A set of transgenic lines enables spatiotemporal control of gene expression in Arabidopsis.
Abstract
Understanding the context-specific role of gene function is a key objective of modern biology. To this end, we generated a resource for inducible cell type-specific transactivation in Arabidopsis (Arabidopsis thaliana) based on the well-established combination of the chimeric GR-LhG4 transcription factor and the synthetic pOp promoter. Harnessing the flexibility of the GreenGate cloning system, we produced a comprehensive set of transgenic lines termed GR-LhG4 driver lines targeting most tissues in the Arabidopsis shoot and root with a strong focus on the indeterminate meristems. When we combined these transgenic lines with effectors under the control of the pOp promoter, we observed tight temporal and spatial control of gene expression. In particular, inducible expression in F1 plants obtained from crosses of driver and effector lines allows for rapid assessment of the cell type-specific impact of an effector with high temporal resolution. Thus, our comprehensive and flexible method is suitable for overcoming the limitations of ubiquitous genetic approaches, the outputs of which often are difficult to interpret due to the widespread existence of compensatory mechanisms and the integration of diverging effects in different cell types.
The key to the evolutionary success of multicellularity, which arose independently in plants and animals, is the division of labor between highly specialized cell types. This requires the robust specification of cell fate through epigenetic and transcriptional programming, despite the identical genetic makeup of each cell. In plants, cell fate acquisition is based largely on positional information, which depends on cell-to-cell communication and medium- to long-distance morphogenetic signals that cooperate in organ patterning (Efroni, 2018). Conversely, individual genes, pathways, and metabolites can have diverse or even opposing roles depending on the tissue context. A prominent example for the context dependency of a fundamental patterning process is given by the interplay of the auxin and cytokinin phytohormones (Furuta et al., 2014; Greb and Lohmann, 2016; Truskina and Vernoux, 2018). In the shoot apical meristem, harboring the stem cell niche ultimately responsible for most aboveground plant organs, cytokinin signaling is associated with maintaining a pluripotent, undifferentiated state, whereas auxin signaling promotes differentiation. In marked contrast, auxin is required for stem cell maintenance in the root apical meristem (RAM; Pacifici et al., 2015; Weijers and Wagner, 2016). Therefore, the global effects of genetic lesions or of knockins can dilute and mask specific functions and often are difficult to interpret.
Routinely, stable genetic gain- and loss-of-function mutants remain the main pillar of the reductionist approach to biology, and the phenotypes of such mutants are assessed to deduce a function of the mutated locus in the wild type. However, the function of many gene products is context specific; thus, the phenotypes of mutants or transgenic lines can be complex. In addition, mutant organisms can undergo life-long adaptation, impeding the interpretation of their phenotype. Moreover, transgenic and mutational approaches can interfere with plant vitality, precluding an in-depth analysis.
Many of these problems can be overcome by inducible, cell type-specific expression mediated by two-component transcription activation systems (Moore et al., 2006). An expression cassette is constructed using a heterologous or synthetic promoter and, hence, is silent unless a cognate transcription factor is present. An efficient approach is to generate transgenic plants called driver lines that express the transcription factor in a spatially and temporally controlled manner and a responder line carrying the effector construct. After crossing of the two lines, expression can be induced and the phenotypic consequences of the effector can be studied. In the abstract, these expression systems are highly valuable because they ideally enable cell type-specific or stage-specific complementation or knockdown, facilitate time-resolved monitoring of the response to a given cue, can overcome the lethality of constitutive expression, and allow the study cell-autonomous and non-cell-autonomous effects with high temporal and spatial resolution. However, the considerable effort and time requirements for DNA cloning and the generation of stable transgenic plants are a major bottleneck curtailing their use to date. For the same reason and because distinct tissue-specific promoters were not always available in the past, attention is usually given to one tissue or cell type of interest at a time, and unbiased approaches targeting a larger spectrum of individual tissues are rarely followed.
Here, we report on the generation of a comprehensive set of Arabidopsis (Arabidopsis thaliana) driver lines suited for tissue-specific transactivation of an effector cassette in a wide range of cell types and with the possibility to monitor gene activation in space and time by a fluorescent promoter reporter. To ensure rapid, stable induction with minimal adverse effects on plant growth caused by the inducer, our system takes advantage of the widely used LhG4/pOp system (Moore et al., 1998; Craft et al., 2005; Samalova et al., 2005) combined with the ligand-binding domain of the rat glucocorticoid receptor (GR; Picard, 1993; Craft et al., 2005). LhG4 is a chimeric transcription factor consisting of a mutant version of the Escherichia coli lac repressor with high DNA-binding affinity (Lehming et al., 1987) and the transcription activation domain of yeast Gal4p (Moore et al., 1998). N-terminal fusion with the GR ligand-binding domain renders the transcription factor inactive in the cytosol through sequestration by HEAT SHOCK PROTEIN90 in the absence of the inducer. Nuclear import after treatment with the synthetic glucocorticoid dexamethasone (Dex; Picard, 1993) results in the transcriptional activation of expression cassettes that are under the control of the synthetic Op 5′ regulatory region consisting of a cauliflower mosaic virus (CaMV) 35S minimal promoter and two upstream lac operators (Moore et al., 1998; Craft et al., 2005). Combining multiple interspersed repeats of the operator elements (pOp4 and pOp6) and localized expression of LhG4 enables strong overexpression of a target gene in a cell type-specific manner (Craft et al., 2005).
Our work builds on these seminal studies by creating 19 well-characterized and stable driver lines targeting most cell types in Arabidopsis with a focus on the three main meristems of the plant, the RAM, the shoot apical meristem (SAM), and the cambium. Of note, for several cell types such as the pith in the inflorescence stem or the xylem pole pericycle cells in the root, inducible expression systems are not available so far. The driver lines were generated employing the fast and flexible GreenGate cloning system (Lampropoulos et al., 2013) but are compatible with any vector/transgenic line in which the expression of an effector is under the control of derivatives of the pOp promoter element (Moore et al., 1998). An important feature of our driver lines is the presence of a fluorescent reporter amenable to live imaging, which allows monitoring the spatiotemporal dynamics of gene induction and may serve as a readout for any effect on the respective tissue identity. Similarly, it allows us to assess whether the expression of the effector has an impact on the transcriptional circuitries targeting the promoter it is expressed from. The material described here allows testing the effect of genetic perturbations in a broad repertoire of individual tissues on a distinct developmental or physiological process. As transactivation efficiently occurs in the presence of the inducer in F1 plants derived from a cross between a driver and an effector line, the effect of a given expression cassette can be assessed relatively quickly in a wide range of cell types, demonstrating the usefulness of this resource for the broader research community.
RESULTS
Design of Driver Lines with Cell Type-Specific Expression of GR-LhG4
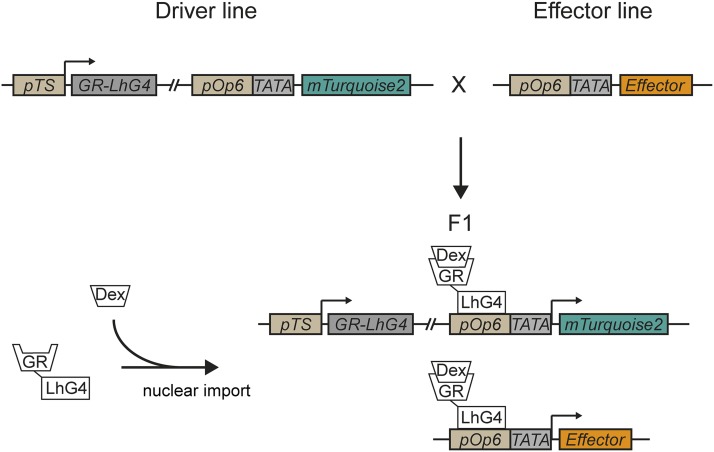
To generate a comprehensive set of driver lines expressing the chimeric GR-LhG4 transcription factor under the control of cell type-specific promoters, we made use of the Golden Gate-type GreenGate cloning system, which enables quick, modular, and scarless assembly of large constructs (Engler et al., 2008; Lampropoulos et al., 2013). Our design included, on the same T-DNA, the coding sequence for an mTurquoise2 fluorescent reporter (Goedhart et al., 2012) targeted to the endoplasmic reticulum (ER) through translational fusion with an N-terminal signal peptide from sweet potato (Ipomoea batatas) Sporamin A (SP; Lampropoulos et al., 2013) and the ER retention motif His-Asp-Glu-Leu (HDEL) under the control of pOp6 and a minimal cauliflower mosaic virus 35S promoter (pOp6:SP-mTurquoise2-HDEL; Fig. 1). In our setup, the GR-LhG4 transcription factor is expressed under the control of a tissue- or cell type-specific promoter. Consequently, GR-LhG4 activates the expression of the mTurquoise2 reporter and any other effector downstream of a pOp promoter after Dex treatment specifically in those tissues (Fig. 1). We anticipate that the most utility can be obtained from this system if lines harboring effector cassettes are crossed with driver lines and analyses are performed with F1 plants. However, other modes such as direct transformation of multiple driver lines or introgression into different (mutant) backgrounds also are conceivable. Notably, even though the mTurquoise2 reporter is expressed from the same T-DNA as GR-LhG4, there is no mechanistic difference from the activation of an effector in trans (Fig. 1).
