Abstract
Transmembrane protein domains often contain interfacial aromatic residues, which may play a role in the insertion and stability of membrane helices. Residues such as Trp or Tyr, therefore, are often found situated at the lipid-water interface. We have examined the extent to which the precise radial locations of interfacial Trp residues may influence peptide helix orientation and dynamics. To address these questions, we have modified the GW5,19ALP23 (acetyl-GGALW5(LA)6LW19LAGA-[ethanol]amide) model peptide framework to relocate the Trp residues. Peptide orientation and dynamics were analyzed by means of solid-state nuclear magnetic resonance (NMR) spectroscopy to monitor specific 2H- and 15N-labeled residues. GW5,19ALP23 adopts a defined, tilted orientation within lipid bilayer membranes with minimal evidence of motional averaging of NMR observables, such as 2H quadrupolar or 15N-1H dipolar splittings. Here, we examine how peptide dynamics are impacted by relocating the interfacial Trp (W) residues on both ends and opposing faces of the helix, for example by a 100° rotation on the helical wheel for positions 4 and 20. In contrast to GW5,19ALP23, the modified GW4,20ALP23 helix experiences more extensive motional averaging of the NMR observables in several lipid bilayers of different thickness. Individual and combined Gaussian analyses of the 2H and 15N NMR signals confirm that the extent of dynamic averaging, particularly rotational “slippage” about the helix axis, is strongly coupled to the radial distribution of the interfacial Trp residues as well as the bilayer thickness. Additional 2H labels on alanines A3 and A21 reveal partial fraying of the helix ends. Even within the context of partial unwinding, the locations of particular Trp residues around the helix axis are prominent factors for determining transmembrane helix orientation and dynamics within the lipid membrane environment.
Introduction
The lipid bilayers of cell membranes host a variety of membrane proteins for which essential protein-lipid interactions influence the management of biological function. Indeed, a better understanding of protein-lipid interactions will be crucial for more complete elucidation of the molecular control mechanisms that underlie cell signaling and other actions mediated by membrane proteins. Transmembrane domains of proteins are often anchored within their lipid environment by flanking aromatic or charged residues (1). For example, four Trp (W) residues at positions 9, 11, 13, and 15 govern the folding and anchor the subunits of gramicidin A (gA) ion channels produced by Bacillus brevis, wherein the channel protein consists of two monomeric peptides that dimerize at the N-terminal in the lipid bilayer (2). The subunit anchoring is a result of the Trp indole ring’s affinity for the membrane interface and is also what prevents each subunit from crossing the bilayer (3). Therefore, the peptide must be added to each opposing bilayer leaflet to form channels. Whereas tryptophans 9 and 11 in gA are essential for efficient channel formation, replacing residues 13 and 15 in gA with Phe interestingly allows the peptides to cross the membrane in a putative double-stranded conformation and then form channels (4, 5). The different outcomes for modified gA sequence isomers (having Phe 9, 11 as opposed to Phe 13, 15) are particularly intriguing in that the differing properties are caused by different relative locations of interfacial aromatic Trp and Phe residues.
Indeed, interfacial Trp residues are important for the functioning of a number of membrane proteins. For example, conserved Trp residues provide membrane anchoring and govern helix dynamics and association to form functional integrin αIIbβ3 heterodimers (6), wherein it is noted that particular Trp residues at different helix locations interact with heterogeneous lipids and cannot necessarily be functionally replaced by Tyr or Phe. Additionally, the switching between the kinase and phosphatase activities of a bacterial thermosensor is linked not only to temperature, membrane thickness, and the hydrophobic length of a helix but also to whether Trp or Phe occupies a particular location on the transmembrane helix (7). For larger and more complex membrane proteins (for example, NADH:ubiquinone oxidoreductase, which has multiple transmembrane helices with interfacial Trp and Tyr residues (8)), it becomes more difficult to characterize, individually or collectively, the local or global impact exerted by the aromatic residues. However, fundamental and broadly applicable insights may be obtained at the single-residue level from investigations of model peptide-lipid systems that incorporate defined molecular features. In particular, examinations of how individual interfacial residues affect the orientation and dynamic behavior of a transmembrane helix are feasible using model peptide systems. Within such model systems, direct sequence modifications can be made and compared such that resulting changes in the peptide-lipid interaction can be investigated in detail. Varying the membrane composition, lipid acyl chain length, or peptide helix length may offer additional insights into these types of interactions (9, 10). To match the hydrophobic lengths of different lipids, variable numbers of repeating pairs of core helix-forming residues, such as leucine-alanine (LA)n repeats, may be used within a peptide’s central helical sequence (11), with aromatic or charged residues flanking each end of the principal helix.
The original acetyl-GWW(LA)nLWWA-amide (WALP) family peptides were based on a sequence that included four interfacial Trp residues, acetyl-GWW-(LA)n-LWWA-[ethanol]amide (Fig. 1; Table 1) (11). These peptides proved useful for examining the fundamentals associated with membrane partitioning by interfacial tryptophan residues and the modulation of lipid phase behavior when the hydrophobic length of the peptide helix was shorter than the thickness of the lipid bilayer (11). Although the original concept envisioned a transmembrane helix normal to the bilayer, the WALP peptides subsequently were discovered to adopt a defined tilted orientation within lipid bilayers, with “apparently” only a “minor” dependence on lipid acyl chain length (12, 13). Indeed, a nonzero tilt angle between the helix axis and the bilayer normal has by now been universally observed with many peptides and has been rationalized in terms of the favorable precession entropy that accompanies a finite tilt angle (14). The minor apparent response to bilayer thickness subsequently was attributed to motional averaging of the solid-state nuclear magnetic resonance (NMR) observables (15, 16), primarily in the form of rotational “slippage” about the helix axis (16, 17). The motion furthermore has been correlated with the presence of four interfacial tryptophan residues, two at each end of the core sequence, potentially competing among themselves for preferential orientations at the lipid-water interface (17, 18, 19, 20). The pairs of tryptophan residues were mutated to other aromatic or charged residues (21, 22) to observe how different pairs of identical residues could impact the transmembrane peptide helix behavior.
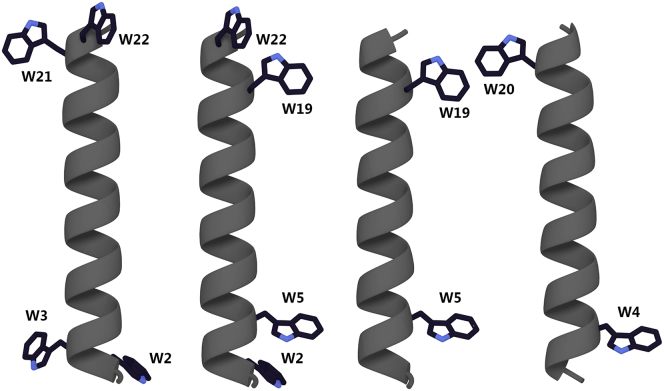
Figure 1.
Molecular models of GWALP-like peptides. From left to right, W2,3,21,22ALP23, W2,22W5,19ALP23, GW5,19ALP23, and GW4,20ALP23 are shown. For amino acid sequences, see Table 1. The model for GW4,20ALP23 was rotated 90°. To see this figure in color, go online.
Table 1.
Sequences of GW5,19ALP23 and High Dynamic Averaging GWALP-like Peptides
| Name | Sequence | Reference |
|---|---|---|
| GW5,19ALP23 | acetyl-GGALW5LALALALALALALW19LAGA-amide | (15) |
| W2,22W5,19ALP23 | acetyl-GW2ALW5LALALALALALALW19LAW22A-amide | (4) |
| W2,3,21,22ALP23 | acetyl-GW2W3LALALALALALALALALW21W22A-amide | (28) |
| Y4,5GW19ALP23 | acetyl-GGAY4Y5LALALALALALALW19LAGA-amide | (11) |
| GW4,20ALP23 | acetyl-GGAW4ALALALALALALALAW20AGA-amide | this work |
It was eventually determined that a single tryptophan residue at each end of the core helix is sufficient to minimize aggregation, regulate a hydrophobic peptide helix within a lipid membrane, and define its orientation, as seen with GW5,19ALP23 (acetylGGALW-(LA)6-LWLAGA-amide) (23, 24). Furthermore, it was discovered that fraying of helix ends may be crucially important for stabilizing the defined transmembrane orientation (25). The presence of fewer tryptophans tends to diminish the motional averaging and increase the sensitivity of the peptide helix to changes in lipid bilayer thickness. Indeed, GW5,19ALP23 is observed to adopt a well-defined tilt angle that scales with lipid bilayer thickness (26). The limited averaging exhibited by transmembrane GW5,19ALP23 stands in stark contrast to the behavior of transmembrane WALP peptides (27, 28).
The effects of repositioning individual Trp residues have been tested by following placements that were shifted by ±200° from positions 5,19 to positions 3, 21 (outward) or positions 7, 17 (inward), thereby increasing or decreasing the length of the hydrophobic core and the Trp-Trp distance along the helix axis while maintaining an analogous radial separation of the flanking tryptophans on one face of the helix (27). GW3,21ALP23 adopts a similar tilt distribution to that of GW5,19ALP23 within various lipid bilayer membranes, with low to moderate levels of motional averaging, whereas GW7,17ALP23 orients with smaller average tilt angles because of its shorter hydrophobic core helix.
Bearing in mind the extensive differences in motional averaging observed with different numbers of interfacial Trp residues, a principal aim of the research described here is to examine the influence of individual Trp residue radial locations on the transmembrane helix properties when two Trp residues are present, one at each end of the core helix. To this end, we moved W5 and W19 of GW5,19ALP23 each 100° outward to positions 4 and 20. As a consequence, the radial separation of the two aromatic residues changed from −40 to +160°. Whereas W5 and W19 are located on the same helix face, W4 and W20 are located on opposite faces of the helix, raising new questions: how will these Trp residue placements affect the peptide dynamics? What role might helix-terminal unwinding play now that fewer residues are available for fraying beyond each aromatic residue? Answering such questions will provide a framework to better understand how the interactions between transmembrane domains and their lipid environments are governed by specific residues within their sequences.
