Abstract
In eukaryotic chromatin, islands of histone tail acetylation are found near transcription start sites and enhancers, whereas linker histones (LHs) are localized in intergenic regions with wild-type (WT) histone tails. However, the structural mechanisms by which acetylation, in combination with LH binding, modulates chromatin compaction and hence transcription regulation are unknown. To explore the folding propensity by which these features may govern gene expression, we analyze 20 kb fibers that contain regularly spaced acetylation islands of two sizes (2 or 5 kb) with various LH levels by mesoscale modeling. Specifically, we investigate the effect of acetylating each histone tail (H3, H4, H2A, and H2B) individually, in combination (H3 and H4, or all tails), and adding LH to WT regions. We find that fibers with acetylated H4 tails lose local contacts (<1 kb) and fibers with all tails acetylated have decreased long-range contacts in those regions. Tail interaction plots show that this opening of the fiber is driven by the loss of tail-tail interactions in favor of tail-parent core interactions and/or increase in free tails. When adding LH to WT regions, the fibers undergo hierarchical looping, enriching long-range contacts between WT and acetylated domains. For reference, adding LH to the entire fiber results in local condensation and loss of overall long-range contacts. These findings highlight the cooperation between histone tail acetylation and regulatory proteins like LH in directing folding and structural heterogeneity of chromatin fibers. The results advance our understanding of chromatin contact domains, which represent a pivotal part of the cell cycle, diseased states, and differentiation states in eukaryotic cells.
Introduction
Chromatin, the DNA/protein complex polymer that stores genetic information in eukaryotic organisms, plays many roles in control of gene expression. At the most basic level, ∼147 base pairs wrap ∼1.75 times around eight core histone proteins (two copies each of H2A, H2B, H3, and H4) to form the nucleosome core particle (1). Each histone protein within the nucleosome contains a segment of intrinsically disordered protein at the N-terminal end that extends away from the nucleosome into the adjoining solvent (2). These histone tails can be chemically modified in a myriad of ways during transcription, often changing the intrinsic dynamic nature of each tail. Nucleosomes are regularly spaced along the genome of eukaryotic organisms and connected to one another by linker DNA. Such chains of DNA wrapped around nucleosomes can be visualized using microscopy techniques. At low salt, such nucleosome chains resemble irregular “beads-on-a-string” arrays (3, 4). At physiological salt, such chains undergo additional condensation to form secondary structures, the details of which are not precisely known (5). Although chromatin fibers reconstituted in vitro using DNA sequences designed specifically to stabilize nucleosome formation lead to straight fibers with a well-defined 30-nm radius (6), such evidence is more difficult to find in vivo (7, 8, 9), and state-of-the-art electron microscopy images instead report fibers with variable diameters that range from 5 to 25 nm (8, 10, 11). The condensation/decondensation of these various chromatin fiber secondary structures depends subtly on auxiliary proteins like linker histone (LH), remodeling proteins, and epigenetic modulation.
LH proteins are essential components of chromatin in eukaryotic cells. They are recognized to be important for the stability of higher-order chromatin structure and are usually associated with repressed chromatin (12, 13, 14, 15, 16). LH is composed of three domains: a small neutrally charged N-terminal domain, a well-folded globular head that binds to the major groove of nucleosomal DNA within the entry/exit dyad (13), and a long, positively charged C-terminal domain (CTD) thought to be crucial to chromatin structure. The CTD is intrinsically disordered, similar to the nucleosomal histone tails (17). The CTD binds linker DNA in a salt-dependent manner, in which low salt favors a bidente configuration (CTD binds only one strand of linker DNA) and physiological salt encourages a tridente configuration (CTD binds both exiting/entering DNA) (18, 19). Similarly, symmetric as well as off-symmetric binding motifs are now recognized in crystallographic studies (20). LH proteins are known to bind transiently, residing on a specific nucleosome for seconds to minutes before dissociating into the solvent (21). The relatively short residence time makes the study of LH difficult in vivo, but preliminary studies show that LH localizes in specific regions of chromatin fibers, often anticorrelated with specific posttranslational modifications (PTMs) such as acetylation (22). Here we use the term condense or decondense to refer to changes in linear compaction of the chromatin fiber, such as we observe when LH is added (condensation) or all tails are acetylated (decondensation). Long-range contacts refer to interactions between nonadjacent nucleosomes at least 1 kb apart.
PTMs often mark for genetically active or inactive chromatin (23) and are associated with many human diseases (24). However, the structural mechanisms by which PTMs control transcription, what their interplay is with LH, and to what degree they locally decondense chromatin are not well understood. Acetylation involves the addition of an acetyl group, which contains a negatively charged oxygen, to a lysine residue of protein side chains. This oxygen can then be a receptor for hydrogen bonding of other protein side chains, increasing the possibility of the protein forming well-folded secondary structures. Very broadly, acetylation marks are associated with active chromatin and often cluster in regions near transcriptional start sites or enhancer sites, forming small islands that range from 1 to 5 kb in size (25, 26). In particular, in vitro work on reconstituted chromatin fibers has demonstrated the effects of tail acetylation on fiber compaction. Acetylation of reconstituted chromatin fibers at H4K16 (i.e., Lysine residue 16 on tail H4) leads to a lower sedimentation coefficient than nonacetylated fibers, suggesting a significantly less compact fiber (27). This has been corroborated by electron-microscopy-based images of these fibers, in which H4K16Ac fibers are visibly less compact (6, 28), and by all-atom molecular dynamics simulations of tail/nucleosome interactions (29, 30). In these simulations, acetylated tails exhibit prolonged residence times of local protein secondary structures (29, 30) and fewer interactions with nonparental nucleosomes through association with an acidic patch near the center of the surface of each nucleosome (30, 31).