Figure 1.
Overview of the Dex-inducible GR-LhG4/pOp system. In driver lines, expression of the synthetic transcription factor LhG4 is controlled by a tissue-specific promoter (pTS), whereas translational fusion with the ligand-binding domain of rat GR prevents nuclear translocation in the absence of the inducer (Dex). After crossing with an effector line harboring a transcriptional cassette under the control of a pOp element and a TATA box-containing minimal 35S promoter and the addition of Dex, GR-LhG4 drives the expression of the effector as well as the mTurquoise2 reporter encoded by the driver line.
To establish a rather comprehensive set of driver lines, we first selected respective tissue-specific promoters based on literature reports and our own expression data (Table 1). Subsequently, we generated stable transgenic driver lines in the Arabidopsis Columbia-0 (Col-0) background using 19 specific promoters that cover most cell types in the RAM, the SAM, and the cambium. Several of the promoters have been shown previously to work robustly in cell type-specific misexpression approaches (Nakajima et al., 2001; Weijers et al., 2006; Mustroph et al., 2009; Miyashima et al., 2011; Roppolo et al., 2011; Vatén et al., 2011; Naseer et al., 2012; Cruz-Ramírez et al., 2013; Ohashi-Ito et al., 2014; Wang et al., 2014; Chaiwanon and Wang, 2015; Serrano-Mislata et al., 2015; Vragović et al., 2015; Marquès-Bueno et al., 2016; Siligato et al., 2016; Doblas et al., 2017). Next, we generated T3 lines in which the resistance to the selective agent sulfadiazine appeared homozygous after segregating as a single locus in the T2 generation based on resistance or standard addition quantitative real-time PCR (SA-qPCR) analyses (Huang et al., 2013).
Table 1. Overview of promoters utilized in this study.
| Promoter | Gene | Expression | Reference |
|---|---|---|---|
| pSCR | SCARECROW | Endodermis, quiescent center (QC) in RAM, starch sheath in stem | Di Laurenzio et al. (1996); Wysocka-Diller et al. (2000) |
| pATHB-8 | HOMEOBOX GENE8 | Procambium, xylem precursors and columella in RAM | Baima et al. (1995) |
| pXPP | XYLEM POLE PERICYCLE | Xylem pole pericycle cells | Andersen et al. (2018) |
| pAHP6 | HISTIDINE PHOSPHOTRANSFER PROTEIN6 | Protoxylem precursors, pericycle, organ primordia in the SAM | Mähönen et al. (2006); Besnard et al. (2014) |
| pPXY | PHLOEM INTERCALATED WITH XYLEM | (Pro)cambium | Fisher and Turner (2007) |
| pTMO5 | TARGET OF MONOPTEROS5 | Xylem precursors | Schlereth et al. (2010); De Rybel et al. (2013) |
| pSMXL5 | SMAX1-LIKE5 | Phloem (precursors) | Wallner et al. (2017) |
| pCASP1 | CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN1 | Endodermis | Roppolo et al. (2011) |
| pVND7 | VASCULAR RELATED NAC-DOMAIN PROTEIN7 | Protoxylem (differentiating) in root, vessels in stem | Kubo et al. (2005) |
| pAPL | ALTERED PHLOEM DEVELOPMENT | Phloem (differentiating) | Bonke et al. (2003) |
| pNST3 | NAC SECONDARY WALL THICKENING PROMOTING3 | Fibers | Mitsuda et al. (2007) |
| pWOX4 | WUSCHEL RELATED HOMEOBOX4 | (Pro)cambium | Hirakawa et al. (2010) |
| pLTP1 | LIPID TRANSFER PROTEIN1 | Epidermis in stem | Thoma et al. (1994) |
| pAT2G3830 | Pith | Valério et al. (2004) | |
| pML1 | MERISTEM LAYER1 | L1 layer, epidermis | Lu et al. (1996) |
| pCLV3 | CLAVATA3 | SAM stem cells | Fletcher et al. (1999) |
| pREV | REVOLUTA | SAM central zone | Otsuga et al. (2001) |
| pUFO | UNUSUAL FLOWER ORGANS | SAM peripheral zone | Levin and Meyerowitz (1995) |
| pCUC2 | CUP-SHAPED COTYLEDON2 | Boundaries in SAM and leaf | Aida et al. (1997) |
Validation of the Specificity of Driver Lines
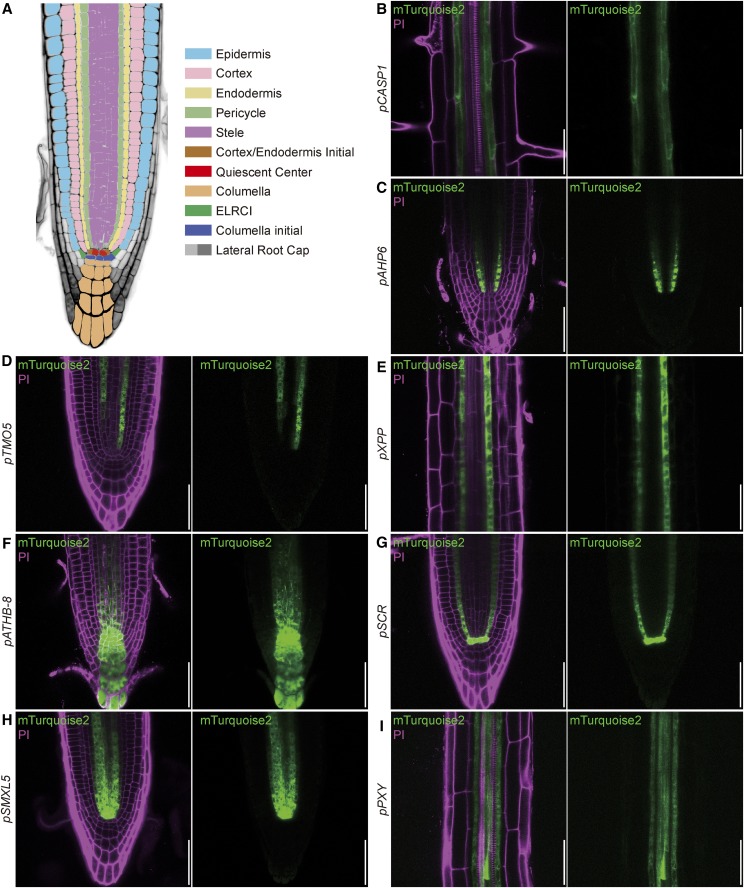
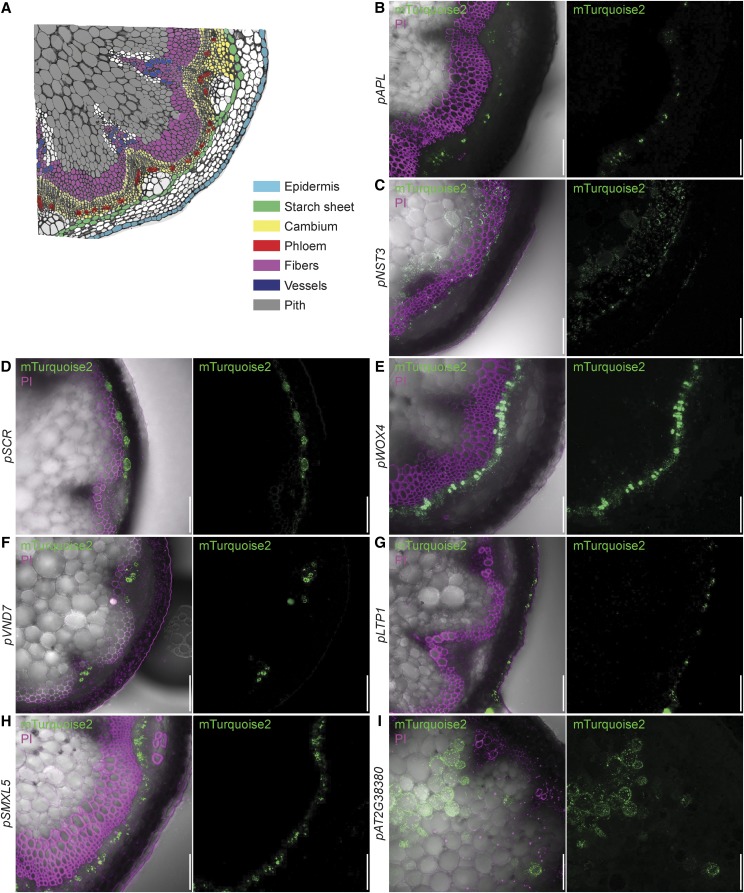
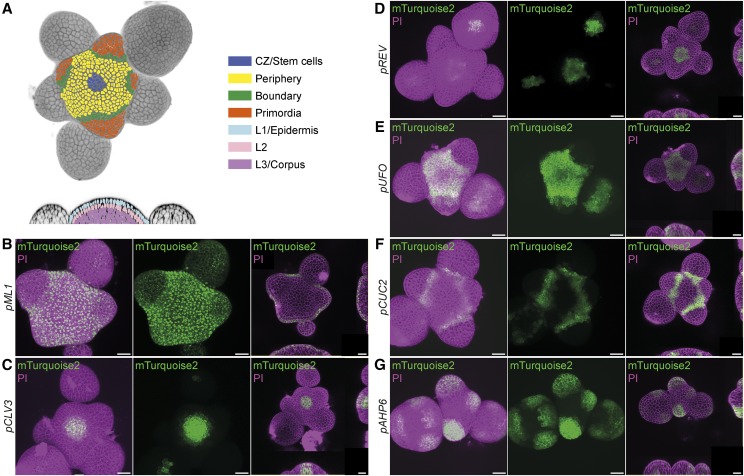