Materials and Methods
2H-labeled peptide synthesis
The sequence of the host peptide GW5,19ALP23 was altered by changing tryptophans 5 and 19 to alanines while changing leucines 4 and 20 to tryptophans to yield the sequence for GW4,20ALP23 (Table 1). Peptides were synthesized using solid-phase FastMoc chemistry on a 0.1 mmol scale, as previously described (18), using a model 433 A Applied Biosciences synthesizer by Life Technologies (Foster City, CA). L-alanine-d4 was purchased from Cambridge Isotope Laboratories (Andover, MA) and modified to Fmoc-L-alanine-d4, as previously described (29). Successful synthesis of Fmoc-L-alanine-d4 was verified by 1H NMR. Each peptide was labeled with two Ala-d4 residues in different isotope abundances. Peptides were purified using reversed-phase high-performance liquid chromatography with an octyl silica column (Zorbax Rx-C8, 9.4 × 250 mm, 5 μm particle size; Agilent Technologies, Santa Clara, CA) and with a gradient of 92–96% methanol (with 0.1% trifluoroacetic acid) over 32 min. Peptide purity of >95% and molar mass were confirmed by means of reversed-phase high-performance liquid chromatography and matrix-assisted laser desorption/ionization mass spectrometry, respectively, as indicated in Figs. S1 and S2.
To confirm peptide α-helicity, circular dichroism (CD) samples were prepared by combining 62.5 nM peptide with 3.75 μM lipid (1/60). Mixtures were initially dissolved in methanol/chloroform; the solvents were removed with a stream of N2 gas. The peptide/lipid films were further dried under vacuum for 48 h, hydrated to the final concentrations noted above, and then sonicated to form lipid vesicles with incorporated peptide. Each experiment consisted of 15 scans that were recorded with a Jasco (Easton, MD) J-1500 CD/fluorescence spectropolarimeter at 22°C using a 1 mm cell path length, 1.0 nm bandwidth, 0.1 nm slit, and a scan speed of 20 nm/min.
Solid-state 2H NMR spectroscopy
For mechanically aligned samples, peptides were incorporated into lipid bilayers, as previously described (12), with a peptide/lipid molar ratio of 1:60 using dilauroylphosphatidylcholine (DLPC), dimyristoylphosphatidylcholine (DMPC), or dioleoylphosphatidylcholine (DOPC) from Avanti Polar Lipids (Alabaster, AL) and a final hydration of 45% w/w using deuterium-depleted water from Cambridge Isotope Laboratories. Bilayer alignment in the liquid-crystalline samples was confirmed by 31P NMR using a Bruker (Billerica, MA) Avance 300 spectrometer with broadband 1H decoupling. Sample orientations at both β = 0° (bilayer normal parallel to the magnetic field) and β = 90° were tested. A quadrupole echo pulse sequence was utilized with full phase cycling (30) at 50°C at both sample orientations. The quadrupole echo delay was 4.5 μs, and the recycle delay was 90 ms. Between 0.7 and 1.5 million scans were acquired for each 2H NMR experiment, and the spectra were processed with 150 Hz line broadening.
For experiments in which magnetically aligned bicelle samples containing 2H-labeled peptides were examined, the samples were prepared as described below.
Solid-state 15N NMR spectroscopy
Magnetically aligned bicelle samples (1:105, peptide/total lipid) were prepared by combining 61 μmol DMPC, 19 μmol ether-DHPC (dihexanoylphosphatidylcholine) (q = 3.2, long lipid/short lipid) from Avanti Polar Lipids, and 0.76 μmol peptide. Peptide and DMPC were mixed, dried down under nitrogen flow, and then placed under vacuum for 48 h to remove residual organic solvent. Separate aliquots of ether-linked-DHPC were dried down in the same manner. The peptide/DMPC mixture and the ether-linked-DHPC were hydrated separately with 100 and 75 μL of 2H-depleted water for a minimum of 4 h with intermittent vortexing. The ether-linked-DHPC was then added to the peptide/DMPC mixture, and the combined sample was subjected to multiple freeze-thaw cycles, alternating the temperature between 0 and 42°C with gentle intermittent vortexing until the sample remained clear at 0°C. While still cold, the sample liquid was transferred to a 5 mm NMR tube (New Era Enterprises, Vineland, NJ) and sealed.
For 15N-detected separated local field experiments, GW4,20ALP23 was synthesized with five 15N-labeled residues at positions 13–17. Fmoc-L-Ala-15N and Fmoc-L-Leu-15N were purchased from Cambridge Isotope Laboratories. 15N chemical shifts and 15N/1H dipolar coupling signals were recorded using a 700 MHz Bruker Avance spectrometer with a Magnex magnet and room temperature shims. A home-built, “low-E” 15N/1H double resonance probe with a 5 mm modified Alderman-Grant coil was used to minimize sample heating (31). The GW4,20ALP23 sample in DMPC/ether-linked-DHPC bicelles was equilibrated in the magnetic field at 42°C (just below the critical temperature for DMPC/DHoPC (1,2-O-dihexyl-sn-glycero-3-phosphocholine) bicelle structural transformation at q = 3.2) for 30 min before initiating the NMR measurements. Previously, we have found that the peptide order parameter in plated samples remains essentially constant from 40 to 60°C; optimal spectral resolution is observed at 50°C (17, 32). To confirm this feature, in Fig. S3, 2H NMR spectra for a plated DMPC sample at 42 vs. 50°C show only a ∼0.1 kHz difference in quadrupolar splittings after 800,000 acquisitions. Agreeing with previous results, better spectral resolution is observed for plate samples at 50°C as opposed to the lower temperature. SAMPI-4 (33) separated local field spectra were recorded with 256 scans, 64 t1 increments, and a recycle delay of 6.5 s. The t1 evolution was preceded by a 1 ms cross-polarization mismatch-optimized I to S transfer cross-polarization (34), which compensates for the power mismatch. The 1H irradiation at all times was 46.3 kHz, and the composite pulse small phase incremental alternation, with 16 steps was applied for 1H heteronuclear decoupling during the 10 ms acquisition time (35, 36). The 1H carrier frequency of ∼9 ppm is optimal for transmembrane helices in perpendicular, magnetically oriented bilayers (37).
The 15N NMR data sets were processed and displayed using the programs NMRPipe/NMRDraw (38) and Sparky (39) using a dipolar coupling scaling factor of 0.61 for SAMPI-4 evolution (33). The chemical shifts were externally referenced to 15N-labeled solid ammonium sulfate set to 26.8 ppm, corresponding to the signal from liquid ammonia at 0 ppm (40). The 15N data for W2,22W5,19ALP23 and GW5,19ALP23 (Table 1) had been previously processed with line broadening, with zero filling applied to both dimensions, and with apodization via a 36° shifted sine-squared bell function applied to the indirect dimension with a first point scaling factor of 0.5 (17, 18). To account for truncation of data in both data sets (primarily in t1), the W2,22W5,19ALP23 and GW5,19ALP23 15N data presented in this study were reprocessed by omitting line broadening and instead applying linear prediction, zero filling, and a 36° shifted sine-squared bell function without first point adjustment to both dimensions.
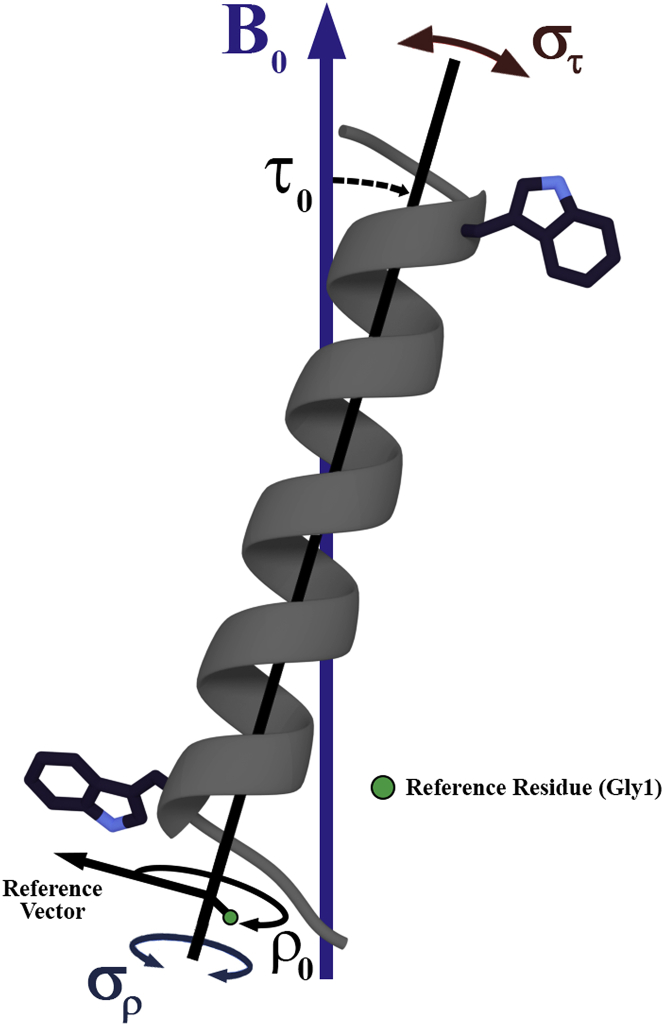
Data analysis
Preferred peptide helical orientations were assessed by several independent or combined analytical methods. The patterns for the recorded 2H quadrupolar splittings were evaluated by a geometric analysis of labeled alanines (“GALA” (geometric analysis of labeled alanines)), as previously described (12). The semistatic GALA method is based on three adjustable parameters: the apparent azimuthal rotation (or tilt direction) ρ0 with respect to the α-carbon of Gly1, the apparent average tilt τ0 of the helix axis relative to the bilayer normal, and a principal order parameter Szz. The whole body dynamics of the peptide can be described by analyzing the oscillations about the helix τ0 or ρ0; Gaussian treatments of 2H quadrupolar splittings, 15N chemical shifts, and 15N/1H dipolar couplings were employed to further analyze the dynamics, following the method of Vostrikov (17). The Gaussian analysis of helix dynamics relies on four adjustable parameters: ρ0, τ0, a distribution σρ (rotational “slippage”), and a distribution στ (helix “wobble”). For this particular study, the Gaussian calculations were performed as previously described (17) over the ranges of 0–30° for στ, 0–200° for σρ, 0–90° for τ0, and 0–359° for ρ0 in 1° increments. For comparative purposes, the 2H quadrupolar splittings and the 15N chemical shifts with 15N/1H dipolar couplings were subjected to individual Gaussian analyses (2H “only” or 15N “only”) as well as to combined analysis of the full data sets.
