The inhibition of the growth regulator TARGET OF RAPAMYCIN (TOR) negatively regulates growth and development; during periods of nutrient scarceness, a repressed activity of TOR helps surmount these conditions and improve survival and development.
Abstract
Upon illumination, etiolated seedlings experience a transition from heterotrophic to photoautotrophic growth. During this process, the tetrapyrrole biosynthesis pathway provides chlorophyll for photosynthesis. This pathway has to be tightly controlled to prevent the accumulation of photoreactive metabolites and to provide stoichiometric amounts of chlorophyll for its incorporation into photosynthetic protein complexes. Therefore, plants have evolved regulatory mechanisms to synchronize the biosynthesis of chlorophyll and chlorophyll-binding proteins. Two phytochrome-interacting factors (PIF1 and PIF3) and the DELLA proteins, which are controlled by the gibberellin pathway, are key regulators of this process. Here, we show that impairment of TARGET OF RAPAMYCIN (TOR) activity in Arabidopsis (Arabidopsis thaliana), either by mutation of the TOR complex component RAPTOR1B or by treatment with TOR inhibitors, leads to a significantly reduced accumulation of the photoreactive chlorophyll precursor protochlorophyllide in darkness but an increased greening rate of etiolated seedlings after exposure to light. Detailed profiling of metabolic, transcriptomic, and physiological parameters revealed that the TOR-repressed lines not only grow slower, they grow in a nutrient-saving mode, which allows them to resist longer periods of low nutrient availability. Our results also indicated that RAPTOR1B acts upstream of the gibberellin-DELLA pathway and its mutation complements the repressed greening phenotype of pif1 and pif3 after etiolation.
Light is one of the major environmental factors that regulate plant growth and development. After germinating in soil, seedlings grow heterotrophically in the dark via a process referred to as skotomorphogenesis (etiolation; Von Arnim and Deng, 1996). Etiolated seedlings lack chlorophyll but accumulate the chlorophyll precursor protochlorophyllide (Pchlide) within the prolamellar body (PLB) of etioplasts (Sperling et al., 1998). Once the emerging seedlings are illuminated by sunlight, their genetic program switches to photomorphogenesis (deetiolation), inducing chlorophyll biosynthesis and, thus, greening of the cotyledons. This process is mediated by the light-activated NADPH:protochlorophyllide oxidoreductase (POR), which catalyzes the conversion of Pchlide into chlorophyllide (Chlide), and the later is esterified subsequently by chlorophyll synthase with phytyl pyrophosphate to synthesize chlorophyll a (Reinbothe et al., 1996). The amount of Pchlide that accumulates in etiolated tissue is correlated with the level of POR. A deregulated chlorophyll synthesis pathway would cause excessive levels of free Pchlide, which could produce large amounts of reactive oxygen species (ROS) upon light exposure, leading to severe photobleaching of cotyledons or even cell death (Reinbothe et al., 1996; Sperling et al., 1998; Meskauskiene et al., 2001; Lee et al., 2003; Buhr et al., 2008). Therefore, a tight regulation of Pchlide formation and its subsequent conversion to chlorophyll is particularly important for plants during the transition from heterotrophic to photoautotrophic growth.
Plants have developed multiple levels of regulation to efficiently and safely produce chlorophyll and the pigment-binding proteins of photosynthesis, including by fine-tuning of enzymatic activities, control of protein stability and suborganelle localization, modulation of protein complex formation, and transcriptional regulation (Brzezowski et al., 2015). The transcriptional regulation of tetrapyrrole biosynthesis during greening is mediated by LONG HYPOCOTYL5, PHYTOCHOROME-INTERACTING FACTORS (PIFs), GOLDEN2-LIKE transcription factors, FAR-RED ELONGATED HYPOCOTYL3, REVEILLE1, and scarecrow-like transcription factors (Kobayashi and Masuda, 2016). Among these regulators, PIFs are important transcription factors that inhibit chlorophyll biosynthesis by controlling the expression of a number of biosynthetic and ROS-responsive genes (Leivar et al., 2009; Stephenson et al., 2009; Chen et al., 2013; Liu et al., 2013; Toledo-Ortiz et al., 2014). The GA-regulated DELLA (GA-INSENSITIVE [GAI], REPRESSOR OF GA1-3 [RGA], RGA-LIKE1 [RGL1], RGL2, and RGL3) proteins are important for etiolated seedlings to cope with the transition to photoautotrophic growth (Cheminant et al., 2011). GA relieves the DELLA-mediated repression of gene expression by forming a complex with its receptor GA-INSENSITIVE DWARF1 (GID1) and DELLAs, which subsequently stimulates the proteasome-dependent degradation of the DELLA proteins (Dill et al., 2004; Fu et al., 2004; Griffiths et al., 2006; Willige et al., 2007). DELLAs accumulate in etiolated cotyledons to derepress chlorophyll biosynthesis by repressing the transcriptional activity of PIFs and simultaneously up-regulating POR expression in a PIF-independent manner (Cheminant et al., 2011). Although ga1-3 mutants, which accumulate DELLAs, have more Pchlide under dark conditions, the mutants have high rates of greening after transfer to light, implying that they also have elevated POR protein activity (Cheminant et al., 2011).
Alongside transcriptional regulation, several metabolites are involved directly or indirectly in the precise control of chlorophyll biosynthesis in plants. The accumulation of Pchlide negatively regulates its own biosynthesis in darkness. After binding to Pchlide, both PORB and PORC can interact with FLUORESCENT IN BLUE (FLU) and CHL27, a protein of the Mg protoporphyrin monomethylester cyclase complex, which is involved in the inhibition of glutamyl-tRNA reductase (GluTR) in the absence of light (Kauss et al., 2012). Similarly, heme, another end product of the tetrapyrrole biosynthesis pathway, inhibits 5-aminolevulinic acid synthesis (Vothknecht et al., 1998). These two paths provide sufficient negative feedback regulation within the tetrapyrrole pathway to prevent plants from overaccumulating toxic photoreactive precursors of chlorophyll. Hence, mutants lacking FLU accumulate excessive amounts of Pchlide in the dark due to perturbation of the negative feedback inhibition of GluTR and do not survive day/night cycles (Meskauskiene et al., 2001; Kauss et al., 2012). Etiolated leaves and the PLBs contain carotenoids, which are essential photoprotective and antioxidant pigments synthesized by all photosynthetic organisms (Shumskaya and Wurtzel, 2013). In particular, carotenoids are documented to convert excess excitation energy from chlorophyll into heat in order to protect plants against photooxidative damage (Park et al., 2002). Intriguingly, the induction of carotenoid biosynthesis in the dark ultimately improves greening upon illumination (Rodríguez-Villalón et al., 2009).
TARGET OF RAPAMYCIN (TOR) is an evolutionarily conserved eukaryotic regulator of growth that modulates protein synthesis (Ma and Blenis, 2009) and metabolism in animals (González and Hall, 2017; Saxton and Sabatini, 2017) and plants (Xiong and Sheen, 2014; Dobrenel et al., 2016). In animal cells, TOR typically forms two distinct complexes with specific regulatory functions (Wullschleger et al., 2006). The three central components of TOR complex 1 (TORC1), namely TOR, REGULATORY ASSOCIATED PROTEIN OF TOR (RAPTOR), and LETHAL with SEC THIRTEEN8 (LST8), also have been described in several plant species (Dobrenel et al., 2016). Interactions between TOR and RAPTOR (Mahfouz et al., 2006), as well as between TOR and LST8 (Moreau et al., 2012), occur in Arabidopsis (Arabidopsis thaliana), indicating the existence of a functional TORC1 in plants. Interestingly, contrary to yeast (Saccharomyces cerevisiae; Heitman et al., 1991), the Arabidopsis genome contains only one TOR isoform, and mutations in this gene result in embryo lethality (Menand et al., 2002).
The TOR complex is a key regulator for an array of cellular processes (Dobrenel et al., 2016). More specifically, TOR is implicated in the control of cell cycle activity within the root meristem and during the transition from heterotrophic to photoautotrophic growth of young Arabidopsis seedlings (Xiong et al., 2013). This effect is different in root and shoot apices, with auxin biosynthesis being required for the activation of TOR in the latter but not in the former (Li et al., 2017). Furthermore, TOR signaling promotes the accumulation of the brassinosteroid transcription factor BZR1 (Zhang et al., 2016) and BRASSINOSTERIOD INSENSITIVE2 (Xiong et al., 2017), which modulate growth through complex hormonal and environmental signals.
In contrast to the single TOR gene present in most plant species (Dobrenel et al., 2016), there are two homologous RAPTOR isoforms (RAPTOR1A and RAPTOR1B) in Arabidopsis (Anderson and Hanson, 2005; Deprost et al., 2005) and other plant species (Dobrenel et al., 2016). Disruption of the major RAPTOR isoform, RAPTOR1B, causes a wide range of developmental defects, whereas mutants of the lowly expressed RAPTOR1A do not show any major phenotype (Anderson et al., 2005; Deprost et al., 2005; Salem et al., 2017). Accordingly, it was shown that a deficiency of RAPTOR1B alters the hormone and metabolite compositions of Arabidopsis seeds and causes severe alterations in seed morphology, viability, and germination potential (Salem et al., 2017). Moreover, the disruption of RAPTOR1B alters the hormonal composition of plants in their vegetative growth phase, which led to morphological phenotypes of shoot and root tissue, delays in developmental transitions, and decreased carbon fixation rates (Salem et al., 2018). Recent evidence also linked TOR and the major cellular energy sensor SnRK1 in plants (Broeckx et al., 2016; Nukarinen et al., 2016). This interplay seems to be mediated through an interaction between SnRK1 and RAPTOR in both animal and plant cells, placing RAPTOR in a central position to regulate TOR function (Hindupur et al., 2015; Baena-González and Hanson, 2017).