To confirm the expected expression patterns in the root, driver lines were germinated on medium containing 30 µm Dex or DMSO and analyzed with confocal laser scanning microscopy (CLSM) 5 d after germination (DAG). In each case, we recorded mTurquoise2-derived fluorescence in longitudinal optical sections of the root meristem (Fig. 2; Supplemental Fig. S1) and, where appropriate, in cross sections through the meristem or the differentiation zone (Supplemental Fig. S2). To visualize expression in the shoot, lines were grown on soil in long-day conditions and the aerial part of plants with 15-cm-tall inflorescence stems were dipped either in tap water containing 10 µm Dex (Fig. 3) or only the solvent DMSO (Supplemental Fig. S3). After 24 h, free-hand sections of the stem were stained with PI to highlight xylem elements and analyzed by confocal microscopy. To analyze expression in the SAM, inflorescence meristems of 15-cm-tall plants were treated with Dex 48 h before being dissected and imaged with CLSM, again using PI as a cell wall counterstain (Fig. 4). Reporter gene activities were consistent with the expected patterns and strictly dependent on the presence of Dex (Supplemental Figs. S1, S3, and S4). In addition, the complete absence of reporter activity in tissues adjacent to cells in which activity was expected suggested that the chimeric GR-LhG4 protein does not move between cells. We did not observe any negative effect of Dex treatment on plant growth (Supplemental Fig. S5).
Figure 2.
Analysis of induced driver lines in seedling roots. A, Schematic representation of root tissue layers. B to I, Induced driver line roots displaying fluorescence from propidium iodide (PI)-stained cell walls and the mTurquoise2 reporter (Fig. 1; Table 1). The indicated promoters mediate expression in the differentiating endodermis (B; pCASPARIAN STRIP MEMBRANE DOMAIN PROTEIN1 [pCASP1]), phloem precursor cells and adjacent pericycle cells (C; pHISTIDINE PHOSPHOTRANSFER PROTEIN6 [pAHP6]), xylem precursor cells (D; pTARGET OF MONOPTEROS5 [pTMO5]), xylem pole pericycle cells (E; pXYLEM POLE PERICYCLE [pXPP]), stele initials, cortex/endodermis initial (CEI), and columella initials (F; pHOMEOBOX GENE8 [pATHB-8]), endodermis, CEI, and QC ([G]; pSCARECROW [pSCR]), stele initials, phloem, and procambial cells (H; pSMAX1-LIKE5 [pSMXL5]), and procambial cells (I; pPHLOEM INTERCALATED WITH XYLEM [pPXY]). PI fluorescence is false colored in magenta, and mTurquoise2 fluorescence is false colored in green. Bars = 50 μm.
Figure 3.
Analysis of induced driver lines in the stem. A, Schematic representation of inflorescence stem tissue layers. B to I, Induced driver line stems displaying fluorescence from PI-stained cell walls and the mTurquoise2 reporter (Fig. 1; Table 1). The promoters mediate expression in differentiated phloem (B; pALTERED PHLOEM DEVELOPMENT [pAPL]), xylem fibers and interfascicular fibers (C; pNAC SECONDARY WALL THICKENING PROMOTING3 [pNST3]), starch sheath (D; pSCR), cambium (E; pWUSCHEL RELATED HOMEOBOX4 [pWOX4]), xylem vessels (F; pVASCULAR RELATED NAC DOMAIN PROTEIN7 [pVND7]), epidermal cells (G; pLIPID TRANSFER PROTEIN1 [pLTP1]), the incipient phloem (H; pSMXL5), and pith (I; pAT2G38380). PI fluorescence is false colored in magenta, and mTurquoise2 fluorescence is false colored in green. Bars = 50 μm.
Figure 4.
Analysis of induced driver lines in the SAM. A, Schematic representation of cell identity domains in the SAM. B to G, Induced driver line stems displaying fluorescence from PI-stained cell walls and the mTurquoise2 reporter (Fig. 1; Table 1). The left and middle images are maximum projections of confocal stack, and the right images consist of a single median confocal xy section and xz and yz views of the stack. The indicated promoters mediate expression in the L1 layer/epidermis (B; pMERISTEM LAYER1 [pML1]), the stem cell domain (C; pCLV3), the central zone (D; pREVOLUTA [pREV]), the peripheral zone (E; pUNUSUAL FLOWER ORGANS [pUFO]), the boundary domain (F; pCUP-SHAPED COTYLEDON [pCUC2]), and organ primordia (G; pAHP6). PI fluorescence is false colored in magenta, and mTurquoise2 fluorescence is false colored in green. Bars = 20 µm.
