Abstract
Bait fusion proteins with a glycosyl-phosphatidylinositol signal sequence anchor enable effective split ubiquitin screening for interactions with otherwise soluble membrane proteins.
Protein-protein interactions play critical roles in a wide range of cellular functions. The need to analyze these interactions has spawned a variety of biophysical and biochemical techniques, including coimmunoprecipitation, yeast two-hybrid, bimolecular fluorescence complementation, Förster resonance energy transfer, and split-luciferase methods (Xing et al., 2016). Nonetheless, there remains a high demand for rapid, reliable, and cost-effective methods for studying protein interactions, especially those occurring at membrane surfaces.
Techniques based on heterologous expression of bait and prey proteins in yeast have gained much popularity because they are quick, easy to perform, and inexpensive. Compared with the conventional yeast two-hybrid system, commonly used for screening interactors from a prey library, the yeast mating-based split-ubiquitin system (mbSUS; Grefen et al., 2007, 2009; Horaruang and Zhang, 2017) has proven effective, especially for studying interactions between membrane proteins. The technology incorporates several advantages over conventional yeast two-hybrid approaches, including mating-based recombination, which greatly enhances transformation efficiencies, and bait suppression by Met, which improves false positive detection (Grefen et al., 2007, 2009; Horaruang and Zhang, 2017). mbSUS is particularly stringent and has proven effective even in identifying the individual amino acid residues that define interaction motifs essential for binding with the prey (Honsbein et al., 2009; Grefen et al., 2010, 2015; Hachez et al., 2014; Zhang et al., 2015, 2017). The mbSUS technology relies on fusion of the bait with a transcriptional activator that, on prey interaction, is cleaved from the bait and moves into the nucleus to activate the reporter gene (Fig. 1A). Thus, only the transcriptional activator needs transit to the nucleus. As a consequence, mbSUS methods work well with large, full-length proteins, which often is not possible using yeast two-hybrid approaches (Xing et al., 2016).
Figure 1.
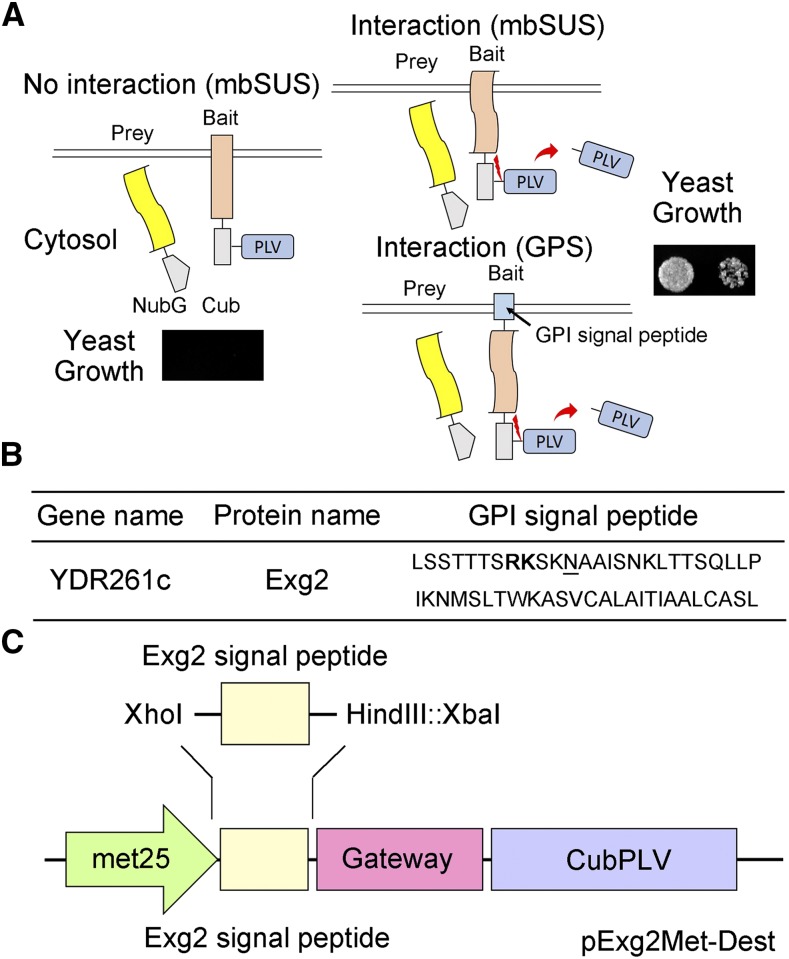
The Mating Based Split-Ubiquitin System (mbSUS). A, Schematic of the split-ubiquitin system. As in conventional SUS methods, ubiquitin is split between the N-terminal half (Nub) and the C-terminal half (Cub), and the latter is fused with the transcriptional activator complex Protein A-LexA-VP16 (PLV). Mutating the isoleucine at position 13 of the wild-type Nub (NubI or NubWt) to Gly yields the NubG, which blocks spontaneous reassembly of ubiquitin. The NubG-Prey and Bait-CubPLV fusion constructs are transformed into mating strains of yeast that require differing amino-acid supplementation for growth. After mating, interaction of the two proteins expressed in the diploid yeast is tested by growth on selective media as in the standard SUS assay (Grefen et al., 2010; Zhang et al., 2015, 2017). Interaction between bait and prey reassembles a functional ubiquitin and leads to