In this study, we provide several lines of evidence suggesting that the impairment of TOR complex activity, either by mutation of its component protein RAPTOR1B or by administering drugs that inhibit the TOR kinase, results in increased greening of seedlings after extended etiolation. This improved greening response was primed by a decrease of the chlorophyll precursor Pchlide, a reduction in ROS levels, enhanced potential of POR activity, and greater amounts of primary and lipophilic metabolites as well as pyrimidine nucleotides. We further found that TOR activity was dependent on a functional GA-sensing and -signaling pathway during greening.
RESULTS
Inhibition of the TOR Complex Promotes the Greening of Etiolated Seedlings
TOR is crucial for the control of cell cycle activity within the root meristem during the heterotrophic-to-photoautotrophic transition of young light-grown Arabidopsis seedlings (Xiong et al., 2013). This observation prompted us to investigate whether TOR also is important for the dark-to-light transition of etiolated seedlings. As tor mutants are embryo lethal (Menand et al., 2002), we decided to use four independent T-DNA insertion mutants of RAPTOR1B (Supplemental Fig. S1, A and B). These lines were named, as in our previous articles (Salem et al., 2017, 2018), rb for raptor1b, followed by the first two nonzero digits of the SALK line number (Alonso et al., 2003). As can be seen from Supplemental Figure S1C, the rb lines had minute levels of RAPTOR1B transcripts, while the RAPTOR1A transcript showed a 10% to 50% increase in the rb mutant background.
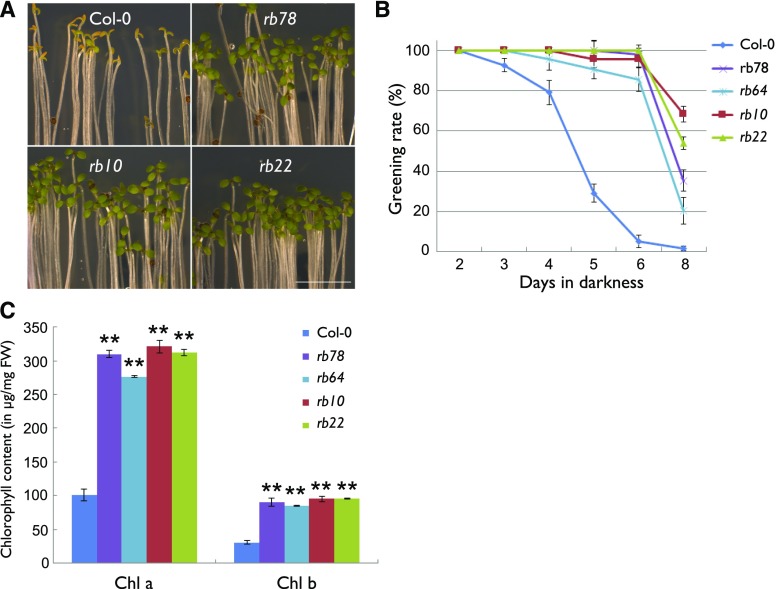
In an initial experiment, the four rb lines and the control (Columbia-0 [Col-0], referred to further as the wild type) were grown in darkness for 2, 3, 4, 5, 6, and 8 d on agar plates before they were transferred to light for another 2 d. Unexpectedly, all the rb seedlings showed significantly higher greening rates compared with the wild type after illumination (Fig. 1A). Less than 6% of 6-d-old etiolated wild-type seedlings were able to turn green after exposure to light. In contrast, more than 85% of the rb seedlings developed green cotyledons under the same conditions (Fig. 1B). This greening behavior correlated well with the extractable levels of chlorophyll, which were significantly higher in the rb seedlings compared with the wild type (Fig. 1C).
Figure 1.
Mutation of RAPTOR1B promotes the greening of etiolated seedlings. A, Representative images of 6-d-old etiolated seedlings after exposure to light for 2 d. Bar = 5 mm. B, Greening rates of etiolated seedlings grown in darkness for the indicated number of days before exposure to light for 2 d. Data are means ± se (n = 3, more than 80 seedlings for each biological replicate). C, Chlorophyll content (μg g−1 fresh weight [FW]) of seedlings grown in darkness for 5 d and exposure to light for 2 d. Data are means ± se (n = 3). Asterisks represent values from rb plants that are significantly different from those from wild-type plants (**, P < 0.01 by Student’s t test).
Since rb seeds showed morphological and metabolic changes compared with the wild type (Salem et al., 2017), we performed an additional control experiment, using two previously described inhibitors of the active site of the TOR kinase (AZD-8055 and Torin2; Montané and Menand, 2013), to determine whether the inhibition of TOR in etiolated wild-type seedlings also leads to enhanced greening upon light exposure. Plants were germinated for 2, 4, and 6 d on plates in the dark before transferring them for another 2 d to the light. Similar to the results obtained for the rb mutants, the greening rate of the TOR-inhibited plants was elevated significantly compared with the wild-type plants treated with the drug vehicle only and phenotypically resembled the rb seedlings (Fig. 1, A and B; Supplemental Fig. S2, A and B). These data demonstrate that the repression of TOR activity during etiolation improved the greening efficiency upon illumination.
rb Seedlings Accumulate Lower Amounts of Tetrapyrrole Precursors, Show Increased POR Expression, and Have an Increased PLB Size
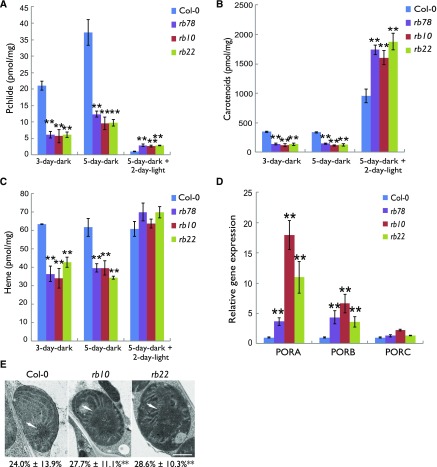
Chlorophyll, one of the final products of the tetrapyrrole biosynthesis pathway (Tanaka et al., 2011), was increased significantly in rb seedlings after a 2-d light exposure (Fig. 1C). Therefore, we asked what happened to other products or intermediates of the tetrapyrrole biosynthesis pathway in the rb lines. To answer this question, we initially determined the level of Pchlide, which is the last product of the chlorophyll-synthesizing branch of the tetrapyrrole pathway in angiosperms grown in the dark (Tanaka et al., 2011). Contrary to the observed increase in chlorophyll in the rb seedlings after the 2-d light treatment, we detected only 25% to 30% of the wild-type Pchlide levels in the 3- and 5-d-old etiolated rb seedlings (Fig. 2A). Similarly, Pchlide levels were decreased significantly in wild-type etiolated seedlings that were exposed to the TOR inhibitors AZD-8055 and Torin2 (Supplemental Fig. S2C). Surprisingly, the level of Pchlide, which was almost depleted in the wild-type seedlings after 2 d of light exposure, was significantly higher in the light-exposed rb seedlings (Fig. 2A).
Figure 2.
The rb seedlings accumulate altered amounts of Pchlide, heme, and carotenoids. A to C, Absolute quantification of Pchlide (A), carotenoids (B), and heme (C) in 3-d-old etiolated, 5-d-old etiolated, and 5-d-old etiolated plus 2-d light-grown seedlings.The data for each histogram are means ± SE (n = 4). D, Analysis of PORA/B/C gene expression by RT-qPCR in 5-d-old etiolated seedlings. The expression level of UBQ10 was used as an internal standard. Error bars represent sd (n = 3). E, Transmission electron microscopy images of cotyledon etioplasts from 5-d-old etiolated seedlings. The arrows indicate the PLB. The PLB area as a percentage of host etioplast area is shown under each image and represents the mean ± se (n > 300). Asterisks represent values that are statistically different from that of the wild type sample at the same condition (**, P < 0.01 by Student’s t test).
Similar to the situation observed for Pchlide, we detected significantly lower concentrations of carotenoids in the rb seedlings prior to illumination and increased levels after illumination as compared with the wild type (Fig. 2B). This was especially interesting, since carotenoids are able to dissipate the excess energy of triplet excited chlorophylls and, therefore, reduce the harmful formation of singlet oxygen (Niyogi, 2000).
Another major product of tetrapyrrole biosynthesis is heme, a product of the iron branch (Tanaka et al., 2011). Because we detected significant changes in chlorophyll biosynthesis, we asked if other products of this pathway also were affected in rb seedlings. Indeed, we found an approximately 50% decrease in the steady-state level of heme in the 5-d etiolated mutants compared with the wild type (Fig. 2C). These data are in agreement with the Pchlide and carotenoid data outlined above. However, contrary to the chlorophyll levels, which were increased significantly in the light-exposed rb seedlings (Fig. 1C), the heme content did not change significantly in the rb lines upon light exposure (Fig. 2C).
Based on the reduced Pchlide levels in the rb seedlings in the dark (Fig. 2A) and the increased chlorophyll levels in the mutants after exposure to light (Fig. 1C), we decided to determine if the abundance of POR, the main enzyme that catalyzes the conversion of Pchlide to Chlide (Tanaka et al., 2011), was quantitatively changed in these seedlings. Reverse transcription quantitative PCR (RT-qPCR) revealed that the expression of PORA and PORB was significantly higher in 5-d etiolated rb seedlings as compared with the wild type (Fig. 2D). The bulk of POR protein is found within the membranes of the PLB; hence, the PLB size correlates to the amount of POR in etiolated seedlings (Sperling et al., 1998). To determine if PLB size is different in the rb plants, we examined the ultrastructure of cotyledon etioplasts of 5-d-old seedlings and compared these with the corresponding wild type. To correct the variable size of PLB resulting from the angle of the section for transmission electron microscopy, we calculated the PLB area as a percentage of host etioplast area. Indeed, the PLB size was slightly but significantly larger in the rb lines as compared with the wild type (Fig. 2E), which is consistent with the higher POR expression in the rb seedlings.
