Summary
The olfactory system must recognize and discriminate among highly diverse chemicals in the environment. To contend with such diversity, insects have evolved a unique family of odorant-gated ion channels composed of a highly conserved co-receptor (Orco) and a divergent odorant receptor (OR) that confers chemical specificity. Here, we present the single-particle cryo-electron microscopy structure of an Orco homomer at 3.5 Å resolution, providing the first structural insight into this receptor family. Orco possesses a novel channel architecture, with four subunits symmetrically arranged around a central pore that diverges into four lateral conduits that open to the cytosol. The Orco tetramer has few inter-subunit interactions within the membrane and is bound together by a small cytoplasmic anchor domain. The minimal sequence conservation among ORs maps largely to the pore and anchor domain, suggesting how the unique architecture of this receptor family accommodates their remarkable sequence diversity and facilitates the evolution of odour tuning.
Introduction
Insects are the most diverse group of multicellular organisms on Earth, representing over half of all identified animal species on the planet1. The success of insects reflects their remarkable capacity to adapt to a wide range of ecological niches. The rapid evolution of insect olfactory receptors is thought to contribute to this adaptation2, endowing each insect species with the ability to selectively detect volatile chemicals associated with their specialized habitat and lifestyle.
The olfactory systems of insects and mammals share a similar logic for odour detection and discrimination3,4. Each olfactory sensory neuron generally expresses just one member of a large family of receptors, an organizational principle that allows odours to be encoded by the combinatorial activation of different sensory neuron ensembles5,6. However, insect olfactory receptors are unrelated to the G protein-coupled chemoreceptors present in other animals. Instead, they have been proposed to form a unique class of heteromeric cation channels7,8 composed of two related heptahelical subunits: a divergent odorant receptor (OR) subunit that confers odour specificity, and a highly conserved co-receptor (Orco) subunit. Most species express just one Orco and a distinct complement of ORs9, ranging from just four members in the damselfly10 to more than 350 in some ants11. Variation in receptor number is paralleled by their striking sequence diversity, with an average of only ~20% amino-acid identity shared between ORs, either within or across species12. Indeed, orthologous ORs are rarely apparent between insect orders, highlighting how different species have evolved unique repertoires of receptors suited to their specific chemical environments.
Orco was initially identified as a member of the OR family in Drosophila13. However, in contrast to other ORs, Orco is broadly expressed in olfactory sensory neurons and is almost invariant in sequence across distant insect lineages, underscoring its essential role in olfactory transduction. ORs cannot assemble, traffic, or function in the absence of Orco14 and the loss of this single receptor results in dramatically impaired olfactory behaviours14–17. Orco is present in the olfactory sensory neurons of evolutionarily basal insects that lack ORs9, suggesting that it may represent the ancestral form of the olfactory receptor complex. Indeed, in the absence of ORs, Orco forms autonomous cation channels that can be activated by synthetic agonists18.
As insect olfactory receptors lack homology to any other protein family, many of their most elementary functional and structural characteristics have remained elusive, including their stoichiometry and how odor binding is transduced to ion flux. Moreover, in the absence of a structural model, how a single Orco can assemble with such a wide array of highly divergent ORs has remained unclear. Here, we present the structure of an Orco homomer from the parasitic fig wasp Apocrypta bakeri19. Orco forms a tetrameric channel comprised of four loosely assembled transmembrane domains, surrounding a central ion-conduction pathway, and a small intracellular anchor domain through which most inter-subunit interactions are formed. The minimal sequence conservation across ORs is largely localized to the pore and anchor domain, revealing how diverse Orco-OR heterotetramers can assemble. The structure of Orco thus defines the architecture of an archetypal insect olfactory receptor and provides insight into how this large family can rapidly diversify, allowing insects to adapt to different chemical landscapes.
Structure determination of an Orco-Fab complex
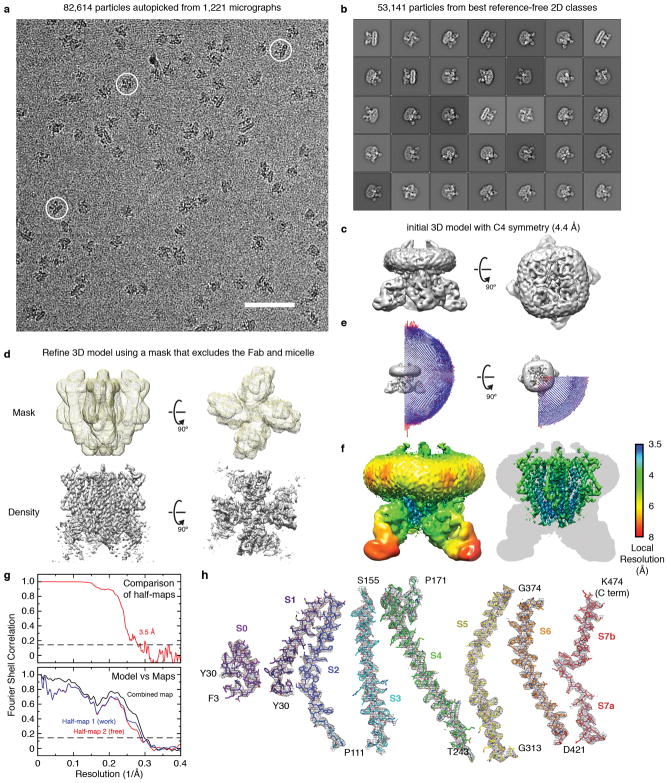
Insect olfactory receptors function as obligate multimers20. We therefore screened Orco orthologs by size-exclusion chromatography to find those that stably formed higher-order complexes, and identified the Orco from A. bakeri as a promising candidate. A. bakeri Orco19 exhibits the characteristic sequence and functional conservation of Orcos: it shares >60% sequence identity with Orco orthologs spanning the majority of insect orders and could couple to ORs from evolutionary distant species to mediate odor-gated signaling21 (Extended Data Fig. 1). Furthermore, when expressed independently, A. bakeri Orco formed a cation channel activated by the agonist VUAA118 (Fig. 1a,b; Extended Data Fig. 2). Chemical cross-linking suggested that Orco assembles into a tetramer, a stoichiometry further supported by its migration on native gels (Extended Data Fig. 3a–d).
Fig. 1. Channel activity, cryo-EM density and model of the Orco-Fab complex.
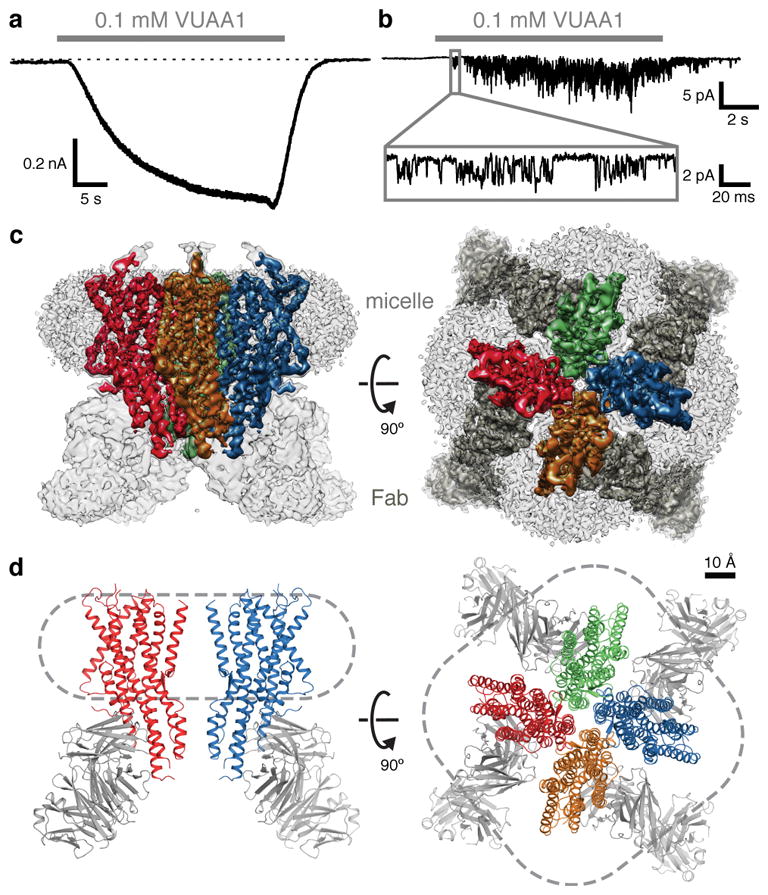
a, Whole-cell voltage-clamp recording from a cell expressing Orco with local perfusion of VUAA1 (holding potential of −80 mV). Dotted line represents 0 pA. b, Outside-out patch recording at −80 mV. Inset shows single channel openings. c, Cryo-EM density of the Orco-Fab complex shown from the side (within the plane of the membrane; left) and from the top (the extracellular surface; right). Density of each Orco subunit (contoured at 6σ) is coloured differently. In the top view, Fab density is dark grey. Density of the detergent micelle (contoured at 4σ) is light grey. d, Ribbon diagrams of Orco corresponding to views shown in c. Two subunits are shown in the side view (left) while four are present in the top view (right). Dashed lines indicate micelle outlines. The Fabs were not modeled and are included in d for illustrative purposes only.