Thus, H4K16ac decondenses chromatin fiber conformations globally by the loss of crucial internucleosome interactions rather than by charge modulation per se. The multiscale mechanism described in (30) involves more rigid tails in the acetylated regions that decrease interactions between nucleosomes, which in turn destabilize and unfold the chromatin fiber globally. Atomic modeling of acetylation on other histone tails shows a similar increase in propensity for those tails to fold into defined secondary structures, and incorporating them into a mesoscale model results in the dramatic decrease of local fiber contacts, in line with experimental results (30). Such roles as epigenetic markers for gene activation are also suggested by common histone acetylases, such as CREB binding protein or the closely related P300 histone acetylase, that acetylate all four histone tail types (23). Furthermore, such decondensation mechanisms can work cooperatively with chromatin domain aggregation, as we showed recently in (32). Specifically, we showed that alternating wild-type (WT)/acetylated fibers lead to distinct aggregation patterns such as those seen by chromatin conformation capture techniques (Hi-C) without protein cofactors (32). These intrinsic folding and segregation trends fit well with the observation that regions of active and inactive gene expression tend to spontaneously cosegregate in living tissue, where histone tail acetylations are associated with active chromatin contact domains (33).
Other PTM marks on reconstituted chromatin fibers do not show significant effects on fiber compaction directly in vitro, but some act to structurally affect fibers through secondary mechanisms in vivo. For example, H3K27me3 compacts chromatin in living systems through recruitment and expression of heterochromatin protein 1 and LH (23, 34), which is known to help condense chromatin fibers in vitro by other mechanisms (28). However, LH can in some cases promote transcription despite not being found in gene encoding regions or near transcription start sites (15, 22).
An outstanding question, therefore, concerns whether LH proteins and epigenetic marks act together to direct chromatin fiber folding, in particular with regards to long-range nucleosome interactions that are important in cellular signaling. To that end, we present mesoscale simulations of 20 kb chromatin fragments with a nucleosome repeat length of 200 bp and regularly spaced acetylation islands separated by WT histone tails. The islands are either 5 kb (25 nucleosomes) or 2 kb (10 nucleosomes) long. We call these alternating constructs AC1 and AC2, respectively. We systematically investigate the cumulative effects of acetylating each histone tail individually and in combination in AC1 fibers and the role of LH in both fibers. First, regarding selective acetylations, we find that although acetylating histone tails H3, H2A, or H2B individually does not significantly affect the overall fiber folding, acetylating the H4 tails alone diminishes local contacts (<1 kb) in a dramatic decondensation. Acetylating all tail types simultaneously has the strongest affect on disrupting long-range contacts (≥1 kb) between distal acetylated regions. Our tail-contact probabilities indicate that although WT regions naturally aggregate because of tail-tail interactions, acetylated regions are more open because of a loss of tail-tail interactions and gain of tail-parent nucleosome interactions and/or free tails.
Second, regarding the interaction of LH and acetylated islands, we present the effects of adding LH either uniformly (+LH) or specifically to WT regions (wtLH) in AC1 and AC2 fibers. Although the uniform addition of LH throughout the fiber removes all long-range contacts because of structural condensation, unexpectedly, the addition of LH in the WT regions encourages long-range contacts between regions with LH and acetylated histone tails. In summary, our results highlight the cooperative role that both LH and acetylation islands play in folding and unfolding global chromatin structures. Both factors, when applied individually, unfold the fiber, but together, when placed strategically, can direct the folding of the fiber.
Methods
Mesoscale model and energy parameters
Our mesoscale chromatin model (35, 36, 37) is composed of four distinct coarse-grained components: linker DNA beads, derived from a worm-like chain polymer treatment; electrostatic partial charges determined by the Discrete Surface Charge Optimization algorithm (38); coarse-grained beads for flexible histone tails (39); and LH beads based on a united all-atom protein subunit model (18). The LH globular head is modeled with eight beads, and the CTD is modeled with 28 beads. Charges and connectivity for the LH model are based on the rat LH variant H1.4 (18). Each coarse-grained object interacts with all other coarse-grained objects through bonded and/or nonbonded interactions governed by the topology of the fiber. Bonded interactions consist of bending and twisting terms with associated parameters for bending rigidity, persistence length, and twisting constant. Nonbonded interactions consist of an excluded-volume term modeled via a Lennard-Jones 10–12 potential and an electrostatic term modeled via a Debye-Hückel approximation. For a full treatment of the model and parameters, see (40).
Chromatin system parameters
We built four systems, each with 100 nucleosomes (∼20 kb) and a nucleosome repeat length of 200 bp, starting from idealized zigzag coordinates (see Fig. 1). The WT system consists of 100 nucleosomes with all-WT tails, and the all-acetylated control consists of all 100 nucleosomes, all with acetylated tails. We also consider two alternating constructs: “AC1,” which has 2× (25 acetylated, 25 WT) cores, and “AC2,” which has 5× (10 acetylated, 10 WT) cores, representing two generalized arrangements of acetylated regions in living chromatin (25). The acetylation in AC1 was considered with each histone individually (H3, H4, H2A, or H2B), H3 and H4 tails combined, or all tails. To test for cooperation between tail acetylation and LH, additional AC1 and AC2 systems were prepared, with LHs placed either uniformly throughout the fiber (+LH) or selectively in regions with WT tails (wtLH) similar to patterns observed in ChIP-Seq data in mouse embryonic stem cells (22). AC1 and AC2 fibers were simulated with 50 independent trajectories, and all other systems were simulated by 24 independent trajectories. All trajectories were simulated for over 40 million Monte Carlo (MC) steps. Internucleosome contacts were calculated and reported every 10,000 steps during the simulation, in which a contact was reported if any two constituents were found to be within 2 nm of one another. Contact maps for each trajectory are normalized by the maximal number of contacts seen throughout the trajectory, and the resulting normalized frequencies are summed together. Considering that the contacts near the diagonal are an order of magnitude larger than those at far ranges, it can be difficult to visualize small differences in intensity for long-range contacts while showing short-range contacts simultaneously. For this reason, we adopt a method similar to those commonly used in presenting Hi-C results. We present contact maps in which the color map (i.e., the Z axis) is truncated by 10% of the maximal Z value, which only effects values of contacts along the diagonal (<1 kb). Internucleosome contact frequencies (presented as one-dimensional contact probabilities) are calculated by combining the final 100 frames (∼10 M steps) of each trajectory. The resulting contact frequencies are calculated across the entire concatenated trajectory and then normalized. Fiber images are rendered from a single representative trajectory.
