Abstract
Nitric oxide radical (NO) is a signaling molecule involved in several physiological and pathological processes and a new nitrate-nitrite-NO pathway has emerged as a physiological alternative to the "classic" pathway of NO formation from L-arginine. Since the late 1990s, it has become clear that nitrite can be reduced back to NO under hypoxic/anoxic conditions and exert a significant cytoprotective action in vivo under challenging conditions. To reduce nitrite to NO, mammalian cells can use different metalloproteins that are present in cells to perform other functions, including several heme proteins and molybdoenzymes, comprising what we denominated as the "non-dedicated nitrite reductases". Herein, we will review the current knowledge on two of those "non-dedicated nitrite reductases", the molybdoenzymes xanthine oxidoreductase and aldehyde oxidase, discussing the in vitro and in vivo studies to provide the current picture of the role of these enzymes on the NO metabolism in humans.
Abbreviations: AO, aldehyde oxidase; AFR, activity-to-flavin ratio; Cb, cytoglobin; Cc, cytochrome c; CcO, cytochrome c oxidase; DAF-FM, 4-amino-5-methylamino-2′,7′-difluorofluorescein; DAF-FM DA, DAF-FM diacetate; DPI, diphenyleneiodonium chloride; EPR, electron paramagnetic resonance; Hb, hemoglobin; HepG2, human epithelial cells from liver carcinoma; HL, human liver; HMEC, human microvascular endothelial cells; L-NAME, Nω-nitro-L-arginine methyl ester hydrochloride; mARC, mitochondrial amidoxime reducing component; Mb, myoglobin; MGD2-Fe, iron-N-methyl-D-glucamine dithiocarbamate; Nb, neuroglobin; NO, nitric oxide radical (•NO); NOS, NO synthase; RL, rat liver; SO, sulfite oxidase; SOD, superoxide dismutase; XD, xanthine dehydrogenase; XO, xanthine oxidase; XOR, xanthine oxidoreductase
Keywords: Nitric oxide, Nitrite, Xanthine oxidoreductase, Aldehyde oxidase, Oxygen availability, Molybdenum
Graphical abstract
Highlights
-
•
Xanthine oxidoreductase (XOR) and aldehyde oxidase (AO) catalyze the nitrite reduction to •NO.
-
•
Molybdoenzymes-dependent •NO formation is relevant to virtually all forms of life.
-
•
Acidosis greatly amplifies the •NO formation by XOR and AO.
-
•
Dioxygen availability controls the rate of •NO formation by XOR and AO.
-
•
Independent studies support the XOR and AO involvement in the •NO formation in vivo.
1. Introduction
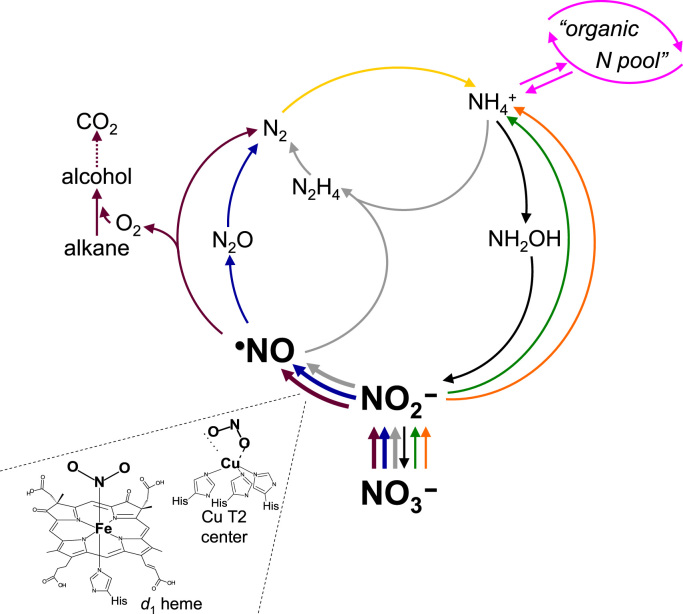
Nitrite is presently recognized as a relevant source of nitric oxide radical (•NO, herein abbreviated as NO) for human cell signaling and survival under challenging conditions [1], [2], [3], [4], [5], [6], [7]. In plants and bacteria, nitrite is also gaining grounds as a source of signaling NO (reviewed in [8], [9], [10]), suggesting that nitrite-derived NO would be relevant in all forms of life! This (at first sight surprising) ubiquity of the nitrite-derived NO is not unexpected at all: the so-called "human" nitrate-nitrite-NO pathway is, in fact, part of ancient "respiratory" prokaryotic pathways of the biogeochemical cycle of the nitrogen (Fig. 1) [8], [11], [12], [13]. Hence, the nitrite reduction to NO can be thought as a heritage from a distant pre-aerobic past, that has been reused every since (evolutionary convergence).
Fig. 1.
Overview of the biogeochemical cycle of nitrogen. The steps common to the human nitrate-nitrite-NO pathway are highlighted with thicker lines and bold characters. The catalytic centers (d1 heme and copper T2 center) of the enzymes responsible for the nitrite reduction to NO in those steps are also presented. Denitrification (a "respiratory" pathway), blue arrows; anaerobic ammonium oxidation (AnAmmOx; "respiratory" pathway), grey arrows; "denitrification/intra-aerobic methane oxidation" ("respiratory" pathway that links the nitrogen and carbon cycles), violet arrows; dinitrogen fixation (nitrogen assimilatory pathway), yellow arrow; assimilatory ammonification (nitrogen assimilatory pathway), orange arrows; "organic nitrogen pool", pink arrows; dissimilatory nitrate reduction to ammonium ("respiratory" pathway), green arrows; nitrification and complete ammonium oxidation (ComAmmOx) ("respiratory" pathways), black arrows. Adapted from Ref. [8] with permission.
2. "Classic" pathways of NO formation
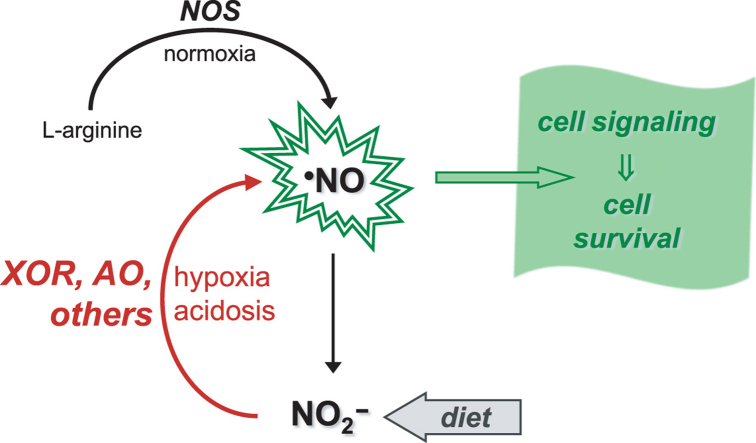
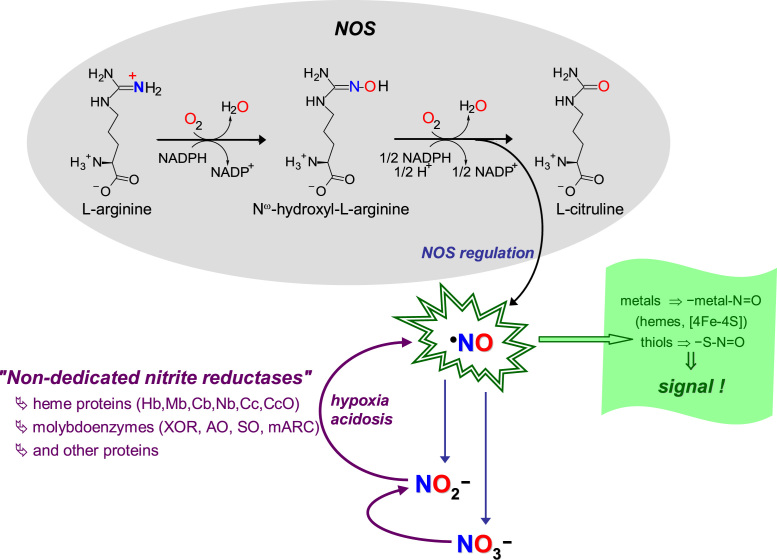
In mammals, NO controls a plethora of functions, including vasodilation (through the well-known activation of guanylate cyclase), neurotransmission, platelet aggregation, apoptosis, gene expression, immune response, and mediates a wide range of both anti-tumor and anti-microbial activities [14]. In humans (Fig. 2), three tissue-specific isoforms of NO synthase (NOS; neuronal, endothelial and inducible NOS) catalyze the formation of NO from L-arginine and dioxygen [15], [16], [17]. Because of this dioxygen dependency, the onset of hypoxia/anoxia hampers the NOS catalytic activity and the NO formation can become compromised. The specificity of the NO signaling is guaranteed by the NOS tight regulation and by the limited NO life time, which is achieved through its rapid oxidation to nitrate (by the well known reaction with oxy-hemoglobin and oxy-myoglobin [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29]) and to nitrite (by ceruloplasmin [30], cytochrome c oxidase [31] or dioxygen [32], [33], [34]).
Fig. 2.
"Classic" and novel pathways of NO formation. The "classic" pathways of NO formation (black arrows, grey shadowed area) are catalyzed by NOS, complex homodimeric enzymes, constituted by one flavinic reductase C-terminal domain and one hemic oxygenase N-terminal domain. During catalysis, NO is formed from one oxygen atom of dioxygen (printed in red) and from the guanidinium nitrogen atom of L-arginine (in blue), and three reducing equivalents are consumed (in the form of NADPH). The electrons from NADPH are transferred through the reductase domain to the b heme iron of the oxygenase domain; on the heme, the dioxygen is activated to hydroxylate L-arginine; the Nω-hydroxy-L-arginine formed is, then, oxidized to yield L-citrulline and NO. To control the specificity of NO signaling (indigo arrows and text), and also to limit the NO toxicity, NOS are tightly regulated and the NO life time is controlled through its rapid oxidation to nitrate and nitrite. The novel pathways of NO formation (violet arrows and text) are reductive in nature (contrary to the oxidative NOS-catalyzed pathways) and are dependent on the nitrite reduction under hypoxic and anoxic conditions. These pathways are catalyzed by "non-dedicated nitrite reductases", metalloproteins that are present in cells to perform other functions, including several heme proteins and molybdoenzymes. The NO biological effects are accomplished (green arrows and text), mainly, by post-translational modification of cysteine residues and other thiols and of transition metal centers, mostly labile [4Fe-4S] centers and hemes (as is the case of the well known activation of guanylate cyclase), to yield nitrosothiol (-S-N=O) and nitrosyl (-metal-N=O) derivates, respectively.
3. Nitrite-derived NO
At the same time as our knowledge about the physiological roles of NO in humans was growing exponentially, nitrate and nitrite were ignored and considered "useless" end-products of NO metabolism. This dogma changed in the early XXI century, when it became clear that nitrite can be reduced back to NO under hypoxic conditions (Eq. (1)) and it was re-discovered that nitrite administration can be cytoprotective during in vivo ischemia and other pathological conditions (Fig. 2) [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69] (interestingly, the physiological action of nitrite had already been described in 1880 [70]). Since then, a novel concept emerged and nitrite began to be thought as a "storage form" of NO, that can be used to maintain the NO formation and ensure cell functioning under conditions of hypoxia/anoxia, precisely when the dioxygen-dependent NOS activity is impaired and a "rescue" pathway would be needed to form NO. Through the nitrite/NO "recycling" pathway, an organ under ischemia can maintain (or even increase) the blood flow, modulate the dioxygen distribution and the reactive oxygen species formation and, at the same time, maintain an anti-inflammatory and anti-apoptotic environment.
| NO2- + 1e- + 2H+ ——→ •NO + H2O | (1) |
4. "Who" is reducing nitrite to NO?
Simultaneously to the emergence of the new paradigm of nitrite-derived NO, the search for the human (mammalian) protein(s) responsible for the nitrite reduction to NO began. (Following the studies in mammals, more recently, a similar quest began in plants [8], [9], [10]).
The reduction of nitrite to NO is known for a long time in prokaryotic organisms, where it is catalyzed by copper-containing or heme-containing nitrite reductases, in "respiratory" pathways, as part of the biogeochemical cycle of the nitrogen (Fig. 1) [8], [11], [12]. However, to date, no "dedicated" (true) mammalian nitrite reductase was ever identified (what definitively contributed to consider nitrite as a "useless" molecule in the earlier years). Hence, the scientific community searched for the nitrite reductase activity in proteins that perform other (already well known) functions in the cell.
In the recent years, several mammalian metalloproteins, with different molecular features, subcellular localization and cellular roles (enzymes, metabolite transporters and electron transfers), were shown to be able to reduce nitrite to NO -comprising what we denominated as the "non-dedicated nitrite reductases" [8] (Fig. 2). The long list of "non-dedicated nitrite reductases" includes all the known mammalian molybdenum-containing enzymes (xanthine oxidoreductase (XOR), aldehyde oxidase (AO), sulfite oxidase (SO) [71] and mitochondrial amidoxime reducing component (mARC) [72]), and a growing number of heme-containing proteins, where hemoglobin (Hb) and myoglobin (Mb) stands out in number of publications, but including also neuroglobin (Nb) [73], cytoglobin (Cb) [74], cytochrome c (Cc) [75], cytochrome P450 [76], cytochrome c oxidase (CcO) [77], [78], [79], and several other proteins [80], [81], [82], [83]. At this pace, other mammalian nitrite reductases will probably be identified in the next years. In addition, some protein-independent pathways were also proposed for the nitrite reduction in ischemic tissues, stomach and brain [84], [85], [86], [87], [88], [89], [90], [91], [92], [93]. Noteworthy, the protein-independent nitrite reduction to NO in the stomach (the pioneering work of Lundberg's and Benjamin's groups [84], [85], [87]) and in ischemic tissues (Zweier's group work [86], [88]) were the first pathways through which nitrite was suggested to be a relevant source of bioactive NO, in the 1990s.
In all those pathways, the nitrite reduction to NO was described to occur under hypoxic or anoxic and acidic conditions, but the level of characterization of each pathway is very dissimilar. So far, only Mb and XOR have been demonstrated to be directly involved in the cytoprotective action of nitrite in vivo or ex vivo [38], [45], [94], [95], [96], but only the XOR nitrite reductase activity was thoroughly characterized. The nitrite reductase activity of Hb, on it is turn, has been extensively characterized in vitro, with several mechanisms being proposed to explain how it would be possible for NO to escape being trapped by the heme (reviewed, e.g., in [8]). The characterization of the other mammalian nitrite reductases is more limited. Yet, regardless of the knowledge so far accumulated, the nitrite "recycling" to NO is still a complex subject, overshadowed by several (bio)chemical constrains, of which we highlight: (i) In the case of enzymes, how can nitrite compete with the "classic" oxidizing substrates? (ii) In the case of the heme proteins, how can the formed NO avoid being rapidly trapped by the heme itself? (iii) How can we reconcile the in vivo observed nitrite effects with the in vitro knowledge of nitrite reduction through those diverse pathways? (iv) How are all those pathways orchestrated in vivo? Are all equally relevant? Are tissue-specific? Have different triggering levels/conditions?