2D-class averages calculated from an initial cryogenic electron microscopy (cryo-EM) dataset of purified Orco failed to show coherent structural features (data not shown). To increase the effective molecular mass of the protein particle (210 kDa), we raised monoclonal antibodies against Orco and purified a 1:1 complex, in which each of the four Orco subunits was bound by an antigen-binding fragment (Fab). Isothermal titration calorimetry (ITC) experiments confirmed that the Orco-Fab complex retained its binding affinity for VUAA1 (Extended Data Fig. 3e,f). A vitrified sample of the purified Orco-Fab complex generated homogenous and mono-dispersed particles with a tetrameric organization that was immediately apparent from the raw cryo-EM images and 2D-class averages (Extended Data Fig. 4a,b). 3D reconstruction using ~53,000 particles and imposing C4 symmetry yielded a density map with ~4 Å overall resolution. Further refinement after masking out the Fab and micelle regions improved the resolution to 3.5 Å (Fig. 1c; Extended Data Fig. 4c–g; Extended Data Table 1). Side-chain density was clearly resolved for most of the Orco channel and 82% of the protein could be accurately modeled, with the exception of the second extracellular loop (Val156–Ile170) and second intracellular loop (Leu244–Asn312). Density for the Fab was generally weaker, especially for the constant region, and was not modeled.
Architecture of the Orco homotetramer
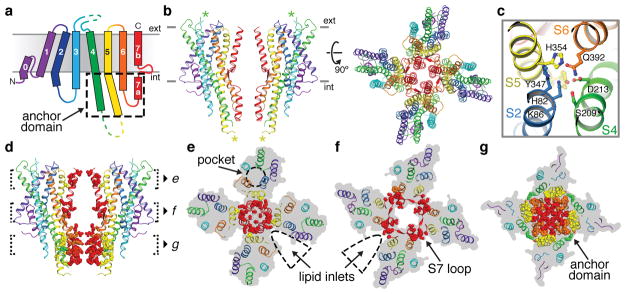
The architecture of Orco represents a novel fold. Viewed from the extracellular surface, Orco forms a tetrameric pinwheel 100 Å in diameter and 80 Å axially, with four subunits encircling a central pore (Fig. 1d). The majority of the protein resides within the micelle, with only short loops projecting from the extracellular surface and a small intracellular domain extending below. We term this protruding cytoplasmic helical bundle the ‘anchor domain’, as it contains the majority of inter-subunit interactions as described below and thus “anchors” the four loosely packed subunits within the micelle or lipid membrane.
Each Orco subunit has seven membrane-spanning helical segments (S1–S7), with an intracellular amino terminus and an extracellular carboxy terminus (Fig. 2a,b), a topology proposed by previous studies20. A short helix (S0) contributes to a re-entrant loop at the amino terminus that packs underneath S4 at the outer perimeter of the channel. Multiple helical segments (S1, S2, S4, S5) traverse the membrane at an angle (~30° relative to the membrane normal), facilitated by kinks in S2 and S5 at the intracellular membrane surface that serve to transform the largely parallel helical bundle of the anchor domain to the tilted helices within the membrane. S7 resides nearest the central four-fold axis and is divided into two helices—a cytoplasmic segment (S7a) and transmembrane segment (S7b)—separated by a 15-residue β-hairpin loop. S7b lines the central pore, while S7a forms the core of the anchor domain. The S4, S5, and S6 helices extend well beyond the membrane, projecting up to 40 Å into the cytosol where they surround S7a to complete the anchor domain.
Fig. 2. Architecture of the Orco homotetramer.
a, Topology of an Orco subunit. b, Ribbon representation of Orco shown from the side (left, two subunits) and top (right, four subunits). Asterisks represent residues that were not modeled. Grey lines indicate membrane boundaries. c, Hydrophilic network in the intracellular leaflet. d, Side view of two Orco subunits with residues within 5 Å from another subunit shown as spheres. e–g, 15 Å cross-sections from d.
The transmembrane domain of each subunit is stabilized by multiple charged and polar amino acids in S2, S4, S5 and S6 that coalesce to form a dense network of hydrophilic interactions within the intracellular membrane leaflet (Fig. 2c). Several of these interactions, including the salt bridge between Lys86 and Asp213, and the hydrogen bond between Asp213 and Gln392, were previously identified as amino acid pairs with high co-evolutionary coupling across ORs22.
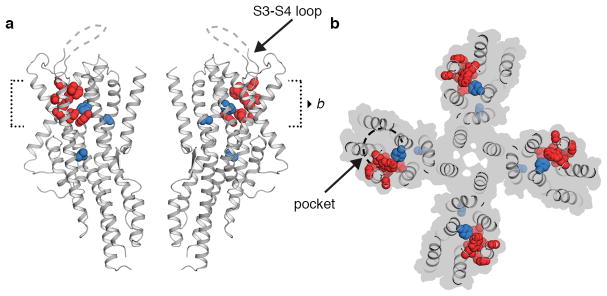
Within the extracellular leaflet, the S1–S6 helices splay apart to form a 10-Å deep crevice, approximately 20 Å long. Several residues implicated in determining Orco sensitivity to VUAA1 line this pocket23, suggesting that it may serve as a binding site for ligands that gate the channel (Extended Data Fig. 5). Mutations that alter odorant specificity in ORs24–27 also map to residues within this pocket, pointing to a potentially shared structural locus for ligand binding in Orcos and ORs. In the Orco structure, an ordered section of the S3–S4 extracellular loop restricts access to the pocket, which may prevent odorant binding to Orco, thus preserving the specificity of odour tuning in Orco-OR complexes.
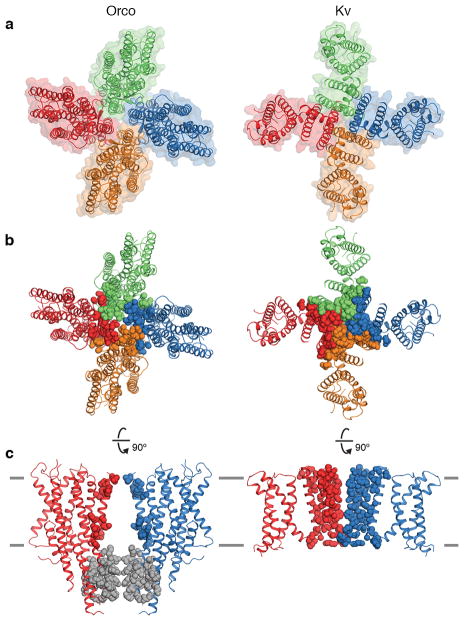
Within the membrane, the S1–S6 helices of each subunit are narrowly tethered to S7b and separated from their neighboring subunits by deep inlets that would be filled with lipid within the membrane (Fig. 2e,f). Thus interactions between subunits are largely confined to the pore-forming helices while the peripheral portions of each subunit are isolated from each other, an arrangement reminiscent of the more extensively studied superfamily of tetrameric cation channels28,29 (Extended Data Fig. 6a). However, while each subunit of a tetrameric cation channel contributes multiple helices to the central ion pathway, creating extensive contacts that intertwine the subunits together, the Orco pore is comprised of just a single helix (S7b), with far fewer amino acids linking the tetramer within the plane of the membrane (Fig. 2d–f; Extended Data Fig. 6b,c). The limited interactions between Orco subunits result in a small 7-Å gap between neighboring subunits suggesting that the lipid membrane forms an integral part of the inter-subunit interface that completes the structural boundary of the pore.
In contrast to the minimal contacts between Orco subunits within the membrane, the helices of the cytosolic anchor domain are densely packed, forming an interface that buries 1740 Å2 of surface area, accounting for nearly 70% of the contacts between subunits (Fig. 2g). Orco can therefore be coarsely divided into two domains—four loosely attached peripheral transmembrane domains and a single tightly packed central anchor domain. The elongated helices within each Orco subunit link these domains. In particular, the S4 and S5 helices are each more than 50 residues long, forming linchpins that moor the transmembrane domains to the cytoplasmic anchor.
The channel pore
At the center of the Orco tetramer, the S7b segment from each subunit lines the ion-conduction pathway (Fig. 3a). The pore is narrowest near the extracellular end, where it is tapered to 2 Å in diameter by a pair of hydrophobic residues, Leu473 and Val469 (Fig. 3b,c). As this narrowing is too small to allow hydrated ions to pass, the structure of Orco appears to represent a closed state. From this extracellular constriction, the pore opens to a large aqueous vestibule lined largely by polar residues. The anchor domain occludes the vestibule at the intracellular face of the membrane, presenting a barrier to the flow of ions into the cytosol. One possibility is that upon channel opening, the extensive inter-subunit interactions within the anchor domain rupture to generate a continuous ~80 Å pathway along the central four-fold axis of the channel. However, this structural rearrangement is not required for ion permeation: four lateral conduits formed at the interfaces between subunits provide a continuous passageway for ions between the central vestibule and the cytosol (Fig. 3). Each lateral conduit is lined by the structured S7 loop and the S5 and S6 segments from adjacent subunits within the anchor domain to form a 6-Å-long channel that runs approximately parallel to the plane of the membrane. The diameter of this lateral pathway is greater than 5 Å throughout its length, which would allow partially hydrated cations to pass unhindered. Thus, the pore of Orco appears to exhibit a quadrivial architecture with a single extracellular entryway that diverges to four equivalent intracellular outlets.
Fig. 3. The ion-permeation pathway.