cleavage and release of the PLV complex by ubiquitin-specific proteases. Critical to this technology, the bait must be a membrane-bound protein, although the prey may be either membrane-bound or soluble. In the GPS system, the bait protein is fused at the N terminus with an Exg2 GPI signal peptide-anchor sequence, which anchors it in the membrane. B, The C-terminal GPI signal sequence of the Exg2 protein of yeast. The dibasic Arg-Lys (RK) residue motif is shown in bold, and the ω-site is underlined. C, Schematic of the pExg2Met-Dest vector used to express the bait fusion protein, including the N-terminal fusion with the GPI signal sequence (see also Supplemental Fig. S1).
There are two caveats to the mbSUS technology. First, membrane anchoring of the bait fusion protein is vital, as a soluble bait could transit to the nucleus and activate reporter gene expression even in the absence of prey binding. Second, constructions normally require fusion of the transcriptional activator with the C terminus of the bait to ensure correct assembly of the two halves of the ubiquitin moiety and cleavage of the transactivator. These caveats mean that the bait must normally be anchored with its C terminus free in the cytosol. So work with Type II, C-terminally anchored membrane proteins, for example, is generally limited to their use as prey. Here, we describe a method that circumvents some of these difficulties through synthetic N-terminal anchoring of potential bait proteins. By incorporating a hydrophobic signal sequence from a glycosyl-phosphatidylinositol (GPI) protein, we show that the mbSUS method can be extended to test these interactions as well as those of small, soluble bait proteins.
We made use of a bait fusion with a GPI signal peptide derived from glucan-1,3-β-glucosidase 2 (Exg2), which is normally anchored in the plasma membrane and involved in cell wall β-glucan assembly (Nebreda et al., 1986; Cid et al., 1995). GPI-anchored proteins (GPI proteins) are expressed in all eukaryotic cells and transported from endoplasmic reticulum to the cell surface by vesicle traffic (Hamburger et al., 1995). GPI proteins have an endoplasmic reticulum import signal sequence at the N terminus and a GPI anchor sequence at the C terminus. This C-terminal sequence is composed of a dibasic residue motif and attachment site (ω-site; Caro et al., 1997). The C-terminal sequence is recognized by the GPI transamidase for lipid attachment, after which the GPI complex is inserted into the plasma membrane, thereby anchoring the protein to the membrane surface (Pittet and Conzelmann, 2007). We introduced the gene-synthesized Exg2 GPI signal peptide sequence (Fig. 1B) in front of the Gateway cassette of the bait vector pMetOYC-Dest of the CytoSUS system (Karnik et al., 2015) using unique restriction sites XhoI and XbaI (GenScript USA; HindIII also was introduced for future cloning work), replacing a sequence encoding an OST4 leader, to obtain the vector pExg2Met-Dest for the Exg2 GPI signal peptide-anchored mbSUS (GPS) system (Fig. 1C; see Supplemental Fig. S1 for the Exg2 GPI signal peptide-anchored sequence in the vector pExg2Met-Dest).