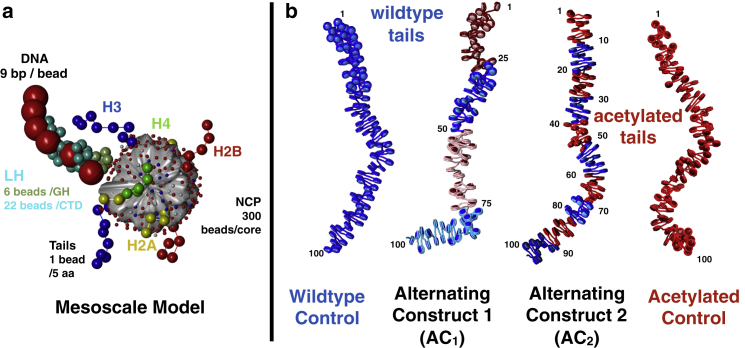
Figure 1.
Chromatin model and fiber systems. (a) Our mesoscale model building block, which consists of linker DNA beads, a rigid nucleosome core with discrete charges, flexible histone tail beads, and flexible LH beads. (b) Starting configurations for four chromatin fiber systems studied with varying levels of acetylation are shown. From left to right: wild-type (WT) system, Alternating Construct 1 (AC1) system consisting of alternating WT (blue) and acetylated regions (magenta or pink) with 25 nucleosome segments, Alternating Construct 2 (AC2) with 10 nucleosome segments, and the all-acetylated control (red). Histone tails are not rendered here for clarity. To see this figure in color, go online.
Sampling methods
Five types of MC moves are implemented for local and global sampling, namely, a global pivot move, a configurationally biased regrow routine to simulate the rapid movement of histone tails (41), a fold-swap move to mimic histone tail acetylation (described below), and local translation and rotation moves. The global pivot move chooses a random position along the fiber and then rotates the shorter section of the bisected chain around a randomly chosen axis running through that point (37); the resulting coordinates are subject to Metropolis acceptance/rejection criteria (42). In the regrow MC move, a chosen tail is “regrown,” starting with the bead closest to the core according to the Rosenbluth method (41, 43). This process is then repeated 10–12 times, and the tail configuration with lowest resulting energy is subject to a Metropolis accept/reject criterion. All DNA and LH beads are also subject to translation/rotation moves with Metropolis accept/reject criteria.
Modeling acetylated histone tails
To model acetylation content in the mesoscale regime, we developed a routine inspired by the procedure of reference (30). In that study, copies of each histone tail were acetylated at known acetylation sites and modeled using several force fields at all-atom resolution with Replica Exchange MD. Coordinates from these runs representing folded tails were chosen and coarse-grained using the united atom coarse-graining approach, and the resulting coordinates are used as “folded” tail coordinates in our mesoscale model.
For our study, we also utilize the folded-tail coordinates and equilibrium values from (30) to represent a highly acetylated tail. To mimic acetylation/deacetylation as a steady state phenomenon, we introduce a “swap-fold” MC move. In this move, tails that are eligible to fold are specified by the user on startup, and during the simulation an eligible tail is chosen at random and its fold state is swapped. In other words, if the chosen tail is currently unfolded, its coordinates and equilibrium values are swapped with those of the folded version of that tail (i.e., a “fold” move), and if the chosen tail is already folded, then the coordinates are swapped for those of the unfolded version of that tail (i.e., an “unfold” move). The new coordinates are then subject to a standard Metropolis MC acceptance/rejection criterion based on the changes in local electrostatic energy. When tails are folded, they do not interact with their parent cores, nor do they interact sterically with one another, and are not subject to the Rosenbluth regrow routine. Throughout the simulation, the total concentration of folded tails is held near a target value by biasing the probability of choosing a “fold” versus “unfold” move. The swap-fold move is attempted on average every 200–400 MC steps, which results in eligible tails folding/unfolding less often than the average frequency of core rotation/translation. Swap-fold frequencies and the rates of tail sampling were adjusted for systems 4× larger than those previously studied in (30), specifically by increasing the frequency of Rosenbluth tail-regrow steps and decreasing the average step size and frequency of global steps. Finally, for all systems, the fold-swap move is only invoked after several million steps of equilibration have been performed.