Characterization of Gene Activation
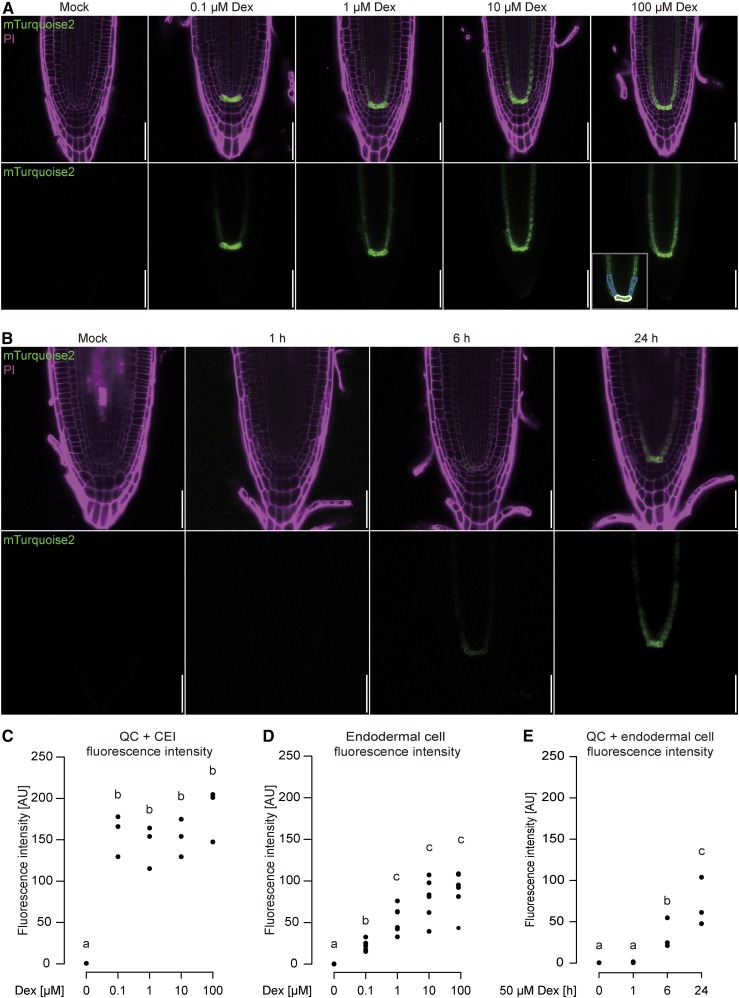
We next tested whether the dose-response and induction dynamics observed previously with the GR-LhG4 system (Craft et al., 2005) were recapitulated in our setup. To this end, we germinated the pSCR driver line mediating GR-LhG4 expression in the QC and the endodermis (Di Laurenzio et al., 1996; Wysocka-Diller et al., 2000) on plates containing solvent or 0.1, 1, 10, or 100 µm Dex. Visualizing reporter fluorescence 5 DAG indeed revealed increasing reporter activity with increasing Dex concentrations (Fig. 5A), arguing for the possibility to fine-tune gene expression by adjusting the levels of the inducer. We noticed that QC cells showed markedly stronger fluorescence compared with the endodermis, putatively reflecting higher promoter GR-LhG4/reporter stability in the QC, as this was not observed with previously published lines using the same promoter fragment (Gallagher et al., 2004; Heidstra et al., 2004; Cruz-Ramírez et al., 2013). Therefore, we quantified fluorescence separately in the QC cells and the endodermal initials (Fig. 5, C and D). Whereas the QC did not show a significant difference in fluorescence intensity between any of the treatments (Fig. 5C), the endodermis fluorescence intensity correlated with the concentration of the inducer until saturation appeared to be reached between 10 and 100 µm Dex (Fig. 5D). Consequently, we concluded that, to fine-tune gene expression by applying different Dex concentrations, the appropriate concentration range has to be determined for each promoter and cell type individually.
Figure 5.
Dose-response and time-course analyses of driver line seedling roots. A, The pSCR driver line was grown on 0, 0.1, 1, 10, and 100 µm Dex and imaged 5 DAG. B, Time course of pSCR driver line induction for 1, 6, and 24 h with 10 µm Dex. Bars = 50 µm. C, Quantification of the mTurquoise2 fluorescence intensity dose response in QC cells and CEI (cells outlined in white in A). D, Quantification of mTurquoise2 fluorescence intensity of the first three endodermal cells after the CEI (cells outlined in blue in A). E, Quantification of the induction time course (B) in QC cells, CEI, and the first three endodermal cells. Significant differences in C to E are based on the results of a two-tailed Student’s t test with P < 0.05 (a), P < 0.01 (b), and P < 0.001 (c), n = 3 to 6 roots each. AU, arbitrary units.
To further assess induction kinetics, the pSCR driver line was germinated on plates with control medium and transferred onto plates containing 50 µm Dex after 5 d. As expected, a time-dependent increase of reporter activity was observed over a period of 24 h (Fig. 5B). Combined quantification of fluorescence in the QC and the endodermis initials detected reporter activity 6 h after induction (Fig. 5E), and the activity values were close to the values of constitutive Dex treatment after 24 h (Fig. 5, D and E). These observations suggested that 6 h are sufficient to allow the nuclear import of GR-LhG4, the induction of gene transcription, and initial protein translation and that, within 24 h, protein levels reached a steady-state level. In addition, 5-d-old roots that were induced at 2, 3, or 4 DAG showed similar reporter activities, demonstrating that responsiveness to the inducer is sustainable (Supplemental Fig. S6). To assess the kinetics of reporter expression after removal of the inducer, we germinated the pSCR>GR>mTurquoise2 line on Dex-containing medium and transferred the seedlings to Dex-free medium 2 DAG. Quantifying reporter fluorescence revealed that, 1 d after transfer, fluorescence intensity was indistinguishable from that in control plants transferred to inducer-containing plates but declined over the course of the next 2 d to hardly detectable levels (Supplemental Fig. S7).
To estimate the level of transcription mediated by the GR-LhG4/pOp system, we employed a line expressing PECTIN METHYLESTERASE INHIBITOR5 (PMEI5; Wolf et al., 2012) under the control of the strong and nearly ubiquitous 35S promoter (p35S:PMEI5). When comparing roots from the p35S:PMEI5 line with roots from a Dex-treated GR-LhG4/pOp line conferring expression of the same PMEI5 coding sequence in xylem pole pericycle (XPP) cells (designated as pXPP>GR>PMEI5; Craft et al., 2005), we observed PMEI5 transcript levels similar to or slightly exceeding those in the p35S:PMEI5 line (Supplemental Fig. S8). This was despite the fact that the XPP expression domain contains only approximately six cell files in the young root (Supplemental Fig. S2). Thus, we concluded that, although activating transcription in a very local manner, the GR-LhG4/pOp system can lead to strong expression in the respective cell types.
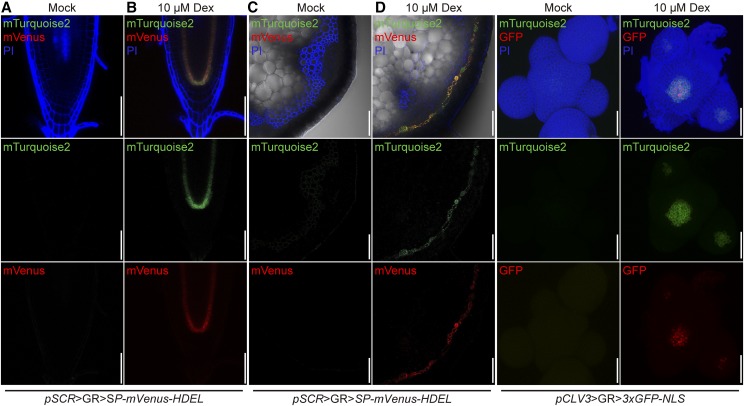
The ER-localized mTurquoise2 reporter present in our driver lines is transcribed from the same T-DNA that harbors the GR-LhG4 module (Fig. 1). To analyze the response of an independent T-DNA insertion carrying the pOp6 element in trans, we generated a transgenic line carrying an ER-targeted mVenus reporter under the control of the pOp6 promoter (pOp6:SP-mVenus-HDEL) and crossed it with the pSCR driver line. The resulting F1 plants did not show any reporter activity when grown on plates without Dex (Fig. 6), again confirming that the GR-LhG4/pOp system is fully Dex dependent. After Dex induction, we visualized both mTurquoise2 and mVenus fluorescence in the root and the stem and observed a complete congruence of both reporter activities (Fig. 6). Likewise, transgenic lines expressing a nucleus-targeted triple GFP fusion protein under the control of the pOp6 promoter were generated and crossed with the pCLAVATA3 (CLV3) driver line mediating expression in stem cells of the SAM (Fletcher et al., 1999). As expected, upon Dex induction, the 3xGFP-NLS signal was observed in a narrow domain at the tip of the SAM, which also expressed the mTurquoise2 marker (Fig. 6). Together, these observations confirmed the robust and specific transactivation of transgenes in F1 plants.
Figure 6.
Induction of mTurquoise2 and mVenus/3xGFP fluorescence in the root, stem, and SAM of F1 plants from a driver line-effector line cross. Cells are counterstained with PI (which, in the stem, highlights lignified vessel elements and fibers). Fluorescence channels are false colored. Bars = 50 µm for the root and the stem and 40 µm for the SAM.