Previously, we used the N-terminal leader of the yeast oligosaccharyl transferase complex (OST4p; Möckli et al., 2007) to anchor bait proteins to a membrane surface. Although the complex in yeast normally resides on the luminal face of the endoplasmic reticulum, this system was sufficient for work with the soluble protein SEC11, a member of the Sec1/Munc18 protein family (Südhof and Rothman, 2009). Using the OST4p anchor with the SEC11 bait, we confirmed its interaction with the Q-SNARE SYP121, which is anchored by its C terminus to the plasma membrane (Karnik et al., 2015). However, efforts to isolate the binding domain failed when SEC11 truncations, including the N-terminal peptide SEC11Δ149 (Karnik et al., 2015), were used as baits with the OST4p anchor. These studies showed yeast growth even with the negative (NubG) control as a prey. Possible explanations include nonspecific cleavage of the protein and, given the low molecular mass of SEC11Δ149 (∼15 kD), an inability of the OST4p anchor to prevent transit of the uncleaved bait to the nucleus. Attempts with the signal sequences of three other membrane proteins, notably yeast aspartic proteinase 3 (Yap3), 1,3-β-glucanosyltransferase (Gas2), and killer toxin resistant 1 (Kre1; Caro et al., 1997), also were unsuccessful, suggesting that the hydrophobicity of such leader sequences per se is not sufficient as a general indicator of their utility.
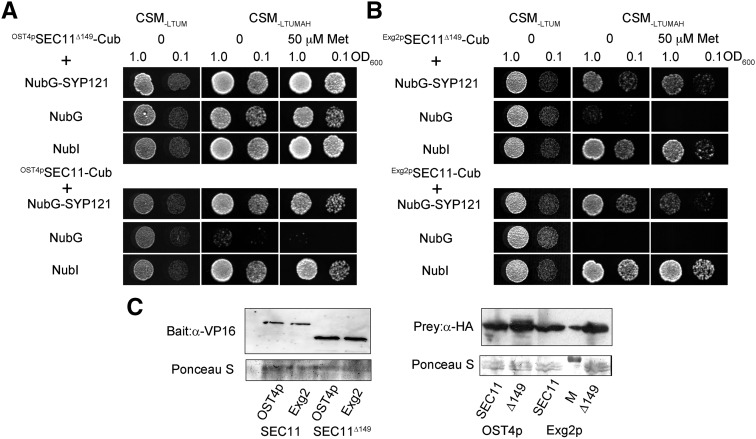
To test the new GPS system, we used Exg2 GPI signal peptide-anchored SEC11 and SEC11Δ149 as baits. Initial tests were carried out with the full-length Q-SNARE SYP121, anchored by its C-terminal transmembrane domain, as prey and with NubG and NubI (NubWt) as negative and positive controls, respectively. For comparison, we carried out parallel assays with the same pairs of proteins using the original OST4p-anchored system (Karnik et al., 2015). Diploid yeast were obtained and grown as before (Grefen et al., 2010; Zhang et al., 2015, 2017). Figure 2 shows the mbSUS assay and supporting immunoblot data from one of three independent experiments, each yielding similar results. A reliable mbSUS assay depends on a mating control as well as the positive and negative controls noted above. Diploid yeast should grow on synthetic medium (CSM) lacking Leu, Thr, Ura, and Met (−LTUM). Growth should also be evident on synthetic medium additionally lacking Ade and His (−LTUMAH), but only when bait and prey proteins interact. Thus, on CSM−LTUMAH, growth should be evident with the positive (NubI) and absent with the negative (NubG) controls. Finally, a strong and selective interaction should be evident even when bait expression is suppressed by including Met in the growth medium.
Figure 2.
The GPS system is suitable for analyzing protein-protein interactions of the soluble SEC11Δ149. A, Diploid yeast expressing OST4pSEC11-CubPLV or OST4pSEC11Δ149-CubPLV as bait and a NubG-X fusion of SYP121 and the controls (negative, NubG; positive, NubI) as prey were spotted onto different media as indicated. Growth on CSM−LTUM was used to verify the presence of both bait and prey vectors in the diploid yeast. Growth on CSM−LTUMAH was used to verify interaction, and additions of different concentrations of Met were used to suppress bait expression as a test for interaction strength. Diploid yeast was dropped at 1.0 and 0.1 OD600 in each case. Incubation time was 24 h on CSM−LTUM and 72 h on CSM−LTUMAH. B, Diploid yeast expressing Exg2 GPI signal peptide-fused Exg2pSEC11-CubPLV and Exg2pGPI-SEC11Δ149-CubPLV as bait, with NubG-SYP121 fusion and the controls (negative, NubG; positive, NubI) as prey spotted onto different media as indicated. Growth was as in A. C, Immunoblot analysis of the diploid yeast carried out with commercial anti-HA antibody for the prey fusions and anti-VP16 antibody for the bait fusions. Ponceau S stains were used for blotting/loading control.