Results
Fiber conformations in alternating constructs depend on length of acetylation island
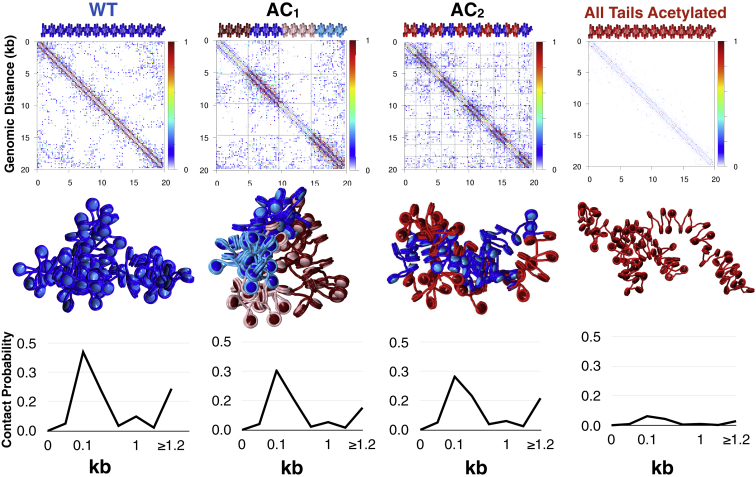
In living systems, acetylation marks are often found in small islands around transcription start sites or enhancer regions, ranging from 1 to 5 kb in size (25). To see how acetylation islands of this size affect long-range structure, we tested two alternating constructs, AC1 and AC2 with 2× (25 acetylated, 25 WT nucleosomes) and 5× (10 acetylated, 10 WT nucleosomes), respectively. The WT control consists of 100 nucleosomes with WT tails, and the “all-acetylated” control consists of 100 nucleosomes with acetylated tails. As we see in Fig. 2, control fibers adopt expected configurations: a loose globule with the hierarchical looping motif we observed for condensed chromatin with WT tails (left) (10), and open structures with few long-range contacts for the fully acetylated fiber (right). Hierarchical loops form when two loops are stacked together in three-dimensional space, leading to a compact, unknotted fiber similar to rope flaking used in mountain climbing (44).
Figure 2.
Probability contact maps, fiber renderings, and short-range internucleosome contact probability profiles for WT, AC1, AC2 and acetylated control fibers (see Fig. 1). Regions of acetylated histone tails (which are more tightly folded) are rendered in red or pink, whereas WT regions are colored blue. To see this figure in color, go online.
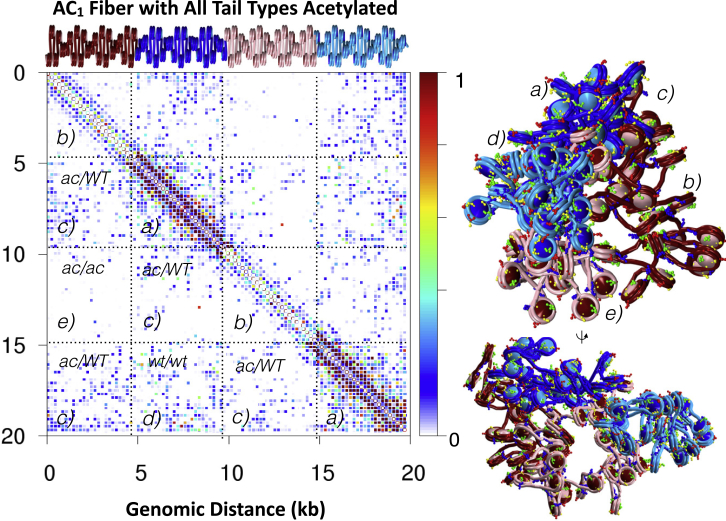
AC1 and AC2 fibers form globules with hierarchical looping, similar to WT fibers. However, acetylation regions are visibly more open. Significantly, in the AC1 fibers, the acetylated nucleosomes (red) spontaneously segregate from WT nucleosomes (blue). AC2 fibers also show aggregation of WT regions, but not aggregation of acetylated regions. The contact probability matrices and internucleosome contact profiles in Fig. 2 reveal the total internal contacts of each system ensemble. Both local and global contacts decrease near acetylated regions, whereas contacts between WT regions dominate long-range contacts. Hierarchical loops are evident in the contact matrices as regions of increased intensity parallel to the main diagonal, as opposed to hairpins, associated with regions perpendicular to the diagonal. In Fig. 3, we annotate the contact map for AC1 to highlight five interaction types: WT/WT local, WT/WT long-range, WT/acetylated, acetylated/acetylated local, and acetylated/acetylated long-range. The fiber rendering shows that these contacts arise from the overall stacking of loops in three-dimensional space.
Figure 3.
Structural analysis of AC1 fibers with all tail types acetylated within the noted 25-nucleosome segment. The contact map (left) is divided into WT/WT local contacts (a), acetylated/acetylated local contacts (b), WT/acetylated contacts (c), WT/WT long-range contacts (d), and acetylated/acetylated long-range contacts (e). The fiber, here rendered with histone tails (right), shows a representative conformation in two views. To see this figure in color, go online.