Cell Type-Specific Induction of VND7 Demonstrates the Efficacy of Transactivation
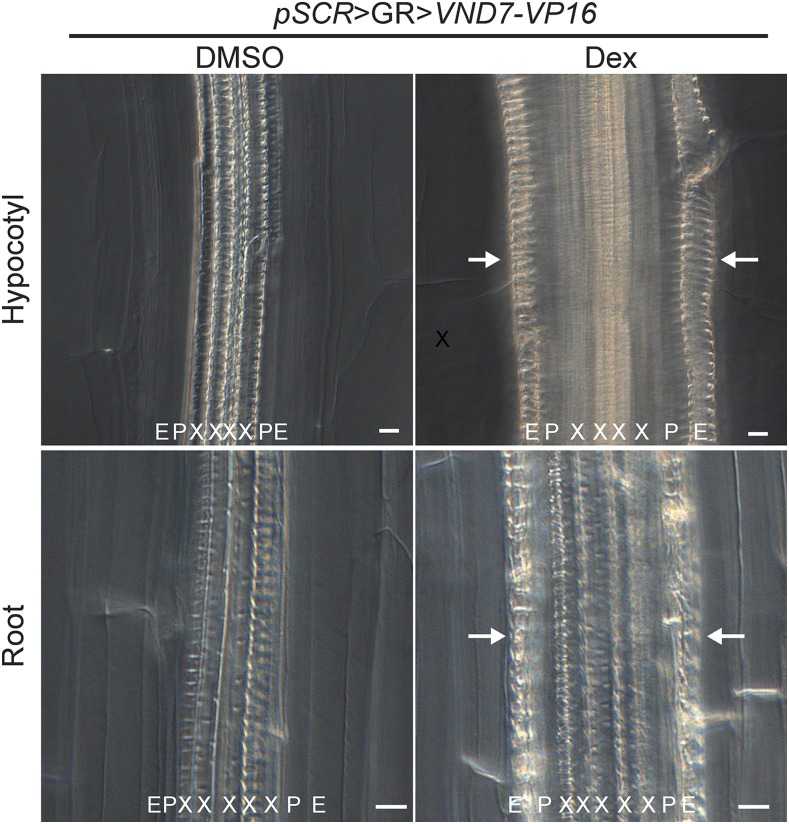
To explore the potential of our lines to mediate the expression of a biologically active effector, we crossed the pSCR driver line with a line harboring the VND7 effector fused to the VP16 activation domain able to induce the formation of xylem vessels in a broad range of cell types (Kubo et al., 2005; Yamaguchi et al., 2010). F1 plants were grown on control medium for 5 d and then transferred to medium containing either 10 µm Dex or solvent. Five days later, fully differentiated vessel-like elements could be observed in the endodermis of the root and hypocotyl (Fig. 7), whereas in DMSO-treated controls, xylem elements were clearly restricted to the stele. These results demonstrate that this resource for cell type-specific and inducible transactivation can be used to study gene function with high spatiotemporal resolution.
Figure 7.
Cell type-specific induction demonstrates the efficacy of transactivation. Plants expressing VND7-VP16 as an effector in the endodermal cells (pSCR>GR>VND7-VP16) show ectopic vessel formation (white arrows) after 5 d of Dex induction in both root and hypocotyl endodermis, in contrast to DMSO-treated plants. The spiral secondary cell wall thickening was observed after fixing and clearing the samples and visualized by differential interference contrast microscopy. E, Endodermis; P, pericycle; X, xylem. Bars = 20 μm.
DISCUSSION
In this study, we combined the proven efficacy of the well-established GR-LhG4/pOp expression system (Craft et al., 2005; Rutherford et al., 2005; Samalova et al., 2005) with the ease of cloning enabled by the GreenGate system (Lampropoulos et al., 2013) to provide a comprehensive toolbox for inducible, cell type-specific expression in Arabidopsis. The driver lines described here cover a large proportion of the known cell types in the three main meristems of the plant, the RAM, the SAM, and the cambium. Our analysis demonstrates that this system achieves nonleaky, adjustable, and robust transactivation of effectors in the F1 generation after crossing with effector-carrying plants. Therefore, generating a line harboring an effector cassette under the control of the pOp6 promoter should enable users to rapidly assess a battery of different expression regimes for a wide range of applications. In most cases, the effector might be the coding region of a gene one may want to misexpress in a spatially and temporally controlled manner, but other uses are conceivable, such as adjustable (pulsed) expression of reporters, domain-specific knockdown through artificial microRNAs, cell type-specific complementation studies, the acquisition of cell type-specific transcriptomes/translatomes/proteomes/epigenomes, or the local induction of genome editing, for example through the expression of Cre recombinase or CRISPR/Cas9 modules (Birnbaum et al., 2003; Brady et al., 2007; Dinneny et al., 2008; Gifford et al., 2008; Mustroph et al., 2009; Deal and Henikoff, 2011; Hacham et al., 2011; Iyer-Pascuzzi et al., 2011; Petricka et al., 2012; Fridman et al., 2014; Adrian et al., 2015; Vragović et al., 2015; Efroni et al., 2016; Kang et al., 2017). Thus, this system should be a valuable tool for the generation of inducible genetic perturbations to overcome the limitations of end-point genetics and to study genetic activities in specific tissue contexts.
Design of the Transactivation System
Two-component transactivation and chemically induced gene expression systems have been used widely by plant biologist in the past. For example, a large collection of enhancer-trapping lines based on the yeast Gal4 transcription factor (Haseloff, 1999; Engineer et al., 2005) are an invaluable tool for constitutive, tissue-specific transactivation in Arabidopsis (Aoyama and Chua, 1997; Sabatini et al., 2003; Weijers et al., 2003, 2005; Swarup et al., 2005). In addition, an inducible system based on Gal and cognate upstream activation sequence has been devised (Aoyama and Chua, 1997) but appears to induce unspecific growth defects (Kang et al., 1999). Transactivation based on LhG4 (Moore et al., 1998) shows only minimal detrimental effects on plant development, is thoroughly characterized and optimized (Moore et al., 1998, 2006; Baroux et al., 2005; Craft et al., 2005; Rutherford et al., 2005; Samalova et al., 2005), and has been used by the plant community in a number of studies (Schoof et al., 2000; Baroux et al., 2001; Eshed et al., 2001; Hay and Tsiantis, 2006; Nodine and Bartel, 2012; Sauret-Güeto et al., 2013; Hazak et al., 2014; Serrano-Mislata et al., 2015; Jiang and Berger, 2017). Parallel to the development of these tools for cell type-specific expression, a number of inducible systems have been conceived to enable temporal control of gene expression (Gatz et al., 1992; Weinmann et al., 1994; Caddick et al., 1998; Zuo et al., 2000). Subsequently, combining and optimizing the available technology has succeeded in generating tools to mediate inducible expression in a cell type-specific manner (Deveaux et al., 2003; Laufs et al., 2003; Maizel and Weigel, 2004; Craft et al., 2005).