Figure 2 shows that, with SEC11 as bait, the mbSUS assay yielded growth both with the OST4p (Fig. 2A) and Exg2 GPI signal peptide anchors (Fig. 2B), and in each case no growth was recovered in the negative controls. However, with SEC11Δ149 as the bait, the OST4p anchor showed growth both in the positive and negative controls under all conditions, precluding any meaningful analysis. By contrast, with Exg2 GPI signal peptide-anchored SEC11Δ149 as the bait, no growth was recovered in the negative control, and yeast growth was recovered with SYP121 as the prey, consistent with published pull-down studies (Karnik et al., 2015). Immunoblot analysis of each bait and prey protein is included in Figure 2C, confirming that the Exg2 signal peptide GPI fusion did not have an appreciable effect on bait protein expression.
To confirm the broader utility of the GPS system, we tested the iLOV protein as GPI-bait fusion with soluble SEC11 as the prey. We also used the truncated fragment of the R-SNARE VAMP721, VAMP721Δ127, as a GPI-bait fusion with the full-length Kv channel protein KAT1 as prey (Supplemental Fig. S2). iLOV is a soluble phototropin protein (∼13 kD) that, when expressed, distributes throughout the cytosol and nucleus of Arabidopsis (Arabidopsis thaliana) and of human embryonic kidney cells (Chapman et al., 2008). iLOV does not bind with SEC11 in pull-down assays (Karnik et al., 2013). VAMP721Δ127 (∼14 kD) comprises the cytosolic N-terminal (so-called longin) domain of R-SNARE, and its interaction with KAT1 has been verified in vitro and in vivo (Zhang et al., 2017). With the OST4p anchor, both iLOV and VAMP721Δ127 showed growth in the negative controls, preventing further analysis. By contrast, with the GPS system, both bait fusion proteins yielded sensible results with the positive (NubI) and negative (NubG) controls. The GPS system also showed interaction of VAMP721Δ127 with KAT1 and no interaction between iLOV and SEC11. These results, again, highlight the efficiency of the GPS system for mbSUS analysis, including in work with low-molecular-weight, soluble proteins as baits.
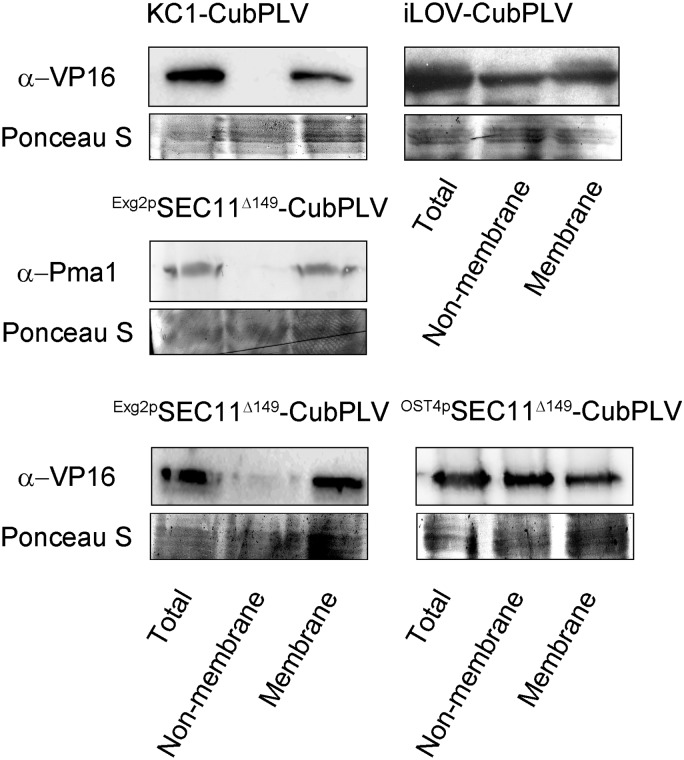
To assess the localization of the bait proteins, we harvested yeast expressing SEC11Δ149 with each of the membrane anchors. For comparison, we also transformed and expressed the integral plasma membrane Kv channel, KC1, which interacts with the SNARE proteins (Grefen et al., 2010; Zhang et al., 2015), and the iLOV protein, without an OST4p or Exg2 GPI signal peptide anchor, using the bait vector (pMetYC-Dest) of the original mbSUS system (Grefen et al., 2009). After mating with yeast transformed with SYP121 as the prey, the diploid yeast was harvested. Total, soluble, and membrane protein fractions were isolated as described before (Rosa and Correia, 1991) and analyzed by immunoblot (Fig. 3). To confirm fraction separations, the anti-Pma1 antibody (ab4656; Abcam) to the H+-ATPase (Pma1p; Martínez-Muñoz and Kane, 2008) was used to verify the distribution of the yeast plasma membrane. As expected, a band corresponding to Pma1p was recovered in the total and membrane fractions, but not in the soluble (nonmembrane) fraction. Similarly, KC1 was evident by immunoblot only in the total sample and membrane fractions. The iLOV protein, by contrast, was detected in all three fractions. We