Herein, we will review our current knowledge about the nitrite reductase activity of the mammalian molybdoenzymes XOR and AO, discussing the in vitro and in vivo studies to provide the best possible current picture of the role of these enzymes on the NO metabolism.
5. Human XOR and AO
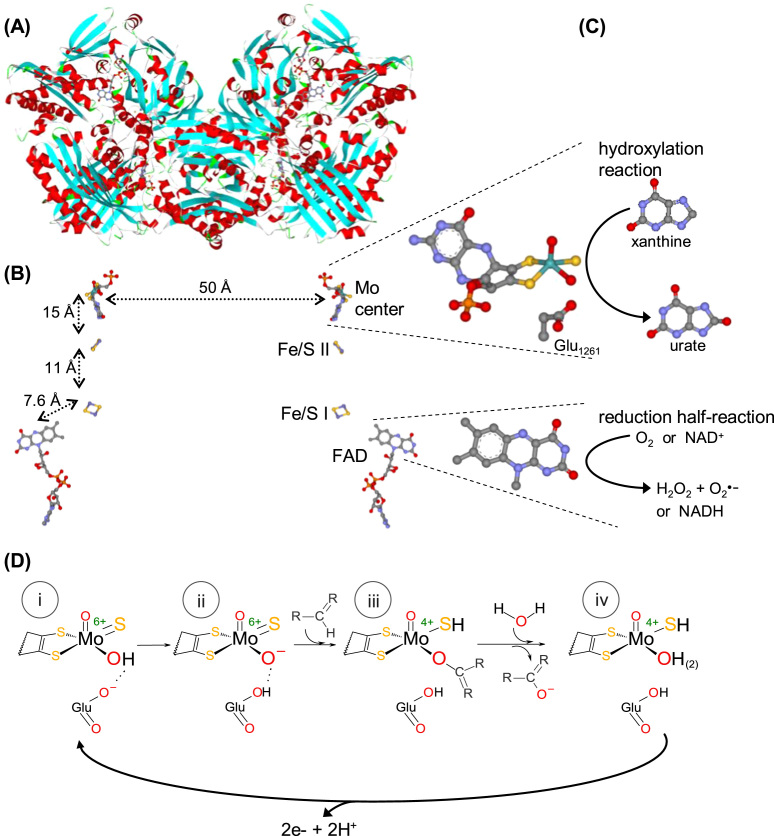
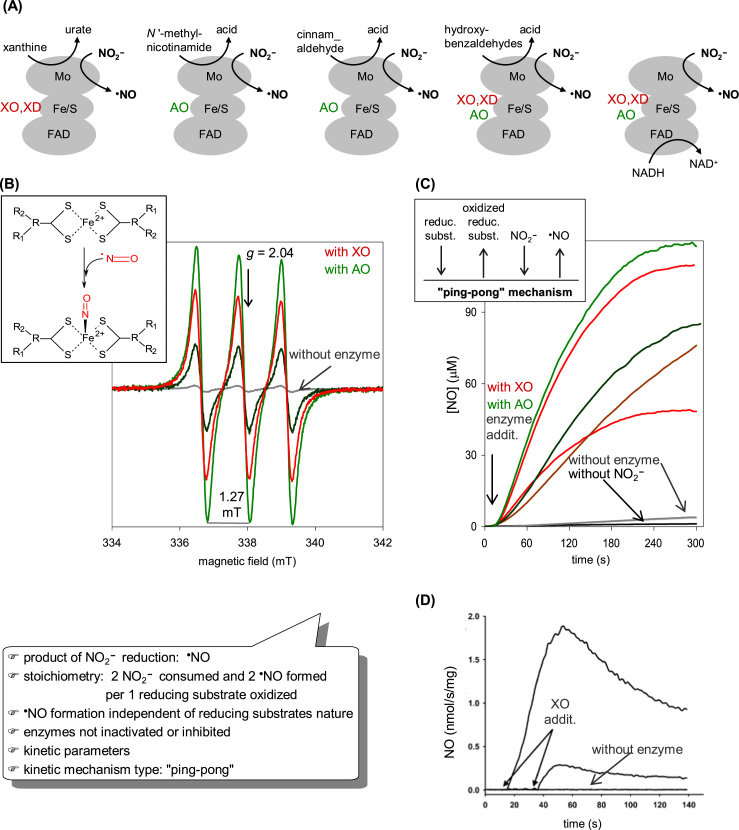
XOR is a key enzyme in purine catabolism, where it catalyzes the oxidation of both hypoxanthine and xanthine to the terminal metabolite, urate [97], [98], [99], [100], [101], [102]. AO catalyzes the oxidation of aldehydes into the respective carboxylates and, although its physiological function remains a matter of debate, it seems to be a probable partner in the metabolism of some neurotransmitters and retinoic acid [103], [104], [105], [106], [107], [108]. Both enzymes contribute also to the xenobiotic metabolism (due to their low substrate specificity) and are allegedly involved in signaling (physiological conditions) and oxidative stress-mediated pathological conditions (due to their ability to form reactive oxygen species, superoxide anion radical and hydrogen peroxide) [109], [110], [111], [112], [113], [114], [115], [116], [117], [118], [119], [120], [121], [122], [123], [124], [125], [126], [127], [128], [129], [130], [131], [132], [133], [134], [135], [136], [137], [138], [139], [140], [141], [142], [143], [144]. In vivo, AO exists exclusively as an oxidase (reduces dioxygen; EC 1.2.3.1; Eq. (2)), whereas XOR exists predominantly as a NAD+-dependent dehydrogenase, named xanthine dehydrogenase (XD; EC 1.17.1.4; Eq. (3)) [97], [98], [99], [100], [101], [102], [104], [106], [108], [145], [146]. Yet, XD can be rapidly converted into a "strict" oxidase form that reduces dioxygen instead of NAD+ − the very well documented xanthine oxidase (XO; EC 1.17.3.2; Eq. (4)). (Note1 gives more details for readers interested in the nature of the two XOR forms.) Overall, XO, XD and AO catalyze the transfer of one oxygen atom from a water molecule to a carbon center of the substrate (as indicated by the red oxygen atoms in Eqs. (2), (3), (4); Fig. 3(D)), through a reaction mechanism that is identical in all enzymes, with dioxygen or NAD+ acting as electron acceptors [97], [98], [99], [100], [101], [102], [152], [153].
Fig. 3.
XOR structure and reactivity. (A) Three-dimensional structure view of the bovine milk XO homodimer. (B) Arrangement of the four redox-active centers shown in the same orientation as in (A). The four centers are identified on the monomer on the right, and the distances between adjacent centers are shown on the monomer on the left. The molybdenum center, together with the conserved glutamate residue, and the FAD isoalloxazine ring are zoomed on the right. (C) The oxidation and reduction half-reactions are represented next to the centers where they are catalyzed. (D) The molecular reaction mechanism of XOR and AO-catalyzed hydroxylation reactions is presently well established [97], [98], [99], [100], [101], [102] and is here exemplified with a R2-C-H molecule that is hydroxylated to R2-C-OH to represent both heterocyclic compounds and linear aldehydes. The hydroxylation catalysis is initiated with the activation of the molybdenum hydroxyl ligand (Mo6+-OH) by a neighboring conserved deprotonated glutamate residue, to form an oxidized Mo6+-O-(=S) core (base-assisted catalysis) (i→ii). It follows the nucleophilic attack of Mo6+-O- on the carbon atom to be hydroxylated, with the simultaneous hydride transfer from substrate to the sulfo ligand (Mo6+=S → Mo4+-SH), resulting in the formation of a covalent intermediate, Mo4+-O-C-R2(-SH) (ii→iii). The subsequent hydrolysis of the Mo4+-O bond releases the product hydroxylated and yields a Mo4+-OH(2)(-SH) core (oxidation half-reaction) (iii→iv). Finally, the two electrons transferred from the substrate to the molybdenum are rapidly transferred, via the Fe/S centers, to the FAD, where the dioxygen or NAD+ reduction takes place (reduction half-reaction) (iv→i). In the now oxidized molybdenum center, the sulfo group is deprotonated and the initial Mo6+-OH(=S) core is regenerated and the catalytic cycle can be reinitiated (i). The structures shown ((A),(B)) are based on the PDB file 1FO4; in (A), α helices and β sheets are shown in red and turquoise, respectively; in (B) and (C), atoms are color coded as follows: carbon, grey; nitrogen, blue; oxygen, red; sulfur, gold; phosphorous, orange; iron, dark gold; molybdenum, cyan.
Structurally, XOR (Fig. 3(A), (B)) and AO are also very similar. Both are complex homodimeric molybdoenzymes that harbor (per monomer) one identical molybdenum center, where the hydroxylation reactions occur, two [2Fe-2S] centers and one FAD, responsible for the reduction of dioxygen (XO, XD, AO) and NAD+ (XD) (Fig. 3(C)) [97], [98], [99], [100], [101], [102], [147], [153], [154], [155]. The protein three-dimensional structure of the molybdenum and iron/sulfur domains of XD and XO is identical; only the protein conformation at the FAD domain is different in XD and XO (see note1). In accordance, the two enzyme forms are virtually identical in respect to the binding and catalysis of substrates at the molybdenum center, as is the case of oxidation of xanthine and other heterocyclic compounds and aldehydes.
| (2) |
| (3) |
| (4) |
6. XOR and AO-catalyzed nitrite reduction: formation of NO
In the quest for the new pathways of NO formation, the potential nitrite reductases must meet some criteria.
6.1. NO is formed
It is mandatory to guarantee that NO is really being formed and released, and not other possible products of nitrite reduction (e.g., NO- or NH4+). Hence, the use of indirect methods, e.g., quantification of cGMP, should be avoided, because they do not allow the unambiguous identification of NO. Also the use of most fluorescent probes is inadequate, because the NO detection is made indirectly, through the reaction of amines with an oxidation product of NO (detection by deamination or triazole ring formation) [156], [157], [158], [159].
Because NO is a radical, electron paramagnetic resonance (EPR) spectroscopy, using a spin-trap (to "stabilize" the radical and increase its life time), is one of the methodologies of choice. The EPR spectrum provides valuable information regarding the nature, structure and environment of the paramagnetic species and, therefore, unequivocal information about the identity of the radical species. In addition, the EPR spectroscopy can be used to quantify the NO formed and, thus, follow its kinetics of formation. Chemiluminescence and polarographic methodologies are other good options. In the former, NO is reacted with ozone (after being purged to a gas phase chamber) to produce an excited state, NO2, that generates light; in the second methodology, NO is oxidized at a Clark-type electrode surface (poised at ≈ 0.8 V vs Ag/AgCl) enclosed by a gases-only-permeable membrane (that ensures the electrochemical measurements selectivity).
That NO is the product of XOR and AO-catalyzed nitrite reduction was unequivocally demonstrated by EPR, using the spin-trap iron-N-methyl-D-glucamine dithiocarbamate ((MGD)2-Fe), by chemiluminescence and with the NO-selective electrode (further confirmed by the rapid and complete inhibition of NO generation after the addition of hemoglobin, an effective NO scavenger) (Fig. 4) [133], [139], [160], [161], [162], [163], [164], [165], [166], [167]. Using the NO-selective electrode, it was also demonstrated that approximately one NO molecule is formed per nitrite molecule and that approximately two NO molecules are formed per reducing substrate molecule consumed [133], [139] (as would be expected, since the XOR and AO reducing substrates are oxidized by two electrons and NO is formed by the one-electron reduction of nitrite).
Fig. 4.
Formation of NO during XOR and AO catalyzed nitrite reduction. Different assays, with various proportions of enzyme, nitrite and reducing substrate are shown to illustrate how the NO formation depends on those three elements. (A) The different reducing substrates used to demonstrate the XOR and AO ability to catalyze the nitrite reduction to NO are represented, indicating in which enzyme redox center each substrate reacts. (B) EPR assays using the spin-trap iron-N-methyl-D-glucamine dithiocarbamate ((MGD)2-Fe), whose structure is shown on the left panel. In the presence of this spin-trap, NO gives rise to a mononitrosyl-iron complex ((MGD)2-Fe-NO), which exhibits a characteristic EPR triplet signal at g ≈ 2.04, with a hyperfine splitting of 1.27mT. Data adapted with permission from Ref. [137]. Copyright 2015 American Chemical Society. (C) Assays with the NO-selective electrode. The kinetic mechanism type, "ping-pong", is schematically indicated on the left panel. In both methodologies, the grey lines refer to curves without enzyme; black line, without nitrite; red and dark red lines, with XOR; green and dark green lines, with AO. Data adapted with permission from Ref. [137]. Copyright 2015 American Chemical Society. (D) Assays of NO formation catalyzed by XO performed using a chemiluminescence NO analyzer. Reprinted with permission from Ref. [161].
6.2. Independence on co-substrates nature
If the nitrite reduction to NO is an intrinsic property of a protein, then the NO formation should be independent on the reagent used to reduce that protein and should be feasible with physiologically available reducers.
The XOR and AO-catalyzed NO generation is dependent on the simultaneous presence of enzyme, nitrite and one reducing substrate (there is no NO formation in the absence of any of these three elements) and the rate of NO formation is a function of the concentration of protein, nitrite and reducing substrate (a Michaelis-Menten function) (Fig. 4) [133], [139], [160], [161], [162], [163], [164], [165], [166]. The NO generation can be triggered in the presence of several reducing substrates, with different chemical natures and sites of reaction with the enzymes: heterocyclic compounds (such as xanthine (for XOR) and N '-methyl-nicotinamide (for AO)) and aldehydes (both XOR and AO), that react at the enzymes molybdenum center, and also NADH (both XOR and AO), that reacts at the FAD center.
6.3. Sustainable NO formation
To be a viable source of NO, the potential nitrite reductase should not be inactivated/inhibited by NO. This is particularly important in XOR and AO, because NO can modify the enzymes cysteine residues and metal centers (molybdenum and iron/sulfur) and, thus, affect the enzyme activity. Another concern is related with the fact that the NO formation from nitrite is a one-electron reduction process (Eq. (1)) and the molybdoenzymes need a two-electron reduction process to regenerate the catalytically competent enzyme form (Mo4+ → Mo6+; Fig. 3). Hence, if the molybdenum center is trapped in an intermediate oxidation state (Mo5+), it would not be able to continue the catalysis (formation of dead-end forms).
The modifications of most enzymes redox-active centers can be accessed with EPR spectroscopy (and UV–visible spectroscopy in some cases). Comparing the spectra of the enzymes during nitrite reduction turnover and "classic" turnover (xanthine oxidation, in the case of XOR) could provide evidence regarding possible modifications and presence of dead-end intermediates. Follow the entire time-course of NO formation is also a good strategy to detect possible deviations from the expected enzymatic behavior, what can be easily achieved with a NO-selective electrode. The form of the time courses provides evidence, e.g., of enzyme inactivation and inhibition or of concurrent non-enzymatic processes (associated with time courses where the NO concentration sharply and rapidly diminishes after an initial, brief, "burst"). Comparing the time-courses curves of nitrite reduction and reducing substrate oxidation (xanthine oxidation, in the case of XOR) can be particularly informative, providing also information about the stoichiometry and extent of reaction.