Acetylation of tail types cooperatively diminishes long-range contacts
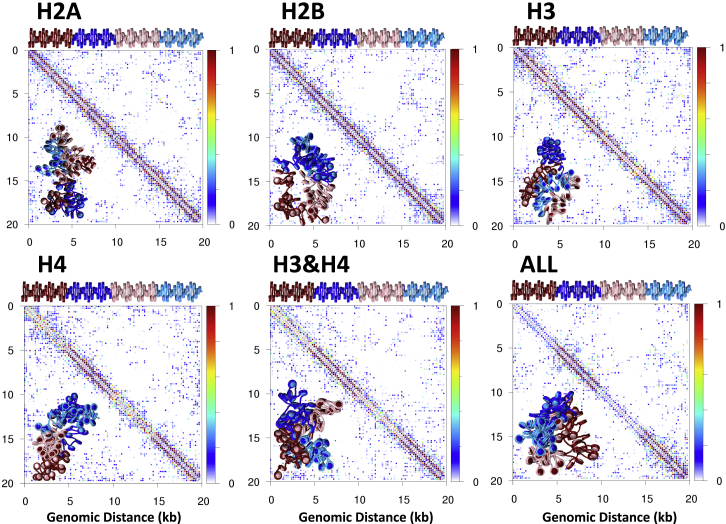
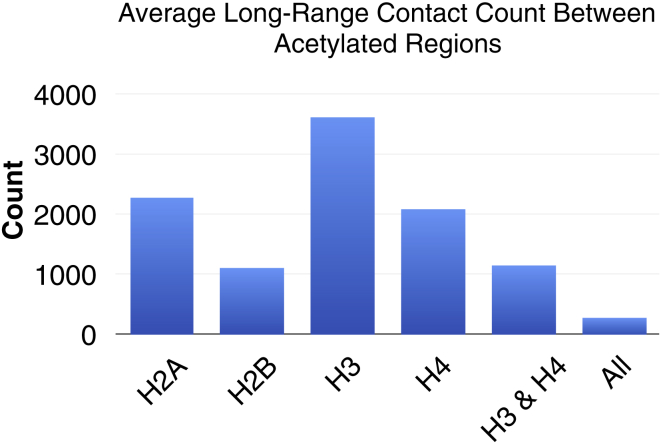
Common histone acetyltransferases such as CREB binding protein or P300 can acetylate all four histone tail types at various locations, but the specific effects of these acetylations on mid-long-range contacts is still poorly understood. Previously we have shown that acetylating all histone tails simultaneously can result in spontaneous segregation of acetylated regions from WT regions (32), but the role of each tail acetylation in this segregation was not explored. Studies performed in vitro show that H4 tail acetylation alone is sufficient for local opening of chromatin fibers. Our series of simulations of individual tail acetylation is designed to dissect the effects of acetylating specific tail types. Fig. 4 shows the contact probability matrices for AC1 fibers (with 25 nucleosome subregions) with H3, H4, H2A, or H2B tails acetylated individually, as well as all tails acetylated in the specified region. Representative fiber renderings are shown in the bottom left of each matrix. The contact maps show that although H4 folding is sufficient to disrupt local contacts near the diagonal, only fibers with all tails acetylated in the specified region disrupt long-range contacts. The ability of the H4 tails to modulate local contacts along the diagonal is in good agreement with the fact that H4 tails interact heavily with the acidic patch of nonparent nucleosomes (45, 46), stabilizing interactions between near-neighbor nucleosomes. This is clearer with the quantitative data in Fig. 5, which shows the total number of long-range interactions between acetylated/acetylated contact regions for each selective tail acetylation. Contact counts show that H3 acetylation has the least effect on long-range contacts, whereas H2A, H2B, H4, and H3 and H4 result in decreased long-range contacts between the islands. Significantly, acetylation of all tail types results in the least number of acetylation/acetylation contacts, showing cooperation between tail types in modulating chromatin structure.
Figure 4.
Fiber systems with various tail segments for acetylation: Probability contact maps and fiber renderings for AC1 with H2A, H2B, or H3 only (top left to right, respectively), and H4, H3 and H4, or all tails acetylated (bottom left to right, respectively). To see this figure in color, go online.
Figure 5.
Average long-range contact count between acetylated/acetylated regions for AC1 fibers with H2A, H2B, H3, and H4, H3 and H4, or all tail types acetylated. A contact is defined when any two elements are within 2 nm of one another in space. To see this figure in color, go online.
Acetylation diminishes tail/tail and tail/nonparent DNA contacts in favor of parental core interactions and free tails
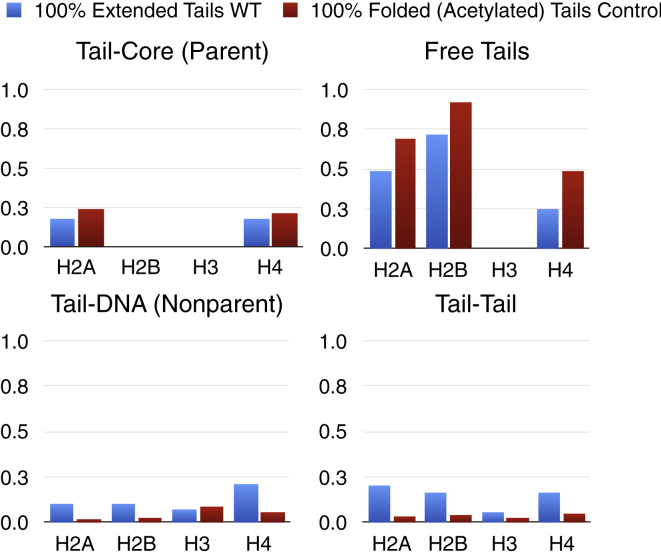
To further investigate the role of each tail type in contact formation, we quantified the tail contact behavior of all structures, counting whether each tail was found in contact with nonparental DNA, the parental nucleosome core, another tail, or free from all contacts. Fig. 6 compares contact frequencies between WT and acetylated control fibers. Acetylated tails show a significant decrease in tail-tail interactions compared to WT fiber, with an increase of free tails and tail-parental core interactions. Thus, we can explain the loss of long-range contacts between acetylated regions of the chromatin fiber through an exchange of tail interaction type, in which acetylation favors free or parental-core interactions and WT tails favor tail-tail interactions. H2A and H2B tails lose the most tail-tail interactions upon acetylation, and H2A and H4 tails gain the most tail-parental core interactions, whereas H2A, H2B, and H4 show the largest increase in free tails.
Figure 6.
Tail-contact analysis for WT (blue) and acetylated control fibers (red). Normalized contact counts are given for tail-tail, tail-parent DNA, free tails, and tail-parent core interactions. A contact is defined when any two elements are within 2 nm of one another in space. To see this figure in color, go online.