suspect that iLOV appeared in the membrane fraction because, when heterologously expressed in yeast, the protein is mistargeted and sequestered within organelles (Bassham and Raikhel, 2000; Vögtle et al., 2012). When expressed as a fusion with the Exg2 GPI signal peptide anchor, SEC11Δ149 was found only in the total and membrane fractions, but when anchored with OST4p, SEC11Δ149 was distributed across all three fractions. We conclude, therefore, that the OST4p anchor was insufficient to prevent SEC11Δ149 release from the membrane, whereas the Exg2 GPI signal peptide anchor was.
Figure 3.
SEC11Δ149 is expressed in the membrane fraction in the GPS system. Yeast membrane fractions were prepared as described before (Rosa and Correia, 1991). Twenty milliliters of yeast containing the constructs were cultured until 0.6–0.8 OD600 and harvested by centrifugation at 4°C. The pellets were quickly frozen (−70°C) after the addition of 5 mL of 100 mm Tricine, 5 mm EDTA, and 2 mm DTT. The samples were thawed, dispersed by vortexing with 1.5-mm-diameter glass beads, and diluted with 5 mL of 0.33 m Suc, 0.1 m Tris, 5 mm EDTA, and 2 mm DTT, adjusted to pH 8. After centrifugation for 3 min at 900g, the supernatants were decanted and centrifuged for 45 min at 40,000g at 4°C. The new supernatants were used as the nonmembrane fractions, and the pellets were resuspended in 20% glycerol, 10 mm Tris, 0.1 mm EDTA, and 0.1 mm DTT, adjusted to pH 7.5, and comprised the membrane fractions. Samples harvested directly from the yeast (total), the nonmembrane fraction, and the membrane fraction were loaded and separated by gel electrophoresis at 20 μg of protein per lane. Immunoblot analysis of the samples was carried out with commercial anti-Pma1 antibody and anti-VP16 antibody for the bait fusions. Ponceau S staining was used for blotting/loading control.
Clearly, incorporating an Exg2 GPI signal peptide sequence at the N terminus can be used as a strategy for anchoring otherwise soluble proteins to the membrane in yeast and is an efficient approach in applications of the yeast mbSUS assay for protein-protein interactions. Interestingly, although the GPI signal sequence is normally found near the C terminus of membrane-anchored proteins and has been used for cell-surface display of proteins in yeast (Fujita et al., 2002, 2004; Yamada et al., 2011; Inokuma et al., 2014), we were able to employ the Exg2 GPI signal peptide as an N-terminal membrane anchor. At present, we cannot be certain why the GPS anchor should prove superior to the OST4p anchor, but we note that the Exg2 signal peptide incorporates a hydrophobic domain like other GPI protein signal peptides. Such domains enhance GPI integration into the membrane (Galian et al., 2012) and may be sufficient for anchoring of protein fusions. Consequently, the early cleavage and relocation to the nucleus observed in the OST4p-anchored mbSUS assays are less likely to occur in the GPS system. Regardless of the explanation, we conclude that the Exg2 GPS system is a superior and promising tool with which to explore the interactions of proteins that would otherwise be difficult to assay using the standard mbSUS technology or other means.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. iLOV does not interact with SEC11 while VAMP721Δ127 interacts with KAT1 in the GPS system.