We subsequently determined whether the larger PLB resulted in higher POR activity (i.e. the light-dependent reduction of Pchlide to Chlide) when etiolated rb seedlings were exposed to light. For this purpose, we measured the light-induced conversion of protochlorophyllide to chlorophyllide using a spectrofluorometric assay. Interestingly, even though rb seedlings contained reduced Pchlide levels at the end of the 5-d etiolation period (Fig. 2A; Supplemental Fig. S3A), the Chlide content, as indicated by the fluorescence peak at 670 nm, increased substantially more after light treatment in the rb than in the wild-type seedlings (Supplemental Fig. S3B). Therefore, we hypothesize that the rb seedlings have higher POR activity, or a higher capacity for Pchlide reduction to Chlide, than the wild-type seedlings, as they can produce more Chlide from a smaller pool of Pchlide upon illumination.
rb Seedlings Produce Low Levels of Singlet Oxygen
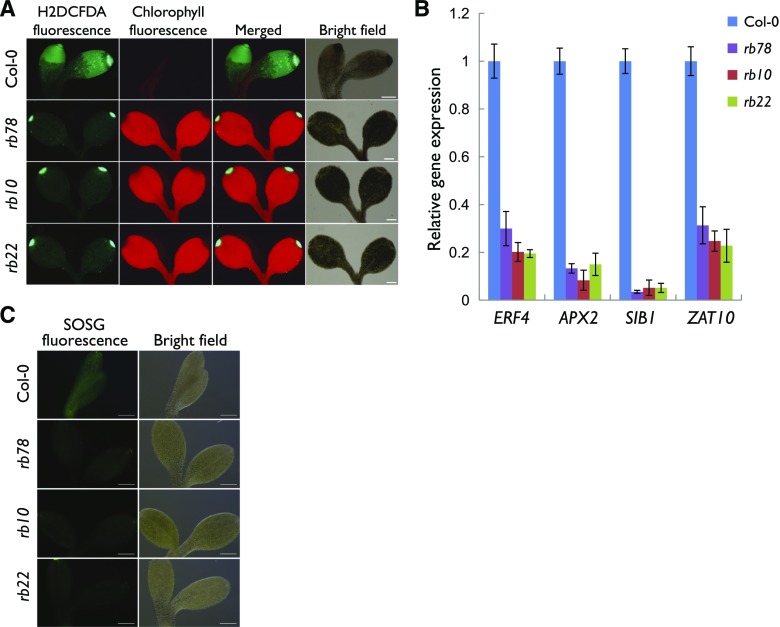
Based on the increased Pchlide concentrations (Fig. 2A) and increased levels of carotenoids (Fig. 2B) in rb upon exposure to light, we reasoned that rb lines might produce lower levels of ROS than the wild type. Therefore, we determined cellular ROS levels in wild-type and rb seedlings using the ROS-sensitive dye 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), which is sensitive to most oxidizing species in the cell. Consistent with the observed greening phenotype, 5-d-old etiolated rb cotyledons displayed much weaker H2DCFDA fluorescence compared with wild-type seedlings after 2 d of light exposure (Fig. 3A). In addition, after transfer of the etiolated seedlings to white light for 1 h, the expression levels of ROS-responsive genes, including the two singlet oxygen-responsive genes ETHYLENE RESPONSIVE TRANSCRIPTION FACTOR4 and SIGMA FACTOR BINDING PROTEIN1, were decreased significantly in rb seedlings when compared with the wild type (Fig. 3B).
Figure 3.
Mutation of RAPTOR1B leads to decreased accumulation of ROS after exposure to light. A, Fluorescence images of ROS (as indicated by H2DCFDA) and chlorophyll in the cotyledons of 5-d-old etiolated seedlings followed by 2 d of white light. Bars = 200 µm. B, Analysis of ROS-responsive gene expression by RT-qPCR of 5-d-old etiolated seedlings after exposed to white light for 1 h. The expression level of UBQ10 was used as an internal standard. Error bars represent sd (n = 3). C, Singlet oxygen (as indicated by SOSG) production in the cotyledons of 5-d-old etiolated seedlings. Bars = 200 µm.
To specifically dissect the ROS response, we further investigated the production of singlet oxygen using the specific fluorescent probe Singlet Oxygen Sensor Green (SOSG). As shown in Figure 3C, rb cotyledons displayed weaker SOSG fluorescence than wild-type seedlings, indicating that less singlet oxygen was released in the mutants upon light irradiation. By contrast, the levels of two other ROS species, superoxide and hydrogen peroxide, were higher in rb as compared with wild-type seedlings (assessed by nitroblue tetrazolium and diaminobenzidine staining, respectively; Supplemental Fig. S4).
TOR Signaling Functions Upstream of the GA-DELLA Signaling Pathway, and Disruption of RAPTOR1B Complements the Greening Defects of PIF Mutants
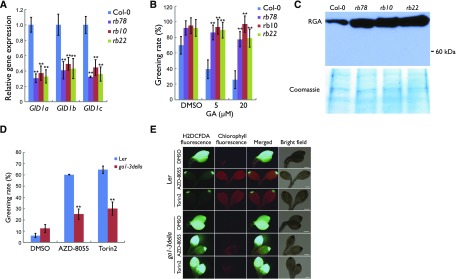
DELLA proteins are known to promote POR expression and PLB formation in the dark, which protects seedlings from photooxidative damage during deetiolation (Cheminant et al., 2011). Therefore, we hypothesized that the GA signaling pathway might be impaired in rb lines, which, in turn, could contribute to the higher expression level of POR genes in the rb seedlings. To assess this, we analyzed the expression levels of several GA signaling pathway genes in rb by RT-qPCR. We found that all three GA receptors (GID1A/B/C) were down-regulated significantly in rb (Fig. 4A). In agreement with the impaired expression of GA receptors, rb plants also were less sensitive to exogenous GA treatment. While the greening rate of 4-d-old etiolated wild-type seedlings decreased significantly in a GA dose-dependent manner, the greening rate of the rb seedlings was significantly less affected by GA (Fig. 4B). These data support a role for RAPTOR1B in the regulation of the greening process by affecting the expression of GA receptors. Moreover, the amount of RGA, one of the major DELLA proteins, which is degraded in a GA-dependent process (Achard and Genschik, 2009), was significantly higher in rb than in the wild type (Fig. 4C), confirming that GA signaling is reduced in the rb seedlings.
Figure 4.
The rb seedlings have an impaired GA signaling pathway, and mutation of DELLAs suppresses the Tor inhibition-caused greening phenotype. A, Analysis of GID1A/B/C gene expression by RT-qPCR in 5-d-old etiolated seedlings. The expression level of UBQ10 was used as an internal standard. Error bars represent sd (n = 3). B, Greening rates of 4-d-old etiolated seedlings followed by 2 d of white light with or without exogenous GA treatment. Data are means ± se (n = 3, more than 80 seedlings for each biological replicate). DMSO, Dimethyl sulfoxide. C, Western blot of DELLA proteins in 5-d-old etiolated seedlings. A protein gel stained with Coomassie Blue is shown as a loading control. D, Greening rates of etiolated seedlings grown in the dark for 5 d before exposure to light for 2 d. Data are means ± se (n = 3, more than 80 seedlings for each biological replicate). Ler, Landsberg erecta. E, Fluorescence images of ROS (as indicated by H2DCFDA) and chlorophyll in the cotyledons of 5-d-old etiolated seedlings followed by 2 d of white light. Bars = 200 μm.
To confirm that this phenotype was specific to inactivation of the TOR complex, wild-type and ga1-3della mutant seedlings were treated with the ATP-competitive TOR inhibitors AZD-8055 and Torin2 (Montané and Menand, 2013) and their greening rates were assessed. Both inhibitors dramatically increased the greening rate (Fig. 4D) and reduced the ROS level (Fig. 4E) in wild-type seedlings. However, the effects were lessened in ga1-3della mutants compared with wild-type plants (Fig. 4, D and E).
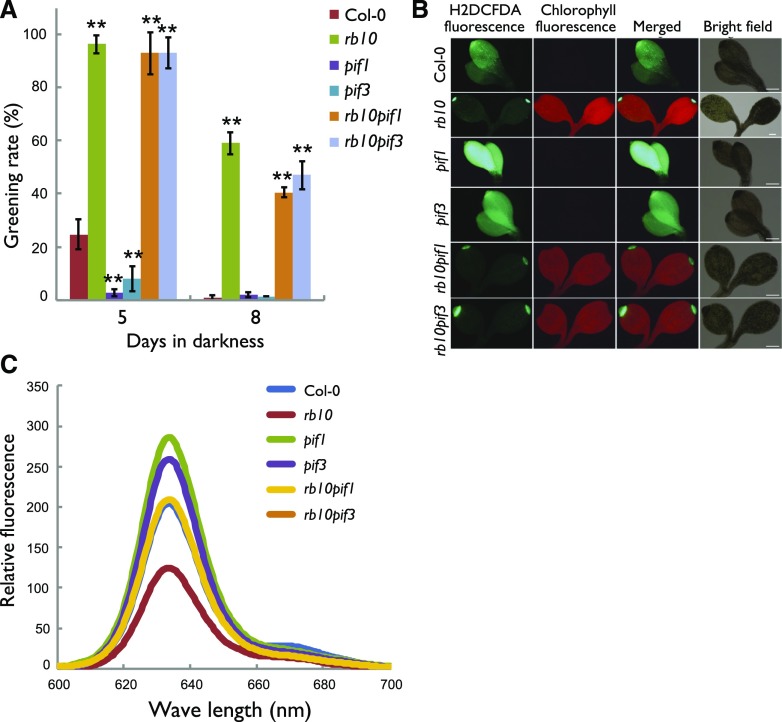
Similar to the GA signaling pathway, PIF transcription factors are critical negative regulators of chlorophyll biosynthesis (Stephenson et al., 2009). To determine whether the TOR complex-mediated repression of cotyledon greening depends on PIFs, we assessed possible genetic interactions between PIF1 or PIF3 and RAPTOR1B. For this purpose, we made reciprocal crosses between rb10 and pif1 or pif3 and analyzed the behavior of the progeny during the greening process after extended etiolation. Mutations in RAPTOR1B substantially increased the greening rate upon light irradiation in both the pif1 and pif3 backgrounds (Fig. 5A).