Results
Model transmembrane peptides such as GW5,19ALP23 and its analogs exhibit α-helical secondary structure in lipid-bilayer membranes. The high stability of the repeating Leu-Ala core helix is manifested in the increased strength of backbone hydrogen bonds within a hydrophobic lipid environment (41, 42). Relocating the Trp residues in GW5,19ALP23 to positions 4 and 20, as in GW4,20ALP23, increases the length of the hydrophobic core sequence from (LA)6L (13 residues) to A(LA)7 (15 residues). To characterize the secondary structure, CD spectra were recorded at 22°C for GW4,20ALP23 incorporated within DLPC, DMPC, and DOPC vesicles with a P/L ratio of 1:60 (Fig. 2). Indeed, an α-helical folding motif is confirmed in each lipid bilayer sample by the spectral minima observed at 208 and 222 nm. The relative ellipticity ratio (ϵ222/ϵ208) is greater than 0.91 in each lipid. When DOPC is the host lipid, the spectral noise observed below 200 nm is due to the absorbance of ultraviolet radiation by the double bonds present in the acyl chains of DOPC.
Figure 2.
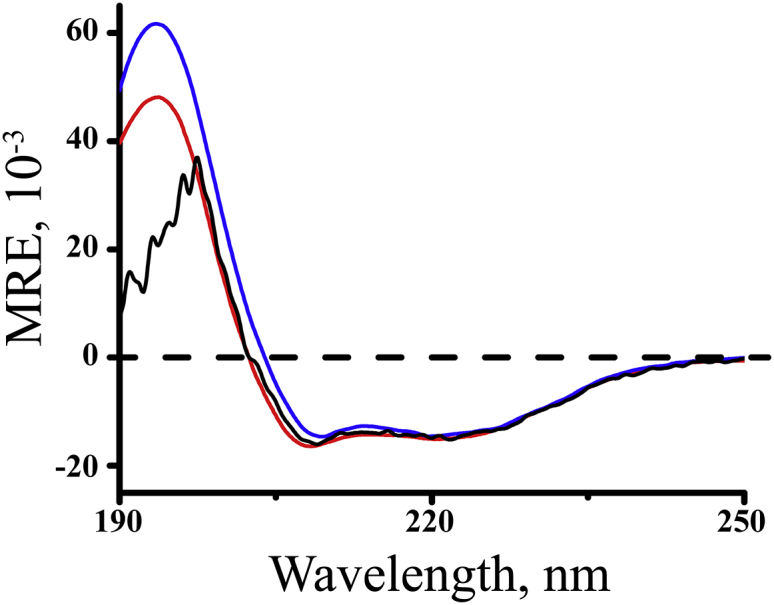
Circular dichroism spectra for GW4,20ALP23 in DLPC (red), DMPC (blue), and DOPC (black) vesicles. To see this figure in color, go online.
Solid-state NMR techniques can be utilized to determine the extent of a helix and the average peptide and lipid orientations and dynamics within lipid bilayers. The 31P NMR spectra confirmed the presence of oriented lipids within the mechanically aligned bilayers and magnetically aligned bicelle samples. The peptide-lipid bilayer samples (1:60, peptide:lipid) were set so that the membrane normal was either perpendicular (β = 90°) or parallel (β = 0°) to the applied magnetic field (Fig. 3 A). For the β = 90° spectra, a single peak was typically observed near −16 ppm; for the β = 0° spectra, a major peak was observed near +30 ppm with a minor peak at −16 ppm, indicating small amounts of unoriented lipid. Peptide-bicelle samples (1:80, peptide:DMPC; or 1:105, peptide:(DMPC+DHoPC), q = 3.2) were observed to align properly giving β = 90° with respect to the applied magnetic field (Fig. 3 B). In Fig. 3 B, the major 31P peak results from the DMPC head groups in the bicelles, and the minor peak is from the ether-linked-DHPC head groups of the shorter lipids.
Figure 3.
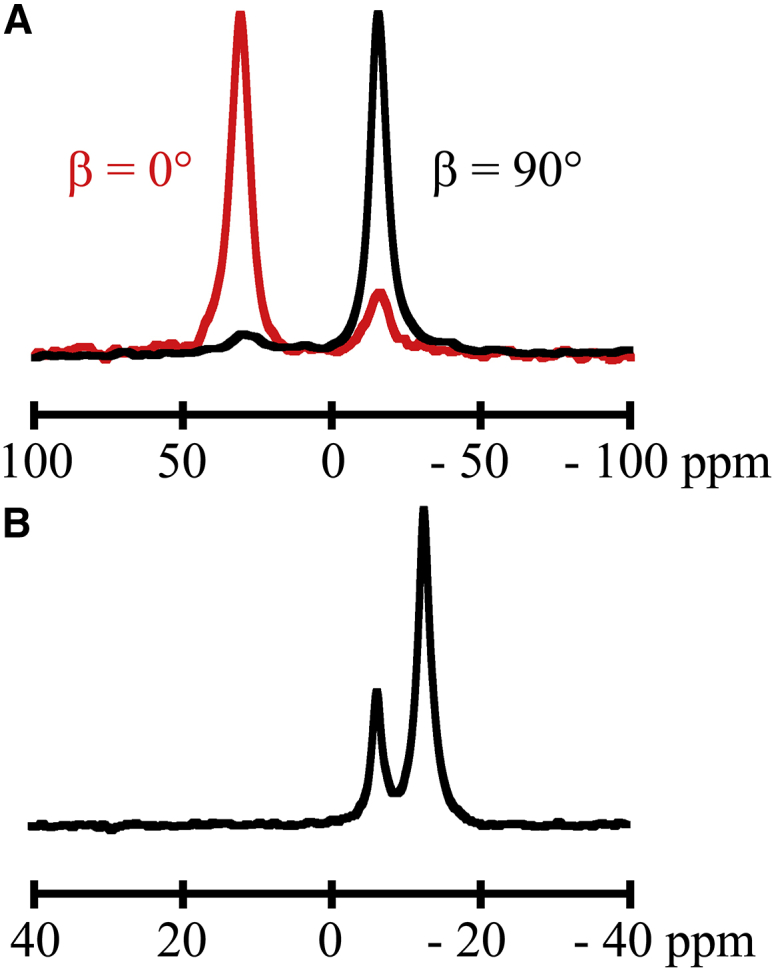
31-Phosphorous NMR spectra for GW4,20ALP23 in (A) oriented DMPC bilayers and (B) DMPC/DHPC bicelles. To see this figure in color, go online.
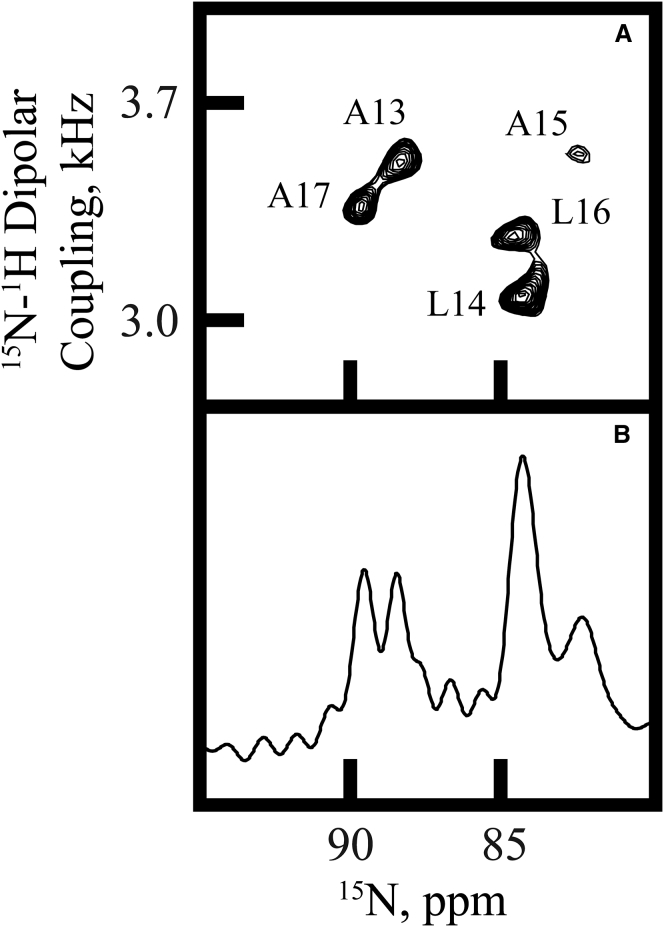
Insights into the orientation and dynamics of the GW4,20ALP23 transmembrane helix were gained from 15N and 2H solid-state NMR experiments. The 15N NMR spectrum of GW4,20ALP23 in DMPC/DHoPC bicelles reveals five 15N amide resonances, two of which overlap (Fig. 4 B). The 15N chemical shift frequencies span the narrow range between 82 and 90 ppm (Table 2), providing evidence of substantial motional averaging. Additional assignments (shown in Fig. 4 A) indicate that the 15N resonances from leucines 14 and 16 overlap in Fig. 4 B. The two-dimensional 15N-detected SAMPI-4 spectra reveal a “PISA” (polarity index slant angle) wheel composed of the five signals corresponding to the five 15N-labeled core residues 13–17 (Fig. 4 A). In similar fashion to the 15N chemical shifts, the 15N/1H splittings also lie within a narrow range (3.1–3.6 kHz) (Fig. 4 A; Table 2). The signals in Fig. 4 A were assigned by comparison with 2H NMR data and a subsequent combined analysis (see below). Position L16 produced an “aberrant” dipolar coupling value that moved the peak somewhat “inside” the PISA wheel, probably also reflecting dynamic averaging; because of the peak location in Fig. 4 A, the L16 dipolar coupling was not used in the analysis described below. The averaging observed for the 15N signals suggests that the GW4,20ALP23 helix may experience extensive motion about its apparent average orientation in a bicelle environment.
Figure 4.
Separated local-field PISEMA spectra. (A) 15N/1H-separated local-field spectrum for GW4,20ALP23 is shown. The sample is oriented in DMPC/DHPC bicelles and contains 15N-labeled residues 13–17 (assigned as depicted). (B) A one-dimensional 15N NMR spectrum for GW4,20ALP23 is shown.
Table 2.