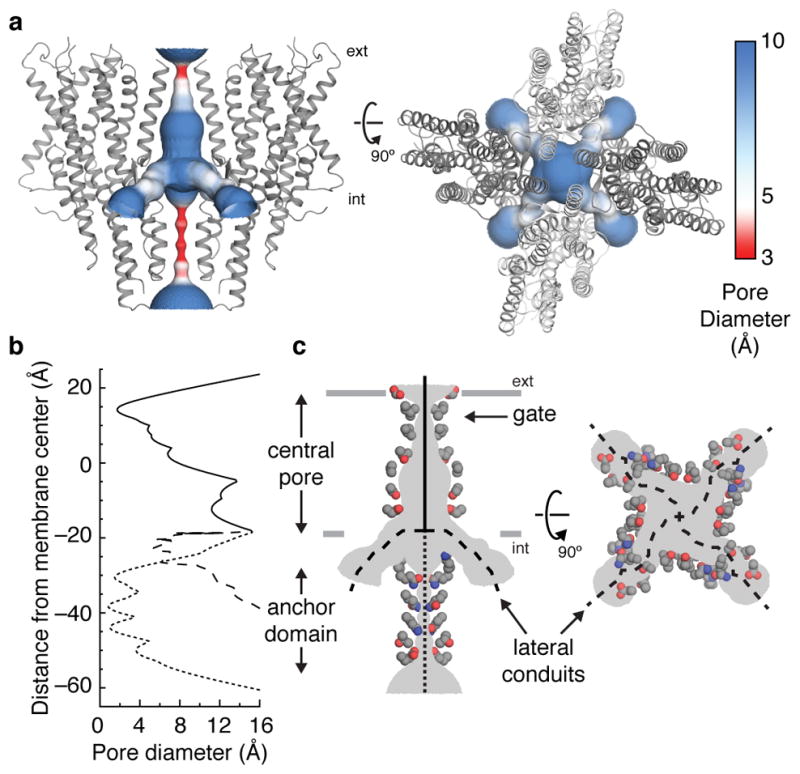
a, The channel pore, coloured according to pore diameter, is shown from the side (left) and top (right). Orco subunits are shown as grey ribbons. b, The diameter along the central pore (solid line), anchor domain (dotted line) and lateral conduits (dashed line) are plotted along the membrane normal. c, Side chains of residues that line the ion-conduction pathway are shown as coloured spheres (grey carbons, blue nitrogens and red oxygens).
The branched nature of Orco’s ion pathway bears resemblance to acid-sensing ion channels (ASIC)30 and ATP-gated P2X channels31. The loosely packed pores of these unrelated channels are also stabilized by soluble domains at the membrane surface that must be circumnavigated to allow ions to pass. ASIC and P2X channels therefore display a similarly complex permeation pathway, in which ions access the central pore through lateral portals formed at the interfaces between subunits.
Ion selectivity and the extracellular gate
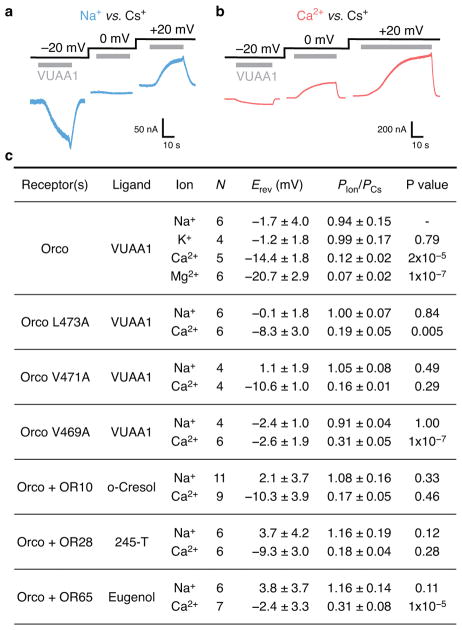
To examine how amino acids lining the Orco pore contribute to ion-conduction properties (Fig. 4a), we heterologously expressed the channel in mammalian cells and assessed the relative permeability of different cations. Under bi-ionic conditions, the measured reversal potential (Erev) of ionic currents through a channel will shift towards that of its most permeant ion. In the presence of intracellular Cs+ and extracellular Na+ or K+, we found the Erev for VUAA1-evoked currents to be near 0 mV, suggesting all three cations pass through Orco equivalently (Fig. 4b, Extended Data Fig. 7). In contrast, in the presence of extracellular Ca2+ or Mg2+, VUAA1-evoked currents reversed at more negative potentials32, indicating that these ions do not pass through the pore as readily as Cs+, likely as a consequence of their larger hydrated radii.
Fig. 4. A hydrophobic gate contributes to ion selectivity.
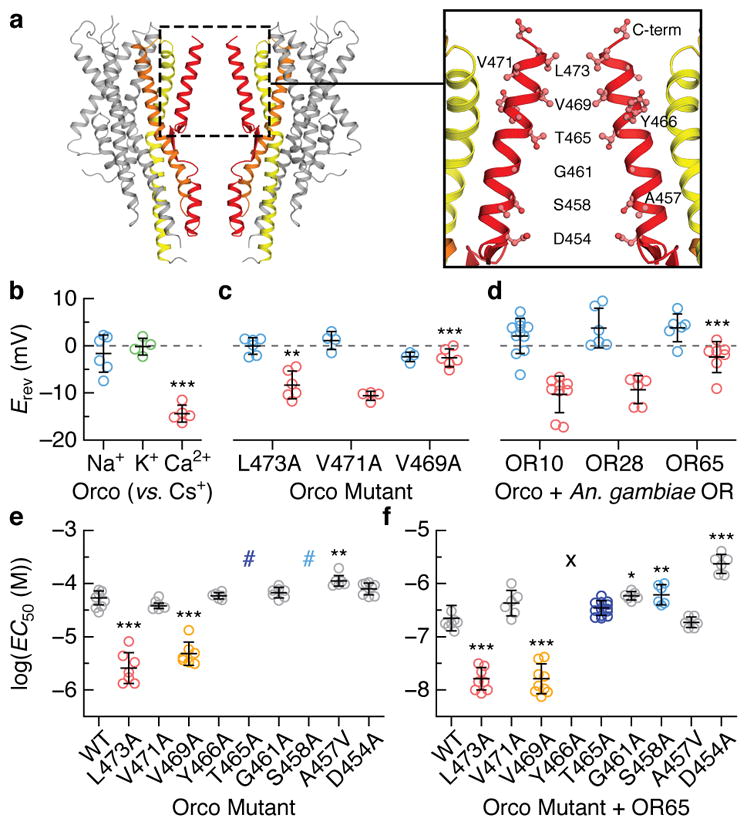
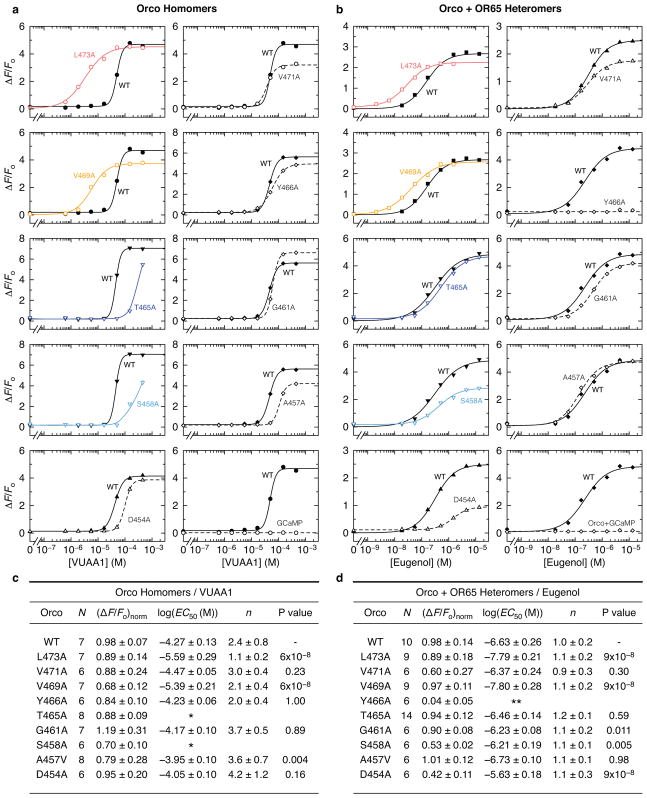
a, Location of residues lining the central pore. b–d, Reversal potentials (Erev; independent replicates and mean ± s.d) for wild-type and mutant Orcos alone or with An. gambiae ORs (intracellular CsCl with extracellular NaCl (blue), KCl (green) or CaCl2 (red)). Asterisks represent Erev that are significantly different from wild-type Orco with the same ions. e,f, Average fitted concentrations for half-maximal response (EC50; independent replicates and mean ± s.d.) for (e) wild-type (WT) and mutant Orco alone or (f) with OR65. Dose-response curves that did not saturate are indicated by #; a lower-bound EC50 estimate is ~3×10−4 M. No binding observed indicated by the X. Asterisks indicate EC50 that are significantly different from wild-type Orco or Orco-OR. In b–f, statistical significance was determined using one-way ANOVA tests followed by Tukey-Kramer multiple comparison tests; P value <0.05 (*), <0.01 (**), <0.001(***). See Extended Data Figs. 7–8 for additional statistical information.
Mutation of either Val469 or Leu473 to alanine resulted in channels that were more permissive toward Ca2+, suggesting that a smaller hydrophobic amino acid at either position generates a larger ion-conduction pathway (Fig. 4c). In contrast, mutation of the nearby Val471, which lies on the opposite face of S7b and points into the lipid membrane, had no affect on ion selectivity. We found that the Ca2+ permeability of Orco-OR complexes depended on the identity of the OR (Fig. 4d), consistent with the notion that both subunits contribute to the ion pathway in the heteromeric receptor32,33. Although Val469 and Leu473 are highly conserved among Orcos, a broader distribution of hydrophobic residues is observed among ORs, which could underlie the distinct permeation properties of different heteromers.
To assess whether Val469 and Leu473 also contribute to gating of the ion pathway, we compared signaling in wild-type and alanine-mutant receptors by co-expressing a genetically encoded Ca2+ indicator, GCaMP6s, in mammalian cells. Mutation of either Val469 or Leu473 enhanced the apparent sensitivity of Orco homomers to VUAA1 10-fold (Fig. 4e; Extended Data Fig. 8). A similar shift was evident when these mutant Orcos were co-expressed with OR65 from the mosquito Anopheles gambiae and activated by odorant (Fig. 4f). Since Val469 and Leu473 line the ion-conduction pathway, it is unlikely these residues directly bind VUAA1, but rather shape the energetics of gating by modulating the relative stability of the open versus closed states.