LH rescues long-range contacts when bound to acetylated regions
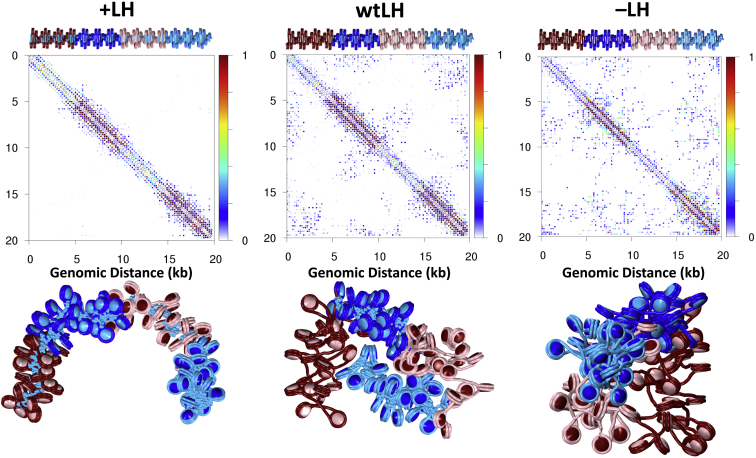
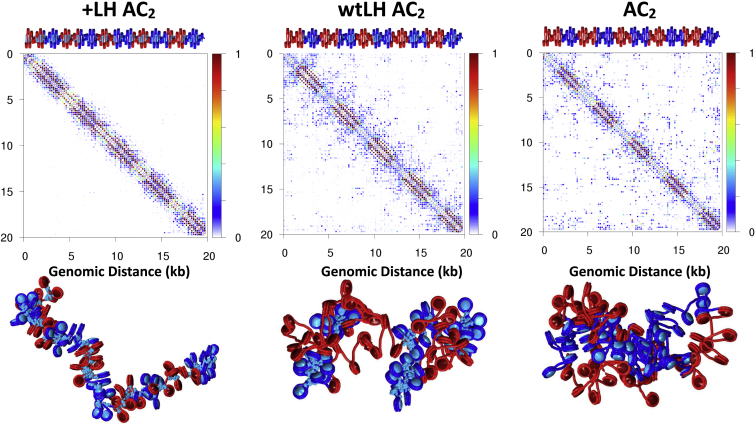
LH is known to bind regions near TSSs and enhancer regions, but the effects of LH binding on chromatin fiber structure near sites of high acetylation is not well understood. To examine the effects of LH near acetylated regions, we added LH either uniformly to the entire fiber (+LH) or selectively to regions with WT tails (wtLH) in both the AC1 and AC2 fibers. As shown in Figs. 7 and 8, +LH AC1 fibers form relatively stiff fibers with few long-range contacts, whereas wtLH AC1 fibers fold into short hairpins that maximize interactions between LH-saturated and acetylated regions. Similarly, contact maps show that uniform LH distribution leads to a loss of long-range contacts (left), whereas selective LH addition to WT tail areas encourages contacts between LH-rich and acetylated regions. In the case of +LH AC2 fibers, saturation of LH similarly results in a straighter fiber, whereas wtLH AC2 fibers form a series of hairpins, maximizing interaction between LH and WT regions.
Figure 7.
Probability contact maps with LH in AC1 fibers and representative conformations. Contact maps (top), and fiber renderings (bottom) for AC1 fibers with all tail types acetylated are shown, with uniformly distributed LH (+LH), with LH placed only in WT regions (wtLH), and no LH (−LH), on the left, middle, and right, respectively. The LH protein, which condenses fibers locally, also acts to attract regions dense with acetylation. To see this figure in color, go online.
Figure 8.
Probability contact maps with LH in AC2 fibers and representative conformations. Contact maps (top), and fiber renderings (bottom) for AC2 fibers with all tail types acetylated are shown, with uniformly distributed LH (+LH), with LH placed only in WT regions (wtLH), and no LH (−LH), on the left, middle, and right, respectively. To see this figure in color, go online.
Discussion
Contact domains in living systems can be divided into two general sizes: topologically associating domains (TADs) that span kilobases and compartment domains that typically span megabases of DNA. Recent work has shown that TAD formation depends on cohesin and operates independently from the formation of compartment domains (32). Compartments with actively transcribed chromatin are also known as A domains, whereas domains with low levels of transcription are referred to as B domains. These compartments form spontaneously without cohesin (as opposed to TADs) and correlate well with certain PTM patterns, prompting many suggestions that PTMs may impart the physical characteristics that lead to segregation/aggregation between active and inactive chromatin compartments (47, 48, 49). Because Hi-C data are typically averaged across millions of cells or lack nucleosome resolution, detailed effects of how these marks may lead to compartment domains are unclear.
Previously, we have shown that regularly repeated acetylation islands in mesoscale simulations of chromatin fibers can result in the loss of local and long-range contacts, resembling aspects of chromatin compartment domains. The typical size of acetylated regions within compartment domains range from 1 to 5 kb (25), but the density and spacing of such islands can vary widely (50). Therefore, both the AC1 and AC2 systems studied here represent realistic representations of possible living systems, but they are far from a complete representation of possible configurations. Of course, more work is needed to tie those features to the larger contact domains observed in living cells (32, 51). Such connections are particularly important when considering techniques that utilize ENCODE ChIP-Seq assays to computationally predict TADs or contact domains based on learning algorithms (52). Although these algorithms may be successful in their predictions, those predictions cannot be used to infer the physical mechanisms involved.
Our studies provide insight into such mechanisms. Here, we show that the structural effects of histone tail acetylation are cumulative across tail type; thus, acetylation of all tail types diminishes long-range contacts most effectively. On the contrary, short-range contacts can be diminished by H4 tail acetylation alone. These findings fit well with results from in vitro assays showing that H4 tail acetylation at lysine 16 is sufficient to open chromatin fibers (27). This also agrees with the observation that H4K16Ac, H3K27Ac, and the acetylated H2A.Z variant (H2A.ZAc) are all commonly found in regions of active chromatin (by a combination of ChIP-Seq and chromatin-conformation capture techniques) (32, 53). However, ChIP-Seq assays for H2A and H2B acetylation are not commonly reported for human cell lines. Our studies suggest that H2A and H2B tail acetylation likely play a structural role in demarcating active from inactive regions.
