The GNC and GLK families of transcription factors act cooperatively and independently to regulate chloroplast biogenesis and photosynthetic activity.
Abstract
Fundamental questions regarding how chloroplasts develop from proplastids remain poorly understood despite their central importance to plant life. Two families of nuclear transcription factors, the GATA NITRATE-INDUCIBLE CARBON-METABOLISM-INVOLVED (GNC) and GOLDEN TWO-LIKE (GLK) families, have been implicated in directly and positively regulating chloroplast development. Here, we determined the degree of functional overlap between the two transcription factor families in Arabidopsis (Arabidopsis thaliana), characterizing their ability to regulate chloroplast biogenesis both alone and in concert. We determined the DNA-binding motifs for GNC and GLK2 using protein-binding microarrays; the enrichment of these motifs in transcriptome datasets indicates that GNC and GLK2 are repressors and activators of gene expression, respectively. ChIP-seq analysis of GNC identified PHYTOCHROME INTERACTING FACTOR and brassinosteroid activity genes as targets whose repression by GNC facilitates chloroplast biogenesis. In addition, GNC targets and represses genes involved in ERECTA signaling and thereby facilitates stomatal development. Our results define key regulatory features of the GNC and GLK transcription factor families that contribute to the control of chloroplast biogenesis and photosynthetic activity, including areas of independence and cross talk.
Chloroplasts are one of the defining features of plants. Many of the unique properties of plants are related to the chloroplast’s capacity for photosynthesis, including the subcellular and cellular organization of the leaf, the leaf shape, and leaf movements (Sakamoto et al., 2008; Jarvis and López-Juez, 2013). Although most generally known for their ability to convert light energy into chemical energy to sustain plant growth (Waters and Langdale, 2009), chloroplasts also play a pivotal role in plant metabolism, being involved in the biosynthesis of amino acids, fatty acids, and phytohormones (Neuhaus and Emes, 2000). Despite their central importance to plant life, fundamental aspects of chloroplast development remain poorly understood (Sakamoto et al., 2008; Jarvis and López-Juez, 2013).
The chloroplasts of vascular plants develop from a nonphotosynthetic progenitor, the proplastid, which is maintained in meristematic cells (Waters and Langdale, 2009). Proplastids are colorless and contain limited amounts of internal membranes but can differentiate into a variety of plastid types with specialized activities, such as amyloplasts in the roots for starch storage, leucoplasts for lipid storage, chromoplasts for pigment accumulation, etioplasts in dark-grown shoots, and chloroplasts in light-grown shoots. The different types of plastids are interconvertible in response to developmental and environmental changes, with light and the phytohormone cytokinin playing pronounced roles in the induction of chloroplast development from proplastids (Stetler and Laetsch, 1965; Mullet, 1988; Chory et al., 1994; Sakamoto et al., 2008; Waters and Langdale, 2009; Jarvis and López-Juez, 2013). Based on the profoundly different phenotypes found in chloroplasts compared to proplastids and plastids from nonphotosynthetic tissues, the existence of chloroplast-specific “master controllers” has been proposed (López-Juez, 2007). Such master controllers would serve as switches to coordinately control the expression of suites of genes involved in the development of the chloroplast. Two families of nuclear transcription factors—the GATA NITRATE-INDUCIBLE CARBON-METABOLISM-INVOLVED (GNC) and GOLDEN TWO-LIKE (GLK) families—have been implicated in the direct, positive regulation of chloroplast biogenesis and thus are candidates for such master controllers.
The GNC family of GATA transcription factors is implicated in the control of chloroplast development in Arabidopsis (Arabidopsis thaliana) based primarily on the analysis of GNC and CYTOKININ-RESPONSIVE GATA1 (CGA1)/GNC-LIKE. These transcription factors regulate the development of the chloroplast from the proplastid and control chloroplast growth and division (Bi et al., 2005; Mara and Irish, 2008; Richter et al., 2010; Köllmer et al., 2011; Bastakis et al., 2018). The gnc and gnc cga1 mutants exhibit a reduction in chlorophyll levels (Bi et al., 2005; Mara and Irish, 2008; Hudson et al., 2011; Chiang et al., 2012), while overexpression increases chlorophyll and chloroplast production (Richter et al., 2010; Hudson et al., 2011; Köllmer et al., 2011; Chiang et al., 2012). Ectopic expression results in an expanded zone for chloroplast production to roots and hypocotyls, and perhaps most significantly for chloroplast production, to the epidermis of the hypocotyls and the epidermal pavement cells of the cotyledons and leaves (Chiang et al., 2012).
The GLK family has also been implicated in chloroplast development (Rossini et al., 2001; Fitter et al., 2002; Yasumura et al., 2005; Waters et al., 2008, 2009; Powell et al., 2012). Arabidopsis contains two GLK family members (GLK1 and GLK2) with Myb-like DNA-binding domains. The glk1 glk2 double mutant is pale green and shows a severe reduction in thylakoids and grana (Fitter et al., 2002; Waters et al., 2008). The GLK transcription factors promote the expression of many nuclear-encoded photosynthetic genes that are associated with chlorophyll biosynthesis and light-harvesting functions (Waters et al., 2009). In addition, ectopic expression of GLKs induces an increase in the production of chloroplasts in nongreen tissues such as roots and callus of Arabidopsis and rice (Oryza sativa; Nakamura et al., 2009; Kobayashi et al., 2012, 2013), as well as the chloroplast number per cell in tomato (Solanum lycopersicum; Nguyen et al., 2014), emphasizing their role in plastid development.
Consistent with their postulated roles as master controllers of chloroplast biogenesis, the expression of these transcription factors is regulated by signals involved in the control of chloroplast development, in particular light and cytokinin (Fitter et al., 2002; Rashotte et al., 2006; Naito et al., 2007; Chiang et al., 2012; Kobayashi et al., 2012; Martín et al., 2016). In addition, their expression is repressed under conditions where greening is inhibited (Mara and Irish, 2008; Richter et al., 2010, 2013b; Kobayashi et al., 2012; Zhang et al., 2013; Martín et al., 2016). These data suggest the existence of a transcriptional network in which the GNC and GLK families function to regulate chloroplast development, growth, and division. In this study, we first determined the degree of functional overlap of the GNC and GLK transcription factor families, characterizing these alone and in concert for their ability to regulate chloroplast biogenesis. Second, we determined DNA-binding motifs for GNC and GLK2 using protein-binding microarrays. The enrichment of these motifs in transcriptomic datasets supports GNC and GLK2 as repressors and activators of gene expression, respectively. Third, we performed a chromatin immunoprecipitation/DNA sequencing (ChIP-seq) analysis for GNC and identified targets whose repression by GNC would facilitate chloroplast biogenesis as well as stomatal development to coordinate photosynthesis. Results from these analyses define key regulatory features of these transcription factor (TF) families in the control of chloroplast biogenesis and photosynthetic activity, including areas of independence and cross talk.
RESULTS
Characterization of Loss-of-Function Mutants Involving the GNC and GLK Families of Transcription Factors
The GNC and GLK transcription factor families are both implicated in the regulation of chloroplast biogenesis. We therefore examined the degree of functional overlap of the GNC and GLK families, characterizing these alone and in concert for their ability to regulate chloroplast biogenesis. To functionally characterize the roles of the GNC and GLK families of transcription factors, we made use of the double mutants gnc cga1 and glk1 glk2, these loss-of-function mutants having previously been demonstrated to impact chloroplast development in Arabidopsis (Fitter et al., 2002; Bi et al., 2005; Mara and Irish, 2008; Waters et al., 2008; Hudson et al., 2011; Chiang et al., 2012). We also generated a gnc cga1 glk1 glk2 quadruple mutant (hereafter referred to as quad), making use of these same T-DNA insertion lines, to functionally characterize the role of both families together.
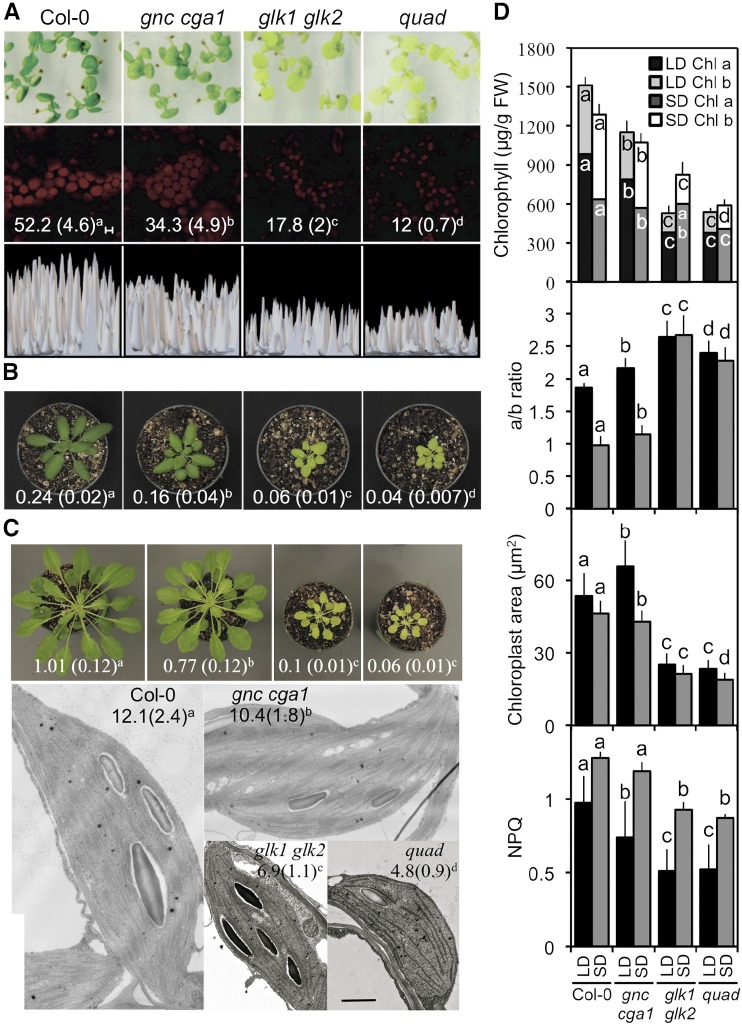
The mutants were compared to the wild type in terms of plant growth, chlorophyll levels, chloroplast size and ultrastructure, photosynthetic parameters, and gene expression. Plants were grown under both long- and short-day conditions (16/8-h and 8/16-h day/night cycles, respectively), because day length can have significant effects on photosynthetic activity (Walters and Horton, 1994; Lepistö and Rintamäki, 2012). Both the gnc cga1 and glk1 glk2 mutants exhibited reduced shoot growth compared to the wild type under either long or short days, this effect being most pronounced in the glk1 glk2 mutant (Fig. 1, A–C). The quad mutant was similar to glk1 glk2, indicating that the GNC family mutations have little or no additive effect in concert with the GLK family mutations. In addition, mutations in the GNC and GLK families resulted in early flowering and consequently fewer rosette leaves than wild type (Supplemental Fig. S1, A and B).
Figure 1.
Effect of GNC and GLK family mutants on photosynthetic and chloroplast-related phenotypes. Wild type, gnc cga1, glk1 glk2, and gnc cga1 glk1 glk2 (quad) loss-of-function mutants were characterized. Statistical analysis was performed using ANOVA with post-hoc Holm multiple-comparison calculation (P < 0.05); same letters indicate no significant difference. A, Ten-day-old seedlings (first row) showing a representative field of chloroplast fluorescence from palisade cells of the cotyledons (second row; scale bar, 5 µm) and the relative fluorescent intensity of the chloroplasts (third row). Chlorophyll a content (ng/mm2 ± sd; n = 30) of the cotyledons is given in the second row. B, Shoot phenotypes of 28-d-old plants grown under long-day conditions. Numbers give rosette weight (g; ±sd); n = 18. C, Shoot and chloroplast phenotypes of 45-d-old plants grown under short-day conditions. Numbers give rosette weight (g; ±sd); n = 15. Representative transmission electron microscopy images of chloroplasts are shown, with numbers giving thylakoid density (number thylakoid membranes per 500 nm; ±sd); n ≥ 20. Scale bar, 500 nm. D, Chloroplast-related parameters. Chlorophyll a and b content (n = 18), chlorophyll a/b ratio (n = 18), mesophyll chloroplast size (n = 100), and nonphotochemical quenching (NPQ; n = 20) were measured for plants grown under long- or short-day conditions. All parameters were measured for mature plants, except mesophyll chloroplast size under long-day conditions, which was determined for 10-d-old seedlings. Statistical analysis was performed independently on genotypes grown under long- and short-day conditions. Statistical analysis was performed for both chlorophyll a and b levels. Error bars indicate ± sd.
