Abstract
Anti-tuberculosis (TB) drugs possess diverse abilities to penetrate the different host tissues and cell types in which infecting Mycobacterium tuberculosis bacilli are located during active disease. This is important since there is increasing evidence that the respective “lesion-penetrating” properties of the front-line TB drugs appear to correlate well with their specific activity in standard combination therapy. In turn, these observations suggest that rational efforts to discover novel treatment-shortening drugs and drug combinations should incorporate knowledge about the comparative abilities of both existing and experimental anti-TB agents to access bacilli in defined physiological states at different sites of infection, as well as avoid elimination by efflux or inactivation by host or bacterial metabolism. However, while there is a fundamental requirement to understand the mode of action and pharmacological properties of any current or experimental anti-TB agent within the context of the obligate human host, this is complex and, until recently, has been severely limited by the available methodologies and models. Here, we discuss advances in analytical models and technologies which have enabled investigations of drug metabolism and pharmacokinetics (DMPK) for new TB drug development. In particular, we consider the potential to shift the focus of traditional pharmacokinetic-pharmacodynamic analyses away from plasma to a more specific “site of action” drug exposure as an essential criterion for drug development and the design of dosing strategies. Moreover, in summarizing approaches to determine DMPK data for the “unit of infection” comprising host macrophage and intracellular bacillus, we evaluate the potential benefits of including these analyses at an early stage in the preclinical drug development algorithm.
INTRODUCTION
Tuberculosis (TB) remains a leading cause of death owing to an infectious disease despite the existence of multiple front-line and second-line drugs that are active against Mycobacterium tuberculosis (Mtb), its etiological agent (1,2). A critical limitation of the current anti-TB regimen is that it involves a minimum six months combination therapy, a requirement thought to reflect the inability of the existing drugs to sterilize bacilli located in different host micro-environments and in variable metabolic states, often resulting in clearance of bacilli from sputum followed by subsequent disease relapse (3,4). Consequently, rational approaches to addressing this problem are urgently needed as they might offer the prospect of elucidating (at least partially) the reasons for the often described difficulty in translating compound potency in vitro into drug efficacy in vivo (5) – and, in turn, could inform the choices of chemical properties and screening assays to be prioritized in the critical-path algorithms that drive medicinal chemistry efforts as part of new TB drug discovery (6).
A seminal review (7) highlighted the absence of knowledge about the distributions of widely used anti-TB agents into the pulmonary lesions in which infecting bacilli are sequestered and, therefore, the need to understand how individual compounds distribute into the different host cell environments (8,9). In order to reach its mycobacterial target, an anti-TB antibiotic must penetrate complex host lesions and lesion compartments comprising multiple cell types (6,7,10). For intracellular bacilli, the drug must also overcome the barrier of the host cell membrane and, in many cases, an encapsulating vesicular membrane, as well as potential sequestration by the different organelles and intracellular bodies within different cell types (11,12). The occupation of discrete host loci (13–15) presents an additional confounder (Figure 1), in that individual bacteria from a clonal infecting population are characterized by different physiological states, and this can impact drug susceptibility (11,16,17) and the ability of the drug to penetrate the complex mycobacterial cell wall (18), as well as subverting the active compound’s activity against a metabolic target that might be essential only under specific conditions (19,20). As if that weren’t already sufficiently complex, host-mediated (21,22) and/or mycobacterium-mediated (23,24) biotransformation might further complicate the passage of drug from ingestion by the patient to its intrabacillary target (7). Although some guiding principles have been inferred from both preclinical and clinical observations in TB as well as other diseases, the physico-chemical and pharmacological properties which enable drugs to navigate this complex delivery pathway, avoid host metabolism, and penetrate the bacillus, remain poorly understood (6,7,25). It is not surprising, therefore, that active compounds selected (and optimized) for potency in a handful of in vitro assays often fail to demonstrate activity in vivo in the infected host (26).
Figure 1.
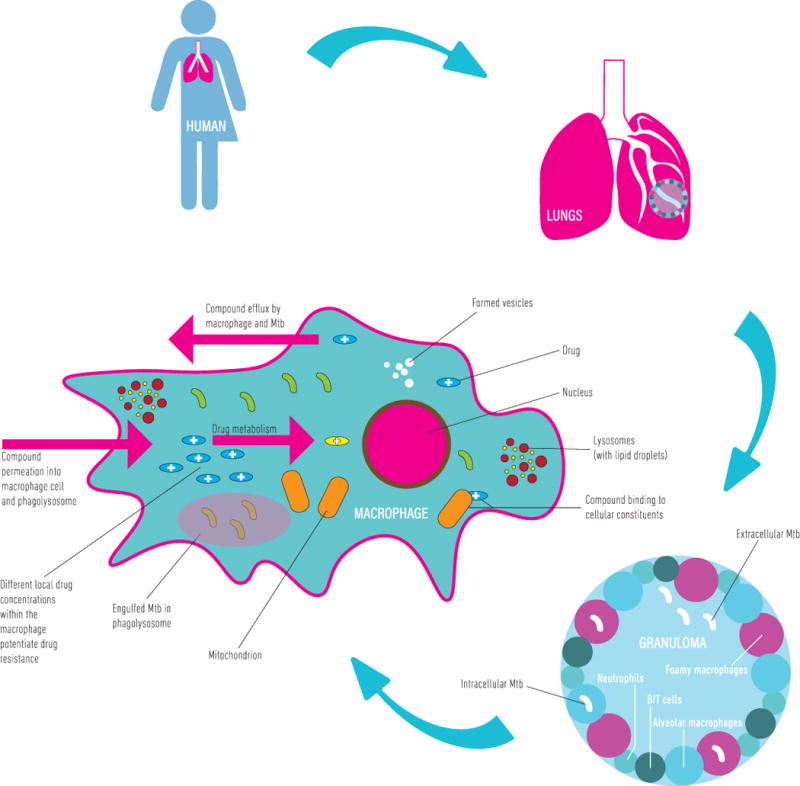
The complexity of targeting M. tuberculosis bacilli in an infected host. After administration of drug to a patient, the drug passes from the bloodstream into the lung tissues, diffusing into the lung fluid. The drug must then penetrate the granulomatous structure and enter the various cellular microenvironments in which bacilli can be sequestered. There are a number of processes that affect drug efficacy in this microenvironment including, but not limited to: (i) drug metabolism and biotransformation, (ii) binding to various cellular components and lipids, (iii) differences in local cellular drug concentrations giving rise to potential drug resistant bacteria, (iv) compound specific differences in the permeation of drug into the macrophage or cellular environment, (v) bacterial and cell mediated drug efflux, and (vi) differences in intracellular pH leading to in-/activation of the drug.
Fortunately, the last decade has seen considerable efforts towards developing accurate methods (Table 1) to measure drug levels directly in Mtb-infected tissues (27–29). Through the application of advanced chromatographic, mass spectrometry, and imaging techniques including HPLC coupled to tandem mass spectrometry (LC–MS/MS) and MALDI mass spectrometry imaging (MALDI–MSI), the quantification of tissue and lesion distribution of known and experimental anti-TB drugs in animal models and clinical samples has become increasingly attainable – primarily through the work of Veronique Dartois and colleagues (4,7,8,28,30) – thus offering the prospect of informing a systems pharmacology approach to the design of antimycobacterial therapy (31). In addition to the well-established appreciation that TB drugs can be differentially active against bacilli in different metabolic states (32–34), these new analytical techniques have demonstrated that those same drugs can also possess distinct lesion-penetrating abilities (6,7,9,18). Moreover, in many cases, tissue penetration appears to correlate well with the demonstrated activity of specific drugs in vivo (9); for example, the ability of pyrazinamide to diffuse through the necrotic areas at the center of mature granulomas is consistent with its sterilizing ability in the TB regimen (7). This is a critical observation, which prompted the prediction that “the next major step towards curing TB and preventing the development of resistance will come from a combination of complementary drugs, each of which preferentially distributes in the lesion or lesion compartment where its most vulnerable target bacterial population resides” (7). That is, future combination therapies for TB should be based on multidrug regimens optimized according to the specific activities and tissue distributions of each constituent drugs.
Table 1.
Methods to measure intracellular drug concentrations
| Measurement method | Detection | Advantages | Limitations |
|---|---|---|---|
| Fluorescence microscopy (1–5) | Combination of optical imaging microscopy with computational analysis to quantify fluorescence signals from molecules or compounds | Allows temporal and physiological studies of drugs in various microenvironments | Few drugs fluoresce above background detection limit Incorporation of fluorescent tag or “click chemistry” enabled fluorescent derivatization not always feasible |
| Raman microscopy (6–8) | Light scattering through change in polarisation potential or vibrational energy | Shortened data collection times for high-throughput analyses and provide indications of cellular state of Mtb and host cells | Can only be applied to biological systems with lower energy excitation |
| Nuclear microscopy (9–11) | Uses ion microbeam with particle-induced X-ray emission | Used as a quantitative standard to complement MRI in animal or human studies where tissues may be removed after imaging | Not widely accessible technology and limited to metal containing drugs/compounds |
| Microautoradiography (12,13) | Exposure or tagging using radiolabel and confocal microscopy | coupling technique to fluorescence in situ hybridisation technique allows more in depth single cell analysis | Resolution limitations; requires radioactive material and extensive processing time; often semi-quantitative |
| PET imaging (14–16) | Emitted positron collides with local electron, produce photons and these are detected by ƴ-detectors | Can be coupled to other techniques such as micro dialysis to allow receptor site PK studies | Expensive owing to necessity of radio-labelled ligands, difficulty overcoming resolution issues |
| Analysis by MS or HPLC (17–21) | Mass spectrometric (MS) analysis coupled to high performance liquid chromatography (HPLC) | Allows for intracellular drug concentrations to be calculated using pharmacokinetic approach | Long sample processing times and extensive optimisation steps, bulk analysis can cause loss of spatial information (cellular compartments) |
| MALDI-MSI (22–27) | Laser desorption based ionisation technique | Allows temporal and spatial resolution of drug distribution in different organs | Difficulty in determining whether analyte truly absent or simply below limit of detection |
Mandal S, Zhou Y, Shibata A, Destache CJ. Confocal fluorescence microscopy: An ultra-sensitive tool used to evaluate intracellular antiretroviral nano-drug delivery in HeLa cells. AIP Adv [Internet]. AIP Publishing LLC; 2015 Aug 8;5(8):84803. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4499042/
Lammertink BHA, Deckers R, Derieppe M, De Cock I, Lentacker I, Storm G, et al. Dynamic Fluorescence Microscopy of Cellular Uptake of Intercalating Model Drugs by Ultrasound-Activated Microbubbles. Mol Imaging Biol [Internet]. New York: Springer US; 2017 Feb 17;19(5):683–93. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5574961/
Manning AJ, Ovechkina Y, McGillivray A, Flint L, Roberts DM, Parish T. A high content microscopy assay to determine drug activity against intracellular Mycobacterium tuberculosis. Methods [Internet]. The Authors; 2017;127:3–11. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1046202316303619
Scarabelli S, Tan KT, Griss R, Hovius R, D’Alessandro PL, Vorherr T, et al. Evaluating Cellular Drug Uptake with Fluorescent Sensor Proteins. ACS Sensors. 2017;2(8):1191–7.