In particular, we propose a role for LH in mediating and selecting long-range contacts within chromatin compartment domains. We hypothesize that these contacts form because of an increase of available negatively charged DNA in these regions, where the folded acetylated tails are less effective as electrostatic screens for the negatively charged DNA polyelectrolyte compared to WT histone tails. Furthermore, it is known that LH can be selectively driven to specific regions of the chromatin fiber (22), possibly through complex mechanisms such as PARP ribosylation of linker DNA (54).
This role for LH as a domain agent/catalyst is particularly important in the context of establishing mechanistic models for active versus inactive chromatin compartments often used for genome-wide structural analysis utilizing coarse polymer models. In these models, active chromatin is generally considered one domain with a specific mixing coefficient, and inactive chromatin is considered as another domain with a different mixing coefficient (47, 48, 49). However, such models assume, a priori, that well-defined separations between compartments can be driven through the binding of protein cofactors and PTM concentrations, which would require segregation of active/inactive compartments, aggregation of active compartments, and aggregation of inactive compartments. Our data suggest that such a model requires more specificity in promoting/blocking long-range contacts than can be imparted with acetylation and LH binding alone. That is, despite the fact that acetylation is capable of segregating two otherwise similar regions, acetylation alone is not a good mechanism for aggregation of active regions. This role may instead be played by low levels of LH or other protein cofactors such as transcription factors, whereas aggregation of inactive regions may be accomplished by high levels of LH and general aggregating proteins such as heterochromatin protein 1. Our work suggests that a simple active/inactive domain model, in which active domains contain exclusively activating marks and inactive domains contain exclusively inactivating marks, may reflect an oversimplification of the data. A more detailed picture, in particular one that includes other structural proteins like CTCF, transcription factors, and long-noncoding RNA, is necessary to fully understand aggregation/segregation mechanisms that are more apparent at megabase scales.
Conclusions
The effects demonstrated in this study, namely, that simultaneous acetylation of all histone tails decreases long-range contacts and that LH can rescue such contacts when localized to specific regions of the fiber, are important to a wide range of applications. Although chromatin structure has enjoyed an explosion of attention in the last decade, this work has raised as many questions as it has solved. We now know that TADs and chromatin contact domains form and operate under independent principles, by which the former is facilitated by cohesin and the latter is spontaneously formed in solution (32). Both structures are crucial for understanding gene expression, but the link between each and the efficiency of transcription are still hazy. We know that active genes tend to colocalize and that these regions are enriched with acetylation and other specific PTMs, but how these PTMs interact to affect the polymer is not well characterized.
Here we suggest general principles for directed folding that can help describe the underlying structure of living chromatin. Namely, histone tail acetylation operates cooperatively along with LH across all tail types to unfold the chromatin polymer on both local and global levels. This is not, however, sufficient to explain the formation of distinct active and inactive compartment domains in living tissue.
This is an exciting time for chromatin structure investigators. As epigenetic profiles for various cell types are becoming available, the underlying structural characteristics will help establish and predict new interventional techniques for guiding gene expression in the future.
Author Contributions
T.S. and G.D.B. planned and designed the work. G.D.B. performed the modeling and stimulations. G.D.B. and T.S. performed data analysis and wrote the manuscript.
Acknowledgments
We dedicate this study to the memory of Jörg Langowski, a pioneer in chromatin simulation who made seminal contributions to the field. His passion for DNA was highly contagious and always brought a smile to our faces.
We thank R. Collepardo-Guevara for sharing folded-tail coordinates from a previous study and discussing the procedure for modeling acetylated tails.
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences awards R01-GM055264 and R35-GM122562, and Phillip-Morris USA and Phillip-Morris International to T.S. Computing was performed on the New York University High Performance Computing cluster Prince.
Editor: Andrzej Stasiak.