Figure 5.
The greening phenotype of rb is epistatic to PIF1 and PIF3. A, Greening rates of etiolated seedlings grown in darkness for the indicated number of days before exposure to light for 2 d. Data are means ± se (n = 3, more than 80 seedlings for each biological replicate). Asterisks represent values that are statistically different from that of the wild-type sample at the same condition (**, P < 0.01 by Student’s t test). B, Fluorescence images of ROS (as indicated by H2DCFDA) and chlorophyll in the cotyledons of 5-d-old etiolated seedlings followed by 2 d of white light. Bars = 200 µm. C, Relative fluorescence of Pchlide in 5-d-old etiolated seedlings.
Consistent with the improved greening rate of the double mutants, both the ROS level after light treatment (Fig. 5B) and the Pchlide level in etiolated seedlings (Fig. 5C) were decreased significantly in rb10pif1 and rb10pif3 double mutants as compared with the pif1 and pif3 single knockout mutants. Notably, even though the Pchlide levels in the double mutants were reduced significantly, they were still higher than the levels observed in the rb mutant lines, reaching wild-type levels (Fig. 5C). This result indicates that the decrease in the Pchlide levels was not necessarily solely responsible for the reduced ROS levels and the improved greening rates of the rb10pif mutants (Fig. 5A). Accordingly, we can conclude that rb10pif1 and rb10pif3 double mutants not only regained partial control of tetrapyrrole biosynthesis but also showed improved antioxidant capacity leading to substantially reduced ROS levels (Fig. 5B).
RAPTOR1B Deficiency Influences the Sugar and Amino Acid Levels of Etiolated and Deetiolated Seedlings
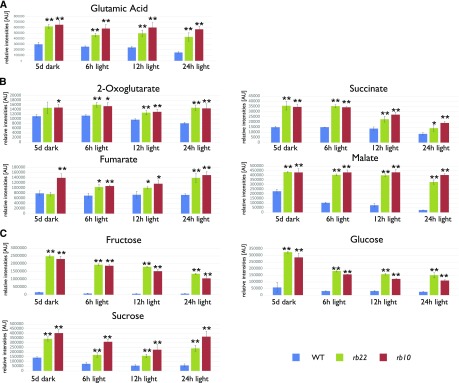
Because most of the tetrapyrrole biosynthesis pathway intermediates were reduced significantly at the end of the dark phase and increased after illumination, we asked how defects in RAPTOR1B function affect global metabolite levels. As the first committed precursor for the synthesis of tetrapyrroles is 5-aminolevulinic acid, which is synthesized from Glu (Tanaka et al., 2011), we decided to initially focus our analysis on the amino acids to assess if the decreased Pchlide levels in the rb seedlings were due to a decreased availability of Glu. For this purpose, we grew rb mutants and the wild type for 5 d in the dark before exposing them to light for 6, 12, and 24 h.
Interestingly, we found that the wild-type samples were depleted of almost all 18 detected amino acids during the 5-d etiolation period (Supplemental Fig. S5; Supplemental Table S1). The levels of 10 of the 18 amino acids were elevated in the rb lines throughout the whole 24-h illumination period, indicating that these compounds were either generated at higher levels or consumed at a slower pace than in the wild type. Only two amino acids (Pro and Arg) were reduced upon light exposure in the rb lines, while the levels of the remaining six amino acids did not differ between the wild type and the mutant lines (Supplemental Fig. S5). Glu was one of the 10 amino acids that showed a high level at the end of the dark phase as well as in response to light in the rb mutants (Fig. 6A).
Figure 6.
rb seedlings show considerably altered primary metabolism. Histograms show the relative levels of metabolites extracted from Col-0, rb10, and rb22 seedlings. A, Glu is an essential precursor for tetrapyrrole biosynthesis. B and C, Tricarboxylic acid cycle intermediates (B) and carbohydrates from primary metabolism (C). All seedlings were grown in the dark for 5 d and then transferred to the light. Samples were collected immediately prior to light exposure (5 d of dark) or after defined periods of light exposure (6, 12, or 24 h of light). For each genotype and time point, three independent biological samples were harvested. The histograms show means of three replicates ± sd (n = 3). Asterisks represent values that are statistically different from that of the wild-type sample at the same condition (*, P < 0.05 and **, P < 0.01 by Student’s t test). The normalized data of all measured primary metabolites can be obtained from Supplemental Table S1. AU, Arbitrary units; WT, wild type.
More surprisingly, the levels of all the detected tricarboxylic acid cycle intermediates (2-oxoglutarate, succinate, fumarate, and malate) were elevated significantly in the etiolated rb lines in comparison with the wild type and were maintained at similar levels throughout the illumination period (Fig. 6B; Supplemental Table S1). By contrast, the level of pyruvate, the cytosolic precursor of the tricarboxylic acid cycle, did not differ between the rb and wild-type seedlings across the time points analyzed (Supplemental Table S1).
In agreement with the increased levels of amino acids and tricarboxylic acid cycle intermediates, the levels of the most abundant sugars (Glc, Fru, and Suc) also were elevated massively in the mutant lines as compared with the wild type (Fig. 6C; Supplemental Table S1).
Taken together, our metabolic analyses indicate that there are substantial changes in the production and utilization of essential biosynthetic precursors responsible for the efficient and successful conversion from heterotrophic to photoautotrophic growth, implying that the fluxes through these pathways were significantly different in the rb seedlings in comparison with the wild type.
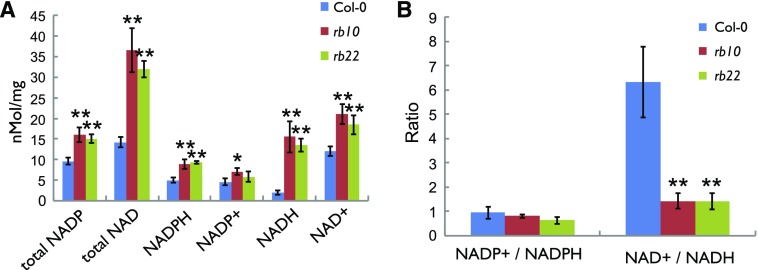
rb Seedlings Maintain Higher Levels of Pyrimidine Nucleotides during Etiolation Compared with the Wild Type
The synthesis of tetrapyrroles not only requires Glu and the Glu-tRNA synthetase but also consumes ATP and NADPH to produce the direct precursor of 5-aminolevulinic acid, Glu 1-semialdehyde (Tanaka et al., 2011). Given the elevated levels of amino acids, organic acids from the tricarboxylic acid cycle, and their sugar respiratory precursors, we decided to evaluate the levels of pyrimidine nucleotides (NAD+, NADP+, NADH, and NADPH) in the rb mutants. Because the measurement of these energy-rich metabolites is particularly arduous and given that the relative changes in the levels of metabolites were largely the same irrespective of the etiolation state, we chose to perform these measurements only at the end of day 5, prior to the start of illumination. Surprisingly, the sum of the reduced and oxidized pyrimidine nucleotides was significantly higher in the etiolated rb seedlings, perhaps implying that these reduction equivalents are consumed less during etiolation in the rb lines than in the wild type (Fig. 7A). We found a 2-fold increase in the total NAD levels in the rb plants compared with the wild type. In addition, the amount of NADP nucleotides was increased by approximately 50% in the etiolated rb seedlings (Fig. 7A). When the cellular redox poise of the NAD+/NADH and NADP+/NADPH couples was calculated, the NADP+/NADPH ratio was found to be invariant, while the NAD+/NADH ratio was found to be elevated considerably in the wild type compared with the rb seedlings (Fig. 7B). These results clearly point toward an increased availability of energy in the rb seedlings at the end of the etiolation process as compared with the wild type.
Figure 7.
The mutation of RAPTOR1B leads to increased pyrimidine nucleotide levels. The quantification of pyrimidine levels (A) and the NADP+/NADPH and NAD+/NADH ratios (B) are shown in 5-d-old etiolated seedlings. Data are means ± se (n = 5). Asterisks represent values that are statistically different from those of the wild-type samples (*, P < 0.05 and **, P < 0.01 by Student’s t test).
Mutation of RAPTOR1B Results in Considerable Alterations in Lipophilic Metabolite Levels during the Deetiolation Process
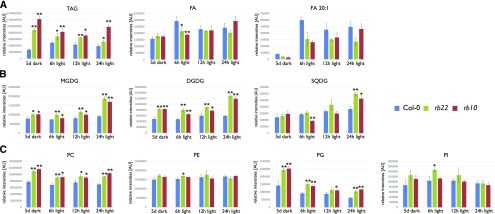
The reduction of pyrimidine nucleotides in nonphotosynthesizing tissues is related directly to the tricarboxylic acid cycle and the oxidative breakdown of storage lipids (Penfield et al., 2006). To validate if changes in the levels of storage lipids, namely triacylglycerols (TAGs), and the levels of structural lipids occur in the rb seedlings, we decided to conduct a lipidomic analysis (Salem et al., 2016) of the same samples used for the metabolic analysis (Supplemental Table S2).