For the generation of this resource, we built on groundbreaking previous work establishing the LhG4 system in combination with the GR ligand-binding domain (Craft et al., 2005), which has since been proven to be a valuable resource (Reddy and Meyerowitz, 2005; Ongaro et al., 2008; Ongaro and Leyser, 2008; Heisler et al., 2010; Jiang et al., 2011; Dello Ioio et al., 2012; Merelo et al., 2016; Caggiano et al., 2017; Tao et al., 2017). For the generation of our driver lines, we exploited the power of the GreenGate cloning system (Lampropoulos et al., 2013). We were able to rapidly assemble a large number of constructs efficiently, circumventing the bottleneck imposed previously by the challenging generation of large DNA constructs with varying promoter elements, coding regions, and terminators. Thus, the limiting factor in generating this resource was plant transformation and obtaining single-insertion, homozygous transgenic lines. As a general workflow, we aimed to generate at least 40 T1 transformants, then scored segregation ratios of antibiotic/herbicide resistance in the T2 generation and maintained lines in which the resistance segregated as a single locus. These lines usually showed similar characteristics concerning the response to the inducer and the expression levels achieved through transactivation (based on fluorescence intensity). Nevertheless, reporter expression in any set of newly generated driver lines should be assessed carefully and compared with the literature and within lines, as genome integration in the vicinity of endogenous promoter and/or enhancer elements might influence the expression pattern. As expected, we occasionally observed widespread silencing in the T2 generation of the driver lines, which did not correlate with any particular DNA element present in multiple constructs
An important feature of our driver lines is the incorporation of a reporter amenable to live imaging, which can be used to monitor the induction and visualize the spatial expression domain. In addition, it allows us to assess whether the expression of the effector has an impact on the transcriptional circuitries of the cell type it is expressed from. For some applications, the internal reporter of the driver lines also might serve as an inducible marker even in the absence of any further effector expression. We chose mTurquoise2 as a fluorescent reporter, since its spectral characteristics make it compatible with more widely used green and red fluorophores and it displays high photostability, fast maturation, and high quantum yield (Goedhart et al., 2012). The fluorescent protein was N-terminally fused with a signal peptide and modified with a C-terminal HDEL motif to mediate retention in the ER, which in our experience is the preferable subcellular localization for a fluorescent reporter when cross sections through the highly differentiated cells of the stem are required.
Transactivation Characteristics
Our system allows stringent temporal control of gene expression, as indicated by the lack of reporter expression in the absence of the inducer Dex. Moreover, the transactivated reporter faithfully reproduced previously described expression patterns associated with the respective 5′ regulatory regions, suggesting that the chimeric GR-LhG4 transcription factor is not cell-to-cell mobile. However, we noticed that, in some cases, transactivation led to slightly different expression patterns as compared with fusions of the same 5′ regulatory region with a reporter gene in cis. For example, expression driven from the CLV3 promoter seemed broader than what was described in pCLV3:XFP lines but consistent with a similarly designed pCLV3-driven transactivation (Serrano-Mislata et al., 2015), possibly because the multiple binding sites of the pOp6 promoter increase expression in cells where the CLV3 promoter is only weakly active. Alternatively, the high protein stability of the chimeric transcription factor, the reporter, or both might cause prolonged activity of these proteins in cells that are already displaced from the stem cell region. This potential issue is less relevant for organs such as the root, where cells of one cell type also largely have the same clonal identity (Kidner et al., 2000; Costa, 2016).
Our experiments, in agreement with previous results, suggested that GR-LhG4/pOp-mediated transactivation can achieve tissue-specific overexpression of the target gene, dependent on the concentration of the inducer. However, the possibility of squelching, the sequestration of general transcription factors required for other processes by the LhG4 activation domain, must be taken into account at very high expression levels. Consistent with previous reports (Craft et al., 2005), our analysis of the pSCR driver line revealed a linear dose response over at least 2 orders of magnitude, but the induction kinetics might be affected by the genomic location of the transgene and, thus, should be determined empirically for each line. It should be noted that the expression of effectors using LhG4/pOp systems can be quenched by adding isopropyl β-d-1-thiogalactopyranoside (Craft et al., 2005), which would allow pulsing experiments. However, we did not test the effect of isopropyl β-d-1-thiogalactopyranoside in our lines.
Distribution of Driver Lines and DNA Constructs
The lines described here, as well as DNA constructs, are available to the community upon request. While GR-LhG4 and the sulfadiazine resistance gene are expressed constitutively, care should be taken to amplify seeds only from noninduced plants to minimize the chance of inducing posttranscriptional gene silencing through the high expression levels of the reporter (Schubert et al., 2004; Abranches et al., 2005).
MATERIALS AND METHODS
Cloning
All constructs were produced by GreenGate cloning (Lampropoulos et al., 2013) using the modules described in Supplemental Table S1. The Eco31I (BsaI) sites of the SCR, PXY, and WOX4 promoters were removed by the QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies) using the primers listed in Supplemental Table S1 following the manufacturer’s instructions. The Eco31I site of the ATHB-8 promoter was removed by amplifying the 5′ part of the promoter up to the endogenous Eco31I restriction site, which was mutated by a single-base exchange in the primer. This primer contained an Eco31I restriction site in the 5′ overhang. The 3′ fragment of the promoter was amplified with a forward primer directed against the region immediately 3′ of the endogenous Eco31I site (containing an Eco31I site in the 5′ overhang) and the reverse primer binding to the region immediately upstream of the ATG. The two fragments were amplified separately, digested with Eco31I, and ligated afterward. As Eco31I is a type II restriction enzyme, the recognition site in the primer overhangs were removed by digestion.
The repetitive sequences of the pOp promoter increase the likelihood of recombination events while amplifying the plasmids. To discriminate against clones with shorter pOp sequences, we designed primers that bind in the short flanking sequences at the beginning and end of pOp6 (pOp6_F, 5′-TGCATATGTCGAGCTCAAGAA-3′; and pOp6_R, 5′-CTTATATAGAGGAAGGGTCTT-3′) for PCR amplification and size assessment through gel electrophoresis. Final constructs were always confirmed by sequencing in Escherichia coli and Agrobacterium tumefaciens. The occasional recombination events were detected only in E. coli.
Plant Material and Growth Conditions
All constructs were transformed by the floral dip method (Clough and Bent, 1998) as modified by Zhang et al. (2006) into Arabidopsis (Arabidopsis thaliana) Col-0. Transformed seeds were selected on half-strength Murashige and Skoog plates containing 1.875 to 3.75 µg mL−1 sulfadiazine or 7.5 µg mL−1 glufosinate ammonium. Only single integration lines based on T2 segregation ratios were propagated to T3, in which plants homozygous for the resistance were selected. All plants were grown in long-day conditions (16 h of light/8 h of dark) at 22°C. For root analysis, plants were grown vertically on half-strength Murashige and Skoog plates containing 1% (w/v) Suc and 0.9% (w/v) plant agar (Duchefa; P1001). For the induction treatments on plates, the seeds were sown on plates containing Dex (Sigma-Aldrich; D4903) at the indicated concentrations while the same volume of DMSO (D139-1; Fisher Scientific) was added for the mock control. For the transactivation experiment, seeds were sown on plates without Dex and seedlings were transferred to Dex-containing plates at 1, 6, and 24 h before imaging 5 DAG. For analysis of the stem, the aerial parts of 15-cm-tall plants were dipped for 30 s in either tap water containing 10 µm Dex with 0.02% (v/v) Silwet L-77 (Kurt Obermeier) or water with the same volume of DMSO with 0.02% (v/v) Silwet. After 24 h, free-hand sections of the stem were performed with a razor blade. Sections were transferred to a small petri dish (35/10 mm; Greiner Bio-One) with 0.25 mg mL−1 PI for 5 min and mounted on microscope slides to be visualized by CSLM. For SAM imaging, the inflorescence meristems of 25- to 30-DAG plants were sprayed with 10 µm Dex, whereas an equal volume of DMSO was added to water sprayed onto the mock controls. At 48 h after the treatment, the inflorescence meristems were dissected by cutting of the stem, flowers, and buds. The SAM was stained in 0.25 mg mL−1 PI (Sigma-Aldrich; P4170) for 5 min, mounted in a 3% (w/v) agarose small petri dish (35/10 mm; Greiner Bio-One), and visualized by CLSM.
Microscopy
Root samples were imaged using a Leica TCS SP5 laser scanning confocal microscope with an HCX PL APO lambda blue 63× water-immersion objective. The mTurquoise2 fluorophore was excited by an argon laser at 458 nm, and emission was collected between 460 and 516 nm. The mVenus fluorophore was excited by 514 nm, and emission was collected between 520 and 580 nm. Cells were counterstained by PI (Sigma-Aldrich; P4170) and imaged with 488 nm for excitation, and emission was collected between 590 and 660 nm.
For stem and SAM samples, we used a Nikon (Minato) A1 confocal microscope with a CFI Apo LWD 25× water-immersion objective. The PI-counterstained cells were imaged with 561 nm for excitation and 570 to 620 nm for emission. mTurquoise2 fluorescence was acquired using excitation at 405 nm, and emission was collected between 425 and 475 nm. For the transactivation experiments, the 3xGFP-NLS signal in the SAM was imaged with 488 nm for excitation and 500 to 550 nm for emission. In the root, mVenus was excited with 514 nm, and the emission was collected between 500 and 550 nm.