The time courses of XOR and AO display the expected exponential form (Fig. 4), providing evidence that XOR and AO are not inactivated or inhibited during the catalysis of NO formation (further confirmed by "classic" activity time-courses curves) [133], [139]. Also the EPR spectra show no alteration of the redox centers (which exhibit characteristic EPR signals) during and after the nitrite reduction catalysis [133], [168].
6.4. Kinetically relevant NO formation
One last criterion regards the rates of NO formation. The rate of NO formation of the potential nitrite reductase has to fit in a relatively tight window, complying with the characteristics of a local signaling molecule: a too slow rate of NO formation could be physiologically irrelevant and a two fast rate could lead to deleterious effects (as the ones observed in some reactive nitrogen species-mediated diseases or during the immune response). Therefore, it is not reasonable to search/aim for a pathway that generates NO at micromolar concentrations. Also the concentration of nitrite and reducing substrate needed to promote the NO formation should be physiologically relevant. A glance of the fulfillment of this criterion can be obtained through the determination of the kinetic parameters, with which a kinetic model can be built to predict how relevant the reaction could be in vivo - discussed in the point below.
7. XOR and AO-catalyzed nitrite reduction: magnitude and kinetics of NO formation
The nitrite reduction to NO has been characterized, under anaerobic conditions, in purified enzymes from bovine milk (XO) [133], [160], [161], [162], [163], [164], [166], rat liver (RL) and human liver (HL) (XD, XO and AO) [139], and also in a bacterial aldehyde oxidoreductase (an enzyme structurally and functionally very similar to the mammalian ones, that can be used as a model) [133].
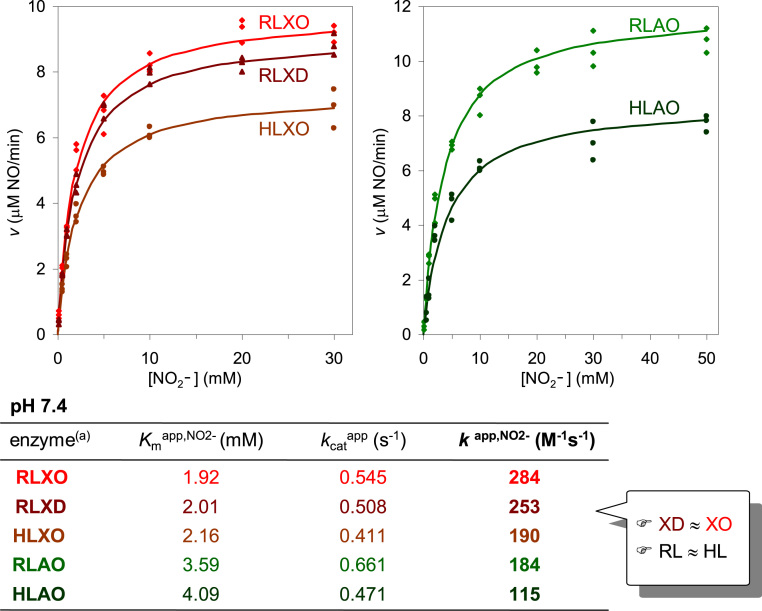
The kinetic parameters of the mammalian enzymes for the nitrite reduction were found to be dependent on the pH (described below) and on the reducing substrate used. The pseudo-first order rate constant values (k = kcat/Km) clearly follow the trend observed for the semi-reaction of enzyme reduction by the reducing substrate: NADH (the slowest to reduce XO, XD and AO) gives rise to the lowest k- values of NO formation (87 M−1 s−1, with 1 mM NADH), while xanthine and N '-methyl-nicotinamide (the fastest to reduce XO/XD and AO, respectively) produce the highest values (774 and 996 M−1 s−1, with 10 μM xanthine and 40 μM methyl-nicotinamide, respectively) [139]. (Exhaustive tables summarizing kinetic parameters with different reducing substrates can be found in [9], [139].) To make the direct comparison between XO/XD and AO more rational, a reducing substrate with a similar rate of reduction of all enzymes -an aldehyde- was also studied. Under such "standardized" conditions (similar reducing power and nitrite and enzyme concentrations), the amount of NO formed by XO, XD and AO is similar, with the Kmapp,NO2- values ranging from 1.9 to 4.1 mM and the kcatapp,NO2- values varying from 0.4 and 0.7 s−1 at pH 7.4 (Fig. 5) [139]. Moreover, and most important, the kinetic studies showed that the rat and human liver enzymes have very similar kinetic parameters (Fig. 5) [139], clearly supporting the utilization of the rat liver enzymes (whose tissue is easier to obtain) as a suitable model of the human counterparts. Also noteworthy is the observation that XO and XD display identical kinetic parameters (within the experimental error; Fig. 5) [139]. Although this is the expected conclusion (because the molybdenum domain of the two XOR forms is identical (see note1) and the nitrite reduction occurs at the molybdenum center (discussed below)), this confirmation is essential to clearly establish that the potential in vivo significance of XOR-derived NO formation does not depend on the XOR form present, XD or XO (note that the XOR predominant form present under different physiological and pathological conditions is still a matter of debate).
Fig. 5.
Kinetics of XO, XD and AO-catalyzed, aldehyde-dependent, nitrite reduction to NO at pH 7.4. (a) Enzyme source: HL, human liver; RL, rat liver. Adapted with permission from Ref. [137]. Copyright 2015 American Chemical Society.
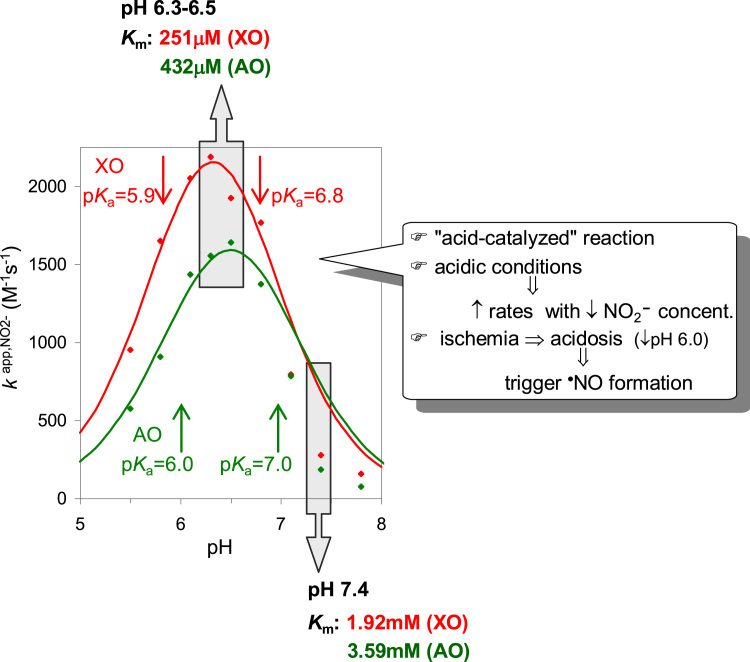
The kinetic parameters dependence on the pH is well exemplified by the variation of the pseudo-first order rate constant of NO formation (note that this is the constant that would govern the reaction rate under physiological conditions, because the nitrite concentration physiologically available (< 20 μM [169], [170], [171]) is much lower than the KmNO2- values reported). The resulting bell-shaped curves (Fig. 6) are characterized by apparent pKa values of ≈ 6 and ≈ 7 and clearly demonstrate that the nitrite reduction is favored under lower pH values: when the pH is decreased from the normal 7.4 to 6.3, the enzymes-catalyzed nitrite reduction to NO is increased ≈ 8 times (k app,NO2- = 2.20 × 103 M−1 s−1 (XO) and 1.64 × 103 M−1 s−1 (AO) at pH 6.3; Fig. 6) [139]. Moreover, these increased rate constants of NO formation are mainly due to a variation in the KmNO2- values, that decrease ≈ 8 times when the pH is decreased from 7.4–6.3 (Kmapp,NO2- = 251 μM (XO) and 432 μM (AO) at pH 6.3; Fig. 6) [139].
Fig. 6.
pH dependence of the kinetic parameters of XO and AO-catalyzed nitrite reduction to NO. Adapted with permission from Ref. [137]. Copyright 2015 American Chemical Society.
These key "kinetic" conclusions show that the XO, XD and AO are able to trigger (amplify) the NO formation, to respond to a decrease in the pH, as the one expected upon an ischemic event, when the pH can drop to values as low as 6.0–5.5 (acidosis) [172], [173], [174]. The parallel decrease in the KmNO2- values shows that the enzymes are also able to cope with the constrain imposed by the low nitrite availability (< 20 μM [169], [170], [171]). The millimolar order of the KmNO2- values determined at pH 7.4 (compared with the nitrite availability) was one of the strongest arguments raised against the participation of these enzymes in the in vivo NO formation. Yet, the KmNO2- values are strongly pH-dependent and a decrease in the pH boosts the enzymes efficiency towards nitrite. For example, a rate of 1.6–2.2 nM NO/s can be obtained at pH 6.3, with 10 μM nitrite and 30 μg enzyme/mL (to simulate 30 μg enzyme/g tissue), rates that compare well with the 1 nM/s described for the constitutive NOS [175]. The available reducing substrates is perhaps the most undefined factor in this prediction (namely because of the substrate promiscuity of these enzymes); yet, under ischemia, for example, the resulting reducing substrates accumulation would certainly "charge" the enzymes with the electrons necessary to reduce nitrite and produce NO. Hence, the magnitude and kinetics of NO formation by XO, XD and AO support that these enzymes can contribute to the in vivo NO generation under acidic conditions, as the ones created upon an ischemic event and other pathological conditions. The potential contribution of XO and XD, as well as, of AO, is expected to be very similar (under similar nitrite and enzyme concentrations and reducing power), suggesting that both XOR and AO can play role in the NO metabolism. Comparatively, the contribution of the other two mammalian molybdenum-containing enzymes, mARC and SO, seems to be extremely small: the predicted rates of NO formation, under the same conditions as described above, are of only 0.011 nM NO/s, for mARC, and 0.028 nM NO/s, for SO (based on a k app,NO2- of 11 M−1 s−1 for the human mARC, at pH 7.4 [72], and a k app,NO2- of 28 M−1 s−1 for the human modified SO, at pH 6.5 [71]).
8. XOR and AO-catalyzed nitrite reduction: molecular mechanism of reaction
Presently, it is clear that the nitrite reduction to NO takes place at the molybdenum center of XOR and AO, as was unequivocally demonstrated by several studies combining EPR and NO electrode assays, using molybdenum-specific inhibitors (allopurinol, BOF-4272 (XOR) [133], [139], [161], [162], [163] or ethyleneglycol (aldehyde oxidoreductase) [133]) and employing native enzymes preparations with different AFR values2 [139]. Additional assays with DPI-inhibited enzymes (a FAD-specific inhibitor) and also with deflavo-XOR and deflavo-AO3 [139] provided the "negative confirmation" (that nitrite reduction is not dependent on the FAD center). Simultaneously, using NADH-reduced desulfo-XO4 it was demonstrated that the molybdenum sulfo group (see Fig. 3(B)) is necessary for the nitrite reductase activity [133], [139], [162], thus, providing further confirmation of the involvement of the molybdenum center. Moreover, and most important, it was also demonstrated that the NO formed during the catalytic cycle does not react with the molybdenum sulfo group, what could lead to its abstraction (in the form of a nitrosothiol) and, consequently, to the enzyme inhibition [133], [139].
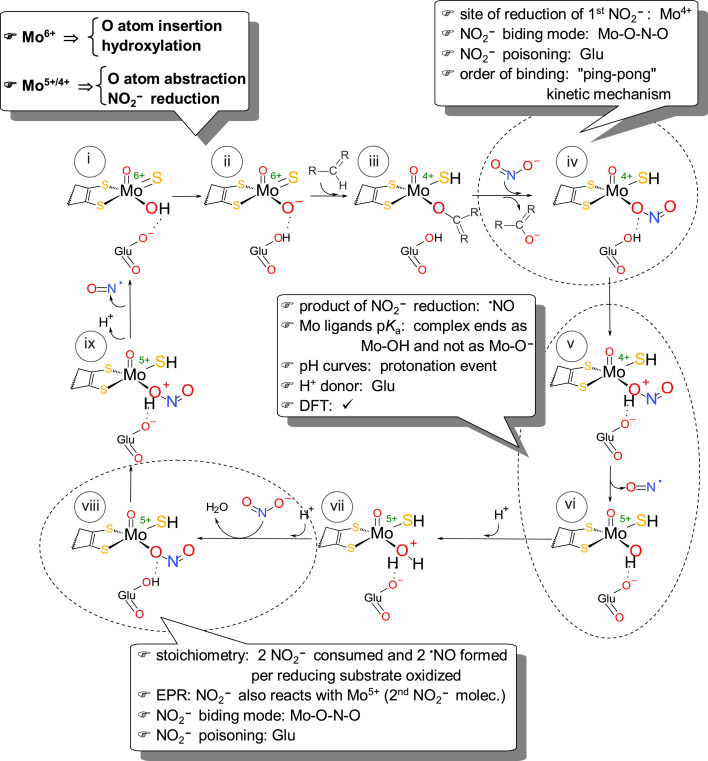
Based on the above kinetic and EPR spectroscopic data and on the general features of these enzymes [97], [98], [99], [100], [101], [102], we have proposed the first and currently accepted molecular mechanism of nitrite reduction by
molybdoenzymes (Fig. 7) [8], [9], [139]. To catalyze the nitrite reduction, the molybdenum center has to be first reduced, by a reducing substrate, in a first part of the catalytic cycle, the oxidation half-reaction (e.g., XOR reduction by xanthine; Fig. 7, i→ii→iii; see Fig. 3(D) for details). After the release of the oxidized reducing substrate (urate, in the example), nitrite binds to the molybdenum atom through one of its oxygen atoms, via the "nitrito" binding mode (Mo4+-O-N-O; Fig. 7, iii→iv). Subsequently, a protonation step, where the nitrite oxygen atom is protonated (Fig. 7, v), triggers the homolyitc O-N bond cleavage, resulting in the formation of one NO molecule and leaving a partially oxidized molybdenum (Mo5+) center (Fig. 7, v→vi). The residue responsible for the protonation step was not yet unambiguously identified, but the conserved glutamate residue that is essential to the hydroxylation half-reaction (Glu1261 in bovine milk XO; Fig. 3(B)), has the adequate and best position inside the active site pocket to act as the proton donor (as demonstrated by theoretical calculations). Accordingly, in the first part of the catalytic cycle (oxidation half-reaction), the deprotonated glutamate functions as a base and assists the Mo-O- nucleophilic attack to the carbon center to be hydroxylated; during the nitrite reduction part (reduction half-reaction), the same glutamate residue, but at this point protonated, functions as a proton donor to facilitate the cleavage of the O-N bond.
Fig. 7.
Molecular reaction mechanism of XOR and AO-catalyzed nitrite reduction to NO. Adapted with permission from Ref. [137].