The effects on shoot growth were associated with reduced chlorophyll levels in the mutants (Fig. 1, A–D), as might be expected given the significant role photosynthesis plays in regulating growth (Kim et al., 2009). The reduction in chlorophyll levels was most pronounced in mutants involving members of the GLK family, which exhibited paler leaves than either wild type or gnc cga1 (Fig. 1, A–C). The glk1 glk2 and gnc cga1 double mutants had altered levels of both chlorophyll a and b, the effects of the quad mutant being similar to those of glk1 glk2; an additive effect of gnc cga1 on glk1 glk2 in the quad was only observed under short days (Fig. 1D). Of particular significance was the effect of the glk1 glk2 double mutation, both alone and in the quad mutant, on the chlorophyll a/b ratio (Fig. 1D). This ratio normally decreases in wild type under short-day compared to long-day growth conditions as a compensatory response to decreased light (Walters and Horton, 1994; Lepistö et al., 2009; Lepistö and Rintamäki, 2012). However, in mutants involving glk1 glk2, the ratio was elevated and unresponsive to day length. Although glk1 glk2 was still capable of increasing chlorophyll levels somewhat in response to decreased day length, this ability was lost in the quad mutant, pointing to a role for the GNC family in the regulation of this process (Fig. 1D).
Chloroplast size followed the same general trend as rosette size and chlorophyll levels, with gnc cga1 exhibiting a slight reduction under short-day growth conditions, glk1 glk2 exhibiting a substantial decrease, and quad being similar to glk1 glk2 in chloroplast size. Nevertheless, a slight additive effect of the gnc cga1 mutant was observed in quad under short-day conditions (Fig. 1, C and D). The reduction in chloroplast size is due, at least in part, to effects on thylakoid density, the glk1 glk2 mutant exhibiting a substantial decrease in grana formation as previously noted (Fitter et al., 2002). Although the effect of the gnc cga1 mutant on thylakoid density is less pronounced than that of the glk1 glk2 mutant, an additive effect is observed in the quad mutant. The decrease in grana is consistent with the decreases in chlorophyll b levels in the mutants, since chlorophyll b is a key component of the light-harvesting complex (LHC), and a reduced LHC results in reduced thylakoid grana abundance (Kim et al., 2009).
Interestingly, despite the significant chlorophyll reduction in the mutants, only modest changes in most photosynthetic parameters were observed (Fig. 1D; Supplemental Table S1). Nonphotochemical energy quenching (NPQ), related to the ability of the photosystems to dissipate excess excitation energy, exhibited a substantial decrease in glk1 glk2 double mutants, although a decrease could also be noted in gnc cga1 under long-day conditions (Fig. 1D). The quad mutant was indistinguishable from glk1 glk2 under both long- and short-day conditions, pointing to the GLK family being the primary modulator of this parameter. The reduction in NPQ is consistent with the reduction in chlorophyll (Chl) b levels, as mutants lacking Chl b in Arabidopsis and barley (Hordeum vulgare) both exhibit reduced NPQ values (Horton et al., 1996; Havaux et al., 2007). In addition, the maximum quantum yield of PSII (Fv/Fm) exhibited a small increase in the glk1 glk2 mutant compared to the wild type under long- but not short-day conditions, with the quad mutant being indistinguishable from glk1 glk2. Increases in Fv/Fm have been previously correlated with reduced Chl b content in rice and oilseed rape (Brassica napus; Zhou et al., 2006; Guo et al., 2007). No significant differences between mutants and the wild type were observed in the flux of electrons through PSII (quantum yield; ΦPSII) or in the proportion of open PSII reaction centers (photochemical quenching) under either long- or short-day conditions (Supplemental Table S1).
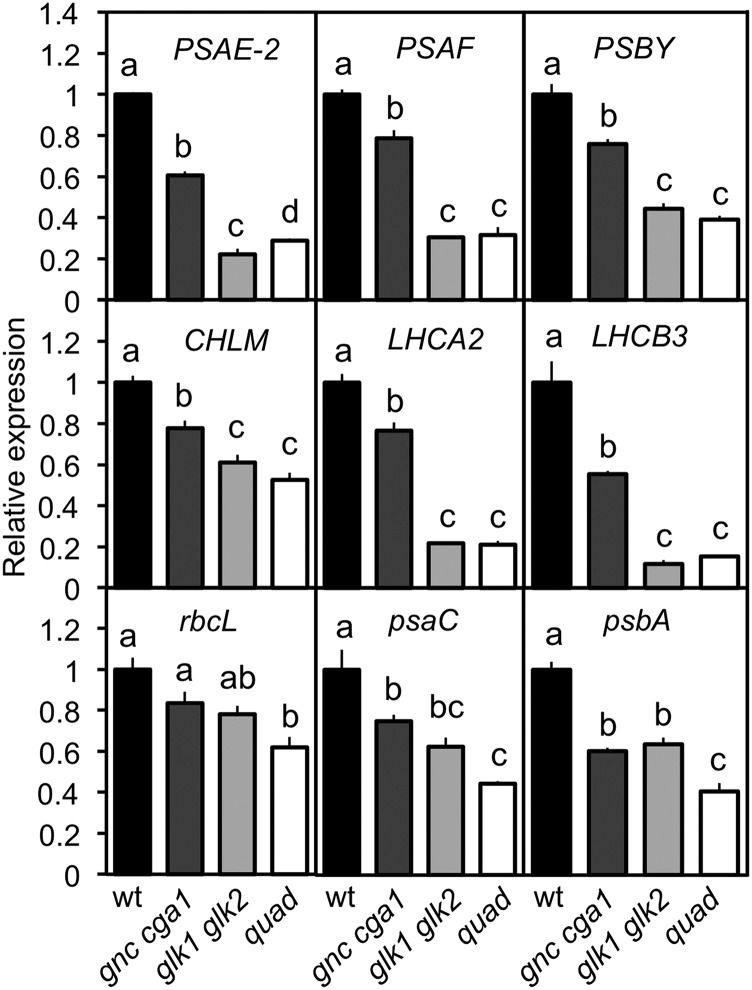
We characterized the mutants for their expression of nuclear-encoded chloroplast-targeted genes as well as plastid-encoded genes (Fig. 2). For this analysis, we selected six nuclear-encoded genes (PSAE-2, PSAF, PSBY, CHLM, LHCA2, and LHCB3) and three plastome-encoded genes (rbcL, psaC, and psbA). CHLM, LHCA2, and LHCB3 were previously identified as direct GLK1 targets (Waters et al., 2009). Expression of the six nuclear-encoded genes followed the same trend observed for rosette size, chlorophyll levels, and chloroplast size, with gnc cga1 exhibiting a partial reduction, glk1 glk2 exhibiting a substantial decrease, and the quad being similar to glk1 glk2. However, the trend was different for the plastome-encoded genes. Similar decreases in their expression were noted in both the gnc cga1 and glk1 glk2 mutants, while an additive effect was observed in the quad mutant. As plastome-encoded genes cannot be direct targets of the GNC or GLK families of transcription factors, the observed effects on expression represent indirect effects arising from their regulation of nuclear gene expression. These physiological and molecular analyses reveal overlapping functions of the GNC and GLK families, with the glk1 glk2 mutants exhibiting stronger effects on the measured parameters than the gnc cga1 mutant.
Figure 2.
GNC and GLK family mutants affect the expression of photosynthetic-related genes. Gene expression was determined by RT-qPCR from 10-d-old wild-type, gnc cga1, glk1 glk2, and gnc cga1 glk1 glk2 (quad) seedlings grown under long-day conditions. Expression was characterized for nuclear-encoded subunits of PSI and PSII (PSAE-2, PSAF, and PSBY; top row), nuclear-encoded chlorophyll biosynthetic enzyme and subunits of light-harvesting complexes (CHLM, LHCA2, and LHCB3; second row), and plastome-encoded subunits of Rubisco, PSI, and PSII (rbcL, psaC, psbA; bottom row). Expression was normalized to the tubulin control and is relative to the wild type. Error bars indicate ± se; statistical analysis was performed using ANOVA with post-hoc Holm multiple-comparison calculation (P < 0.05; n = 3 biological replicates); same letters indicate no significant difference..
Effects of Ectopic Overexpression of GNC and GLK2 on Photosynthesis and Chloroplast Biogenesis
Ectopic overexpression of GNC was previously found to induce chloroplast production in the epidermal cells of cotyledons and leaves, as well as the root cortex and pericycle (Richter et al., 2010; Hudson et al., 2011; Köllmer et al., 2011; Chiang et al., 2012). Ectopic overexpression of GLK1 or GLK2 also enhances chloroplast production in Arabidopsis roots (Kobayashi et al., 2012, 2013). We directly compared the ability of both families to induce chloroplast biogenesis in novel cell types by establishing transgenic Cauliflower mosaic virus (CaMV) 35S:GNC and CaMV 35S:GLK2 lines, examining the effects of these transgenes both alone and in concert. In addition, we examined the interdependence of the TF families by assessing the ability of the CaMV 35S:GNC and CaMV 35S:GLK lines to induce expanded chloroplast production in the complementary gnc cga1 and glk1 glk2 mutant backgrounds.
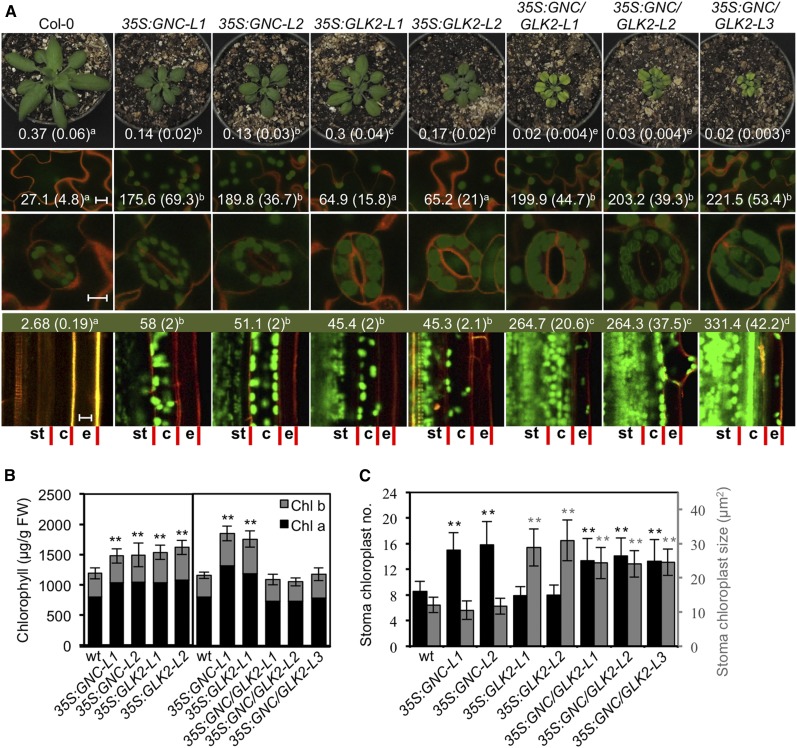
Ectopic overexpression of GNC results in reduced shoot growth but increased leaf chlorophyll levels compared to wild type (Fig. 3, A and B; Supplemental Fig. S2). Analysis of photosynthetic parameters reveals that NPQ was higher in the 35S:GNC lines than in the wild type, the opposite of what is observed with the gnc cga1 loss-of-function mutant, and the flux of electrons through PSII (ΦPSII) was slightly lower than in the wild type (Supplemental Table S1). The 35S:GNC lines matured more slowly than did the wild type, such that there was an increase in the number of days to bolting even though the plants exhibited a slight reduction in rosette leaf number. In terms of chloroplast biogenesis, the 35S:GNC lines resulted in significantly increased chloroplast production in both epidermal pavement and stomatal guard cells (Fig. 3, A and C). Although the number of chloroplasts was higher in the stomatal guard cells of the 35S:GNC lines, these chloroplasts were not significantly different in size from those of the wild type (Fig. 3C). Overexpression of GNC also resulted in a significant enhancement of chloroplast development in roots, particularly in the cortex and pericycle (Fig. 3A).