Stone MRL, Butler MS, Phetsang W, Cooper MA, Blaskovich MAT. Fluorescent Antibiotics: New Research Tools to Fight Antibiotic Resistance. Trends Biotechnol [Internet]. Elsevier; 2018 Apr 9; Available from: http://dx.doi.org/10.1016/j.tibtech.2018.01.004
Baron VO, Chen M, Clark SO, Williams A, Dholakia K, Gillespie SH. Detecting Phenotypically Resistant Mycobacterium tuberculosis Using Wavelength Modulated Raman Spectroscopy BT -. In: Gillespie SH, editor. Gillespie S (eds) Antibiotic Resistance Protocols Methods in Molecular Biology, vol 1736 Humana Press, New York, NY [Internet]. New York, NY: Springer New York; 2018. p. 41–50. Available from: https://doi.org/10.1007/978-1-4939-7638-6_4
Yokoyama M, Nishimura T, Yamada K, Ohno Y, Sakurai Y. Proposal of estimation method for drug concentration in blood by Raman spectroscopy of tear fluids. In: The 6th 2013 Biomedical Engineering International Conference. 2013. p. 1–4.
Chuchuen O, Henderson MH, Sykes C, Kim MS, Kashuba ADM, Katz DF. Quantitative Analysis of Microbicide Concentrations in Fluids, Gels and Tissues Using Confocal Raman Spectroscopy. PLoS One [Internet]. Public Library of Science; 2014 Dec 30;8(12):e85124. Available from: https://doi.org/10.1371/journal.pone.0085124
Ilinski P, Lai B, Cai Z, Yun W, Legnini D, Talarico T, et al. The Direct Mapping of the Uptake of Platinum Anticancer Agents in Individual Human Ovarian Adenocarcinoma Cells Using a Hard X-ray Microprobe. Cancer Res [Internet]. 2003 Apr 15;63(8):1776 LP-1779. Available from: http://cancerres.aacrjournals.org/content/63/8/1776.abstract
Sakurai H, Okamoto M, Hasegawa M, Satoh T, Oikawa M, Kamiya T, et al. Direct visualization and quantification of the anticancer agent, cis-diamminedichloro-platinum(II), in human lung cancer cells using in-air microparticle-induced X-ray emission analysis. Cancer Sci. 2008;99(5):901–4.
Rajendran R, Ronald JA, Ye T, Minqin R, Chen JW, Weissleder R, et al. Nuclear Microscopy: A Novel Technique for Quantitative Imaging of Gadolinium Distribution within Tissue Sections. Microsc Microanal [Internet]. 2009 Aug;15(4):338–44. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2802450/
Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol [Internet]. Nature Publishing Group; 2008 Mar 1;6:199. Available from: http://dx.doi.org/10.1038/nrmicro1838
Solon EG. Autoradiography techniques and quantification of drug distribution. Cell Tissue Res [Internet]. 2015;360(1):87–107. Available from: https://doi.org/10.1007/s00441-014-2093-4
Mammatas LH, Verheul HMW, Hendrikse NH, Yaqub M, Lammertsma A a, Menke-van der Houven van Oordt CW. Molecular imaging of targeted therapies with positron emission tomography: the visualization of personalized cancer care. Cell Oncol (Dordr) [Internet]. 2015;38(1):49–64. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25248503
Matthews PM, Rabiner EA, Passchier J, Gunn RN. Positron emission tomography molecular imaging for drug development. Br J Clin Pharmacol [Internet]. Oxford, UK: Blackwell Science Inc; 2012 Feb 25;73(2):175–86. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3269576/
Ankrah AO, van der Werf TS, de Vries EFJ, Dierckx RAJO, Sathekge MM, Glaudemans AWJM. PET/CT imaging of Mycobacterium tuberculosis infection. Clin Transl Imaging. Springer Milan; 2016;4(2):131–44.
Bhat J, Narayan A, Venkatraman J, Chatterji M. LC-MS based assay to measure intracellular compound levels in Mycobacterium smegmatis: Linking compound levels to cellular potency. J Microbiol Methods [Internet]. The Authors; 2013;94(2):152–8. Available from: http://dx.doi.org/10.1016/j.mimet.2013.05.010
Gordon LJ, Allen M, Artursson P, Hann MM, Leavens BJ, Mateus A, et al. Direct Measurement of Intracellular Compound Concentration by RapidFire Mass Spectrometry Offers Insights into Cell Permeability. J Biomol Screen [Internet]. SAGE Publications Inc STM; 2015 Sep 3;21(2):156–64. Available from: https://doi.org/10.1177/1087057115604141
Chen C, Gardete S, Jansen RS, Shetty A, Dick T, Rhee KY, et al. Verapamil targets membrane energetics in Mycobacterium tuberculosis. Antimicrob Agents Chemother [Internet]. 2018;AAC.02107-17. Available from: http://aac.asm.org/lookup/doi/10.1128/AAC.02107-17
Sarathy JP, Zuccotto F, Hsinpin H, Sandberg L, Via LE, Marriner GA, et al. Prediction of Drug Penetration in Tuberculosis Lesions. ACS Infect Dis [Internet]. 2016 Aug 12;2(8):552–63. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5028112/
Sarathy JP, Via LE, Weiner D, Blanc L, Boshoff H, Eugenin EA, et al. Extreme drug tolerance of Mycobacterium tuberculosis in caseum. Antimicrob Agents Chemother [Internet]. 2017;(December):AAC.02266-17. Available from: http://aac.asm.org/lookup/doi/10.1128/AAC.02266-17
Karlsson O, Hanrieder J. Imaging mass spectrometry in drug development and toxicology. Arch Toxicol. Springer Berlin Heidelberg; 2017;91(6):2283–94.
Prentice BM, Chumbley CW, Caprioli RM. Absolute Quantification of Rifampicin by MALDI Imaging Mass Spectrometry Using Multiple TOF/TOF Events in a Single Laser Shot. J Am Soc Mass Spectrom [Internet]. Journal of The American Society for Mass Spectrometry; 2017;28(1):136–44. Available from: http://dx.doi.org/10.1007/s13361-016-1501-2
Fisher GL, Bruinen AL, Ogrinc Potočnik N, Hammond JS, Bryan SR, Larson PE, et al. A New Method and Mass Spectrometer Design for TOF-SIMS Parallel Imaging MS/MS. Anal Chem [Internet]. American Chemical Society; 2016 Jun 21;88(12):6433–40. Available from: http://dx.doi.org/10.1021/acs.analchem.6b01022
Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O’Brien P, et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med [Internet]. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2015 Sep 7;21:1223. Available from: http://dx.doi.org/10.1038/nm.3937
Prideaux B, Dartois V, Staab D, Weiner DM, Goh A, Via LE, et al. High-Sensitivity MALDI-MRM-MS Imaging of Moxifloxacin Distribution in Tuberculosis-Infected Rabbit Lungs and Granulomatous Lesions. Anal Chem [Internet]. 2011 Mar 15;83(6):2112–8. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3158846/
Irwin SM, Prideaux B, Lyon ER, Zimmerman MD, Brooks EJ, Schrupp CA, et al. Bedaquiline and Pyrazinamide Treatment Responses Are Affected by Pulmonary Lesion Heterogeneity in Mycobacterium tuberculosis Infected C3HeB/FeJ Mice. ACS Infect Dis [Internet]. 2016;2(4):251–67. Available from: http://pubs.acs.org/doi/abs/10.1021/acsinfecdis.5b00127
Reynolds J, Heysell SK. Understanding pharmacokinetics to improve tuberculosis treatment outcome. Expert Opin Drug Metab Toxicol [Internet]. 2014 Jun 6;10(6):813–23. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4112565/
Sloan DJ, McCallum AD, Schipani A, Egan D, Mwandumba HC, Ward SA, et al. Genetic determinants of the pharmacokinetic variability of rifampin in Malawian adults with pulmonary tuberculosis. Antimicrob Agents Chemother. 2017;61(7):1–9.
Wright DFB, Winter HR, Duffull SB. Understanding the time course of pharmacological effect: a PKPD approach. Br J Clin Pharmacol [Internet]. Blackwell Science Inc; 2011 Jun 12;71(6):815–23. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3099368/
Upton RN, Mould DR. Basic Concepts in Population Modeling, Simulation, and Model-Based Drug Development: Part 3—Introduction to Pharmacodynamic Modeling Methods. CPT Pharmacometrics Syst Pharmacol [Internet]. Nature Publishing Group; 2014 Jan 2;3(1):e88. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3917320/
Lyons MA, Lenaerts AJ. Computational pharmacokinetics/pharmacodynamics of rifampin in a mouse tuberculosis infection model. J Pharmacokinet Pharmacodyn [Internet]. 2015 Aug 31;42(4):375–89. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4506877/
Probing the unit of infection: the Mtb-infected macrophage
Developing comprehensive rules for targeted TB drug delivery necessitates concerted research efforts to elucidate the various factors that influence host tissue penetration and bacillary permeation. From a drug discovery perspective, the ability to assess rapidly whether a putative hit compound possesses the required characteristics would profoundly impact the efficiency of development pipelines by ensuring that medicinal chemistry, pharmacology, and biology resources and expertise are applied only to the most promising candidates. However, as noted elsewhere (7,35), this is critically dependent on the development of medium-throughput in vitro assays for intralesional (e.g., necrotic foci or caseum; (11), intrabacillary (33), and intra-macrophage (36)) pharmacokinetic (PK) determinations that can be readily integrated into lead-compound discovery and lead-compound optimization campaigns.
Mtb is sequestered in multiple different host compartments during infection (3,9,29), including cellular granulomas, in which bacilli are located predominantly within macrophages but also in some extracellular niches; necrotic granulomas, where they are extracellular in the caseous centres and considered metabolically quiescent (37); and the inner surface of open cavities, where bacilli occur within multiple cell types while some are extracellular (38,39), an environment in which they are protected from the immune system and able to replicate freely (40). There is even evidence that bacilli can reside in cells in distal loci of the lung (41) and in other organs (42–44). These observations corroborate the suggestion that TB disease should be treated as a polymicrobial infection (45), and simultaneously reinforce the complexities inherent in determining the factors that might be essential to ensure optimal drug exposure for extended durations. From a drug development perspective, they also highlight a major obstacle to defining the optimal pre-clinical assays to be used in selecting compounds for advancement through the discovery pipeline.
How does the micro environment in which Mtb persists influence compound penetration and permeation? Current anti-TB chemotherapy may cause bacilli to revert to a drug-tolerant phenotype, which may explain the difficulty in achieving complete bacterial clearance (46,47). Therefore, more research is needed to understand the mechanism(s) by which exposure to these compounds might force the exposed bacilli into a different metabolic state (48). It is well-established that a biphasic reduction in bacterial load is observed under current TB therapy (32), a phenomenon which has been explained by two models: (i) the killing of actively dividing bacilli in an initial rapid killing phase, followed by the gradual decrease in the persistent population of bacteria, or (ii) the lack of treatment of bacterial populations due to the inaccessibility of bacteria within granulomas (49). Adding a further layer of complexity is the fact that the bacteria exist within both an intracellular and extracellular environment (50), an observation which is exacerbated by the knowledge that these lesions are very heterogeneous, often differing even within the same patient (4,14).