Immediately below the hydrophobic gate, conserved threonine (Thr465) and serine (Ser458) residues form two rings of hydroxyls that point into the pore, where they may stabilize permeant cations (Fig. 4a). Mutation of either Thr465 or Ser458 to alanine strongly attenuated VUAA1 responses in Orco homomers (Fig. 4e). However, co-expression of OR65 restored the functional sensitivity of these mutants (Fig. 4f), demonstrating that Orco and OR subunits can compensate for each other in the heteromeric receptor. Conversely, mutation of S7b residues that reside at the interface between neighboring subunits (Tyr466 and Asp454), resulted in functional Orco homomers but significantly impaired heteromeric channels. Mutation of this tyrosine in Drosophila Orco has been shown to differentially impact in-vivo odour signaling depending on the identity of the OR33, suggesting that interactions between subunits are not strictly conserved across different Orco-OR assemblies. Mutation of nearby hydrophobic residues that either point toward the lipid (Val471 and Ala457) or the aqueous cavity of the pore (Gly461) did not strongly alter channel gating, either alone or in combination with OR65. Together, these results highlight how Orco and OR subunits work in concert to transduce odorant binding into ion flux within the heteromeric complex.
OR conservation maps to key interaction motifs
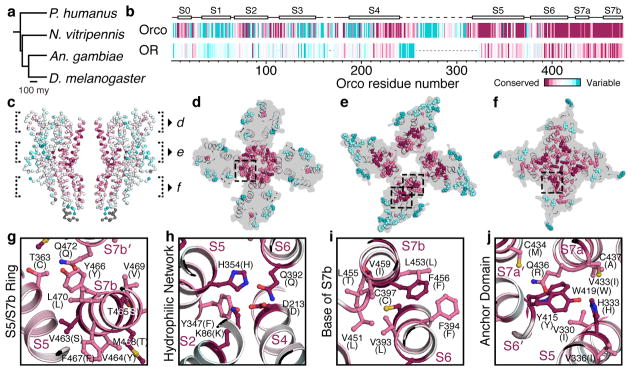
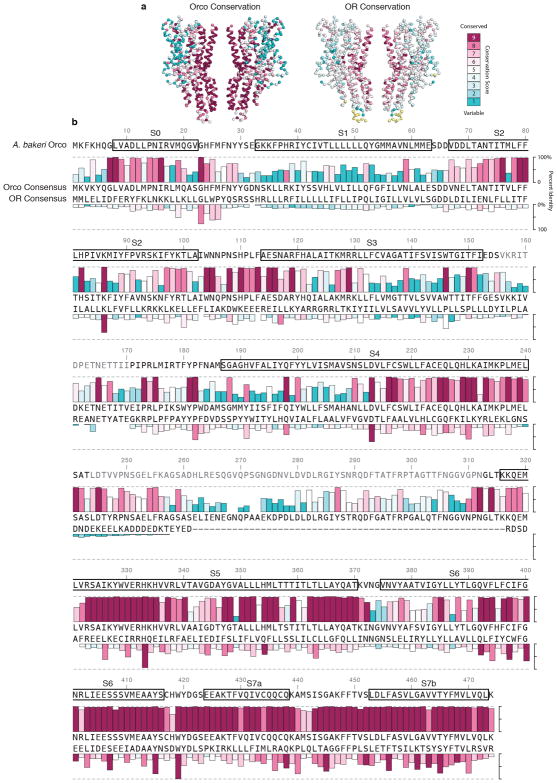
An Orco subunit from one species can interact with ORs from another21, yielding thousands, if not millions, of possible Orco-OR heteromeric channels. How can a single Orco assemble with such a diverse array of ORs? To address this question, we aligned OR sequences from four distantly related insect species separated by nearly 400 million years of evolution and defined whether each residue was evolving more slowly or rapidly than the overall average amino acid substitution rate (Fig. 5a,b; Extended Data Fig. 9)34. While the ORs from these four species exhibit the low sequence identity characteristic of this receptor family (average ~20%), the pattern of relative sequence conservation resembled that in Orco (Fig. 5b).
Fig. 5. OR conservation maps to key interaction domains in Orco.
a, Phylogenetic tree of insects whose OR sequences were used in the alignment. b, Residue conservation among 176 Orcos from 174 species (top) and 371 ORs from the species in a (bottom), calculated using ConSurf34, are plotted using a colour scale (far right) and aligned to A. bakeri Orco. Orco and OR conservation scores are independently normalized. Orco secondary structure is indicated above the plot. c, OR conservation scores from b mapped onto the structure of Orco. d–f, 15 Å cross-sections through c. The most conserved and variable amino acids are shown as spheres. g–j, Selected regions from d–f shown in greater detail. Residues are labeled according to A. bakeri Orco, with the most common OR amino acid at that position indicated in parentheses.
When mapped onto the structure of Orco, residues with higher conservation principally line the pore and cluster within the anchor domain (Fig. 5c–f). In the extracellular leaflet, conserved residues from S7b and S5 form a ring that encompasses most of the inter-subunit contacts stabilizing the central pore (Fig. 5d,g). Near the intracellular surface, conserved residues in S7b form an extensive hydrophobic interface with residues lining S6, securing the base of the S7b pore helix (Fig. 5i). Likewise, the anchor domain contains an intricate network of many conserved residues. Notably, highly conserved tyrosine (Tyr415) and tryptophan (Trp419) residues from the cytosolic end of S6 project into a pocket formed by a conserved histidine (His333) and hydrophobic residues from S5 and S7a of a neighboring subunit (Fig. 5j). Thus, OR sequence conservation preferentially maps to inter-subunit interfaces that could serve to stabilize interactions between Orco and ORs in the heteromer. However, most ORs possess only a subset of these conserved residues suggesting a basis for why ORs are incapable of assembling in the absence of Orco.
As in other membrane proteins, more variable residues tend to be distributed along the protein surface exposed to the lipid membrane34. Indeed, the most rapidly evolving OR residues map to the perimeter of the channel at the extensive protein/lipid interface surrounding each subunit. Residues within the transmembrane regions of S1–S5 are generally poorly conserved, except for a subset that comprises the hydrophilic network and bridges S2, S4, S5, and S6; residues equivalent to Lys86, Asp213, His354 and Gln392 are among the most highly conserved in ORs (Fig. 5h). The conservation of this hydrophilic network suggests it forms an important structural motif that maintains the organization of the transmembrane helices within OR subunits.
Viewed through the lens of the Orco structure, the limited OR sequence conservation can be understood as largely concentrated at key structural and functional nodes in the channel. The striking correspondence between OR sequence conservation and Orco structural elements highlights their evolutionary relatedness and suggests that the Orco homotetramer can serve as a structural template for highly divergent Orco-OR receptor complexes. We thus propose that heteromeric insect olfactory receptors adopt the same overall architecture as the Orco homomeric channel, with one or more (most likely two20) Orco subunits replaced by an OR.
Discussion
Olfactory detection poses a unique challenge. For example, while only a few photoreceptors are necessary to detect the entire visible spectrum, large repertoires of olfactory receptors are required to discriminate among the myriad of molecularly distinct chemicals in the environment3,4. Diverse species, from insects to mammals, have evolved families of tens to thousands of receptors dedicated to the task of odorant detection. Our data provide structural and functional corroboration that insect olfactory receptors form a novel class of heteromeric ligand-gated ion channels, structurally and mechanistically distinct from other chemoreceptors.
Our work supports a model in which Orco and OR subunits assemble into a heterotetramer with a central shared ion-conduction pathway32,33. Each subunit contributes only a single helix, S7b, to a central pore, which is encircled by four loosely tethered S1–S6 transmembrane domains. We propose a simple gating mechanism in which either VUAA1 binding to Orco or odorant binding to the OR dilates the hydrophobic aperture at the extracellular end of S7b to allow passage of cations through the quadrivial pore. This small conformational change presents a relatively low energetic barrier to channel opening, consistent with the weak affinity of VUAA1 and most odorants35. One potential site for odorant binding is the extracellular pocket – a distinctive crevice formed by the loose packing of S1–S6 helices that incorporates amino acids required for VUAA1 sensitivity23. Residues lining this pocket are variable across ORs and have been previously implicated in defining odorant specificity24–27. However, most ORs bind multiple structurally distinct odorants with varying affinities35, a biophysical property integral to the combinatorial coding of odors by the olfactory system. The extensive sequence diversity of ORs may reflect the existence of additional odorant-binding sites distributed throughout the receptor.
A hallmark of insect olfactory receptors is their inordinate diversity within and across insect lineages. Several features of the architecture of Orco provide a framework to understand how this receptor family accommodates such diversity. First, the organization of the homotetrameric Orco channel offers an explanation for how a single Orco can assemble and function with an array of OR subunits. The majority of inter-subunit interactions are localized to the pore and anchor domain, leaving the majority of residues relatively unconstrained and free to diversify. Second, Orco affords flexibility to ORs by contributing highly conserved structural and functional elements to the heteromeric complex. Orco can complement and compensate for OR diversity, thereby relaxing evolutionary constraints on the ORs. Since a single Orco must assemble and function with up to hundreds of distinct OR partners within a given species, the strict conservation of Orco is likely required to preserve functionality across these diverse complexes. Thus, conservation and variability are largely segregated to separate protein subunits in the heteromer, allowing for the modular assembly of an enormous number of receptors with distinct chemical tuning.
Our work offers the first structural insight into likely the largest family of ion channels found in nature, with many hundreds of thousands of different variants distributed across the hundreds of thousands of insect species. The structure of Orco provides an inroad to consider how protein variation can be selected through evolution to perceive the vast chemical world.