Figure 3.
Effects of ectopically overexpressed GNC and GLK2 on chlorophyll accumulation and chloroplast biogenesis. Wild-type (Col-0), two 35S:GNC, two 35S:GLK2, and three 35S:GNC 35S:GLK2 lines were characterized. A, Representative images of 28-d-old plants (top row; rosette fresh weight in grams indicated ± sd, n = 18), pavement cotyledon cells (second row; scale bar, 10 µm; number of chloroplasts per 0.1024 mm2 area ± sd indicated, n = 10), stomata guard cells (third row; scale bar, 10 µm), and root sections (bottom row; scale bar, 10 µm; chlorophyll content in µg/g fresh weight [FW] indicated, n = 6). Chloroplasts are visualized by chlorophyll autofluorescence (green) and cell walls by propidium iodide (orange). Root cell files for stele and endodermis (st), cortex (c), and epidermis (e) are indicated. Statistical analysis performed using ANOVA with post-hoc Holm multiple-comparison calculation; P < 0.05. B, Chlorophyll a and b content of rosette leaves from lines with ectopically overexpressed GNC and GLK2 (±sd, n = 10); plants were grown as in A. Statistical analysis performed on total chlorophyll for comparison of CaMV 35S lines with wild type, using one-way ANOVA with post-hoc Holm multiple-comparison calculation; *P < 0.05, **P < 0.01. C, Effect of ectopically overexpressed GNC and GLK2 on chloroplast production in stomata guard cells. Number of chloroplasts per stoma (black, ±sd, n = 50) and stoma chloroplast size (gray, ±sd, n = 100) were examined. Statistical analysis performed as in B.
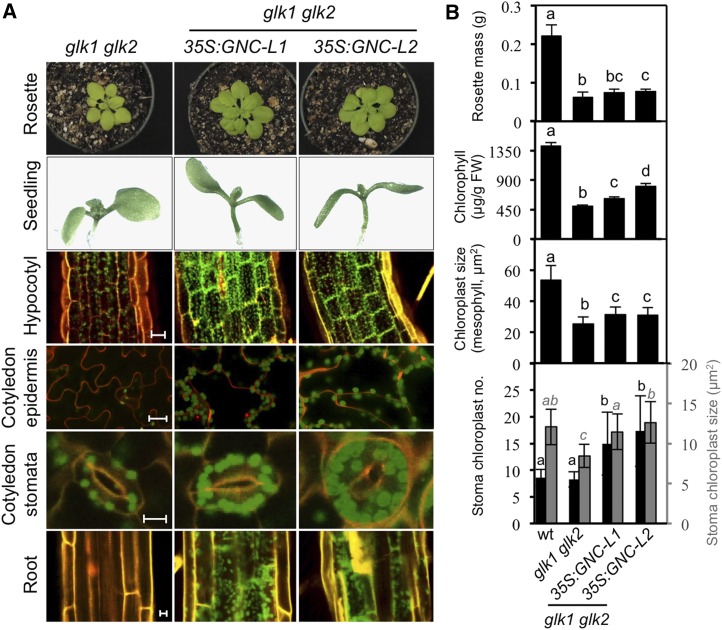
When expressed in a glk1 glk2 mutant background, 35S:GNC partially rescued the mutant phenotype in terms of rosette mass, mesophyll chloroplast size, and chlorophyll levels, although these parameters remained substantially reduced compared to the wild type (Fig. 4, A and B; Supplemental Fig. S2). As in the wild-type background, 35S:GNC induced an increase in NPQ and a decrease in the flux of electrons through PSII (ΦPSII). Significantly, ectopic overexpression of GNC still resulted in enhanced chloroplast production in both epidermal pavement and stomata guard cells as well as in hypocotyls and roots, similarly to what was observed in the wild-type background (Fig. 4, A and B). Thus, the ability of 35S:GNC to induce chloroplast production in novel cell types, as well as its effects on photosynthetic parameters, functions independently from the GLK family.
Figure 4.
Analysis of 35S:GNC expressed in the glk1 glk2 background. A, Effect of 35S:GNC expression on chloroplast development in hypocotyl, epidermis, and roots. Two independent 35S:GNC lines in the glk1 glk2 background were analyzed and compared to glk1 glk2. Representative rosettes of 28-d-old plants (top row), 7-day-old seedlings, (second row), chloroplasts in the hypocotyls of 3-day-old seedlings (third row; scale bar, 25 µm), chloroplasts in the cotyledon adaxial pavement cells (fourth row; scale bar, 25 µm) and stoma guard cells (fifth row; scale bar, 10 µm) of 10-d-old seedlings, and chloroplasts in roots of 7-day-old seedlings (bottom row; scale bar, 10 µm) are shown. Young seedlings were grown under constant white light. Chloroplasts were visualized by chlorophyll autofluorescence (green) and cell walls by staining with propidium iodide (orange). B, Ability of 35S:GNC expression to rescue the glk1 glk2 phenotypes. Rosette mass (n = 18), total chlorophyll in mature leaves (n = 16), chloroplast size in mesophyll cotyledon cells of 10-d-old seedlings (n = 100), and stoma chloroplast number and size (black columns, n = 50; and gray columns, n = 100, respectively) were measured for Col-0, glk1 glk2, and two independent 35S:GNC lines in the glk1 glk2 background. Error bars indicate ± sd. Statistical analysis was performed using ANOVA with post-hoc Holm multiple-comparison calculation; P < 0.05, same letters indicate no significant difference..
As with GNC, ectopic overexpression of GLK2 resulted in reduced shoot growth, increased chlorophyll levels, increased NPQ, and a decrease in the flux of electrons through PSII (Fig. 3, A and B; Supplemental Table S1). Additionally, the 35S:GLK2 lines exhibited a late-flowering phenotype based on rosette leaf number as well as days to bolting (Supplemental Fig. S1, C and D), consistent with previous studies (Waters et al., 2008). The 35S:GLK2 lines, like 35S:GNC, exhibited chloroplast development in the root cortex and pericycle, as previously noted (Kobayashi et al., 2012, 2013). In contrast to the 35S:GNC lines, the stomatal guard cells of the 35S:GLK2 lines did not exhibit an increase in chloroplast number compared to the wild type. Nevertheless, the guard cell chloroplast area in 35S:GLK2 lines was approximately 2.5-fold greater than those of wild type or the 35S:GNC lines (Fig. 3, A and C), a phenotype opposite to that observed in glk1 glk2 mutants and consistent with a role in promoting development of the light-harvesting complex (Fitter et al., 2002; Kim et al., 2009). GLK overexpression also induced a slight but significant increase in the size of the pavement cell chloroplasts, although this effect was substantially reduced compared to its effect in stoma cells (Supplemental Fig. S3), along with a slight trend toward increased chloroplast number in the pavement cells (Fig. 3). Interestingly, GLK2 could be overexpressed in the wild-type background, but we were unable to obtain overexpression in the gnc cga1 background (Supplemental Fig. S4). This was not a simple case of silencing, as GLK2 transcript was detected, most of it arising from the transgene, but the levels were similar to those in the wild type. Thus, the expression level of GLK2 appears to be dependent on the GNC family; by contrast, GNC expression does not require the GLK family, although the GLK family may play a minor role in regulating the maximal level of GNC expression (Supplemental Figs. S2 and S4).
Ectopic overexpression of GNC and GLK2 together resulted in severe growth reduction in the shoot (Fig. 3A). Maturation of the plants, based on the number of days to bolting, was substantially slowed. However, the number of rosette leaves at bolting falls in between that found for 35S:GNC and 35S:GLK2 (i.e. higher than in 35S:GNC but lower than in 35S:GLK2), suggesting independent effects on this phenotype (Supplemental Fig. S1). These growth defects coincided with substantial decreases in photosynthetic parameters related to the quantum yield of PSII (Fv/Fm and ΦPSII) and both photochemical and nonphotochemical quenching (Supplemental Table S1). The 35S:GNC 35S:GLK2 plants had chlorotic interveinal spaces, although overall leaf chlorophyll levels were similar to those in wild type (Fig. 3, A and B). An examination of the mesophyll chloroplasts revealed reduced sizes compared to wild type, although they still exhibited strong chlorophyll fluorescence (Supplemental Fig. S5). The chlorosis observed in 35S:GNC 35S:GLK2 plants is potentially due to oxidative stress, as we observed enhanced expression of marker genes ZAT12 and BAP1 (Nitschke et al., 2016), primarily due to the action of 35S:GNC (Supplemental Fig. S6). The veinal regions of leaves were still dark green (Fig. 3A), similar to what we sometimes observed in 35S:GNC lines and which may relate to the ability of GNC to induce increased chloroplast production in endodermis and bundle sheath cells, tissues that normally exhibit reduced chloroplast production (Chiang et al., 2012).
The expression of 35S:GLK2 did not affect the ability of 35S:GNC to induce chloroplasts in epidermal pavement cells, but resulted in additional phenotypes in the stomatal guard cells and roots (Fig. 3, A and C). In stomatal guard cells, the chloroplasts increased in number, as found in 35S:GNC lines, and also increased in size, as found in 35S:GLK2 lines (Fig. 3, A and C). Simultaneous overexpression of GNC and GLK2 stimulated chloroplast production in root tissues even further, resulting in chloroplast production in the epidermis, which was not observed when the transcription factors were ectopically overexpressed individually (Fig. 3A). These data emphasize similarities and differences between the two families and their role in chloroplast biogenesis, as well as the need to maintain careful control of their expression due to the possibly severe photosynthetic defects that may arise when both are misregulated, potentially due to photo-oxidative stress (Sakuraba et al., 2010). This misregulation was most pronounced in the mesophyll cells, which already produce substantial numbers of active chloroplasts, but was not apparent in cells to which chloroplast production ectopically expanded in the transgenic lines.