15N Chemical Shifts and 1H-15N Dipolar Splittings for GW4,20ALP23 in DMPC/DHPC Bicelles
| DMPC/DHPC | Position |
||||
|---|---|---|---|---|---|
| 13 | 14 | 15 | 16 | 17 | |
| 15N chemical shift (ppm) | 88.4 | 84.1 | 82.4 | 84.5 | 89.7 |
| 1H-15N coupling (kHz) | 3.53 | 3.15 | 3.55 | 3.33a | 3.38 |
The aberrant 1H-15N coupling for residue 16 was omitted from future data analyses.
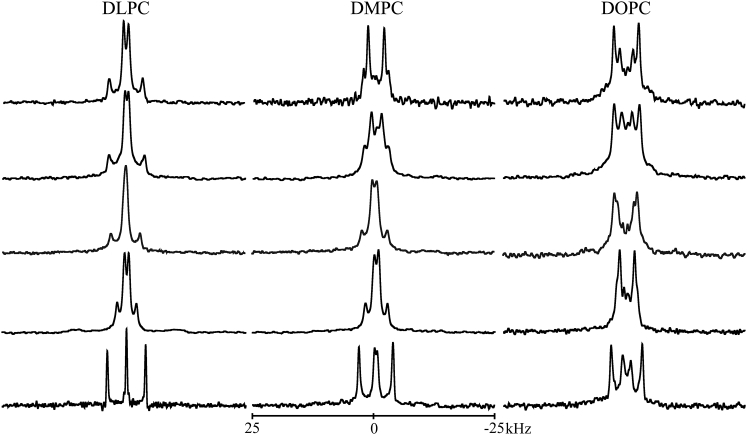
Additional solid-state NMR experiments, including 2H NMR measurements, can be used to analyze further the nature of helical peptides in oriented lipid bilayer membranes. To this end, 2H-labeled alanine residues were incorporated into the core helix of GW4,20ALP23, and solid-state NMR was used to monitor the peptide’s behavior in aligned lipid bilayers of increasing thickness (DLPC < DMPC < DOPC). For these spectra, mechanically aligned bilayer samples produce solid-state NMR observables that are similar to those from magnetically aligned bicelle samples (17). GW5,19ALP23 was previously observed to adopt well-defined orientations relative to bilayer thickness (23, 26), and similar responses were seen when the interfacial tryptophan residues were relocated by ± two sequence positions to effectively increase or decrease the length of the Leu-Ala core sequence (27). The helix of GW4,20ALP23 (Fig. 5) behaves differently.
Figure 5.
Deuterium quadrupolar splittings for GW4,20ALP23 in oriented DLPC, DMPC, and DOPC bilayers for samples oriented with β = 90°. The 2H-labeled alanine identities are, from top to bottom, (5,7), (9,11), (13,15), (17,19), and (3,21), with the first alanine of each pair 100% deuterated and the second alanine 50% deuterated.
Each 2H-labeled core alanine residue in GW4,20ALP23 yields a unique deuterium quadrupolar splitting when the lipid-peptide samples are aligned with β = 90° (Fig. 5) or β = 0° (Fig. S4). The variations among the core Ala-CD3 Δνq magnitudes (Fig. 5; Table 3) indicate that GW4,20ALP23 is tilted away from the bilayer normal because each value would be the same if the peptide helix were parallel to the membrane normal (12). The Δνq magnitudes for the core labels are observed, nevertheless, to span a frequency range from ∼0.5 to 16.0 kHz in all three lipids. The range is indeed narrow when compared to that of the parent transmembrane GW5,19ALP23 helix, for which the core alanines display 2H |Δνq| values ranging from 1.0 to 27 kHz (26). The large extent of motional averaging of the 2H Δνq values for GW4,20ALP23 correlates with the narrow ranges of resonance frequencies and dipolar splittings observed in the 15N-based experiments (Fig. 4), again suggesting the presence of additional motions in the GW4,20ALP23 helix; these may involve “wobble” about the helix tilt angle or “slippage” around the helix axis (17, 19, 28, 43).
Table 3.
Quadrupolar Splitting Magnitudes for Labeled Alanine CD3 Groups in GW4,20ALP23
| Lipid(s) | Alanine CD3 Position/ Δνq (kHz) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 5 | 7 | 9 | 11 | 13 | 15 | 17 | 19 | 21 | |
| DLPC | 15.7 | 2.1 | 14.0 | 1.4 | 14.6 | 0.8 | 12.2 | 1.8 | 8.2 | 0.1 |
| DMPC | 14.5 | 6.6 | 10.3 | 4.1 | 9.9 | 1.8 | 10.3 | 1.6 | 9.0 | 1.0 |
| DOPC | 13.1 | 10.2 | 5.3 | 10.4 | 4.2 | 9.4 | 6.8 | 2.9 | 6.2 | 3.3 |
| DMPC/DH(o)PC | 12.8 | 5.4 | 11.0 | 3.0 | 9.2 | 1.9 | 11.2 | 1.9 | 8.2 | 6.4 |
Quadrupolar splittings (|Δνq|, in kHz) are reported for samples oriented with β = 0° and are twice the magnitude observed when β = 90°. The core alanine residues 5, 7, 9, 11, 13, 15, 17, and 19 sit between W4 and W20 in the sequence, whereas the juxtaterminal alanines 3 and 21 are near the ends. The DMPC/DH(o)PC lipid mixture indicates bicelle samples with q = 3.2.
Overlaying the 15N PISA wheel of GW5,19ALP23 (Fig. 6 A) onto the spectrum of GW4,20ALP23 (Fig. 6 B) puts the extent of motional averaging into a visual context. The 15N backbone signals (black contours) for GW4,20ALP23 are clustered to one side of the wheel produced by GW5,19ALP23 (red contours, black ring). The 15N resonances of GW4,20ALP23 converge around essentially the same location as the extensively averaged signals from the corresponding labels in residues 13–17 in W2,22W5,19ALP23 (blue contours in Fig. 6 B). Neither set of peptide 15N resonances converges toward the value calculated for τ0 = 0° (4.1 kHz, 79.3 ppm, when β = 90°; Fig. 6), and they instead remain within the larger GW5,19ALP23 PISA wheel, which implies that the PISA signal convergence and 2H signal averaging are likely due to dynamics (17). We note that the data set presented here for W2,22W5,19ALP23 has been reprocessed to isolate previously unresolved peaks; see Materials and Methods. The PISA wheel signal convergence suggests that the GW4,20ALP23 helix undergoes extensive motional averaging in DMPC/DHoPC bicelles. In Fig. S5, we additionally show several examples of PISA wheels that illustrate the different trends for changing the helix tilt as opposed to changing the helix motion. Fitting the polarization inversion with spin exchange at magic angle (PISEMA) spectra for GW4,20ALP23 and W2,22W5,19ALP23 (Fig. 6) requires extensive motional averaging.
Figure 6.
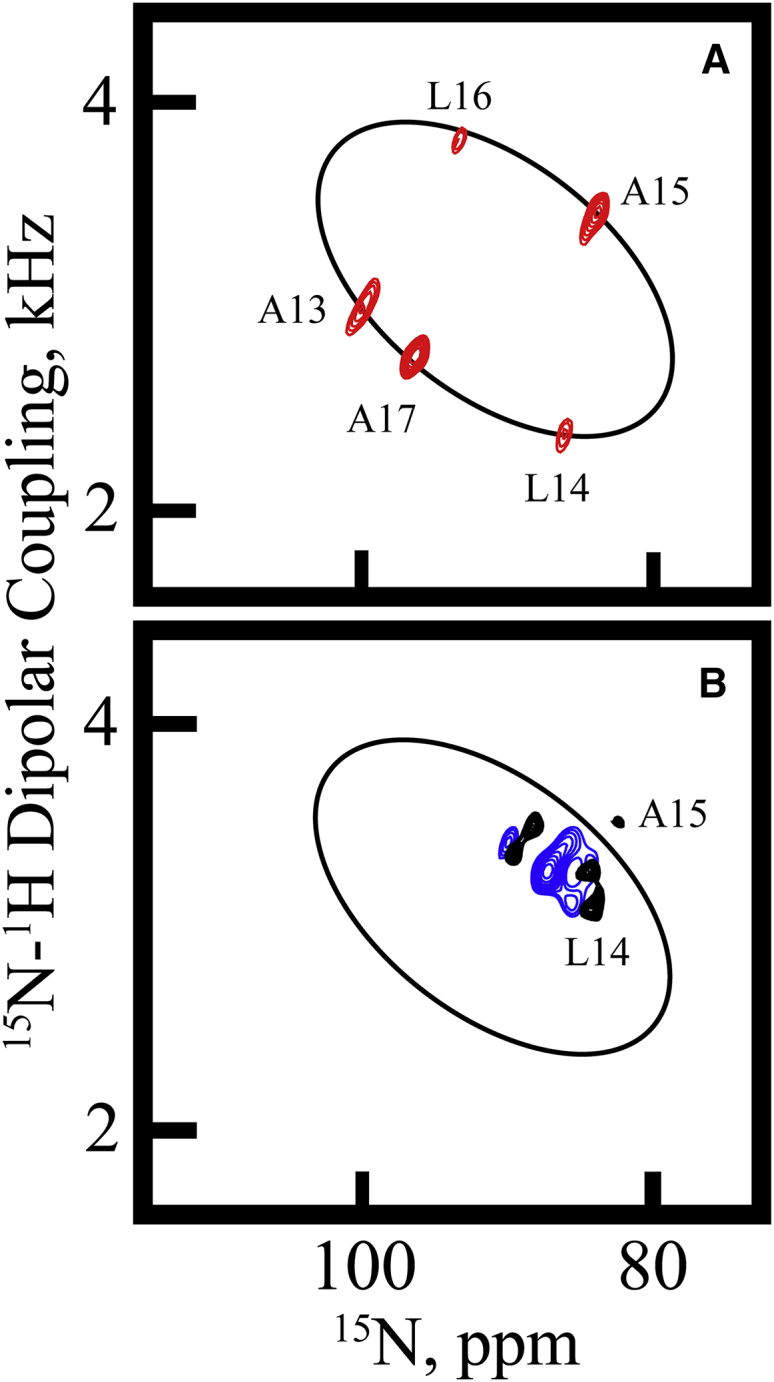
Helix dynamics illustrated by separated local-field PISEMA spectra. (A) Data and PISA wheel for GW5,19ALP23 are shown. (B) The PISA wheel from (A) is repeated along with data showing extensive motional averaging for W2,22W5,19ALP23 (blue contours) and for GW4,20ALP23 (black contours). Each sample is oriented in DMPC/DHoPC bicelles with β = 90° and is 15N-labeled in residues 13–17 (assigned as indicated). The spectra for GW5,19ALP23 and W2,22W5,19ALP23 were recorded previously (17, 18). To see this figure in color, go online.