As shown in Figure 8A, we found that the total amounts of TAGs, which are the major source of storage carbon in germinating Arabidopsis seedlings (Graham, 2008; Nguyen et al., 2015), were reduced significantly in the wild-type samples at all four time points compared with the rb seedlings. These results were surprising because the TAG levels in the mature seeds of the wild type and the rb mutants were almost identical (Salem et al., 2017). Nevertheless, these results indicate that the etiolated wild-type seedlings seem to utilize significantly larger amounts of this lipid-derived energy resource during the etiolation period (Fig. 8A). Indeed, these data are in full agreement with the decreased concentration of NADH in the wild type (Fig. 7A), which is one of the main products of the β-oxidation of fatty acids and is derived from the breakdown of TAG (Graham, 2008).
Figure 8.
The mutation of RAPTOR1B has considerable effects on lipid metabolism. Histograms show the relative levels of the indicated lipid classes extracted from Col-0, rb10, and rb22 seedlings. A, Levels of storage lipids (TAGs) and their breakdown products (FAs). B and C, Plastidial lipids (B) and the main lipids from endomembranes (C). All seedlings were grown in darkness for 5 d and then transferred to the light. Samples were collected immediately prior to light exposure (5 d of dark) or after defined periods of light exposure (6, 12, or 24 h of light). The histograms show means of the sums of the individual lipid species of each lipid class ± sd (n = 3). Asterisks represent values that are statistically different from that of the wild-type sample at the same condition (*, P < 0.05 and **, P < 0.01 by Student’s t test). The normalized data of all individual lipid species can be obtained from Supplemental Table S2. AU, Arbitrary units.
Contrary to the decreased production of chemical energy during the 5-d etiolation, the rb lines can utilize their energy resource more efficiently to synthesize plastidic membrane lipids in response to light. This is highlighted in Figure 8B, as the levels of monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and sulfoquinovosyldiacylglycerol (SQDG) do not increase substantially in the wild type during the 24-h illumination period, while the levels were doubled in the rb seedlings. This result is in agreement with the data obtained from the chlorophyll measurements (Fig. 1C), indicating that the rb seedlings efficiently build up their chloroplasts to effectively transit from heterotrophic to photoautotrophic growth.
Beyond the massive changes in the levels of the storage and plastidic lipids, the phospholipids in the wild type and mutant seedlings showed significant differences only for the phospatidylcholines, which are abundantly present in the chloroplast envelope (Figure 8C). Accordingly, these results point towards a differential allocation of energy and nutrients for the synthesis of plastidic membranes and endomembranes between the two genotypes (Li-Beisson et al., 2013).
Mutation of RAPTOR1B Leads to Considerable Alterations in Gene Expression
To complement the metabolite measurements, we analyzed transcriptomic changes in the wild-type and rb seedlings using microarrays. The data were obtained from rb10 and the wild-type control at the end of the 5-d etiolation process and after 1 h of illumination, respectively. The 1-h time point was included to trap early regulatory events.
The most obvious result from our global analysis was the increased expression of ROS-responsive genes after light exposure in the wild-type samples, while the induction of ROS-marker genes was reduced in rb10 (Fig. 9A). This result, which is consistent with the RT-qPCR analysis of the singlet oxygen-responsive genes (Fig. 3B) and the chromatographic data showing increased levels of carotenoids in the rb lines (Fig. 2B), suggested that rb10 seedlings might experience less ROS stress than wild-type seedlings during the beginning of the deetiolation process.
Figure 9.
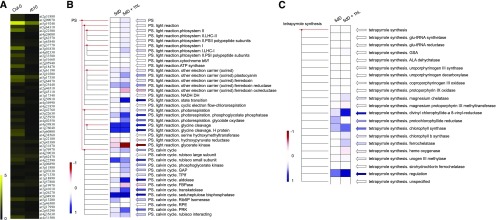
The mutation of RAPTOR1B has considerable effects on the expression of ROS-responsive genes and genes coding for proteins involved in photosynthesis and tetrapyrrole biosynthesis. Seedlings were grown in the dark for 5 d and then transferred to light. Samples were collected immediately prior to light exposure (5dD) or 1 h after illumination (5dD + 1hL). A, Heat map showing the fold changes of the expression of ROS-responsive genes in Col-0 and rb10 after exposure to light for 1 h with respect to samples without light treatment (5dD + 1hL versus 5dD). B and C, PageMan heat map showing fold changes of transcripts (rb10 versus the wild type) in different functional categories of photosynthesis (B) and tetrapyrrole synthesis (C). The means of the data points (log2 values) in each bin were used for the heat map.
To gain further insights into the transcriptional regulation of greening, we next investigated how the differentially expressed genes in the two genotypes mapped in terms of metabolic pathways using PageMan (Usadel et al., 2006) and MapMan (Thimm et al., 2004). Notably, there was a prominent increase in the expression of PSII components, other electron carriers of the photosynthetic light reactions (with the exception of ferredoxin oxidoreductase, which was decreased), and genes encoding proteins involved in state transition, photorespiration, and Gly cleavage of the photosynthetic light reaction in the samples of illuminated rb10 compared with wild-type seedlings (Fig. 9B). These changes were paralleled with an up-regulation of genes encoding components of the Calvin-Benson-Basham cycle (with the exception of FBPase; Fig. 9B). For tetrapyrrole biosynthesis, the expression of several genes was elevated in the rb10 illuminated samples, including genes encoding divinylchlorophyllide-a 8-vinyl-reductase, chlorophyll synthase, ferrochelatase, and enzymes involved in the regulation of tetrapyrrole synthesis (Fig. 9C). Similar to our metabolic and lipidomic results, these transcriptional changes support an elevation in the biosynthetic activity associated with the chloroplast in the rb seedlings during the deetiolation process.
Alongside these changes, considerable alterations were observed in transcripts coding for proteins involved in other processes, including glycolysis, gluconeogenesis, cell wall synthesis/modification, nitrogen metabolism, sulfur assimilation, and secondary metabolism (Supplemental Fig. S6, A–C). Notably, the differences in transcript profiles between rb and wild-type seedlings were similar in dark and light conditions; however, the differences were more pronounced when the seedlings were illuminated, supporting the hypothesis that the rb lines are able to induce the transcriptional programs required for successful greening, while the nutritional and energetically depleted wild type encounters difficulties in doing so.
Suc Supplementation of the Medium Rescues the Low Greening Rates of Wild-Type Seedlings after Extended Etiolation
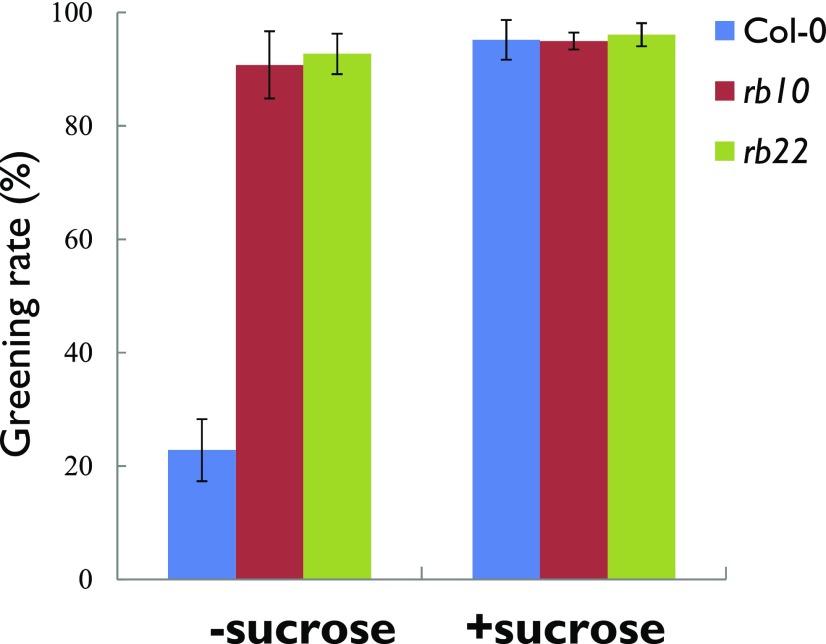
Our data implied that metabolic depletion might be a major cause for the poor greening efficiency of the etiolated wild-type seedlings. To test this, we repeated the greening assay on medium supplemented with 1% (w/v) Suc. As can be seen in Figure 10, the Suc supplementation led to almost 100% greening events of 5-d etiolated wild-type seedlings. This is in contrast to the greening rate on medium without Suc, where less than 30% of wild-type seedlings displayed greening (Fig. 1B).
Figure 10.
Suc can rescue the greening rate of etiolated wild-type seedlings. The greening rate is shown for rb and wild-type seedlings grown in darkness for 5 d before exposure to light for 2 d. Plants were grown on medium with or without 1% Suc. Data are means ± se (n = 3, more than 80 seedlings for each biological replicate).
DISCUSSION
During the process of germination, the plant embryo develops from heterotrophy, relying on the nutrient resources deposited in the seed, into a photoautotrophic organism, synthesizing all required compounds from light, water, and CO2. As germination under natural conditions takes place in the soil, in the absence of light, the efficient germination and etiolation of the seedling provide an improved chance of developmental success for the plant. Nevertheless, being efficient and possibly faster than the other seeds is not the only criterion for success. The passage from skotomorphogenesis to photomorphogenesis involves several challenges, including the synthesis of photosynthetic pigments and the development of functional chloroplasts (Pogson and Albrecht, 2011), which hold the photosynthetic machinery. During this process, the coordinated regulation of the synthesis of proteins, lipids, photosynthetic pigments, and photoprotective compounds is crucial.