For visualization of the xylem, plants were germinated on half-strength Murashige and Skoog plates and transferred 5 DAG to either 10 µm Dex- or mock solution-containing half-strength Murashige and Skoog plates. To visualize ectopic xylem formation, plants were collected 5 d after induction and fixed overnight in a 1:3 acetic acid:ethanol solution. Then, they were cleared in a 8:1:2 chloral hydrate:glycerol:water solution for at least 3 h. Samples were mounted on microscope slides containing 50% (v/v) glycerol solution, and bright-field images were obtained using an Axioimager M1 microscope equipped with an AxioCamHRc (Carl Zeiss).
qPCR and SA-qPCR Analyses
Analysis of PMEI5 expression by qPCR was performed as described (Wolf et al., 2012). For SA-qPCR, plant DNA extraction was performed as described by Allen et al. (2006) and SA-qPCR was performed as described (Huang et al., 2013). Quadruplicate qPCR was performed in a final volume of 12.5 µL, including 6.25 µL of ABsolute qPCR SYBR Green Mix (Thermo Scientific), 0.25 µL of each primer (10 µm), and 2 µL of genomic DNA (1.6 ng µL−1) with different amounts (0, 1, or 3 µL) of plasmid (0.1 pg µL−1) as a reference. The SulfR resistance gene was amplified with primers SulfR_Fwd (5′-GCATGATCTAACCCTCTGTCTC-3′) and SulfR_Rvs (5′-GAAGTCACTCGTTCCCACTAG-3′), and the plasmid target sequence was amplified with PL_Fwd (5′-GCCGTACTAAACCTCTCATCG-3′) and PL_Rvs (5′-CTGACCGGAAAGTTTGTTATTCG-3′).
Accession Numbers
The Arabidopsis Genome Initiative numbers of genes used in this study are as follows: SCR (AT3G54220), ATHB-8 (AT4G32880), XPP (At4g30450), AHP6 (AT1G80100), PXY (AT5G61480), TMO5 (AT3G25710), SMXL5 (AT5G57130), CASP1 (AT2G36100), VND7 (AT1G71930), APL (AT1G79430), NST3 (AT1G32770), WOX4 (AT1G46480), PMEI5 (AT2G31430), LTP1 (AT2G38540), ML1 (AT4G21750), CLV3 (AT2G27250), REV (AT5G60690), UFO (AT1G30950), and CUC2 (AT5G53950).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Analysis of DMSO-treated mock controls for driver line seedling root induction 5 DAG.
Supplemental Figure S2. Analysis of induced driver lines in 5-DAG seedling root.
Supplemental Figure S3. Analysis of DMSO-treated driver lines in the stem.
Supplemental Figure S4. Analysis of DMSO-treated driver lines in the SAM.
Supplemental Figure S5. Growth on 50 µm Dex does not impair root growth of Col-0.
Supplemental Figure S6. Reporter activation in the pSCR>GR>mTurquoise2 line is sustainable.
Supplemental Figure S7. Kinetics of pSCR>GR>mTurquoise2 reporter activity after removal of inducer.
Supplemental Figure S8. Quantification of GR-LhG4-mediated transactivation.
Supplemental Table S1. List of primers used and DNA constructs generated in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the members of the Greb, Wolf, and Lohmann laboratories, Alexis Maizel (all at COS, Heidelberg University, Germany), and Joop Vermeer (University of Zürich, Switzerland) for discussion and support.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (Grants WO 1660/2-1 and WO 1660/6-1 to S.W. and Grant GR 2104/4-1 to T.G.) and a European Research Council consolidator grant (PLANTSTEMS 647148) to T.G.
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Abranches R, Shultz RW, Thompson WF, Allen GC (2005) Matrix attachment regions and regulated transcription increase and stabilize transgene expression. Plant Biotechnol J 3: 535–543 [DOI] [PubMed] [Google Scholar]
- Adrian J, Chang J, Ballenger CE, Bargmann BO, Alassimone J, Davies KA, Lau OS, Matos JL, Hachez C, Lanctot A, et al. (2015) Transcriptome dynamics of the stomatal lineage: birth, amplification, and termination of a self-renewing population. Dev Cell 33: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1: 2320–2325 [DOI] [PubMed] [Google Scholar]
- Andersen TG, Naseer S, Ursache R, Wybouw B, Smet W, De Rybel B, Vermeer JEM, Geldner N (2018) Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature 555: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua NH (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G (1995) The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121: 4171–4182 [DOI] [PubMed] [Google Scholar]
- Baroux C, Blanvillain R, Moore IR, Gallois P (2001) Transactivation of BARNASE under the AtLTP1 promoter affects the basal pole of the embryo and shoot development of the adult plant in Arabidopsis. Plant J 28: 503–515 [DOI] [PubMed] [Google Scholar]
- Baroux C, Blanvillain R, Betts H, Batoko H, Craft J, Martinez A, Gallois P, Moore I (2005) Predictable activation of tissue-specific expression from a single gene locus using the pOp/LhG4 transactivation system in Arabidopsis. Plant Biotechnol J 3: 91–101 [DOI] [PubMed] [Google Scholar]
- Besnard F, Refahi Y, Morin V, Marteaux B, Brunoud G, Chambrier P, Rozier F, Mirabet V, Legrand J, Lainé S, et al. (2014) Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 505: 417–421 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Bonke M, Thitamadee S, Mähönen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426: 181–186 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Caddick MX, Greenland AJ, Jepson I, Krause KP, Qu N, Riddell KV, Salter MG, Schuch W, Sonnewald U, Tomsett AB (1998) An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat Biotechnol 16: 177–180 [DOI] [PubMed] [Google Scholar]
- Caggiano MP, Yu X, Bhatia N, Larsson A, Ram H, Ohno CK, Sappl P, Meyerowitz EM, Jönsson H, Heisler MG (2017) Cell type boundaries organize plant development. eLife 6: e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY (2015) Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol 25: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costa S. (2016) Cell identity: a matter of lineage and neighbours. New Phytol 210: 1155–1158 [DOI] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I (2005) New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J 41: 899–918 [DOI] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Wachsman G, Du Y, Arteága-Vázquez M, Zhang H, Benjamins R, Blilou I, Neef AB, Chandler V, et al. (2013) A SCARECROW-RETINOBLASTOMA protein network controls protective quiescence in the Arabidopsis root stem cell organizer. PLoS Biol 11: e1001724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff S (2011) The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc 6: 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V, Scheres B, Sabatini S, Baulcombe D, Maini PK, et al. (2012) A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr Biol 22: 1699–1704 [DOI] [PubMed] [Google Scholar]
- De Rybel B, Möller B, Yoshida S, Grabowicz I, Barbier de Reuille P, Boeren S, Smith RS, Borst JW, Weijers D (2013) A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell 24: 426–437 [DOI] [PubMed] [Google Scholar]
- Deveaux Y, Peaucelle A, Roberts GR, Coen E, Simon R, Mizukami Y, Traas J, Murray JA, Doonan JH, Laufs P (2003) The ethanol switch: a tool for tissue-specific gene induction during plant development. Plant J 36: 918–930 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N (2017) Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355: 280–284 [DOI] [PubMed] [Google Scholar]
- Efroni I. (2018) A conceptual framework for cell identity transitions in plants. Plant Cell Physioll 59: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD (2016) Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineer CB, Fitzsimmons KC, Schmuke JJ, Dotson SB, Kranz RG (2005) Development and evaluation of a Gal4-mediated LUC/GFP/GUS enhancer trap system in Arabidopsis. BMC Plant Biol 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL (2001) Establishment of polarity in lateral organs of plants. Curr Biol 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Fisher K, Turner S (2007) PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr Biol 17: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Fridman Y, Elkouby L, Holland N, Vragović K, Elbaum R, Savaldi-Goldstein S (2014) Root growth is modulated by differential hormonal sensitivity in neighboring cells. Genes Dev 28: 912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta KM, Hellmann E, Helariutta Y (2014) Molecular control of cell specification and cell differentiation during procambial development. Annu Rev Plant Biol 65: 607–638 [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN (2004) Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 14: 1847–1851 [DOI] [PubMed] [Google Scholar]
- Gatz C, Frohberg C, Wendenburg R (1992) Stringent repression and homogeneous de-repression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J 2: 397–404 [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TW Jr, Royant A (2012) Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun 3: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Lohmann JU (2016) Plant stem cells. Curr Biol 26: R816–R821 [DOI] [PubMed] [Google Scholar]