The proposal of the protonation step is supported by different lines of evidence (Fig. 7). (a) The kinetic characterization of the pH effect (described in the previous section) shows that the nitrite reduction is greatly accelerated under acid conditions and that the nitrite affinity is significantly increased. (b) The pKa values of the molybdenum coordinated ligands change dramatically with the oxidation state and the lower oxidation states hold highly protonated ligands [176], [177], [178], [179], [180]. For this reason, in the Mo5+ complex, both terminal oxygen and sulfur atoms should be protonated.5 Therefore, if nitrite is protonated before it is converted into NO, the resulting molybdenum complex would be in a more stable form, Mo5+-OH(-SH) (Fig. 7, vi), than if it is as a Mo5+-O-(-SH) complex. (c) Also theoretical calculations support this reasoning [181]. (d) This mechanistic approach has a precedent in the bacterial copper-containing nitrite reductase that follows a similar mechanistic strategy (see note6).
At this stage (Fig. 7, vi), one molecule of NO is already formed and released. However, because the oxidation half-reaction of the catalytic cycle is a two-electron process, the molybdenum center still has one electron to reduce a second nitrite molecule to NO. The reaction is suggested to proceed with the formation of a good leaving group, a water molecule (Mo5+–OH2; Fig. 7, vi→vii), which is subsequently, displaced by nitrite (Fig. 7, vii→viii). After a second cycle of nitrite reduction/molybdenum oxidation, triggered by a protonation step, a second NO molecule is released (Fig. 7, viii→i). The molybdenum is now in a 6+ oxidation state, which would favor the deprotonation of its ligands, and the center is ready to start a new catalytic cycle.
Overall, the ability of the molybdoenzymes to reduce nitrite to NO arises from two main key features. (a) The molybdenum unique chemistry - This chemistry makes the molybdenum centers excellent "oxygen atom exchangers", as long as the thermodynamics of the reactions is favorable [176], [177], [178], [179], [180]. As a result, organisms developed numerous molybdoenzymes to catalyze different oxotransfer reactions (both oxo-abstractions and oxo-insertions) in the carbon, sulfur and nitrogen metabolism (reviewed in [99], [101]) -this is precisely what is needed to convert nitrite into NO, an oxygen atom abstraction reaction (Eq. (1)). The Mo6+ cores act as competent oxo group donors (catalyzing, e.g., hydroxylation reactions), while the Mo4+ cores act as oxo group acceptors (catalyzing the nitrate and nitrite reduction and other reductions) [99], [101]. Hence, the nitrite reductase activity is just another manifestation of the oxotransfer reactivity of the molybdenum cores. (b) The protonation step - This is thought to be crucial for the O-N bond homolysis with the molybdoenzymes and also with the copper-containing enzymes.
9. XOR and AO-catalyzed nitrite reduction: inhibition of NO formation by dioxygen
After the above discussion, it is clear that XO, XD and AO can catalyze the formation of NO. The magnitude and kinetics of the nitrite reduction reaction, as well as the feasibility of the reaction mechanism, unequivocally demonstrate it. However, as mentioned, the kinetic characterization was carried out under anaerobic conditions.
As can be anticipated, the enzymes "classic" oxidizing substrates -dioxygen and NAD+- should act as strong competitive inhibitors of the NO formation, both in vitro and in vivo. The "classic" oxidizing substrates are efficiently oxidized by the enzymes, consuming the electrons needed to reduce nitrite (Eqs. (2), (4) versus Eq. (1); in other words, in the presence of the "classic" oxidizing substrates, the concentration of reduced enzymes molecules, the ones that react with nitrite, is smaller). Hence, the potential role of the enzymes in the NO metabolism can not be properly evaluated without taking into account the effect of dioxygen; the NAD+ inhibition of the XD-dependent NO formation should also be considered, but this issue was not yet studied.
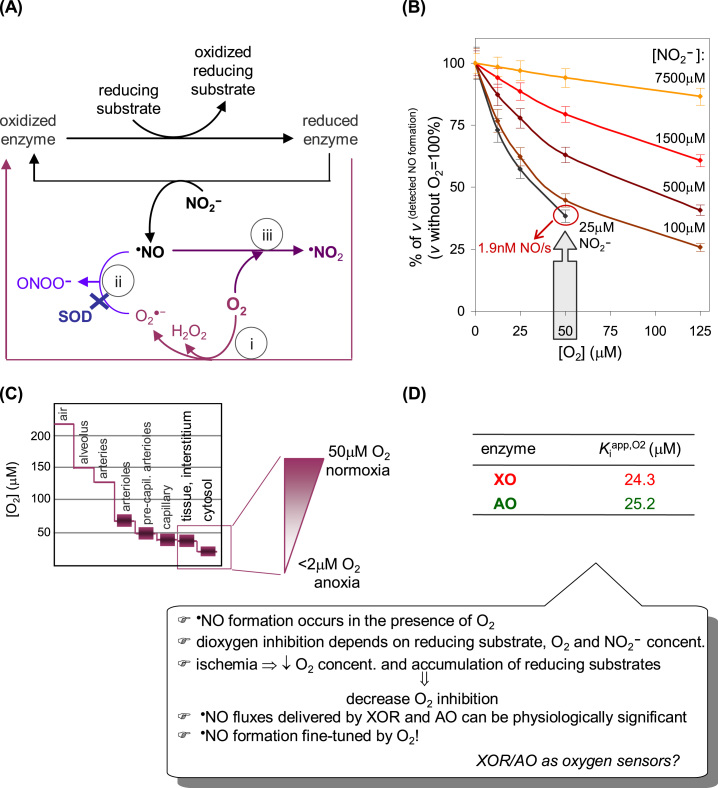
Dioxygen interferes with NO at different levels, decreasing both the amount of NO formed (dioxygen inhibition of NO formation; Fig. 8(A), i) and the amount of NO available (available to exert its in vivo actions and to be detected; Fig. 8(A), ii, iii). (The simultaneous occurrence of these effects complicates the interpretation of the dioxygen effect on the nitrite reduction/NO formation, what probably explains some of the inconsistencies found in literature).
Fig. 8.
Effects of dioxygen on the XOR and AO-dependent NO status. (A) Dioxygen interferes with NO at different levels. (i) Dioxygen is efficiently reduced at the enzymes FAD center (Fig. 3) and rapidly consumes the electrons derived from the reducing substrates. Consequently, dioxygen readily decreases the concentration of enzyme molecules with reduced molybdenum available to react with nitrite, thus decreasing (inhibiting) the NO formation. Theoretically, it can be demonstrated that the dioxygen competitive inhibition constant (Kiapp,O2) is formally equal to the Kmapp,O2 of the hydroxylation reaction. (ii) At the same time, the superoxide anion radical formed (Eqs. (2), (4)) reacts with NO to yield peroxynitrite, in a diffusion controlled reaction (k ≈ 109–1010 M−1 s−1). This effect can be counteracted by the presence of superoxide dismutase (k ≈ 2 × 109 M−1 s0.1), although, in this case, the dioxygen is partially restored in the system. (iii) In addition, dioxygen can also react directly with NO, but in a comparatively slow reaction (k ≈ 106–107 M−2 s−1), to yield different products, including •NO2. This dioxygen effect can not be experimentally counteracted. (B) Data from XO-catalyzed NO formation at pH 6.3; adapted with permission from Ref. [137]. Copyright 2015 American Chemical Society. (C) Scheme modified from Ref. [180]. (D) Data pH 6.3; adapted with permission from Ref. [137]. Copyright 2015 American Chemical Society.
The direct reaction of NO with dioxygen (Fig. 8(A), iii) was evaluated using the deflavo-XO and deflavo-AO [139]. These enzymes forms, lacking the flavin, can not reduce dioxygen (see note3; Fig. 3) and, consequently, both the dioxygen inhibition (Fig. 8(A), i) and the NO consumption by superoxide anion radical (Fig. 8(A), ii) are abolished, while the nitrite reduction is not affected (as discussed above). Using the deflavo-enzymes, it was shown that the effect of the NO/O2 reaction on the detected rates of NO formation in vitro can be disregarded [139]. However, in vivo, under non-anoxic conditions, this may not be the case. Nevertheless, this dioxygen effect (Fig. 8(A), iii) is also exerted in the "classic" NOS systems, which must catalyze the NO formation in the presence of dioxygen (one of the reaction substrates; Fig. 2). The effect of dioxygen related with the NO consumption by superoxide in the absence of superoxide dismutase (SOD) (Fig. 8(A), ii) can be substantial [139]. Yet, the XO and AO-dependent NO can be detected in the absence SOD, with the higher amounts of NO being obtained at saturating reducing substrate concentrations. These results highlight the physiological relevance of SOD to achieve a net flux of NO from these enzymes (and other systems), as well as, to avoid the formation of the deleterious peroxynitrite.
The dioxygen inhibition of NO formation (Fig. 8(A), i) was assessed with the native enzymes, in the presence of SOD to eliminate the NO consumption by superoxide radical anion (Fig. 8(A), ii), a reaction that would lead to false dioxygen inhibitions (erroneous lower NO formation rates) [139], [162], [164], [166]. The inhibition of NO formation by dioxygen is dependent not only on the dioxygen concentration, but also on the reducing substrate concentration, with a higher reducing power giving rise to lower inhibitions (because the Kiapp,O2 values increase with increasing reducing substrate concentrations) [139]. Hence, the ischemia-induced accumulation of reducing substrates is expected to decrease the inhibition imposed by dioxygen. Lower dioxygen inhibitions are also obtained with higher nitrite concentrations (Fig. 8(B); because the Kmapp,NO2- values increase with increasing dioxygen concentrations, while the kcatapp,NO2- values do not change significantly, feature of competitive inhibition) [139]. However, even without extravagant nitrite concentrations, a reasonable NO formation can be obtained: with 25 μM nitrite, a rate of 1.3–1.9 nM NO/s [139] can be observed in the presence of 50 μM dioxygen, that is, under the normal (normoxic) tissue dioxygen concentrations [182]; under hypoxia, e.g., 25 μM dioxygen, the values increase to 2.0–2.9 nM NO/s [139] (all values at pH 6.3, with 30 μg enzyme/mL, to simulate 30 μg enzyme/g tissue). These rate values compare very well with the 1 nM/s described for the constitutive NOS [175] and show that the amount of NO delivered by XO, XD and AO can be physiologically significant, even in the presence of dioxygen. Moreover, and most significant for the potential physiological role of these enzymes, the Kiapp,O2 values of XO (24 μM) and AO (25 μM) (Fig. 8(D)) [139] are within the in vivo tissue dioxygen concentrations, going from normoxia to hypoxia (Fig. 8(C)). These relatively high Kiapp,O2 values clearly refute the view argued by some authors that "there is no NO formation by these enzymes in the presence of dioxygen ". Furthermore, these values suggest that the in vivo NO formation by XO and AO could be fine-tuned by the dioxygen availability, thus providing a straightforward mechanism through which the "classic" hydroxylating activity versus nitrite reductase activity of these enzymes could be "automatically" regulated: (i) under normoxic conditions, the enzymes display the hydroxylating activity and the NO formation is considerably hindered; (ii) as the dioxygen concentration decreases towards hypoxic and anoxic conditions, the dioxygen inhibition on the NO formation is relieved and the NO generation is amplified, sustained by the accumulation of reducing substrates and decrease in the cellular pH.
10. XOR and AO-catalyzed nitrite reduction: models to envisage the potential NO formation in vivo
Measuring the NO formation in vivo is a challenging task and several approaches have been used, all with their own disadvantages. Most of the NO-specific electrodes or chemiluminescence (that measures NO only in the gas phase) only detect NO after cell disruption or after NO has diffused out of the cell/tissue (so, measure the surplus NO that didn't reacted inside the cell). Therefore, they do not measure the functional NO present inside a living cell nor can provide spatial information about the site where NO is being formed. A microelectrode can overcome this problem, but limited to a certain scale (μm), and they are very difficult to operate. On the other hand, a non-toxic cell-permeable probe, such as a spin-trap (EPR) or a fluorescence probe, can provide measures of the NO present inside a living cell, but they can induce alterations on the biological system. In addition, in the case of most fluorescence probes, they do not detect NO directly (instead, they react with NO oxidation product(s)) and are not selective in the biological milieu [156], [157], [158], [183], [184]. Hence, to confirm that the measured NO is really meaningful in a given physiological context, it is valuable to have different approaches to measure the NO formation.
To evaluate the potential physiological role of XOR and AO in the generation of NO, different models have been explored, such as models of ischemia/reperfusion and hypoxic injury in heart [38], [39], [45], liver [39], [40], [185], kidney [186], [187], [188], blood vessels and cells (models of inflammation) [189], [190], [191], [192], [193], [194], [195], pulmonary hypertension [55], [65], [196], [197], and several other, that include the "less physiological" homogenates (of heart, aorta, liver, lung, kidney) (Fig. 9) [38], [55], [65], [164], [165], [186], [190], but also animal models [38], [45], [65]. In this respect, it should be borne in mind that some caution must be taken when extrapolating results from animal models to humans, as the enzymes tissue distribution, relative expression and presence of different forms (see notes1 to 4) can be different in diverse animal species.
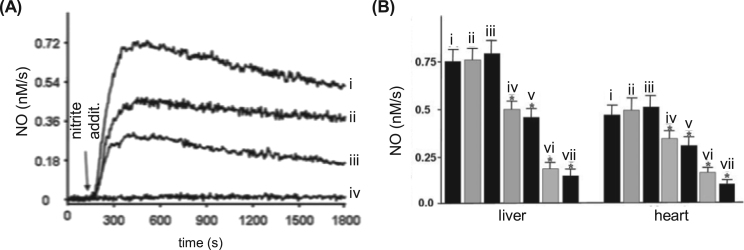
Fig. 9.
Nitrite-dependent NO formation in liver, heart, aorta, and blood. (A) Curves (i), (ii), (iii) and (iv) show the nitrite-dependent NO formation by liver, heart, aorta and blood, respectively (1 g of tissue or 5 mL of blood with 100 μM nitrite). (B) The contribution of or complex I (ii), cytochrome P450 (iii), XOR (iv), AO (v) and XOR plus AO (vi) to the measured NO, in liver and heart, was accessed using the inhibitors rotenone (ii), clotrimazole (iii), oxypurinol (iv), raloxifene (v) and oxypurinol plus raloxifene (vi), respectively (comparatively to an assay without inhibitors (i)). The general enzymatic contribution was accessed by the decreased NO formation caused by preheated the tissue (liver or heart) at 95 °C for 5 min (vii). NO measurements were performed using a chemiluminescence NO analyzer. *, significant inhibition, p < 0.05, compared with the respective control (bar i). Adapted from reference 163 with permission.