In this study, we showed that the greening efficiency of rb seedlings, which are mutants of an essential component of the TOR complex (Dobrenel et al., 2016), is enhanced substantially in comparison with that of wild-type seedlings if both are kept in the dark for several days (Fig. 1). This observation was startling, because not only was TOR described to act as a positive regulator of chlorophyll biosynthesis (Li et al., 2015; Sun et al., 2016), but it also was shown that the mutation of RAPTOR1B leads to substantially delayed greening and germination of seedlings in the light (Salem et al., 2017). These two controversial observations, namely the delayed and slowed germination (Salem et al., 2017) and the improved greening after etiolation (Fig. 1), indicate substantial differences in the impact of TOR activity on germination, chlorophyll biosynthesis, and greening. While pharmacological TOR inhibitors (Montané and Menand, 2013) and the repression of TOR using RNA interference (Deprost et al., 2007; Caldana et al., 2013) led to leaf bleaching, the RAPTOR1B mutants used here, which repress TOR activity to a lesser extent (Salem et al., 2018), show neither reduction in chlorophyll concentrations nor decreased photosynthetic efficiency (Salem et al., 2018). Taken together, these results indicate that repressing the same gene or protein under different developmental conditions does not necessarily lead to the same phenotype. Accordingly, rb plants exhibit several growth phenotypes that are reminiscent of those observed in TOR-repressed plants as well as rb-specific ones (Salem et al., 2018). Interestingly, most of the observed growth phenotypes of the rb plants (Salem et al., 2018) and seeds (Salem et al., 2017) were accompanied by massive alterations in hormone levels. In particular, the levels of abscisic acid and GAs were altered massively in the rb mutants, and these changes were causally responsible for many of the observed phenotypes (Salem et al., 2017, 2018).
Consistent with the previous results showing altered hormone levels in the rb mutants, we found that the etiolated rb seedlings were less sensitive to GA treatment (Fig. 4D), which is known to negatively impact tetrapyrrole biosynthesis and POR expression and activity (Achard and Genschik, 2009). This phenotypic connection was supported further by a significant decrease in transcript levels of the GA receptor GID1 (Fig. 4A) and by increased levels of the GA-controlled DELLA transcription factor RGA (Fig. 4C). Mechanistically, the binding of GA to its receptor GID1 triggers the degradation of DELLAs through the ubiquitin-proteasome pathway (Dill et al., 2004; Fu et al., 2004; Griffiths et al., 2006; Willige et al., 2007). Therefore, we postulate that the decrease in GID1 expression (Fig. 4A) and the increase of the DELLA protein RGA (Fig. 4C) could account for the reduced sensitivity of the rb mutants to exogenous GA as compared with the wild type (Fig. 4D).
DELLAs are critical modulators of cotyledon greening in dark-grown seedlings (Cheminant et al., 2011). They accumulate in etiolated seedlings and regulate the greening process through two opposing paths. On the one hand, DELLAs reduce the binding affinity of the PIF1 transcription repressor to the promoters of chlorophyll biosynthesis genes, thus promoting the expression of these genes and eventually the accumulation of phototoxic Pchlide in the dark. On the other hand, DELLAs induce the expression of the photoprotective enzymes PORA and PORB by a mechanism that is independent of PIF1 (Cheminant et al., 2011). In our data, the rb seedlings not only have a higher amount of the DELLA protein RGA at the end of the 5-d etiolation phase (Fig. 4C) but also show higher expression levels of PORA and PORB, and increased PLB size, where POR and Pchlide are stored (Fig. 2B). As mutations in DELLAs suppress the greening phenotype conferred by TOR inhibitors, we hypothesize that the DELLAs act at least partially downstream of the TOR complex for the regulation of POR gene expression.
As mentioned above, the PIF transcription factors are negative regulators of chlorophyll biosynthesis in etiolated seedlings (Huq et al., 2004; Moon et al., 2008; Shin et al., 2009; Stephenson et al., 2009; Tang et al., 2012; Liu et al., 2013). Several genes that encode proteins involved in chlorophyll biosynthesis, like PORC, HEMA1, CHLH, and HEMB1, are major PIF targets that contribute to the photobleaching phenotype in pif mutants (Shin et al., 2009; Stephenson et al., 2009; Tang et al., 2012; Liu et al., 2013). Here, we found that the inhibition of greening in pif plants is largely suppressed in rb10pif double mutants (Fig. 5A). This phenotype, on the one hand, is accompanied by a reduction of Pchlide levels in the double mutants, as compared with the pif1 and pif3 mutants (Fig. 5C). On the other hand, the rb10pif double mutants also show massively reduced ROS production, which has been shown to be the main reason for the decreased greening efficiency of pif3 (Stephenson et al., 2009). Accordingly, it is difficult to judge from these data if TOR repression independently down-regulates tetrapyrrole biosynthesis through a PIF-independent pathway (Fig. 5C), which, in turn, leads to improved greening of the double mutants (Fig. 5A), or if the rb10pif double mutants only have substantially increased antioxidative capacities, which constitute the improved greening.
Even though our results imply that TOR may have a regulatory role in the tetrapyrrole pathways, metabolic analyses of the etiolated and deetiolated rb seedlings indicate that the greening phenotype might be derived from a more indirect, possibly pleiotropic, effect of the reduced TOR activity. In our metabolomics data set, we showed that many essential metabolites, required for the growth and biosynthesis of essential cellular components, are depleted substantially in wild-type seedlings after 5 d of etiolation, while the slower growing rb seedlings maintain higher levels of the corresponding metabolites. Hence, under these conditions, the reduced TOR activity would lead to an energy- and nutrient-sparing growth behavior and, therefore, allow the mutant to maintain higher levels of nutrient and energy resources, enabling the successful transition from heterotrophic to phototrophic growth (Fig. 1). This exhaustion of nutritional resources in the wild type is best reflected by the massive depletion of NADs and by the significantly reduced ratio of the reduced species (Fig. 7). NADH and NADPH, which are produced sufficiently during oxygenic photosynthesis in light-grown plants, can only be obtained in the dark from the oxidative breakdown of highly reduced nutrients like storage lipids or proteins (Penfield et al., 2006). As a consequence, the dark-grown seedlings rely mostly on the sugar, protein, and storage lipid resources in their seeds. Notably, the storage lipids, which provide the substrates for the β-oxidation (Graham, 2008), are largely depleted in the etiolated wild-type seedlings, while the rb seedlings still contain approximately 5-fold higher concentrations (Fig. 8A). This differential resource availability therefore may fuel the costly conversion of etioplasts into fully functional chloroplasts in the rb seedlings after etiolation (Pogson and Albrecht, 2011). Consequently, the etiolated rb seedlings managed to synthesize significant amounts of essential plastidic components, including chlorophyll and the thylakoid membrane lipids MGDG, DGDG, and SQDG (Fig. 8B), within the first 24 h of deetiolation. Similarly, rb seedlings manage to produce substantial amounts of ROS-scavenging carotenoids (Fig. 2B), while wild-type seedlings fail to do so. This capacity to save energy and nutrients during etiolation and use these resources for the substantial synthesis of ROS-scavenging molecules also might explain the substantially improved greening of the rb10pif double mutant as compared with the pif seedlings (Stephenson et al., 2009).
Contrary to the complementation of the greening phenotype of the pif mutants, our metabolic data showed that the most likely explanation for the failure of the wild-type seedlings to efficiently transit from skotomorphogenesis to photomorphogenesis relies on nutritional depletion (Figs. 6–8). This assumption is fully supported by the fact that the supplementation of the growth medium with 1% Suc was sufficient to reestablish maximal greening rates in the wild type (Fig. 10). Still, even though this explanation is the most likely, it should be kept in mind that applying 1% Suc to the growth medium not only provides additional nutritional supply but also distorts the transcriptional and metabolic network in these plants, which also might contribute partially to the complementation of the greening phenotype.
Taken together, our results show that dealing with complex phenotypes of mutant plants, affecting essential processes like chloroplast biogenesis, central metabolism or translation, requires special caution in dissecting the cause and the order of the events leading to the observed phenotypes. Even though strong evidence exists that TOR is a global regulator of multiple processes, some of the observed phenotypes might be derived frompleiotropic effects, which need to be dissected from the direct targets of TOR in future studies.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) RAPTOR1B T-DNA insertion mutants SALK_078159 (named rb78 here), SALK_006431 (named rb64 here), SALK_101990 (named rb10 here), and SALK_022096 (named rb22 here) as well as the pif1 (SALK_072677) and pif3 (SALK_030753) mutants are in the Col-0 ecotype and were obtained from the European Arabidopsis Stock Centre. The ga1-3della mutant (ga1-3; gai-t6; rga-t2; rgl1-1; rgl2-1) is in the Landsberg erecta ecotype. For the greening experiments, if not specified, more than 60 plants were grown on one-half-strength Murashige and Skoog medium without Suc in darkness for the indicated time period and transferred into continuous white light (120 μmol m−2 s−1) for 2 d before calculating the greening rate. For the Suc supplementation experiment, 1% (w/v) Suc was added. For the TOR inhibitor treatment, 0.5 μm AZD-8055 and 0.4 μm Torin2 (Montané and Menand, 2013) were used.
Fluorescence Imaging of ROS and Singlet Oxygen
After exposure to continuous white light for 2 d, seedlings were incubated with 10 μm H2DCFDA (Invitrogen) in 10 mm Tris-HCl (pH 7.2) for 30 min at room temperature and washed for 30 min with 10 mm Tris-HCl (pH 7.2). To image singlet oxygen, dark-grown seedlings were incubated with 10 μm SOSG (Invitrogen) in one-half-strength Murashige and Skoog liquid medium for 1 h in darkness followed by light exposure for 15 h. The fluorescence images were captured by a Leica MZ16 FA microscope equipped with a Leica DFC300 FX camera.