References
- 1.Davey C.A., Sargent D.F., Richmond T.J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 2.Luger K., Richmond T.J. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 3.Olins A.L., Olins D.E. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 4.Hansen J.C. Human mitotic chromosome structure: what happened to the 30-nm fibre? EMBO J. 2012;31:1621–1623. doi: 10.1038/emboj.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grigoryev S.A., Arya G., Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. USA. 2009;106:13317–13322. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li G., Margueron R., Reinberg D. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol. Cell. 2010;38:41–53. doi: 10.1016/j.molcel.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fussner E., Ching R.W., Bazett-Jones D.P. Living without 30nm chromatin fibers. Trends Biochem. Sci. 2011;36:1–6. doi: 10.1016/j.tibs.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Eltsov M., Maclellan K.M., Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. USA. 2008;105:19732–19737. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeshima K., Hihara S., Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr. Opin. Cell Biol. 2010;22:291–297. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Grigoryev S.A., Bascom G., Schlick T. Hierarchical looping of zigzag nucleosome chains in metaphase chromosomes. Proc. Natl. Acad. Sci. USA. 2016;113:1238–1243. doi: 10.1073/pnas.1518280113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ou H.D., Phan S., O’Shea C.C. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:1–13. doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y.B., Gerchman S.E., Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 14.Thomas J.O. Histone H1: location and role. Curr. Opin. Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 15.Fan Y., Nikitina T., Skoultchi A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 16.Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 17.Caterino T.L., Hayes J.J. Structure of the H1 C-terminal domain and function in chromatin condensation. Biochem. Cell Biol. 2011;89:35–44. doi: 10.1139/O10-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luque A., Collepardo-Guevara R., Schlick T. Dynamic condensation of linker histone C-terminal domain regulates chromatin structure. Nucleic Acids Res. 2014;42:7553–7560. doi: 10.1093/nar/gku491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang H., Wei S., Hayes J.J. Chromatin structure-dependent conformations of the H1 CTD. Nucleic Acids Res. 2016;44:9131–9141. doi: 10.1093/nar/gkw586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fyodorov D.V., Zhou B.R., Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat. Rev. Mol. Cell Biol. 2018;19:192–206. doi: 10.1038/nrm.2017.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misteli T., Gunjan A., Brown D.T. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 22.Geeven G., Zhu Y., de Laat W. Local compartment changes and regulatory landscape alterations in histone H1-depleted cells. Genome Biol. 2015;16:289. doi: 10.1186/s13059-015-0857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012;13:343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teif V.B., Vainshtein Y., Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- 26.Roh T.Y., Wei G., Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q., Yang R., Nordenskiöld L. Regulation of nucleosome stacking and chromatin compaction by the histone H4 N-terminal tail-H2A acidic patch interaction. J. Mol. Biol. 2017;429:2075–2092. doi: 10.1016/j.jmb.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Li G., Reinberg D. Chromatin higher-order structures and gene regulation. Curr. Opin. Genet. Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potoyan D.A., Papoian G.A. Energy landscape analyses of disordered histone tails reveal special organization of their conformational dynamics. J. Am. Chem. Soc. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 30.Collepardo-Guevara R., Portella G., Orozco M. Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: a multiscale computational study. J. Am. Chem. Soc. 2015;137:10205–10215. doi: 10.1021/jacs.5b04086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R., Erler J., Langowski J. Histone acetylation regulates chromatin accessibility: role of H4K16 in inter-nucleosome interaction. Biophys. J. 2017;112:450–459. doi: 10.1016/j.bpj.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao S.S.P., Huang S.C., Aiden E.L. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao S.S., Huntley M.H., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bryan L.C., Weilandt D.R., Fierz B. Single-molecule kinetic analysis of HP1-chromatin binding reveals a dynamic network of histone modification and DNA interactions. Nucleic Acids Res. 2017;45:10504–10517. doi: 10.1093/nar/gkx697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collepardo-Guevara R., Schlick T. Chromatin fiber polymorphism triggered by variations of DNA linker lengths. Proc. Natl. Acad. Sci. USA. 2014;111:8061–8066. doi: 10.1073/pnas.1315872111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arya G., Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. USA. 2006;103:16236–16241. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perišić O., Collepardo-Guevara R., Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J. Mol. Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beard D.A., Schlick T. Modeling salt-mediated electrostatics of macromolecules: the discrete surface charge optimization algorithm and its application to the nucleosome. Biopolymers. 2001;58:106–115. doi: 10.1002/1097-0282(200101)58:1<106::AID-BIP100>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 39.Arya G., Zhang Q., Schlick T. Flexible histone tails in a new mesoscopic oligonucleosome model. Biophys. J. 2006;91:133–150. doi: 10.1529/biophysj.106.083006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bascom G.D., Sanbonmatsu K.Y., Schlick T. Mesoscale modeling reveals hierarchical looping of chromatin fibers near gene regulatory elements. J. Phys. Chem. B. 2016;120:8642–8653. doi: 10.1021/acs.jpcb.6b03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arya G., Schlick T. Efficient global biopolymer sampling with end-transfer configurational bias Monte Carlo. J. Chem. Phys. 2007;126:044107. doi: 10.1063/1.2428305. [DOI] [PubMed] [Google Scholar]
- 42.Metropolis N., Ulam S. The Monte Carlo method. J. Am. Stat. Assoc. 1949;44:335–341. doi: 10.1080/01621459.1949.10483310. [DOI] [PubMed] [Google Scholar]
- 43.Rosenbluth M.N., Rosenbluth A.W. Monte Carlo calculation of the average extension of molecular chains. J. Chem. Phys. 1955;23:356–359. [Google Scholar]
- 44.Bascom G., Schlick T. Linking chromatin fibers to gene folding by hierarchical looping. Biophys. J. 2017;112:434–445. doi: 10.1016/j.bpj.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voltz K., Trylska J., Langowski J. Unwrapping of nucleosomal DNA ends: a multiscale molecular dynamics study. Biophys. J. 2012;102:849–858. doi: 10.1016/j.bpj.2011.11.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalashnikova A.A., Porter-Goff M.E., Hansen J.C. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface. 2013;10:20121022. doi: 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barbieri M., Chotalia M., Nicodemi M. Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. USA. 2012;109:16173–16178. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jost D., Carrivain P., Vaillant C. Modeling epigenome folding: formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 2014;42:9553–9561. doi: 10.1093/nar/gku698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Pierro M., Zhang B., Onuchic J.N. Transferable model for chromosome architecture. Proc. Natl. Acad. Sci. USA. 2016;113:12168–12173. doi: 10.1073/pnas.1613607113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rotem A., Ram O., Bernstein B.E. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015;33:1165–1172. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagano T., Lubling Y., Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Pierro M., Cheng R.R., Onuchic J.N. De novo prediction of human chromosome structures: epigenetic marking patterns encode genome architecture. Proc. Natl. Acad. Sci. USA. 2017;114:12126–12131. doi: 10.1073/pnas.1714980114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdés-Mora F., Gould C.M., Clark S.J. Acetylated histone variant H2A.Z is involved in the activation of neo-enhancers in prostate cancer. Nat. Commun. 2017;8:1346. doi: 10.1038/s41467-017-01393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnakumar R., Gamble M.J., Kraus W.L. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]