The pollen sequestrome is found to be a highly dynamic compartment for the storage of translationally repressed mRNAs, and nontranslating monosomes are proposed to be the primary mediators of mRNA sequestration in Nicotiana tabacum.
Abstract
Reproduction success in angiosperm plants depends on robust pollen tube growth through the female pistil tissues to ensure successful fertilization. Accordingly, there is an apparent evolutionary trend to accumulate significant reserves during pollen maturation, including a population of stored mRNAs, that are utilized later for a massive translation of various proteins in growing pollen tubes. Here, we performed a thorough transcriptomic and proteomic analysis of stored and translated transcripts in three subcellular compartments of tobacco (Nicotiana tabacum), long-term storage EDTA/puromycin-resistant particles, translating polysomes, and free ribonuclear particles, throughout tobacco pollen development and in in vitro-growing pollen tubes. We demonstrated that the composition of the aforementioned complexes is not rigid and that numerous transcripts were redistributed among these complexes during pollen development, which may represent an important mechanism of translational regulation. Therefore, we defined the pollen sequestrome as a distinct and highly dynamic compartment for the storage of stable, translationally repressed transcripts and demonstrated its dynamics. We propose that EDTA/puromycin-resistant particle complexes represent aggregated nontranslating monosomes as the primary mediators of messenger RNA sequestration. Such organization is extremely useful in fast tip-growing pollen tubes, where rapid and orchestrated protein synthesis must take place in specific regions.
The male gametophyte, a highly organized haploid organ, offers a unique chance to analyze the development and differentiation of a single haploid cell, cell-cell interaction, and the recognition between the pollen grain and the stigma/transmitting tissue. Furthermore, cellular polarity and pollen tube tip growth can be studied easily. Continuous completion of genome sequencing, which has been accomplished in an increasing number of plant species, facilitates transcriptomic and proteomic studies in the male gametophyte (for review, see Fíla et al., 2017) and, in several cases, also of particular pollen developmental stages and the progamic phase. Analysis has been performed in species such as Arabidopsis (Arabidopsis thaliana; Honys and Twell, 2004; Wang et al., 2008; Qin et al., 2009; Ge et al., 2011), tomato (Solanum lycopersicum; Chaturvedi et al., 2013; Paul et al., 2016), tobacco (Nicotiana tabacum; Hafidh et al., 2012a, 2012b; Bokvaj et al., 2014; Ischebeck et al., 2014; Conze et al., 2017), and rice (Oryza sativa; Dai et al., 2006, 2007; Wei et al., 2010). Transcriptomic studies clearly showed that male gametophyte development is under the control of two subsequent developmental programs, early and late, accompanied by stage-specific gene expression patterns (Honys and Twell, 2004; Wei et al., 2010). Although the inhibition of transcription by actinomycin D did not influence early pollen tube growth (Lafleur and Mascarenhas, 1978), the translation inhibition by cycloheximide had an immediate and fatal impact on pollen germination and pollen tube growth (Čapková-Balatková et al., 1980; Honys and Twell, 2004). This clearly demonstrates the requirement for protein synthesis during pollen tube growth. Interestingly, pollen germination in some species is not affected by protein synthesis inhibitors, and de novo protein synthesis is only needed for later pollen tube growth and fertilization (Fernando, 2005). Accordingly, posttranscriptional regulation has been demonstrated to play a role in the fine spatial and temporal modulation of male gametophytic gene expression (Borges et al., 2008; Grant-Downton et al., 2009a, 2009b).
Therefore, it is no surprise that a number of pollen mRNAs were shown to accumulate in pollen-stored ribonucleoprotein (RNP) particles, which remain intact even in a buffer comprising strong detergents. This novel class of detergent-resistant RNP particles was annotated as EDTA/puromycin-resistant particles or EPPs (Honys et al., 2000). The identification of the EPPs in the tobacco male gametophyte highlighted the presence of germ cell-like granules in flowering plants. In analogy to the role played by Drosophila melanogaster germinal granules in delivering maternal mRNAs during the initial stages of embryogenesis (Schisa, 2012), in our previous study (Honys et al., 2009), we isolated and characterized the proteome of EPPs and proposed that, during pollen maturation, the storage EPPs represent preloaded complex machinery devoted to mRNA processing, transport, subcellular localization, and protein synthesis (Honys et al., 2009). Discovery of the EPPs also supported the long-hypothesized presence of stored messenger ribonucleoprotein (mRNP) particles in developing tobacco male gametophytes (Honys et al., 2000, 2009). The physiological advantage that the EPPs convey is that, being composed of mRNA sets that are stored and translationally silenced at earlier stages of development, they enable an immediate activation of translation of selected mRNAs during germination of the pollen grain and subsequent pollen tube growth (Honys et al., 2009). This potential role of EPPs particularly resembles that of the growing axons of the human neuronal cells. The directional growth in neurons is facilitated by the transport of sequestered mRNAs by neuronal granules to the synaptic surfaces for translation (for review, see Buchan, 2014). Similar to EPPs, neuronal granules also are preloaded with translational machinery and represent the mediators of nerve cell networking, which deposit transcripts to the growing tip and catalyze their efficient translation, thereby promoting directional growth (Elvira et al., 2006; Hirokawa, 2006). During the progamic phase, an intense molecular dialogue occurs between male and female reproductive tissues (for review, see Vogler et al., 2016) that, among others, involves proteins secreted by growing pollen tubes (Hafidh et al., 2016). In Arabidopsis, a genome-wide study of the in vivo pollen tube translatome revealed that transcripts encoding pollen-secreted proteins are involved in pollen guidance (Lin et al., 2014).
Here, we present a thorough transcriptomic and proteomic analysis of stored and translated transcripts and their storage ribonucleoprotein particles throughout tobacco pollen development. Analysis was performed on microspores and early-late bicellular pollen as well as mature pollen grains and pollen tubes. We were able to define a pollen sequestrome, the entity of stored and translationally repressed transcripts, and its dynamics and propose that the nontranslating monosomes might serve as primary mediators of selective mRNA sequestration.
RESULTS
Definition of the Pollen Sequestrome
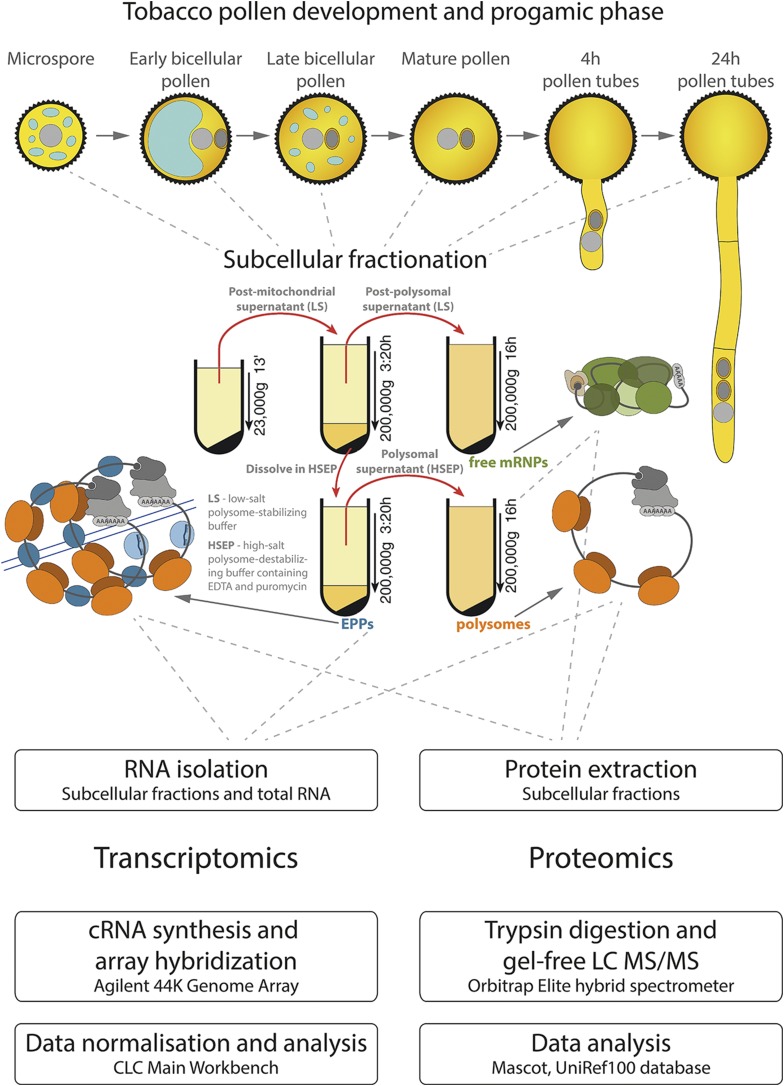
The objective of this work was the characterization of the extent and dynamics of translational regulation during tobacco male gametophyte development and the subsequent functional progamic phase. First, we aimed to demonstrate the different sets of transcripts comprising the three types of mRNA-containing ribonucleoprotein particles, polysomal RNPs (hereafter referred to as polysomes [POL]), free mRNPs (RNPs), and EPPs cosedimenting with polysomes (EPP complexes [EPP]). The ribonucleoprotein complexes were separated by two-step Suc gradient centrifugation in low-salt and high-salt gradient buffers (Fig. 1; Honys et al., 2000, 2009), and total RNA was isolated from each fraction. Based on the function of ribonucleoprotein particles (Honys et al., 2000, 2009) present in each fraction, their transcriptomes were marked as follows (Fig. 1): sequestrome (mRNA sequestered/long-term stored in the form of EPPs [EPP]; Honys et al., 2009), translatome (actively translated transcripts on polysomes; Lin et al., 2014), and mRNPome (transcripts present in free mRNP particles).
Figure 1.
Schematic representation of the experimental workflow.
For the analyses, six developmental stages of pollen development were used: uninucleate microspores (UNM), early bicellular pollen (eBCP), late bicellular pollen (lBCP), mature pollen (MPG), and in vitro-cultivated pollen tubes for 4 h (PT4) and 24 h (PT24; Fig. 1). The purity of the isolated spore populations was evaluated microscopically and was published in our earlier works (Hafidh et al., 2012a, 2012b; Bokvaj et al., 2014). In an effort to understand the nature and diversity of transcripts associated with the isolated fractions, we applied tobacco microarray hybridization (Imaxio) and performed comparative transcriptomic analysis. Normalized expression values are presented in Supplemental Table S1. For each sample, two biological replicates were used, and each value represents the respective mean. For systematic comparison of the tissue samples, only a subset of expressed genes was considered for analysis (see “Materials and Methods”). These genes had the detection call of present in both biological replicates in all three subcellular fractions as well as in the independent total transcriptome. Therefore, only the overlap of genes identified as being expressed in the total transcriptome and in all three subcellular fractions were considered further (Fig. 2A). Although there were differences between total RNA and pooled fractions, the affected genes were only those with weak expression signals. In all samples, overlapping genes comprised 70% to 90% of all genes expressed in at least one fraction or in the total transcriptome (Supplemental Fig. S1B). Similarly, principal component analysis (PCA) always grouped both samples of the same developmental stage (Supplemental Fig. S1C). Independently applied hierarchical clustering to visualize the relationship among the subcellular fractions in mature pollen (Fig. 2G) and correlation evaluation of all data sets (Supplemental Figs. S1D and S2) again demonstrated the close relationship of the corresponding samples. All these analyses unequivocally showed the consistency between the total transcriptome and pooled subcellular fractions. Moreover, the obtained normalized expression arrays were confirmed as being of good quality for downstream analyses.
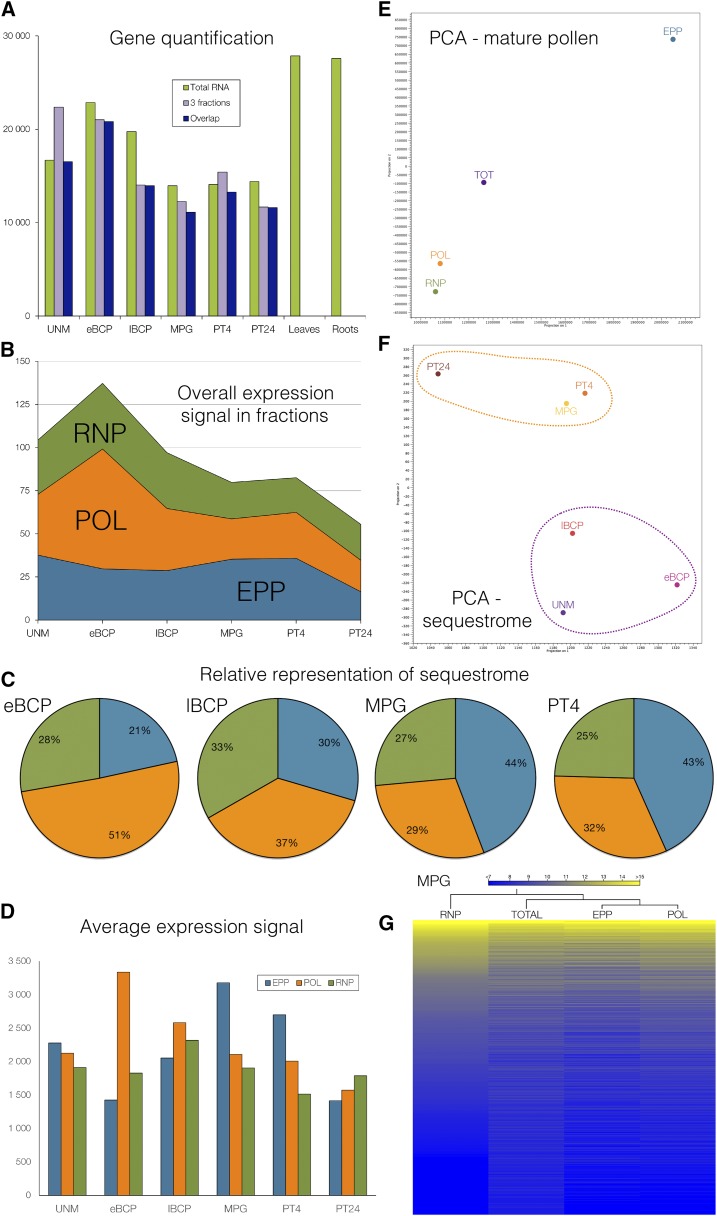
Figure 2.
Definition of the pollen sequestrome. A, Quantification of expressed transcripts in total RNA, transcripts expressed in all three fractions, and the overlap of both sets. B, Overall expression signal in all three fractions during pollen development and the progamic phase. C, Relative distribution of the expression signal between subcellular fractions in four stages of pollen development and the progamic phase. D, Average expression signal in each subcellular fraction during pollen development and the progamic phase. E, PCA of transcripts present in all three subcellular fractions and total RNA in MPG. F, PCA of transcripts forming the sequestrome fraction during pollen development and the progamic phase. G, Hierarchical clustering of transcripts present in all three subcellular fractions and in total RNA in mature pollen.
The quantification of expressed genes (Fig. 2A; Supplemental Fig. S1A) confirmed the general trends of general reduction of the total transcriptome complexity during pollen maturation followed by its slight increase after pollen germination. PCA and correlation analyses revealed the developmental shift in the very last period of pollen maturation, since the transcriptomes of immature pollen formed one group different from the transcriptomes of mature pollen and pollen tubes (Supplemental Fig. S3, D and E). In developing pollen, there were smaller differences between the transcriptomes of the subcellular fractions than later, in mature pollen and during the early progamic phase (MPG and PT4). The latter showed higher divergence of translationally active and stored transcript populations, as also confirmed by PCA showing the dynamics of the transcriptomes of the subcellular fractions in the male gametophyte and the relationship of the total and fraction transcriptomes of all six developmental stages (Supplemental Fig. S3C). We also selected five candidate genes with different expression profiles to verify their microarray expression by reverse transcription quantitative PCR (RT-qPCR) in mature pollen in three subcellular fractions (Supplemental Fig. S4).