As noted elsewhere (51), the immunological lifecycle (52) of TB disease involves multiple stages at which the interaction between host macrophage and invading pathogens might be critical to the outcome of infection (3,37,53). That is, the ability of Mtb to survive and replicate within the macrophage is a defining feature of mycobacterial pathogenesis (54). This observation has motivated a longstanding interest in understanding the dynamics of the host-pathogen interaction within this phagocytic cell (30,50,53,55), as well as efforts to understand the factors which might undermine therapeutic outcomes (17,56). It has also underpinned the use of alternative screens to identify compounds active against bacilli in this intracellular environment (57–60), as well as approaches to understand the impact of the intracellular environment on drug partitioning and how this knowledge might be exploited for rationale drug and drug regimen design (36). The perceived centrality of the host macrophage in infection outcomes (61–63) has also been key to the use of standard mouse models as distinct from the “Kramnik” or C3HeB/FeJ model; (29,64) in pre-clinical efficacy assessments (65). Therefore, while cultured cells in vitro do not fully recapitulate the specialist functional properties of differentiated macrophages in vivo (3,66) – with their diverse ontogenies and differential trajectories of activation and development (67),– the utility of the macrophage model in inferring disease-relevant mycobacterial physiological and metabolic adaptations, as well as innate host defence strategies, seems convincing (3,51,68). This notion is perhaps best summarized in the concept that the Mtb-infected macrophage represents the “minimal unit of infection” (50), a term which encapsulates critical concepts in immunometabolism (69) and pathometabolism (70).
The concentration which any drug achieves within its target (myco)bacterial cell is a function of multiple factors, including passive or active uptake, pathogen-mediated metabolism, active drug efflux, and cell growth (71). Mycobacterial drug uptake is generally encapsulated in the concept of “permeation”. The complex mycobacterial cell wall is thought to function as an impermeable barrier to most compounds and undergoes dynamic architectural modifications during infection that correlate with a switch from active replication to a persistent state (18,72). Moreover, the mechanisms which enable many of the known anti-TB drugs (in particular, small hydrophilic compounds like isoniazid, ethambutol, and cycloserine) to permeate the lipid-rich mycobacterial cell wall remain unknown (73). Mtb also possesses an expanded complement of efflux pumps (25,74,75) which have been implicated in intrinsic resistance to applied drugs in vitro (76,77) and in experimental models of infection (78). Moreover, growth within THP-1 and J774 macrophages has been shown to induce Mtb efflux pump activity (79), resulting in tolerance to RIF and other drugs mediated by the efflux transporter, Rv1258c (79,80). As highlighted elsewhere (7), this is an important observation since it supports the need for in vitro assays to determine (and, ultimately, predict) drug distributions within immune cells such as macrophages, as well as the sub-cellular organelles in which the bacterium might be contained.
The prevailing drug discovery paradigm
It is generally acknowledged that, in order to understand how a drug will respond in the human body, the pharmacokinetic/pharmacodynamic (PK/PD) parameters in various tissues and cells must be understood fully (4,30,45). The propensity for Mtb to occupy different microenvironments – and at the same time – within an infected host (13–15), makes it essential that new combination regimens comprise partner drugs that are active in the various micro environments and, potentially, have the ability to modulate permeability of the mycobacterial cell (81).
Anti-TB drug discovery often begins with a basic screen against replicating Mtb, followed by subsequent cytotoxicity screens to determine the compound selectivity against the pathogen (82). This allows high-throughput screening whilst identifying potential candidates with initial activity against replicating bacteria (82), but neglects aspects such as intracellular compound activity, membrane permeability, involvement of efflux, and metabolism of the compound (83). The use of a standard growth medium alone ignores the metabolic changes that Mtb undergoes when in the host cell environment, and could influence compound efficacy (84): the evidence is strong that the metabolic status of the bacterium is a function of the host environment (51,85). Therefore, the active compounds that emerge from in vitro screens are likely to be active only under specific conditions (86). For example, amikacin displayed potent activity in initial screens against extracellular bacteria, but little to no activity against intracellular Mtb (87).
Next, the PK parameters of the compounds are assessed using in vitro or in vivo absorption, distribution, metabolism, and excretion (ADME) studies. In the simplest sense, the ADME parameters for a compound in combination with the dose of the drug determine the time course and concentration in serum and, consequently, the tissues and fluids. The pharmacodynamic (PD) parameters relate drug concentration with the observed antimicrobial effect. The PK/PD characteristics of a drug have implications for dosing, clinical efficacy, and the combinatorial drug classes which can be used to treat TB (88). Summary exposure parameters, such as area under the concentration-time curve (AUC) and peak concentration (Cmax), are often used in combination with PD parameters, such as MIC (BOX 1). Importantly, drugs display varied and distinctive patterns of PK/PD relationships which can influence their activity (89,90). The main patterns include bacterial killing based on maximum drug concentration (Cmax/MIC), total exposure over a certain time period (AUC/MIC), or time above MIC (T>MIC) (91). Notably, these parameters have been experimentally determined using multiple approaches, including the ex vivo hollow fibre model, which allows for the recapitulation of the different bacterial populations and their corresponding PK/PD responses. Applying this technique, Gumbo and colleagues were able to predict the inability of moxifloxacin (MXF) used at WHO recommended doses to improve clinical outcomes significantly in a series of drug trials (92–97).
Box 1. PK/PD relationship of TB drugs.
The PK/PD relationship for TB drugs is established in the literature between AUC or Cmax (adjusted for MIC) and/or time above a certain concentration threshold (T>MIC), with many of the first-line drugs described by AUC/MIC and some of the newer compounds best described by T>MIC (28,29).
Convincing arguments can be made to justify using AUC>MIC to allow for penetration in the lesions or other “hard to reach” compartments/tissues in the body, or saturation of efflux pumps. Similar arguments can be made for the use of T>MIC to avoid the concentration falling below the threshold at which Mtb is able to start replicating, possibly leading to resistance if low drug concentrations are present.
As discussed, for TB the PK/PD relationship is complicated by the fact that the PK is generally measured in plasma in clinical trials, but the majority of bacilli are sequestered in other compartments into which the drugs may not readily penetrate. Therefore, the PK profile in these regions is likely to be different from that measured in plasma. A reasonable PK/PD modelling approach in such cases is to use a hypothetical “effect compartment” (30,31) mimicking the concentration at the site of action which, while dependent on the concentration in plasma, is “delayed” by the drug traversing through the numerous layers of biological tissue, thus significantly smoothing out peak and trough concentrations. Also, the time over a certain concentration would be very different in this effect compartment. The best proxy for the concentration at the site of action is plasma AUC since the ratio between the average concentrations would remain the same, and AUC is closely related to average concentrations. In the literature, some results from clinical data report the superiority of Cmax>MIC versus AUC>MIC for prediction of clinical efficacy, however this should be interpreted with care since it is very difficult to discriminate which of these PK exposure parameter is most closely related to outcome - especially when the only available data is derived from observational clinical studies where all patients are on a similar dosing regimen. Without targeted studies of dose fractionation (when the same total dose is given in a single or multiple dosing events), Cmax and AUC are generally strongly correlated and difficult to separate (32). Comparing the two parameters, AUC data can also be variable, even on a daily basis, and this should also be considered when interpreting PK/PD results. The variability will result in a Cmax which fluctuates considerably as the determination of this parameter is based on a single sample. AUC may also change but is generally more stable due to the parameter being determined by an entire PK profile. Therefore, the use of AUC/MIC might better predict the outcome of clinical efficacy in diseases such as TB which have sites of action that are disparate from the plasma. Although the best PK/PD values would be derived from target site concentrations, these are often difficult to access.
Mandal S, Zhou Y, Shibata A, Destache CJ. Confocal fluorescence microscopy: An ultra-sensitive tool used to evaluate intracellular antiretroviral nano-drug delivery in HeLa cells. AIP Adv [Internet]. AIP Publishing LLC; 2015 Aug 8;5(8):84803. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4499042/
Lammertink BHA, Deckers R, Derieppe M, De Cock I, Lentacker I, Storm G, et al. Dynamic Fluorescence Microscopy of Cellular Uptake of Intercalating Model Drugs by Ultrasound-Activated Microbubbles. Mol Imaging Biol [Internet]. New York: Springer US; 2017 Feb 17;19(5):683–93. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5574961/
Manning AJ, Ovechkina Y, McGillivray A, Flint L, Roberts DM, Parish T. A high content microscopy assay to determine drug activity against intracellular Mycobacterium tuberculosis. Methods [Internet]. The Authors; 2017;127:3–11. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1046202316303619
Scarabelli S, Tan KT, Griss R, Hovius R, D’Alessandro PL, Vorherr T, et al. Evaluating Cellular Drug Uptake with Fluorescent Sensor Proteins. ACS Sensors. 2017;2(8):1191–7.
Stone MRL, Butler MS, Phetsang W, Cooper MA, Blaskovich MAT. Fluorescent Antibiotics: New Research Tools to Fight Antibiotic Resistance. Trends Biotechnol [Internet]. Elsevier; 2018 Apr 9; Available from: http://dx.doi.org/10.1016/j.tibtech.2018.01.004
Baron VO, Chen M, Clark SO, Williams A, Dholakia K, Gillespie SH. Detecting Phenotypically Resistant Mycobacterium tuberculosis Using Wavelength Modulated Raman Spectroscopy BT -. In: Gillespie SH, editor. Gillespie S (eds) Antibiotic Resistance Protocols Methods in Molecular Biology, vol 1736 Humana Press, New York, NY [Internet]. New York, NY: Springer New York; 2018. p. 41–50. Available from: https://doi.org/10.1007/978-1-4939-7638-6_4
Yokoyama M, Nishimura T, Yamada K, Ohno Y, Sakurai Y. Proposal of estimation method for drug concentration in blood by Raman spectroscopy of tear fluids. In: The 6th 2013 Biomedical Engineering International Conference. 2013. p. 1–4.
Chuchuen O, Henderson MH, Sykes C, Kim MS, Kashuba ADM, Katz DF. Quantitative Analysis of Microbicide Concentrations in Fluids, Gels and Tissues Using Confocal Raman Spectroscopy. PLoS One [Internet]. Public Library of Science; 2014 Dec 30;8(12):e85124. Available from: https://doi.org/10.1371/journal.pone.0085124
Ilinski P, Lai B, Cai Z, Yun W, Legnini D, Talarico T, et al. The Direct Mapping of the Uptake of Platinum Anticancer Agents in Individual Human Ovarian Adenocarcinoma Cells Using a Hard X-ray Microprobe. Cancer Res [Internet]. 2003 Apr 15;63(8):1776 LP-1779. Available from: http://cancerres.aacrjournals.org/content/63/8/1776.abstract
Sakurai H, Okamoto M, Hasegawa M, Satoh T, Oikawa M, Kamiya T, et al. Direct visualization and quantification of the anticancer agent, cis-diamminedichloro-platinum(II), in human lung cancer cells using in-air microparticle-induced X-ray emission analysis. Cancer Sci. 2008;99(5):901–4.
Rajendran R, Ronald JA, Ye T, Minqin R, Chen JW, Weissleder R, et al. Nuclear Microscopy: A Novel Technique for Quantitative Imaging of Gadolinium Distribution within Tissue Sections. Microsc Microanal [Internet]. 2009 Aug;15(4):338–44. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2802450/
Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol [Internet]. Nature Publishing Group; 2008 Mar 1;6:199. Available from: http://dx.doi.org/10.1038/nrmicro1838
Solon EG. Autoradiography techniques and quantification of drug distribution. Cell Tissue Res [Internet]. 2015;360(1):87–107. Available from: https://doi.org/10.1007/s00441-014-2093-4
Mammatas LH, Verheul HMW, Hendrikse NH, Yaqub M, Lammertsma A a, Menke-van der Houven van Oordt CW. Molecular imaging of targeted therapies with positron emission tomography: the visualization of personalized cancer care. Cell Oncol (Dordr) [Internet]. 2015;38(1):49–64. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25248503
Matthews PM, Rabiner EA, Passchier J, Gunn RN. Positron emission tomography molecular imaging for drug development. Br J Clin Pharmacol [Internet]. Oxford, UK: Blackwell Science Inc; 2012 Feb 25;73(2):175–86. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3269576/
Ankrah AO, van der Werf TS, de Vries EFJ, Dierckx RAJO, Sathekge MM, Glaudemans AWJM. PET/CT imaging of Mycobacterium tuberculosis infection. Clin Transl Imaging. Springer Milan; 2016;4(2):131–44.