Supplementary Methods
Expression and purification of Orco
A synthetic construct consisting of residues Lys2–Lys474 (the native carboxy terminus) of A. bakeri Orco (also known as Or219) was cloned into a pEG BacMam vector36 along with an amino-terminal Strep II tag, superfolder GFP37 and an HRV 3C protease site. Baculovirus containing the Orco-coding sequence was created in Sf9 cells (ATCC CRL-1711) and used to infect HEK293S GnTi− cells (ATCC CRL-3022)36. Cells were293 ibco) supplemented with 2% (v/v) fetal bovine serum (FBS; Gibco) with 8% (v/v) carbon dioxide until they reached a density of ~3×106 cells/mL and then infected at a multiplicity of infection of 1. After 12 h, 10 mM sodium butyrate (Sigma-Aldrich) was added to the medium and the temperature was reduced to 30 °C. The cells were harvested ~48 h later by centrifugation and washed once in phosphate-buffered saline (pH 7.5; Gibco). Cell pellets were frozen in liquid nitrogen and stored at −80 °C until needed.
Cell pellets were thawed on ice and resuspended in 10 mL of lysis buffer per gram of cells. Lysis buffer was composed of 50 mM HEPES/NaOH (pH 7.5), 375 mM NaCl, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 μg/mL pepstatin A, 1 mM phenylmethylsulfonyl fluoride (all from Sigma-Aldrich). Orco was extracted by adding 0.25% (w/v) lauryl maltose neopentyl glycol (LMNG; Anatrace) with 0.05% (w/v) cholesterol hemisuccinate (CHS; Sigma-Aldrich) for 2 h at 4 °C. The mixture was clarified by centrifugation at 90,000 g and the supernatant was added to 0.2 mL StrepTactin Sepharose resin (GE Healthcare) per gram of cells and rotated at 4 °C for 1 h. The resin was collected, washed with 10 column volumes (cv) of 20 mM HEPES/NaOH (pH 7.5), 150 mM NaCl (HBS) with 0.01% (w/v) LMNG, 0.002% (w/v) CHS, and then with 10 cv of HBS with 0.05% (w/v) digitonin (Sigma-Aldrich). Orco was eluted by adding 2.5 mM desthiobiotin (DTB; Sigma-Aldrich) to the digitonin buffer. The Strep-GFP tag was cleaved by HRV 3C protease (Novagen), added at 10 U/mg of Orco, overnight at 4 °C. DTB was removed by using a PD-10 column (GE Healthcare) and the desalted sample was reapplied to the StrepTactin Sepharose resin (regenerated and re-equilibrated following the manufacturers instructions) to remove uncut material. Fab was added to Orco at a 1.5-fold molar excess and incubated on ice for 30 min. Orco and Fab concentrations were calculated from their absorbances at 280 nm assuming extinction coefficients (ε280) of 57.3 and 73.1 mM−1 cm−1, respectively (calculated by ProtParam38). The complex was concentrated to ~5 mg/mL (Amicon Ultra; 50 kDa cutoff) and injected onto a Superose 6 Increase column (GE Healthcare) equilibrated in HBS with 0.05% (w/v) digitonin.
Purification of Fab
Monoclonal antibodies were produced by culturing hybridoma line 9G11/C7 in hybridoma serum-free medium (Gibco) supplemented with 1% (v/v) ultra-low IgG FBS (Gibco) and 1% (v/v) Nutridoma-SP (Roche) using CELLine disposable bioreactors (Argos Technologies). Medium supernatant was dialyzed against 10 mM Tris/HCl (pH 7.5), 10 mM NaCl (Spectra/Por 6; 10 kDa cutoff) overnight at 4 °C. Precipitates were removed by centrifugation and the supernatant was passed through a 0.2 μm filter and onto a 5 mL HiTrap Q HP column (GE Heathcare) equilibrated in 10 mM Tris/HCl (pH 7.5), 10 mM NaCl. The antibody was eluted from the column using a linear gradient to 10 mM Tris/HCl (pH 7.5), 0.3 M NaCl. Fractions containing the antibody were pooled and digested for 3 h at 37 °C by papain (Worthington) at a 1:50 (w:w) papain:antibody ratio with L-cysteine and ethylenediaminetetraacetic acid (EDTA) added at 10 mM each (Sigma-Aldrich). The digestion was terminated by the addition of 10 mM iodoacetamide (Sigma-Aldrich) for 20 min. The mixture was dialyzed twice against 10 mM Tris/HCl (pH 7.5), 50 mM NaCl overnight at 4 °C and applied to a HiTrap Q HP equilibrated in the same buffer. Fab was collected from the flow-through and concentrated to ~10 mg/mL (Amicon Ultra; 10 kDa cutoff).
Cryo-EM sample preparation and data acquisition
Peak fractions containing the Orco-Fab complex were concentrated to 4–5 mg/mL (assuming ε280 = 130 mM−1 cm−1). Cryo-EM grids were frozen using a Vitrobot Mark IV (FEI) as follows: 3 μL of the concentrated sample was applied to a glow-discharged Quantifoil R1.2/1.3 holey carbon 400 mesh gold grid, blotted for 3–4 s in >90% humidity at room temperature, and plunge frozen in liquid ethane cooled by liquid nitrogen.
Cryo-EM data were recorded on a Titan Krios (FEI) operated at 300 kV, equipped with a Gatan K2 Summit camera. SerialEM39 was used for automated data collection. Movies were collected at a nominal magnification of 29,000× in super-resolution mode resulting in a calibrated pixel size of 0.5 Å/pixel, with a defocus range of approximately −0.8 to −2.0 μm. Fifty frames were recorded over 15 s of exposure at a dose rate of 1.6 electrons per Å2 per frame.
Movie frames were aligned and binned over 2×2 pixels using MotionCor240 and the contrast transfer function parameters for each motion-corrected image were estimated using CTFFIND441. Particles were auto-picked using Gautomatch42 using templates derived from averages that were generated from an initial data set of 5,000 particles from 116 images. A total of 82,614 particles from 1,221 images were extracted into 384×384-pixel boxes, binned over 3×3 pixels, and subjected to 2D classification using RELION-2.043. After removal of junk particles, the remaining 53,141 particles were re-extracted without binning and subjected to 3D refinement in RELION with C4 symmetry imposed, using, as initial reference, a 3D map that was calculated in EMAN244 from a subset of 2D-class averages. The four-fold symmetry was immediately evident from the individual particles and 2D-class averages, and refinement imposing no symmetry produced an equivalent map. Subsequent 3D classification into four classes showed a single dominant class containing 47,934 particles (90% of the dataset), suggesting that the Orco-Fab complex was highly homogeneous with limited structural variability. The orientation parameters of the particles from all four classes were further refined using Frealign45 employing a soft mask that excluded the Fab and micelle. The map was sharpened using a B-factor of −160 Å2 yielding a final resolution of 3.5 Å, estimated using the Fourier shell correlation (FSC) = 0.143 cutoff criterion46. The images in Fig. 1d were created using Chimera47.
Model building
The 3.5-Å density map was of sufficient quality for de novo atomic model building. A poly-alanine model for Orco was built in Coot48 and subsequent amino-acid assignments were made based on side-chain densities. Since the entire Fab was masked out during particle alignment, density for the Fab was much weaker and therefore was not modeled. The Orco tetramer was refined using real-space refinement implemented in PHENIX49 with four-fold non-crystallographic symmetry applied. The structure was compared to existing structures in the Protein Data Bank using the Dali server50 and no significant hits were obtained suggesting that Orco adopts a novel fold. All images of the model were created using PyMOL51.
Structure Analyses
Residues at subunit interfaces were identified using PyMOL as any residue within 5 Å of a neighbouring subunit (Fig. 2d–g). The inter-subunit surface area was calculated as the solvent-accessible surface area (SA) that would be occluded upon tetramer formation (SA of a single Orco subunit minus one-fourth the SA of the Orco tetramer).
The pore diameters along the central axis and lateral conduits were calculated using the program HOLE52 (Fig. 3a). Two calculations were performed: one along the central four-fold axis (central pore) and another between subunits near the cytosolic membrane interface (lateral conduits). Both pores overlapped in the central vestibule.
Isothermal titration calorimetry (ITC)
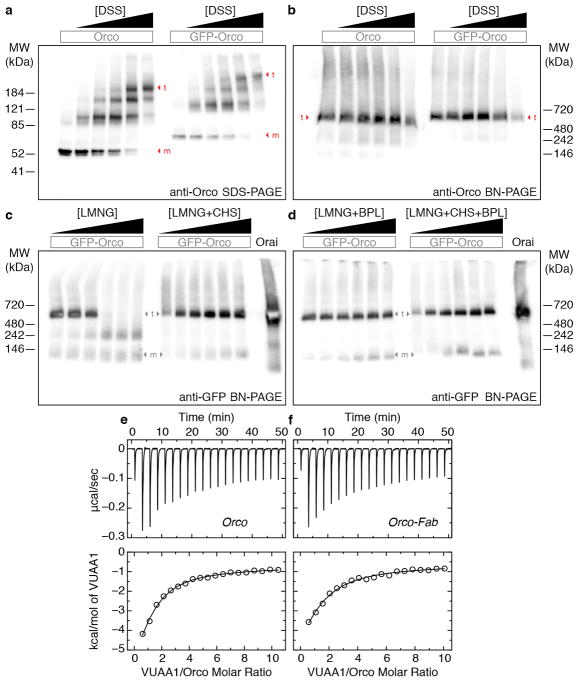
Samples of Orco and Orco-Fab complex were expressed and purified as described above, and concentrated to ~10 μM (monomer). A VUAA1 (Princeton Biomedical Research) stock was prepared in dimethylsulfoxide (DMSO; Sigma-Aldrich) at 100 mM and diluted to 0.5 mM in the same buffer as Orco. 0.5% (v/v) DMSO was added to Orco and buffer samples to match the amount of DMSO originating from the VUAA1 stock solution. ITC experiments were performed using a MicroCal Auto-iTC200 (Malvern) at 25 °C. Each experiment began with a single injection of 0.4 μL followed by 19 injections of 2 μl each (at 0.5 μL/s, 150 s apart) into a 0.2-mL Orco sample. The experiments were repeated using Orco samples obtained from independent purifications (biological replicates).