Identification of DNA-Binding Motifs for GNC and GLK2
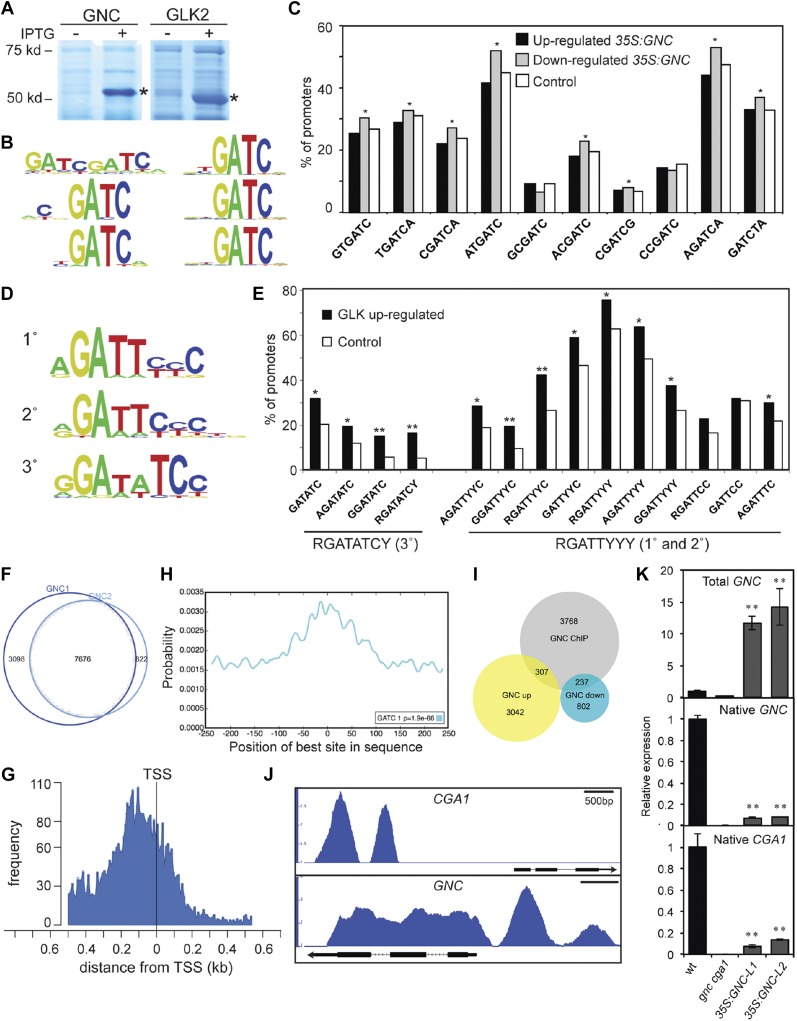
To characterize the DNA-binding motifs for GNC and GLK2, we employed PBM11 protein-binding microarrays (Fig. 5, A–E; Godoy et al., 2011; Franco-Zorrilla et al., 2014). GNC bound with high affinity to DNA sequences containing a core GATC motif, and this motif appeared when either 8-mer or 6-mer sequences were analyzed for enrichment (Fig. 5B). This GATC DNA-binding motif has been found for other TFs of the GATA family in plants (Chang et al., 2013; Franco-Zorrilla et al., 2014; Weirauch et al., 2014). We analyzed published transcriptomic data from CaMV 35S:GNC plants for overrepresentation of the GNC DNA-binding motif (Richter et al., 2010). The top 10 scoring 6-mers, all of which contain the GATC motif, were evaluated for their representation in promoters of genes up- or down-regulated in the GNC overexpressing plants (Richter et al., 2010). We found that GATC-containing 6-mers are overrepresented in promoters of down-regulated genes, but not of up-regulated genes (Fig. 5C), suggesting that GNC primarily functions as a transcriptional repressor.
Figure 5.
Genomic analysis of GNC and GLK2. A, Expression of GNC and GLK2 DNA-binding domains as MBP fusions in E. coli. SDS-PAGE protein profiles from IPTG-induced (+) and uninduced (−) cultures are shown. B, Consensus 8-mer (left) and 6-mer (right) DNA-binding motifs for GNC based on PBM analysis. C, GNC-binding motifs are enriched in promoters for genes repressed in a 35S:GNC line. The top 10 scoring 6-mers were evaluated for overrepresentation (*P < 0.05) in genes whose expression was up- or down-regulated 3-fold or more (Richter et al., 2010). D, Consensus DNA-binding motifs for GLK2 based on PBM analysis. E, GLK2-binding motifs are enriched in genes up-regulated in response to GLK1 and GLK2 induction (*P < 0.05; **P < 0.01; Waters et al., 2009). F, Proportional Venn diagram of summit regions in each GNC ChIP-seq sample with 7,676 summit regions being common to the two samples, the common summits being used for subsequent analyses. G, GNC-binding sites are enriched near transcriptional start sites (TSS). H, The GATC DNA-binding motif for GNC is enriched in summit regions of ChIP-binding peaks based on analysis with CentriMo (E value = 1.9e-66; Bailey and Machanick, 2012). I, Proportional Venn diagram for GNC candidate gene targets based on ChIP-seq with genes whose expression is up- or down-regulated in 35S:GNC (Richter et al., 2010). J, GNC ChiP-seq binding profiles for CGA1 and GNC based on IGB viewer. Fold enrichment (IP/input) was computed by MACS using bdgcmp and ranges from 1 (IP = input) to 3 (CGA1) or 4 (GNC). K, Expression of native CGA1 and GNC genes in the 35S:GNC lines based on RT-qPCR analysis (**P < 0.01).
GLK2, which contains a Myb-like DNA-binding domain, recognized two types of DNA motifs (Fig. 5D). The type-1 motif corresponds to the first and second 8-mer motifs identified and has a consensus sequence RGATTYYY (R = A/G; Y = C/T), whereas the type-2 motif corresponds to the third 8-mer motif and has a consensus sequence RGATATCY. Both sequences contain RGAT, but the type-2 motif is a perfect palindrome, unlike the type-1 motif. Both type-1 and type-2 motifs are enriched in the promoters of genes up-regulated in response to GLK1 or GLK2 expression (Fig. 5E; Waters et al., 2009). These data are consistent with GLK2 functioning as a transcriptional activator. We note that very few genes are down-regulated following induction of GLK1 or GLK2, precluding analysis for motif enrichment; this result is also consistent with members of the GLK family functioning as transcriptional activators.
Genome-Wide Identification of GNC-Binding Sites
To identify potential targets by which GNC enhances chloroplast development in the 35S:GNC lines, we performed ChIP-seq with two independent lines, taking advantage of the GFP tag for immunoprecipitation. GNC-binding peaks were identified by model-based analysis of ChIP-seq (MACS), and the summits of these peaks were defined as points of local maximum read density (Zhang et al., 2008). Substantial overlap was found for the DNA regions bound in the two GNC samples, with 7,676 summits in common (Fig. 5F). The maximum peak frequency occurred near the transcription start sites (TSSs; Fig. 5G), consistent with a classic regulatory role for GNC in the control of gene expression. Furthermore, the GNC DNA-binding motif GATC was significantly enriched at the summits of the binding peaks (Fig. 5H), reinforcing the significance of the DNA-binding motif identified via PBM.
We identified 4,312 candidate gene targets based on their proximity to the GNC-binding peaks (Supplemental Data Set 1). We compared these GNC candidate gene targets to the up-regulated and down-regulated genes in GNC overexpressing plants (Fig. 5I; Supplemental Data Set S2; Richter et al., 2010). Down-regulated genes were significantly overrepresented (P < 0.001; Z-score = 10.6) whereas up-regulated genes were significantly underrepresented (P < 0.001; Z-score = −5.3) in the GNC targets. These data are consistent with GNC functioning as a transcriptional repressor. Similar results were observed when examining for the enrichment of the GATC-binding motif in the same dataset. Prior studies suggest an autoregulatory role with members of the GNC family serving to inhibit their own expression (Richter et al., 2010; Ranftl et al., 2016; Xu et al., 2017). We identified GNC-binding sites by ChIP-seq in the promoters of GNC and CGA1, consistent with direct inhibitory regulation, and confirmed that expression of both CGA1 and the native GNC gene is inhibited in the 35S:GNC lines (Fig. 5, J and K).
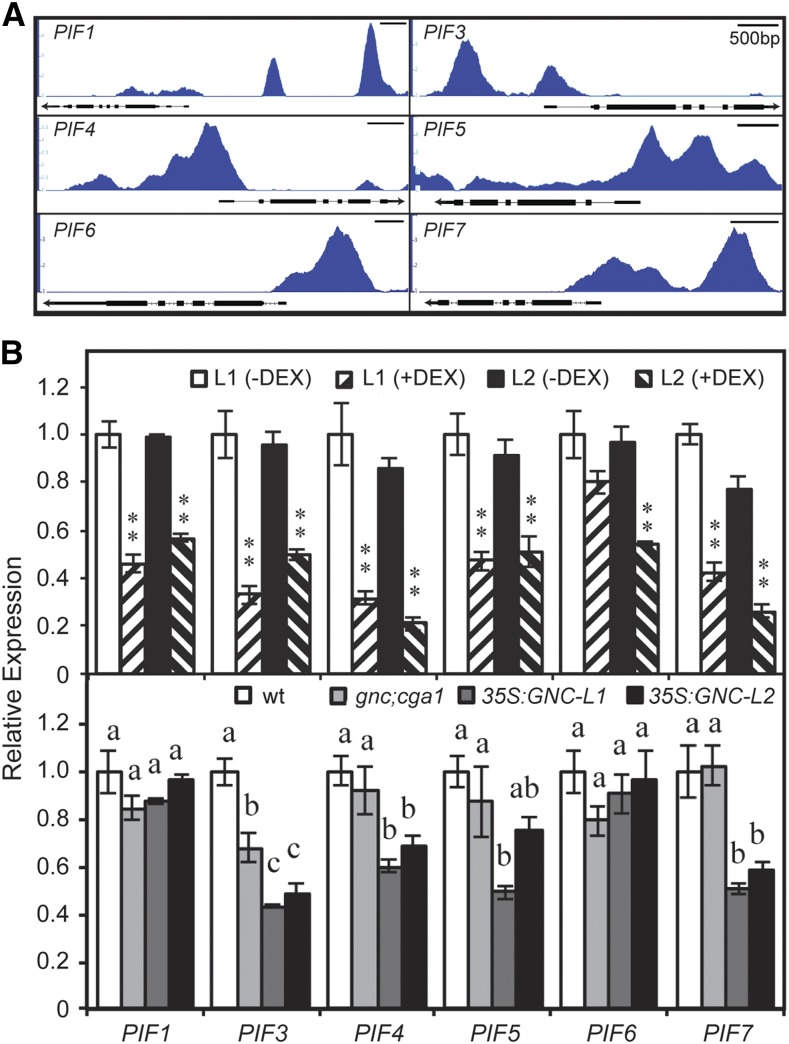
GNC Inhibits Expression of PIF and Brassinosteroid Activity Genes to Facilitate Chloroplast Biogenesis
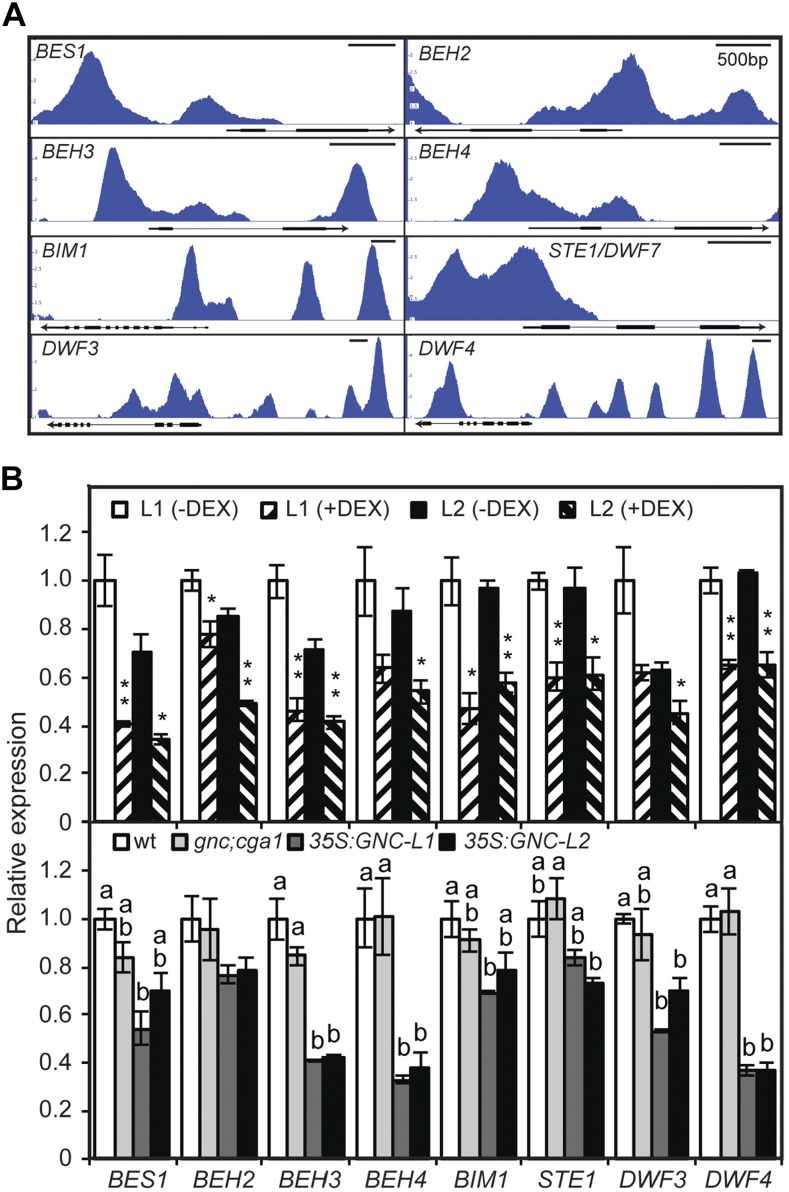
The predicted role of GNC as a transcriptional repressor suggested that it might counteract pathways that negatively regulate chloroplast development. We therefore explored the GNC candidate gene targets for those that could play such a role. GO analysis of the candidate targets indicates enrichment for response to light stimulus (1.7e-9), of particular note being GNC-binding peaks associated with the six members of the PHYTOCHROME INTERACTING FACTOR (PIF) transcription factor family that negatively regulate photomorphogenesis and repress chloroplast development (Stephenson et al., 2009; Leivar and Quail, 2011; Liu et al., 2017; Fig. 6A). Gene Ontology (GO) analysis of the candidate targets also indicated enrichment for various hormones, including brassinosteroids (BR; 3.47e-6), BRs being a class of hormones that also negatively regulate photomorphogenesis and chloroplast development (Wang et al., 2012). GNC-binding peaks are associated with genes that encode enzymes involved in BR biosynthesis (STE1/DWF7, DWF3, and DWF4) and that encode BR-activated transcription factors (BES1, BEH2, BEH3, BEH4, and BIM1; Fig. 7A).