Bhat J, Narayan A, Venkatraman J, Chatterji M. LC-MS based assay to measure intracellular compound levels in Mycobacterium smegmatis: Linking compound levels to cellular potency. J Microbiol Methods [Internet]. The Authors; 2013;94(2):152–8. Available from: http://dx.doi.org/10.1016/j.mimet.2013.05.010
Gordon LJ, Allen M, Artursson P, Hann MM, Leavens BJ, Mateus A, et al. Direct Measurement of Intracellular Compound Concentration by RapidFire Mass Spectrometry Offers Insights into Cell Permeability. J Biomol Screen [Internet]. SAGE Publications Inc STM; 2015 Sep 3;21(2):156–64. Available from: https://doi.org/10.1177/1087057115604141
Chen C, Gardete S, Jansen RS, Shetty A, Dick T, Rhee KY, et al. Verapamil targets membrane energetics in Mycobacterium tuberculosis. Antimicrob Agents Chemother [Internet]. 2018;AAC.02107-17. Available from: http://aac.asm.org/lookup/doi/10.1128/AAC.02107-17
Sarathy JP, Zuccotto F, Hsinpin H, Sandberg L, Via LE, Marriner GA, et al. Prediction of Drug Penetration in Tuberculosis Lesions. ACS Infect Dis [Internet]. 2016 Aug 12;2(8):552–63. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5028112/
Sarathy JP, Via LE, Weiner D, Blanc L, Boshoff H, Eugenin EA, et al. Extreme drug tolerance of Mycobacterium tuberculosis in caseum. Antimicrob Agents Chemother [Internet]. 2017;(December):AAC.02266-17. Available from: http://aac.asm.org/lookup/doi/10.1128/AAC.02266-17
Karlsson O, Hanrieder J. Imaging mass spectrometry in drug development and toxicology. Arch Toxicol. Springer Berlin Heidelberg; 2017;91(6):2283–94.
Prentice BM, Chumbley CW, Caprioli RM. Absolute Quantification of Rifampicin by MALDI Imaging Mass Spectrometry Using Multiple TOF/TOF Events in a Single Laser Shot. J Am Soc Mass Spectrom [Internet]. Journal of The American Society for Mass Spectrometry; 2017;28(1):136–44. Available from: http://dx.doi.org/10.1007/s13361-016-1501-2
Fisher GL, Bruinen AL, Ogrinc Potočnik N, Hammond JS, Bryan SR, Larson PE, et al. A New Method and Mass Spectrometer Design for TOF-SIMS Parallel Imaging MS/MS. Anal Chem [Internet]. American Chemical Society; 2016 Jun 21;88(12):6433–40. Available from: http://dx.doi.org/10.1021/acs.analchem.6b01022
Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O’Brien P, et al. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med [Internet]. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2015 Sep 7;21:1223. Available from: http://dx.doi.org/10.1038/nm.3937
Prideaux B, Dartois V, Staab D, Weiner DM, Goh A, Via LE, et al. High-Sensitivity MALDI-MRM-MS Imaging of Moxifloxacin Distribution in Tuberculosis-Infected Rabbit Lungs and Granulomatous Lesions. Anal Chem [Internet]. 2011 Mar 15;83(6):2112–8. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3158846/
Irwin SM, Prideaux B, Lyon ER, Zimmerman MD, Brooks EJ, Schrupp CA, et al. Bedaquiline and Pyrazinamide Treatment Responses Are Affected by Pulmonary Lesion Heterogeneity in Mycobacterium tuberculosis Infected C3HeB/FeJ Mice. ACS Infect Dis [Internet]. 2016;2(4):251–67. Available from: http://pubs.acs.org/doi/abs/10.1021/acsinfecdis.5b00127
Reynolds J, Heysell SK. Understanding pharmacokinetics to improve tuberculosis treatment outcome. Expert Opin Drug Metab Toxicol [Internet]. 2014 Jun 6;10(6):813–23. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4112565/
Sloan DJ, McCallum AD, Schipani A, Egan D, Mwandumba HC, Ward SA, et al. Genetic determinants of the pharmacokinetic variability of rifampin in Malawian adults with pulmonary tuberculosis. Antimicrob Agents Chemother. 2017;61(7):1–9.
Wright DFB, Winter HR, Duffull SB. Understanding the time course of pharmacological effect: a PKPD approach. Br J Clin Pharmacol [Internet]. Blackwell Science Inc; 2011 Jun 12;71(6):815–23. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3099368/
Upton RN, Mould DR. Basic Concepts in Population Modeling, Simulation, and Model-Based Drug Development: Part 3—Introduction to Pharmacodynamic Modeling Methods. CPT Pharmacometrics Syst Pharmacol [Internet]. Nature Publishing Group; 2014 Jan 2;3(1):e88. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3917320/
Lyons MA, Lenaerts AJ. Computational pharmacokinetics/pharmacodynamics of rifampin in a mouse tuberculosis infection model. J Pharmacokinet Pharmacodyn [Internet]. 2015 Aug 31;42(4):375–89. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4506877/
Potential for a paradigm shift?
Many of the anti-TB drugs were discovered before the routine use of PK/PD analyses, a problem that continues to impair correct drug dosage today. This is exemplified by rifampicin (RIF) (98), for which recent studies have highlighted the benefits of using a larger dose than has been employed for decades. Although plasma and tissue PK analyses are routinely used in drug discovery (99), they may not reveal the entire picture owing to the complex nature of TB lesions and the difficulty faced for a drug to access, and then penetrate, the infecting bacilli. This is further complicated by the avascular nature of necrotic lesions, where the relationship between plasma and target site drug concentrations is even more difficult to predict (7). PK within the human setting can be spatially unique and the heterogeneous nature of both the bacterial and lesion phenotypes leads to increased complexity (100).
This complexity is shown in the use of RIF in first-line anti-TB therapy. The current dosing of RIF (10 mg/kg daily) is sub-optimal for TB, with some studies suggesting that significantly higher doses are needed for more effective treatment (101). Moreover, the problem persists in vulnerable population groups, with Sub-Saharan individuals (primarily in South Africa and Malawi) displaying reduced drug exposure (102,103). This often places individuals below the recommended target concentrations, potentially exposing them to increased burden of disease, the development of resistance, and risk of mortality (104,105). Many of the problems associated with RIF stem from a poor underlying understanding of dosing requirements in combination with other first-line treatments. Interestingly, MXF has been shown to be antagonistic with most antibiotics, including RIF and pretomanid (106,107). This knowledge, in conjunction with the deficiency of MXF in penetrating different lesions types (27), could explain the recent failed drug trials (108,109). Given the cellular concentrations of these first-line drug combinations, it may have been possible to predict their efficacy before commencement of the trials.
A series of studies conducted between 1950 and the 1980’s showed that concentrations of isoniazid (INH) and RIF are far lower in TB lesion homogenate than in the plasma of infected patients (110,111), a finding which was confirmed in MALDI studies (27). Despite the increasing appreciation that the activity of antibiotics depends on their ability to reach and accumulate in lesions, there is a continued reliance on blood and plasma levels to drive drug discovery (112,113). In various other diseases, target areas and plasma levels are often quite closely related, which leads to predictable and reliable plasma PK/PD values, even when using plasma exposure as PK target (114). However, owing to the complex nature of TB pathology, blood supply is often absent and free drug has difficulty in entering the different lesion compartments homogeneously. Drug PK/PD relationships are therefore extremely difficult to determine, particularly for newer drugs. So, while plasma PK measures still offer some value for many diseases, these determinations need to be repositioned in the TB drug discovery pipeline and factored in with new PK analyses at the target site.
Using the technologies available today (Table 1), it is possible to evaluate PK/PD properties at cellular level (100). The use of in vitro macrophage culture to represent Mtb’s intracellular environment has shown some success with the identification of compound Q203 (86). Similar assays have been undertaken in various cell types including mouse-derived macrophages and epithelial cells, with a limited number of studies having used cell lines such as the human-derived THP-1 macrophage-like cells (60,115,116). Progress in the use of metabolomics to obtain PK/PD information from treated mycobacterial cells (117,118) has been especially useful in determining the systems-level impact of drug treatment on Mtb physiology, and has the capacity to provide key insights into the mechanisms of action of new (117,119) and even known (118,120,121) anti-TB drugs. However, while the effective intrabacterial concentration is critical to efficacy, for most drugs the identity of the active metabolite(s) remains unknown. It is likely, therefore, that, in addition to revealing intracellular modes of action of known drugs, analyses of active drug metabolites generated via host- or Mtb-mediated biotransformation could identify new targets (7).
Metabolism alone does not always account for the disappearance of drug during PK/PD analyses: instead, this is can be a multifactorial phenomenon, involving the binding of drugs to macromolecules such as plasma proteins (122) and binding to lower molecular weight targets such as the oxidative stress protectants, glutathione and mycothiol (123). The phagolysosome in which these molecules must function itself undergoes significant pH changes in response to IFNƴ-dependent macrophage activation, dropping from a pH of 6.2 to approximately 4.5 (124,125), a shift which has the potential to limit the activities of acid-labile drugs while elevating the efficacy of drugs such as pyrazinamide which function optimally at low pH (126).
Drug discovery at the target site
The importance of determining lesion-specific drug concentrations stems from various sources. Mitchison and Coates (2004) described a model which explained the relationship between the different microenvironments in TB and current first-line TB drug efficacy. In their model, actively growing bacilli were killed by INH, semi-dormant bacilli were killed by RIF, intracellular bacilli were targeted by pyrazinamide (PZA) in the acidic phagolysosome, while dormant persisters were found in hypoxic environments, making them harder to treat using standard therapies (127,128).
Pioneering work by Dartois et al. in 2012 allowed scientists in the TB drug discovery environment to start looking at the target site of pulmonary TB in far more detail (8). Using a combination of New Zealand White rabbits, imaging mass spectrometry, three first-line anti-TB drugs (INH, RIF, and PZA), and the fluoroquinolone MXF, the group demonstrated that drug plasma concentration was indeed a poor proxy for drug concentration in TB lesions (8). The lack of data on how this might translate into humans combined with the absence of cellular protein binding data were acknowledged as weaknesses by the authors; nevertheless, this work prompted a growing appreciation of the need to include permeation studies in their drug development pipeline (16,129,130).
In a later study, Prideaux et al. (2015) provided a compelling extension of this work by taking advantage of a small set of resected lung samples from drug-resistant TB cases (131). In this case, the patients received a steady-state dose of RIF, INH, MXF, and PZA at intervals ranging from 2-24 hours pre-surgery, and lung samples were analysed using imaging mass spectrometry. This revealed that RIF and PZA were able to penetrate lesions, with RIF accumulating to steady-state levels in the caseum (27). Although multiple dosing was not investigated, patients had been receiving these drugs for several weeks/months. A controlled trial involving infected, drug-naïve patients would be necessary to confirm these findings and control for drug levels. This work could potentially correlate clinical trial data with lack of efficacy and motivate earlier inclusion of such studies in a pre-clinical setting (132).