The raw heat evolutions were baseline-corrected and a single binding site model was fit to the integrated data using AFFINImeter (https://www.affinimeter.com), excluding the first injection. The number of binding sites per monomer was fixed to be 1 and a dissociation constant (Kd), enthalpy of binding (ΔH) and heat of sample dilution (ΔQdil) were fit. Separate experiments injecting VUAA1 into buffer alone showed no significant heats of dilution and were not subtracted from the Orco data prior to fitting.
Electrophysiological experiments
Experiments on wild-type Orco alone were conducted with a Flp-In T-Rex 293 cell line (Invitrogen) with a stably integrated GFP-tagged Orco cloned into pcDNA6/TR. Constructs used for mutant Orco or Orco-OR experiments were cloned into pEG BacMam vectors as described above, except that OR proteins were tagged with mCherry. HEK293 cells were maintained in high-glucose DMEM supplemented with 10% (v/v) FBS (Gibco), 1% (v/v) MEM (Sigma-Aldrich) and 1% (v/v) GlutaMAX (Gibco) plus the appropriate antibiotics. Cells were plated on 12 mm poly-D-lysine-coated coverslips (Corning) 24–72 h before recording, and transfected or infected with the appropriate constructs 24 h before recording. Electrodes were drawn from borosilicate patch glass (Sutter Instruments) and polished (MF-83, Narishige Co.) to a resistance of 3–6 MΩ when filled with pipette solution. Analog signals were filtered at 2 kHz using the built-in 4-pole Bessel filter of a Multiclamp 700B patch-clamp amplifier (Molecular Devices) in patch mode and digitized at 20 kHz (Digidata 1440A, Molecular Devices). Signals were further filtered offline at 1 kHz for analysis and representations.
Whole-cell and single-channel recordings in Fig. 1 and Extended Data Fig. 2 were performed using an extracellular (bath) solution composed of 135 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM glucose, 10 mM HEPES-Na/HCl (pH 7.3, 310 mOsm/kg) and an intracellular (pipet) solution composed of 150 mM KCl, 10 mM NaCl, 1 mM EDTA-Na, 10 mM HEPES-Na/HCl (pH 7.45, 310 mOsm/kg). Single-channel recordings were done in excised outside-out mode. VUAA1 (100 mM in DMSO) was diluted to the final concentrations using the extracellular solution. Solutions were locally perfused using a microperfusion system (ALA Scientific Instruments).
For the ion-selectivity studies in Fig. 4, the intracellular (pipet) solution consisted of 150 mM CsCl, 10 mM HEPES-Cs (pH 7.3). The extracellular solutions for monovalent cations (X = Na, K) consisted of 150 mM XCl, 10 mM HEPES-X/HCl (pH 7.3). The extracellular solutions for divalent cations (X = Ca, Mg) consisted of 100 mM XCl2 10 mM HEPES-Cs/HCl (pH 7.3). VUAA1 and odorants were diluted to the appropriate concentration in extracellular solution. Wild-type and mutant Orco alone used 0.1 mM VUAA1, while Orco with An. gambiae OR10, OR28 and OR65 used 0.3 mM o-cresol, 0.3 mM 2,4,5-trimethylthiazole, and 10 μM eugenol, respectively (odors were from Sigma-Aldrich). Solutions were perfused locally in a bath composed of 150 mM NaCl, 10 mM HEPES-Na/HCl (pH 7.3). Motility differences between the cesium-filled pipette and the sodium-filled bath were measured to be 3 mV and the measured reversal potentials (Erev) were corrected for this liquid junction potential. The ion-permeability ratios (PX/PCs) in Extended Data Fig. 7 were calculated from Erev using the simplified Goldman-Hodgkin-Katz equations:
Cell-based calcium sensor fluorescence assay
Constructs used in this assay were cloned into a modified pME18s vector. HEK293 cells were maintained in FluoroBrite DMEM (Gibco) supplemented with 10% (v/v) FBS, 1% MEM, and 1% (v/v) GlutaMAX. For each transfection, 0.5 μg of GCaMP6s (Addgene #4075353) and 1.5 μg of the appropriate construct(s) were diluted in 250 μL Opti-MEM, 1% GlutaMAX (Gibco), mixed with an equal volume of medium containing 7 μL lipofectamine 2000 (Invitrogen), and incubated for 20 min at room temperature. HEK293 cells were detached with trypsin and resuspended at 1×106 cells/mL, mixed with the transfection solution and 42 μL added to 2×16 wells in a 384-well plate (Greiner CELLSTAR). After 4 h, the transfection medium was replaced with fresh medium, and after another 24 h, the medium was replaced with 20 μL of reading buffer composed of 1x HBSS (Gibco) supplemented with 20 mM HEPES/NaOH (pH 7.4), 5 mM CaCl2, 1 mM MgSO4, and 3 mM Na2CO3. The fluorescence emission at 527 nm (excited at 480 nm) was continuously recorded by a Hamamatsu FDSS plate reader set to 37 °C. After 34 s, 20 μL of VUAA1 or odorant solution, diluted in reading buffer, was added and recording was continued for 2 min. All solutions were pre-warmed to 37 °C prior to use.
Seven VUAA1 or odour concentrations were used (plus one without ligand); each repeated four times on the plate covering 2×16 wells per construct and together considered a single replicate. The baseline fluorescence (Fo) was the average fluorescence of the 30 s prior to VUAA1 or odour delivery. ΔF was the fluorescence difference from baseline. The Hill equation was fitted to ΔF/Fo values using GraphPad Prism. In Extended Data Fig. 8 (ΔF/Fo)norm is the fitted maximum ΔF/Fo value relative to a separate wild-type Orco (or Orco-OR) control experiment from the same plate. In Extended Data Fig. 1 and Extended Data Fig. 8, averages of the four replicates at each ligand concentration are plotted.
Sequence alignment of Orco and OR proteins
For the sequence alignment of Orco proteins, 176 Orco sequences from 174 different organisms were aligned using Clustal Omega54,55 with minimal manual adjustment. For the sequence alignment of OR proteins, 361 sequences were included from four insect species: Anopheles gambiae (72/79 ORs56) Drosophila melanogaster (61/62 ORs57), Nasonia vitripennis (221/301 ORs58) and Pediculus humanus (7/10 ORs59). Not all OR sequences from each insect were used as those with large insertions or deletions, or with regions of unknown residues were removed. The alignment was initially generated using MAFFT60,61 with subsequent manual adjustment, primarily to reduce the number of gaps, using the Orco alignment as a guide. The sequence alignments are included as Supplementary Data. Sequence alignments were analyzed independently using ConSurf34,62.
Data Availability
The 3D cryo-EM density map of the Orco-Fab complex has been deposited in the Electron Microscopy Data Bank under accession number EMD-7352. The coordinates of the atomic model of Orco have been deposited in the Protein Data Bank under accession number 6C70.
Extended Data
Extended Data Fig. 1. Ligand-gated signaling of A. bakeri and An. gambiae Orcos.
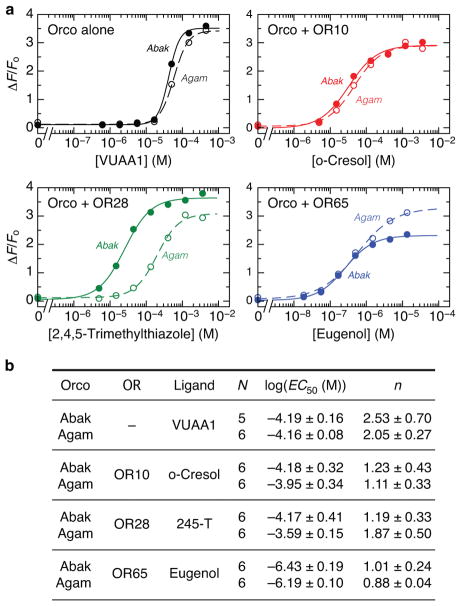
a, Fluorescence changes in HEK293 cells transfected with a genetically encoded Ca2+ indicator, GCaMP6s, and Orco from A. bakeri (Abak; closed circles) or An. gambiae (Agam; open circles) alone, or with an An. gambiae OR. Dose-response curves were obtained by titrating with VUAA1 or the cognate odour of the OR. b, Average fitted Hill equation parameters from N number of independent replicates. EC50 is the concentration for half-maximal response and n is the Hill coefficient (both mean ± s.d.). 245-T is 2,4,5-trimethylthiazole.
Extended Data Fig. 2. Electrophysiological characterization of Orco.
