A/MYC protein expression, which promotes cancer cell proliferation. Becides, KDM2B recruits PRC and Nsg2 to enhance cell migration and controls self-renewal of cancer stem cells. On the other hand, KDM2B inhibits ribosomal RNA genes, MYC protein, and promots c-Fos ubiquitylation, which led to a decrease of cancer cell proliferation.
Up to now, the research of KDM2B in cancer is just on the way. The role of KDM2B in cancer may be more complex than originally believed. Now, most studies focus on the phenotype changes by KDM2B overexpression or knockdown. Future studies should seek to clarify the long-term effect of KDM2B in vivo studies. In addition, several questions still need to be addressed. For instance, what upstream signals trigger the expression of KDM2B, and why KDM2B has different function in cancers? It is under what conditions that KDM2B may act on H3K36me2 or H3K4me3. Besides, why the KDM2B displays multiaspect roles in the same cancer? This paradoxical phenomenon implies us whether the regulation of KDM2B changes during different stages of tumor. Moreover, it would be a challenge to identify potential inhibitors of KDM2B. Obviously, more research is required to further elucidate these questions. Here, we present evidence from multiple papers that may contribute to have a better understanding of KDM2B in tumor development. It may open new therapeutic approaches to suppress cancer progression.
Acknowledgements
We thank all laboratory members for advice and encouragement; We also acknowledge generous support from the National Natural Science Foundation of China (Grant NO: 81273202, 81671541) and the Clinical Medicine Science & Technology Project of Jiangsu province of China (Grant NO: BL2013024).
Disclosure of conflict of interest
None.
References
- 1.Lal G, Bromberg JS. Epigenetic mechanisms of regulation of Foxp3 expression. Blood. 2009;114:3727–3735. doi: 10.1182/blood-2009-05-219584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanwal R, Gupta S. Epigenetics and cancer. J Appl Physiol (1985) 2010;109:598–605. doi: 10.1152/japplphysiol.00066.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elbjeirami WM. DNA methylation in the inflammatory response and relevance to chronic kidney disease. J Periodontol. 2008 [Google Scholar]
- 4.Ducasse M, Brown MA. Epigenetic aberrations and cancer. Mol Cancer. 2006;5:60. doi: 10.1186/1476-4598-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 7.Celano M, Mio C, Sponziello M, Verrienti A, Bulotta S, Durante C, Damante G, Russo D. Targeting post-translational histone modifications for the treatment of non-medullary thyroid cancer. Mol Cell Endocrinol. 2017;469:38–47. doi: 10.1016/j.mce.2017.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Rondelet G, Wouters J. Human DNA (cytosine-5)-methyltransferases: a functional and structural perspective for epigenetic cancer therapy. Biochimie. 2017;139:137–147. doi: 10.1016/j.biochi.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Koschmann C, Nunez FJ, Mendez F, Brosnan-Cashman JA, Meeker AK, Lowenstein PR, Castro MG. Mutated chromatin regulatory factors as tumor drivers in cancer. Cancer Res. 2017;77:227–233. doi: 10.1158/0008-5472.CAN-16-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Amamoto Y, Aoi Y, Nagashima N, Suto H, Yoshidome D, Arimura Y, Osakabe A, Kato D, Kurumizaka H, Kawashima SA, Yamatsugu K, Kanai M. Synthetic posttranslational modifications: chemical catalyst-driven regioselective histone acylation of native chromatin. J Am Chem Soc. 2017;139:7568–7576. doi: 10.1021/jacs.7b02138. [DOI] [PubMed] [Google Scholar]