The influence of caseum binding on the permeation of compounds into the deeper recesses of the granuloma was the next object of study (11). Exploiting a very large sample set which covered the molecular space using 64 parameters and over 200 compounds, it was observed that the compound’s ClogP value best describing penetration into the granuloma (11). The use of surrogate caseum was also investigated: while it was acknowledged that the surrogate failed to reproduce the full composition of the in vivo material, and so required further development to mimic patient-derived lesions more accurately, the results supported the prospect for this assay to be standardised, thereby allowing more in-depth in vitro assays to take advantage of this methodology.
Another recent study investigated the mystery of ethambutol’s (EMB) clinical efficacy despite its modest potency against non-replicating Mtb (30). Using the TB-infected rabbit model, along with microdissection and LC-MS/MS, the group determined that EMB partitions into the caseous lesions with great efficiency, potentially explaining its efficacy and again motivating for permeation studies to be conducted in early pre-clinical drug development (30). This work was followed up by a further study using the infected rabbit model, where ex vivo MBC measurements were taken in caseum (MBCcaseum) for several first-line TB drugs (16). The Wayne (133) and Loebel (134) models for non-replicating persistence failed to predict the extent to which resistance was being generated in the caseous environment, whilst also indicating that PZA has activity in the caseum, a fact which in the current non-replicating models would have missed. This motivates for the use of an in vitro assay which closely mimics the in vivo situation.
Challenges and perspectives
Incorporating findings from these recent studies into the model of Mitchison and Coates (32) has revealed interesting correlations with, and departures from, their original ideas. For example, Prideaux et al. (2015) suggested that drug accumulation within granulomas may not simply follow the binary ‘in/out’ dichotomy (131). Instead, four distinct patterns of drug accumulation were identified including: (i) rapid and homogeneous distribution with no accumulation appearing over time (INH/linezolid), which may explain the predominant killing of extracellular bacteria by INH; (ii) rapid and heterogeneous distribution with accumulation in the cellular rim rather than the caseum (fluoroquinolones and oxazolidinones); (iii) slow distribution with gradual accumulation of drug over time (RIF) explaining the intracellular killing of Mtb; and (iv) rapid distribution with massive accumulation in the cellular layers and poor diffusion into the caseum (clofazimine and bedaquiline) (27).
From a translational perspective, knowledge about the permeation of current first-line drugs and novel drug candidates into TB microenvironments is clearly essential: in addition to elucidating reasons for therapeutic failure that do not simply invoke patient non-adherence, the potential to inform combinations inferred from high-order drug interactions (107) with these data promises a route to mitigate the criticisms often levelled at in vitro-derived compound synergies. That is, coupling penetration and potentiation might enable design of new regimens that incorporate information about drug permeation into the heterogeneous TB microenvironments. Towards this end, a number of factors may have to be considered in ensuring clinical relevance (BOX 2). In this context, it is worth noting the parallel development and application to Mtb infection studies of increasingly sophisticated systems for three-dimensional cell culture (135,136) since these might offer a useful intermediate in bridging the in vitro/in vivo divide.
Box 2. Determining the clinical relevance of in vitro and in vivo assays.
Is there a quantifiable and agreed measure of “cure” that can be evaluated in an in vivo model?
What host-pathogen relationships can be effectively modelled in a non-clinical setting?
Is there a way to model bacterial physiology, pathogenesis, and drug susceptibility effectively to allow better understanding of these aspects in patients?
What critical PK/PD relationships can be modelled using in vitro/in vivo systems and how might these be applied in clinical trial settings?
Can dosing regimens be altered according to data emerging from in vitro/in vivo experiments employed in the pre-clinical setting?
In summary, there is increasing evidence that the incorporation of the efficacy of drugs in the microenvironments must be assessed early on in pre-clinical development to allow correct dosing in further in vivo experiments. A better understanding of target site drug concentrations, rather than plasma concentrations, is needed, particularly for a complex, multi-faceted disease such as TB. The aim of novel drug discovery programs should be to target populations of bacilli with the most effective drugs, which should be most proficient at reaching relevant sites in the human body. In driving this concept of target site drug concentrations, it is hoped that the translational link between the lab bench and clinic can be strengthened, allowing expedited and “smarter” drug discovery and, by implication, enhanced therapeutic regimens.
Acknowledgments
We acknowledge the financial support of the Department of Science and Technology (DST) and National Research Foundation (NRF) of South Africa, the Strategic Health Innovation Partnerships (SHIP) initiative the South African Medical Research Council, the University of Cape Town, and the US National Institute of Allergy and Infectious Diseases (NIAID) R21AI115993. We also acknowledge the support of the Research Council of Norway (INTPART, AMR-PART) for work on antimicrobial resistance in M. tuberculosis.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Kyu HH, Maddison ER, Henry NJ, Mumford JE, Barber R, Shields C, et al. Lancet Infect Dis [Internet] 3. Vol. 18. Elsevier; 2016. Mar 2, The global burden of tuberculosis: results from the Global Burden of Disease Study 2015; pp. 261–84. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallis RS, Maeurer M, Mwaba P, Chakaya J, Rustomjee R, Migliori GB, et al. Tuberculosis—advances in development of new drugs, treatment regimens, host-directed therapies, and biomarkers. Lancet Infect Dis [Internet] 2016 Apr; doi: 10.1016/S1473-3099(16)00070-0. [cited 2017 Sep 16];16(4):e34–46 Available from: http://linkinghub.elsevier.com/retrieve/pii/S1473309916000700. [DOI] [PubMed]
- 3.Russell DG, Barry CE, Flynn JL. NIH Public Access. Russell J Bertrand Russell Arch. 2010;328(5980):852–6. doi: 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry CE, Boshoff H, Dartois V, Dick T, Ehrt S, Flynn J, et al. The spectrum of latent tuberculosis: rethinking the goals of prophylaxis. Nat Rev Microbiol. 2009;7(12):845–55. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knudson SE, Kumar K, Awasthi D, Ojima I, Slayden RA. In vitro-in vivo activity relationship of substituted benzimidazole cell division inhibitors with activity against Mycobacterium tuberculosis. Tuberculosis [Internet] Elsevier Ltd. 2014;94(3):271–6. doi: 10.1016/j.tube.2014.03.007. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dartois V, Barry CE. Bioorganic Med Chem Lett [Internet] 17. Vol. 23. Elsevier Ltd; 2013. A medicinal chemists’ guide to the unique difficulties of lead optimization for tuberculosis; pp. 4741–50. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dartois V. Nat Rev Microbiol [Internet] 3. Vol. 12. Nature Publishing Group; 2014. The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells; pp. 159–67. Available from: http://www.nature.com/doifinder/10.1038/nrmicro3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kjellsson MC, Via LE, Goh A, Weiner D, Low KM, Kern S, et al. Pharmacokinetic evaluation of the penetration of antituberculosis agents in rabbit pulmonary lesions. Antimicrob Agents Chemother. 2012 Jan;56(1):1. 446–57. doi: 10.1128/AAC.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dartois V, Barry CE. Curr Clin Pharmacol [Internet] 2. Vol. 5. The Novartis Institute for Tropical Diseases; Biopolis, Singapore, Singapore: 2010. Clinical pharmacology and lesion penetrating properties of second- and third-line antituberculous agents used in the management of multidrug-resistant (MDR) and extensively-drug resistant (XDR) tuberculosis; pp. 96–114. veronique.dartois@novartis.com. Available from: http://europepmc.org/abstract/MED/20156156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenaerts A, Barry CE, Dartois V. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev. 2015;264(1):288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarathy JP, Zuccotto F, Hsinpin H, Sandberg L, Via LE, Marriner GA, et al. Prediction of Drug Penetration in Tuberculosis Lesions. ACS Infect Dis [Internet] 2016 Aug 12;2(8):552–63. doi: 10.1021/acsinfecdis.6b00051. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5028112/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carryn S, Van Bambeke F, Mingeot-Leclercq MP, Tulkens PM. Comparative intracellular (THP-1 macrophage) and extracellular activities of β-lactams, azithromycin, gentamicin, and fluoroquinolones against Listeria monocytogenes at clinically relevant concentrations. Antimicrob Agents Chemother. 2002;46(7):2095–103. doi: 10.1128/AAC.46.7.2095-2103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin PL, Ford CB, Coleman MT, Myers AJ, Gawande R, Ioerger T, et al. Nat Med [Internet] Vol. 20. Nature Publishing Group a division of Macmillan Publishers Limited; 2013. Dec 15, Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing; p. 75. All Rights Reserved. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadena AM, Fortune SM, Flynn JL. Nat Rev Immunol [Internet] Nature Publishing Group; 2017. Heterogeneity in tuberculosis; pp. 1–12. Available from: http://www.nature.com/doifinder/10.1038/nri.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin CJ, Cadena AM, Leung VW, Lin L, Maiello P, Hicks N, et al. crossm Tuberculosis. MBio. 2017;8(3):1–12. doi: 10.1128/mBio.00312-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarathy JP, Via LE, Weiner D, Blanc L, Boshoff H, Eugenin EA, et al. Extreme drug tolerance of Mycobacterium tuberculosis in caseum. Antimicrob Agents Chemother [Internet] 2017 Dec; doi: 10.1128/AAC.02266-17. AAC.02266-17 Available from: http://aac.asm.org/lookup/doi/10.1128/AAC.02266-17. [DOI] [PMC free article] [PubMed]