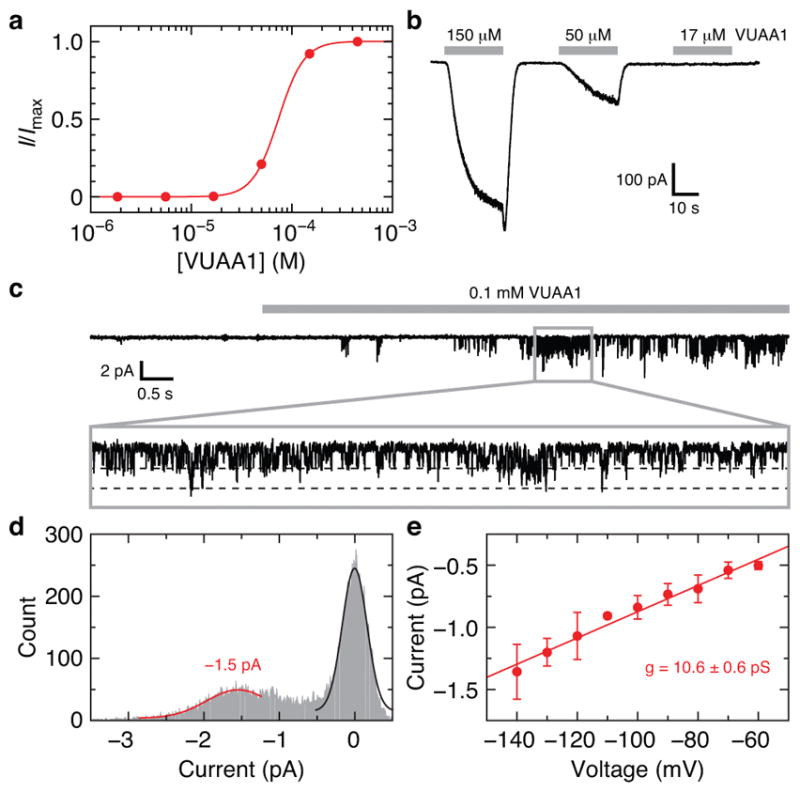
a, Example VUAA1 dose-response curve using whole-cell current from a HEK293 cell expressing Orco. I/Imax is the measured current relative to the maximum current obtained at the highest concentration of VUAA1. The titration experiment was repeated using four independent cells with equivalent results. The average fitted Hill equation parameters are (mean ± s.d.): EC50 = 64 ± 7 μM, n = 3.5 ± 0.5. b, Inward whole-cell current for a subset of the VUAA1 concentrations used in a (held at −80 mV). c, VUAA1-evoked Orco currents recorded from an outside-out membrane patch from a HEK293 cell expressing Orco (held at −140 mV). Inset (0.9 s) highlights single channel openings. d, Amplitude histogram determined from the inset in c. Fitted Gaussian distributions for single-channel Orco current (red) and baseline current (black) are shown. e, Amplitude histograms were obtained at multiple voltages and the single-channel conductance of 10.6 ± 0.6 pS was determined from the slope of the current-voltage plot (mean ± s.d. for 2–6 data points per voltage from 9 patches). The specific numbers of replicates at each voltage were: 5 (−140 mV), 4 (−130 mV), 5 (−120 mV), 3 (−110 mV), 6 (−100 mV), 4 (−90 mV), 4 (−80 mV), 3 (−70 mV), 2 (−60 mV).
Extended Data Fig. 3. Stoichiometry and ligand binding of the Orco homotetramer.
a, Western blot of cross-linked Orco in transfected HEK293 cells. SDS-PAGE showing a ladder of four bands that appears with both GFP-tagged and untagged Orco after treatment with increasing concentration of the amine cross-linker disuccinimidyl suberate (DSS). Monomer (m) and tetramer (t) bands are indicated. DSS concentrations were (μM): 0, 25, 75, 125, 250, 2500. b, Western blot of a Blue Native (BN)-PAGE63 gel of the same samples as in a showing that tetrameric Orco is present in all samples and higher-order aggregates are not induced by cross-linking. c,d Western blot of Orco extracted with increasing concentrations of detergent (LMNG) showed gradual loss of the tetrameric species. Addition of (c) CHS, (d) porcine brain polar lipid extract (BPL; Anatrace) or the combination of the two stabilized the Orco tetramer. LMNG concentrations were (% w/v): 0.01, 0.05, 0.1, 0.25, 0.5, 1. Concentration of CHS, BPL and the sum of CHS + BPL were added at 1/5 that of LMNG. GFP-labeled Orai (55 kDa)64 was used as a molecular weight marker as it is a hexamer with a similar total size as the Orco-GFP tetramer (340 kDa). The larger apparent molecular weights observed in BN-PAGE gels (b–d) reflect the additional mass of the micelle. Primary antibodies used were: anti-Orco clone 20F7 and anti-GFP (Life Technologies). Each experiment in this figure was repeated three times with similar results. The molecular weight markers on the native gels are approximate: they are from a separate gel run under the same conditions (see Supplementary Data). e,f, Representative baseline-corrected isothermal titration calorimetry (ITC) data for (e) Orco (11 μM) and (f) Orco-Fab complex (10 μM) titrated with VUAA1. Integrated heats and fitted single-site binding isotherms are shown at the bottom. The number of binding sites per monomer was fixed at 1 and the dissociation constant (Kd), enthalpy of binding (ΔH), and heat associated with sample dilution (ΔQdil) were fit. The experiments were repeated three times each using Orco samples obtained from independent purifications. The average fitted thermodynamic parameters are as follows (mean ± s.d.). Orco: Kd = 13 ± 1 μM, ΔH = −8.3 ± 0.6 kcal/mol, ΔQdil = −1.0 ± 0.1 kcal/mol. Orco-Fab: Kd = 18 ± 2 μM, ΔH = −9.7 ± 0.4 kcal/mol, ΔQdil = −1.0 ± 0.2 kcal/mol.
Extended Data Fig. 4. Cryo-EM data analysis.
a, A representative motion-corrected micrograph showing the distribution of Orco-Fab single particles (three particles are circled). Scale bar is 50 nm. b, 2D averages of classes selected for further processing. c, Initial density map from 3D refinement in RELION using all of the particles in b with C4 symmetry imposed. d, Soft mask and final density map after refinement in Frealign. e, Fourier shell correlation (FSC) curves for the final cryo-EM density maps. The horizontal dashed line represents the 0.143 cutoff value. f, Orientation distribution of the particles included in the final 3D map of the Orco-Fab complex in d (as reported by RELION). g, Approximate local resolution of the entire Orco-Fab density map (left) and only Orco (right). h, Cryo-EM densities for the modeled regions are shown as grey mesh. Orco models are drawn as sticks with carbon atoms coloured according to Fig. 2, and oxygen, nitrogen and sulfur atoms coloured red, blue and yellow, respectively.
Extended Data Fig. 5. Potential extracellular facing odour-binding pocket.
a, Side view of Orco highlighting the location of residues in ORs that, when mutated, alter ligand binding specificity. Orco residues that are equivalent to point mutations that alter odour specificity in Helicoverpa assaulta OR14b24, An. gambiae OR1525, D. melanogaster OR85b26 or Ostrinia furnacalis OR327 are shown as red spheres. Residues required for VUAA1 sensitivity in Orco23 are shown as blue spheres. In Orco, the S3–S4 extracellular loop is positioned above the pocket. b, A 15-Å cross-section through the pocket from a.
Extended Data Fig. 6. Inter-subunit interactions in Orco and Kv channels.
a, Top views of Orco (left) and Kv10.1 (Eag1)65 (right) highlighting the overall organization of these tetrameric cation channels. In this Kv channel, the pore and voltage-domains are not domain-swapped and so it more closely resembles the quaternary structure of Orco compared to other Kv channels. b, Top and c, side views showing inter-subunit interactions. Residues within 5 Å of a different subunit within the transmembrane region are shown as coloured spheres (16 residues in Orco, 58 residues in Kv10.1). In Orco, residues at subunit interfaces in the anchor domain are grey spheres (35 residues). The extracellular and intracellular domains of Kv10.1 are not shown.
Extended Data Fig. 7. Reversal potentials and ion-permeability ratios.
a,b, VUAA1-evoked whole-cell current from HEK293 cells expressing Orco with 150 mM intracellular CsCl and 150 mM extracellular NaCl (a, blue), or 100 mM extracellular CaCl2 (b, red). c, Summary of reversal potentials (Erev) and permeability ratios (Pion/PCs) for wild-type and mutant Orco and Orco-OR complexes measured under bi-ionic conditions (mean ± s.d). Erev were measured using N independent cells and corrected for the measured junction potential. Pion/PCs were calculated from Erev using Goldman-Hodgkin-Katz equations (see Methods) with their errors determined by propagation of the standard deviations. Three one-way ANOVA tests were performed using these Erev data followed by Tukey-Kramer multiple comparison tests. (1) Orco selectivity of Na+, K+, Ca2+ and Mg2+ (P values are from comparisons to Na+ Erev). (2) Wild-type and mutant Orco Na+ and Ca2+ selectivity (P values are from comparisons to wild-type Orco with the same ion, Na+ or Ca2+, as appropriate). (3) Orco homomer and Orco+OR heteromer Na+ and Ca2+ selectivity (P values are from comparisons to the Orco homomer with the same ion, Na+ or Ca2+, as appropriate). 245-T is 2,4,5-trimethylthiazole.
Extended Data Fig. 8. Cell-based GCaMP assay parameters.
a,b, Relative fluorescence changes in HEK293 cells transfected with GCaMP plus wild-type (WT) or mutant Orco alone (a; titrated with VUAA1) or in the presence of An. gambiae OR65 (b; titrated with eugenol). Plots using the same symbols were collected on the same day. c,d, Fitted Hill equation parameters for Orco and Orco-OR65 (mean ± s.d). The assays were repeated N number of independent times to obtain the maximum ΔF/Fo response, concentration for half-maximal response (EC50) and Hill coefficient (n). (ΔF/Fo)norm is the fitted maximum ΔF/Fo response relative to a wild-type (WT) control Orco or Orco-OR65 experiment from the same plate (for mutations that could not be accurately fit, the measured maximum ΔF/Fo response was used). Two one-way ANOVA tests were performed using these EC50 data followed by Tukey-Kramer multiple comparison tests. (1) Wild-type and mutant Orco homomers (P values are from comparisons to wild-type Orco) (2) Wild-type and mutant Orco co-transfected with OR65 (P values are from comparisons to wild-type Orco with OR65). *Fitted parameters for Orco T465A or S458A homomers + VUAA1 could not be obtained as the dose-response curves did not saturate: a lower-bound estimate for their EC50 are ~3×10−4 M. **No binding was observed for Orco Y466A in the presence of OR65.