- 12.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 13.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 14.Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 16.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 17.Dimitrova E, Turberfield AH, Klose RJ. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015;16:1620–1639. doi: 10.15252/embr.201541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaniskan HU, Martini ML, Jin J. Inhibitors of protein methyltransferases and demethylases. Chem Rev. 2017 doi: 10.1021/acs.chemrev.6b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi YG, Tsukada Y. The discovery of histone demethylases. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams ST, Walport LJ, Hopkinson RJ, Madden SK, Chowdhury R, Schofield CJ, Kawamura A. Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates. Epigenetics. 2014;9:1596–1603. doi: 10.4161/15592294.2014.983381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou H, Yu H. Structural insights into histone lysine demethylation. Curr Opin Struct Biol. 2010;20:739–748. doi: 10.1016/j.sbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood. 2011;117:3869–3880. doi: 10.1182/blood-2010-10-312736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janzer A, Stamm K, Becker A, Zimmer A, Buettner R, Kirfel J. The H3K4me3 histone demethylase Fbxl10 is a regulator of chemokine expression, cellular morphology, and the metabolome of fibroblasts. J Biol Chem. 2012;287:30984–30992. doi: 10.1074/jbc.M112.341040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, Koseki H, Brockdorff N, Ponting CP, Kessler BM, Klose RJ. KDM2B links the polycomb repressive complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musselman CA, Kutateladze TG. PHD fingers: epigenetic effectors and potential drug targets. Mol Interv. 2009;9:314–323. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc Natl Acad Sci U S A. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, Wong KK, Bardeesy N. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–33069. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiu WT, Huang YF, Tsai HY, Chen CC, Chang CH, Huang SC, Hsu KF, Chou CY. FOXM1 confers to epithelial-mesenchymal transition, stemness and chemoresistance in epithelial ovarian carcinoma cells. Oncotarget. 2015;6:2349–2365. doi: 10.18632/oncotarget.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNiel EA, Tsichlis PN. Analyses of publicly available genomics resources define FGF-2-expressing bladder carcinomas as EMT-prone, proliferative tumors with low mutation rates and high expression of CTLA-4, PD-1 and PD-L1. Signal Transduct Target Ther. 2017:2. doi: 10.1038/sigtrans.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohde M, Sievers E, Janzer A, Willmann D, Egert A, Schorle H, Schule R, Kirfel J. Overexpression of histone demethylase Fbxl10 leads to enhanced migration in mouse embryonic fibroblasts. Exp Cell Res. 2016;348:123–131. doi: 10.1016/j.yexcr.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 33.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49:1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Yu F, Shen H, Deng HW. Systemic analysis of osteoblast-specific DNA methylation marks reveals novel epigenetic basis of osteoblast differentiation. Bone Rep. 2017;6:109–119. doi: 10.1016/j.bonr.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SM, Lee J, Noh KM, Choi WY, Jeon S, Oh GT, Kim-Ha J, Jin Y, Cho SW, Kim YJ. Intragenic CpG islands play important roles in bivalent chromatin assembly of developmental genes. Proc Natl Acad Sci U S A. 2017;114:E1885–E1894. doi: 10.1073/pnas.1613300114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 38.He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15:373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konuma T, Nakamura S, Miyagi S, Negishi M, Chiba T, Oguro H, Yuan J, Mochizuki-Kashio M, Ichikawa H, Miyoshi H, Vidal M, Iwama A. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp Hematol. 2011;39:697–709. e695. doi: 10.1016/j.exphem.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Wang JJ, Dong R, Wang LP, Wang JS, Du J, Wang SL, Shan ZC, Fan ZP. Histone demethylase KDM2B inhibits the chondrogenic differentiation potentials of stem cells from apical papilla. Int J Clin Exp Med. 2015;8:2165–2173. [PMC free article] [PubMed] [Google Scholar]
- 41.Inagaki T, Iwasaki S, Matsumura Y, Kawamura T, Tanaka T, Abe Y, Yamasaki A, Tsurutani Y, Yoshida A, Chikaoka Y, Nakamura K, Magoori K, Nakaki R, Osborne TF, Fukami K, Aburatani H, Kodama T, Sakai J. The FBXL10/KDM2B scaffolding protein associates with novel polycomb repressive complex-1 to regulate adipogenesis. J Biol Chem. 2015;290:4163–4177. doi: 10.1074/jbc.M114.626929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yun Y, Gao R, Yue H, Guo L, Li G, Sang N. Sulfate aerosols promote lung cancer metastasis by epigenetically regulating the Epithelial-to-Mesenchymal Transition (EMT) Environ Sci Technol. 2017;51:11401–11411. doi: 10.1021/acs.est.7b02857. [DOI] [PubMed] [Google Scholar]