Figure 6.
GNC enhances chloroplast biogenesis by inhibiting expression of PIF family genes. A, GNC ChiP-seq binding profiles based on IGB viewer. Fold enrichment for each sample (IP/input) was computed by MACS using bdgcmp. The range starts at 1 (IP = input), and the maximum varies per panel (5 for PIF1 and PIF5; 4 for PIF3, PIF4, PIF6, and PIF7). B, Gene expression determined by RT-qPCR on 6-d-old green seedlings for two independent DEX-inducible GNC lines, as well as wild-type, gnc cga1, and two independent 35S:GNC lines. Statistical analysis was performed using one-way ANOVA with post-hoc Holm multiple-comparison calculation for comparison of DEX-treated to -untreated for each line (*P < 0.05, **P < 0.01) and using ANOVA with post-hoc Holm multiple-comparison calculation for comparison of mutants to the wild type (P < 0.05; same letters indicate no significant difference); n = 3 biological replicates. Error bars indicate ± se.
Figure 7.
GNC enhances chloroplast biogenesis by inhibiting expression of target genes involved in brassinosteroid biosynthesis and signaling. BR-activated transcription factors encoded by BES1, BEH2, BEH3, BEH4, BIM1, and enzymes involved in BR biosynthesis encoded by STE1/DWF7, DWF3, and DWF4 were characterized. A, GNC ChiP-seq binding profiles based on IGB viewer. Fold enrichment for each sample (IP/input) was computed by MACS using bdgcmp. The range starts at 1 (IP = input), and the maximum varies per panel (5 for BES1, BEH3, and DWF4; 4 for DWF3; 3.5 for BEH2 and BIM1; 3 for BEH4 and STE1). B, Gene expression determined by RT-qPCR on 6-d-old green seedlings for two independent DEX-inducible GNC lines, as well as wild-type, gnc cga1, and two independent 35S:GNC lines. Statistical analysis was performed as in Figure 6.
We tested the ability of GNC to regulate expression of these candidate targets by employing an inducible version of GNC that is controlled by application of the steroid hormone dexamethasone (DEX; Chiang et al., 2012). For this purpose, seedlings were treated with 10 µm DEX for 4 h, treatment conditions that we previously determined resulted in good induction of GNC (Chiang et al., 2012). The short time of DEX induction should facilitate the identification of GNC-mediated changes in primary target expression while minimizing secondary indirect or homeostatic effects on gene expression. A similar inducible approach was successful in identifying GLK-regulated genes for chloroplast development, which were missed when transcriptome analysis was performed using constitutive GLK overexpression (Waters et al., 2009). DEX-induction of GNC represses expression of all six PIF genes, as well as genes for BR biosynthesis and BR-activated transcription factors (Figs. 6B and 7B). The expression of many of these genes is also reduced in 35S:GNC lines (Figs. 6B and 7B). For example, three out of the six PIF genes (PIF3, PIF4, and PIF7) exhibit lower expression levels than in the wild type in two independent 35S:GNC lines. In contrast, gene expression was similar to the wild type for the gnc cga1 mutant (Figs. 6B and 7B). Taken together, the data from DEX induction, 35S:GNC lines, and ChIP analysis support an ability of GNC to regulate chloroplast development by directly repressing multiple genes of the PIF family as well as those that mediate BR activity.
The GNC Family Inhibits Expression of ER and EPF Gene Families to Enhance Stomatal Development
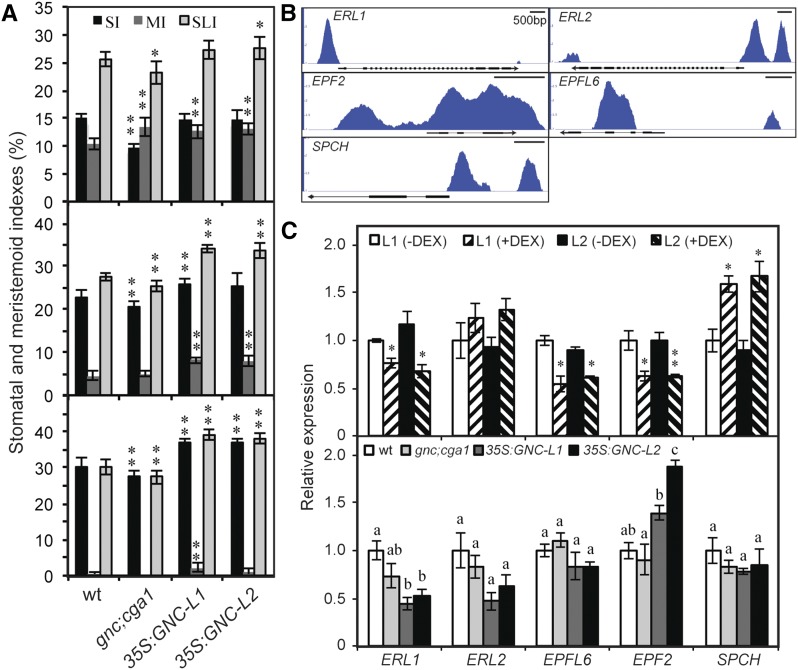
Stomata control the uptake of carbon dioxide needed for photosynthesis and their development, like that of chloroplasts, is responsive to light (Casson et al., 2009; Lepistö et al., 2009; Casson and Hetherington, 2010; Lepistö and Rintamöki, 2012). In particular, increased light intensity or photoperiod results in an increased stomatal index (percentage stomata out of the total number of epidermal cells plus stomata) in leaves. Members of the GNC family were recently implicated in the regulation of stomatal development, loss-of-function mutants and overexpression lines exhibiting decreased or increased numbers of stomata, respectively (Klermund et al., 2016). Genetic analysis indicated that the GNC family functioned upstream of several stomata formation regulators, including SPCH, which was identified as a direct target of CGA1.
We focused on the role of the GNC family in stomatal development in the cotyledon, the primary focus of the GNC-family study by Klermund et al. (2016) having been on the hypocotyl. The choice of the cotyledon allowed us to determine key developmental parameters such as stomatal index (SI; number of stomata/total number of stomata + nonstomatal epidermal cells × 100), meristemoid index (MI; number of meristemoids/total number of stomata + nonstomatal epidermal cells × 100), and stomatal lineage index (SLI; number of stomata + meristemoids/total number of stomata + nonstomatal epidermal cells × 100; Fig. 8A). We examined cotyledons of three ages (3, 6, and 10 d old) because stomatal development parameters change over time, MI being highest in young cotyledons and SI being highest in older cotyledons (Fig. 8A). The SLI is reduced in gnc cga1 seedlings but increased in the 35S:GNC seedlings, demonstrating a positive role for the GNC family in stomatal development (Fig. 8A). The gnc cga1 seedlings exhibit reduced SI at all developmental times, although they have higher MI than the wild type at 3 d old, suggesting that their stomatal development may be delayed. In contrast, the increased MI of the 35S:GNC lines at all time points examined translates into enhanced SI. Thus, overall, the 35S:GNC lines produce more meristemoids than the wild type, and these mature into stomata, resulting in higher SI and SLI. We also observed pairs of stomata in direct contact (one to three times per cotyledon in the 6- and 10-d-old seedlings) in the 35S:GNC lines, breaking the “one-cell-spacing rule” found in the wild type and indicating that 35S:GNC affects the formation of satellite meristemoids. We also examined stomata-related indexes of the glk1 glk1 mutant and 35S:GLK2 lines but found no alterations in stomata development (Supplemental Fig. S7), indicating that this function is specific to the GNC family.
Figure 8.
The GNC family regulates stomatal development in cotyledons. A, Effect on stomata index (SI), meristemoid index (MI), and stomata lineage index (SLI) in cotyledons of Col-0, gnc cga1, and two independent 35S:GNC lines. Parameters were determined for 3-d-old (top), 6-d-old (middle), and 10-d-old seedlings (bottom). For statistical analysis, one-way ANOVA with post-hoc Holm multiple-comparison calculation was used to compare the mutant lines to the wild type (*P < 0.05, **P < 0.01, n = 10). Error bars indicate ± sd. B, GNC ChiP-seq binding profiles based on IGB viewer. Fold enrichment for each sample (IP/input) was computed by MACS using bdgcmp. The range starts at 1 (IP = input), and the maximum varies per panel (4 for ERL1 and ERL2; 3 for EPF2 and EPFL6; 2.5 for SPCH). C, GNC regulates expression of genes involved in stomatal development and patterning. Gene expression determined by RT-qPCR on two independent DEX-inducible GNC lines, as well as wild-type, gnc cga1, and two independent 35S:GNC lines. Statistical analysis was performed as in Figure 6.
We explored the GNC candidate gene targets for those that could play a role in stomata development. GNC-binding peaks were identified in the promoters for ERECTA LIKE1 (ERL1) and ERL2, which encode members of the ERECTA family of receptor kinases, and in the intronic regions of EPIDERMAL PATTERNING FACTOR2 (EPF2) and EPIDERMAL PATTERNING FACTOR LIKE6 (EPFL6) genes, which encode Cys-rich secretory peptides that serve as ligands for the ERECTA family (Fig. 8B). The ERECTA family and their ligands inhibit stomata development, EPF2 having also been shown to regulate satellite meristemoid formation and thus assist in maintaining the “one-cell-spacing” rule (Shimada et al., 2011; Lee et al., 2012). We also identified two binding peaks for GNC in the SPEECHLESS (SPCH) promoter (Fig. 8B), a positive regulator of stomata development previously proposed to be a direct target of the GNC family (Klermund et al., 2016). The DEX induction of GNC significantly down-regulated the expression of ERL1, EPF2, and EPFL6, but up-regulated expression of SPCH (Fig. 8C). ERL1 and ERL2 were also down-regulated in 35S:GNC lines, indicating long-term effects of GNC on their expression; the expression of the other stomata-related genes was either similar to the wild type or increased, as in the case of EPF2 in the 35S:GNC lines. These results demonstrate that GNC plays a positive role in regulating stomatal development by inhibiting the activity of the ERECTA pathway as well as by inducing SPCH expression.