On the basis of the selected criteria, 37,610 probes (85.9% of 43,803 probes present on the array) had a positive signal in at least one gametophytic or sporophytic data set and, thus, were considered expressed. Of them, 25,034 probes (66.6% of expressed genes) were present in the male gametophyte in at least one developmental stage. Similarly, a previous analysis of the tobacco transcriptome that utilized 40K custom-designed Affymetrix microarrays reported reliable expression of 76% of the probe sets in 19 different tobacco tissues, including leaves and roots (Edwards et al., 2010). The highest number of genes was expressed in early bicellular pollen (Fig. 2A; Supplemental Fig. S1A). This also was the developmental stage with the highest proportion of actively translated transcripts present in the polysomal fraction (Fig. 2, B and C) that gradually decreased later on. On the contrary, the proportion of the sequestrome increased during the final phases of pollen maturation and reached its maximum in mature pollen and 4-h cultivated pollen tubes (Fig. 2, B and C). The dominance of the sequestrome in MPG and PT4 was not only relative but also absolute (Fig. 2D), regardless of the fact that these two developmental stages expressed the lowest number of genes. Of them, the most abundant transcripts were stored in the sequestrome fraction (Supplemental Table S1).
The uniqueness of the total male gametophyte versus sporophyte transcriptomes was already demonstrated (Bokvaj et al., 2014; Supplemental Fig. S3, D and E). We extended the PCA to visualize the relationship of all individual total and fraction transcriptomes in pollen (Fig. 2E) that highlighted the sequestrome as the most unique fraction in mature pollen. The close proximity of all pairs of replicates confirmed the high reproducibility among replicates (Supplemental Fig. S3, B, E, and F). This distribution also was similar at other developmental stages (data not shown). However, the sequestrome dynamics resembled that of total RNA, being formed by a set of transcripts diverting from the sporophytic transcriptomes especially in the late progamic phase (Fig. 2F; Supplemental Fig. S3D). From the observed distribution, we concluded (1) a low degree of posttranscriptional regulation between free mRNPs and polysomes and (2) a great distance between two presumed RNA storage compartments, free mRNPs and EPPs. This suggests a complex and precise regulation of transcript distribution between these two compartments. Free mRNPs probably harbor transcripts that are on their way to becoming associated with polysomes, whereas EPPs were shown to more likely contain long-term-stored transcripts without immediate relation to translation, as already indicated by previously published results (Honys et al., 2009).
Analyses of transcriptome characteristics of individual transcripts not only showed great variation of their expression profiles at the level of absolute abundance but also allowed us to observe their distribution among three subfractions and their dynamics throughout pollen development and the progamic phase that is linked to their translational and storage status. This was highlighted in 17 clusters of transcripts grouped according to their storage and translation status throughout pollen development (Supplemental Fig. S5; Supplemental Table S2). These results confirmed the previous finding that the eBCP and lBCP stages contained the highest proportion of translated transcripts. Moreover, the mRNA storage in EPPs begins in bicellular pollen, and the transcripts stored there are preferably either short term stored (stored in eBCP and lBCP only) or long term stored (from eBCP to PT24; Supplemental Fig. S5).
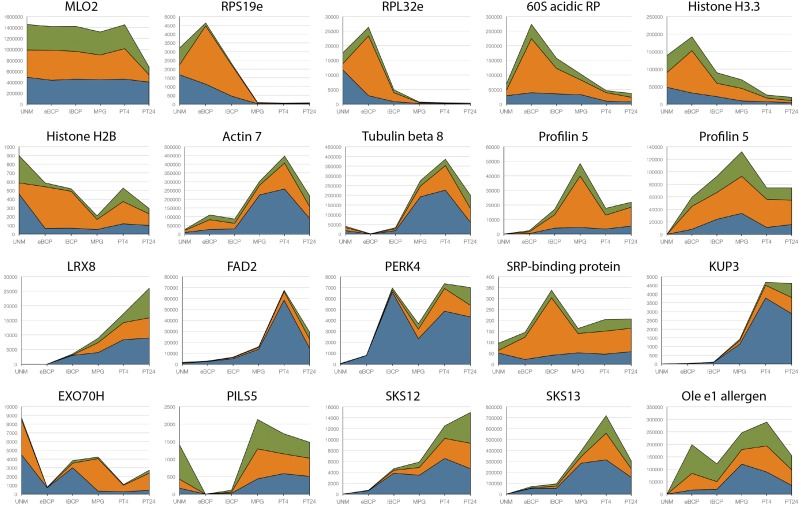
A few transcripts were selected to demonstrate the large variability of expression and translation/storage profiles among pollen-expressed transcripts (Fig. 3; Supplemental Table S3). The integral membrane Mildew resistance locus o family protein MLO2 (Devoto et al., 2003) exemplifies transcripts with uniform distribution among individual fractions throughout the whole male gametophyte development and progamic phase. Several transcripts encoding ribosomal proteins (RPs) and histones share early expression profiles being associated predominantly with polysomes. On the contrary, the abundance of transcripts encoding cytoskeletal proteins increases later, but their expression profiles differ. Actin7 and β-tubulin8 are the most abundant in growing pollen tubes and are likely to be subject to stronger translational regulation, whereas profilin5 isoforms are peaking in mature pollen, mostly in the polysomal fraction. Similar late expression profiles are characteristic for proteins involved in pollen tube growth and cell-cell communication. For example, the cell wall enzyme Nicotiana tabacum pollen-specific glycoprotein303 (NTP303) exemplified by its two paralogs, SKU5-similar12 (SKS12) and SKS13 (Sedbrook et al., 2002), Leu-rich repeat/extensin8 (LRX8), the K+ uptake transporter KUP3, and Pro-rich receptor-like kinase4 are putatively translationally regulated, whereas PIN FORMED (PIN)-like5 (Barbez et al., 2012; Dal Bosco et al., 2012) and Ole e1 allergen (de Dios Alché et al., 2004) are not. Interestingly, in Arabidopsis, LRX8 is among several pollen-expressed members of the LRX family that localize in the cell wall and were shown recently to be important for Arabidopsis pollen grain and pollen tube cell wall development and integrity (Fabrice et al., 2018; Sede et al., 2018). In addition, their deficiency negatively affects pollen germination (Wang et al., 2018). Moreover, LXR proteins interact with the small rapid alkalinization factors RALF4 and RALF19 to control pollen tube growth (Mecchia et al., 2017). Since RALFs and their regulatory potential were discovered in tobacco (Pearce et al., 2001) and then studied in several species (Murphy and De Smet, 2014), the observed presence, late expression profile, and translational regulation of the LXR8 tobacco homolog in pollen tubes are not surprising. The massive translation and protein secretion mainly during the early phases of pollen tubes are reflected by the strong translation of the signal recognition particle (SRP)-binding protein and exocyst subunit Exo70. Finally, FATTY ACID DESATURASE2 (FAD2) is expressed almost exclusively in pollen tubes but is not translated at all. Fatty acid desaturases are hypothesized to be involved in stress signaling and pathogen detection in plants (Walley et al., 2013), so their presence in the EPP fraction may be part of the fast-acting late pollen tube guidance/communication network or thermotolerance mechanisms. The presented charts show only a very limited selection of male gametophytic transcripts; the full list is presented in Supplemental Table S1.
Figure 3.
Expression profiles of selected transcripts in six stages of pollen development and the progamic phase. In each chart, relative expression signals are shown for each subcellular fraction. Blue represents the sequestrome, orange the translatome, and green the mRNPome. Expression profiles and accession numbers of selected genes are shown in Supplemental Table S3.
The Variability of the Pollen Sequestrome Is Accompanied by the Dynamics of Associated Proteins
The mRNA fate in the cytoplasm is controlled primarily by associated RNA-binding proteins. Considering the same being true for mRNA storage and activation in growing pollen tubes, we employed a proteomic approach to identify and analyze proteins present in all subcellular fractions in the corresponding stages of the progamic phase (Fig. 4A). Isolated proteins were subjected to label-free liquid chromatography-tandem mass spectrometry (LC-MS/MS) that resulted in the identification of 9,317 protein groups across all samples and replicates (Supplemental Tables S4 and S5). We compared three subcellular fractions (EPP, POL, and RNP) in mature pollen and in two progamic phase time points (MPG, PT4, and PT24), each sample in two biological replicates, each of which was represented by three technical replicates. For quantitative analyses, only proteins identified by five or more peptides in all biological and technical replicates of the particular sample were considered as reliably present. This reduced the number of robustly identified proteins to 2,089 that were identified reliably in at least one subcellular fraction of at least one developmental stage (Supplemental Table S5).
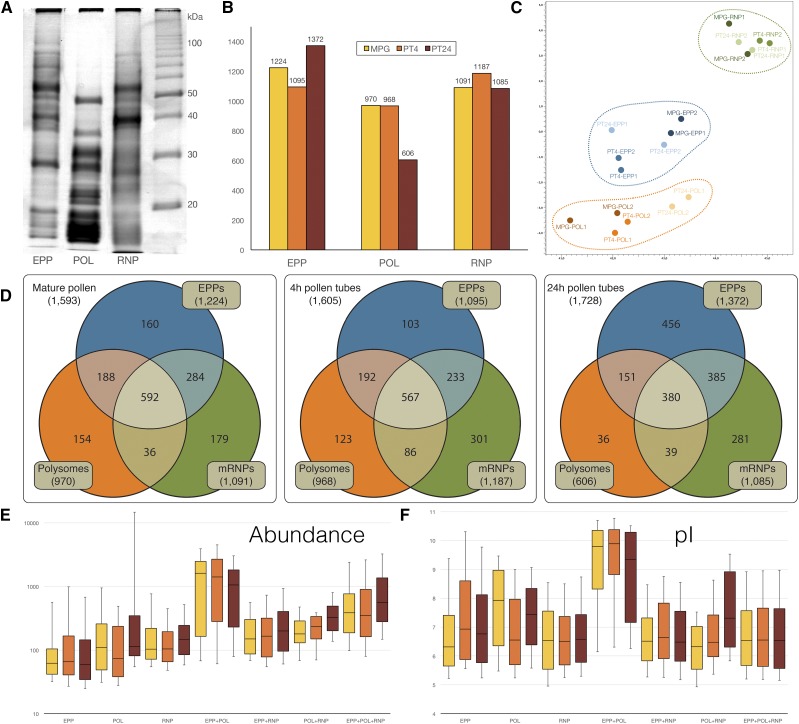
Figure 4.
Quantification of fraction proteomes. A, 1D SDS-PAGE electrophoretogram, 12.5% polyacrylamide gel, with Coomassie Brilliant Blue R250 staining. B, Number of proteins identified in each fraction in each MPG and two stages of the progamic phase (PT4 and PT24). C, PCA of proteins present in all three fractions in MPG, PT4, and PT24. D, Venn diagrams showing the number of unique and overlapping proteins in all fractions of MPG, PT4, and PT24. E, Average expression signal (area) of proteins unique for each fraction and proteins shared by two or all three fractions (see C). F, Distribution of expression signals of data presented in D. In E and F, the bottom whiskers show the 9th percentile and the top whiskers mark the 91st percentile.
Of the total 9,317 proteins identified in the three subcellular fractions, 3,814 proteins were present (the identification of which was achieved according to at least one peptide) also in at least one of the stages of the previously published male gametophyte total proteome (Ischebeck et al., 2014), which was reanalyzed from raw files in this study. However, it should be mentioned that, for the subsequent comparison analyses, only mature pollen grains were fully comparable between Ischebeck’s and our data sets, since Ischebeck’s pollen tubes were cultivated for 5 h instead of 4 or 24 h in our case. Consequently, further comparisons between our data describing the subcellular fractions and Ischebeck’s total proteomic data were performed exclusively on MPGs (Supplemental Table S5). There were 7,329 proteins (of the total 9,317) present in at least one MPG subfraction. Only 2,185 proteins from the 7,329 proteins present in MPG subcellular fractions were reflected in the total MPG proteome (Ischebeck et al., 2014). On the other hand, 264 proteins were exclusive for Ischebeck’s total MPG proteome and did not appear in our MPG subcellular fractions. Besides the differences of sample handling and extraction protocols used in our and Ischebeck’s study, the lack of several proteins in our data set likely was caused by the fact that they were not present in our subcellular fractions and were lost during the subcellular fractionation. The appearance of proteins exclusively in our data set that were absent from Ischebeck’s data can be explained by the fact that low-expressed proteins might not reach the detection limits at the level of the whole proteome, whereas subcellular fractionation enabled their enrichment and, hence, detection.
To further compare Ischebeck’s total proteomic data with our subcellular fraction proteomes, two protein groups were selected, namely ribosomal proteins and poly(A)-binding proteins (PABPs; Supplemental Table S6). In our data set, 170 proteins were identified according to at least five peptides in three technical replicates of at least one sample. This highly stringent criterion was chosen in order to work further with only the most reliable candidates. From the 170 ribosomal proteins, 33 proteins were found only in our MPG subcellular fractions and not in Ischebeck’s MPG total proteome. Moreover, 121 ribosomal proteins were at least 5 times more abundant in all MPG subcellular fractions together compared with the total in Ischebeck’s MPG proteome. Taken together, there were 154 ribosomal proteins that were more abundant or exclusive to our subcellular fractions in MPG out of 170 proteins (i.e. 91% ribosomal proteins). These data strongly support the fact that the subcellular fractions presented in this study contain ribosomal subunits and that the protocol used enabled the enrichment of the acquired samples for ribosomes.
The second candidate group selected were the PABPs. In total, eight PABPs were identified by at least five peptides in three technical replicates of at least one sample. Six of these PABPs were present in at least one of our MPG subcellular fractions but, on the contrary, did not appear in Ischebeck’s total MPG proteome. Moreover, the remaining PABPs were at least twice more abundant in MPG subcellular fractions compared with Ischebeck’s MPG proteome. These results further supported the feasibility of our fractionation protocol for the enrichment of translation-related ribonucleoprotein complexes. PABPs bind the poly(A) tail of mRNAs and, thus, should be present in the RNA-rich RNP complexes (Gorgoni and Gray, 2004). Interestingly, PABP5 and PABP7 were more abundant in the male gametophyte compared with the other tissues, and in our subcellular fraction proteomes, they were enriched in polysomes both at MPG (represented by 47%–70%) and PT4 (56%–69%) stages, whereas the remaining PABPs were found predominantly in nontranslated fractions (EPP and RNP), and polysomes contained a maximum of 15% (MPG) or 19% (PT4) PABPs. The abundance of all PABPs in PT24 polysomes was lower (a maximum of 25%), which was consistent with the overall lower rate of translation after 24 h of pollen tube growth.