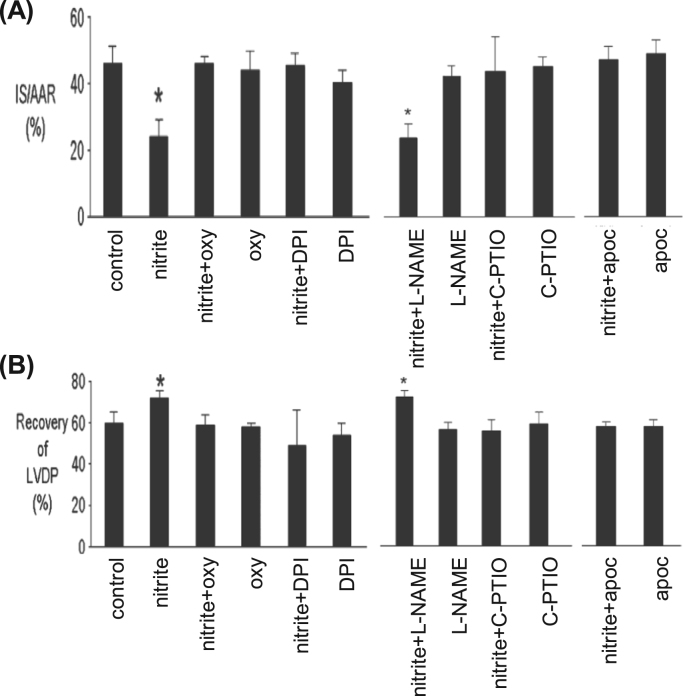
The ground-breaking work in this context is probably the one of Webb et al. [38] that demonstrated the nitrite and XOR-dependent NO formation in an isolated rat heart under ischemic conditions. This NO generation is NOS-independent (L-NAME was used to eliminate the NOS activity) and largely dependent on XOR (as assessed by the inhibition with allopurinol and BOF-4272, two XOR specific inhibitors; NO was detected by chemiluminescence). Noteworthy, the nitrite administration was shown to be cardioprotective (reduced infarct size and improved recovery of left ventricular function) and, most significant, no nitrite-dependent NO formation was observed in hearts that did not experienced the ischemic insult. Corroborating results were also obtained in rat and human heart homogenates, where the NO formation is also pH, nitrite and dioxygen concentration-dependent [38]. Hence, Webb et al. showed that during myocardial ischemia, the hypoxic and acidic environment generated promotes the reduction of nitrite to cardioprotective NO, in a process that is XOR-dependent, and the functionally relevant nitrite-dependent NO formation occurs selectively within ischemic regions. Similar conclusions were later obtained by Baker et al. [45] that also described the nitrite-dependent cardioprotection following ischemia/reperfusion in intact and isolated rat heart (reduction of myocardial necrosis and decline in ventricular function). This protective effect is nitrite concentration-dependent, NOS-independent and associated with XOR, as well as NADPH oxidase and KATP channels (whose inhibition abolishes the cardioprotection) (Fig. 10). Baliga et al. [65] observed the pulmonary protection with dietary nitrite (and nitrate) in a scenario of pulmonary hypertension (reduced the right ventricular pressure and hypertrophy and pulmonary vascular remodeling). This favorable pharmacodynamic profile is dependent on the endothelial NOS and XOR (as assessed with mice lacking endothelial NOS or treated with the allopurinol; NO was detected by chemiluminescence).
Fig. 10.
Nitrite-dependent cardioprotection. Rat hearts were perfused with and without nitrite for 15 min prior to 30 min regional ischemia and 180 min of reperfusion, and percentage of infarction (A) and recovery of left ventricular developed pressure (B) were evaluated (IS, infarct size; LVDP, left ventricular developed pressure). Oxypurinol (oxy, another molybdenum-specific inhibitor, metabolite of allopurinol) or DPI or L-NAME or C-PTIO (a NO oxide scavenger) or apocynin (apoc, an inhibitor of NADPH oxidase) was present in the coronary perfusate for 30 min prior to ischemia, where indicated. * , significant inhibition, p < 0.05, compared with control. Adapted from Ref. [45], Copyright (2007), with permission from Elsevier.
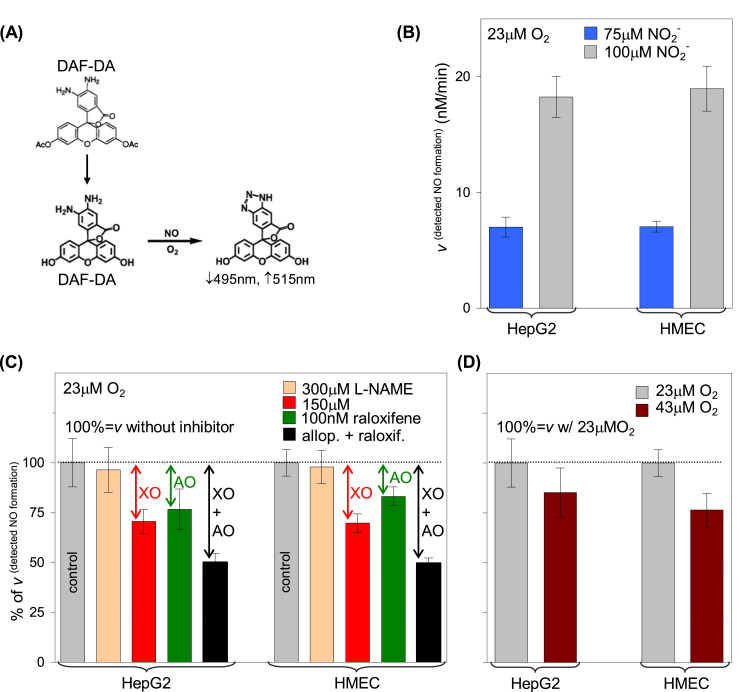
More recently, a different approach was assayed by Maia et al. [139] that used human epithelial cells from liver carcinoma (HepG2) and human microvascular endothelial cells (HMEC) to assess the role of both XOR and AO in the NO generation under hypoxic (23 μM dioxygen) and relative normoxic (43 μM dioxygen) conditions. Those two simple cellular systems generate nitrite-dependent NO in a similar extent and in a manner that is pH, nitrite and dioxygen concentration-dependent (NO was detected with a fluorescence probe) (Fig. 11). NOS, as well as the mitochondrial complex I, do not contribute to the NO formation (L-NAME and rotenone do not significantly inhibit the NO formation), while XOR and AO can account for a remarkable ≈ 50% of the measured NO in these systems (as assessed by the inhibition with allopurinol (XOR) and raloxifene (AO)). Noteworthy, the NO formation is only slightly decreased (14–23%) when these systems are passed from hypoxic (23 μM dioxygen) to relative normoxic conditions (43 μM dioxygen) [139]. This small effect, although in opposition to what Webb et al. described in rat heart, is in accordance with the in vitro kinetic characterization of the enzymes (see above) and maybe due to specificities of each system (cell involved) and pH and dioxygen availability.
Fig. 11.
Nitrite-dependent NO formation by HepG2 and HMEC. (A) The in situ NO formation was followed by spectrofluorimetry, in the presence of the cell-permeant fluorescence probe 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM DA); after internalization, the DAF-FM DA acetate groups are hydrolyzed, to yield DAF-FM, which will then react with the NO oxidation products (intracellular conversion of DAF-FM DA to DAF-FM). (B) The intracellular NO formation by HepG2 and HMEC is dependent on the nitrite concentration (D) and also on the dioxygen availability. (C) The contribution of XOR, AO and NOS to the measured NO was accessed using the inhibitors, allopurinol, raloxifene and L-NAME, respectively. Adapted with permission from Ref. [137]. Copyright 2015 American Chemical Society.
All these studies point to the same conclusion: the nitrite-dependent, XOR and AO-catalyzed NO generation under acidic and hypoxic conditions can be physiologically relevant. Nevertheless, it should be here noted that some authors failed to obtain similar compelling results and, instead, support the involvement of other proteins, such as the Mb and Hb [4], [39], [94], [95], [96], [198], [199].
11. XOR and AO-catalyzed nitrite reduction: outline
The participation of the molybdoenzymes XOR and AO in the mammalian nitrite-derived NO pathway is highly probable. The molybdenum centers are excellent "oxygen atom exchangers", a requirement to convert nitrite into NO, an oxygen atom abstraction reaction (Eq. (1)), making the nitrite reduction to NO by a molybdoenzyme a feasible reaction from the chemical perspective. Moreover, numerous in vitro studies support that the mammalian XOR and AO can contribute to the NO generation. However, the in vivo XOR and AO-dependent NO formation would be modulated by several factors. (i) Availability of reducing substrates - first, and obviously, because they provide the electrons needed to reduce nitrite, but also because they modulated the extension of dioxygen inhibition. Hence, the ischemia-induced reducing substrates accumulation is expected to "fuel" the enzymes with reducing equivalents and also to decrease the inhibition imposed by dioxygen on the NO formation by the low physiological nitrite concentrations. (ii) Dioxygen availability - with Ki values within the transition from normoxia to hypoxia, dioxygen would fine-tune the nitrite-dependent NO formation, being the probable key factor that regulates the NO formation by these enzymes. (iii) Presence of SOD - necessary to achieve a net NO production under non-anoxic conditions. (iv) NAD+ - NAD+ inhibition was not yet studied, but it probably has a marked impact on the XD-dependent NO formation. (v) Acidic conditions - since cellular pH values lower than 6.8 greatly amplifies the nitrite reduction. (vi) And, of course, the nitrite availability. In accordance, several in situ and in vivo studies support that XOR and AO-dependent NO formation can, in fact, occur in vivo, even though the NO fluxes through these enzymes are most certainly modulated by the dioxygen and nitrite availability (competition as oxidizing substrates for XOR and AO).
12. Wrapping up – an outlook on the nitrite/no metabolism
Nitrite is well described as one of the players of the biogeochemical cycle of nitrogen, participating in key prokaryotic pathways crucial to the planetary "recycling" of nitrogen (Fig. 1) and, consequently, to life on Earth. More recently, nitrite was also recognized as a molecule relevant to cell signaling and survival, under challenging conditions, in virtually all forms of life, from bacteria to humans. In spite of those diverse biological functions, in different organism types, nitrite reduction to NO is remarkably similar in all cases: it involves the one-electron reduction of nitrite by a redox active metalloprotein, typically an iron or copper or molybdenum-containing protein, and requires just protons and an electron donor to reduce the metal.
To generate nitrite-dependent signaling NO, organisms seem to be able to use different metalloproteins, present in the cells to accomplish other functions, all apparently dependent on or linked with the cellular redox status and/or dioxygen availability (as is the case of XOR, AO, SO, mARC, Hb, Mb, Nb, Cb, Cc, CcO and many other proteins). In this way, variations on the cellular redox status and/or dioxygen availability can directly "switch" the activity of the metalloproteins, from the "classic" activity to a new nitrite reductase ("non-dedicated nitrite reductase") that produces NO. From a chemical point of view, the organisms are just using the redox chemistry of an available redox system (a heme protein or a molybdoenzyme, e.g.) and doing a "substrate adaptation" to generate NO -thus saving cellular resources, while eliciting a prompt response to the triggering event. From a biological point of view, the use of a single protein to accomplish more than one function (the "classic" one and the nitrite reduction) is no novelty introduced with the nitrite/NO metabolism. This is a well recognized phenomenon -moonlighting- with important implications for Systems Biology and, in particular, for human physiology and pathology (see, e.g., [200]). The existence of two functions within the same protein allows the cell to create regulatory/signaling points, from where the metabolism can be modulated/adapted to properly respond to the event that triggered the activity "switch". In this context, the nitrite reduction to NO can be thought as an universal, conserved mechanism, inherited from a distant pre-aerobic past, that "translates" the cellular redox status and/or dioxygen availability into a differentiated flux of NO; the differentiated flux of NO is, then, "translated" into a biological response to circumvent/overcome/correct the alterations imposed by the redox and/or dioxygen variations. Noteworthy, a pathway inherited from the anaerobic world is a perfect solution for the human cells to cope with hypoxic/anoxic conditions! This exciting hypothesis suggests that nitrite is not only a source of NO, but also a redox status and/or an oxygen sensing molecule. Furthermore, this hypothesis suggests that each individual "non-dedicated nitrite reductase" could be activated when the dioxygen concentration decreases below its own threshold of oxygen-dependent "classic" activity. So, different pathways would be triggered by different dioxygen concentrations/redox conditions. In this way, all "non-dedicated nitrite reductases" could act in a concerted and self-regulated manner, with each individual pathway being relevant under different conditions and in different tissues.
Overall, during the last two decades, it is becoming clear that mammals (and humans in particular) can use two distinct pathways, that operate under opposite conditions, to generate NO: (i) an oxidative pathway that is mediated by specific, tissue-dependent, heme NOS enzymes and depends on dioxygen, (ii) and a reductive pathway that is mediated by (apparently several) "non-dedicated nitrite reductases", depends on nitrite and is favored under low dioxygen concentrations conditions. With these two pathways, cells can maintain the NO formation under the entire dioxygen gradient, from normoxia to anoxia. In humans, these nitrite-dependent pathways are creating new therapeutic opportunities for the management of several pathological conditions, including ischemia, cardiovascular dysfunctions, myocardial infarction, stroke, pulmonary hypertension, infection and also in solid organ transplantation. This is particularly interesting given the known safety of nitrite: it is an anion naturally occurring in our diet and an already FDA-approved therapeutic (for cyanide poisoning). As such, the characterization of potential mammalian nitrite reductases is of most interest and the present review aimed to provide the current picture of our knowledge on the role of the molybdoenzymes XOR and AO on the NO metabolism.
Acknowledgments
We acknowledge Fundação para a Ciência e Tecnologia (FCT/MCTES) for the financial support granted to the Associate Laboratory for Green Chemistry - LAQV, which is financed by national funds from FCT/MCTES (UID/QUI/50006/2013) and co-financed by the ERDF under the PT2020 Partnership Agreement (POCI-01-0145-FEDER - 007265). LBM also thanks to FCT/MCTES for the fellowship grant (SFRH/BPD/111404/2015, which is financed by national funds and co-financed by FSE).
Footnotes
EC according to International Union of Biochemistry and Molecular Biology, Enzyme Nomenclature Committee (www.chem.qmul.ac.uk/iubmb).
Mammalian XO and XD are two forms of the same protein (same gene product). Mammalian XOR enzymes are synthesized as a NAD+-dependent dehydrogenase form, the XD, and are believed to exist mostly as XD under normal physiological conditions [97–102]. However, the XD form can be readily converted into a "strict" oxidase form, the XO. This conversion can be either reversible, through oxidation of Cys535 and Cys992, or irreversible, by proteolysis after Lys551 or Lys569 (bovine XOR numbering) [146], [147], [148], [149], [150], [151]. The only "functional" distinction between XD and XO lies in the electron acceptor used by each form: while XD transfers the electrons preferentially to NAD+, XO fails to react with NAD+ and uses exclusively dioxygen. During the XD into XO conversion process, the protein conformation at the FAD center is modified and this conformational alteration is responsible for the differentiated oxidizing substrate specificity displayed by XO and XD [146–153] (note that both dioxygen and NAD+ react at the FAD center). On the other hand, the protein structure at the iron/sulfur and molybdenum centers is not changed during the conversion and, in accordance, the two enzyme forms are virtually identical in respect to the binding and catalysis of substrates at the molybdenum center, as is the case of oxidation of xanthine and other heterocyclic compounds and aldehydes [97–102]. For these reasons, XO and XD can be considered as one unique enzyme in what concerns the overall structural organization of the molybdenum domain and the molybdenum center reactivity.