- 43.Ayob AZ, Ramasamy TS. Cancer stem cells as key drivers of tumour progression. J Biomed Sci. 2018;25:20. doi: 10.1186/s12929-018-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gearhart MD, Corcoran CM, Wamstad JA, Bardwell VJ. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol Cell Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong SJ, Gearhart MD, Taylor AB, Nanyes DR, Ha DJ, Robinson AK, Artigas JA, Lee OJ, Demeler B, Hart PJ, Bardwell VJ, Kim CA. KDM2B recruitment of the polycomb group complex, PRC1.1, requires cooperation between PCGF1 and BCORL1. Structure. 2016;24:1795–1801. doi: 10.1016/j.str.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao W, Li Q, Ayers S, Gu Y, Shi Z, Zhu Q, Chen Y, Wang HY, Wang RF. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell. 2013;152:1037–1050. doi: 10.1016/j.cell.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 50.Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, Pei D. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Z, Yang X, He J, Liu J, Wu F, Yu S, Liu Y, Lin R, Liu H, Cui Y, Zhou C, Wang X, Wu J, Cao S, Guo L, Lin L, Wang T, Peng X, Qiang B, Hutchins AP, Pei D, Chen J. Kdm2b regulates somatic reprogramming through variant PRC1 complex-dependent function. Cell Rep. 2017;21:2160–2170. doi: 10.1016/j.celrep.2017.10.091. [DOI] [PubMed] [Google Scholar]
- 52.Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer. 2017;16:104. doi: 10.1186/s12943-017-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Almozyan S, Colak D, Mansour F, Alaiya A, Al-Harazi O, Qattan A, Al-Mohanna F, Al-Alwan M, Ghebeh H. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. 2017;141:1402–1412. doi: 10.1002/ijc.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C, Zhu M, Lou X, Liu C, Chen H, Lin X, Ji W, Li Z, Su C. Transcriptional factor OCT4 promotes esophageal cancer metastasis by inducing epithelial-mesenchymal transition through VEGF-C/VEGFR-3 signaling pathway. Oncotarget. 2017;8:71933–71945. doi: 10.18632/oncotarget.18035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen YP, Zhu WF, Chen LF, Lu JP, He TM, Fu WD, Xu CW, Chen G. Clinicopathologic features and expression of OCT4 protein in testicular diffuse large B cell lymphoma. Zhonghua Bing Li Xue Za Zhi. 2017;46:383–387. doi: 10.3760/cma.j.issn.0529-5807.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Goyama S, Kitamura T. Epigenetics in normal and malignant hematopoiesis: an overview and update 2017. Cancer Sci. 2017;108:553–562. doi: 10.1111/cas.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kottakis F, Foltopoulou P, Sanidas I, Keller P, Wronski A, Dake BT, Ezell SA, Shen Z, Naber SP, Hinds PW, McNiel E, Kuperwasser C, Tsichlis PN. NDY1/KDM2B functions as a master regulator of polycomb complexes and controls self-renewal of breast cancer stem cells. Cancer Res. 2014;74:3935–3946. doi: 10.1158/0008-5472.CAN-13-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, Wong KK, Lan F, Trojer P, Park PJ, Bardeesy N. KDM2B promotes pancreatic cancer via polycomb-dependent and -independent transcriptional programs. J Clin Invest. 2013;123:727–739. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hong X, Xu Y, Qiu X, Zhu Y, Feng X, Ding Z, Zhang S, Zhong L, Zhuang Y, Su C, Hong X, Cai J. MiR-448 promotes glycolytic metabolism of gastric cancer by downregulating KDM2B. Oncotarget. 2016;7:22092–22102. doi: 10.18632/oncotarget.8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, Olofsson T, Rade J, Fontes M, Porwit-Macdonald A, Behrendtz M, Hoglund M, Johansson B, Fioretos T. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 61.Andricovich J, Kai Y, Peng W, Foudi A, Tzatsos A. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J Clin Invest. 2016;126:905–920. doi: 10.1172/JCI84014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN. Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci U S A. 2008;105:1907–1912. doi: 10.1073/pnas.0711865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van den Boom V, Maat H, Geugien M, Rodriguez Lopez A, Sotoca AM, Jaques J, Brouwers-Vos AZ, Fusetti F, Groen RW, Yuan H, Martens AC, Stunnenberg HG, Vellenga E, Martens JH, Schuringa JJ. Non-canonical PRC1.1 targets active genes independent of H3K27me3 and is essential for leukemogenesis. Cell Rep. 2016;14:332–346. doi: 10.1016/j.celrep.2015.12.034. [DOI] [PubMed] [Google Scholar]