The number of proteins associated with individual fractions ranged from 606 to 1,372 (Fig. 4B). The protein quantification in fractions highlighted the reduced translational activity in older pollen tubes (reduction of polysome-associated proteins from 968 to 606) as well as the higher number of proteins associated with the long-term storage of sequestered transcripts that was most apparent in EPPs in PT24. PCA (Fig. 4C) grouped the proteomes of all individual fractions together regardless of the developmental stage. It confirmed that the fraction proteomes throughout development were much more uniform than fraction transcriptomes, suggesting the similar regulatory mechanisms associated with transcript storage. In this respect, EPP and polysomal proteomes were more similar to each other than to the RNP proteome. It likely reflected the presence of the set of ribosomal proteins in both fractions (Honys et al., 2009). Categorization of proteins present in all fractions revealed that the proportion of proteins shared by all three fractions (Fig. 4D) decreased during the progamic phase from 37% (MPG) to 22% (PT24). Accordingly, the proportion of proteins associated with nontranslated transcripts increased gradually, from 13% (MPG) to 33% (PT24) in EPPs and from 16% (MPG) to 26% (PT24) in the RNP fraction. This trend was not followed in the polysomal fraction, where the proportion of fraction-specific proteins decreased from 16% (MPG) to 6% (PT24). On the contrary, proteins forming the polysomal fraction were the most abundant not only among the polysome fraction-exclusive proteins (Fig. 4E) but especially among proteins shared by POL/EPP and POL/EPP/RNP fractions, as documented by their median expression signal (Fig. 4E) as well as the highest and even gradually increasing values in quartiles 2 and 3 of their relative abundance box plot profiles. Accordingly, these polysomal proteins also were by far the most basic, with the median pI value ranging between 9 and 10 (Fig. 4F). Moreover, although the proportion of proteins shared by the POL/EPP and POL/EPP/RNP fractions decreased during the progamic phase, quartiles 2 and 3 increased in abundance, showing that the most abundant proteins formed the core of these fraction proteomes. The proteins shared by all three subcellular fractions also were identified by the highest number of peptides in all three developmental stages (Supplemental Fig. S6).
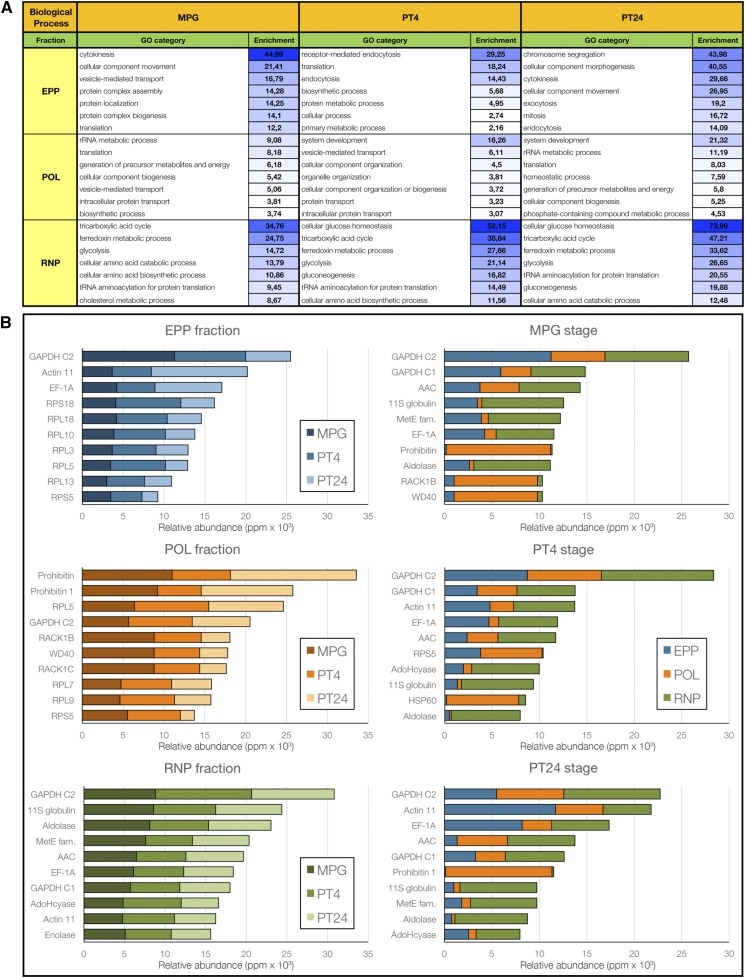
Gene enrichment analysis also was applied to the fraction proteomes (Fig. 5A; Supplemental Table S7) and highlighted the GO terms overrepresented in each fraction as well as the individual GO categories overrepresented in different developmental stages. Alongside the identification of overrepresented GO categories, we exemplified the GO categorization by identifying the most abundant proteins in individual fractions and developmental stages (Fig. 5B). In the EPP fraction, eight of the 10 most abundant proteins (ppm) were indeed related to translation. There were seven ribosomal protein of large (RPL) and small (RPS) subunits (RPL3, RPL5, RPL10, RPL13, RPL18, RPS5, and RPS18) and elongation factor GTP-binding EF-Tu/EF-1A. However, the most abundant proteins in EPP complexes were the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) subunit C and Actin11. Subunit C of GAPDH was among the most abundant protein components in all fractions. In the polysomal fraction, four ribosomal proteins, RPL5, RPL7, RPL9, and RPS5, were accompanied by three Trp-Asp-repeat proteins (two of them were receptors for activated C kinase1 [RACK1B and RACK1C], both ribosomal proteins) and two prohibitins. Two of the identified Trp-Asp-repeat proteins, receptors for RACK1B and RACK1C, also were annotated as ribosomal proteins (Kakehi et al., 2015). Taken together, both ribosomal subunit-containing fractions included a significant proportion of highly abundant ribosomal proteins and other proteins associated with translation and the cytoskeleton (nine in EPPs and six in POL out of 10 each). On the contrary, the top 10 proteins in the RNP fraction were the most diverse, containing several cytosolic metabolic enzymes accompanied by the seed storage protein legumin and previously identified GAPDH C proteins, elongation factor TU/EF-1A, and Actin11. The representation of translation-related proteins was thus much lower than in the previous two fractions.
Figure 5.
Gene Ontology (GO) summary of fraction proteomes. A, Top seven categories in each fraction (EPP, POL, and RNP) in all three developmental stages (MPG, PT4, and PT24). B, Expression dynamics of the 10 most abundant proteins present in each subcellular fraction and in each developmental stage.
The top 10 lists in all three developmental stages were very similar to the corresponding lists in individual fractions, differing mainly in the order of individual proteins. This is not surprising, since proteins shared by all three fractions were among the most abundant ones. Proteins highly present in the RNP fraction were the main contributors; however, the most abundant proteins in each stage were the strongest also in the EPP complexes.
The categorization of the proteomes of individual fractions and their developmental dynamics throughout the progamic phase not only confirmed the abundant presence of proteins related to translation in both polysomes and RNA storage EPPs but also the higher heterogeneity of the RNP fraction.
Preferential Distribution of Ribosomal Proteins in the Sequestrome
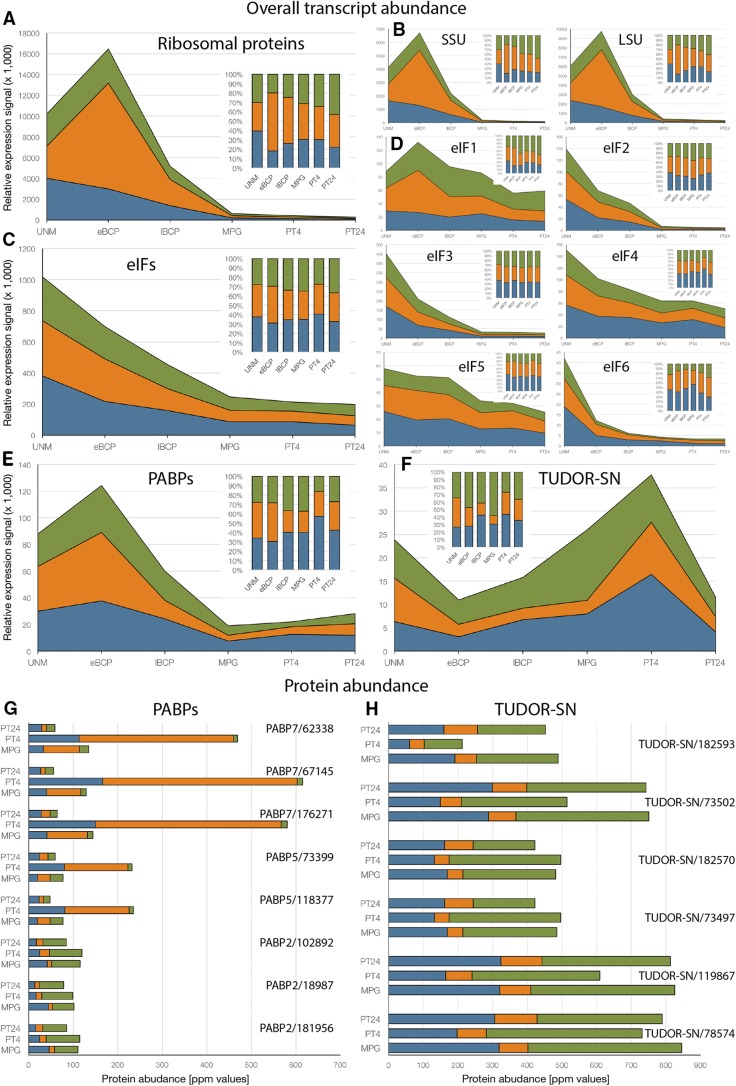
The broad presence of ribosomal proteins, which were among the most abundant proteins in both EPP and POL fractions, inspired our subsequent analysis of the dynamics of ribosomal proteins throughout pollen development as well as the distribution of their encoding transcripts. Therefore, we prepared a transcriptomic table containing only RP transcripts (Supplemental Table S8) by a subtraction from Supplemental Table S1. We identified 321 RP transcripts showing a reliable expression signal in at least one stage of pollen development. From previous studies, it was known that mRNAs encoding RPs were abundant at early stages of pollen development and strongly declined in abundance after pollen mitosis I (PMI; Honys and Twell, 2004). This finding also was confirmed in tobacco (Bokvaj et al., 2014), where the abundance of RP transcripts peaked soon after PMI (Fig. 6A); however, the inclusion of fraction transcriptomes also revealed that the majority of RP transcripts (62%) were actively translated, leaving the remaining RP mRNAs in EPP (18%) and RNP (20%) mRNA storage compartments. Later, the overall abundance of RP transcripts dropped dramatically (by 96%) until MPG (Fig. 6A). This expression profile and drop in abundance were similar for both 40S and 60S ribosomal subunits (Fig. 6B). In addition, we extracted other proteins (Supplemental Table S9) involved in translation regulation, namely eIFs (Fig. 6C) and supplemented them with examples of proteins known to affect mRNA cellular fate, metabolism, translation, and storage, PABP (Fig. 6E) and proteins containing Tudor and staphylococcal/micrococcal-like nuclease (SN) domains, TSN (Fig. 6, E and F) transcripts. PABPs bind the poly(A) tail of mRNA at its 3′ end and play multiple roles in mRNA processing, nuclear export, translation initiation, and degradation (for review, see Goss and Kleiman, 2013). Similarly, TSN proteins are emerging to play a complex role in mRNA catabolism and storage (Gutierrez-Beltran et al., 2015; Chou et al., 2017). We compared their expression profiles with those of RP transcripts. Unlike RP transcripts, eIFs in general were up-regulated during the time of PMI, although individual gene families showed different expression patterns (Fig. 6D). For example, eIF1 showed the least apparent decline in abundance with time. eIF1 transcripts together with those encoding eIF4 and eIF5 proteins persisted in 24-h pollen tubes in reasonable amounts: 45% of their abundance in eBCP for eIF1s, 49% for eIF4s, and 48% for eIF5s. Most of the eIF transcripts also showed a higher degree of translational regulation than RP transcripts themselves. On the contrary, the abundance of PABP and TSN transcripts increased following pollen germination. TSN transcripts even peaked in PT4, before PMII, with only 30% of them being actively translated.
Figure 6.
Dynamics of RPs, eukaryotic initiation factors (eIFs), PABPs, and TUDOR-SN (TSN) proteins during pollen development and the progamic phase. A, Overall abundance of RP transcripts. B, Abundance of RP transcripts present in 60S and 40S subunits. C, Overall abundance of eIF transcripts. D, Abundance of transcripts forming individual eIFs. E, Overall abundance of PABP transcripts. F, Overall abundance of TSN transcripts. G, Expression dynamics of seven protein groups associated with PABPs in all subcellular fractions during the progamic phase. H, Expression dynamics of six protein groups associated with TSN proteins in all subcellular fractions during the progamic phase. Transcriptomic data (A–F) were plotted as a sum of expression signals, and relative visualization of the same data is shown in the respective inlet. Proteomic data are plotted as protein abundance values (ppm).
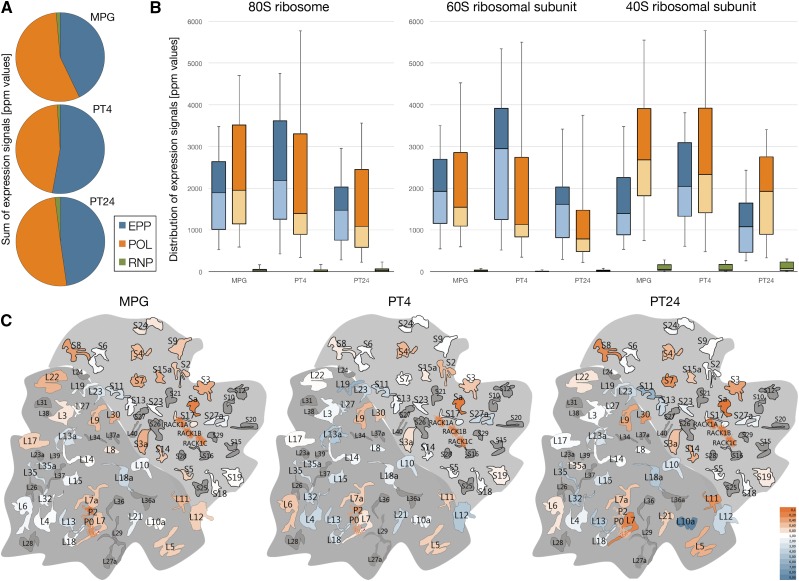
Previously, we demonstrated that EPPs contained both 60S and 40S ribosomal subunits (Honys et al., 2009). Therefore, we aimed to elucidate how the developmental dynamics in the synthesis of RPs is reflected by their distribution among the subcellular fractions during the progamic phase (Supplemental Table S6). In general, the vast majority of RPs were present in the EPP and POL fractions, leaving only negligible 1.08% (PT4) to 1.98% (PT24) of the overall expression signal in free RNPs. The distribution of the overall expression signal between the two dominant fractions was almost uniform, ranging from 43% to 53% in EPPs and between 46% and 56% in polysomes (Fig. 7A). A more detailed view showed that there were no significant differences in the abundance of RPs during the progamic phase (Fig. 7B), with the sole exception of higher quartile 3 in both fractions at early stages of pollen tube growth (PT4), demonstrating the greater variability among more abundant RPs at this stage. On the contrary, there were differences between both ribosomal subunits. In all developmental stages, the majority of 60S RPs based on the expression signal were associated with EPPs, whereas the most 40S RPs were found in polysomes. This shift was more apparent in mature pollen and especially in the early phase of pollen tube growth (PT4). The most detailed view, at the level of individual RPs (Fig. 7C; Supplemental Fig. S7), confirmed this observation, since 63% of 60S RPs were more abundant in EPPs, whereas 65% of 40S RPs were associated predominantly with polysomes. Supplemental Table S6 and corresponding Figure 7C and Supplemental Figure S7 cover only RPs identified by five or more peptides in all biological and technical replicates. Therefore, not all proteins less reliably expressed are shown, although they were present in the complete proteomic data set (Supplemental Table S5). More interestingly, the association of individual RPs with EPP complexes or polysomes was highly dynamic throughout the progamic phase. Within the small subunit, RPS7 and RPS8 were among the most variable, whereas RPS4 and RPS19 showed the most stable distribution pattern. RPS19 was almost uniformly distributed between both fractions, whereas RPS4 was more abundant in polysomes. Here, the acidic ribosomal protein RPSA represented the protein most significantly associated with translating polysomes. The dynamics of proteins within the large subunit was generally larger than within the small subunit. There, RPL7a and RPL9 were the most stably distributed, both associated more abundantly with polysomes. On the contrary, RPL10a and RPL12 were the most variable, showing the highest rate of accumulation in EPPs, especially in later phases of the progamic phase. In fact, there were several 60S RPs that accumulated in EPPs in PT4 and PT24: RPL10a, RPL12, RPL13, RPL18A, RPL19, RPL23, RPL32, and RPL35. In the 40S subunit, only RPS11 and RPS13 followed this distribution pattern. Taken together, our observations confirmed and extended the previous findings that both EPP and POL fractions contained RPs (Honys et al., 2009) and exhibited heterogeneity in the composition of ribosomal subunits, showing different dynamics of the expression and distribution patterns of individual RPs.