Western-Blot Analysis
Western-blot analysis was performed as described with anti-GAI antibody, which recognizes GAI, RGA, and RGL2 (Piskurewicz et al., 2009).
Transmission Electron Microscopy
Five-day-old dark-grown cotyledons were high pressure frozen in 3-mm-diameter B-type sample carriers with 1-hexadecene as a cryoprotectant using a Leica HPM 100. Cryofixed samples were freeze substituted for 5 d at −85°C in 2% (w/v) osmium tetroxide and 8% (v/v) dimethoxypropane in acetone using a Leica AFS2, then embedded in Spurr’s resin, sectioned, and poststained as described (McFarlane et al., 2008). Samples were viewed using a Zeiss EM910 at 80 kV, and images were collected using an Olympus Quemesa CCD camera. PLB area was measured, relative to the area of the whole etioplast, using ImageJ.
Pchlide, Chlide, Heme, and Carotenoid Analysis
Seedlings were homogenized in liquid nitrogen, and 1 mL of 80% (v/v) acetone was added to 50-mg samples. Pchlide and Chlide were extracted at 4°C in the dark overnight. Room temperature fluorescence emission spectra were acquired with an excitation wavelength of 440 nm and an emission wavelength of 600 to 700 nm by a fluorescence spectrofluorometer (PekinElmer; LS55). Carotenoids were extracted with cold acetone:0.1 m NH4OH (9:1, v/v) in a three-step cycle of resuspension and centrifugation using 500 μL of the same solution. Supernatants were combined and analyzed by HPLC using authentic standards as reported previously (Kim et al., 2013). Noncovalently bound heme was extracted from the pellet after pigment extraction using 100 μL of acetone/HCl/DMSO (10:0.5:2, v/v/v) in the same three-step protocol. HPLC analysis of heme was performed essentially as described (Scharfenberg et al., 2015).
Metabolite Extraction
For the metabolite extraction, three independent pools of samples were ground to a fine powder, and 50 mg of ground tissue of each independent pool was extracted in 1 mL of a precooled (∼15°C) mixture of methyl-tert-butyl ether (MTBE; 3:1:1, v/v/v) as described previously (Giavalisco et al., 2011). MTBE buffer was added to the frozen powder and vortexed until the tissue was fully resuspended. The sample was incubated for 30 min at 4°C on an orbital shaker before incubating it for another 10 min in an ultrasonication bath with ice. Then, 700 μL of ultra-performance liquid chromatography (UPLC)-grade water:methanol (3:1, v/v) was added. The tubes were then centrifuged at 20,800g at room temperature for 5 min, and 700 μL of the upper MTBE phase, containing the lipids, was transferred to a fresh 1.5-mL Eppendorf tube and dried down in a Speed-Vac. The remaining organic phase was then removed completely from the extract, and 150 μL of the lower, polar fraction was transferred to a fresh 1.5-mL Eppendorf tube and used for gas chromatography (GC) analysis after drying.
GC-Time of Flight-Mass Spectrometry Analysis
Prior to GC-time of flight-mass spectrometry analysis, the samples were derivatized. A variation of the two-stage technique used by Roessner et al. (2001) was employed as described previously (Krueger et al., 2011; Salem et al., 2016). First, the carbonyl moieties were protected via methoximation in a 90-min reaction at 30°C using 5 µL of 40 mg mL−1 methoxyamine hydrochloride (Sigma) in pyridine (Merck), followed by the derivatization of acidic protons via a 30-min reaction at 37°C with the addition of 45 µL of N-methyl-N-trimethylsilyltrifluoroacetamide (Macherey-Nagel). Forty microliters of a mixture of retention time standards (fatty acid methyl esters), containing 3.7% (w/v) heptanoic acid, 3.7% (w/v) nonanoic acid, 3.7% (w/v) undecanoic acid, 3.7% (w/v) tridecanoic acid, 3.7% (w/v) pentadecanoic acid, 7.4% (w/v) nonadecanoic acid, 7.4% (w/v) tricosanoic acid, 22.2% (w/v) heptacosanoic acid, and 55.5% (w/v) hentriacontanoic acid dissolved in tetrahydrofuran at 10 mg mL−1 total concentration, was added prior to trimethylsilylation. One microliter of the derivatized sample was injected onto the column, and analysis was commenced in nonsplit mode. The analysis was performed under the following temperature program: 5 min of isothermal heating at 70°C, followed by a 5°C min−1 increase in oven temperature to 350°C, and a final 5 min of heating at 330°C. The samples were run in an Agilent 7683 series autosampler (Agilent) coupled to an Agilent 6890 gas chromatograph coupled to a LecoPegasus 2 time-of-flight mass spectrometer (Leco). Chromatograms were exported from LecoChroma TOF software (version 3.25) to R software (www.r-project.org). Peak detection, retention time alignment, and library matching were performed using the TargetSearch R package (Cuadros-Inostroza et al., 2009). Metabolites were quantified by the peak intensity of a compound-specific mass.
Lipid Analysis
For lipid analysis, which was performed as described previously (Hummel et al., 2011; Salem et al., 2016), the dried pellets were resuspended in 250 µL of a mixture of acetonitrile:isopropanol (7:3 v/v), thoroughly vortexed, and centrifuged. Then, 100-µL aliquots were transferred into glass vials and 2 µL was injected and separated on an Acquity UPLC system (Waters) using a reverse-phase C8 column. The UPLC solvents (A = water with 1% [v/v] of a solution of 1 m ammonium acetate and 0.1% [v/v] acetic acid; B = 70% [v/v] acetonitrile and 30% [v/v] isopropanol with 1% [v/v] of a solution of 1 m ammonium acetate and 0.1% [v/v] acetic acid). The gradient separation was performed at a flow rate of 400 µL min−1 as follows: 1 min at 45% A, a 3-min linear gradient from 45% A to 35% A, an 8-min linear gradient from 25% A to 11% A, and a 3-min linear gradient from 11% A to 1% A. After washing the column for 3 min with 1% A, the buffer was returned to 45% A and the column was reequilibrated for 4 min (22-min total run time). The samples were measured in either positive or negative ion mode. The mass spectra were acquired using an Exactive high-resolution mass spectrometer (Thermo Fisher). The spectra were recorded using alternating full-scan and all-ion fragmentation scan mode, covering a mass range from 100 to 1,500 m/z The resolution was set to 10,000, with 10 scans per second, restricting the Orbitrap loading time to a maximum of 100 ms with a target value of 1 E6 ions. The capillary voltage was set to 3 kV, with a sheath gas flow of 60 and an auxiliary gas flow of 35 (values are arbitrary units). The capillary temperature was set to 150°C, and the drying gas in the heated electrospray source was set to 350°C. The skimmer voltage was held at 25 V, and the tube lens was set to a value of 130 V. The spectra were recorded from 1 to 20 min of the UPLC gradients. The obtained raw chromatograms were processed further using Excalibur software version 2.10 (Thermo Fisher) or Refiner MS software version 6.0 (Gene-Data). Peaks from raw chromatograms were first determined, then aligned by their parent masses, chemical noise was subtracted, and a final alignment file of all chromatograms was the output, which contains information about m/z, retention times, and retention time deviations for each annotated peak. The peaks were assigned using the chemical formula database GoBioSpace (gmd.mpimp-golm.mpg.de) to compounds with sum formulas, according to retention time and m/z, and finally were corrected manually (Hummel et al., 2011).
RNA Preparation, RT-qPCR, and Transcript Expression Profiling
Seedlings were treated as indicated in the text, and total RNA was purified by using the RNase Plant Mini Kit (Qiagen). Reverse transcription was performed by using SuperScript II (Invitrogen), and RT-qPCR assays used the Power SYBR Green RT-PCR Reagents Kit (Applied Biosystems). Primer sequences used for RT-qPCR can be found in Supplemental Table S3.
Microarray analysis was performed using Affymetrix Gene 1.0 ST Array (Thermo Fisher) with three biological replicates of wild-type and rb10 samples. Total RNA was isolated from pools of seedlings, and labeling was performed using 1 μg of RNA as described previously. Affymetrix Gene 1.0 ST Array hybridizations were performed by Atlas Biolabs. Raw CEL files were analyzed using Bioconductor software (Gentleman et al., 2004) for R, and the GC Robust Multiarray Average expression estimation was obtained using the gcrma package (Wu et al., 2004). Expression data obtained for rb and the wild type were submitted to the NCBI Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo). Significant overrepresentation of functional annotations was analyzed using PageMan with Benjamini-Hochberg correction (Usadel et al., 2006). Gene clusters of functional categories were visualized by heat maps or using the BiNGO 2.3 plugin tool in Cytoscape version 2.6 with MapMan bin files (Maere et al., 2005; Usadel et al., 2005). Overrepresented MapMan bins were identified using a hypergeometric test with a significance threshold of 0.05 after Benjamini-Hochberg false discovery rate correction (Benjamini and Hochberg, 1995).
Pyridine Nucleotide Measurements
The procedure of extraction and assay of NADs was performed according to the method described by Schippers et al. (2008).
Chlorophyll Measurements
Chlorophyll was measured as described previously (Caldana et al., 2013).
Statistical Analysis
All statistical analyses were performed using R version 2.12.2 (www.r-project.org). Metabolite data obtained from GC-time of flight-mass spectrometry analysis were normalized by fresh weight, followed by sample total ion count and global outlier replacement, as described previously (Giavalisco et al., 2011; Huege et al., 2011; Hummel et al., 2011). Lipophilic and polar fractions analyzed by liquid chromatography-high-resolution mass spectrometry were normalized in the same manner, except using internal standards (phosphatidylcholine 34:0 and ampicillin, respectively) instead of the total ion content. The significance of differences was tested for each metabolite by a paired Student’s t test (P < 0.05) comparing genotypes.