- 64.Ueda T, Nagamachi A, Takubo K, Yamasaki N, Matsui H, Kanai A, Nakata Y, Ikeda K, Konuma T, Oda H, Wolff L, Honda Z, Wu X, Helin K, Iwama A, Suda T, Inaba T, Honda H. Fbxl10 overexpression in murine hematopoietic stem cells induces leukemia involving metabolic activation and upregulation of Nsg2. Blood. 2015;125:3437–3446. doi: 10.1182/blood-2014-03-562694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuang Y, Lu F, Guo J, Xu H, Wang Q, Xu C, Zeng L, Yi S. Histone demethylase KDM2B upregulates histone methyltransferase EZH2 expression and contributes to the progression of ovarian cancer in vitro and in vivo. Onco Targets Ther. 2017;10:3131–3144. doi: 10.2147/OTT.S134784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Agthoven T, Sieuwerts AM, Meijer D, Meijer-van Gelder ME, van Agthoven TL, Sarwari R, Sleijfer S, Foekens JA, Dorssers LC. Selective recruitment of breast cancer anti-estrogen resistance genes and relevance for breast cancer progression and tamoxifen therapy response. Endocr Relat Cancer. 2010;17:215–230. doi: 10.1677/ERC-09-0062. [DOI] [PubMed] [Google Scholar]
- 67.Galbiati A, Penzo M, Bacalini MG, Onofrillo C, Guerrieri AN, Garagnani P, Franceschi C, Trere D, Montanaro L. Epigenetic up-regulation of ribosome biogenesis and more aggressive phenotype triggered by the lack of the histone demethylase JHDM1B in mammary epithelial cells. Oncotarget. 2017;8:37091–37103. doi: 10.18632/oncotarget.16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 69.Han XR, Zha Z, Yuan HX, Feng X, Xia YK, Lei QY, Guan KL, Xiong Y. KDM2B/FBXL10 targets c-Fos for ubiquitylation and degradation in response to mitogenic stimulation. Oncogene. 2016;35:4179–4190. doi: 10.1038/onc.2015.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu X, Wang J, Wu J, Shi Y. A systematic study of the cellular metabolic regulation of Jhdm1b in tumor cells. Mol Biosyst. 2015;11:1867–1875. doi: 10.1039/c5mb00166h. [DOI] [PubMed] [Google Scholar]
- 71.Zhao E, Tang C, Jiang X, Weng X, Zhong X, Zhang D, Hou J, Wang F, Huang M, Cui H. Inhibition of cell proliferation and induction of autophagy by KDM2B/FBXL10 knockdown in gastric cancer cells. Cell Signal. 2017;36:222–229. doi: 10.1016/j.cellsig.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 72.Satterfield L, Shuck R, Kurenbekova L, Allen-Rhoades W, Edwards D, Huang S, Rajapakshe K, Coarfa C, Donehower LA, Yustein JT. miR-130b directly targets Arhgap1 to drive activation of a metastatic CDC42-PAK1-AP1 positive feedback loop in Ewing sarcoma. Int J Cancer. 2017;141:2062–2075. doi: 10.1002/ijc.30909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakatsuka T, Tateishi K, Kudo Y, Yamamoto K, Nakagawa H, Fujiwara H, Takahashi R, Miyabayashi K, Asaoka Y, Tanaka Y, Ijichi H, Hirata Y, Otsuka M, Kato M, Sakai J, Tachibana M, Aburatani H, Shinkai Y, Koike K. Impact of histone demethylase KDM3A-dependent AP-1 transactivity on hepatotumorigenesis induced by PI3K activation. Oncogene. 2017;36:6262–6271. doi: 10.1038/onc.2017.222. [DOI] [PubMed] [Google Scholar]