Figure 7.
Dynamics of ribosomal proteins in fraction proteomes during the progamic phase. A, Sum of expression signals of ribosomal proteins in all fractions. B, Distribution of expression signals in 80S ribosomes as well as individually in small and large ribosomal subunits. C, Dynamics of individual small and large subunit ribosomal proteins between EPP and POL fractions during the progamic phase. The heat map intensity shows the predominant presence of the respective protein in the polysomal (orange) or EPP (blue) fraction.
Stored Transcripts Contain Longer Untranslated Regions Harboring More Upstream Open Reading Frames
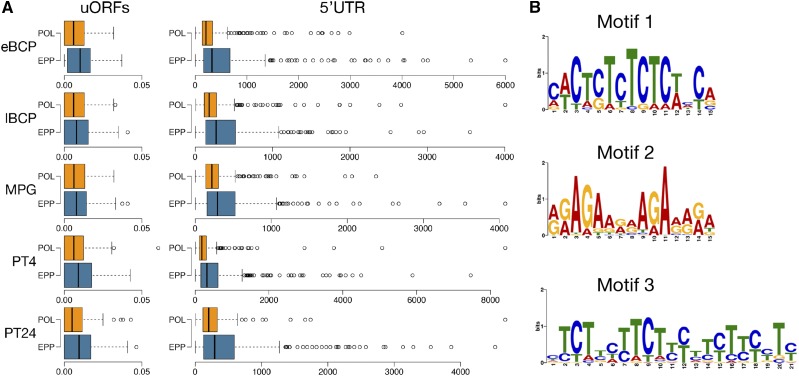
Pollen germination and the initiation of pollen tube growth was shown previously to be accompanied by the activation of the translation apparatus. In previous sections, we demonstrated the dynamics of the pollen sequestrome that was reflected by the massive redistribution of transcripts from the storage compartment (EPPs) to translationally active polysomes. Therefore, we were interested in the comparison of known regulatory features of stored transcripts, namely their untranslated regions (UTRs): 5′UTR and 3′UTR. Therefore, we collected known UTR sequences of stored and translated tobacco transcripts for subsequent analyses. In all stages observed, untranslated regions of stored transcripts were longer than those of translated transcripts (Fig. 8A). The difference was more dramatic in 5′UTRs, and, with the exception of PT4, the 5′UTR length increased with time. The mean 5′UTR length of transcripts associated with EPPs remained stable throughout the progamic phase, ranging from 149 to 171 nucleotides, whereas the mean length of the 5′UTR of actively translated transcripts was shorter, in a range of 55 to 145 bases. Accordingly, the highest margin between the 5′UTR of stored and translated transcripts was observed in long-term stored mRNAs in PT24. On the contrary, the length difference in 3′UTRs, although observed, was not significant.
Figure 8.
5′UTR and upstream open reading frames (uORF) analysis. Sequences were extracted from the Nicotiana tabacum TN90 cDNA database (Sierro et al., 2013, 2014). A, Box plots comparing 5′UTR length and number of uORFs between POL and EPP resident transcripts. All observed differences in the number of uORFs and 5′UTR length (with the exception of the uORF comparison at MPG) were statistically significant. B, Top three common motifs identified in the 5′UTRs of stored transcripts at the PT24 time point.
A motif search of upstream sequences revealed several sequence motifs overrepresented in 5′UTRs of stable transcripts. The most prominent motifs are shown in Figure 8B. Interestingly, GO categorization of transcripts harboring these motifs in their 5′UTRs showed their bias toward male and female gametophyte-associated categories (Supplemental Table S9). Alongside sequence similarity analyses, we also observed higher complexity and the overall higher presence of stable secondary structures in 5′UTRs of stored transcripts. 5′UTRs of EPP-derived transcripts showed, on average, −25.32 kcal mol−1 minimal folding energy (Zuker, 2003) lower than the average of POL-derived 5′UTR at all stages (data not shown). uORFs were shown previously to be associated with transcript storage and activation under specialized conditions (von Arnim et al., 2014). The association of uORFs with several subunits of eIF3 was found to be important for their sequestration and controlled activation. However, this regulatory module has never been associated with the male gametophyte. Here, we identified the higher occurrence of uORFs in transcripts stored in all developmental stages of pollen development (Fig. 8A), and again, the enrichment of 5′UTRs with uORFs was observed in stable transcripts stored in pollen tubes, most abundantly in PT24.
A Subset of Translationally Repressed Transcripts Are Sperm Cell Expressed
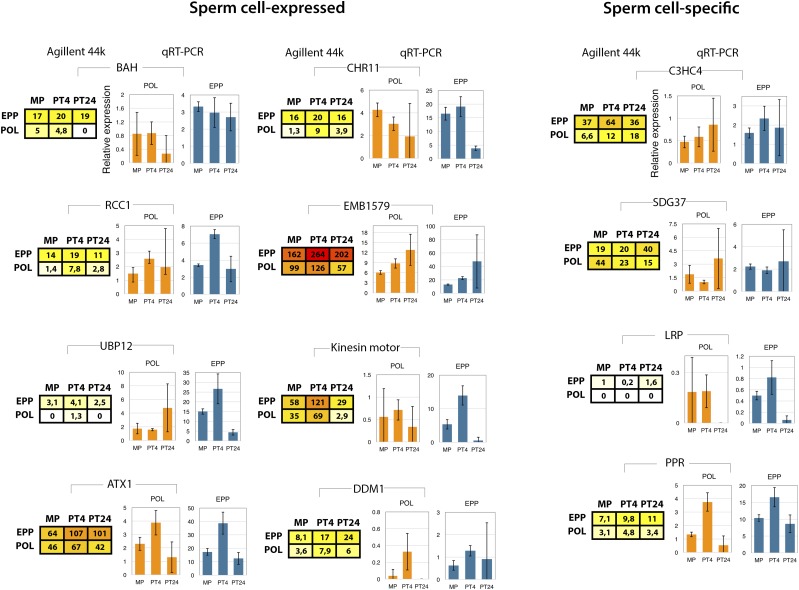
We hypothesized that a likely function of some of the stored transcripts after 24 h of pollen tube growth would be in promoting embryogenesis; thus, they are likely to be delivered via sperm cells. We mapped direct homologs of tobacco to the Arabidopsis sperm cell transcriptome (Borges et al., 2008). We identified 151 (5.9%) tobacco homologs from the 2,562 genes expressed in Arabidopsis sperm cells as stored transcripts present at greater than 50% in the EPP fraction at PT24 (Fig. 9; Supplemental Table S10). Their distribution in POL and EPP fractions at PT24 also was investigated by RT-qPCR analysis (Fig. 9). We found that five out of 12 genes correlated well with the RT-qPCR results in both fractions (EPP and POL) and at all time points (MPG/PT4/PT24), six out of 12 genes also correlated in one of the fractions at all time points, and only one gene (CHR11) showed no correlation with the RT-qPCR profile (Fig. 9). Of the tested genes, ARABIDOPSIS HOMOLOG OF TRITHORAX (ATX1), REGULATOR OF CHROMOSOME CONDENSATION (RCC1), BROMO-ADJACENT HOMOLOGY (BAH) DOMAIN-CONTAINING PROTEIN, and DECREASED DNA METHYATION1 (DDM1) all showed a steady predominant distribution in the EPP fraction from MPG to PT24. For others, such as CHROMATIN REMODELLING FACTOR11 (CHR11), KINESIN MOTOR, and UBP12, their transcripts were utilized during pollen tube growth but still remained predominant in the EPP fraction at PT24. By contrast, EMBRYO DEFECTIVE1579 (EMB1579) showed increased accumulation in the EPP fraction toward PT24 (Fig. 9).
Figure 9.
Dynamics of stored transcripts from sperm cell-expressed and sperm cell-specific genes between POL and EPP up to 24 h of in vitro pollen tube growth. Data were derived from an Agilent 44K array chip and validated by qPCR. Error bars represent sd among replicates.
We also mapped 26 tobacco homologs of Arabidopsis sperm cell-specific genes (absent call in pollen and seedlings; Borges et al., 2008), which are stored at PT24 (Fig. 9; Supplemental Table S11). The profile distribution in POL and EPP fractions of four genes, C3HC4-TYPE RING FINGER, SET DOMAIN GROUP37 ZINC ION BINDING (SDG37), LEUCINE-RICH REPEAT PROTEIN (LRP), and PENTATRICOPEPTIDE REPEAT CONTAINING PROTEIN (PPR), was verified by RT-qPCR, and all four genes showed steady storage in the EPP fraction (Fig. 9). To gain insight into the various functions of the stored mRNAs that are expressed in sperm cells, we performed GO analysis. Among the top 10 enriched GO terms were cell division (GO:0051301), microtubule-based movement (GO:0003777), DNA duplex unwinding (GO:0032508), pollen development (GO:0009555), embryo sac development (GO:0009553), and cell fate specification (GO:0001708). A complete GO list can be found in Supplemental Table S12. This analysis revealed that top activities of the stored sperm cell-expressed genes are associated with chromatin remodeling, transcription activation, development postfertilization, and cell fate specification. Our results suggest a likely conserved posttranscriptional fate of Arabidopsis and tobacco sperm cell-expressed transcripts with a possible function before and after fertilization (Huanca-Mamani et al., 2005; Chen et al., 2009; Robert et al., 2009; Tsugeki et al., 2009; Zhao et al., 2017).
Monosomes Dominate in Dehisced Pollen Grains
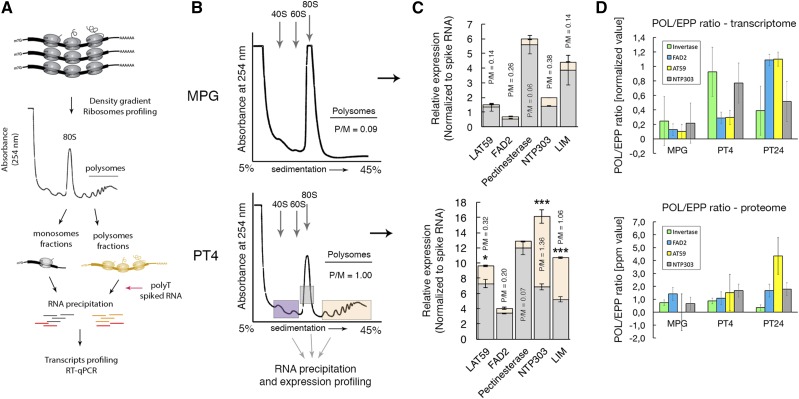
Our results thus far have supported the findings that pollen and the pollen tube sequestrome constitute diverse mRNA populations that are highly dynamic between POL and EPP subfractions in the course of pollen development and pollen tube growth (Figs. 2 and 3). The efficiency of translation of a given mRNA at a given developmental stage can be assessed from its distribution in the polysomal profile. Generally speaking, the more the transcript shifts from monosomal to polysomal fractions, the higher the translation rates can be expected, with the exception of mRNAs containing inhibitory uORFs, short ORFs, low abundance regulatory proteins, and targets of nonsense-mediated decay such as unspliced transcripts, all of which can be found in monosomal fractions (Heyer and Moore, 2016). A totally different class of fully assembled monosomes is represented by stalled 80S couples at the initiating AUG codon on various mRNAs, which may serve as one of the means of translational control. Therefore, we wished to understand the identity of monosomes in the male gametophyte. To do that, we focused on the translation status of five mRNAs encoding genes from the tobacco pollen sequestrome with orthologs in Arabidopsis upon pollen hydration and pollen tube growth, in particular a Pectate lyase ortholog of the tomato Lat59 protein (AT59; AT1G14420), FAD2 (AT3G12120), PLANT INVERTASE FAMILY PROTEIN (AT5G27870), POLLEN-SPECIFIC CELL WALL GLYCOPROTEIN (NTP303 ortholog SKU5 Similar12; AT1G55570), and POLLEN-SPECIFIC LIM DOMAIN CONTAINING PROTEIN (WLIM2B; AT3G55770). Note that all selected mRNAs were associated predominantly with the EPP fraction at the MPG stage (Supplemental Table S1). To follow their translation status, we applied high-velocity centrifugation in a linear Suc density gradient (5%–45%) to two developmental stages of the male gametophyte, MPG and PT4, and separated free ribosomal subunits from monosomes and polysomes by Teledyne ISCO subcellular fractionation (Fig. 10). We profiled mRNAs associated with polysomes relative to those occurring in monosomes as a relative measure of translation efficiency (Fig. 10A). As expected, polysomal profiling confirmed that monosomes vastly dominated in MPG with practically no polysomes, as illustrated by the calculated polysome:monosome ratio (P/M = ∼0.09; Fig. 10B). In contrast, there was a robust increase in polysomal fractions at the expense of monosomes at the PT4 stage of pollen tube growth (P/M = ∼1; Fig. 10B). This clearly illustrates a rapid change from a nonactive to an active translational status that must inevitably occur in the growing pollen tubes.
Figure 10.
Polysome profiling revealed that monosomes dominate at MPG. A, Experimental workflow. B, Polysome profiles from a subcellular fractionation-coupled Suc density gradient showing a dominant monosome peak in the MPG sample and only traces of polysomes. At the PT4 stage, the increase in polysome abundance is likely associated with high translation activities in pollen tubes. P/M represents a ratio of polysomes versus monosomes calculated per area of occupancy on the plot by the two fractions. C, Quantification of transcript occupancy in monosomes versus polysomes of five selected genes at the MPG stage and after 4 h of in vitro pollen tube growth (PT4). The P/M ratio represents the translatability rate of each transcript from the respective gene at each stage. Asterisks report a statistically significant difference (Welch’s t test, P < 0.05) between the P/M ratio at MPG versus PT4 as a metric of induced translation. Error bars represent sd among replicates. D, Quantification of the POL/EPP ratio for selected transcripts and the respective proteins during the progamic phase. Error bars represent sd among replicates.