AFR, activity-to-flavin ratio, is a measure of the number of XOR and AO molecules with an intact, active molybdenum center, per FAD center. Only XOR and AO molecules with an intact molybdenum center are able to catalyze hydroxylation reactions [97], [98], [99], [100], [101], [102]. Therefore, the hydroxylation activity assay constitutes a suitable way to measure the number of XOR and AO molecules with the molybdenum center intact. Enzyme preparations with a higher amount of damaged molybdenum centers display low hydroxylation activities, relatively to the FAD content, and, consequently, have lower AFR values. Hence, if the nitrite reduction also occurs in the molybdenum center, than the values of nitrite reductase activity would follow the same tendency as the AFR values -what was, in fact, demonstrated [139].
Enzyme form whose FAD center was chemically removed. If the nitrite reduction occurred at the FAD center, than the DPI-inhibited enzymes, as well as, the deflavo-forms would not display nitrite reductase activity.
Enzyme form whose molybdenum sulfo group (Fig. 3(B)) was chemically removed.
This Mo5+-OH (-SH) complex would give rise to the rapid type EPR signal, with two interacting protons [168].
The bacterial copper-containing nitrite reductase displays a similar pH dependence, with pKa values of 5 and 7, and theoretical calculations have suggested that it is the proton transfer from a key neighboring aspartate residue that triggers the electron transfer from copper to nitrite (reviewed in [8]). Moreover, also in CuNiR, the previous nitrite protonation results in the formation of a more stable metal complex, Cu-OH instead of Cu-O-, and the choice of the proton donor, one aspartate in CuNiR and a glutamate residue in XOR/AO, is also similar. In addition, both metal centers share the same square pyramidal geometry and have a redox active HOMO on the xy plane.
References
- 1.Lundberg J.O., Weitzberg E., Gladwin M.T. Nat. Rev. Drug Dis. 2008;7:156. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 2.Lundberg J.O., Gladwin M.T., Ahluwalia A., Benjamin N., Bryan N.S., Butle R.A., Cabrales P., Fago A., Feelisch M., Ford P.C., Freeman B.A., Frenneaux M., Friedman J., Kelm M., Kevil C.G., Kim-Shapiro D.B., Kozlov A.V., Lancaster Jr. J.R., Lefer D.J., McColl K., McCurry K., Patel R.P., Petersson J., Rassaf T., Reutov V.P., Richter-Addo J.B., Schechter A., Shiva S., Tsuchiya K., Faassen E.E., Webb A.J., Zuckerbraun B.S., Zweier J.L., Weitzberg E. Nat. Chem. Biol. 2009;5:865. doi: 10.1038/nchembio.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castiglione N., Rinaldo S., Giardina G., Stelitano V., Cutruzzola F. Antioxid. Redox Signal. 2012;17:684. doi: 10.1089/ars.2011.4196. [DOI] [PubMed] [Google Scholar]
- 4.Shiva S. Redox Biol. 2013;1:40. doi: 10.1016/j.redox.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim-Shapiro D.B., Gladwin M.T. Nitric Oxide. 2014;38:58. doi: 10.1016/j.niox.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan N.S., Ivy J.L. Nutr. Res. 2015;35:643. doi: 10.1016/j.nutres.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Khatri J., Mills C.E., Maskell P., Odongerel C., Webb A.J. Br. J. Clin. Pharmacol. 2017;83:129. doi: 10.1111/bcp.12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maia L.B., Moura J.J.G. Chem. Rev. 2014;114:5273. doi: 10.1021/cr400518y. [DOI] [PubMed] [Google Scholar]
- 9.Maia L.B., Moura J.J.G. J. Biol. Inorg. Chem. 2015;20:403. doi: 10.1007/s00775-014-1234-2. [DOI] [PubMed] [Google Scholar]
- 10.Chamizo-Ampudia A., Sanz-Luque E., Llamas A., Galvan A., Fernandez E. Trends Plant Sci. 2017;22:163. doi: 10.1016/j.tplants.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Maia L., Moura J.J.G. Lessons from denitrification for the human metabolism of signalling nitric oxide. In: Moura I., Moura J.J.G., Pauleta S.R., Maia L., editors. Chapter 17. The Royal Society of Chemistry; Camb: 2017. pp. 419–443. (Metalloenzymes in Denitrification: Applications and Environmental Impacts). [Google Scholar]
- 12.Moura I., Maia L., Pauleta S.R., Moura J.J.G. In: Moura I., Moura J.J.G., Pauleta S.R., Maia L., editors. Chapter 1. The Royal Society of Chemistry; Camb: 2017. pp. 1–10. (A Bird’s-Eye View of Denitrification in Relation to the Nitrogen Cycle. In Metalloenzymes in Denitrification: Applications and Environmental Impacts). [Google Scholar]
- 13.Kuypers M.M.M., Marchant H.K., Kartal B. Nat. Rev. Microbiol. 2018;16:263. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 14.Moncada S., Palmer R.M.J., Higgs E.A. Pharmacol. Rev. 1991;43:109. [PubMed] [Google Scholar]
- 15.Pfeiffer S., Mayer B., Hemmens B. Angew. Chem. Int. Ed. Engl. 1999;38:1714. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1714::AID-ANIE1714>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Stuehr D.J. Biochim. Biophys. Acta. 1999;1411:217. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 17.Alderton W.K., Cooper C.E., Knowles R.G. Biochem. J. 2001;357:593. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle M.P., Hoekstra J.W. J. Inorg. Biochem. 1981;14:351. doi: 10.1016/s0162-0134(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 19.Sharma V.S., Taylor T.G., Gardiner R. Biochemistry. 1987;26:3837. doi: 10.1021/bi00387a015. [DOI] [PubMed] [Google Scholar]
- 20.Eich R.F., Li T., Lemon D.D., Doherty D.H., Curry S.R., Aitken J.F., Mathews A.J., Johnson K.A., Smith R.D., Jr., Olson J.S. Biochemistry. 1996;35:6976. doi: 10.1021/bi960442g. [DOI] [PubMed] [Google Scholar]
- 21.Liu X., Miller M.J.S., Joshi M.S., Sadowska-Krowicka H., Clark D.A., Lancaster J.R. J. Biol. Chem. 1998;273:18709. doi: 10.1074/jbc.273.30.18709. [DOI] [PubMed] [Google Scholar]
- 22.Gow A.J., Luchsinger B.P., Pawloski J.R., Singel D.J., Stamler J.S. Proc. Natl. Acad. Sci. USA. 1999;96:9027. doi: 10.1073/pnas.96.16.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunori M. Trends Biochem. Sci. 2001;26:21. doi: 10.1016/s0968-0004(00)01698-4. [DOI] [PubMed] [Google Scholar]
- 24.Flögel U., Merx M.W., Gödecke A., Decking U.K.M., Schrader J. Proc. Natl. Acad. Sci. USA. 2001;98:735. doi: 10.1073/pnas.011460298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang K.-T., Han T.H., Hyduke D.R., Vaughn M.W., Herle H.V., Hein T.W., Zhang C., Kuo L., Liao J.C. Proc. Natl. Acad. Sci. USA. 2001;98:11771. doi: 10.1073/pnas.201276698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herold S., Exner M., Nauser T. Biochemistry. 2001;40:3385. doi: 10.1021/bi002407m. [DOI] [PubMed] [Google Scholar]
- 27.Witting P.K., Douglas D.J., Mauk A.G. J. Biol. Chem. 2001;276:3991. doi: 10.1074/jbc.M005758200. [DOI] [PubMed] [Google Scholar]
- 28.Joshi M.S., Ferguson T.B., Jr., Han T.H., Hyduke D.R., Liao J.C., Rassaf T., Bryan N., Feelisch M., Lancaster J.R., Jr. Proc. Natl. Acad. Sc. USA. 2002;99:10341. doi: 10.1073/pnas.152149699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner P.R., Gardner A.M., Brashear W.T., Suzuki T., Hvitved A.N., Setchell K.D.R., Olson J.S. J. Inorg. Biochem. 2006;100:542. doi: 10.1016/j.jinorgbio.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 30.Shiva S., Wang X., Ringwood L.A., Xu X., Yuditskaya S., Annavajjhala V., Miyajima H., Hogg N., Harris Z.L., Gladwin M.T. Nat. Chem. Biol. 2006;2:486. doi: 10.1038/nchembio813. [DOI] [PubMed] [Google Scholar]
- 31.Brunori M., Giuffre A., Forte E., Mastronicola D., Barone M.C., Sarti P. Biochim. Biophys. Acta. 1655;2004:365. doi: 10.1016/j.bbabio.2003.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Wink D.A., Darbyshire J.F., Nims R.W., Saavedra J.E., Ford P.C. Chem. Res. Toxicol. 1993;6:23. doi: 10.1021/tx00031a003. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein S., Czapski G. J. Am. Chem. Soc. 1995;117:12078. doi: 10.1021/ja00269a020. [DOI] [PubMed] [Google Scholar]
- 34.Liu X., Liu Q., Gupta E., Zorko N., Brownlee E., Zweier J.L. Nitric Oxide. 2005;13:68. doi: 10.1016/j.niox.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Johnson G., 3rd, Tsao P.S., Mulloy D., Lefer A.M. J. Pharmacol. Exp. Ther. 1990;252:35. [PubMed] [Google Scholar]
- 36.Demoncheaux E.A., Higenbottam T.W., Foster P.J., Borland C.D., Smith A.P., Marriott H.M., Bee D., Akamine S., Davies M.B. Clin. Sci. (Lond.) 2002;102:77. [PubMed] [Google Scholar]
- 37.Hunter C.J., Dejam A., Blood A.B., Shields H., Kim-Shapiro D.B., Machado R.F., Tarekegn S., Mulla N., Hopper A.O., Schechter A.N., Power G.G., Gladwin M.T. Nat. Med. 2004;10:1122. doi: 10.1038/nm1109. [DOI] [PubMed] [Google Scholar]
- 38.Webb A., Bond R., McLean P., Uppal R., Benjamin N., Ahluwalia A. Proc. Natl. Acad. Sci. USA. 2004;101:13683. doi: 10.1073/pnas.0402927101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duranski M.R., Greer J.J., Dejam A., Jaganmohan S., Hogg N., Langston W., Patel R.P., Yet S.F., Wang X., Kevil C.G., Gladwin M.T., Lefer D.J. J. Clin. Investig. 2005;115:1232. doi: 10.1172/JCI22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu P., Liu F., Yao Z., Wang C.Y., Chen D.D., Tian Y., Zhang J.H., Wu Y.H. Hepatobiliary Pancreas Dis. Int. 2005;4:350. [PubMed] [Google Scholar]
- 41.Lundberg J.O., Weitzberg E. Arterioscler. Thromb. Vasc. Biol. 2005;25:915. doi: 10.1161/01.ATV.0000161048.72004.c2. [DOI] [PubMed] [Google Scholar]
- 42.Pluta R.M., Dejam A., Grimes G., Gladwin M.T., Oldfield E.H. JAMA. 2005;293:1477. doi: 10.1001/jama.293.12.1477. [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya K., Kanematsu Y., Yoshizumi M., Ohnishi H., Kirima K., Izawa Y., Shikishima M., Ishida T., Kondo S., Kagami S., Takiguchi Y., Tamaki T. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H2163. doi: 10.1152/ajpheart.00525.2004. [DOI] [PubMed] [Google Scholar]
- 44.Jung K.-H., Chu K., Ko S.-Y., Lee S.-T., Sinn D.-I., Park D.-K., Kim J.-M., Song E.-C., Kim M., Roh J.K. Stroke. 2006;37:2744. doi: 10.1161/01.STR.0000245116.40163.1c. [DOI] [PubMed] [Google Scholar]
- 45.Baker J.E., Su J., Fu X., Hsu A., Gross G.J., Tweddell J.S., Hogg N. J. Mol. Cell. Cardiol. 2007;43:437. doi: 10.1016/j.yjmcc.2007.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bryan N.S., Calvert J.W., Elrod J.W., Gundewar S., Ji S.Y., Lefer D.J. Proc. Natl. Acad. Sci. USA. 2007;104:19144. doi: 10.1073/pnas.0706579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dezfulian C., Raat N., Shiva S., Gladwin M.T. Cardiovasc. Res. 2007;75:327. doi: 10.1016/j.cardiores.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldfield E.H., Cannon R.O., 3rd., Schechter A.N., Gladwin M.T. Circulation. 2007;116:1821. doi: 10.1161/CIRCULATIONAHA.107.712133. [DOI] [PubMed] [Google Scholar]
- 49.Shiva S., Sack M.N., Greer J.J., Duranski M., Ringwood L.A., Burwell L., Wang X., MacArthur P.H., Shoja A., Raghavachari N., Calvert J.W., Brookes P.S., Lefer D.J., Gladwin M.T. J. Exp. Med. 2007;204:2089. doi: 10.1084/jem.20070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bryan N.S., Calvert J.W., Gundewar S., Lefer D.J. Free Radic. Biol. Med. 2008;45:468. doi: 10.1016/j.freeradbiomed.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez F.M., Shiva S., Vincent P.S., Ringwood L.A., Hsu L.Y., Hon Y.Y., Aletras A.H., Cannon R.O., 3rd, Gladwin M.T., Arai A.E. Circulation. 2008;117:2986. doi: 10.1161/CIRCULATIONAHA.107.748814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maher A.R., Milsom A.B., Gunaruwan P., Abozguia K., Ahmed I., Weaver R.A., Thomas P., Ashrafian H., Born G.V., James P.E., Frenneaux M.P. Circulation. 2008;117:670. doi: 10.1161/CIRCULATIONAHA.107.719591. [DOI] [PubMed] [Google Scholar]
- 53.Sinha S.S., Shiva S., Gladwin M.T. Trends Cardiovasc. Med. 2008;18:163. doi: 10.1016/j.tcm.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Raat N.J., Shiva S., Gladwin M.T. Adv. Drug. Deliv. Rev. 2009;61:339. doi: 10.1016/j.addr.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Zuckerbraun B.S., Shiva S., Ifedigbo E., Mathier M.A., Mollen K.P., Rao J., Bauer P.M., Choi J.J., Curtis E., Choi A.M., Gladwin M.T. Circulation. 2010;121:98. doi: 10.1161/CIRCULATIONAHA.109.891077. [DOI] [PubMed] [Google Scholar]