- 17.Liu Y, Tan S, Huang L, Abramovitch RB, Rohde KH, Zimmerman MD, et al. J Exp Med [Internet] 5. Vol. 213. The Rockefeller University Press; 2016. May 2, Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo; pp. 809–25. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4854729/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal-Mutalik R, Nikaido H. Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc Natl Acad Sci [Internet] 2014 Apr 1;111(13):4958 LP–4963. doi: 10.1073/pnas.1403078111. Available from: http://www.pnas.org/content/111/13/4958.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant SS, Kawate T, Nag PP, Silvis MR, Gordon K, Stanley SA, et al. ACS Chem Biol [Internet] 10. Vol. 8. American Chemical Society; 2013. Oct 18, Identification of Novel Inhibitors of Nonreplicating Mycobacterium tuberculosis Using a Carbon Starvation Model; pp. 2224–34. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, et al. Tryptophan biosynthesis protects mycobacteria from CD4 T cell-mediated killing. Cell [Internet] 2013 Dec 5;155(6):1296–308. doi: 10.1016/j.cell.2013.10.045. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3902092/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Boogaard J Den, Kibiki GS, Kisanga ER, Boeree MJ, Aarnoutse RE. New drugs against tuberculosis: Problems, progress and evaluation of agents in clinical development. Antimicrob Agents Chemother. 2009;53(3):849–62. doi: 10.1128/AAC.00749-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds J, Heysell SK. Understanding pharmacokinetics to improve tuberculosis treatment outcome. Expert Opin Drug Metab Toxicol [Internet] 2014 Jun 6;10(6):813–23. doi: 10.1517/17425255.2014.895813. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4112565/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awasthi D, Freundlich JS. Trends Microbiol [Internet] xx. Elsevier Ltd; 2017. Antimycobacterial Metabolism : Illuminating Mycobacterium tuberculosis Biology and Drug Discovery; pp. 1–12. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warrier T, Kapilashrami K, Argyrou A, Ioerger TR, Little D, Murphy KC, et al. <em>N</em>-methylation of a bactericidal compound as a resistance mechanism in <em>Mycobacterium tuberculosis</em>. Proc Natl Acad Sci [Internet] 2016 Aug 2;113(31):E4523 LP–E4530. doi: 10.1073/pnas.1606590113. Available from: http://www.pnas.org/content/113/31/E4523.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarathy JP, Dartois V, Lee EJD. Pharmaceuticals [Internet] 11. Vol. 5. MDPI; 2012. Nov 9, The Role of Transport Mechanisms in Mycobacterium Tuberculosis Drug Resistance and Tolerance; pp. 1210–35. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3816664/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. Nature [Internet] 7331. Vol. 469. Nature Publishing Group; 2011. The challenge of new drug discovery for tuberculosis; pp. 483–90. Available from: http://www.nature.com/doifinder/10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 27.Prideaux B, Via LE, Zimmerman MD, Eum S, Brien PO, Chen C, et al. Into Tuberculosis. Lesions. 2016;21(10):1223–7. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prideaux B, Dartois V, Staab D, Weiner DM, Goh A, Via LE, et al. High-Sensitivity MALDI-MRM-MS Imaging of Moxifloxacin Distribution in Tuberculosis-Infected Rabbit Lungs and Granulomatous Lesions. Anal Chem [Internet] 2011 Mar 15;83(6):2112–8. doi: 10.1021/ac1029049. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3158846/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenaerts A, Barry CE, Dartois V. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev [Internet] 2015 Mar 1;264(1):288–307. doi: 10.1111/imr.12252. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimmerman M, Lestner J, Prideaux B, O’Brien P, Freedman I, Chen C, et al. Ethambutol partitioning in tuberculous pulmonary lesions explains its clinical efficacy. Antimicrob Agents Chemother [Internet] 2017 Jul; doi: 10.1128/AAC.00924-17. AAC. 00924-17. Available from: http://aac.asm.org/lookup/doi/10.1128/AAC.00924-17. [DOI] [PMC free article] [PubMed]
- 31.Pienaar E, Sarathy J, Prideaux B, Dietzold J, Dartois V, Kirschner DE, et al. Comparing efficacies of moxifloxacin, levofloxacin and gatifloxacin in tuberculosis granulomas using a multi-scale systems pharmacology approach [Internet] PLOS Computational Biology. 2017:e1005650. doi: 10.1371/journal.pcbi.1005650. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28817561%0Ahttp://dx.plos.org/10.1371/journal.pcbi.1005650. [DOI] [PMC free article] [PubMed]
- 32.Mitchison D, Davies G. The chemotherapy of tuberculosis: past, present and future. Int J Tuberc Lung Dis [Internet] 2012 Jun;16(6):724–32. doi: 10.5588/ijtld.12.0083. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3736084/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarathy J, Dartois V, Dick T, Gengenbacher M. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2013;57(4):1648–53. doi: 10.1128/AAC.02202-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z, Siddiqi N, Rubin EJ. Antimicrob Agents Chemother [Internet] 11. Vol. 49. American Society for Microbiology; 2005. Nov 22, Differential Antibiotic Susceptibilities of Starved Mycobacterium tuberculosis Isolates; pp. 4778–80. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1280169/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gumbo T, Lenaerts AJ, Hanna D, Romero K, Nuermberger E. Nonclinical Models for Antituberculosis Drug Development: A Landscape Analysis. J Infect Dis. 2015 Sep;211:S83–95. doi: 10.1093/infdis/jiv183. [DOI] [PubMed] [Google Scholar]
- 36.Schump MD, Fox DM, Bertozzi CR, Riley LW. Subcellular partitioning and intramacrophage selectivity of antimicrobial compounds against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2017;61(3):1–12. doi: 10.1128/AAC.01639-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MPR. The Immune Response in Tuberculosis. Annu Rev Immunol [Internet] 2013 Mar 21;31(1):475–527. doi: 10.1146/annurev-immunol-032712-095939. Annual Reviews. Available from: [DOI] [PubMed] [Google Scholar]
- 38.Eum S-Y, Kong J-H, Hong M-S, Lee Y-J, Kim J-H, Hwang S-H, et al. Chest [Internet] 1. Vol. 137. American College of Chest Physicians; 2010. Jan 11, Neutrophils Are the Predominant Infected Phagocytic Cells in the Airways of Patients With Active Pulmonary TB; pp. 122–8. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2803122/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe DM, Redford PS, Wilkinson RJ, O’Garra A, Martineau AR. Trends Immunol [Internet] 1. Vol. 33. Elsevier; 2012. Mar 2, Neutrophils in tuberculosis: friend or foe? pp. 14–25. Available from: [DOI] [PubMed] [Google Scholar]
- 40.Kaplan G, Post FA, Moreira AL, Wainwright H, Kreiswirth BN, Tanverdi M, et al. Infect Immun [Internet] 12. Vol. 71. American Society for Microbiology; 2003. Dec 18, Mycobacterium tuberculosis Growth at the Cavity Surface: a Microenvironment with Failed Immunity; pp. 7099–108. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC308931/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernández-Pando R, Jeyanathan M, Mengistu G, Aguilar D, Orozco H, Harboe M, et al. Lancet [Internet] 9248. Vol. 356. Elsevier; 2000. Mar 2, Persistence of DNA from Mycobacterium tuberculosis in superficially normal lung tissue during latent infection; pp. 2133–8. Available from: [DOI] [PubMed] [Google Scholar]
- 42.Barrios-Payán J, Saqui-Salces M, Jeyanathan M, Alcántara-Vazquez A, Castañon-Arreola M, Rook G, et al. Extrapulmonary Locations of Mycobacterium tuberculosis DNA During Latent Infection. J Infect Dis [Internet] 2012 Oct 15;206(8):1194–205. doi: 10.1093/infdis/jis381. Available from: [DOI] [PubMed] [Google Scholar]
- 43.Reece ST, Vogelzang A, Tornack J, Bauer W, Zedler U, Schommer-Leitner S, et al. Mycobacterium tuberculosis-Infected Hematopoietic Stem and Progenitor Cells Unable to Express Inducible Nitric Oxide Synthase Propagate Tuberculosis in Mice. J Infect Dis [Internet] 2018 Feb;17:jiy041–jiy041. doi: 10.1093/infdis/jiy041. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beigier-Bompadre M, Montagna GN, Kühl AA, Lozza L, Weiner J, Kupz A, et al. Mycobacterium tuberculosis infection modulates adipose tissue biology. PLoS Pathog. 2017;13(10) doi: 10.1371/journal.ppat.1006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evangelopoulos D, da Fonseca JD, Waddell SJ. Int J Infect Dis [Internet] Vol. 32. International Society for Infectious Diseases; 2015. Understanding anti-tuberculosis drug efficacy: Rethinking bacterial populations and how we model them; pp. 76–80. Available from: [DOI] [PubMed] [Google Scholar]
- 46.Cohen J. Approval of Novel TB Drug Celebrated-With Restraint. Science (80-) [Internet] 2013;339(6116):130–130. doi: 10.1126/science.339.6116.130. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23307714%5Cnhttp://www.sciencemag.org/cgi/doi/10.1126/science.339.6116.130. [DOI] [PubMed] [Google Scholar]
- 47.Saito K, Warrier T, Somersan-Karakaya S, Kaminski L, Mi J, Jiang X, et al. Rifamycin action on RNA polymerase in antibiotic-tolerant <em>Mycobacterium tuberculosis</em> results in differentially detectable populations. Proc Natl Acad Sci [Internet] 2017 Jun 13;114(24):E4832 LP–E4840. doi: 10.1073/pnas.1705385114. Available from: http://www.pnas.org/content/114/24/E4832.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, et al. Dynamic Persistence of Antibiotic-Stressed Mycobacteria. Science (80-) [Internet] 2013;339(6115):91–5. doi: 10.1126/science.1229858. Available from: http://www.sciencemag.org/cgi/doi/10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 49.Horsburgh CR, Barry CE, Lange C. Treatment of Tuberculosis. N Engl J Med [Internet] 2015;373(22):2149–60. doi: 10.1056/NEJMra1413919. Available from: http://www.nejm.org/doi/10.1056/NEJMra1413919. [DOI] [PubMed] [Google Scholar]
- 50.Vanderven BC, Huang L, Rohde KH, Russell DG. HHS Public Access. Microbiol Spectr. 2017;4(6):1–26. doi: 10.1128/microbiolspec.TBTB2-0025-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrt S, R K. Mycobacterium Tuberculosis Metabolism and Host Interaction: Mysteries and Paradoxes. Curr Top Microbiol Immunol. 2013;374(1):163–88. doi: 10.1007/82_2012_299. [DOI] [PubMed] [Google Scholar]
- 52.Srivastava S, Ernst JD, Desvignes L. Beyond macrophages: The diversity of mononuclear cells in tuberculosis. Immunol Rev. 2014;262(1):179–92. doi: 10.1111/imr.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohde K, Yates RM, Purdy GE, Russell DG. Mycobacterium tuberculosis and the environment within the phagosome. Immunol Rev. 2007;219(1):37–54. doi: 10.1111/j.1600-065X.2007.00547.x. [DOI] [PubMed] [Google Scholar]
- 54.Russell DG. The evolutionary pressures that have molded Mycobacterium tuberculosis into an infectious adjuvant. Curr Opin Microbiol [Internet] 2013 Feb 3;16(1):78–84. doi: 10.1016/j.mib.2012.11.007. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3637961/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fortune SM, Rubin EJ. Cell Host Microbe [Internet] 1. Vol. 2. Elsevier; 2007. Mar 2, The Complex Relationship between Mycobacteria and Macrophages: It’s Not All Bliss; pp. 5–6. Available from: [DOI] [PubMed] [Google Scholar]
- 56.Raffetseder J, Pienaar E, Blomgran R, Eklund D, Brodin VP, Andersson H, et al. Replication rates of mycobacterium tuberculosis in human macrophages do not correlate with mycobacterial antibiotic susceptibility. PLoS One. 2014;9(11):1–10. doi: 10.1371/journal.pone.0112426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, et al. High Content Screening Identifies Decaprenyl-Phosphoribose 2′ Epimerase as a Target for Intracellular Antimycobacterial Inhibitors. In: Bishai W, editor. PLoS Pathog [Internet] 10. Vol. 5. San Francisco, USA: Public Library of Science; 2009. Oct 30, p. e1000645. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2763345/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brodin P, Christophe T. Curr Opin Chem Biol [Internet] 4. Vol. 15. Elsevier Ltd; 2011. High-content screening in infectious diseases; pp. 534–9. Available from: [DOI] [PubMed] [Google Scholar]