DISCUSSION
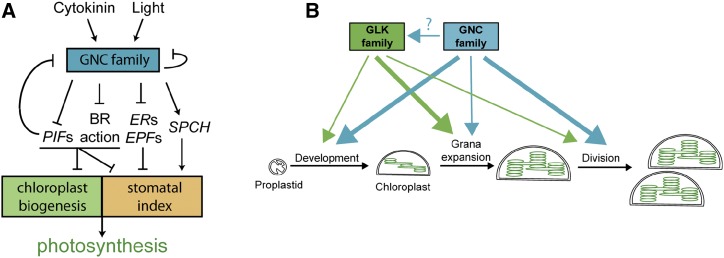
In this study, we took an integrative approach to analyze the roles of two transcription factor families proposed to function as master regulators of chloroplast development. Our results shed light on the mechanism of action for GNC and GLK family members based on the use of protein-binding microarrays to identify DNA-binding motifs, ChIP-seq analysis of GNC to identify binding sites and candidate targets for gene regulation, and the integration of these results with transcriptomic data. Our results highlight the independent and overlapping roles of the GNC and GLK family members in the regulation of chloroplast development and photosynthesis and identify direct transcriptional targets by which GNC regulates chloroplast and stomatal development. We propose models illustrating these regulatory mechanisms (Fig. 9). Below, we discuss the mechanism of action of the GNC and GLK families and their roles in regulating chloroplast development and photosynthesis.
Figure 9.
Models for the regulation of chloroplast biogenesis and photosynthesis. A, Regulation of photosynthesis by the GNC family. Expression of the GNC family is stimulated by light and cytokinin. The GNC family regulates expression of genes controlling chloroplast biogenesis and stomatal index, both of which contribute to photosynthesis. B, Overlapping and independent roles of the GNC and GLK families in chloroplast biogenesis in the shoot. The thickness of the arrows indicates the relative contribution of each gene family to the development of chloroplasts from the proplastid, to grana expansion, and to chloroplast division.
Mechanism of Action
Our results demonstrate that GNC binds to DNA sequences containing a GATC motif, primarily functions as a repressor, and functions within several cross-repressive regulatory circuits that modulate its expression. GATA-family TFs were originally named due to vertebrate family members recognizing and binding to DNA motifs containing the sequence GATA, although it was subsequently discovered that some of their zinc fingers recognize the target sequence GATC instead (Ko and Engel, 1993; Newton et al., 2001). Based on protein binding microarray analysis, we found that GNC binds exclusively to GATC-containing motifs, with no evidence of binding to GATA-containing motifs. The finding that the GATC motif is enriched at GNC-binding peaks identified by ChIP-seq is consistent with this analysis. Furthermore, the GATC-binding motif of GNC is consistent with what has been found for other plant GATA family members, including GATA4 (Chang et al., 2013), GATA12 (Franco-Zorrilla et al., 2014), and GATAs 1, 3, 6, 7, 8, 9, 10, 11, 15, 26, and 27 (Weirauch et al., 2014). Prior studies on GNC-family members have sometimes operated under the assumption that plant GATA family TFs will bind to GATA motifs in the genome (Hudson et al., 2011; Richter et al., 2013a; Ranftl et al., 2016), but, based on what is now known about GNC and other plant GATA-family members, the focus should be on the GATC motif.
GNC primarily functions as a repressor based on (1) the enrichment of its GATC-binding motif in genes repressed in a 35S:GNC line, (2) repressed genes being significantly overrepresented whereas induced genes were significantly underrepresented in GNC targets identified by ChIP-seq, and (3) the suppression of representative targets following short-term DEX induction of GNC (Fig. 9A). Prior studies have generally assumed that GNC will serve as an activator, thus informing the search for target genes (Hudson et al., 2011; Klermund et al., 2016; Xu et al., 2017), although a repressive function in the control of SOC1 for flowering time was uncovered (Richter et al., 2013a). Even though the evidence suggests that GNC primarily functions as a repressor, this does not preclude its functioning in some instances as a transcriptional activator, as we confirm that induction of GNC stimulates expression of the target gene SPCH, consistent with a prior study (Klermund et al., 2016).
The repressor activity of GNC plays a substantive role in modulating various cross-repressive interactions with other transcription factors, including members of the GNC family, the PIF family, and SOC1 to regulate flowering time (Fig. 9A). Prior studies have indicated that members of the GNC family repress their own gene expression (Richter et al., 2010; Ranftl et al., 2016; Xu et al., 2017). Our study defines novel GNC-binding sites in the promoters of GNC and CGA1, not identified by previous studies, which often focused on potential GATA rather than GATC motifs. These results confirm that GNC can inhibit expression of both itself and CGA1. GNC is also involved in other cross-repressive interactions. We find that GNC directly represses multiple members of the PIF family, which promote skotomorphogenesis and regulate shade responses, prior data indicating that PIFs also negatively regulate GNC family expression (Fig. 9A; Richter et al., 2010). A similar negative regulatory circuit also appears to function in the regulation of flowering time, with GNC directly repressing expression of SOC1 but SOC1 also directly repressing GNC and CGA1 expression (Richter et al., 2013a). Our ChIP-seq analysis supports multiple GNC-binding sites within the SOC1 promoter, rather than in the first intron of the 5′ UTR, as previously proposed. Taken together, these studies establish multiple regulatory circuits that modulate the expression of GNC family members and point to the importance of feedback inhibition in this process. The more subtle changes in gene expression observed in the gnc cga1 mutant may arise in part due to buffering by these regulatory circuits.
Our results also suggest that the GNC family may play a role in regulating the expression of GLK2, based on our ability to overexpress GLK2 in the wild-type background but not in the gnc cga1 background. This was not a simple case of silencing because we could detect the GLK2 transcript, most of this arising from the transgene but not at levels above those in the wild type. The role of the GNC family in regulating GLK2 expression is likely to be indirect, as ChIP analysis did not reveal GNC-binding sites within the portion of the GLK2 sequence used in the 35S:GLK2 construct. It may be that GNC represses the expression of other transcriptional repressors, with one of these serving to regulate expression of GLK2; the cross-repressive transcriptional regulation mediated by GNC, as described earlier, points to the potential complexity of such regulation. It may also be that the expression of GLK2 is a target for retrograde signaling by the chloroplast and is affected by the alterations in chloroplast development arising in the gnc cga1 mutant. Retrograde signaling from the chloroplast has been found to regulate gene expression through mechanisms involving transcriptional repressors, chromatin remodeling, and alternative splicing (Jarvis and López-Juez, 2013; Petrillo et al., 2014; Feng et al., 2016). We note that even though the CaMV 35S promoter was used to drive overexpression GLK2, the genomic GLK2 sequence was used in the construct; therefore, its introns provide potential noncoding sites for regulation.
Our analysis also provides new information on the mechanism of action of the GLK family. Prior analysis of GLK1 supports its role as a transcriptional activator, with its immediate targets being primarily associated with chlorophyll biosynthesis and light harvesting functions of the chloroplast (Waters et al., 2009; Franco-Zorrilla et al., 2014). By use of protein-binding microarrays with GLK2, we confirmed a DNA-binding motif previously identified for GLK1 as well as uncovered a new binding motif. Both binding motifs are enriched in genes up-regulated in response to expression of GLK1 or GLK2 (Waters et al., 2009). Thus, of the two transcription factor families proposed to function as master regulators in chloroplast development, members of the GLK family primarily function as transcriptional activators, while members of the GNC family primarily function as transcriptional repressors.
Regulation of Chloroplast Development and Photosynthesis
Proplastids are colorless and contain limited amounts of internal membranes, but they can differentiate into a variety of plastid types with specialized activities, including chloroplasts in photosynthetically active tissues (Fig. 9B), light, and the phytohormone cytokinin playing pronounced roles in the induction of chloroplast development from proplastids (Stetler and Laetsch, 1965; Mullet, 1988; Chory et al., 1994; Sakamoto et al., 2008; Waters and Langdale, 2009; Jarvis and López-Juez, 2013). The activity of the GNC and GLK families of transcription factors is regulated by light and cytokinin, and these proteins have been proposed to be master regulators that coordinately control the expression of gene suites involved in the development of the chloroplast (López-Juez, 2007). Our results confirm and extend upon previous findings.
Based on the analysis of loss-of-function mutants, the GNC and GLK families exhibit overlapping functions in the control of rosette size, flowering time, chlorophyll levels, NPQ, chloroplast size, thylakoid density, and the regulation of chloroplast-targeted genes. Both the GNC and GLK mutants affect these phenotypes, but the effect of the GLK mutants is greater, and the quad mutant is often similar to the glk1 glk2 mutant. The similarity of the quad mutant phenotypes to those of the glk1 glk2 mutant suggests that the GLK family may operate downstream of the GNC family or that a stronger perturbation of the response may have occurred in the glk1 glk2 mutant. Even though the glk1 glk2 mutant generally exhibits greater physiological defects than the gnc cga1 mutant, in some cases additive effects are observed in the quad mutant (e.g. thylakoid density; effect on plastome-encoded gene expression), demonstrating that the GNC family can, to some extent, regulate these phenotypes independently of the GLK family. Additional phenotypes, such as stomatal development, are affected by the GNC family and not by the GLK family.
The chloroplast-related phenotypes found in shoots of the GLK loss- and gain-of-function mutants are consistent with the previously identified role of the GLK family in positively regulating gene expression for components of the light-harvesting antenna (Fig. 9B; Waters et al., 2009). It was proposed that the GLK family may play a role in adaptation to changing light conditions (Waters et al., 2009), and our functional analysis under short- and long-day conditions confirms this hypothesis. The chlorophyll a/b ratio is normally lower in the wild type under short-day compared to long-day growth conditions, the increase in the antenna complex and its associated chlorophyll b likely being a compensatory response to the decreased light and resulting in more grana stacking (Walters and Horton, 1994; Lepistö et al., 2009; Lepistö and Rintamäki, 2012). However, in mutants involving glk1 glk2, the chlorophyll a/b ratio is elevated and unresponsive to day length. GLK mutants also exhibit a reduction in NPQ, consistent with the reduction in Chl b levels, as mutants that lack Chl b exhibit reduced NPQ values (Horton et al., 1996; Havaux et al., 2007).
Our understanding of the role of the GNC family in chloroplast development largely originates from the analysis of overexpression phenotypes (Richter et al., 2010; Hudson et al., 2011; Köllmer et al., 2011; Chiang et al., 2012). In particular, ectopic overexpression leads to the induction of chloroplast production, growth, and division in cell types where chloroplast production is normally reduced or absent compared to leaf mesophyll cells (Fig. 9B). Our results demonstrate that GNC promotes chloroplast biogenesis independently of the GLK family, doing so in part by repressing the expression of PIFs and genes for BR activity (Fig. 9A). The PIFs are negative regulators of chloroplast development, pif mutants exhibiting enhanced chloroplast development in the dark (Stephenson et al., 2009; Liu et al., 2017). PIFs directly bind to the promoters of LHCA and LHCB and suppress their expression; since these genes encode key components of the light-harvesting complex, this mechanism inhibits chloroplast development (Liu et al., 2017). BR is another key negative regulator of chloroplast development, BR deficiency resulting in increased accumulation of chlorophyll and photosynthetic proteins (Wang et al., 2012). Furthermore, members of the PIF and BZR/BES/BEH family interact to regulate gene expression, including for the repression of GLK1 (Oh et al., 2012; Wang et al., 2012), emphasizing the significance of GNC in regulating both signaling pathways in the control of chloroplast development.
Interactions between 35S:GNC and 35S:GLK2 in the enhancement of chloroplast development vary depending on the tissue and cell type. In stoma cells, which normally produce small chloroplasts, GNC and GLK2 exhibit independent and additive effects. GNC induces an increase in chloroplast number, consistent with its role in stimulating chloroplast division; GLK2 induces an increase in chloroplast size, consistent with its role in enhancing formation of the antenna complex; and together they induce an increase in both chloroplast number and size. In epidermal pavement cells of cotyledons and leaves, GNC still exhibits a strong ability to induce chloroplast production, and the effects of GLK2 on chloroplast size and number are minimal either alone or in concert with GNC. In the root, both GNC and GLK2 can individually induce the formation of chloroplasts to a similar extent in stele and cortex cells; when both are expressed together, both chloroplast number and their production zone increases, such that chloroplasts are also detected in the root epidermis. The effect of GNC is therefore fairly consistent in all these tissues/cell types, involving its roles in stimulating chloroplast production and division. In contrast, the effect of GLK2 appears to be dependent on other cell-specific factors. Its role in stimulating grana/antenna formation occurs primarily in shoot tissues adapted to photosynthesis, but outside of these tissues, it has either minimal effect, such as in epidermal pavement cells, or plays a greater role in chloroplast proliferation than in grana/antenna modulation, such as in roots or the tomato fruit (Nguyen et al., 2014).