- 56.Alef M.J., Vallabhaneni R., Carchman E., Morris S.M., Jr., Shiva S., Wang Y., Kelley E.E., Tarpey M.M., Gladwin M.T., Tzeng E., Zuckerbraun B.S. J. Clin. Investig. 2011;121:1646. doi: 10.1172/JCI44079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blood A.B., Schroeder H.J., Terry M.H., Merrill-Henry J., Bragg S.L., Vrancken K., Liu T., Herring J.L., Sowers L.C., Wilson S.M., Power G.G. Circulation. 2011;123:605. doi: 10.1161/CIRCULATIONAHA.110.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilchrist M., Shore A.C., Benjamin N. Cardiovasc. Res. 2011;89:492. doi: 10.1093/cvr/cvq309. [DOI] [PubMed] [Google Scholar]
- 59.Kevil C.G., Kolluru G.K., Pattillo C.B., Giordano T. Free Radic. Biol. Med. 2011;51:576. doi: 10.1016/j.freeradbiomed.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larsen F.J., Schiffer T.A., Borniquel S., Sahlin K., Ekblom B., Lundberg J.O., Weitzberg E. Cell Metab. 2011;13:149. doi: 10.1016/j.cmet.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Lundberg J.O., Carlstrom M., Larsen F.J., Weitzberg E. Cardiovasc. Res. 2011;89:525. doi: 10.1093/cvr/cvq325. [DOI] [PubMed] [Google Scholar]
- 62.Murillo D., Kamga C., Mo L., Shiva S. Nitric Oxide. 2011;25:70. doi: 10.1016/j.niox.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pattillo C.B., Bir S., Rajaram V., Kevil C.G. Cardiovasc. Res. 2011;89:533. doi: 10.1093/cvr/cvq297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sindler A.L., Fleenor B.S., Calvert J.W., Marshall K.D., Zigler M.L., Lefer D.J., Seals D.R. Aging Cell. 2011;10:429. doi: 10.1111/j.1474-9726.2011.00679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baliga R.S., Milsom A.B., Ghosh S.M., Trinder S.L., Macallister R.J., Ahluwalia A., Hobbs A.J. Circulation. 2012;125:2922. doi: 10.1161/CIRCULATIONAHA.112.100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sparacino-Watkins C.E., Lai Y.C., Gladwin M.T. Ther. Circ. 2012;125:2824. doi: 10.1161/CIRCULATIONAHA.112.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bueno M., Wang J., Mora A.L., Gladwin M.T. Antioxid. Redox Signal. 2013;18:1797. doi: 10.1089/ars.2012.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghosh S.M., Kapil V., Fuentes-Calvo I., Bubb K.J., Pearl V., Milsom A.B., Khambata R., Maleki-Toyserkani S., Yousuf M., Benjamin N., Webb A.J., Caulfield M.J., Hobbs A.J., Ahluwalia A. Hypertension. 2013;61:1091. doi: 10.1161/HYPERTENSIONAHA.111.00933. [DOI] [PubMed] [Google Scholar]
- 69.Omar S.A., Webb A. J. Mol. Cell Cardiol. 2014;73:57. doi: 10.1016/j.yjmcc.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 70.Reichert E.T., Mitchell S.W. Am. J. Med. Sci. 1880;159:158. [Google Scholar]
- 71.Wang J., Krizowski S., Fischer-Schrader K., Niks D., Tejero J., Sparacino-Watkins C., Wang L., Ragireddy V., Frizzell S., Kelley E.E., Zhang Y., Basu P., Hille R., Schwarz G., Gladwin M.T. Antioxid. Redox Signal. 2015;23:283. doi: 10.1089/ars.2013.5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sparacino-Watkins C.E., Tejero J., Sun B., Gauthier M.C., Thomas J., Ragireddy V., Merchant B.A., Wang J., Azarov I., Basu P., Gladwin M.T. J. Biol. Chem. 2014;289:10345. doi: 10.1074/jbc.M114.555177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tiso M., Tejero J., Basu S., Azarov I., Wang X., Simplaceanu V., Frizzell S., Jayaraman T., Geary L., Shapiro C., Ho C., Shiva S., Kim-Shapiro D.B., Gladwin M.T. J. Biol. Chem. 2011;286:18277. doi: 10.1074/jbc.M110.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H., Hemann C., Abdelghany T.M., El-Mahdy M.A., Zweier J.L. J. Biol. Chem. 2012;278:36623. doi: 10.1074/jbc.M112.342378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basu S., Azarova N.A., Font M.D., King S.B., Hogg N., Gladwin M.T., Shiva S., Kim-Shapiro D.B. J. Biol. Chem. 2008;283:32590. doi: 10.1074/jbc.M806934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Liu X., Cui H., Chen Y.R., Cardounel A.J., Zweier J.L. J. Biol. Chem. 2006;281:12546. doi: 10.1074/jbc.M511803200. [DOI] [PubMed] [Google Scholar]
- 77.Castello P.R., Woo D.K., Ball K., Wojcik J., Liu L., Poyton R.O. Proc. Natl. Acad. Sci. USA. 2008;105:8203. doi: 10.1073/pnas.0709461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benamar A., Rolletschek H., Borisjuk L., Avelange-Macherel M.-H., Curien G., Mostefai H.A., Andriantsitohaina R., Macherel D. Biochim. Biophys. Acta. 1777;2008:1268. doi: 10.1016/j.bbabio.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 79.Castello P.R., David P.S., McClure T., Crook Z., Poyton R.O. Cell Metab. 2006;3:277. doi: 10.1016/j.cmet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 80.Badejo A.M., Jr., Hodnette C., Dhaliwal J.S., Casey D.B., Pankey E., Murthy S.N., Nossaman B.D., Hyman A.L., Kadowitz P. J. Am. J. Physiol. Heart Circ. Physiol. 2010;299:H819. doi: 10.1152/ajpheart.00959.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kozlov A.V., Staniek K., Nohl H. FEBS Lett. 1999;454:127. doi: 10.1016/s0014-5793(99)00788-7. [DOI] [PubMed] [Google Scholar]
- 82.Gautier C., van Faassen E., Mikula I., Martasek P., Slama-Schwok A. Biochem. Biophys. Res. Commun. 2006;341:816. doi: 10.1016/j.bbrc.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 83.Vanin A.F., Bevers L.M., Slama-Schwok A., van Faassen E.E. Cell Mol. Life Sci. 2006;64:96. doi: 10.1007/s00018-006-6374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benjamin N., O’Driscoll F., Dougall H., Duncan C., Smith L., Golden M. Nature. 1994;368:502. doi: 10.1038/368502a0. [DOI] [PubMed] [Google Scholar]
- 85.Lundberg J.O.N., Weitzberg E., Lundberg J.M., Alving K. Gut. 1994;35:1543. doi: 10.1136/gut.35.11.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zweier J.L., Wang P., Samouilov A., Kuppusamy P. Nat. Med. 1995;1:804. doi: 10.1038/nm0895-804. [DOI] [PubMed] [Google Scholar]
- 87.McKnight G.M., Smith L.M., Drummond R.S., Duncan C.W., Benjamin N. Gut. 1997;40:211. doi: 10.1136/gut.40.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zweier J.L., Samouilov A., Kuppusamy P. Biochim. Biophys. Acta. 1999;1411:250. doi: 10.1016/s0005-2728(99)00018-3. [DOI] [PubMed] [Google Scholar]
- 89.Gago B., Lundberg J.O., Barbosa R.M., Laranjinha J. Free Radic. Biol. Med. 2007;43:1233. doi: 10.1016/j.freeradbiomed.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 90.Rocha B.S., Gago B., Barbosa R.M., Laranjinha J. Toxicology. 2009;265:41. doi: 10.1016/j.tox.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 91.Pereira C., Ferreira N.R., Rocha B.S., Barbosa R.M., Laranjinha J. Redox Biol. 2013;9:276. doi: 10.1016/j.redox.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rocha B.S., Nunes C., Pereira C., Barbosa R.M., Laranjinha J. Food Funct. 2014;5:1646. doi: 10.1039/c4fo00124a. [DOI] [PubMed] [Google Scholar]
- 93.Rocha B.S., Lundberg J.O., Radi R., Laranjinha J. Redox Biol. 2016;8:407. doi: 10.1016/j.redox.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hendgen-Cotta U.B., Merx M.W., Shiva S., Schmitz J., Becher S., Klare J.P., Steinhoff H., Goedecke A., Schrader J., Gladwin M.T., Kelm M., Rassaf T. Proc. Natl. Acad. Sci. USA. 2008;105:10256. doi: 10.1073/pnas.0801336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shiva S., Huang Z., Grubina R., Sun J., Ringwood L.A., MacArthur P.H., Xu X., Murphy E., Darley-Usmar V.M., Gladwin M.T. Circ. Res. 2007;100:654. doi: 10.1161/01.RES.0000260171.52224.6b. [DOI] [PubMed] [Google Scholar]
- 96.Rassaf T., Flögel U., Drexhage C., Hendgen-Cotta U., Kelm M., Schrader J. Circ. Res. 2007;100:1749. doi: 10.1161/CIRCRESAHA.107.152488. [DOI] [PubMed] [Google Scholar]
- 97.Hille R. Arch. Biochem. Biophys. 2005;433:107. doi: 10.1016/j.abb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 98.Okamoto K., Kusano T., Nishino T. Curr. Pharm. Des. 2013;19:2606. doi: 10.2174/1381612811319140010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hille R., Hall J., Basu P. Chem. Rev. 2014;114:3963. doi: 10.1021/cr400443z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Battelli M.G., Polito L., Bortolotti M., Bolognesi A. Curr. Med. Chem. 2016;23:4027. doi: 10.2174/0929867323666160725091915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maia L., Moura I., Moura J.J.G. Molybdenum and tungsten-containing enzymes: an overview. In: Hille R., Schulzke C., Kirk M.L., editors. Chapter 1. Royal Society of Chemistry; Cambridge: 2017. pp. 1–80. (Molybdenum and Tungsten Enzymes). [Google Scholar]
- 102.Nishino T., Okamoto K., Leimkuhler S. Enzymes of the xanthino oxidase family. In: Hille R., Schulzke C., Kirk M.L., editors. Chapter 6. Royal Society of Chemistry; Cambridge: 2017. pp. 192–239. (Molybdenum and Tungsten Enzymes). [Google Scholar]
- 103.Huang D.Y., Furukawa A., Ichikawa Y. Arch. Biochem. Biophys. 1999;364:264. doi: 10.1006/abbi.1999.1129. [DOI] [PubMed] [Google Scholar]
- 104.Garattini E., Fratelli M., Terao M. Cell. Mol. Life Sci. 2008;65:1019. doi: 10.1007/s00018-007-7398-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pryde D.C., Dalvie D., Hu Q., Jones P., Obach R.S., Tran T.D. J. Med. Chem. 2010;53:8441. doi: 10.1021/jm100888d. [DOI] [PubMed] [Google Scholar]
- 106.Garattini E., Terao M. Drug Metab. Rev. 2011;43:374. doi: 10.3109/03602532.2011.560606. [DOI] [PubMed] [Google Scholar]
- 107.Swenson T.L., Casida J.E. Toxicol. Sci. 2013;133:22. doi: 10.1093/toxsci/kft066. [DOI] [PubMed] [Google Scholar]
- 108.Terao M., Romão M.J., Leimkuhler S., Bois M., Fratelli M., Coelho C., Santos-Silva T., Garattini E. Arch. Toxicol. 2016;90:753. doi: 10.1007/s00204-016-1683-1. [DOI] [PubMed] [Google Scholar]
- 109.Granger D.N., Rutili G., McCord J.M. Gastroenterology. 1981;81:22. [PubMed] [Google Scholar]
- 110.McCord J.M. N. Engl. J. Med. 1985;312:159. doi: 10.1056/NEJM198501173120305. [DOI] [PubMed] [Google Scholar]
- 111.Lieber C.S. N. Engl. J. Med. 1988;319:1639. doi: 10.1056/NEJM198812223192505. [DOI] [PubMed] [Google Scholar]
- 112.Zweier J.L., Kuppusamy P., Lutty G.A. Proc. Natl. Acad. Sci. USA. 1988;85:4046. doi: 10.1073/pnas.85.11.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cederbaum A.I. Free Radic. Biol. Med. 1989;7:537. doi: 10.1016/0891-5849(89)90029-4. [DOI] [PubMed] [Google Scholar]
- 114.Kato S., Kawase T., Alderman J., Inatomi N., Lieber C. Gastroenterology. 1990;98:203. doi: 10.1016/0016-5085(90)91311-s. [DOI] [PubMed] [Google Scholar]
- 115.Nordmann R., Ribière C., Rouach H. Free Radic. Biol. Med. 1992;12:219. doi: 10.1016/0891-5849(92)90030-k. [DOI] [PubMed] [Google Scholar]
- 116.Terada L.S., Dormish J.J., Shanley P.F., Leff J.A., Anderson B.O., e Repine J.E. Am. J. Physiol. 1992;263:L394. doi: 10.1152/ajplung.1992.263.3.L394. [DOI] [PubMed] [Google Scholar]
- 117.Zweier J.L., Broderick R., Kuppusamy P., Thompson-Gorman S., Lutty G.A. J. Biol. Chem. 1994;269:24156. [PubMed] [Google Scholar]
- 118.Mira L., Maia L., Barreira L., Manso C.F. Arch. Biochem. Biophys. 1995;318:53. doi: 10.1006/abbi.1995.1203. [DOI] [PubMed] [Google Scholar]
- 119.Weinbroum A., Nielsen V.G., Tan S., Gelman S., Matalon S., Skinner K.A., Bradley E., Jr., Parks D.A. Am. J. Physiol. 1995;268:G988. doi: 10.1152/ajpgi.1995.268.6.G988. [DOI] [PubMed] [Google Scholar]
- 120.Harrison R. Biochem. Soc. Trans. 1997;25:786. doi: 10.1042/bst0250786. [DOI] [PubMed] [Google Scholar]
- 121.Nishino T., Nakanishi S., Okamoto K., Mizushima J., Hori H., Iwasaki T., Nishino T., Ichimori K., Nakazawa H. Biochem. Soc. Trans. 1997;25:783. doi: 10.1042/bst0250783. [DOI] [PubMed] [Google Scholar]
- 122.Wright R.M., Repine J.E. Biochem. Soc. Trans. 1997;25:799. doi: 10.1042/bst0250799. [DOI] [PubMed] [Google Scholar]
- 123.Beckman K.B., Ames B.N. Physiol. Rev. 1998;78:547. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 124.Suzuki H., Delano F.A., Parks D.A., Jamshidi N., Granger D.N., Ishii H., Suematsu M., Zweifach B.W., Schmid-Schonbein G.W. Proc. Natl. Acad. Sci. USA. 1998;95:4754. doi: 10.1073/pnas.95.8.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wright R.M., McManaman J.L., Repine J.E. Free Radic. Biol. Med. 1999;26:348. doi: 10.1016/s0891-5849(98)00204-4. [DOI] [PubMed] [Google Scholar]
- 126.Harrison R. Free Radic. Biol. Med. 2002;33:774. doi: 10.1016/s0891-5849(02)00956-5. [DOI] [PubMed] [Google Scholar]
- 127.Stirpe F., Ravaioli M., Battelli M.G., Musiani S., Grazi G.L. Am. J. Gastroenterol. 2002;97:2079. doi: 10.1111/j.1572-0241.2002.05925.x. [DOI] [PubMed] [Google Scholar]