For further analysis, collected fractions representing monosomes were used as individual fractions, and all polysomal fractions were pooled together (four to five fractions) to represent the entire polysomal area (Fig. 10). Total RNA was precipitated from these monosomal and polysomal fractions using SDS:phenol:chloroform and chloroform:isopropanol. Quantification of transcripts by RT-qPCR revealed that the vast majority of mRNAs that were identified as being stored in the EPP were associated with monosomes at the MPG stage, showing a P/M ratio ranging between 0.14 and 0.38 (Fig. 10C). At the PT4 stage, INVERTASE showed a minor shift toward polysomes with the P/M ratio increasing by 1.16-fold (from 0.06 to 0.07; P > 0.98, Welch’s t test), whereas FAD2 remained strongly associated with monosomes showing even a small shift toward monosomes (the P/M ratio decreased by ∼1.3-fold, from 0.26 to 0.2; P > 0.09, Welch’s t test), suggesting a continuous stable transcript storage even after 4 h of pollen tube growth (Fig. 10C). Conversely, AT59 showed a medium shift toward polysomes with a 2.2-fold increase of the P/M ratio (from 0.14 to 0.32; P < 0.0018, Welch’s t test) at the PT4 stage (Fig. 10C), whereas NTP303 and WLIM2B transcripts showed the greatest increases by 3.6-fold (from 0.38 to 1.36; P < 0.0001, Welch’s t test) and 7.6-fold (from 0.14 to 1.06; P < 0.0001, Welch’s t test), respectively (Fig. 10C). Remarkably, LC-MS/MS proteome data analysis of the EPP and POL fractions revealed an almost perfect correlation between the monosome-to-polysome shift of the selected transcripts and the increase in protein abundance in POL fractions between MPG and PT4 stages, with the exception of WLIM2B. INVERTASE showed an increase in the translation rate from MPG to PT4 by 1.16-fold, whereas FAD2 showed a decrease by 1.3-fold (Fig. 10D). AT59 then showed a 1.5-fold increase in the efficiency of translation, whereas NTP303 translation was enhanced by more than 2.5-fold at the PT4 stage (Fig. 10D). These results suggest that the observed dynamic changes in the association of various mRNAs with monosomes versus polysomes in pollen and pollen tubes likely reflect their translatability that varies among different developmental stages. Unexpectedly, the proteome data obtained for WLIM2B at the MPG stage was not reliable between the replicates and, therefore, could not be used further. Taken together, our results reveal that there are numerous mRNAs whose translational activity remains poor even in mature pollen grains with a reactivated translational apparatus. Interestingly, this suggests that mRNA-bound monosomes could represent the pollen sequestrome pool of stored mRNAs awaiting its activation at a specific developmental stage by a yet-to-be-elucidated mechanism.
DISCUSSION
Sequestrome Dynamics during Pollen Development and the Progamic Phase
Translation regulation has gradually received increased attention as an important regulatory level playing an essential role in plant development following developmental cues as well as in complex stress responses (for review, see Van Der Kelen et al., 2009; Muench et al., 2012; Roy and von Arnim, 2013; Salomé, 2017; Mills et al., 2018). Several studies have aimed to identify subsets of actively translated transcripts, termed the translatome, using either polysome profiling (Kawaguchi et al., 2004) or translating ribosome affinity purification using a FLAG-tagged ribosomal protein L18 (FLAG-RPL18; Zanetti et al., 2005). Polysome profiling has been used to characterize translatomes during photomorphogenesis (Liu et al., 2012, 2013) or under heat stress (Yángüez et al., 2013). Translating ribosome affinity purification has been applied to identify condition- or cell-specific translatomes for the characterization of translation modulation during plant immune responses (Meteignier et al., 2017) or in in vivo-growing pollen tubes (Lin et al., 2014). The latter study provided interesting insight into selective translation during the progamic phase by the identification of 519 transcripts selectively loaded onto polysomes during pollen tube growth in vivo as a result of contact between the male gametophyte and female reproductive tissues.
Considering the importance of selective translation activation for pollen tube growth, we aimed to get a deeper insight into the phenomenon of long-term transcript storage and translation in pollen and pollen tubes. We wished to extend our previous analyses (Honys et al., 2000, 2009) by characterizing the dynamics of translational regulation during tobacco male gametophyte development and the subsequent functional progamic phase. Therefore, we separated the ribonucleoprotein complexes by two-step Suc gradient centrifugation (Fig. 1; Jurečková et al., 2017) to isolate the three types of mRNA-containing ribonucleoprotein particles: POL, RNP, and EPP. We defined the EPP transcriptome as the pollen sequestrome, a specific and highly dynamic compartment for stored, translationally repressed transcripts clearly distinct from other RNA-containing fractions (Fig. 2, E–G).
The quantification of expressed genes (Fig. 2A; Supplemental Fig. S1A) confirmed the general trends of previously published profiles of male gametophytic gene expression in tobacco (Hafidh et al., 2012a, 2012b; Bokvaj et al., 2014), Arabidopsis (Honys and Twell, 2004; Wang et al., 2008; Qin et al., 2009), and rice (Wei et al., 2010) as well as the developmental shift during late stages of pollen development (Honys and Twell, 2004). Despite the reduction of transcriptome complexity toward pollen maturity that was reflected in the similar reduction of the translatome, we observed an increased abundance of transcripts forming the sequestrome (Fig. 2, C and D). The sequestrome peaked in mature pollen, and this observation was consistent with the general concept of reduced translation activity accompanied by the storage of presynthesized transcripts in this quiescent stage (Mascarenhas, 1993; Štorchová et al., 1994; Honys et al., 2000). For example, the tobacco highly abundant pollen-specific transcript NTP303 (Weterings et al., 1992) encoding cell wall glycoprotein (Čapková et al., 1987), shown previously to be translationally repressed (Wittink et al., 2000), was found predominantly in the sequestrome but gradually redistributed to the polysomal fraction following pollen germination (Fig. 3). In fact, NTP303 was the seventh most abundant sequestered transcript in mature pollen. A similar expression and translation profile was observed for the other NTP303 homologs SKS12 (Fig. 3; Sedbrook et al., 2002) and NTP805 (Supplemental Table S1; Weterings et al., 1995).
Recently, SKS14, one of the Arabidopsis orthologs of NTP303, was localized to processing bodies (P-bodies) in tobacco pollen (Scarpin et al., 2017). Analogous to other organisms, P-bodies were speculated in that study to represent the mRNA storage compartment in tobacco pollen. However, the observed occurrence of NTP303 in P-bodies in pollen was unexpectedly low (Scarpin et al., 2017) and corresponded neither to the previously published high abundance of NTP303 transcript in tobacco pollen and pollen tubes (Štorchová et al., 1994; Wittink et al., 2000) nor to the accumulation of the majority of this abundant transcript in the EPPs (this study). Furthermore, our protein composition analysis also did not confirm the suggested hypothesis that EPPs may correspond to P-bodies. The DCP1 and VCS protein markers of P-bodies (Xu et al., 2006) were absent from EPPs in the original proteomic study (Honys et al., 2009) as well as in this study (we identified VCS associated with nonpolysomal particles only in 24-h pollen tubes). Last but not least, P-bodies in contrast to EPPs are not considered to contain ribosomal subunits and translation factors (Chantarachot and Bailey-Serres, 2018). Therefore, although the NTP303 mRNA partially colocalized with P-bodies in the aforementioned study, it seems unlikely that P-bodies represent the main storage compartment for NTP303 as well as for many other quiescent mRNAs, as suggested (Scarpin et al., 2017). It is possible that these two assemblies coexist in pollen while having distinct functions for its maturation and development.
Ribosomal or ribosome-associated proteins occupied eight of the 10 most abundant proteins in the EPP fraction and six out of the 10 most abundant proteins in the polysomal fraction (Fig. 5B). The most abundant and the most basic proteins, often RPs, were shared by EPP and POL fractions (Fig. 4, E and F). Interestingly, the most abundant protein in most of the fractions was GAPDH subunit C (Zeng et al., 2016). The main function of this enzyme is in the glycolytic pathway (Sirover, 2011). GAPDH is not only a cytosolic protein; in human and animal cells, it also was localized to the membrane, the nucleus, polysomes, the endoplasmic reticulum, and the Golgi (Sirover, 2012). Moreover, GAPDH was shown to bind RNA (Nicholls et al., 2012). Therefore, alongside this canonical metabolic activity, GAPDH has been implicated also in other functions, including transcription, translation, and endoplasmic reticulum-to-Golgi protein transport (Tristan et al., 2011; Sirover, 2012). If the presence of GAPDH subunit C did not result from a contamination, one could speculate that this protein might fulfill one of the above-mentioned roles also in pollen. It was even proposed that cytosolic GAPDH might function as a sensor for redox signals in yeast, plants, and mammals and as an information hub to transduce the stress signals for appropriate adaptive responses (Hildebrandt et al., 2015).
In our study, we confirmed the dramatic decline in the abundance of transcripts encoding cytosolic ribosomal proteins in late pollen development (Fig. 6), a phenomenon observed previously in Arabidopsis and tobacco pollen (Honys and Twell, 2004; Bokvaj et al., 2014). This decline was similar for both ribosomal subunits. The massive synthesis of ribosomal proteins in young pollen grains, soon after the completion of PMI, was demonstrated not only by the high abundance of RP transcripts in the cell (Bokvaj et al., 2014) but also by their prevalence in association with polysomes (Fig. 6, A and B), demonstrating their active translation at this stage and persistence in vast amounts for the whole progamic phase (Fig. 7). Translation plays an important role during the progamic phase, since its inhibition causes a dramatic suppression of pollen tube growth (Čapková-Balatková et al., 1980; Hao et al., 2005). Therefore, it was not surprising that such a transcript down-regulation pattern was not entirely followed by other players involved in translation, namely some translation initiation factors and various forms of the poly(A)-binding proteins (Fig. 6, C–E), as also observed previously in Arabidopsis (Honys and Twell, 2004). Whereas the expression of transcripts encoding eIF2 and eIF3 subunits as well as the eIF6 factor also declined in late pollen development, eIF1, eIF4A, eIF4E, eIF4G, eIFiso4E, eIFiso4G, eIF5, and eIF5C were actively transcribed also during the progamic phase, and PABP transcripts were even up-regulated in pollen tubes. Since all of the listed eIFs are critically required for cap-dependent translation initiation (for review, see Valášek, 2012; Browning and Bailey-Serres, 2015; Valášek et al., 2017), we believe that the observed differences in expression profiles simply reflect the varying protein stability of these eIFs. Trimetric eIF2 and multimeric eIF3 complexes are known to be very stable, which may markedly reduce the demand for the de novo transcription of mRNAs encoding their subunits. Interestingly, eIF4B, eIF4G, eIFiso4F, and eIF5B, which also were identified in the EPP proteome, were among the proteins that were phosphorylated in tobacco mature pollen and dephosphorylated immediately after pollen germination as a consequence of pollen activation (Fíla et al., 2016). The most abundant eIF in pollen and pollen tube fraction proteomes, eIF4A, was constitutively phosphorylated both in the mature pollen and throughout the onset of the progamic phase (Fíla et al., 2016). Even though more factor-focused analyses are required to understand the phosphorylation status changes of individual eIFs, these data hint at another level of regulation that could occur specifically during pollen development.
As expected given the stable composition of EPPs, despite the sharp decline in the transcript levels, ribosomal proteins persisted in large amounts in growing pollen tubes (Fig. 7). In particular, the abundance of ribosomal proteins increased during the first 4 h of pollen tube growth and then finally decreased in 24-h pollen tubes. This pattern closely followed the rate of tobacco pollen tube growth (Čapková et al., 1987; Hafidh et al., 2012b). A substantial fraction (around 50% in general) of ribosomal proteins remained associated with EPPs even during pollen tube growth. This finding further confirmed our previously published observations that EPPs occurring in the growing pollen tube still contain both ribosomal subunits (Honys et al., 2009). Unexpectedly, the abundance of ribosomal proteins was not uniform during the progamic phase, and neither was their distribution among polysomes and EPP complexes. Especially, the EPP-POL ratio varied not only for the entire subunits (Fig. 7A) but also for individual ribosomal proteins (Fig. 7C) throughout the progamic phase. Here, the large ribosomal proteins were redistributed more profoundly toward the storage EPP complexes in both PT4 and PT24, whereas the small ribosomal proteins remained more often in the polysomal fraction (Fig. 7, B and C). Interestingly, even though the differential accumulation of ribosomal proteins and ribosome remodeling were documented previously as part of the response to environmental cues and stress (Horiguchi et al., 2012; Tiruneh et al., 2013; Wang et al., 2013), these striking observations await further explanation. Of note, the most characteristic distribution pattern was observed for acidic ribosomal proteins (RPP0 and RPP2) as well as for the small ribosomal protein RACK1. The acidic ribosomal protein P0 and phosphoproteins P1 to P3 form a lateral stalk structure in the active site of the 60S ribosomal subunit (Szick et al., 1998). RACK1 is a soluble, plasma membrane-associated or ribosome-bound protein that is found in a variety of signaling complexes, suggesting that it links cell regulation and translation (Gibson, 2012). All these proteins are associated with actively translating ribosomes (Szick et al., 1998; Turkina et al., 2011; Browning and Bailey-Serres, 2015), and in our proteomic data sets, they all remained associated predominantly with the polysomal fraction throughout the progamic phase (Fig. 7C; Supplemental Fig. S7; Supplemental Table S6). PABP5 and PABP7 were enriched in the polysomal fraction, whereas PABP2 was enriched in the RNP data set (Supplemental Table S6). This is consistent with proposed multiple roles of PABP proteins in mRNA storage and translation in plants (Goss and Kleiman, 2013). TSN proteins, besides their well-documented role in mRNA catabolism in stress granules and P-bodies (Gutierrez-Beltran, 2015; Gutierrez-Beltran et al., 2015), have been implicated in the stabilization of stress-induced mRNAs in Arabidopsis (Frei dit Frey et al., 2010) and in mRNA storage and localization in rice endosperm (Chou et al., 2017). Accordingly, the vast majority (87% of protein abundance) of TSN proteins was found outside the polysomal fraction, namely 35% in the EPP fraction and 52% in the RNP fraction (Fig. 6H; Supplemental Table S6).
Collectively, the data presented in this section introduced EPP complexes as a distinguishable compartment for mRNA storage in pollen and pollen tubes characterized by a specific subset of associated proteins and harboring a dynamic yet distinct set of stored stable transcripts, a sequestrome.
Stored Transcript in Sperm Cells
Transcript profiling in late stages of pollen tube growth revealed that approximately 6% (151 genes) of the EPP stored transcripts at the PT24 time point (a more than halfway growth of tobacco pollen tube toward the ovule) are expressed in sperm cells and some are sperm cell specific (Fig. 9; Supplemental Tables S11 and S12). It is likely that not all transcripts associated with sperm cells are utilized for sperm cell-related cellular activities prefertilization. We hypothesized that some of these stored transcripts could be inherited and involved in embryogenesis, for instance. RNA inheritance via gametes in plants and its significance are still little explored; however, recent studies have provided a glimpse of its significance (Autran et al., 2011; Nodine and Bartel, 2012; Del Toro-De León et al., 2014). From the GO analysis of the stored sperm cell transcripts, we identified CHR11 (AT3G06400), a SWI2/SNF2 chromatin-remodeling factor, and a BTB scaffold protein (AT1G05690) as direct regulators of embryo and embryo sac development. CHR11 is involved in haploid nuclear proliferation during megagametogenesis, and chr11 loss of function blocks the completion of the mitotic haploid nuclear divisions and cell expansion in the female gametophyte (Huanca-Mamani et al., 2005). BTB functions redundantly with BT1 and BT2 to regulate male and female gametophyte development (Robert et al., 2009). Two additional sperm cell-expressed genes, RBR1 (AT3G12280) and NO VEIN (NOV; AT4G13750), were identified under the enriched category of cell fate specification. In the male gametophyte, RBR1 is required for the correct differentiation of the male cell types (Chen et al., 2009). It also regulates nuclear proliferation in the female gametophyte and promotes germline entry (Zhao et al., 2017). After fertilization, RBR1 coordinates with MSI1 to activate the expression of the imprinted genes FIS2 and FWA through the repression of MET1. NOV is a plant-specific nuclear factor that promotes cell fate decisions associated with auxin gradients and maxima to establish PIN polarity and patterning (Tsugeki et al., 2009). nov loss-of-function mutations alter auxin gradients and early embryo patterning. Thus, the biological significance of transcript storage in pollen tubes is likely widespread beyond its function in embryogenesis, and ongoing research will highlight those yet unidentified roles.