Several methods were employed to deduce the average helix orientation and quantify the dynamics of GW4,20ALP23. Notably, the availability of eight core alanine data points enabled a full Gaussian analysis (see (17) and (28)) for GW4,20ALP23 using the 2H quadrupolar splitting magnitudes |Δνq| observed for the core helix alanine residues spanning sequence positions 5–19 in bilayers of DLPC, DMPC, and DOPC (Table 4). The Gaussian analysis generates estimates for peptide dynamics by considering the widths of the distributions (στ and σρ) about the helix’s average tilt (τ0) and azimuthal rotation (ρ0). A full Gaussian treatment requires four adjustable parameters (τ0, στ, ρ0, and σρ) while holding a principal order parameter Szz to a fixed value, typically 0.88, as an estimate for the overall isotropic motion of the peptide with respect to its average orientation (16). A modified Gaussian analysis (13, 19) can provide a suitable alternative approach when there are insufficient data points available to treat the four variables independently. While also setting Szz to 0.88, this modified calculation requires only three adjustable parameters (τ0, ρ0, and σρ), as στ has been found to be a less important descriptor (28) that can be assigned a small finite value (19).
Table 4.
Comparison of Orientations and Dynamics Calculated by GALA and Gaussian Analyses Using Ala-CD3 |Δνq| Magnitudes of GWALP23 Family Peptides
| Analysis |
DLPC |
DMPC |
DOPC |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD3 Gaussian | τ0 | ρ0 | σρ | RMSD (kHz) | τ0 | ρ0 | σρ | RMSD (kHz) | τ0 | ρ0 | σρ | RMSD (kHz) |
| W4,20 | 16° | 321° | 85° | 0.49 | 5.0° | 347° | 51° | 0.68 | 9.0° | 129° | 122° | 0.78a |
| Y4,5b | 14° | 259° | >90° | 1.7 | 15° | 321° | 124° | 0.63 | 6.0° | 344° | 72° | 0.9 |
| W5,19b | 23° | 304° | 33° | 0.70 | 13° | 308° | 44° | 1.1 | 9.0° | 321° | 48° | 0.7 |
| GALA | τ0 | ρ0 | Szz | RMSD (kHz) | τ0 | ρ0 | Szz | RMSD (kHz) | τ0 | ρ0 | Szz | RMSD (kHz) |
| W4,20 | 6.0° | 322° | 0.72 | 0.70 | 3.3° | 349° | 0.71 | 0.85 | 1.7° | 133° | 0.81 | 0.80a |
| Y4,5 | 5.0° | 260° | 0.66 | 1.6 | 3.0° | 323° | 0.77 | 0.60 | 3.0° | 359° | 0.82 | 1.1 |
| W5,19 | 20.7° | 305° | 0.71 | 0.66 | 11.7° | 311° | 0.86 | 0.90 | 6.0° | 323° | 0.87 | 0.6 |
The abbreviations refer to peptides based on the locations of selected aromatic residues W4,20 in GW4,20ALP23, Y4,5 in Y4,5GW19ALP23, and W5,19 in GW5,19ALP23. See also Table 1.
For GW4,20ALP23 in DOPC, the 2H |Δνq| value for position 17 was left out of the analysis, as it deviates from the calculated GALA curve (see Fig. 7C).
For the six core Ala-CD3 data points of GW5,19ALP23 and Y4,5GW19ALP23, a modified three-variable Gaussian treatment was used as described by (28) while constraining στ to 5° (19). For the eight core Ala-CD3 data points of GW4,20ALP23, a full Gaussian analysis was used, and the resulting στ-values were 15°, 20°, and 5° in DLPC, DMPC, and DOPC, respectfully.
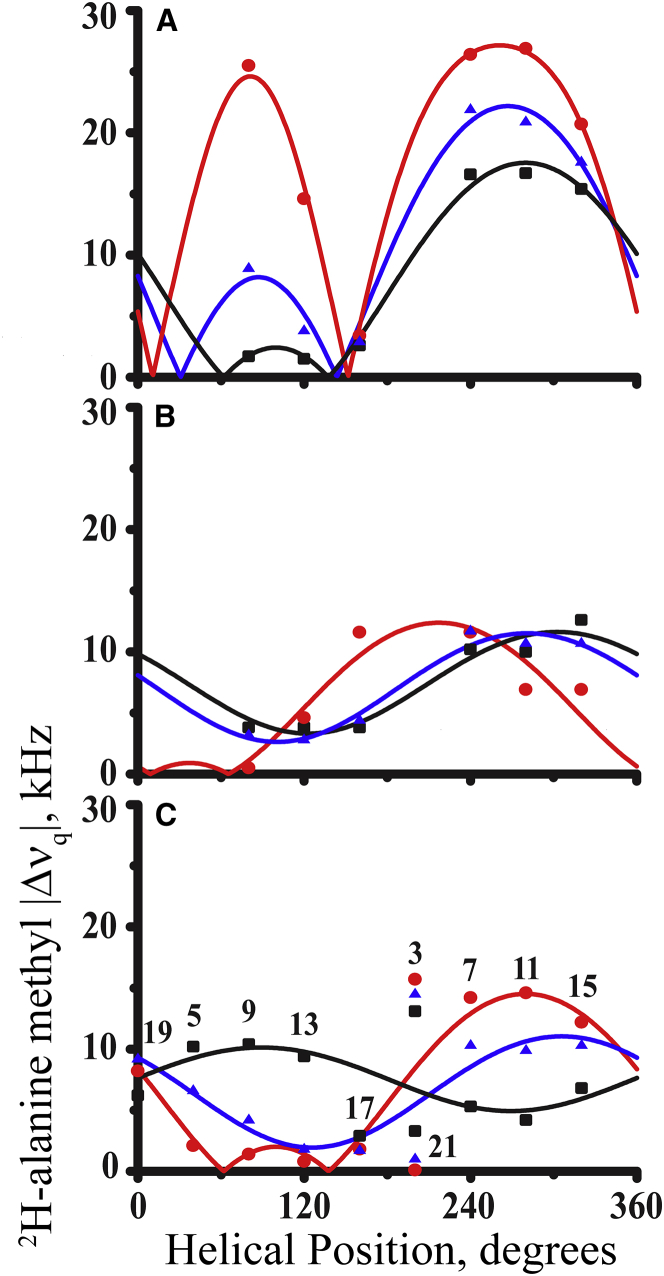
For comparison with GW4,20ALP23, a modified Gaussian analysis was performed for GW5,19ALP23 to represent low dynamic averaging and Y4,5GW19ALP23 to represent high dynamic averaging (18). Each of the latter helices contains only six core alanine data points. The fitted σρ-values that emerge from the full Gaussian treatment of the eight core alanines for GW4,20ALP23 exceed 80° in both DLPC and DOPC bilayers (Table 4). These results are comparable to those calculated for Y4,5GW19ALP23 (>70°). The large σρ-values are consistent with rotational “slippage” about the helix axis of the peptide’s average orientation, leading to the motional averaging observed in the 15N NMR as well as the 2H NMR data. The behavior of the GW4,20ALP23 helix in DOPC and DLPC bilayers differs substantially from the reduced dynamics calculated for GW5,19ALP23 in the same lipids. Indeed, the σρ-values for the parent GW5,19ALP23 helix are less than 50° in all three lipids. Surprisingly, the estimated σρ-value for GW4,20ALP23 in DMPC is much lower (51°) than its corresponding σρ-values in DLPC and DOPC (Table 4). This lower σρ for GW4,20ALP23 in DMPC bilayers is similar to that observed for GW5,19ALP23 in DMPC and DOPC. The reason for this unusual lipid dependence of σρ for GW4,20ALP23 remains to be determined. However, we note that the gel-fluid phase transition temperature of 23°C for DMPC is higher than those of the other lipids used in the experiments, and this result in DMPC contrasts with that for Y4,5GW19ALP23, which is highly dynamic in all three lipids.
The results of the Gaussian analysis were compared to those from a semistatic GALA analysis, which uses a grid search to find the lowest root mean-square deviation (RMSD) for fitting the 2H alanine |Δνq| values based on three adjustable parameters: τ0, ρ0, and Szz. The GALA curves previously calculated for GW5,19ALP23 are shown for comparison in Fig. 7 A (26). For this peptide helix, similar results emerge from the GALA and Gaussian analyses, even though the values of τ0 are slightly lower for the GALA method regardless of the lipid environment (Table 4). Importantly, the dependence of τ0 upon the bilayer thickness is maintained in both methods of analysis. The trends for the GALA (or Gaussian)-derived values of τ0 for GW5,19ALP23 are that τ0 decreases from ∼20.7° (or 23°) in DLPC to 11.7° (or 13°) in DMPC and to 6.0° (or 9°) in DOPC (Table 4). Thus, it is evident that the helix tilt decreases as the lipid bilayer thickness increases. In contrast, when the tryptophans are relocated to positions 4 and 20, increased motional averaging obscures the trend such that the peptide helix no longer exhibits a clear relationship between an apparent τ0 and the bilayer thickness (Fig. 7 B). Instead, according to the GALA analysis, the GW4,20ALP23 helix “appears” to adopt a relatively small τ0-value in all three lipids (≤6.0°). The low τ0 angles in DLPC and DOPC do not match the tilts obtained in the full Gaussian analysis (which would be expected in the case of a peptide undergoing dynamic averaging, as the GALA does not consider the oscillating motions about ρ0 and τ0), yet in DMPC, the τ0-values from both methods of analysis are remarkably similar.
Figure 7.
“Apparent” GALA quadrupolar wave plots for transmembrane peptide helices in DLPC (red circles), DMPC (blue triangles), or DOPC (black squares) oriented bilayer membranes. (A) GW5,19ALP23, (B) Y4,5GW19ALP23, and (C) GW4,20ALP23 are shown. (C) denotes the positions of the deuterium-labeled alanine residues. Only (A) reflects correctly the variation of the helix tilt τ0 and the constant helix azimuthal rotation ρ0 in the different bilayers. The amplitudes and phases of the waves in (B) and (C) reflect excessive dynamic averaging of the 2H quadrupoles. See text for details. See also Table 4. To see this figure in color, go online.