Extended Data Fig. 9. Conservation of Orco and OR sequences.
a, Orco (left) and OR (right) conservation scores mapped onto the structure of Orco. b, Consensus sequences from Orco and OR amino-acid alignments (see Supplementary Data) are aligned to A. bakeri Orco. In total, 176 Orco sequences and 361 OR sequences were used in the alignments, respectively. The percent identities (bar height) and ConSurf62 conservation scores (bar colour) are plotted for each consensus sequence. Only residues that align to A. bakeri Orco are included.
Extended Data Table 1.
Cryo-EM data collection, refinement and model validation statistics.
| Orco-Fab | |
|---|---|
| Data deposition | |
| PDB | 6C70 |
| EMDB | 7352 |
|
| |
| Cryo-EM Data Collection | |
| Voltage (kV) | 300 |
| Magnification (×) | 29,000 |
| Pixel size (Å) | 1.0 |
| Electron exposure (e−/Å2/frame) | 1.6 |
| Defocus range (μm) | −0.8 to −2.0 |
| Number of image stacks | 1,221 |
| Number of frames per stack | 50 |
|
| |
| Cryo-EM Data Processing | |
| Initial number of particles | 82,614 |
| Final number of particles | 53,141 |
| Symmetry imposed | C4 |
| Map sharpening B factor (Å2) | −160 |
| Map resolution (Å) | 3.5 |
| Map resolution range (Å) | 3–8 |
| FSC threshold | 0.143 |
|
| |
| Model Refinement | |
| Number of Orco amino acids | 388 |
| Number of Fab amino acids | 0 |
| Total non-hydrogen atoms | 3092 |
| Average B factor (Å2) | 104 |
| Bond length r.m.s.d. (Å) | 0.01 |
| Bond angle r.m.s.d. (°) | 1.07 |
| Ramachandran Plot: | |
| Favoured (%) | 97.4 |
| Allowed (%) | 2.6 |
| Outliers (%) | 0 |
| Rotamer outliers (%) | 0 |
| MolProbity score | 1.7 |
| MolProbity clash score | 7.6 |
Supplementary Material
Supplementary Data 1 | Full gel images from Extended Data Fig. 3.
Supplementary Data 2 | Sequence alignment of Orco proteins.
Supplementary Data 3 | Sequence alignment of OR proteins.
Acknowledgments
We thank R. Axel, R. MacKinnon, B. Noro and L. Vosshall for advice on the manuscript. A. Siliciano and C. Monnier for technical advice and preliminary biochemistry, other members of the Ruta lab for helpful discussions, and R. Boggavarapu for help with initial negative-stain EM experiments. We are grateful to M. Ebrahim and J. Sotiris at The Rockefeller University Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center for assistance with microscope operation, F. Weis-Garcia and S. L. Bourne at the MSKCC Antibody & Bioresource Core Facility for hybridoma generation, and R. Lavoisier and C. Adura at The Rockefeller University High Throughput & Spectroscopy Resource Center for support on functional assays. This work was supported by a Helmsley Postdoctoral Fellowship (to K.H.K.) and a Sinsheimer Foundation Award, the National Institutes of Health (R01-AI103171-01A1) and New York Stem Cell Foundation Robertson Neuroscience Investigator Award (to V.R.).
Footnotes
Author Contributions
J.A.B. and M.A.K. expressed and purified Orco, screened monoclonal antibody lines and purified Fab fragments. J.A.B. and K.H.K. collected and analyzed cryo-EM data with input from T.W. J.A.B. built and refined the Orco model and performed ITC measurements. J.M. performed electrophysiology experiments. J.A.R. performed calcium imaging assays. V.R. contributed to all aspects of the project and wrote the paper with J.A.B and input from all authors.
The authors declare no competing interests.
Extended data is available for this paper at XXX.
Supplementary information is available for this paper at XXX.
References
- 1.Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. How many species are there on Earth and in the ocean? PLoS Biol. 2011;9:e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansson BS, Stensmyr MC. Evolution of insect olfaction. Neuron. 2011;72:698–711. doi: 10.1016/j.neuron.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Bargmann CI. Comparative chemosensation from receptors to ecology. Nature. 2006;444:295–301. doi: 10.1038/nature05402. [DOI] [PubMed] [Google Scholar]
- 4.Kaupp UB. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat Rev Neurosci. 2010;11:188–200. doi: 10.1038/nrn2789. [DOI] [PubMed] [Google Scholar]
- 5.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 6.Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 7.Sato K, et al. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 8.Wicher D, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- 9.Missbach C, et al. Evolution of insect olfactory receptors. Elife. 2014;3:e02115. doi: 10.7554/eLife.02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ioannidis P, et al. Genomic features of the damselfly Calopteryx splendens representing a sister clade to most insect orders. Genome Biol Evol. 2017;9:415–430. doi: 10.1093/gbe/evx006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenzie SK, Fetter-Pruneda I, Ruta V, Kronauer DJC. Transcriptomics and neuroanatomy of the clonal raider ant implicate an expanded clade of odorant receptors in chemical communication. Proc Natl Acad Sci USA. 2016;113:14091–14096. doi: 10.1073/pnas.1610800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vosshall LB. Olfaction in Drosophila. Curr Opin Neurobiol. 2000;10:498–503. doi: 10.1016/s0959-4388(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 13.Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102:147–159. doi: 10.1016/s0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- 14.Larsson MC, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Yan H, et al. An engineered orco mutation produces aberrant social behavior and defective neural development in ants. Cell. 2017;170:736–747.e9. doi: 10.1016/j.cell.2017.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trible W, et al. orco Mutagenesis causes loss of antennal lobe glomeruli and impaired social behavior in ants. Cell. 2017;170:727–735.e10. doi: 10.1016/j.cell.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeGennaro M, et al. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 2013;498:487–491. doi: 10.1038/nature12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PL, Pask GM, Rinker DC, Zwiebel LJ. Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci USA. 2011;108:8821–8825. doi: 10.1073/pnas.1102425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu B, et al. Expression and evolutionary divergence of the non-conventional olfactory receptor in four species of fig wasp associated with one species of fig. BMC Evol Biol. 2009;9:43. doi: 10.1186/1471-2148-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones WD, Nguyen TAT, Kloss B, Lee KJ, Vosshall LB. Functional conservation of an insect odorant receptor gene across 250 million years of evolution. Curr Biol. 2005;15:R119–21. doi: 10.1016/j.cub.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Hopf TA, et al. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat Commun. 2015;6:6077. doi: 10.1038/ncomms7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corcoran JA, Sonntag Y, Andersson MN, Johanson U, Löfstedt C. Endogenous insensitivity to the Orco agonist VUAA1 reveals novel olfactory receptor complex properties in the specialist fly Mayetiola destructor. Sci Rep. 2018;8:3489. doi: 10.1038/s41598-018-21631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang K, Huang LQ, Ning C, Wang CZ. Two single-point mutations shift the ligand selectivity of a pheromone receptor between two closely related moth species. Elife. 2017;6:155. doi: 10.7554/eLife.29100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes DT, Wang G, Zwiebel LJ, Luetje CW. A determinant of odorant specificity is located at the extracellular loop 2-transmembrane domain 4 interface of an Anopheles gambiae odorant receptor subunit. Chem Senses. 2014;39:761–769. doi: 10.1093/chemse/bju048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols AS, Luetje CW. Transmembrane segment 3 of Drosophila melanogaster odorant receptor subunit 85b contributes to ligand-receptor interactions. J Biol Chem. 2010;285:11854–11862. doi: 10.1074/jbc.M109.058321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leary GP, et al. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc Natl Acad Sci USA. 2012;109:14081–14086. doi: 10.1073/pnas.1204661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long SB, Tao X, Campbell EB, Mackinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 29.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na+-selective channel. Cell. 2014;156:717–729. doi: 10.1016/j.cell.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pask GM, Jones PL, Rützler M, Rinker DC, Zwiebel LJ. Heteromeric Anopheline odorant receptors exhibit distinct channel properties. PLoS ONE. 2011;6:e28774. doi: 10.1371/journal.pone.0028774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakagawa T, Pellegrino M, Sato K, Vosshall LB, Touhara K. Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLoS ONE. 2012;7:e32372. doi: 10.1371/journal.pone.0032372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 36.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 38.Gasteiger E, et al. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [DOI] [Google Scholar]
- 39.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K. Gautomatch. www.mrc-lmb.cam.ac.uk/kzhang/
- 43.Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in RELION. Acta Crystallogr D Struct Biol. 2017;73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Grigorieff N. Frealign: an exploratory tool for single-particle cryo-EM. Meth Enzymol. 2016;579:191–226. doi: 10.1016/bs.mie.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–5. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The PyMOL Molecular Graphics System, Version 2.0. Schrödinger, LLC; [Google Scholar]
- 52.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 53.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goujon M, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hill CA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 57.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 58.Robertson HM, Gadau J, Wanner KW. The insect chemoreceptor superfamily of the parasitoid jewel wasp Nasonia vitripennis. Insect Mol Biol. 2010;19(Suppl 1):121–136. doi: 10.1111/j.1365-2583.2009.00979.x. [DOI] [PubMed] [Google Scholar]
- 59.Pelletier J, Xu P, Yoon KS, Clark JM, Leal WS. Odorant receptor-based discovery of natural repellents of human lice. Insect Biochem Mol Biol. 2015;66:103–109. doi: 10.1016/j.ibmb.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katoh K, Misawa K, Kuma KI, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinformatics. 2017;30:3059. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashkenazy H, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–50. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 64.Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whicher JR, Mackinnon R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science. 2016;353:664–669. doi: 10.1126/science.aaf8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1 | Full gel images from Extended Data Fig. 3.
Supplementary Data 2 | Sequence alignment of Orco proteins.
Supplementary Data 3 | Sequence alignment of OR proteins.
Data Availability Statement
The 3D cryo-EM density map of the Orco-Fab complex has been deposited in the Electron Microscopy Data Bank under accession number EMD-7352. The coordinates of the atomic model of Orco have been deposited in the Protein Data Bank under accession number 6C70.