- Hacham Y, Holland N, Butterfield C, Ubeda-Tomas S, Bennett MJ, Chory J, Savaldi-Goldstein S (2011) Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J. (1999) GFP variants for multispectral imaging of living cells. Methods Cell Biol 58: 139–151 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M (2006) The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hazak O, Obolski U, Prat T, Friml J, Hadany L, Yalovsky S (2014) Bimodal regulation of ICR1 levels generates self-organizing auxin distribution. Proc Natl Acad Sci USA 111: E5471–E5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B (2004) Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev 18: 1964–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, Traas J, Meyerowitz EM (2010) Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8: e1000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H (2010) TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yin X, Zhu C, Wang W, Grierson D, Xu C, Chen K (2013) Standard addition quantitative real-time PCR (SAQPCR): a novel approach for determination of transgene copy number avoiding PCR efficiency estimation. PLoS ONE 8: e53489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, Jackson T, Cui H, Petricka JJ, Busch W, Tsukagoshi H, Benfey PN (2011) Cell identity regulators link development and stress responses in the Arabidopsis root. Dev Cell 21: 770–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Berger F (2017) DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 357: 1146–1149 [DOI] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y (2011) Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet 7: e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Fang Y, Singh KB (1999) A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J 20: 127–133 [DOI] [PubMed] [Google Scholar]
- Kang YH, Breda A, Hardtke CS (2017) Brassinosteroid signaling directs formative cell divisions and protophloem differentiation in Arabidopsis root meristems. Development 144: 272–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner C, Sundaresan V, Roberts K, Dolan L (2000) Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211: 191–199 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate: a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs P, Coen E, Kronenberger J, Traas J, Doonan J (2003) Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development 130: 785–796 [DOI] [PubMed] [Google Scholar]
- Lehming N, Sartorius J, Niemöller M, Genenger G, v Wilcken-Bergmann B, Müller-Hill B (1987) The interaction of the recognition helix of lac repressor with lac operator. EMBO J 6: 3145–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JZ, Meyerowitz EM (1995) UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7: 529–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Porat R, Nadeau JA, O’Neill SD (1996) Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kinoshita K, Törmäkangas K, Ikeda Y, Oka A, Kakimoto T, Helariutta Y (2006) Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 311: 94–98 [DOI] [PubMed] [Google Scholar]
- Maizel A, Weigel D (2004) Temporally and spatially controlled induction of gene expression in Arabidopsis thaliana. Plant J 38: 164–171 [DOI] [PubMed] [Google Scholar]
- Marquès-Bueno MDM, Morao AK, Cayrel A, Platre MP, Barberon M, Caillieux E, Colot V, Jaillais Y, Roudier F, Vert G (2016) A versatile Multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J 85: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merelo P, Ram H, Pia Caggiano M, Ohno C, Ott F, Straub D, Graeff M, Cho SK, Yang SW, Wenkel S, et al. (2016) Regulation of MIR165/166 by class II and class III homeodomain leucine zipper proteins establishes leaf polarity. Proc Natl Acad Sci USA 113: 11973–11978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Koi S, Hashimoto T, Nakajima K (2011) Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138: 2303–2313 [DOI] [PubMed] [Google Scholar]
- Moore I, Gälweiler L, Grosskopf D, Schell J, Palme K (1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 95: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Samalova M, Kurup S (2006) Transactivated and chemically inducible gene expression in plants. Plant J 45: 651–683 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJ, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Naseer S, Lee Y, Lapierre C, Franke R, Nawrath C, Geldner N (2012) Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proc Natl Acad Sci USA 109: 10101–10106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Saegusa M, Iwamoto K, Oda Y, Katayama H, Kojima M, Sakakibara H, Fukuda H (2014) A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr Biol 24: 2053–2058 [DOI] [PubMed] [Google Scholar]
- Ongaro V, Leyser O (2008) Hormonal control of shoot branching. J Exp Bot 59: 67–74 [DOI] [PubMed] [Google Scholar]
- Ongaro V, Bainbridge K, Williamson L, Leyser O (2008) Interactions between axillary branches of Arabidopsis. Mol Plant 1: 388–400 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25: 223–236 [DOI] [PubMed] [Google Scholar]
- Pacifici E, Polverari L, Sabatini S (2015) Plant hormone cross-talk: the pivot of root growth. J Exp Bot 66: 1113–1121 [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, et al. (2012) The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci USA 109: 6811–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. (1993) Steroid-binding domains for regulating the functions of heterologous proteins in cis. Trends Cell Biol 3: 278–280 [DOI] [PubMed] [Google Scholar]
- Reddy GV, Meyerowitz EM (2005) Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310: 663–667 [DOI] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Dénervaud Tendon V, Pfister A, Alassimone J, Vermeer JE, Yamazaki M, Stierhof YD, Beeckman T, Geldner N (2011) A novel protein family mediates Casparian strip formation in the endodermis. Nature 473: 380–383 [DOI] [PubMed] [Google Scholar]
- Rutherford S, Brandizzi F, Townley H, Craft J, Wang Y, Jepson I, Martinez A, Moore I (2005) Improved transcriptional activators and their use in mis-expression traps in Arabidopsis. Plant J 43: 769–788 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samalova M, Brzobohaty B, Moore I (2005) pOp6/LhGR: a stringently regulated and highly responsive dexamethasone-inducible gene expression system for tobacco. Plant J 41: 919–935 [DOI] [PubMed] [Google Scholar]
- Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E (2013) JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biol 11: e1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644 [DOI] [PubMed] [Google Scholar]
- Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16: 2561–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Mislata A, Schiessl K, Sablowski R (2015) Active control of cell size generates spatial detail during plant organogenesis. Curr Biol 25: 2991–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siligato R, Wang X, Yadav SR, Lehesranta S, Ma G, Ursache R, Sevilem I, Zhang J, Gorte M, Prasad K, et al. (2016) MultiSite Gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol 170: 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ (2005) Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat Cell Biol 7: 1057–1065 [DOI] [PubMed] [Google Scholar]
- Tao Z, Shen L, Gu X, Wang Y, Yu H, He Y (2017) Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551: 124–128 [DOI] [PubMed] [Google Scholar]
- Thoma S, Hecht U, Kippers A, Botella J, De Vries S, Somerville C (1994) Tissue-specific expression of a gene encoding a cell wall-localized lipid transfer protein from Arabidopsis. Plant Physiol 105: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truskina J, Vernoux T (2018) The growth of a stable stationary structure: coordinating cell behavior and patterning at the shoot apical meristem. Curr Opin Plant Biol 41: 83–88 [DOI] [PubMed] [Google Scholar]
- Valério L, De Meyer M, Penel C, Dunand C (2004) Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry 65: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. (2011) Callose biosynthesis regulates symplastic trafficking during root development. Dev Cell 21: 1144–1155 [DOI] [PubMed] [Google Scholar]
- Vragović K, Sela A, Friedlander-Shani L, Fridman Y, Hacham Y, Holland N, Bartom E, Mockler TC, Savaldi-Goldstein S (2015) Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc Natl Acad Sci USA 112: 923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner ES, López-Salmerón V, Belevich I, Poschet G, Jung I, Grünwald K, Sevilem I, Jokitalo E, Hell R, Helariutta Y, et al. (2017) Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr Biol 27: 1241–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y (2014) The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Wagner D (2016) Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67: 539–574 [DOI] [PubMed] [Google Scholar]
- Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R (2003) Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol 133: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R (2005) Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17: 2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G (2006) Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10: 265–270 [DOI] [PubMed] [Google Scholar]
- Weinmann P, Gossen M, Hillen W, Bujard H, Gatz C (1994) A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J 5: 559–569 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Höfte H (2012) Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22: 1732–1737 [DOI] [PubMed] [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN (2000) Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development 127: 595–603 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Goué N, Igarashi H, Ohtani M, Nakano Y, Mortimer JC, Nishikubo N, Kubo M, Katayama Y, Kakegawa K, et al. (2010) VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol 153: 906–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]