- 74.Tyagi A, Vishnoi K, Kaur H, Srivastava Y, Roy BG, Das BC, Bharti AC. Cervical cancer stem cells manifest radioresistance: association with upregulated AP-1 activity. Sci Rep. 2017;7:4781. doi: 10.1038/s41598-017-05162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pang JS, Yen JH, Wu HT, Huang ST. Gallic acid inhibited matrix invasion and AP-1/ETS-1-mediated MMP-1 transcription in human nasopharyngeal carcinoma cells. Int J Mol Sci. 2017:18. doi: 10.3390/ijms18071354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X, Zhang L, Yang H, Huang X, Otu H, Libermann TA, DeWolf WC, Khosravi-Far R, Olumi AF. c-Fos as a proapoptotic agent in TRAIL-induced apoptosis in prostate cancer cells. Cancer Res. 2007;67:9425–9434. doi: 10.1158/0008-5472.CAN-07-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ge R, Wang Z, Zeng Q, Xu X, Olumi AF. F-box protein 10, an NF-kappaB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ. 2011;18:1184–1195. doi: 10.1038/cdd.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurt IC, Sur I, Kaya E, Cingoz A, Kazancioglu S, Kahya Z, Toparlak OD, Senbabaoglu F, Kaya Z, Ozyerli E, Karahuseyinoglu S, Lack NA, Gumus ZH, Onder TT, Bagci-Onder T. KDM2B, an H3K36-specific demethylase, regulates apoptotic response of GBM cells to TRAIL. Cell Death Dis. 2017;8:e2897. doi: 10.1038/cddis.2017.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kottakis F, Polytarchou C, Foltopoulou P, Sanidas I, Kampranis SC, Tsichlis PN. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell. 2011;43:285–298. doi: 10.1016/j.molcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren Y, Wu L, Li X, Li W, Yang Y, Zhang M. FBXL10 contributes to the progression of nasopharyngeal carcinoma via involving in PI3K/mTOR pathway. Neoplasma. 2015;62:925–931. doi: 10.4149/neo_2015_112. [DOI] [PubMed] [Google Scholar]
- 81.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 82.Cadigan KM, Peifer M. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb Perspect Biol. 2009;1:a002881. doi: 10.1101/cshperspect.a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu L, Gao Y, Zhang Z, Cao Q, Zhang X, Zou J, Cao Y. Kdm2a/b lysine demethylases regulate canonical wnt signaling by modulating the stability of nuclear beta-catenin. Dev Cell. 2015;33:660–674. doi: 10.1016/j.devcel.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 84.Feng S, Jin Y, Cui M, Zheng J. Lysine-Specific Demethylase 1 (LSD1) inhibitor S2101 induces autophagy via the AKT/mTOR pathway in SKOV3 ovarian cancer cells. Medical Science Monitor. 2016;22:4742–4748. doi: 10.12659/MSM.898825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trifiro P, Cappa A, Brambillasca S, Botrugno OA, Cera MR, Zuffo RD, Dessanti P, Meroni G, Thaler F, Villa M, Minucci S, Mercurio C, Varasi M, Vianello P. Novel potent inhibitors of the histone demethylase KDM1A (LSD1), orally active in a murine promyelocitic leukemia model. Future Med Chem. 2017;9:1161–1174. doi: 10.4155/fmc-2017-0003. [DOI] [PubMed] [Google Scholar]
- 86.Lee HJ, Kim BK, Yoon KB, Kim YC, Han SY. Novel inhibitors of lysine (K)-specific Demethylase 4A with anticancer activity. Invest New Drugs. 2017;35:733–741. doi: 10.1007/s10637-017-0496-2. [DOI] [PubMed] [Google Scholar]
- 87.D’Oto A, Tian QW, Davidoff AM, Yang J. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 2016;1:34–40. [PMC free article] [PubMed] [Google Scholar]
- 88.Kristensen LH, Nielsen AL, Helgstrand C, Lees M, Cloos P, Kastrup JS, Helin K, Olsen L, Gajhede M. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J. 2012;279:1905–1914. doi: 10.1111/j.1742-4658.2012.08567.x. [DOI] [PubMed] [Google Scholar]
- 89.Vinogradova M, Gehling VS, Gustafson A, Arora S, Tindell CA, Wilson C, Williamson KE, Guler GD, Gangurde P, Manieri W, Busby J, Flynn EM, Lan F, Kim HJ, Odate S, Cochran AG, Liu Y, Wongchenko M, Yang Y, Cheung TK, Maile TM, Lau T, Costa M, Hegde GV, Jackson E, Pitti R, Arnott D, Bailey C, Bellon S, Cummings RT, Albrecht BK, Harmange JC, Kiefer JR, Trojer P, Classon M. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol. 2016;12:531–538. doi: 10.1038/nchembio.2085. [DOI] [PubMed] [Google Scholar]
- 90.Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morera L, Lubbert M, Jung M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin Epigenetics. 2016;8:57. doi: 10.1186/s13148-016-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Labadie SS, Dragovich PS, Cummings RT, Deshmukh G, Gustafson A, Han N, Harmange JC, Kiefer JR, Li Y, Liang J, Liederer BM, Liu Y, Manieri W, Mao W, Murray L, Ortwine DF, Trojer P, VanderPorten E, Vinogradova M, Wen L. Design and evaluation of 1,7-naphthyridones as novel KDM5 inhibitors. Bioorg Med Chem Lett. 2016;26:4492–4496. doi: 10.1016/j.bmcl.2016.07.070. [DOI] [PubMed] [Google Scholar]