Accession Number
Sequence data from this article can be found in the NCBI Gene Expression Omnibus repository under accession number GSE114639.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Molecular characterization of rb seedlings.
Supplemental Figure S2. TOR inhibitor treatment promotes the greening of etiolated seedlings.
Supplemental Figure S3. rb seedlings convert Pchlide more efficiently into Chlide.
Supplemental Figure S4. ROS staining of cotyledons.
Supplemental Figure S5. Histograms showing the relative levels of extracted and detected amino acids from Col-0, rb10, and rb22 seedlings.
Supplemental Figure S6. PageMan and MapMan analyses of the transcript profiling data.
Supplemental Table S1. GC-time of flight-mass spectrometry data of primary metabolites.
Supplemental Table S2. UPLC-mass spectrometry data of lipid metabolites.
Supplemental Table S3. Sequences of primers used in RT-qPCR experiments.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Änne Michaels and Gudrun Wolter for excellent technical assistance with the GC- and liquid chromatography-mass spectrometry measurements. Furthermore, we acknowledge Luis Lopez-Molina and Dr. Urszula Piskurewicz (University of Geneva) for sharing the anti-GAI antibody.
Footnotes
Y.Z., S.P., A.R.F., and P.G. are supported by the Max-Planck Society. S.P. is supported by a R@MAP Professorship at the University of Melbourne and an ARC Future Fellowship (FT160100218). Yi.Z. is supported by the Fundamental Research Funds for the Central Universities. H.E.M. is supported by an EMBO Long-Term Fellowship (EMBO ALTF 1246-2013) and an ARC DECRA (DE170100054.
Articles can be viewed without a subscription.
References
- Achard P, Genschik P (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson GH, Hanson MR (2005) The Arabidopsis Mei2 homologue AML1 binds AtRaptor1B, the plant homologue of a major regulator of eukaryotic cell growth. BMC Plant Biol 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GH, Veit B, Hanson MR (2005) The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Hanson J (2017) Shaping plant development through the SnRK1-TOR metabolic regulators. Curr Opin Plant Biol 35: 152–157 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser A 57: 289–300 [Google Scholar]
- Broeckx T, Hulsmans S, Rolland F (2016) The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J Exp Bot 67: 6215–6252 [DOI] [PubMed] [Google Scholar]
- Brzezowski P, Richter AS, Grimm B (2015) Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim Biophys Acta 1847: 968–985 [DOI] [PubMed] [Google Scholar]
- Buhr F, El Bakkouri M, Valdez O, Pollmann S, Lebedev N, Reinbothe S, Reinbothe C (2008) Photoprotective role of NADPH:protochlorophyllide oxidoreductase A. Proc Natl Acad Sci USA 105: 12629–12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P (2013) Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J 73: 897–909 [DOI] [PubMed] [Google Scholar]
- Cheminant S, Wild M, Bouvier F, Pelletier S, Renou JP, Erhardt M, Hayes S, Terry MJ, Genschik P, Achard P (2011) DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25: 1657–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña-Cortés H, Willmitzer L, Hannah MA (2009) TargetSearch: a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics 10: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Truong HN, Robaglia C, Meyer C (2005) An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem Biophys Res Commun 326: 844–850 [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C (2016) TOR signaling and nutrient sensing. Annu Rev Plant Biol 67: 261–285 [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L (2011) Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high-resolution mass spectrometry. Plant J 68: 364–376 [DOI] [PubMed] [Google Scholar]
- González A, Hall MN (2017) Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. (2008) Seed storage oil mobilization. Annu Rev Plant Biol 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909 [DOI] [PubMed] [Google Scholar]
- Hindupur SK, González A, Hall MN (2015) The opposing actions of target of rapamycin and AMP-activated protein kinase in cell growth control. Cold Spring Harb Perspect Biol 7: a019141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huege J, Krall L, Steinhauser MC, Giavalisco P, Rippka R, Tandeau de Marsac N, Steinhauser D (2011) Sample amount alternatives for data adjustment in comparative cyanobacterial metabolomics. Anal Bioanal Chem 399: 3503–3517 [DOI] [PubMed] [Google Scholar]
- Hummel J, Segu S, Li Y, Irgang S, Jueppner J, Giavalisco P (2011) Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci 2: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R (2012) FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg++-branch of this pathway. FEBS Lett 586: 211–216 [DOI] [PubMed] [Google Scholar]
- Kim S, Schlicke H, Van Ree K, Karvonen K, Subramaniam A, Richter A, Grimm B, Braam J (2013) Arabidopsis chlorophyll biosynthesis: an essential balance between the methylerythritol phosphate and tetrapyrrole pathways. Plant Cell 25: 4984–4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Masuda T (2016) Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front Plant Sci 7: 1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S, Giavalisco P, Krall L, Steinhauser MC, Büssis D, Usadel B, Flügge UI, Fernie AR, Willmitzer L, Steinhauser D (2011) A topological map of the compartmentalized Arabidopsis thaliana leaf metabolome. PLoS ONE 6: e17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Lee DW, Apel K (2003) TIGRINA d, required for regulating the biosynthesis of tetrapyrroles in barley, is an ortholog of the FLU gene of Arabidopsis thaliana. FEBS Lett 553: 119–124 [DOI] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Song Y, Wang K, Dong P, Zhang X, Li F, Li Z, Ren M (2015) TOR-inhibitor insensitive-1 (TRIN1) regulates cotyledons greening in Arabidopsis. Front Plant Sci 6: 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y (2017) Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA 114: 2765–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen CY, Wang KC, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y, et al. (2013) PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25: 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane HE, Young RE, Wasteneys GO, Samuels AL (2008) Cortical microtubules mark the mucilage secretion domain of the plasma membrane in Arabidopsis seed coat cells. Planta 227: 1363–1375 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Menand B (2013) ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot 64: 4361–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhu L, Shen H, Huq E (2008) PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA 105: 9433–9438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, et al. (2012) Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TP, Cueff G, Hegedus DD, Rajjou L, Bentsink L (2015) A role for seed storage proteins in Arabidopsis seed longevity. J Exp Bot 66: 6399–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyogi KK. (2000) Safety valves for photosynthesis. Curr Opin Plant Biol 3: 455–460 [DOI] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L, Wurzinger B, Mair A, Landgraf R, Börnke F, Hanson J, Teige M, Baena-Gonzalez E, et al. (2016) Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci Rep 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Pinfield-Wells HM, Graham IA (2006) Storage reserve mobilisation and seedling establishment in Arabidopsis. The Arabidopsis Book 4: e0100, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Turecková V, Lacombe E, Lopez-Molina L (2009) Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J 28: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V (2011) Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol 155: 1545–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Lebedev N, Apel K (1996) PORA and PORB, two light-dependent protochlorophyllide-reducing enzymes of angiosperm chlorophyll biosynthesis. Plant Cell 8: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M (2009) Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J 60: 424–435 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie A (2001) Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem MA, Jüppner J, Bajdzienko K, Giavalisco P (2016) Protocol: a fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods 12: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem MA, Li Y, Wiszniewski A, Giavalisco P (2017) Regulatory-associated protein of TOR (RAPTOR) alters the hormonal and metabolic composition of Arabidopsis seeds, controlling seed morphology, viability and germination potential. Plant J 92: 525–545 [DOI] [PubMed] [Google Scholar]
- Salem MA, Li Y, Bajdzienko K, Fisahn J, Watanabe M, Hoefgen R, Schöttler MA, Giavalisco P (2018) RAPTOR controls developmental growth transitions by altering the hormonal and metabolic balance. Plant Physiol 177: 565–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168: 960–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfenberg M, Mittermayr L, von Roepenack-Lahaye E, Schlicke H, Grimm B, Leister D, Kleine T (2015) Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ 38: 280–298 [DOI] [PubMed] [Google Scholar]
- Schippers JH, Nunes-Nesi A, Apetrei R, Hille J, Fernie AR, Dijkwel PP (2008) The Arabidopsis onset of leaf death5 mutation of quinolinate synthase affects nicotinamide adenine dinucleotide biosynthesis and causes early ageing. Plant Cell 20: 2909–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumskaya M, Wurtzel ET (2013) The carotenoid biosynthetic pathway: thinking in all dimensions. Plant Sci 208: 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling U, Franck F, van Cleve B, Frick G, Apel K, Armstrong GA (1998) Etioplast differentiation in Arabidopsis: both PORA and PORB restore the prolamellar body and photoactive protochlorophyllide-F655 to the cop1 photomorphogenic mutant. Plant Cell 10: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Yu Y, Hu W, Min Q, Kang H, Li Y, Hong Y, Wang X, Hong Y (2016) Ribosomal protein S6 kinase1 coordinates with TOR-Raptor2 to regulate thylakoid membrane biosynthesis in rice. Biochim Biophys Acta 1861: 639–649 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0145, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Wang W, Chen D, Ji Q, Jing Y, Wang H, Lin R (2012) Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 24: 1984–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Johansson H, Lee KP, Bou-Torrent J, Stewart K, Steel G, Rodríguez-Concepción M, Halliday KJ (2014) The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet 10: e1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Arnim A, Deng XW (1996) Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol 47: 215–243 [DOI] [PubMed] [Google Scholar]
- Vothknecht UC, Kannangara CG, von Wettstein D (1998) Barley glutamyl tRNAGlu reductase: mutations affecting haem inhibition and enzyme activity. Phytochemistry 47: 513–519 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Sheen J (2014) The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol 164: 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F, Zhang R, Meng Z, Deng K, Que Y, Zhuo F, Feng L, Guo S, Datla R, Ren M (2017) Brassinosteriod Insensitive 2 (BIN2) acts as a downstream effector of the Target of Rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol 213: 233–249 [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C, Sun Y, Wang W, Wang ZY (2016) TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr Biol 26: 1854–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]