In addition to chloroplasts, photosynthesis also requires stomata for gas exchange, the development of both chloroplasts and stomata being coordinated in response to light (Casson et al., 2009; Lepistö et al., 2009; Casson and Hetherington, 2010; Lepistö and Rintamöki, 2012). We confirmed a positive role for the GNC family in stomatal development (Klermund et al., 2016) and demonstrated that the GNC family enhances meristemoid production and their maturation to increase the SI and SLI of cotyledons. As with chloroplast development, the ability of GNC to repress PIFs and genes for BR activity is likely to play a role in facilitating stomata development (Fig. 9A). BR inhibits stomatal development in cotyledons and leaves by inhibition of a MAPK pathway, although it may have opposing effects in the hypocotyl (Gudesblat et al., 2012; Kim et al., 2012). The role of PIF family members is also somewhat variable, but genetic analysis indicated that PIF6 is a negative regulator of stomatal development in mature leaves (Casson et al., 2009). In addition, PIFs inhibit stomata formation in dark-grown seedlings, a quadruple pif mutant (pif1/3/4/5) developing stomata on its hypocotyl (Klermund et al., 2016). However, GNC also regulates the expression of genes that play more direct roles in regulation of stomatal development (Fig. 9A); this includes acting as a positive regulator for SPCH expression (Klermund et al., 2016) and a negative regulator for members of the ERECTA family and their ligands (Shimada et al., 2011; Lee et al., 2012).
Overall, our results support a more general role for the GNC family in the regulation of photosynthesis than for the GLK family. The GNC family regulates key elements of photomorphogenic development, in particular, chloroplast and stomatal development, both of which require coordination for optimal photosynthesis. The GLK family plays more specific roles in regulating aspects of chloroplast development, but not stomatal development; these roles vary with tissue type and thus additional regulatory elements are likely to modulate GLK function. The quadruple gnc cga1 glk1 glk2 mutant is viable and makes chloroplasts, suggesting the existence of additional “master regulators” for chloroplast development. However, the full extent to which the GNC family regulates chloroplast development is still uncertain because, whereas the GLK family is composed of only the two members found in the glk1 glk2 mutant, the GNC family is potentially composed of six or seven members, the ectopic overexpression of each GNC family member resulting in enhanced chloroplast development (Behringer et al., 2014; Ranftl et al., 2016). Analysis of the full contribution of the GNC family to chloroplast development has been hampered due to a lack of null mutants for the various family members outside of GNC and CGA1 (Ranftl et al., 2016), but the advent of genome editing approaches such as CRISPR-Cas9 analysis should rectify this deficiency in our understanding.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The wild-type and mutant lines of Arabidopsis (Arabidopsis thaliana) were all in the Columbia ecotype. The gnc, cga1, gnc cga1, glk1, glk2, and glk1 glk2 T-DNA insertion mutants have been previously described (Fitter et al., 2002; Chiang et al., 2012). The gnc cga1 and glk1 glk2 double mutants were crossed to create the quadruple mutant and genotyped by PCR using T-DNA and gene-specific primers as listed in Supplemental Table S2. The CaMV 35S:GNC-GFP overexpression lines and dexamethasone (DEX)-inducible GNC-HA lines in wild type and gnc cga1 mutant backgrounds were previously described (Chiang et al., 2012). Seedlings were grown on medium containing 0.8% (w/v) phytoagar (Research Products International), 1× Murashige and Skoog (MS) salts containing Gamborg’s vitamins, 1% (w/v) Suc, and 0.05% (w/v) MES, pH 5.7, as previously described (Chiang et al., 2012). For DEX induction, seedlings were treated with 10 µM DEX or a vehicle control for 4 h in liquid MS media. For analysis of adult phenotypes, seedlings were transferred to soil at one plant per pot and grown under long-day conditions (16 h day/8 h night) or short-day conditions (8 h day/16 h night) at 100 µmol m−2 s−1 light intensity, 22°C. Unless stated otherwise, leaves 9 and 10 from 28-d-old plants grown under long-day conditions or 45-d-old plants grown under short-day conditions were used for further analysis.
Generation of New Ectopic Overexpression Lines
To generate the 35S:GLK2-GFP construct, the genomic sequence of the GLK2 gene was amplified from genomic DNA using primers (5′-ATTCCGATGTTAACTGTTTCTCCGGC-3′ and 5′-AGGAAGAGGAGGAACATTAGAAACTCCT-3′). The PCR product was cloned into pCR8/GW/TOPO (Invitrogen) to generate an entry clone and subsequently recombined into the pGWB405 (Nakagawa et al., 2007) destination binary vector that confers kanamycin resistance to transformed plants. This vector was transformed into Agrobacterium tumefaciens strain GV3101 and than introduced into wild type, gnc cga1 mutant, and 35S:GNC backgrounds by the floral-dip method (Clough and Bent, 1998). A CaMV 35S:GNC-MYC construct was generated by recombining a GNC entry clone (Chiang et al., 2012) with the pGWB20 vector (Nakagawa et al., 2007), which resulted in the fusion of a 10× Myc tag to the C terminus of GNC; this construct was introduced into glk1 glk2 mutant background, also by the floral-dip method.
Gene Expression Analysis
Total RNA was isolated by using the Plant RNA kit (Omega) with the incorporation of a DNase treatment using the TURBO DNA-free kit (Invitrogen). The iScript cDNA Synthesis kit (Bio-Rad) was used for reverse transcription, and real time-quantitative PCR (RT-qPCR) was performed using iTaq Universal SYBR Green Supermix (Bio-Rad), with gene-specific primers (Supplemental Table S2). β-TUBULIN3 (AT5G62700) was used as a control for RT-qPCR. All procedures were performed according the manufacturer’s guidelines. Three biological replicates were used for each RT-qPCR experiment, each biological replicate representing 20–30 seedlings pooled from the separate plates used for their growth.
Chlorophyll and Photosynthetic Parameter Measurements
Spectrophotometric determination of chlorophyll content extracted from leaves was performed as described (Ritchie, 2006). In brief, chlorophyll a and b were extracted from 10-d-old whole seedlings grown on MS media or from leaves 9 and 10 of mature plants. Extraction was made with 100% (v/v) ethanol at 4°C in the dark with shaking for 16 h. At least 10 biological replicates (seedlings or leaves) for each genotype were used in the assays. The relative fluorescent intensity of cotyledon mesophyll cells of 10-d-old seedlings was determined with Nikon A1 confocal microscope.
Photosynthetic parameters were examined for leaves 9 and 10 of 28-d-old plants grown under long-day conditions and for the four largest mature leaves of 45-d-old plants grown under short-day conditions. Ten plants were used for each genotype/condition. Measurements were performed using Pulse-Amplitude-Modulated Mode of FlourCam 800MF (Photon Systems Instruments) according to the manufacturer’s guidelines.
Chloroplast Analysis
Chloroplasts in living tissues of roots, hypocotyls, and cotyledon epidermal cells and stomata were visualized as described (Chiang et al., 2012). A Nikon A1 confocal microscope was used to visualize chloroplasts by autofluorescence and cell walls by propidium iodide staining using 3-d-old hypocotyls, 7-d-old roots, and cotyledons of 10-d-old seedlings.
To determine chloroplast area and number in stomata, images of the best stoma focal plane (i.e. the 26.8-µm-thick plane in which the most chloroplasts appear) were obtained using NIS-Elements 4.00.03 software, and the images then analyzed using ImageJ software (ImageJ64; National Institutes of Health). The number of chloroplasts per stomata was determined based on the analysis of 50 stomata for each genotype; chloroplast area was based on the analysis of 100 chloroplasts. For these measurements, at least five cotyledons harvested from five seedlings were used. The size of mesophyll chloroplasts was determined as described (Chiang et al., 2012). In brief, cotyledons of 10-d-old seedlings were chopped with a razor blade in 0.33 m sorbitol, 2 mm EDTA, and 50 mm Tris-HCl (pH 8.0) to release chloroplasts, and the average area of 100 chloroplasts determined using ImageJ software (ImageJ64). For short-day conditions, leaves 9 and 10 were used.
To examine chloroplast ultrastructure, samples were cut out from the center part of leaf 9, excluding veins, of 45-d-old plants grown in soil under short-day conditions. Chloroplasts were fixed and then visualized on a JEOL TEM 1010 apparatus equipped with an XR-41B AMT digital camera as described (Chiang et al., 2012). Transmission electron microscopy images were used to analyze thylakoid membrane density. For this purpose, three lines were drawn across each chloroplast to create four equal sections. The number of membranes that crossed the lines was counted and calculated as the number of thylakoid membranes per 500 nm.
Stomatal Density Measurements
The numbers of stomata, meristemoids, and nonstomatal epidermal cells were counted on cotyledons of 3-, 6-, and 10-d-old seedlings, with 7 to 10 cotyledons analyzed for each age/genotype, coming from 7 to 10 seedlings (Peterson et al., 2013). Cotyledons were stained with propidium iodide, and images of the epidermis were made using a Nikon A1 confocal microscope as described above. SI (number of stomata/total number of stomata + nonstomatal epidermal cells × 100), MI (number of meristemoids/total number of stomata + nonstomatal epidermal cells × 100), and SLI (number of stomata + meristemoids/total number of stomata + nonstomatal epidermal cells × 100) were calculated.
Flowering Time and Rosette Mass Measurements
Flowering time was determined based on the number of days from sowing to the first open flower, as well as the number of rosette and cauline leaves produced by the apical meristem at this time point. Rosette mass was determined for 28-d-old plants for long-day conditions and 45-d-old plants for short-day conditions.
Protein-Binding Microarrays
The DNA-binding domains from GNC and GLK2 were amplified from Arabidopsis cDNA (Supplemental Table S2), then fused to MBP by cloning into pMAL-c2x using the restriction enzymes Bam HI and Hind III (New England Biolabs). Recombinant protein was expressed in the BL-21 E. coli strain using the Zymo Dual Media Kit (Zymo Research), with induction by 0.1 mm IPTG for 8 h at 37°C, with the soluble protein extracts then analyzed by SDS-PAGE to confirm induction. Analysis of DNA-binding specificities was performed using protein-binding microarrays (PBM11) as described (Godoy et al., 2011; Franco-Zorrilla et al., 2014). Enrichment of the DNA-binding motifs in promoters of published microarray datasets was determined as described (Franco-Zorrilla et al., 2014). For the GNC overexpression line (Richter et al., 2010), genes were selected for comparison based on their being up- or down-regulated 3-fold or more relative to the wild type. For the GLK induction study (Waters et al., 2009), genes were selected based on their being significantly up-regulated in the combined dataset following induction of GLK1 and GLK2 (Supplemental Table S3 of Waters et al., 2009)
ChIP and Analysis
ChIP-seq was performed using 16-d-old shoots from two independent 35S:GNC lines. Seedling growth, tissue cross-linking, chromatin isolation and immunoprecipitation (ab290 anti-GFP antibody, Abcam), and library construction and sequencing were as described (Zubo et al., 2017; Supplemental Table S3). Reads were aligned, peaks called by comparing the IP to the input derived from each sample, and GNC consensus peaks defined based on the overlap between samples as described (Zubo et al., 2017). Enrichment analysis for the GNC DNA-binding motif at consensus peaks was determined using CentriMo, using a set of 6,729 consensus peaks defined by their summits being no more than 50 bp apart, as this gave high confidence for the peak center (Bailey and Machanick, 2012). GNC consensus peaks were associated with genes based on the closest downstream gene from each strand within 0.5 kb of the binding summit, as well as accepting any gene that the peak was internal to; this resulted in 4,331 peaks linked to at least one gene, with 4,312 unique genes or “candidate targets” identified (Supplemental Dataset 1). GO analysis was performed with VirtualPlant 1.3 (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/; Katari et al., 2010). Comparison of the ChIP gene targets to genes up- and down-regulated in GNC overexpressing plants (Richter et al., 2010; Supplemental Dataset 2) was performed in VirtualPlant1.3 using Genesect, which performs a nonparametric randomization test to determine whether the overlap between two gene lists is higher or lower than expected by chance.