Dynamic averaging is manifest in several ways (see Fig. 8). For example, large values of σρ in the Gaussian analysis indicate extensive rotational averaging about the helix axis, as is evident for the Y4,5GW19ALP23 helix in all three lipids and for the GW4,20ALP23 helix in DLPC and DOPC (Table 4). The fitted “apparent” values of ρ0 also reflect the dynamic properties. When σρ is less than ∼50°, the ρ0-values tend to be rigorously consistent in all three lipid membranes, as is seen for GW5,19ALP23 in Fig. 7 A. By contrast, when σρ is very large, then the apparent ρ0-values vary widely, as is seen for GW4,20ALP23 in Fig. 7 B and as similarly observed for Y4,5GW19ALP23 (18). Interestingly, when σρ is large, the extensive dynamics about the peptide’s helix axis has been indicated by consensus from a number of different analytical treatments of not only experimental data but also populations from molecular simulations (13, 15, 26, 42). Indeed, the accompanying ρ0 predictions from the GALA and Gaussian methods tend to agree (Table 4), even though the τ0 predictions from the GALA method are systematically smaller by 5–10°. Extensive motional averaging in the form of rotational slippage can essentially mask the realistic τ0-values, as has been observed previously (15, 44).
Figure 8.
Model to describe transmembrane peptide motions and dynamics with respect to the bilayer normal B0. The apparent average helix tilt and rotation are denoted by τ0 and ρ0, respectively, and the oscillations about these orientations are shown as the helical “wobble” (στ) and rotational “slippage” (σρ). To see this figure in color, go online.
Besides the 2H NMR data, the 15N chemical shifts and 15N/1H dipolar couplings can be used to characterize peptide dynamics with the Gaussian treatment. Indeed, we have compared Gaussian analyses of GW4,20ALP23 in bicelles of DMPC/DHoPC (based on 15N NMR data) with results from mechanically aligned DMPC bilayer samples. Notably, the 15N NMR data sets for the bicelles suggest a value of ρ0 that differs by ∼25° from the consistent predictions from both the GALA and Gaussian analyses of the 2H NMR data for the same helix in mechanically aligned DMPC bilayers (Table 5). The different analytical outcomes for ρ0 may indicate that the properties of the GW4,20ALP23 helix are somewhat different between the lipid environments provided by the bicelle and bilayer samples. We note also that somewhat larger values of τ0 and σρ are calculated from the analysis of the helix properties in bicelles (Table 5). To investigate further, we prepared 2H-labeled samples of the GW4,20ALP23 helix in DMPC/DHoPC bicelles. The measured 2H |Δνq| values are quite similar for the alanine methyl groups of the core helix but somewhat different for the terminal alanines 3 and 21 (Figs. 9 and S6; Table 3). The semistatic analysis furthermore generates similar GALA curves for the core helix in DMPC bilayers and DMPC/DHoPC bicelles (Fig. 9), with a similar value of τ0 (3.7°) and a value of ρ0 (339°) that differs by ∼10° (Fig. 9) from that found for the helix in DMPC bilayers. A Gaussian treatment applied to the bicelle 2H NMR data also predicts a similar 10° difference in ρ0 while fitting τ0 to 9.0° and σρ to 64° (Table 5). The observed σρ of 64° and the 5° difference in τ0 between the semistatic and full Gaussian analyses would suggest somewhat more extensive dynamics for the GW4,20ALP23 helix in bicelles. These variations contrast with the dynamic analysis of the mechanically oriented DMPC samples, for which similar results emerge from the GALA and Gaussian methods.
Table 5.
Comparison of GW4,20ALP23 in DMPC Oriented Samples and DMPC/DHPC Bicelle Dynamics
| Analysis | DMPC Bilayers | DMPC/DH(o)PC Bicelles | ||||||
|---|---|---|---|---|---|---|---|---|
| 1H-15N/15N Gaussiana | τ0 | ρ0 | σρ | RMSD (kHz) | ||||
| 14° | 321° | 80° | 0.59 | |||||
| CD3 Gaussianb | τ0 | ρ0 | σρ | RMSD (kHz) | τ0 | ρ0 | σρ | RMSD (kHz) |
| 5.0° | 347° | 51° | 0.68 | 9.0° | 338° | 64° | 0.82 | |
| Combined Gaussian | τ0 | ρ0 | σρ | RMSD (kHz) | τ0 | ρ0 | σρ | RMSD (kHz) |
| 4.0° | 344° | 42° | 0.85c | 6.0° | 337° | 62° | 1.00d | |
Nine data points consisting of five 15N chemical shifts and four 1H-15N dipolar splittings.
Eight data points from the core alanine side chain CD3 quadrupolar splittings.
Combined Gaussian analysis was performed using 1H-15N dipolar splittings and 15N chemical shifts obtained from 15N-labeled DMPC/DHPC bicelle samples (nine data points) with 2H data from bilayers (eight data points). The resulting στ-value was 16°.
Combined Gaussian analysis was performed using all bicelle data (17 data points). The resulting στ-value was 17°.
Figure 9.
Comparison of the “apparent” GALA quadrupolar wave plots for GW4,20ALP23 in DMPC oriented bilayers (black circles) and DMPC/DHPC bicelles (red triangles). Positions 3 and 21 were left out of both analyses. To see this figure in color, go online.
To further clarify the helix properties in bilayer and bicelle samples, we performed a combined Gaussian analysis (17) using the 15N chemical shifts and 15N/1H dipolar couplings for bicelle samples along with the 2H |Δνq| data recorded for either bicelles or oriented samples. The combined analysis (using 17 data points) for GW4,20ALP23 in DMPC bilayers (with 15N bicelle data) predicts a similar tilt and rotation to that of the bilayer 2H Gaussian data set alone and a lower σρ of ∼42° (Table 5). An additional combined analysis using the 2H and 15N bicelle data for GW4,20ALP23 (also 17 data points) reveals a similar tilt τ0-value (6°) as the analysis of the smaller 2H data set alone (eight data points; Table 4). When comparing the bicelle and bilayer results using either the 2H data alone or all available data points in the combined analysis (Table 5), the prediction for ρ0 seems to differ by ∼10° between bicelle and oriented samples, and σρ is calculated to be ∼20° higher for the helix in bicelles (62 vs. 42° for bilayers). It is possible that the minor variations with respect to helix azimuthal rotation result from differences between bicelle and lipid bilayer curvature and lipid composition (see Discussion).
To illustrate the key links between helix dynamics and Trp radial locations, the RMSD fits for the Gaussian and GALA analyses as functions of τ0 vs. ρ0 and στ vs. σρ for GW4,20ALP23 and GW5,19ALP23 in DMPC/DHoPC bicelles are shown and compared in Fig. S7. Both the GALA and Gaussian methods generate an acceptable solution area for GW5,19ALP23 and agree on both τ0 and ρ0. The GALA analysis of GW4,20ALP23 reveals a larger range of solutions with comparable probabilities for the peptide helix adopting orientations over a range of ρ0-values at low values of τ0. The Gaussian refines this distribution about ρ0 somewhat, yet the similarly low average values for τ0 are predicted. Furthermore, examination of the fits for στ and σρ (see Fig. S7, top) reveals that GW4,20ALP23 generates the lowest RMSD values at relatively high values of στ (∼17°) and moderate values of σρ, indicating much more extensive motional averaging when the Trp radial locations are moved ±100° from sequence positions (5,19) to (4,20).
Discussion
The interfacial tryptophan residues present in many membrane proteins merit detailed investigations because of their profound effects on the structure and dynamics of transmembrane helices, which are the dominant structural element of the helical class of membrane proteins. GWALP-like peptides serve as effective models for the roles of interfacial tryptophan residues that are tractable experimental systems whose sequences can be readily altered, enabling them to provide insights into how membrane proteins are situated and moving within lipid membrane environments.
The positioning of each particular tryptophan residue at the lipid-water interface may adjust the alignment of the parent helix to minimize hydrophobic mismatch between peptide length and lipid thickness (45) and may also maximize favorable interactions with the lipid head groups. This type of “anchoring” behavior along with possible “fraying” of the helix termini (25) is responsible for the relatively modest dynamic averaging and the well-defined helix tilt of GW5,19ALP23, a tilt that indeed increases systematically whereas lipid thickness decreases (26). Nevertheless, when multiple tryptophan residues are present, as seen in the WALP peptides (11, 12, 13), competition may occur between nearby aromatic residues as each one seeks an optimal position at the interface. Such a molecular “tug of war” could contribute to the extensive motional averaging of the transmembrane helix that is observed (15, 44). The helix motion is particularly evident through rotational averaging of the axis of the tilted helix (15, 17, 43). It should be noted that this phenomenon is not restricted to tryptophan residues. Although the replacement of W5 by Y5 in Y5GW19ALP23 resulted in peptide dynamics similar to the original GW5,19ALP23, an additional Tyr residue in Y4,5GW19ALP23 (18) nevertheless led to additional motional averaging, which was reminiscent of the properties of the WALP and other peptides that have more than two interfacial Trp residues. The extent of averaging was much more modest when Y4 and Y5 were replaced with F4 and F5, likely because of the absence of hydrogen-bonding ability of the phenyl rings of F4 and F5 and, consequently, a lower tendency to “seek” interactions with the lipid head groups (19). Replacing these two aromatic side chains by methyl groups in A4,5GW19ALP23 surprisingly also resulted in a stabilized transmembrane helix experiencing low dynamic averaging (25). The complete lack of any side chain with hydrogen-bonding potential near the N-terminal in both F4,5GW19ALP23 and A4,5GW19ALP23 raised questions concerning how a well-defined tilted transmembrane helix orientation could be stabilized and maintained. The unfolding of helix termini may suggest an answer; exposure of backbone groups caused by the unwinding of 3–4 residues at the N- and/or C-termini could serve to stabilize each peptide helix in a preferred transmembrane orientation and minimize the local motions (25). The potential link between helix fraying and orientational stability of a helix in a phospholipid bilayer environment may provide a bridge between the studies of model systems, such as the WALP and GWALP peptides, and membrane proteins with multiple transmembrane helices.