It is important to clarify that transcript sequestration and mRNA storage, irrespective of the mechanism of repression, does not suggest a complete block in mRNA translation; rather, it is a posttranscriptional mechanism that allows the modulation of protein abundance as well as long-term transcript storage to facilitate immediate translation, particularly in tip growth. Therefore, it is unsurprisingly common that stored transcripts are highly dynamic between subcellular compartments in a long- and short-term manner, and their half-life within storage granules varies across development. In pollen tubes, EPPs possess a likely function to facilitate immediate translation during tip growth and the safekeeping of transcripts with late function from cytosolic 5′ to 3′ and 3′ to 5′ exonuclease activities (Łabno et al., 2016).
Monosomes as a Potential Mechanism for Translation Repression in Pollen
To define the ribosome occupancy pattern during pollen dehiscence and pollen tube growth as a measure of the efficiency of translation, we performed polysome profiling analyses and revealed a dominant monosome peak with nearly no polysomes in mature pollen, further underscoring the fact that mature pollen is translationally practically inactive. By contrast, polysomal profiling of the PT4 stage showed a comparable abundance of polysomes to monosomes, clearly demonstrating resumed translational activity at this stage, as expected (Fig. 10). This distinct pollen polysome profile of the male gametophyte is similar to that of the RNA storage in dormant and embedded seeds (Basbouss-Serhal et al., 2015). Furthermore, in neuronal cells of animals, the mRNA-storing granules are cemented together with the complete translation machinery and form heavier fractions peaking beyond the polysomal profile (Krichevsky and Kosik, 2001; Anderson and Kedersha, 2006; Kiebler and Bassell, 2006; Buchan, 2014; Hafidh et al., 2014; El Fatimy et al., 2016). In plants, and in pollen in particular as shown here, or in other tip-growing cell types, monosomes seem to be the compartment for mRNA storage and could represent an efficient mRNA sequestration mechanism facilitating the rapid activation of translation upon various stimuli. EPPs sediment in much lighter fractions in comparison with the neuronal RNA granules; therefore, their commonalities are mainly in their molecular composition, particularly the translation machinery, subspecies of ribosomal proteins, translation elongation factors, cytoskeleton-associated proteins including actin and microtubules, and the nature of mRNAs associated with both fractions, including variable 5′UTR and frequent uORFs (Buchan, 2014). In a parallel observation, stored transcripts associated with EPPs throughout pollen development and in pollen tubes often bear a long 5′UTR with multiple uORFs (Fig. 8); this is a common characteristic of translationally repressed or stored transcripts interfering with ribosome scanning and translation initiation (Liu et al., 2012, 2013; Muench et al., 2012; Buchan, 2014). Indeed, RNA granules are known to be associated with translation repressors; however, these factors are yet to be identified in plants.
In this study, we observed the similarities between EPPs and monosomes particularly in their proteome composition and close correlation of transcript dynamics between EPP-POL and monosome-POL samples. Such dynamics correlated closely with their translational status, exemplified here with four genes, INVERTASE, FAD2, AT59, and NTP303, and their predominant abundance at MPG and reduced accumulation during pollen tube growth. Together, these observations suggest that the isolated EPPs are, in fact, monosomes stalled on late encoded transcripts in a translationally quiescent state. Therefore, we propose that these forms of the sequestered monosome-mRNA species, most likely also bound by initiation and/or elongation factors, represent a very dynamic and regulatable tool for the mRNA storage of late transcripts in pollen and pollen tubes of tobacco.
CONCLUSION
We performed a thorough transcriptomic and proteomic analysis of stored and translated transcripts and their storage ribonucleoprotein particles throughout tobacco pollen development and pollen tube growth. In this way, we defined a pollen sequestrome as a distinct and highly dynamic compartment for the storage of stable, translationally repressed transcripts and demonstrated its dynamics. We have also proposed that EPP complexes in fact represent nontranslating monosomes that are the actual form of mRNA sequestration. Finally, we have demonstrated that a fraction of the tobacco pollen tube-expressed transcripts is regulated continuously at the posttranscriptional level, suggesting the involvement of these long-stored stable transcripts in postfertilization development.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type tobacco plants (Nicotiana tabacum ‘Samsun’) were used to collect tissue samples for all downstream studies. Seeds were sown in a greenhouse under short-day conditions at 22°C to 25°C. Adult plants with a fully developed root system were transplanted to an outdoor greenhouse on ground compost and grown under the natural day/night photoperiod from March to September. Pollen grains were collected throughout the season, from June to September, and their germination rate was monitored.
Collection of Pollen and in Vitro Pollen Tube Cultivation
Tobacco mature pollen was isolated aseptically as described previously (Petrů et al., 1964). Flowers were collected 1 d before anthesis. Stamens were removed from the flowers into a petri dish to dehisce overnight at room temperature. Dry pollen grains were then filtered through a nylon mesh (Miracloth; pore size, 50 μm), weighed, and stored at −20°C. The germination rate of isolated pollen was monitored as described previously (Hafidh et al., 2012b).
For tobacco in vitro pollen tube germination, approximately 10 mg of pollen was resuspended in 10 mL of pollen germination medium [SMM-MES: 0.3 m Suc, 1.6 mm H3BO3, 3 mm Ca(NO3)2·4H2O, 0.8 mm MgSO4·0.7H2O, 1 mm KNO3, and 25 mm MES-KOH buffer, pH 5.9; Tupý and Rhová, 1984] and divided into aliquots into conical Erlenmeyer flasks. The 24- and 48-h pollen tubes were cultivated with SMM-MES medium supplemented with casein (1 mg mL−1). Cultures were incubated in a water-bath shaker at 140 rpm for 2 h and then slowed down to 90 rpm for the remaining cultivation time at 26°C in the dark. Similar procedures were followed for other pollen tube cultures (13, 24, and 48 h of cultivation), although under sterile conditions. Aliquots of the samples were stained with Aniline Blue and 4′,6-diamidino-2-phenylindole and analyzed using a fluorescence microscope. Pollen tubes were vacuum filtered, flash frozen in liquid nitrogen, and stored at −80°C prior to RNA extraction. Sporophytic tissues (leaf discs and roots) were collected from juvenile plants and also from excavated adult plants. Collected samples were frozen immediately in liquid nitrogen.
Subcellular Fractionation
To fractionate POL, EPP, and RNP from developing pollen and in vitro-cultivated pollen tubes, immature pollen grains from 20 anthers at corresponding developmental stages or 150 mg of dehisced pollen grains or pollen tube pellets were homogenized with low-salt buffer (200 mm Tris-HCl, pH 9, 25 mm KCl, 60 mm magnesium acetate, 2 mm DTT, 0.5 mm PMSF, 1% [v/v] PTE, 1 mm cycloheximide, and 250 mm Suc). The homogenates were centrifuged twice (400g, 10 min, 4°C and 23,000g, 15 min, 4°C) to remove cellular debris and organelles from the postmitochondrial supernatant (Fig. 1; Honys et al., 2009).
The collected postmitochondrial supernatant was loaded on a 30% (w/v) Suc cushion with low-salt gradient buffer (40 mm Tris-HCl, pH 8.5, 15 mm KCl, 30 mm magnesium acetate, 2 mm DTT, 0.5 mm PMSF, and 1 mm cycloheximide) and centrifuged (298,400g, 3 h 20 min, 4°C; Jurečková et al., 2017). The supernatant comprised postpolysomal RNP fraction (free mRNPs), whereas the pelleted fraction contained POL and EPP complexes (Fig. 1). The postpolysomal supernatant was centrifuged (258,000g, 18 h, 4°C). The pellet-constituted fraction of free mRNPs was rinsed with RNase-free water and stored at −80°C. To separate POL and EPP complexes, the pelleted mixed fraction (POL and EPP complexes) was resuspended in a high-salt EPP buffer (200 mm Tris-HCl, pH 9, 500 mm KCl, 2 mm magnesium acetate, 2 mm DTT, 0.5 mm PMSF, 1% [v/v] PTE, 50 mm EDTA, pH 8, 0.2 mm puromycin, and 250 mm Suc), loaded on a 30% (w/v) Suc cushion with high-salt EPP gradient buffer (40 mm Tris-HCl, pH 8.5, 200 mm KCl, 1 mm magnesium acetate, 2 mm DTT, 0.5 mm PMSF, 50 mm EDTA, pH 8, and 0.2 mm puromycin), and centrifuged (298,400g, 3 h 20 min, 4°C). The pellet that constituted EPP complexes was collected, rinsed with RNase-free water, and stored at −80°C. The remaining supernatant was centrifuged (258,000g, 18 h, 4°C). The pellet that constituted the fraction of polysomes was rinsed with RNase-free water and stored at −80°C (Fig. 1).
RNA Extraction, Probe Preparation, and Microarray Hybridization
Total RNA was isolated using the Qiagen RNeasy Plant Kit in accordance with the manufacturer’s instructions (Qiagen) and treated with DNaseI (Promega). RNA was quantified using NanoDrop (Thermo Scientific). Prior to shipment, five replicates from each sample were tested using semiquantitative RT-PCR with two marker genes, NteIF5A and a constitutive 18S rRNA, for reproducibility. RNA concentration, purity, and integrity were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) at ImaGenes and Imaxio. Biotinylated target cRNA was prepared from 66 ng of reverse-transcribed total RNA (One-Cycle Target labeling and control reagents; Agilent Technologies). Labeled cRNA was fragmented, and 1.65 μg was used for Agilent 44 K Tobacco Genome Array hybridization. Hybridized chips were scanned on an Agilent High Resolution Microarray Scanner. The data discussed in this article have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE114806 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114806).
Bioinformatics and Statistical Analysis of Agilent 44K Tobacco Genome Array Data
All transcriptomic data sets were normalized using freely available dChip 1.3 software (http://www.dchip.org). The reliability and reproducibility of the analyses were ensured by the inclusion of duplicates in each experiment, the normalization of all arrays to the median probe intensity level, and the use of normalized intensities of all arrays for the calculation of model-based gene expression values based on the Perfect Match-only model (Li and Wong, 2001). For each sample, only probes with the detection call of present in both replicates were considered to be expressed.
For systematic comparison of the tissue samples, only a subset of expressed genes was considered for analysis. These genes had the detection call of present in both biological replicates in all three subcellular fractions as well as in the independent total transcriptome. Therefore, only the overlap of genes identified as expressed in the total transcriptome and in all three subcellular fractions was considered further. The corresponding number of expressed genes identified in previous transcriptomic studies (Supplemental Fig. S1A; Hafidh et al., 2012a, 2012b) and this study supported our confidence that the selected approach was appropriate and left most possible false positives out. To test the quality of the microarray hybridization, we stringently assessed our data sets to justify their reproducibility among replicates and, thus, provided a feasible comparison between individual fractions and the total transcriptome.
To determine the quality of the normalized data set and the correlation between arrays, CLC Genomics Workbench version 4.5.1 (CLC bio) was used to visualize the correlation between samples using PCA as well as independently using hierarchical clustering. To observe the variance of the distribution of the mean expression levels, scatterplots were used for pairwise comparison between samples.
Sequences of probes located on the chip were BLASTed against the Nicotiana tabacum TN90 cDNA database (Sierro et al., 2013, 2014). Transcripts with the best value of score and E value (E ≤ 10−5) were assigned to individual probes. The closest Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum) homologs were inherited from the original TN90 genome annotation.
To identify transcripts enriched in the EPP and POL fractions in each developmental stage, we applied the following workflow: (1) false discovery rate (FDR)-corrected P (using the t test in the CLC Genomics Workbench) was calculated for each probe comparing EPP and POL samples in each stage; (2) the threshold P was set as the first decile FDR-corrected P among all probes for each stage; and (3) transcripts for which all targeting probes gave FDR-corrected P below the threshold were selected for 5′UTR and uORF analyses. Sequences of 5′UTRs and 3′UTRs were determined by the comparison of cDNA and coding sequences of the reference tobacco TN90 data sets. The list of uORFs was obtained by Python script searching through a 5′ UTR list for ATG starting and TAG, TAA, or TGA terminating motifs.
GO Analysis
GO analyses in proteome analyses were performed using the PANTHER classification system (http://www.pantherdb.org/data/; Mi et al., 2005). We used a statistical overrepresentation test with default settings using the gene identifiers of the closest tomato homologs and a tomato reference organism. The top GO-slim biological processes in the hierarchy with P ≤ 0.05 are shown. GO analysis in transcriptome analyses was performed using DAVID (http://david.abcc.ncifcrf.gov; Huang et al., 2009) with an EASE score (a modified Fisher exact test) cutoff of P ≤ 0.05.
Protein Extraction and Filter-Aided Sample Preparation Processing
Proteins were extracted from fraction samples using SDS (2%)- and DTT (100 mm)-containing Tris-HCl (100 mm, pH 7.6) buffer for 30 min at 95°C. Protein concentration was ascertained by fluorescence. Two microliters of protein solutions or Trp standard was added to 400 µL of 8 m urea in 100 mm Tris-HCl, pH 7.6, and analyzed in a 10-mm quartz cuvette using a Cary Eclipse Fluorescence Spectrophotometer. Fluorescence was measured at 295 nm for excitation and 350 nm for emission. The slits were set to 10 for excitation and emission. Protein solutions were processed by the filter-aided sample preparation (FASP) method (Wiśniewski et al., 2009, 2011). Approximately 100 µg of proteins was mixed with 400 µL of 8 m UA buffer (8 m urea in 100 mm Tris-HCl, pH 8.5). Half (200 µL) was loaded onto the Microcon 30-kD filter unit (Millipore; MILLMRCF0R030). After centrifugation (14,000g, 30 min, 20°C), the second part of the mixture was loaded and centrifuged under the same conditions. The retained proteins were washed with 100 μL of UA buffer. After additional centrifugation (14,000g, 30 min, 20°C), the samples were mixed with 100 μL of UA buffer containing 50 mm iodoacetamide and incubated in the dark for 30 min. After the next centrifugation step, the samples were washed three times with 100 μL of UA buffer and three times with 100 μL of 100 mm triethylammonium bicarbonate buffer. Trypsin (sequencing grade; Promega) was added onto the filter, and the mixture was incubated overnight at 37°C. The tryptic peptides were finally eluted by centrifugation followed by two additional elution steps with 100 μL of 50 mm triethylammonium bicarbonate buffer. The peptide mixture was dried under vacuum. Dried peptides in the FASP tube were resuspended in 50 µL of 50% (w/w) acetonitrile (with 2.5% [w/w] formic acid) and transferred to LC-MS vials with already added polyethylene glycol (2.5 µL of 0.01% [w/w] polyethylene glycol; Stejskal et al., 2013). The FASP tube was washed again using the same solution (50 µL) and subsequently twice with 100% acetonitrile (2 × 100 µL). The combined solution was concentrated under vacuum to a volume below 25 µL. Water was used to get 25 µL of peptide solution.