- 59.Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, et al. Identification of Host-Targeted Small Molecules That Restrict Intracellular Mycobacterium tuberculosis Growth. In: Ehrt S, editor. PLoS Pathog [Internet] 2. Vol. 10. San Francisco, USA: Public Library of Science; 2014. Feb 20, p. e1003946. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3930586/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, et al. Novel Inhibitors of Cholesterol Degradation in Mycobacterium tuberculosis Reveal How the Bacterium???s Metabolism Is Constrained by the Intracellular Environment. PLoS Pathog. 2015;11(2):1–20. doi: 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang L, Russell DG. Protective immunity against tuberculosis: what does it look like and how do we find it? Curr Opin Immunol [Internet] 2017;48(Supplement C):44–50. doi: 10.1016/j.coi.2017.08.001. Available from: http://www.sciencedirect.com/science/article/pii/S0952791517300183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leemans JC, Thepen T, Weijer S, Florquin S, van Rooijen N, van de Winkel JG, et al. Macrophages play a dual role during pulmonary tuberculosis in mice. J Infect Dis. 2005;191(1):65–74. doi: 10.1086/426395. [DOI] [PubMed] [Google Scholar]
- 63.Leemans JC, Juffermans NP, Florquin S, van Rooijen N, Vervoordeldonk MJ, Verbon A, et al. Depletion of Alveolar Macrophages Exerts Protective Effects in Pulmonary Tuberculosis in Mice. J Immunol [Internet] 2001;166(7):4604–11. doi: 10.4049/jimmunol.166.7.4604. Available from: http://www.jimmunol.org/content/166/7/4604.full. [DOI] [PubMed] [Google Scholar]
- 64.Kramnik I, Beamer G. Mouse models of human TB pathology: roles in the analysis of necrosis and the development of host-directed therapies. Semin Immunopathol. 2016;38(2):221–37. doi: 10.1007/s00281-015-0538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nuermberger E, Sizemore C, Romero K, Hanna D. Towards an evidence-based non-clinical roadmap for evaluating the efficacy of new TB drug regimens: Proceedings of a Critical Path to TB Drug Regimens (CPTR)-National Institute of Allergy and Infectious Disease (NIAID) in vivo Pharmacology Workshop for Tu. Antimicrob Agents Chemother [Internet] 2016;60(3):1177–82. doi: 10.1128/AAC.02041-15. Available from: http://aac.asm.org/content/60/3/1177.abstract?etoc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fejer G, Wegner MD, Györy I, Cohen I, Engelhard P, Voronov E, et al. Proc Natl Acad Sci U S A [Internet] 24. Vol. 110. National Academy of Sciences; 2013. Jun 11, Nontransformed, GM-CSF–dependent macrophage lines are a unique model to study tissue macrophage functions; pp. E2191–8. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3683787/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. Nat Immunol [Internet] Vol. 17. Nature Publishing Group a division of Macmillan Publishers Limited; 2015. Dec 17, New insights into the multidimensional concept of macrophage ontogeny, activation and function; p. 34. All Rights Reserved. Available from: [DOI] [PubMed] [Google Scholar]
- 68.Flynn JL, Chan J, Lin PL. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol [Internet] 2011 May 23;4(3):271–8. doi: 10.1038/mi.2011.14. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3311958/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Neill LAJ, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nature Reviews Immunology. 2016:553–65. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eisenreich W, Heesemann J, Rudel T. Metabolic Adaptations of Intracellullar Bacterial Pathogens and their Mammalian Host Cells during Infection (“Pathometabolism”) Microbiol Spectr [Internet] 2015 doi: 10.1128/microbiolspec.MBP-0002-2014. Available from: http://www.asmscience.org/content/journal/microbiolspec/10.1128/microbiolspec.MBP-0002-2014%5Cnpapers3://publication/doi/10.1128/microbiolspec.MBP-0002-2014. [DOI] [PubMed]
- 71.Fange D, Nilsson K, Tenson T, Ehrenberg M. Drug efflux pump deficiency and drug target resistance masking in growing bacteria. Proc Natl Acad Sci [Internet] 2009 May 19;106(20):8215 LP–8220. doi: 10.1073/pnas.0811514106. Available from: http://www.pnas.org/content/106/20/8215.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhamidi S, Scherman MS, Jones V, Crick DC, Belisle JT, Brennan PJ, et al. J Biol Chem [Internet] 26. Vol. 286. 9650 Rockville Pike, Bethesda, MD 20814, U.S.A.: American Society for Biochemistry and Molecular Biology; 2011. Jul 1, Detailed Structural and Quantitative Analysis Reveals the Spatial Organization of the Cell Walls of in Vivo Grown Mycobacterium leprae and in Vitro Grown Mycobacterium tuberculosis; pp. 23168–77. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3123084/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. Mycobacterial outer membranes: in search of proteins. Trends Microbiol [Internet] 2010 Mar 8;18(3):109–16. doi: 10.1016/j.tim.2009.12.005. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2931330/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Louw GE, Warren RM, Gey van Pittius NC, McEvoy CRE, Van Helden PD, Victor TC. Antimicrob Agents Chemother [Internet] 8. Vol. 53. American Society for Microbiology (ASM); 2009. Aug 18, A Balancing Act: Efflux/Influx in Mycobacterial Drug Resistance; pp. 3181–9. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2715638/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.te Brake LHM, de Knegt GJ, de Steenwinkel JE, van Dam TJP, Burger DM, Russel FGM, et al. The Role of Efflux Pumps in Tuberculosis Treatment and Their Promise as a Target in Drug Development: Unraveling the Black Box. Annu Rev Pharmacol Toxicol [Internet] 2018;58(1) doi: 10.1146/annurev-pharmtox-010617-052438. annurev-pharmtox-010617-052438. Available from: http://www.annualreviews.org/doi/10.1146/annurev-pharmtox-010617-052438. [DOI] [PubMed] [Google Scholar]
- 76.Balganesh M, Dinesh N, Sharma S, Kuruppath S, Nair AV, Sharma U. Efflux pumps of Mycobacterium tuberculosis play a significant role in antituberculosis activity of potential drug candidates. Antimicrob Agents Chemother. 2012;56(5):2643–51. doi: 10.1128/AAC.06003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dinesh N, Sharma S, Balganesh M. Antimicrob Agents Chemother [Internet] 4. Vol. 57. 1752 N St., N.W., Washington, DC: American Society for Microbiology; 2013. Apr 1, Involvement of Efflux Pumps in the Resistance to Peptidoglycan Synthesis Inhibitors in Mycobacterium tuberculosis; pp. 1941–3. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3623302/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adams KN, Szumowski JD, Ramakrishnan L. J Infect Dis [Internet] 3. Vol. 210. Oxford University Press; 2014. Aug 1, Verapamil, and Its Metabolite Norverapamil, Inhibit Macrophage-induced, Bacterial Efflux Pump-mediated Tolerance to Multiple Anti-tubercular Drugs; pp. 456–66. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4110457/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, et al. Drug Tolerance in Replicating Mycobacteria Mediated by a Macrophage-Induced Efflux Mechanism. Cell [Internet] 2011 Apr 1;145(1):39–53. doi: 10.1016/j.cell.2011.02.022. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3117281/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, Shi W, Zhang S, Cassell G, Maslov D, Shur K, et al. Mutations in efflux pump Rv1258c (Tap) cause resistance to pyrazinamide and other drugs in M. tuberculosis. bioRxiv [Internet] 2018 Jan 1; Available from: http://biorxiv.org/content/early/2018/01/17/249102.1.abstract.
- 81.Rodriguez-Rivera FP, Zhou X, Theriot JA, Bertozzi CR. Visualization of mycobacterial membrane dynamics in live cells. J Am Chem Soc. 2017;139(9):3488–95. doi: 10.1021/jacs.6b12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manning AJ, Ovechkina Y, McGillivray A, Flint L, Roberts DM, Parish T. Methods [Internet] Vol. 127. The Authors; 2017. A high content microscopy assay to determine drug activity against intracellular Mycobacterium tuberculosis; pp. 3–11. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1046202316303619. [DOI] [PubMed] [Google Scholar]
- 83.Mdluli K, Kaneko T, Upton A. Drug Targets. 2015:1–24. doi: 10.1101/cshperspect.a021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Galagan JE, Minch K, Peterson M, Lyubetskaya A, Sweet L, Gomes A, et al. NIH Public Access. 2014;499(7457):178–83. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boshoff HIM, Barry CE., 3rd . Nat Rev Microbiol [Internet] Vol. 3. Nature Publishing Group; 2005. Jan 1, Tuberculosis — metabolism and respiration in the absence of growth; p. 70. Available from: [DOI] [PubMed] [Google Scholar]
- 86.Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat Med [Internet] 2013;19(9):1157–60. doi: 10.1038/nm.3262. Available from: http://www.nature.com/doifinder/10.1038/nm.3262. [DOI] [PubMed] [Google Scholar]
- 87.Wei J, Dahl JL, Moulder JW, Roberts EA, O’Gaora P, Young DB, et al. J Bacteriol [Internet] 2. Vol. 182. American Society for Microbiology; 2000. Jan 3, Identification of a Mycobacterium tuberculosis Gene That Enhances Mycobacterial Survival in Macrophages; pp. 377–84. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC94286/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.P M, Shah W, Junghanns WS. Dose-response relationship of bactericidal activity in E. coli, K. pneumoniae and Staphylococcus aureus. Dtsch med Wochenschr. 1976;101(9):325–8. doi: 10.1055/s-0028-1104083. [DOI] [PubMed] [Google Scholar]
- 89.Craig WA. Interrelationship between pharmacokinetics and pharmacodynamics in determining dosage regimens for broad-spectrum cephalosporins. Diagn Microbiol Infect Dis [Internet] 1995;22(1):89–96. doi: 10.1016/0732-8893(95)00053-d. Available from: http://www.sciencedirect.com/science/article/pii/073288939500053D. [DOI] [PubMed] [Google Scholar]
- 90.Craig WA. State‐of‐the‐Art Clinical Article: Pharmacokinetic/Pharmacodynamic Parameters: Rationale for Antibacterial Dosing of Mice and Men. Clin Infect Dis [Internet] 1998;26(1):1–10. doi: 10.1086/516284. Available from: https://academic.oup.com/cid/article-lookup/doi/10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 91.Vaddady PK, Lee RE, Meibohm B. In vitro pharmacokinetic/pharmacodynamic models in anti-infective drug development: focus on TB. Future Med Chem [Internet] 2010 Aug;2(8):1355–69. doi: 10.4155/fmc.10.224. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3042695/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasipanodya JG, Srivastava S, Gumbo T. Meta-analysis of clinical studies supports the pharmacokinetic variability hypothesis for acquired drug resistance and failure of antituberculosis therapy. Clinical Infectious Diseases. 2012:169–77. doi: 10.1093/cid/cis353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pasipanodya JG, Srivastava S, Gumbo T. Acquired drug resistance because of pharmacokinetic variability in a young child with tuberculosis. Pediatric Infectious Disease Journal. 2014:1205. doi: 10.1097/INF.0000000000000436. [DOI] [PubMed] [Google Scholar]
- 94.Pasipanodya JG, Nuermberger E, Romero K, Hanna D, Gumbo T. Systematic analysis of hollow fiber model of tuberculosis experiments. Clin Infect Dis. 2015;61:S10–7. doi: 10.1093/cid/civ425. [DOI] [PubMed] [Google Scholar]
- 95.Pasipanodya JG, McIlleron H, Burger A, Wash PA, Smith P, Gumbo T. Serum drug concentrations predictive of pulmonary tuberculosis outcomes. J Infect Dis. 2013;208(9):1464–73. doi: 10.1093/infdis/jit352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gumbo T, Louie A, Deziel MR, Parsons LM, Salfinger M, Drusano GL. Selection of a moxifloxacin dose that suppresses drug resistance in Mycobacterium tuberculosis, by use of an in vitro pharmacodynamic infection model and mathematical modeling. J Infect Dis [Internet] 2004;190(9):1642–51. doi: 10.1086/424849. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15478070%0Ahttp://jid.oxfordjournals.org/content/190/9/1642.full.pdf. [DOI] [PubMed] [Google Scholar]
- 97.Dheda K, Lenders LGT. New insights into the pathogenesis of drug-resistant TB and implications for clinical management. keystone Symp Tuberc co-morbidities immunopathogenesis; 2016; Keystone Resort, Keystone, Color USA. 2016; 2016. [Google Scholar]