- 128.Wu D., Cederbaum A.I. Alcohol Res. Health. 2003;27:277. [PMC free article] [PubMed] [Google Scholar]
- 129.Berry C.E., Hare J.M. J. Physiol. 2004;555:589. doi: 10.1113/jphysiol.2003.055913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maia L., Vala A., Mira L. Free Radic. Res. 2005;39:979. doi: 10.1080/10715760500210962. [DOI] [PubMed] [Google Scholar]
- 131.Kelley E.E., Hock T., Khoo N.K., Richardson G.R., Johnson K.K., Powell P.C., Giles G.I., Agarwal A., Lancaster J.R., Jr., Tarpey M.M. Free Radic. Biol. Med. 2006;40:952. doi: 10.1016/j.freeradbiomed.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 132.Maia L., Duarte R.O., Ponces-Freire A., Moura J.J.G., Mira L. J. Biol. Inorg. Chem. 2007;12:777. doi: 10.1007/s00775-007-0229-7. [DOI] [PubMed] [Google Scholar]
- 133.Maia L.B., Moura J.J.G. J. Biol. Inorg. Chem. 2011;16:443. doi: 10.1007/s00775-010-0741-z. [DOI] [PubMed] [Google Scholar]
- 134.Small D.M., Coombes J.S., Bennett N., Johnson D.W., Gobe G.C. Nephrology. 2012;17:311. doi: 10.1111/j.1440-1797.2012.01572.x. [DOI] [PubMed] [Google Scholar]
- 135.Bachschmid M.M., Schildknecht S., Matsui R., Zee R., Haeussler D., Cohen R.A., Pimental D., Loo B. Ann. Med. 2013;45:17. doi: 10.3109/07853890.2011.645498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cantu-Medellin N., Kelley E.E. Redox Biol. 2013;1:353. doi: 10.1016/j.redox.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Madamanchi N.R., Runge M.S. Free Radic. Biol. Med. 2013;61:473. doi: 10.1016/j.freeradbiomed.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kelley E.E. Pharmacol. Rep. 2015;67:669. doi: 10.1016/j.pharep.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maia L.B., Pereira V., Mira L., Moura J.J.G. Biochemistry. 2015;54:685. doi: 10.1021/bi500987w. [DOI] [PubMed] [Google Scholar]
- 140.Nishino T., Okamoto K. J. Biol. Inorg. Chem. 2015;20:195. doi: 10.1007/s00775-014-1210-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Battelli M.G., Polito L., Bortolotti M., Bolognesi A. Curr. Med. Chem. 2016;23:4027. doi: 10.2174/0929867323666160725091915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Garattini E., Terao M. Xanthine Oxidoreductase and Aldehyde Oxidases. In Comprehensive Toxicology. Third ed. Elsevier; 2017. pp. 208–232. [Google Scholar]
- 143.Paragas E.M., Humphreys S.C., Min J., Joswig-Jones C.A., Jones J.P. Biochem. Pharmacol. 2017;145:210. doi: 10.1016/j.bcp.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 144.Kumar R., Joshi G., Kler H., Kalra S., Kaur M., Arya R. Med. Res. Rev. 2018;38:1073. doi: 10.1002/med.21457. [DOI] [PubMed] [Google Scholar]
- 145.Turner N.A., Doyle W.A., Ventom A.M., Bray R.C. Eur. J. Biochem. 1995;232:646. [PubMed] [Google Scholar]
- 146.Nishino T., Okamoto K., Eger B.T., Pai E.F., Nishino T. FEBS J. 2008;275:3278. doi: 10.1111/j.1742-4658.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 147.Enroth C., Eger B.T., Okamoto K., Nishino T., Nishino T., Pai E.F. Proc. Natl. Acad. Sci. USA. 2000;97:10723. doi: 10.1073/pnas.97.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nishino T., Okamoto K., Kawaguchi Y., Hori H., Matsumura T., Eger B.T., Pai E.F., Nishino T. J. Biol. Chem. 2005;280:24888. doi: 10.1074/jbc.M501830200. [DOI] [PubMed] [Google Scholar]
- 149.Tsujii A., Nishino T. Nucleosides Nucleotides Nucleic Acids. 2008;27:881. doi: 10.1080/15257770802146569. [DOI] [PubMed] [Google Scholar]
- 150.Ishikita H., Eger B.T., Okamoto K., Nishino T., Pai E.F. J. Am. Chem. Soc. 2012;134:999. doi: 10.1021/ja207173p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nishino T., Okamoto K., Kawaguchi Y., Matsumura T., Eger B.T., Pai E.F., Nishino T. FEBS J. 2015;282:3075. doi: 10.1111/febs.13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kuwabara Y., Nishino T., Okamoto K., Matsumura T., Eger B.T., Pai E.F., Nishino T. Proc. Natl. Acad. Sci. USA. 2003;100:8170. doi: 10.1073/pnas.1431485100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Okamoto K., Matsumoto K., Hille R., Eger B.T., Pai E.F., Nishino T. Proc. Natl. Acad. Sci. USA. 2004;101:7931. doi: 10.1073/pnas.0400973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Doonan C.J., Stockert A., Hille R., George G.N. J. Am. Chem. Soc. 2005;127:4518. doi: 10.1021/ja042500o. [DOI] [PubMed] [Google Scholar]
- 155.Coelho C., Mahro M., Trincão J., Carvalho A.T.P., Ramos M.J., Terao M., Garattini E., Leimkühler S., Romão M.J. J. Biol. Chem. 2012;287:40690. doi: 10.1074/jbc.M112.390419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kojima H., Nakatsubo N., Kikuchi K., Kawahara S., Kirino Y., Nagoshi H., Hirata Y., Nagano T. Anal. Chem. 1998;70:2446. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- 157.Sasaki E., Kojima H., Nishimatsu H., Urano Y., Kikuchi K., Hirata Y., Nagano T. J. Am. Chem. Soc. 2005;127:3684. doi: 10.1021/ja042967z. [DOI] [PubMed] [Google Scholar]
- 158.Yu H.B., Zhang X.F., Xiao Y., Zou W., Wang L.P., Jin L.J. Anal. Chem. 2013;85:7076. doi: 10.1021/ac401916z. [DOI] [PubMed] [Google Scholar]
- 159.Loas A., Lippard S.J. J. Mater. Chem. B. 2017;5:8929. doi: 10.1039/c7tb02666h. [DOI] [PubMed] [Google Scholar]
- 160.Millar T.M., Stevens C.R., Benjamin N., Eisenthal R., Harrison R., Blake D.R. FEBS Lett. 1998;427:225. doi: 10.1016/s0014-5793(98)00430-x. [DOI] [PubMed] [Google Scholar]
- 161.Zhang Z., Naughton D., Winyard P.G., Benjamin N., Blake D.R., Symons M.C. Biochem. Biophys. Res. Commun. 1998;249:767. doi: 10.1006/bbrc.1998.9226. [DOI] [PubMed] [Google Scholar]
- 162.Godber H.L.J., Doel J.J., Sapkota G.P., Blake D.R., Stevens C.R., Eisenthal R., Harrison R. J. Biol. Chem. 2000;275:7757. doi: 10.1074/jbc.275.11.7757. [DOI] [PubMed] [Google Scholar]
- 163.Li H., Samouilov A., Liu X., Zweier J.L. J. Biol. Chem. 2001;276:24482. doi: 10.1074/jbc.M011648200. [DOI] [PubMed] [Google Scholar]
- 164.Li H., Samouilov A., Liu X., Zweier J.L. J. Biol. Chem. 2004;279:16939. doi: 10.1074/jbc.M314336200. [DOI] [PubMed] [Google Scholar]
- 165.Li H., Cui H., Kundu T.K., Alzawahra W., Zweier J.L. J. Biol. Chem. 2008;283:17855. doi: 10.1074/jbc.M801785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li H., Kundu T.K., Zweier J.L. J. Biol. Chem. 2009;284:33850. doi: 10.1074/jbc.M109.019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Maia L., Moura J.J.G. Methods Mol. Biol. 2016;1424:81. doi: 10.1007/978-1-4939-3600-7_8. [DOI] [PubMed] [Google Scholar]
- 168.Maia L., Moura I., Moura J.J.G. EPR spectroscopy on mononuclear molybdenum-containing enzymes. In: Hanson G., Berliner L.J., editors. Vol. 33. Springer International Publishing; Cham: 2017. pp. 55–101. (Future Directions in Metalloprotein and Metalloenzyme Research, Biological Magnetic Resonance). [Google Scholar]
- 169.Rodriguez J., Maloney R.E., Rassaf T., Bryan N.S., Feelisch M. Proc. Natl. Acad. Sci. USA. 2003;1000:336. doi: 10.1073/pnas.0234600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bryan N.S., Rassaf T., Maloney R.E., Rodriguez C.M., Saijo F., Rodriguez J.R., Feelisch M. Proc. Natl. Acad. Sci. USA. 2004;101:4308. doi: 10.1073/pnas.0306706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shiva S., Gladwin M.T. J. Mol. Cell Cardiol. 2009;46:1. doi: 10.1016/j.yjmcc.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 172.Opie L.H. Circ. Res. 1976;38:152. [PubMed] [Google Scholar]
- 173.Cobbe S.M., Poole-Wilson P.A. J. Mol. Cell. Cardiol. 1980;12:761. doi: 10.1016/0022-2828(80)90078-4. [DOI] [PubMed] [Google Scholar]
- 174.Momomura S., Ingwall J.S., Parker J.A., Sahagian P., Ferguson J.J., Grossman W. Circ. Res. 1985;57:822. doi: 10.1161/01.res.57.6.822. [DOI] [PubMed] [Google Scholar]
- 175.Giraldez R.R., Panda A., Xia Y., Sanders S.P., Zweier J.L. J. Biol. Chem. 1997;272:21420. doi: 10.1074/jbc.272.34.21420. [DOI] [PubMed] [Google Scholar]
- 176.Stiefel E.I. Proc. Natl. Acad. Sci. USA. 1973;70:988. doi: 10.1073/pnas.70.4.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Holm R.H. Chem. Rev. 1987;87:1401. [Google Scholar]
- 178.Harlan E.E., Berg J.M., Holm R.H. J. Am. Chem. Soc. 1986;108:6992. doi: 10.1021/ja00284a073. [DOI] [PubMed] [Google Scholar]
- 179.Holm R.H. Coord. Chem. Rev. 1990;100:183. [Google Scholar]
- 180.Rajapakshe A., Snyder R.A., Astashkin A.V., Bernardson P., Evans D.J., Young C.G., Evans D.H., Enemark J.H. Inorg. Chim. Acta. 2009;362:4603. doi: 10.1016/j.ica.2009.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang J., Giles L.J., Ruppelt C., Mendel R.R., Bittner F., Kirk M.L. J. Am. Chem. Soc. 2015;137:5276. doi: 10.1021/jacs.5b01112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ward J.P.T. Biochim. Biophys. Acta. 2008;1777:1. doi: 10.1016/j.bbabio.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 183.Rumer S., Krischke M., Fekete A., Mueller M.J., Kaiser W.M. Nitric Oxide. 2012;27:123. doi: 10.1016/j.niox.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 184.Wang T.N., Douglass E.F., Fitzgerald K.J., Spiegel D.D. J. Am. Chem. Soc. 2013;135:12429. doi: 10.1021/ja406077j. [DOI] [PubMed] [Google Scholar]
- 185.Cherif-Sayadi A., Ayed-Tka K.H., Bejaoui M., Gdara N.B., Zaouali M.A., Abdennebi H.B. Ann. Transplant. 2016;21:602. doi: 10.12659/aot.899338. [DOI] [PubMed] [Google Scholar]
- 186.Tripatara P., Patel N.S., Webb A., Rathod K., Lecomte F.M., Mazzon E., Cuzzocrea S., Yaqoob M.M., Ahluwalia A., Thiemermann C. J. Am. Soc. Nephrol. 2007;18:570. doi: 10.1681/ASN.2006050450. [DOI] [PubMed] [Google Scholar]
- 187.Liu M., Zollbrecht C., Peleli M., Lundberg J.O., Weitzberg E., Carlström M. Free Radic. Biol. Med. 2015;84:154. doi: 10.1016/j.freeradbiomed.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 188.Gao X., Yang T., Liu M., Peleli M., Zollbrecht C., Weitzberg E., Lundberg J.O., Erik A., Persson G., Carlström M. Hypertension. 2015;65:161. doi: 10.1161/HYPERTENSIONAHA.114.04222. [DOI] [PubMed] [Google Scholar]
- 189.Jansson E.A., Huang L., Malkey R., Govoni M., Nihlen C., Olsson A., Stensdotter M., Petersson J., Holm L., Weitzberg E., Lundberg J.O. Nat. Chem. Biol. 2008;4:411. doi: 10.1038/nchembio.92. [DOI] [PubMed] [Google Scholar]
- 190.Webb A.J., Milsom A.B., Rathod K.S., Chu W.L., Qureshi S., Lovell M.J., Lecomte F.M., Perrett D., Raimondo C., Khoshbin E., Ahmed Z., Uppal R., Benjamin N., Hobbs A.J., Ahluwalia A. Circ. Res. 2008;103:957. doi: 10.1161/CIRCRESAHA.108.175810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang T., Peleli M., Zollbrecht C., Giulietti A., Terrando N., Lundberg J.O., Weitzberg E., Carlström M. Free Radic. Biol. Med. 2015;83:159. doi: 10.1016/j.freeradbiomed.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 192.Ren X., Ding Y., Lu N. Eur. J. Pharmacol. 2016;775:50. doi: 10.1016/j.ejphar.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 193.Zollbrechta C., Perssonb E.G., Lundberga J.O., Weitzberga E., Carlströma M. Redox Biol. 2016;10:119. doi: 10.1016/j.redox.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Peleli M., Zollbrecht C., Montenegro M.F., Hezel M., Zhong J., Persson E.G., Holmdahl R., Weitzberg E., Lundberg J.O., Carlström M. Free Radic. Biol. Med. 2016;99:472. doi: 10.1016/j.freeradbiomed.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 195.Buerk D.G., Liu Y., Zaccheo K.A., Barbee K.A., Jaron D. Front. Physiol. 2017;8:1053. doi: 10.3389/fphys.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Casey D.B., Badejo A.M., Dhaliwal J.S., Murthy S.N., Hyman A.L., Nossaman B.D., Kadowitz P.J. Am. J. Physiol. 2009;296:H524. doi: 10.1152/ajpheart.00543.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sugimoto R., Okamoto T., Nakao A., Zhan J., Wang Y., Kohmoto J., Tokita D., Farver C.F., Tarpey M.M., Billiar T.R., Gladwin M.T., McCurry K.R. Am. J. Transpl.:Off. J. Am. Soc. Transpl. Am. Soc. Transpl. Surg. 2012;12:2938. doi: 10.1111/j.1600-6143.2012.04169.x. [DOI] [PubMed] [Google Scholar]
- 198.Hare M.T.G., Tsui A.K.Y., Crawford J.H., Patel R.P. Redox Biol. 2013;1:65. doi: 10.1016/j.redox.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.C.C. Helms, X. Liu, D.B. Kim-Shapiro, Biol. Chem., vol. 398, 1017, p. 319. [DOI] [PMC free article] [PubMed]
- 200.Copley S.D. Bioessays. 2012;34:578. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]