In these studies, we again find that the helix termini are partially unwound, as seen with the anomalous quadrupolar splittings of residues 3 and 21 in Figs. 7 C and 9. Partial helix unraveling characterized by such a pattern has also been observed in the absence of a terminal Trp residues in A4,5GW19ALP23 (25), and therefore, the quadrupolar splittings of alanines 3 and 21 are not a result of any interference with an adjacent Trp. Even with the partial unwinding present at the helix termini, relocation of the juxtaterminal tryptophan residues from positions W5 and W19 in GW5,19ALP23 to W4 and W20 is sufficient to introduce additional motion into the system. Indeed, GW4,20ALP23 experiences comparable dynamic averaging to that of Y4,5GW19ALP23 (18), with high values of σρ observed in both DLPC and DOPC bilayers. This extensive azimuthal averaging about the helix axis is also responsible for the apparent mismatch of τ0-values between the semistatic and Gaussian analyses. One factor that may influence the azimuthal averaging is the radial positioning of particular interfacial tryptophan residues, which more specifically are located on opposite sides of the GW4,20ALP23 α-helix. This arrangement is similar to that of W2 and W22 found within the highly dynamic peptides W2,22W5,19ALP23 and W2,3,21,22ALP23 (Fig. 1). Although competition between tryptophan residues oriented on opposite helix faces could play a major role in determining the helix dynamics, it is not the only factor to consider. For example, compared to GW5,19ALP23, GW4,20ALP23 lacks leucine residues on either side of the terminal tryptophans such that W4 and W20 have Ala neighbors instead of Leu neighbors, and this may influence the dynamics. GW4,20ALP23 additionally contains a longer hydrophobic core than that of GW5,19ALP23, consisting of two additional residues. The length of the hydrophobic core varies for each face of the helix. After considering both the core length and the competition between W4 and W20, a high “slippage” about the helix axis is likely the most facile way in which the peptide can compensate and minimize hydrophobic mismatch. Therefore, the extensive motional averaging of GW4,20ALP23 observed in DLPC and DOPC bilayers is not surprising. However, the lack of additional motion when the helix is incorporated into DMPC bilayers is unexpected.
Notably, GW4,20ALP23 is seemingly well-behaved with only modest dynamics about the helix average rotation in DMPC bilayers. The core helix adopts a small tilt angle τ0 of 3–5° as determined by both the GALA and Gaussian analysis methods while maintaining a σρ of ∼50°, a value that is comparable to that observed for GW5,19ALP23 in DMPC, DLPC, and DOPC bilayers (Table 4). This behavior of GW4,20ALP23 is different from that of Y4,5GW19ALP23, which retains its high level of dynamics about the helical axis in all three lipids. Additionally, in DMPC bilayers, the lowest RMSD values are fitted to higher values of στ, which suggests that the helical wobble may have an impact. DLPC and DOPC bilayers have hydrophobic thicknesses (excluding the head group region) of 20.8 and ∼26 Å, respectively, at 50°C (46, 47) and may match the hydrophobic length of GW4,20ALP23 less well than does DMPC. A DMPC bilayer of 24.8 Å (at 50°C) (46) is of similar thickness to the intertryptophan distance of 24 Å for the 16 residues in the α-helix between W4 and W20 in GW4,20ALP23. It is plausible, therefore, that DMPC may be an optimal environment for GW4,20ALP23 to adopt a “stabilized” orientation with wobbling about the membrane normal over a low tilt angle without the need for additional motions about the peptide average rotation to further minimize hydrophobic mismatch. The substantial unwinding of the helix at residues 3 and 21 (Fig. 7) confirms a maximal length for the core helix. The behavior of GW4,20ALP23 in DMPC/DHPC bicelles, although arguably somewhat more dynamic, further supports these concepts.
Data points for peptides in DMPC/DHPC bicelles and DMPC bilayers have been used interchangeably in the past because of the almost indistinguishable results when the sample types are compared using either the semistatic or Gaussian analyses (17). Although the quadrupolar wave plots generated by both analyses are similar (Fig. 9), we find that GW4,20ALP23 nevertheless is consistently found to adopt a slightly different preferred rotation corresponding to a 10° difference in ρ0 in bicelles compared to DMPC bilayer samples. The Gaussian analysis agrees with this difference in rotation and furthermore finds a higher rotational distribution reflected by σρ for the peptide within a bicelle environment. The topology of the mixed lipids found in bicelles is somewhat different from that of DMPC bilayers and may play a role in the different rotational minimum and higher level of dynamic averaging that is observed for the helix in bicelles. A bicelle’s discoid shape is composed of a long-chained DMPC lipid bilayer surrounded by a curved edge formed by a short-chained DHPC lipid assembly that protects the longer lipids from the surrounding solvent (48). Hydration is also somewhat higher for bicelle samples and thus can increase the membrane fluidity. The σρ calculated for GW4,20ALP23 in bicelle samples is in between the value calculated in DMPC bilayers and the extensive motion calculated for both DLPC and DOPC bilayers. The size of the 15N PISA wheel (Fig. 6) agrees with the presence of additional motional averaging for the peptide in bicelles, as the signals partially collapse toward a single locus, a behavior also exhibited by the signals from the highly dynamic W2,22W5,19ALP23 helix (Fig. 6). One possibility is that the more fluid bicelle environment may impact the GW4,20ALP23 helix to induce more extensive rotational oscillations (larger σρ) to compensate. The edges of the bicelle at which the DMPC lipids meet the shorter DHPC lipids may also impact the helix properties, which would be seen as a system average. For example, an averaging between central and outer populations of the peptide would influence the NMR spectra. We also note that the τ0 calculated by the combined Gaussian analysis of the complete bicelle data set does not deviate from the semistatic analysis, whereas σρ remains relatively high. The overall results suggest somewhat increased dynamics about the peptide’s average rotation for GW4,20ALP23 in bicelles versus bilayer samples.
Previously, it was reported that transmembrane peptides with low τ0 usually can be fitted with multiple solutions (17) and that by increasing the number of observed 2H or 15N restraints, the array of possible solutions could be significantly decreased. The large pool of data points and interesting dynamics of the GW4,20ALP23 system provide an opportunity to examine how a full Gaussian analysis, using only 2H |Δνq| values, compares to a combined Gaussian analysis that incorporates both the 2H and 15N data sets. For peptides such as GW5,19ALP23, the six data points available from the CD3 side chains of the six 2H-labeled core alanine residues are not sufficient for reliable solutions for the four variables involved in a full Gaussian analysis. If quadrupolar splittings can be observed from some of the backbone Cα deuterons of the labeled alanines, the resulting larger data set can sometimes suffice for a full Gaussian analysis (49). In the absence of additional data points, a three-parameter modified Gaussian analysis proves to be useful. The modified analysis may be implemented by setting στ to a small finite value and varying σρ, ρ0, and τ0 (13, 19). For well-behaved peptides that adopt a relatively large τ0 in lipid bilayers, such as GW5,19ALP23, the limited data set is less of an issue, as the solutions calculated by the modified analysis tend to agree with a semistatic GALA treatment and predict similar dynamics (see Table 6). The limitations of the modified Gaussian calculation can be highlighted when examining all eight 2H data points from the core alanines of GW4,20ALP23 in DMPC bilayers and comparing these results to those of the full Gaussian calculation over the same data set. The solutions calculated by the modified Gaussian analysis may overestimate the peptide dynamics, predicting somewhat higher values for both τ0 and σρ with similar RMSD values (Table 6). The full Gaussian calculation for GW4,20ALP23 in DMPC narrows the solution range to lower values of τ0 and σρ, and provides a value of 20° for στ while also giving a better RMSD fit. Therefore, in this case, minor limitations to the modified Gaussian analysis are revealed, and the advantage of eight data points allows access to the full calculation and a narrower range of solutions for GW4,20ALP23. Notably, the ρ0- and σρ-values obtained from the combined Gaussian analyses, utilizing bicelle data alone or bilayer 2H data and bicelle 15N data, agree with the dynamic properties predicted by full Gaussian analysis (Table 5) and suggest a lower extent of slippage about the helix axis in DMPC bilayer samples compared to bicelles. Although the combined analysis does not resolve the minor discrepancies in ρ0 and σρ observed between the bilayer and bicelle samples, its overall agreement with the full Gaussian calculation further indicates the advantages of a larger experimental data set when dealing with dynamic transmembrane peptide systems such as GW4,20ALP23.
Table 6.
Comparison of Full versus Modified Gaussian Analyses of GWALP23 Family Peptides in DMPC Bilayers Using 2H-Quadrupolar Splittings
| Modifieda |
GW4,20ALP23 (8 Data Points) |
GW5,19ALP23 (6 Data Points) |
||||||
|---|---|---|---|---|---|---|---|---|
| (Fixed) στ | τ0 | ρ0 | σρ | RMSD (kHz) | τ0 | ρ0 | σρ | RMSD (kHz) |
| 5° | 21° | 346° | 148° | 0.98 | 13° | 308° | 44° | 1.1 |
| 10° | 19° | 346° | 138° | 0.98 | 13° | 308° | 42° | 1.2 |
| 15° | 4.0° | 344° | 42° | 0.96 | 16° | 307° | 51° | 1.6 |
| 20° | 5.0° | 348° | 51° | 0.68 | 18° | 306° | 51° | 2.3 |
| Fullb | τ0 | ρ0 | σρ | RMSD (kHz) | ||||
| 5.0° | 347° | 51° | 0.68 | |||||
The radial locations of particular Trp residues about the helix principal axis influence transmembrane helix dynamics. We have performed multiple analyses incorporating 2H NMR and 15N NMR data from solid-state NMR experiments to determine the effects of relocating interfacial tryptophan residues 5 and 19 outward and to opposing faces of the GW5,19ALP23 framework to positions 4 and 20. The extent of motion around the peptide helix axis then becomes quite high in both DLPC and DOPC bilayers and intermediate in DMPC/DHPC bicelles while remaining moderate in DMPC bilayers. An added benefit of having eight core alanine residues available for 2H labeling in the GW4,20ALP23 framework is the opportunity to perform calculations that reveal the complex behavior of this transmembrane helix. In summary, the numbers and precise locations of tryptophan residues are both important determinants of the helix dynamics.
Author Contributions
All authors designed the research. M.J.M., A.N.M., A.A.D.A., and D.V.G. performed the experiments. All authors analyzed the data and wrote the manuscript.