LC-MS/MS Analysis
Tryptic peptide mixtures obtained by FASP were analyzed using the RSLCnano system connected to an Orbitrap Elite hybrid spectrometer (Thermo Fisher Scientific). Prior to LC separation, tryptic digests were inline concentrated and desalted using a trapping column (100 μm × 30 mm) filled with 3.5-μm X-Bridge BEH 130 C18 sorbent (Waters). After washing the trapping column with 0.1% (w/w) fatty acid (FA), the peptides were eluted (300 nL min−1) from the trapping column onto an Acclaim Pepmap100 C18 column (3-µm particles, 75 μm × 500 mm; Thermo Fisher Scientific) by the following gradient program (mobile phase A: 0.1% [w/w] FA in water; mobile phase B: 0.1% [w/w] FA in 80% [w/w] acetonitrile): the gradient elution started at 1% (w/w) mobile phase B and increased from 1% to 56% (w/w) during the first 100 min (14% in the 30th, 30% in the 60th, and 56% in 100th min), then increased linearly to 80% (w/w) mobile phase B in the next 5 min and remained in this state for the next 15 min. Equilibration of the trapping column and the column was done prior to sample injection to the sample loop. The analytical column outlet was connected directly to the Nanospray Flex Ion Source (Thermo Fisher Scientific).
MS data were acquired in a data-dependent strategy selecting up to the top 10 precursors based on precursor abundance in the survey scan (350–2,000 m/z). The resolution of the survey scan was 60,000 (400 m/z) with a target value of 1 × 106 ions, one microscan, and a maximum injection time of 200 ms. Higher energy collisional dissociation (HCD) MS/MS spectra were acquired with a target value of 50,000 and resolution of 15,000 (400 m/z). The maximum injection time for MS/MS was 500 ms. Dynamic exclusion was enabled for 45 s after one MS/MS spectra acquisition, and early expiration was disabled. The isolation window for MS/MS fragmentation was set to 2 m/z.
The analysis of the mass spectrometric raw data files was carried out using the Proteome Discoverer software (Thermo Fisher Scientific; version 1.4) with in-house Mascot (Matrix Science; version 2.6) and Sequest search engine utilization. MS/MS ion searches were done against a protein database downloaded from ftp://ftp.solgenomics.net/genomes/Nicotiana_tabacum/annotation/ (file Ntab-TN90_AYMY-SS_NGS.prot.annot.faa; downloaded February 27, 2015) with additional sequences from the cRAP database (downloaded from http://www.thegpm.org/crap/). Mass tolerances for peptides and MS/MS fragments were 10 ppm and 0.05 D, respectively. Oxidation of Met and deamidation (Asn and Gln) as optional modification, carbamidomethylation of C as fixed modification, and two enzyme (trypsin) missed cleavages were set for all searches. Percolator was used for the postprocessing of Mascot and Sequest search results. Peptides with FDR (q value) < 1%, rank 1, and at least six amino acids were considered. Label-free quantification using protein area calculation in Proteome Discoverer was used.
Data from the dataset of Ischebeck et al. (2014) were reprocessed starting from raw files using the same pipeline except for these differences: selection of 10 or six peaks within a mass window of 100 D (to N peaks filter) in each MS/MS spectrum prior to Mascot or SequestHT search, respectively, and MS and MS/MS mass tolerances of 10 ppm and 0.8 D, respectively.
Protein groups with at least five identified peptides in each biological and technical replicate were considered for downstream quantitative analyses. Next, protein groups associated with the same tomato gene identifier were stacked together, and the maximum abundance of such joint protein groups was used. To consider a protein up-regulated in the EPP, POL, or RNP fraction of a particular stage, its abundance in that fraction should have been at least 2-fold higher than its median abundance in the stage. Using this strategy, nine groups of up-regulated proteins specific to a certain fraction within the stage were selected and analyzed for enriched GO terms.
Polysome Profiling
Polysomes from tobacco mature pollen grains and 4-h in vitro-cultivated pollen tubes were isolated with freshly prepared polysome extraction buffer (0.2 m Tris-HCl, 0.2 m KCl, 0.25 m EGTA, 0.35 m MgCl2, detergent mix [20% (w/w) Brij-35, 20% (w/w) Triton X-100, 20% (w/w) Igepal, and 20% (w/w) Tween 20], 1% (w/w) deoxycholate, 1% (w/w) PTE, 5 μm DTT, 10 μg mL−1 cycloheximide, 5 μg mL−1 chloramphenicol, 1× proteasome inhibitor [catalog no. MG132; Sigma-Aldrich], and 10 μL mL−1 enzymatic cocktail [catalog no. P9599; Sigma-Aldrich]). After pulverization with a mortar and pestle, 3 volumes of polysome buffer was added to each sample and incubated on ice for 10 min. Samples were centrifuged (17,000g, 10 min, 4°C). An additional 10-min centrifugation was performed to ensure debris-free samples. Polysomes were loaded on top of a 5% to 45% (v/w) Suc gradient prepared by BIOCOMP gradient master (BioComp Instruments) containing 1.32 m Suc, 1× Suc salts (0.04 m Tris-HCl, pH 8.4, 0.02 m KCl, and 0.01 m MgCl2), 5 μg mL−1 chloramphenicol, and 10 μg mL−1 cycloheximide. Ultracentrifugation was performed with Beckman Optima XPN-80 using a Beckman SW 41 Ti rotor at 154,300g for 3.5 h at 4°C without braking. Fractions were collected using a peristaltic pump (Brandel) and a Foxy R1 collector (Teledyne ISCO), and polysome profiles were recorded using the UA-6 UV/VIS detection system with a 254-nm filter (Teledyne ISCO). All fractions were flash frozen and stored at −80°C until further use.
RNA Extraction from Polysome Profiling
For RNA extraction from polysomal fractions, frozen samples were equilibrated to room temperature and universal spike RNA (TATAA Biocenter) was added to each sample. SDS was added to a final concentration of 1% (w/w). One volume of phenol:chloroform (2.5:1) was added, and samples were incubated for 10 min at 65°C with occasional vortexing. Samples were cooled on ice for 5 min and spun at 21,900g with a Sigma 2-16K centrifuge for 5 min at 4°C. The aqueous layer was removed, and the phenol:chloroform precipitation was repeated as above. The final aqueous layer was mixed with 1 volume of chloroform:isopropanol (25:1), vortexed for 1 min, incubated for 1 min at room temperature, and spun at 21,900g at 4°C for 2 min. To the aqueous fractions, 300 mm NaOAC and 2 µL of glycogen (Invitrogen) were added and mixed with 2.5 volumes of 100% ethanol. RNA was precipitated overnight at −20°C. Samples were then spun at 21,900g at 4°C for 45 min. RNA pellets were washed with 75% (w/w) ethanol. Dried pellets were resuspended in 20 µL of Tris-HCl, pH 8, and RNA concentrations were determined by Nanodrop (Thermo Fisher Scientific). For RT-qPCR analysis of each isolated fraction, total RNA was DNase treated (Ambion, Thermo Fisher Scientific) and first-strand cDNA was synthesized using the ImPromII Reverse Transcription System (Promega). qPCR measurements were obtained using GoTaq qPCR Master Mix (Promega) on a LightCycler 480 instrument (Roche). Ct values from each fraction were normalized with the TATAA RNA spike crossing point. The list of primers can be found in Supplemental Table S13.
Accession Numbers
Accession numbers are presented as tobacco identifier/Arabidopsis homolog/Agilent probe as follows: NtREF/SRPP (TA12371/AT1G75020/A_95_P178902); NtLAT59 (BP128327/AT3G53190/A_95_P283783); FAD2 (CV016252/AT3G12120/A_95_P102907); hnRNP A3 (DV161566/AT2G33410/A_95_P244767); NTP303/SKS12 (X61146/AT1G55570/A_95_P005151); NtLAT52 (EB425604/AT1G58340/A_95_P129172); LIM domain protein (EB426388/AT3G55770/A_95_P126982); NTF2 (TA17318/AT3G25150/A_95_P149662); NtDKDM (EB446238/AT4G14710/A_95_P116182); NtLEA (DV159065/AT3G15670/A_95_P244692); NtHMA1 (BP530876/AT4G37270/A_95_P090243); PRF5 (X82120/AT2G19770/A_95_P025251); RPS19 (BP526774/AT5G47320/A_95_P075615); U2AF65B (AJ718624/AT1G60900/A_95_P241774); NtARDCP (DV157797/AT1G04780/A_95_P029706); NtRPS19 (BP526774/AT5G47320/A_95_P075615); and NtHMA1 (BP530876/AT4G37270/A_95_P090243). Accession numbers of all the genes mentioned in this work together with accession numbers of their homologs in Arabidopsis and tomato are listed in their corresponding tables as cited in the text. Raw data for microarray gene expression are accessible through GEO Series accession number GSE114806 at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Quantification of transcriptomic data sets.
Supplemental Figure S2. Validation of transcriptomic data.
Supplemental Figure S3. PCA of total and fraction transcriptomes and a sequestrome throughout pollen development and the progamic phase.
Supplemental Figure S4. Validation of microarray data by RT-qPCR in relation to microarray data in all three subcellular fractions demonstrated independently for five selected genes.
Supplemental Figure S5. Clustering of gene expression profiles according to their translation status.
Supplemental Figure S6. Quantification of fraction proteomes.
Supplemental Figure S7. Dynamics of individual small and large subunit ribosomal proteins between EPP and POL fractions during the progamic phase.
Supplemental Table S1. Total and fraction transcriptomes in six stages of pollen development and the progamic phase.
Supplemental Table S2. Clustering of gene expression profiles according to their translation status.
Supplemental Table S3. Expression profiles of transcripts selected in Figure 3 in six stages of pollen development and the progamic phase.
Supplemental Table S4. Annotation of protein groups including the closest Arabidopsis and tomato homologs.
Supplemental Table S5. Quantitative proteome analysis of tobacco pollen total and fraction proteomes.
Supplemental Table S6. Proteomic data of the distribution of ribosomal proteins and other proteins involved in translation within three subcellular fractions and in the total proteome.
Supplemental Table S7. PANTHER GO-slim analysis of genes enriched in three fraction proteomes in tobacco mature pollen and pollen tubes.
Supplemental Table S8. Transcriptomic data of the distribution of ribosomal proteins and other proteins involved in translation within the subcellular fractions.
Supplemental Table S9. Enriched GO terms associated with overrepresented motifs identified at the 5′UTR of stored transcripts.
Supplemental Table S10. List of sperm cell-expressed transcripts.
Supplemental Table S11. List of putative sperm cell-specific transcripts.
Supplemental Table S12. Enriched GO terms associated with sperm cell-expressed and sperm cell-specific transcripts.
Supplemental Table S13. List of primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Cécile Bousquet-Antonelli and Rémy Merret (Université de Perpignan) for the initial consultation on polysome profiling.
Footnotes
This work was supported by the Czech Science Foundation (Grants GA17-23183S and GA18-02448S to D.H., GA17-23203S to S.H., and GA17-06238S to L.S.V.). We also acknowledge Prague Structural Funds, Project No. CZ.2.16/3.1.00/21519. The proteomic part of this work was supported by the project CEITEC 2020 (LQ1601) with financial support from MEYS CR. We acknowledge the CIISB research infrastructure project LM2015043 funded by MEYS CR for supporting the LC-MS/MS measurements at the Proteomics Core Facility. Computational resources were provided by the CESNET LM2015042 and the CERIT Scientific Cloud LM2015085, provided under the program “Projects of Large Research, Development, and Innovations Infrastructures.”.
Articles can be viewed without a subscription.