- 98.Diacon AH, Patientia RF, Venter A, van Helden PD, Smith PJ, McIlleron H, et al. Antimicrob Agents Chemother [Internet] 8. Vol. 51. American Society for Microbiology; 2007. Aug 21, Early Bactericidal Activity of High-Dose Rifampin in Patients with Pulmonary Tuberculosis Evidenced by Positive Sputum Smears; pp. 2994–6. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1932511/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gu HF, Li HZ, Tang YL, Tang XQ, Zheng XL, Liao DF. Nicotinate-curcumin impedes foam cell formation from THP-1 cells through restoring autophagy flux. PLoS One. 2016;11(4):1–15. doi: 10.1371/journal.pone.0154820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vinegoni C, Dubach JM, Thurber GM, Miller MA, Mazitschek R, Weissleder R. Drug Discov Today [Internet] 9. Vol. 20. Elsevier Ltd; 2015. Advances in measuring single-cell pharmacology in vivo; pp. 1087–92. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bekker A, Schaaf HS, Draper HR, van der Laan L, Murray S, Wiesner L, et al. Pharmacokinetics of Rifampin, Isoniazid, Pyrazinamide, and Ethambutol in Infants Dosed According to Revised WHO-Recommended Treatment Guidelines. Antimicrob Agents Chemother [Internet] 2016 Apr;60(4):2171–2179. doi: 10.1128/AAC.02600-15. Available from: http://europepmc.org/articles/PMC4808214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sloan DJ, McCallum AD, Schipani A, Egan D, Mwandumba HC, Ward SA, et al. Genetic determinants of the pharmacokinetic variability of rifampin in Malawian adults with pulmonary tuberculosis. Antimicrob Agents Chemother. 2017;61(7):1–9. doi: 10.1128/AAC.00210-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Denti P, Gonzalez-Martinez C, Winckler J, Bekker A, Zar H, Davies G, et al. Pharmacokinetics of rifampicin in African children-Evaluation of the new WHO dosing guidelines. 10th International workshop on clinical pharmacology of tuberculosis drugs; Atlanta, Georgia. 2017. p. 28. [Google Scholar]
- 104.Thee S, Seddon JA, Donald PR, Seifart HI, Werely CJ, Hesseling AC, et al. Antimicrob Agents Chemother [Internet] 12. Vol. 55. 1752 N St., N.W., Washington, DC: American Society for Microbiology; 2011. Dec 31, Pharmacokinetics of Isoniazid, Rifampin, and Pyrazinamide in Children Younger than Two Years of Age with Tuberculosis: Evidence for Implementation of Revised World Health Organization Recommendations; pp. 5560–7. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3232790/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seddon JA, Schaaf HS. Pneumonia [Internet] Vol. 8. London: BioMed Central; 2016. Nov 24, Drug-resistant tuberculosis and advances in the treatment of childhood tuberculosis; p. 20. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5471710/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nijland HMJ, Ruslami R, Suroto AJ, Burger DM, Alisjahbana B, van Crevel R, et al. Rifampicin Reduces Plasma Concentrations of Moxifloxacin in Patients with Tuberculosis. Clin Infect Dis [Internet] 2007 Oct 15;45(8):1001–7. doi: 10.1086/521894. Available from: [DOI] [PubMed] [Google Scholar]
- 107.Cokol M, Kuru N, Bicak E, Larkins-Ford J, Aldridge BB. Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis. Sci Adv [Internet] 2017;3(10):e1701881. doi: 10.1126/sciadv.1701881. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29026882%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5636204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dawson R, Diacon AH, Everitt D, van Niekerk C, Donald PR, Burger DA, et al. Lancet [Internet] 9979. Vol. 385. Elsevier; 2017. Dec 22, Efficiency and safety of the combination of moxifloxacin, pretomanid (PA-824), and pyrazinamide during the first 8 weeks of antituberculosis treatment: a phase 2b, open-label, partly randomised trial in patients with drug-susceptible or drug-resistant pul; pp. 1738–47. Available from: [DOI] [PubMed] [Google Scholar]
- 109.Warner DF, Mizrahi V. N Engl J Med [Internet] 17. Vol. 371. Massachusetts Medical Society; 2014. Oct 22, Shortening Treatment for Tuberculosis — Back to Basics; pp. 1642–3. Available from: [DOI] [PubMed] [Google Scholar]
- 110.Kislitsyna NA, Kotova NI. Rifampicin and isoniazid concentration in the blood and resected lungs in tuberculosis with combined use of the preparations. Probl Tuberk. 8:63–65. [PubMed] [Google Scholar]
- 111.Canetti G, Parrot R, Porven G, Le Lirzin M. Rifomycin levels in the lung and tuberculous lesions in man. Acta Tuberc Pneumol Belg. 1969;60:315–322. (in French) [PubMed] [Google Scholar]
- 112.Sauermann R, Karch R, Langenberger H, Kettenbach J, Mayer-Helm B, Petsch M, et al. Antibiotic abscess penetration: Fosfomycin levels measured in pus and simulated concentration-time profiles. Antimicrob Agents Chemother. 2005;49(11):4448–54. doi: 10.1128/AAC.49.11.4448-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wagner C, Sauermann R, Joukhadar C. Principles of Antibiotic Penetration into Abscess Fluid. Pharmacology [Internet] 2006;78(1):1–10. doi: 10.1159/000094668. Available from: https://www.karger.com/DOI/10.1159/000094668. [DOI] [PubMed] [Google Scholar]
- 114.Scaglione F, Paraboni L. Expert Rev Anti Infect Ther [Internet] 3. Vol. 4. Taylor & Francis; 2006. Jun 1, Influence of pharmacokinetics/pharmacodynamics of antibacterials in their dosing regimen selection; pp. 479–90. Available from: [DOI] [PubMed] [Google Scholar]
- 115.Stanley SA, Barczak AK, Silvis MR, Luo SS, Sogi K, Vokes M, et al. Identification of Host-Targeted Small Molecules That Restrict Intracellular Mycobacterium tuberculosis Growth. PLoS Pathog. 2014;10(2) doi: 10.1371/journal.ppat.1003946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Queval CJ, Song O-R, Delorme V, Iantomasi R, Veyron-Churlet R, Deboosère N, et al. A Microscopic Phenotypic Assay for the Quantification of Intracellular Mycobacteria Adapted for High-throughput/High-content Screening. J Vis Exp [Internet] 2014;(83):1–16. doi: 10.3791/51114. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24473237. [DOI] [PMC free article] [PubMed]
- 117.de Carvalho LPS, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis Reveals Compartmentalized Co-Catabolism of Carbon Substrates. Chem Biol [Internet] 2010;17(10):1122–31. doi: 10.1016/j.chembiol.2010.08.009. Available from: http://www.sciencedirect.com/science/article/pii/S1074552110003479. [DOI] [PubMed] [Google Scholar]
- 118.Chakraborty P, Kulkarni S, Rajan R, Sainis K. Drug Resistant Clinical Isolates of Mycobacterium tuberculosis from Different Genotypes Exhibit Differential Host Responses in THP-1 Cells. PLoS One. 2013;8(5):1–9. doi: 10.1371/journal.pone.0062966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Halouska S, Fenton RJ, Barletta RG, Powers R. Predicting the in vivo Mechanism of Action for Drug Leads using NMR Metabolomics. ACS Chem Biol [Internet] 2012 Jan 20;7(1):166–71. doi: 10.1021/cb200348m. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3262886/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Halouska S, Fenton RJ, Zinniel DK, Marshall DD, Barletta RG, Powers R. Metabolomics Analysis Identifies D-Alanine-D-alanine Ligase as the Primary Lethal Target of D-cycloserine in Mycobacteria. J Proteome Res [Internet] 2014 Feb 7;13(2):1065–76. doi: 10.1021/pr4010579. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3975674/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prosser GA, de Carvalho LPS. ACS Med Chem Lett [Internet] 12. Vol. 4. American Chemical Society; 2013. Dec 12, Metabolomics Reveal d-Alanine:d-Alanine Ligase As the Target of d-Cycloserine in Mycobacterium tuberculosis; pp. 1233–7. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3903091/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deborah-Ann J, D SK. Biomed Chromatogr [Internet] 6–7. Vol. 20. Wiley-Blackwell; 2006. Jun 15, The efficacy of certain anti‐tuberculosis drugs is affected by binding to α‐1‐acid glycoprotein; pp. 551–60. Available from: [DOI] [PubMed] [Google Scholar]
- 123.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: Defense against host stresses. Cell Microbiol [Internet] 2009 Aug 6;11(8):1170–8. doi: 10.1111/j.1462-5822.2009.01335.x. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3170014/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.MacMicking JD, Taylor GA, McKinney JD. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science (80-) [Internet] 2003;302(5645):654–9. doi: 10.1126/science.1088063. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14576437%5Cnhttp://www.sciencemag.org/content/302/5645/654.abstract%5Cnhttp://www.sciencemag.org/content/302/5645/654.full.pdf. [DOI] [PubMed] [Google Scholar]
- 125.Vandal OH, Nathan CF, Ehrt S. Acid resistance in Mycobacterium tuberculosis. Journal of Bacteriology. 2009:4714–21. doi: 10.1128/JB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: A review. International. Journal of Tuberculosis and Lung Disease. 2003:6–21. [PubMed] [Google Scholar]
- 127.Mitchison DA. The action of antituberculosis drugs in short-course chemotherapy. Tubercle. 1985;66(3):219–25. doi: 10.1016/0041-3879(85)90040-6. [DOI] [PubMed] [Google Scholar]
- 128.Mitchison DA, Coates ARM. Predictive In Vitro Models of the Sterilizing Activity of Anti-Tuberculosis Drugs. Curr Pharm Des [Internet] 2004;10(26):3285–95. doi: 10.2174/1381612043383269. Available from: http://search.ebscohost.com/login.aspx?direct=true&db=aph&AN=14546022&site=ehost-live&scope=site. [DOI] [PubMed] [Google Scholar]
- 129.Hoagland DT, Liu J, Lee RB, Lee RE. Adv Drug Deliv Rev [Internet] Vol. 102. The Authors; 2016. New agents for the treatment of drug-resistant Mycobacterium tuberculosis; pp. 55–72. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schaaf K, Hayley V, Speer A, Wolschendorf F, Niederweis M, Kutsch O, et al. Assay Drug Dev Technol [Internet] 6. Vol. 14. 140 Huguenot Street, 3rd FloorNew Rochelle, NY 10801 USA: Mary Ann Liebert, Inc; 2016. Aug 1, A Macrophage Infection Model to Predict Drug Efficacy Against Mycobacterium Tuberculosis; pp. 345–54. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4991579/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O’Brien P, et al. Nat Med [Internet] Vol. 21. Nature Publishing Group a division of Macmillan Publishers Limited; 2015. Sep 7, The association between sterilizing activity and drug distribution into tuberculosis lesions; p. 1223. All Rights Reserved. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gillespie SH, Crook AM, McHugh TD, Mendel CM, Meredith SK, Murray SR, et al. Four-Month Moxifloxacin-Based Regimens for Drug-Sensitive Tuberculosis. N Engl J Med [Internet] 2014 Oct 23;371(17):1577–87. doi: 10.1056/NEJMoa1407426. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4277680/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–63. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 134.Gengenbacher M, Rao SPS, Pethe K, Dick T. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology. 2010;156(1):81–7. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 135.Bielecka MK, Tezera LB, Zmijan R, Drobniewski F. Culture Platform Integrated with Microfluidics To Address Antimicrobial Resistance in Tuberculosis. 2017;8(1) doi: 10.1128/mBio.02073-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tezera LB, Bielecka MK, Chancellor A, Reichmann MT, Shammari B Al, Brace P, et al. Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. In: Aldridge B, editor. Elife [Internet] Vol. 6. eLife Sciences Publications, Ltd; 2017. p. e21283. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]