Acknowledgments
This work was supported in part by National Science Foundation Molecular and Cellular Biosciences grant 1713242 and by the Arkansas Biosciences Institute. The peptide, NMR, and mass spectrometry facilities at the University of Arkansas are supported in part by National Institutes of Health (NIH) grants GM103429 and GM103450. The Biotechnology Research Center for NMR Molecular Imaging of Proteins at the University of California, San Diego is supported by NIH grant P41 EB002031, and research on membrane-associated peptides and proteins was supported by NIH grant R35 GM122501.
Editor: Michael Brown.
Footnotes
Seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30461-2.
Supporting Material
References
- 1.Yau W.M., Wimley W.C., White S.H. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 2.Koeppe R.E., II, Anderson O.S. Engineering the gramicidin channel. Annu. Rev. Biophys. Biomol. Struct. 1996;25:231–258. doi: 10.1146/annurev.bb.25.060196.001311. [DOI] [PubMed] [Google Scholar]
- 3.O’Connell A.M., Koeppe R.E., II, Andersen O.S. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 1990;250:1256–1259. doi: 10.1126/science.1700867. [DOI] [PubMed] [Google Scholar]
- 4.Gu H., Lum K., Koeppe R.E., II The membrane interface dictates different anchor roles for “inner pair” and “outer pair” tryptophan indole rings in gramicidin A channels. Biochemistry. 2011;50:4855–4866. doi: 10.1021/bi200136e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKay M.J., Afrose F., Greathouse D.V. Helix formation and stability in membranes. Biochim. Biophys. Acta. 2018 doi: 10.1016/j.bbamem.2018.02.010. Published online February 13, 2018. [DOI] [PubMed] [Google Scholar]
- 6.Situ A.J., Kang S.M., Ulmer T.S. Membrane anchoring of α-helical proteins: role of tryptophan. J. Phys. Chem. B. 2018;122:1185–1194. doi: 10.1021/acs.jpcb.7b11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inda M.E., Oliveira R.G., Cybulski L.E. The single transmembrane segment of minimal sensor DesK senses temperature via a membrane-thickness caliper. J. Bacteriol. 2016;198:2945–2954. doi: 10.1128/JB.00431-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baradaran R., Berrisford J.M., Sazanov L.A. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersen O.S., Koeppe R.E., II Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 10.Thibado J.K., Martfeld A.N., Koeppe R.E., II Influence of high pH and cholesterol on single arginine-containing transmembrane peptide helices. Biochemistry. 2016;55:6337–6343. doi: 10.1021/acs.biochem.6b00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Killian J.A., Salemink I., Greathouse D.V. Induction of nonbilayer structures in diacylphosphatidylcholine model membranes by transmembrane alpha-helical peptides: importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry. 1996;35:1037–1045. doi: 10.1021/bi9519258. [DOI] [PubMed] [Google Scholar]
- 12.van der Wel P.C., Strandberg E., Koeppe R.E., II Geometry and intrinsic tilt of a tryptophan-anchored transmembrane alpha-helix determined by (2)H NMR. Biophys. J. 2002;83:1479–1488. doi: 10.1016/S0006-3495(02)73918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strandberg E., Özdirekcan S., Killian J.A. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys. J. 2004;86:3709–3721. doi: 10.1529/biophysj.103.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J., Im W. Transmembrane helix tilting: insights from calculating the potential of mean force. Phys. Rev. Lett. 2008;100:018103. doi: 10.1103/PhysRevLett.100.018103. [DOI] [PubMed] [Google Scholar]
- 15.Ozdirekcan S., Etchebest C., Fuchs P.F. On the orientation of a designed transmembrane peptide: toward the right tilt angle? J. Am. Chem. Soc. 2007;129:15174–15181. doi: 10.1021/ja073784q. [DOI] [PubMed] [Google Scholar]
- 16.Strandberg E., Esteban-Martín S., Ulrich A.S. Orientation and dynamics of peptides in membranes calculated from 2H-NMR data. Biophys. J. 2009;96:3223–3232. doi: 10.1016/j.bpj.2009.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vostrikov V.V., Grant C.V., Koeppe R.E., II On the combined analysis of 2H and 15N/1H solid-state NMR data for determination of transmembrane peptide orientation and dynamics. Biophys. J. 2011;101:2939–2947. doi: 10.1016/j.bpj.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleason N.J., Vostrikov V.V., Koeppe R.E., II Tyrosine replacing tryptophan as an anchor in GWALP peptides. Biochemistry. 2012;51:2044–2053. doi: 10.1021/bi201732e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sparks K.A., Gleason N.J., Koeppe R.E., II Comparisons of interfacial Phe, Tyr, and Trp residues as determinants of orientation and dynamics for GWALP transmembrane peptides. Biochemistry. 2014;53:3637–3645. doi: 10.1021/bi500439x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Planque M.R., Bonev B.B., Killian J.A. Interfacial anchor properties of tryptophan residues in transmembrane peptides can dominate over hydrophobic matching effects in peptide-lipid interactions. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 21.de Planque M.R., Kruijtzer J.A., Killian J.A. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J. Biol. Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 22.de Planque M.R., Boots J.W., Killian J.A. The effects of hydrophobic mismatch between phosphatidylcholine bilayers and transmembrane alpha-helical peptides depend on the nature of interfacially exposed aromatic and charged residues. Biochemistry. 2002;41:8396–8404. doi: 10.1021/bi0257686. [DOI] [PubMed] [Google Scholar]
- 23.Vostrikov V.V., Grant C.V., Koeppe R.E., II Comparison of “Polarization inversion with spin exchange at magic angle” and “geometric analysis of labeled alanines” methods for transmembrane helix alignment. J. Am. Chem. Soc. 2008;130:12584–12585. doi: 10.1021/ja803734k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vostrikov V.V., Hall B.A., Sansom M.S. Changes in transmembrane helix alignment by arginine residues revealed by solid-state NMR experiments and coarse-grained MD simulations. J. Am. Chem. Soc. 2010;132:5803–5811. doi: 10.1021/ja100598e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortazavi A., Rajagopalan V., Koeppe R.E., II Juxta-terminal helix unwinding as a stabilizing factor to modulate the dynamics of transmembrane helices. ChemBioChem. 2016;17:462–465. doi: 10.1002/cbic.201500656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vostrikov V.V., Daily A.E., Koeppe R.E., II Charged or aromatic anchor residue dependence of transmembrane peptide tilt. J. Biol. Chem. 2010;285:31723–31730. doi: 10.1074/jbc.M110.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vostrikov V.V., Koeppe R.E., II Response of GWALP transmembrane peptides to changes in the tryptophan anchor positions. Biochemistry. 2011;50:7522–7535. doi: 10.1021/bi2006459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strandberg E., Esteban-Martín S., Salgado J. Hydrophobic mismatch of mobile transmembrane helices: merging theory and experiments. Biochim. Biophys. Acta. 2012;1818:1242–1249. doi: 10.1016/j.bbamem.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 29.Thomas R., Vostrikov V.V., Koeppe R.E., II Influence of proline upon the folding and geometry of the WALP19 transmembrane peptide. Biochemistry. 2009;48:11883–11891. doi: 10.1021/bi9016395. [DOI] [PubMed] [Google Scholar]
- 30.Davis J.H., Jeffrey K.R., Higgs T.P. Quadrupolar echo deuteron magnetic-resonance spectroscopy in ordered hydrocarbon chains. Chem. Phys. Lett. 1976;42:390–394. [Google Scholar]
- 31.Grant C.V., Yang Y., Opella S.J. A modified Alderman-Grant coil makes possible an efficient cross-coil probe for high field solid-state NMR of lossy biological samples. J. Magn. Reson. 2009;201:87–92. doi: 10.1016/j.jmr.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Wel P.C., Reed N.D., Koeppe R.E., II Orientation and motion of tryptophan interfacial anchors in membrane-spanning peptides. Biochemistry. 2007;46:7514–7524. doi: 10.1021/bi700082v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nevzorov A.A., Opella S.J. Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. J. Magn. Reson. 2007;185:59–70. doi: 10.1016/j.jmr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Levitt M.H., Suter D., Ernst R.R. Spin dynamics and thermodynamics in solid-state NMR cross polarization. J. Chem. Phys. 1986;84:4243–4255. [Google Scholar]
- 35.Fung B.M., Khitrin A.K., Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 36.Sinha N., Grant C.V., Opella S.J. Peptides and the development of double- and triple-resonance solid-state NMR of aligned samples. J. Pept. Res. 2005;65:605–620. doi: 10.1111/j.1399-3011.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 37.De Angelis A.A., Howell S.C., Opella S.J. Structure determination of a membrane protein with two trans-membrane helices in aligned phospholipid bicelles by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2006;128:12256–12267. doi: 10.1021/ja063640w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 39.Goddard, T. D., and D. G. Kneller. SPARKY 3: University of California, San Francisco, CA.
- 40.Wishart D.S., Bigam C.G., Sykes B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 41.Page R.C., Kim S., Cross T.A. Transmembrane helix uniformity examined by spectral mapping of torsion angles. Structure. 2008;16:787–797. doi: 10.1016/j.str.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S., Cross T.A. Uniformity, ideality, and hydrogen bonds in transmembrane alpha-helices. Biophys. J. 2002;83:2084–2095. doi: 10.1016/S0006-3495(02)73969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Esteban-Martín S., Strandberg E., Ulrich A.S. Solid state NMR analysis of peptides in membranes: influence of dynamics and labeling scheme. Biochim. Biophys. Acta. 2010;1798:252–257. doi: 10.1016/j.bbamem.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 44.Esteban-Martín S., Salgado J. Self-assembling of peptide/membrane complexes by atomistic molecular dynamics simulations. Biophys. J. 2007;92:903–912. doi: 10.1529/biophysj.106.093013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Killian J.A., Nyholm T.K. Peptides in lipid bilayers: the power of simple models. Curr. Opin. Struct. Biol. 2006;16:473–479. doi: 10.1016/j.sbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Kučerka N., Nieh M.P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 47.Pan J., Tristram-Nagle S., Nagle J.F. Temperature dependence of structure, bending rigidity, and bilayer interactions of dioleoylphosphatidylcholine bilayers. Biophys. J. 2008;94:117–124. doi: 10.1529/biophysj.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanders C.R., Prosser R.S. Bicelles: a model membrane system for all seasons? Structure. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 49.Vostrikov V.V., Hall B.A., Koeppe R.E., II Accommodation of a central arginine in a transmembrane peptide by changing the placement of anchor residues. J. Phys. Chem. B. 2012;116:12980–12990. doi: 10.1021/jp308182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.