References
- Anderson P, Kedersha N (2006) RNA granules. J Cell Biol 172: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, Lenormand T, Wittig M, Grob S, Steimer A, Barann M, Klostermeier UC, Leblanc O, et al. (2011) Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145: 707–719 [DOI] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, et al. (2012) A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122 [DOI] [PubMed] [Google Scholar]
- Basbouss-Serhal I, Soubigou-Taconnat L, Bailly C, Leymarie J (2015) Germination potential of dormant and nondormant Arabidopsis seeds is driven by distinct recruitment of messenger RNAs to polysomes. Plant Physiol 168: 1049–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokvaj P, Hafidh S, Honys D (2014) Transcriptome profiling of male gametophyte development in Nicotiana tabacum. Genom Data 3: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS, Bailey-Serres J (2015) Mechanism of cytoplasmic mRNA translation. The Arabidopsis Book 13: e0176, doi/10.1199/tab.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR. (2014) mRNP granules: assembly, function, and connections with disease. RNA Biol 11: 1019–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čapková V, Hrabětová E, Tupý J (1987) Protein-changes in tobacco pollen culture: a newly synthesized protein related to pollen-tube growth. J Plant Physiol 130: 307–314 [Google Scholar]
- Čapková-Balatková V, Hrabětová E, Tupý J (1980) Effects of cycloheximide on pollen of Nicotiana tabacum in culture. Biochem Physiol Pflanz 175: 412–420 [Google Scholar]
- Chantarachot T, Bailey-Serres J (2018) Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol 176: 254–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P, Ischebeck T, Egelhofer V, Lichtscheidl I, Weckwerth W (2013) Cell-specific analysis of the tomato pollen proteome from pollen mother cell to mature pollen provides evidence for developmental priming. J Proteome Res 12: 4892–4903 [DOI] [PubMed] [Google Scholar]
- Chen Z, Hafidh S, Poh SH, Twell D, Berger F (2009) Proliferation and cell fate establishment during Arabidopsis male gametogenesis depends on the retinoblastoma protein. Proc Natl Acad Sci USA 106: 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HL, Tian L, Kumamaru T, Hamada S, Okita TW (2017) Multifunctional RNA binding protein OsTudor-SN in storage protein mRNA transport and localization. Plant Physiol 175: 1608–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conze LL, Berlin S, Le Bail A, Kost B (2017) Transcriptome profiling of tobacco (Nicotiana tabacum) pollen and pollen tubes. BMC Genomics 18: 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Li L, Chen T, Chong K, Xue Y, Wang T (2006) Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 6: 2504–2529 [DOI] [PubMed] [Google Scholar]
- Dai S, Chen T, Chong K, Xue Y, Liu S, Wang T (2007) Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol Cell Proteomics 6: 207–230 [DOI] [PubMed] [Google Scholar]
- Dal Bosco C, Dovzhenko A, Palme K (2012) Intracellular auxin transport in pollen: PIN8, PIN5 and PILS5. Plant Signal Behav 7: 1504–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dios Alché J, M’rani-Alaoui M, Castro AJ, Rodríguez-García MI (2004) Ole e 1, the major allergen from olive (Olea europaea L.) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant Cell Physiol 45: 1149–1157 [DOI] [PubMed] [Google Scholar]
- Del Toro-De León G, García-Aguilar M, Gillmor CS (2014) Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514: 624–627 [DOI] [PubMed] [Google Scholar]
- Devoto A, Hartmann HA, Piffanelli P, Elliott C, Simmons C, Taramino G, Goh CS, Cohen FE, Emerson BC, Schulze-Lefert P, et al. (2003) Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J Mol Evol 56: 77–88 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Bombarely A, Story GW, Allen F, Mueller LA, Coates SA, Jones L (2010) TobEA: an atlas of tobacco gene expression from seed to senescence. BMC Genomics 11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fatimy R, Davidovic L, Tremblay S, Jaglin X, Dury A, Robert C, De Koninck P, Khandjian EW (2016) Tracking the Fragile X Mental Retardation Protein in a highly ordered neuronal ribonucleoparticles population: a link between stalled polyribosomes and RNA granules. PLoS Genet 12: e1006192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sánchez-Carbente M, Servant F, Bell AW, Boismenu D, et al. (2006) Characterization of an RNA granule from developing brain. Mol Cell Proteomics 5: 635–651 [DOI] [PubMed] [Google Scholar]
- Fabrice TN, Vogler H, Draeger C, Munglani G, Gupta S, Herger AG, Knox JP, Grossniklaus U, Ringli C (2018) LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol 176: 1981–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando DD. (2005) Characterization of pollen tube development in Pinus strobus (Eastern white pine) through proteomic analysis of differentially expressed proteins. Proteomics 5: 4917–4926 [DOI] [PubMed] [Google Scholar]
- Fíla J, Radau S, Matros A, Hartmann A, Scholz U, Feciková J, Mock HP, Čapková V, Zahedi RP, Honys D (2016) Phosphoproteomics profiling of tobacco mature pollen and pollen activated in vitro. Mol Cell Proteomics 15: 1338–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fíla J, Záveská Drábková L, Gibalová A, Honys D (2017) When simple meets complex: pollen and the -omics. In Obermeyer G, Feijo J, eds, Pollen Tip Growth: From Biophysical Aspects to Systems Biology. Springer, pp 247–292 [Google Scholar]
- Frei dit Frey N, Muller P, Jammes F, Kizis D, Leung J, Perrot-Rechenmann C, Bianchi MW (2010) The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22: 1575–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Song Y, Zhang C, Zhang Y, Burlingame AL, Guo Y (2011) Proteomic analyses of apoplastic proteins from germinating Arabidopsis thaliana pollen. Biochim Biophys Acta 1814: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson TJ. (2012) RACK1 research: ships passing in the night? FEBS Lett 586: 2787–2789 [DOI] [PubMed] [Google Scholar]
- Gorgoni B, Gray NK (2004) The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct Genomics Proteomics 3: 125–141 [DOI] [PubMed] [Google Scholar]
- Goss DJ, Kleiman FE (2013) Poly(A) binding proteins: are they all created equal? Wiley Interdiscip Rev RNA 4: 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant-Downton R, Hafidh S, Twell D, Dickinson HG (2009a) Small RNA pathways are present and functional in the angiosperm male gametophyte. Mol Plant 2: 500–512 [DOI] [PubMed] [Google Scholar]
- Grant-Downton R, Le Trionnaire G, Schmid R, Rodriguez-Enriquez J, Hafidh S, Mehdi S, Twell D, Dickinson H (2009b) MicroRNA and tasiRNA diversity in mature pollen of Arabidopsis thaliana. BMC Genomics 10: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E. (2015) Genome-wide analysis of uncapped mRNAs under heat stress in Arabidopsis. Genom Data 5: 7–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV (2015) Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 27: 926–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafidh S, Breznenová K, Honys D (2012a) De novo post-pollen mitosis II tobacco pollen tube transcriptome. Plant Signal Behav 7: 918–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafidh S, Breznenová K, Růžička P, Feciková J, Capková V, Honys D (2012b) Comprehensive analysis of tobacco pollen transcriptome unveils common pathways in polar cell expansion and underlying heterochronic shift during spermatogenesis. BMC Plant Biol 12: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafidh S, Potěšil D, Fíla J, Feciková J, Čapková V, Zdráhal Z, Honys D (2014) In search of ligands and receptors of the pollen tube: the missing link in pollen tube perception. Biochem Soc Trans 42: 388–394 [DOI] [PubMed] [Google Scholar]
- Hafidh S, Potěšil D, Fíla J, Čapková V, Zdráhal Z, Honys D (2016) Quantitative proteomics of the tobacco pollen tube secretome identifies novel pollen tube guidance proteins important for fertilization. Genome Biol 17: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Li Y, Hu Y, Lin J (2005) Inhibition of RNA and protein synthesis in pollen tube development of Pinus bungeana by actinomycin D and cycloheximide. New Phytol 165: 721–729 [DOI] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ (2016) Redefining the translational status of 80S monosomes. Cell 164: 757–769 [DOI] [PubMed] [Google Scholar]
- Hildebrandt T, Knuesting J, Berndt C, Morgan B, Scheibe R (2015) Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol Chem 396: 523–537 [DOI] [PubMed] [Google Scholar]
- Hirokawa N. (2006) mRNA transport in dendrites: RNA granules, motors, and tracks. J Neurosci 26: 7139–7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Combe JP, Twell D, Čapková V (2000) The translationally repressed pollen-specific ntp303 mRNA is stored in non-polysomal mRNPs during pollen maturation. Sex Plant Reprod 13: 135–144 [Google Scholar]
- Honys D, Rĕnák D, Feciková J, Jedelský PL, Nebesárová J, Dobrev P, Capková V (2009) Cytoskeleton-associated large RNP complexes in tobacco male gametophyte (EPPs) are associated with ribosomes and are involved in protein synthesis, processing, and localization. J Proteome Res 8: 2015–2031 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H (2012) Ribosomes and translation in plant developmental control. Plant Sci 191-192: 24–34 [DOI] [PubMed] [Google Scholar]
- Huanca-Mamani W, Garcia-Aguilar M, León-Martínez G, Grossniklaus U, Vielle-Calzada JP (2005) CHR11, a chromatin-remodeling factor essential for nuclear proliferation during female gametogenesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 17231–17236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Valledor L, Lyon D, Gingl S, Nagler M, Meijón M, Egelhofer V, Weckwerth W (2014) Comprehensive cell-specific protein analysis in early and late pollen development from diploid microsporocytes to pollen tube growth. Mol Cell Proteomics 13: 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurečková JF, Sýkorová E, Hafidh S, Honys D, Fajkus J, Fojtová M (2017) Tissue-specific expression of telomerase reverse transcriptase gene variants in Nicotiana tabacum. Planta 245: 549–561 [DOI] [PubMed] [Google Scholar]
- Kakehi J, Kawano E, Yoshimoto K, Cai Q, Imai A, Takahashi T (2015) Mutations in ribosomal proteins, RPL4 and RACK1, suppress the phenotype of a thermospermine-deficient mutant of Arabidopsis thaliana. PLoS ONE 10: e0117309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ (2006) Neuronal RNA granules: movers and makers. Neuron 51: 685–690 [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS (2001) Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron 32: 683–696 [DOI] [PubMed] [Google Scholar]
- Łabno A, Tomecki R, Dziembowski A (2016) Cytoplasmic RNA decay pathways: enzymes and mechanisms. Biochim Biophys Acta 1863: 3125–3147 [DOI] [PubMed] [Google Scholar]
- Lafleur GJ, Mascarenhas JP (1978) The dependence of generative cell division in Tradescantia pollen tubes on protein and RNA synthesis. Plant Sci Lett 12: 251–255 [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Chen PW, Chuang MH, Juntawong P, Bailey-Serres J, Jauh GY (2014) Profiling of translatomes of in vivo-grown pollen tubes reveals genes with roles in micropylar guidance during pollination in Arabidopsis. Plant Cell 26: 602–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Chen HM, Wu SH (2012) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, Wu SH (2013) Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP. (1993) Molecular mechanisms of pollen tube growth and differentiation. Plant Cell 5: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, Martínez-Bernardini A, Fabrice TN, Ringli C, Muschietti JP, et al. (2017) RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 358: 1600–1603 [DOI] [PubMed] [Google Scholar]
- Meteignier LV, El Oirdi M, Cohen M, Barff T, Matteau D, Lucier JF, Rodrigue S, Jacques PE, Yoshioka K, Moffett P (2017) Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in Arabidopsis. J Exp Bot 68: 2333–2344 [DOI] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33: D284–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills SC, Enganti R, von Arnim AG (2018) What makes ribosomes tick? RNA Biol 15: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench DG, Zhang C, Dahodwala M (2012) Control of cytoplasmic translation in plants. Wiley Interdiscip Rev RNA 3: 178–194 [DOI] [PubMed] [Google Scholar]
- Murphy E, De Smet I (2014) Understanding the RALF family: a tale of many species. Trends Plant Sci 19: 664–671 [DOI] [PubMed] [Google Scholar]
- Nicholls C, Li H, Liu JP (2012) GAPDH: a common enzyme with uncommon functions. Clin Exp Pharmacol Physiol 39: 674–679 [DOI] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP (2012) Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, Chaturvedi P, Selymesi M, Ghatak A, Mesihovic A, Scharf KD, Weckwerth W, Simm S, Schleiff E (2016) The membrane proteome of male gametophyte in Solanum lycopersicum. J Proteomics 131: 48–60 [DOI] [PubMed] [Google Scholar]
- Pearce G, Moura DS, Stratmann J, Ryan CA Jr (2001) RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc Natl Acad Sci USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrů E, Hrabětová E, Tupý J (1964) The technique of obtaining germinating pollen without microbial contamination. Biol Plant 6: 68–69 [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Quint A, Brand D, Vivian-Smith A, Offringa R (2009) BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J 58: 109–121 [DOI] [PubMed] [Google Scholar]
- Roy B, von Arnim AG (2013) Translational regulation of cytoplasmic mRNAs. The Arabidopsis Book 11: e0165, doi/10.1199/tab.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA. (2017) Some like it HOT: protein translation and heat stress in plants. Plant Cell 29: 2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpin MR, Sigaut L, Temprana SG, Boccaccio GL, Pietrasanta LI, Muschietti JP (2017) Two Arabidopsis late pollen transcripts are detected in cytoplasmic granules. Plant Direct 1: e00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schisa JA. (2012) New insights into the regulation of RNP granule assembly in oocytes. Int Rev Cell Mol Biol 295: 233–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook JC, Carroll KL, Hung KF, Masson PH, Somerville CR (2002) The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sede AR, Borassi C, Wengier DL, Mecchia MA, Estevez JM, Muschietti JP (2018) Arabidopsis pollen extensins LRX are required for cell wall integrity during pollen tube growth. FEBS Lett 592: 233–243 [DOI] [PubMed] [Google Scholar]
- Sierro N, Battey JN, Ouadi S, Bovet L, Goepfert S, Bakaher N, Peitsch MC, Ivanov NV (2013) Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol 14: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5: 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirover MA. (2011) On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim Biophys Acta 1810: 741–751 [DOI] [PubMed] [Google Scholar]
- Sirover MA. (2012) Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J Cell Biochem 113: 2193–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stejskal K, Potěšil D, Zdráhal Z (2013) Suppression of peptide sample losses in autosampler vials. J Proteome Res 12: 3057–3062 [DOI] [PubMed] [Google Scholar]
- Štorchová H, Čapková V, Tupý J (1994) A Nicotiana-tabacum messenger-RNA encoding a 69-kDa glycoprotein occurring abundantly in pollen tubes is transcribed but not translated during pollen development in the anthers. Planta 192: 441–445 [Google Scholar]
- Szick K, Springer M, Bailey-Serres J (1998) Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins reveal a distinct protein of higher plant ribosomes. Proc Natl Acad Sci USA 95: 2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruneh BS, Kim BH, Gallie DR, Roy B, von Arnim AG (2013) The global translation profile in a ribosomal protein mutant resembles that of an eIF3 mutant. BMC Biol 11: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristan C, Shahani N, Sedlak TW, Sawa A (2011) The diverse functions of GAPDH: views from different subcellular compartments. Cell Signal 23: 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R, Ditengou FA, Sumi Y, Teale W, Palme K, Okada K (2009) NO VEIN mediates auxin-dependent specification and patterning in the Arabidopsis embryo, shoot, and root. Plant Cell 21: 3133–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tupý J, Rhová L (1984) Changes and growth effect of pH in pollen tube culture. J Plant Physiol 115: 1–10 [DOI] [PubMed] [Google Scholar]
- Turkina MV, Klang Årstrand H, Vener AV (2011) Differential phosphorylation of ribosomal proteins in Arabidopsis thaliana plants during day and night. PLoS ONE 6: e29307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek LS. (2012) ‘Ribozoomin’: translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci 13: 305–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek LS, Zeman J, Wagner S, Beznosková P, Pavlíková Z, Mohammad MP, Hronová V, Herrmannová A, Hashem Y, Gunišová S (2017) Embraced by eIF3: structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res 45: 10948–10968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Kelen K, Beyaert R, Inzé D, De Veylder L (2009) Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol 44: 143–168 [DOI] [PubMed] [Google Scholar]
- Vogler H, Martinez-Bernardini A, Grossniklaus U (2016) Maybe she’s NOT the boss: male-female crosstalk during sexual plant reproduction. Genome Biol 17: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Jia Q, Vaughn JN (2014) Regulation of plant translation by upstream open reading frames. Plant Sci 214: 1–12 [DOI] [PubMed] [Google Scholar]
- Walley JW, Kliebenstein DJ, Bostock RM, Dehesh K (2013) Fatty acids and early detection of pathogens. Curr Opin Plant Biol 16: 520–526 [DOI] [PubMed] [Google Scholar]
- Wang J, Lan P, Gao H, Zheng L, Li W, Schmidt W (2013) Expression changes of ribosomal proteins in phosphate- and iron-deficient Arabidopsis roots predict stress-specific alterations in ribosome composition. BMC Genomics 14: 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XX, Wang K, Liu XY, Liu M, Cao N, Duan Y, Yin G, Gao H, Wang WL, Ge W, et al. (2018) Pollen-expressed leucins-rich-repeat extensins are essential for pollen germination and growth. Plant Physiol 176: 1993–2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LQ, Xu WY, Deng ZY, Su Z, Xue Y, Wang T (2010) Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics 11: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings K, Reijnen W, van Aarssen R, Kortstee A, Spijkers J, van Herpen M, Schrauwen J, Wullems G (1992) Characterization of a pollen-specific cDNA clone from Nicotiana tabacum expressed during microgametogenesis and germination. Plant Mol Biol 18: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Weterings K, Schrauwen J, Wullems G, Twell D (1995) Functional dissection of the promoter of the pollen-specific gene NTP303 reveals a novel pollen-specific, and conserved cis-regulatory element. Plant J 8: 55–63 [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Ostasiewicz P, Mann M (2011) High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J Proteome Res 10: 3040–3049 [DOI] [PubMed] [Google Scholar]
- Wittink F, Knuiman B, Derksen J, Čapková V, Twell D, Schrauwen J, Wullems G (2000) The pollen-specific gene Ntp303 encodes a 69-kDa glycoprotein associated with the vegetative membranes and the cell wall. Sex Plant Reprod 12: 276–284 [Google Scholar]
- Xu J, Yang JY, Niu QW, Chua NH (2006) Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18: 3386–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yángüez E, Castro-Sanz AB, Fernández-Bautista N, Oliveros JC, Castellano MM (2013) Analysis of genome-wide changes in the translatome of Arabidopsis seedlings subjected to heat stress. PLoS ONE 8: e71425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J (2005) Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol 138: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Deng R, Guo Z, Yang S, Deng X (2016) Genome-wide identification and characterization of glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genomics 17: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bramsiepe J, Van Durme M, Komaki S, Prusicki MA, Maruyama D, Forner J, Medzihradszky A, Wijnker E, Harashima H, et al. (2017) RETINOBLASTOMA RELATED1 mediates germline entry in Arabidopsis. Science 356: eaaf6532. [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]