Statistical Analysis
Statistical analysis was performed using the online calculator (http://astatsa.com/OneWay_Anova_with_TukeyHSD/).
Accession Numbers
GNC ChIP-seq sequence data are available from the NCBI Sequence Read Archive under BioProject no. PRJNA435725. Sequence data for genes used in this study can be found in the GenBank/EMBL libraries under the following accession numbers: GNC (AT5G56860), CGA1 (AT4G26150), GLK1 (AT2G20570), GLK2 (AT5G44190), PSAE-2 (AT2G20260), PSAF (AT1G31330), PSBY (AT1G67740), CHLM (AT4G25080), LHCA2 (AT3G61470), LHCB3 (AT5G54270), rbcL (ATCG00490), psaC (ATCG01060), psbA (ATCG00020), PIF1 (AT2G20180), PIF3 (AT1G09530), PIF4 (AT2G43010), PIF5 (AT3G59060), PIF6 (AT3G62090), PIF7 (AT5G61270), BES1 (AT1G19350), BEH2 (AT4G36780), BEH3 (AT4G18890), BEH4 (AT1G78700), BIM1 (AT5G08130), STE1 (AT3G02580), DWF3 (AT5G05690), DWF4 (AT3G50660), ERL1 (AT5G62230), ERL2 (AT5G07180), EPFL6 (AT2G30370), EPF2 (AT1G34245), SPCH (AT5G53210), BAP1 (AT3G61190), ZAT12 (AT5G59820), β,TUB3 (AT5G62700). T-DNA mutant alleles used in this study were SALK_001778 for GNC, SALK_003995 for CGA1, Atglk1.1/CS9805 for GLK1, and Atglk2.1/CS9806 for GLK2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Flowering time of GNC and GLK family mutants.
Supplemental Figure S2. Expression level of GNC in 35S:GNC lines.
Supplemental Figure S3. Effect of ectopically overexpressed GNC and GLK2 on pavement chloroplast size.
Supplemental Figure S4. Ectopic overexpression of GLK2 requires the presence of the GNC family.
Supplemental Figure S5. Effects of ectopically overexpressed GNC and GLK2 on chloroplasts in mesophyll cells.
Supplemental Figure S6. Expression levels of oxidative stress genes in ectopic overexpression lines.
Supplemental Figure S7. GLK family mutants do not affect stomata development.
Supplemental Table S1. Photosynthetic parameters of GNC and GLK family mutants.
Supplemental Table S2. Primers used in the study.
Supplemental Table S3. Sequencing reads information for GNC ChiP-seq experiment.
Supplemental Data Set 1. Candidate gene targets of GNC.
Supplemental Data Set 2. Comparison of genes with altered expression in GNC overexpressing plants (Richter et al., 2010) with the GNC gene targets.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jane Langdale (Oxford, UK) for the glk mutant seed.
Footnotes
This work was supported by the National Science Foundation (IOS-1022053 and IOS-1238051 to G.E.S., J.J.K., and A.E.L., and IOS-1456487 to G.E.S.) and by Spanish Ministry for Science and Innovation Grant BIO2013-44407-R and Fundacion UAM Grant 2015007 (to R.S.). Computational resources for ChIP-seq analysis were provided through SciDAS with funding from the NSF Office of Cypberinfrastructure (OAC-1659300).
Articles can be viewed without a subscription.
References
- Bailey TL, Machanick P (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastakis E, Hedtke B, Klermund C, Grimm B, Schwechheimer C (2018) LLM-domain B-GATA transcription factors play multifaceted roles in controlling greening in Arabidopsis. Plant Cell 30: 582–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer C, Bastakis E, Ranftl QL, Mayer KF, Schwechheimer C (2014) Functional diversification within the family of B-GATA transcription factors through the leucine-leucine-methionine domain. Plant Physiol 166: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi YM, Zhang Y, Signorelli T, Zhao R, Zhu T, Rothstein S (2005) Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J 44: 680–692 [DOI] [PubMed] [Google Scholar]
- Casson SA, Hetherington AM (2010) Environmental regulation of stomatal development. Curr Opin Plant Biol 13: 90–95 [DOI] [PubMed] [Google Scholar]
- Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC, Hetherington AM (2009) phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol 19: 229–234 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YH, Zubo YO, Tapken W, Kim HJ, Lavanway AM, Howard L, Pilon M, Kieber JJ, Schaller GE (2012) Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol 160: 332–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Reinecke D, Sim S, Washburn T, Brenner M (1994) A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins). Plant Physiol 104: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Feng P, Guo H, Chi W, Chai X, Sun X, Xu X, Ma J, Rochaix JD, Leister D, Wang H, et al. (2016) Chloroplast retrograde signal regulates flowering. Proc Natl Acad Sci USA 113: 10708–10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA (2002) GLK gene pairs regulate chloroplast development in diverse plant species. Plant J 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy M, Franco-Zorrilla JM, Pérez-Pérez J, Oliveros JC, Lorenzo O, Solano R (2011) Improved protein-binding microarrays for the identification of DNA-binding specificities of transcription factors. Plant J 66: 700–711 [DOI] [PubMed] [Google Scholar]
- Gudesblat GE, Schneider-Pizoń J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, Boeren S, Zhiponova M, de Vries S, Jonak C, et al. (2012) SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol 14: 548–554 [DOI] [PubMed] [Google Scholar]
- Guo J-W, Guo J-K, Zhao Y, Du L-F (2007) Changes of photosystem II electron transport in the chlorophyll-deficient oilseed rape mutant studied by chlorophyll fluorescence and thermoluminescence. J Integr Plant Biol 49: 698–705 [Google Scholar]
- Havaux M, Dall’osto L, Bassi R (2007) Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol 145: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Ruban AV, Walters RG (1996) Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol 47: 655–684 [DOI] [PubMed] [Google Scholar]
- Hudson D, Guevara D, Yaish MW, Hannam C, Long N, Clarke JD, Bi YM, Rothstein SJ (2011) GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase (GLU1/Fd-GOGAT) expression in Arabidopsis. PLoS One 6: e26765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010) VirtualPlant: a software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EH, Li XP, Razeghifard R, Anderson JM, Niyogi KK, Pogson BJ, Chow WS (2009) The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: a study using two chlorophyll b-less mutants. Biochim Biophys Acta 1787: 973–984 [DOI] [PubMed] [Google Scholar]
- Kim TW, Michniewicz M, Bergmann DC, Wang ZY (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klermund C, Ranftl QL, Diener J, Bastakis E, Richter R, Schwechheimer C (2016) LLM-Domain B-GATA transcription factors promote stomatal development downstream of light signaling pathways in Arabidopsis thaliana hypocotyls. Plant Cell 28: 646–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Engel JD (1993) DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol 13: 4011–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Baba S, Obayashi T, Sato M, Toyooka K, Keränen M, Aro EM, Fukaki H, Ohta H, Sugimoto K, et al. (2012) Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Sasaki D, Noguchi K, Fujinuma D, Komatsu H, Kobayashi M, Sato M, Toyooka K, Sugimoto K, Niyogi KK, et al. (2013) Photosynthesis of root chloroplasts developed in Arabidopsis lines overexpressing GOLDEN2-LIKE transcription factors. Plant Cell Physiol 54: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllmer I, Werner T, Schmülling T (2011) Ectopic expression of different cytokinin-regulated transcription factor genes of Arabidopsis thaliana alters plant growth and development. J Plant Physiol 168: 1320–1327 [DOI] [PubMed] [Google Scholar]
- Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, McAbee JM, Sarikaya M, Tamerler C, Torii KU (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A, Rintamäki E (2012) Coordination of plastid and light signaling pathways upon development of Arabidopsis leaves under various photoperiods. Mol Plant 5: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E (2009) Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol 149: 1261–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Liu R, Li Y, Shen X, Zhong S, Shi H (2017) EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell 29: 3051–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E. (2007) Plastid biogenesis, between light and shadows. J Exp Bot 58: 11–26 [DOI] [PubMed] [Google Scholar]
- Mara CD, Irish VF (2008) Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol 147: 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet JE. (1988) Chloroplast development and gene-expression. Annu Rev Plant Physiol Plant Mol Biol 39: 475–502 [Google Scholar]
- Naito T, Kiba T, Koizumi N, Yamashino T, Mizuno T (2007) Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Muramatsu M, Hakata M, Ueno O, Nagamura Y, Hirochika H, Takano M, Ichikawa H (2009) Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol 50: 1933–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Newton A, Mackay J, Crossley M (2001) The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem 276: 35794–35801 [DOI] [PubMed] [Google Scholar]
- Nguyen CV, Vrebalov JT, Gapper NE, Zheng Y, Zhong S, Fei Z, Giovannoni JJ (2014) Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 26: 585–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke S, Cortleven A, Iven T, Feussner I, Havaux M, Riefler M, Schmülling T (2016) Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28: 1616–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KM, Shyu C, Burr CA, Horst RJ, Kanaoka MM, Omae M, Sato Y, Torii KU (2013) Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JW, Barta A, Kalyna M, et al. (2014) A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AL, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, et al. (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336: 1711–1715 [DOI] [PubMed] [Google Scholar]
- Ranftl QL, Bastakis E, Klermund C, Schwechheimer C (2016) LLM-domain containing B-GATA factors control different aspects of cytokinin-regulated development in Arabidopsis thaliana. Plant Physiol 170: 2295–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ (2006) A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA 103: 11081–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Bastakis E, Schwechheimer C (2013a) Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol 162: 1992–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Zourelidou M, Schwechheimer C (2013b) Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc Natl Acad Sci USA 110: 13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie RJ. (2006) Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynth Res 89: 27–41 [DOI] [PubMed] [Google Scholar]
- Rossini L, Cribb L, Martin DJ, Langdale JA (2001) The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 13: 1231–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Miyagishima SY, Jarvis P (2008) Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. Arabidopsis Book 6: e0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Yokono M, Akimoto S, Tanaka R, Tanaka A (2010) Deregulated chlorophyll b synthesis reduces the energy transfer rate between photosynthetic pigments and induces photodamage in Arabidopsis thaliana. Plant Cell Physiol 51: 1055–1065 [DOI] [PubMed] [Google Scholar]
- Shimada T, Sugano SS, Hara-Nishimura I (2011) Positive and negative peptide signals control stomatal density. Cell Mol Life Sci 68: 2081–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler DA, Laetsch WM (1965) Kinetin-induced chloroplast maturation in cultures of tobacco tissue. Science 149: 1387–1388 [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P (1994) Acclimation of Arabidopsis thaliana to the light environment: Changes in composition of the photosynthetic apparatus. Planta 195: 248–256 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Waters MT, Langdale JA (2009) The making of a chloroplast. EMBO J 28: 2861–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Moylan EC, Langdale JA (2008) GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J 56: 432–444 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Casaretto JA, Bi YM, Rothstein SJ (2017) Genome-wide binding analysis of AtGNC and AtCGA1 demonstrates their cross-regulation and common and specific functions. Plant Direct 1: e00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura Y, Moylan EC, Langdale JA (2005) A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 17: 1894–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Ding L, Wu Z, Liu R, Meyerowitz EM (2013) Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-S, Wu D-X, Shen S-Q, Sun J-W, Shu Q-Y (2006) High photosynthetic efficiency of a rice (Oryza sativa L.) xantha mutant. Photosynthetica 44: 316–319 [Google Scholar]
- Zubo YO, Blakley IC, Yamburenko MV, Worthen JM, Street IH, Franco-Zorrilla JM, Zhang W, Hill K, Raines T, Solano R, et al. (2017) Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc Natl Acad Sci USA 114: E5995–E6004 [DOI] [PMC free article] [PubMed] [Google Scholar]