Supplemental Figure S2. iLOV does not interact with SEC11 while VAMP721Δ127 interacts with KAT1 in the GPS system.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (BB/I024496/1, BB/K015893/1, BB/L001276/1, BB/M01133X/1, BB/M001601/1, and BB/L019205/1 to M.R.B.) and the Royal Society (University Research Fellowship UF150364 to R.K.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Bassham DC, Raikhel NV (2000) Plant cells are not just green yeast. Plant Physiol 122: 999–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro LHP, Tettelin H, Vossen JH, Ram AF, van den Ende H, Klis FM (1997) In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13: 1477–1489 [DOI] [PubMed] [Google Scholar]
- Chapman S, Faulkner C, Kaiserli E, Garcia-Mata C, Savenkov EI, Roberts AG, Oparka KJ, Christie JM (2008) The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci USA 105: 20038–20043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Durán A, del Rey F, Snyder MP, Nombela C, Sánchez M (1995) Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev 59: 345–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Takahashi S, Ueda M, Tanaka A, Okada H, Morikawa Y, Kawaguchi T, Arai M, Fukuda H, Kondo A (2002) Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl Environ Microbiol 68: 5136–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A (2004) Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl Environ Microbiol 70: 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galian C, Björkholm P, Bulleid N, von Heijne G (2012) Efficient glycosylphosphatidylinositol (GPI) modification of membrane proteins requires a C-terminal anchoring signal of marginal hydrophobicity. J Biol Chem 287: 16399–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Lalonde S, Obrdlik P (2007) Split-ubiquitin system for identifying protein-protein interactions in membrane and full-length proteins. Curr Protoc Neurosci 41: 5.27.1–5.27.41 [DOI] [PubMed] [Google Scholar]
- Grefen C, Obrdlik P, Harter K (2009) The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol Biol 479: 217–233 [DOI] [PubMed] [Google Scholar]
- Grefen C, Chen Z, Honsbein A, Donald N, Hills A, Blatt MR (2010) A novel motif essential for SNARE interaction with the K+ channel KC1 and channel gating in Arabidopsis. Plant Cell 22: 3076–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Karnik R, Larson E, Lefoulon C, Wang Y, Waghmare S, Zhang B, Hills A, Blatt MR (2015) A vesicle-trafficking protein commandeers Kv channel voltage sensors for voltage-dependent secretion. Nat Plants 1: 15108. [DOI] [PubMed] [Google Scholar]
- Hachez C, Laloux T, Reinhardt H, Cavez D, Degand H, Grefen C, De Rycke R, Inzé D, Blatt MR, Russinova E, et al. (2014) Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D, Egerton M, Riezman H (1995) Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J Cell Biol 129: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsbein A, Sokolovski S, Grefen C, Campanoni P, Pratelli R, Paneque M, Chen Z, Johansson I, Blatt MR (2009) A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21: 2859–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horaruang W, Zhang B (2017) Mating based split-ubiquitin assay for detection of protein interactions. Bio Protoc 7: e2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuma K, Hasunuma T, Kondo A (2014) Efficient yeast cell-surface display of exo- and endo-cellulase using the SED1 anchoring region and its original promoter. Biotechnol Biofuels 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik R, Grefen C, Bayne R, Honsbein A, Köhler T, Kioumourtzoglou D, Williams M, Bryant NJ, Blatt MR (2013) Arabidopsis Sec1/Munc18 protein SEC11 is a competitive and dynamic modulator of SNARE binding and SYP121-dependent vesicle traffic. Plant Cell 25: 1368–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnik R, Zhang B, Waghmare S, Aderhold C, Grefen C, Blatt MR (2015) Binding of SEC11 indicates its role in SNARE recycling after vesicle fusion and identifies two pathways for vesicular traffic to the plasma membrane. Plant Cell 27: 675–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Muñoz GA, Kane P (2008) Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem 283: 20309–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckli N, Deplazes A, Hassa PO, Zhang Z, Peter M, Hottiger MO, Stagljar I, Auerbach D (2007) Yeast split-ubiquitin-based cytosolic screening system to detect interactions between transcriptionally active proteins. Biotechniques 42: 725–730 [DOI] [PubMed] [Google Scholar]
- Nebreda AR, Villa TG, Villanueva JR, del Rey F (1986) Cloning of genes related to exo-β-glucanase production in Saccharomyces cerevisiae: characterization of an exo-β-glucanase structural gene. Gene 47: 245–259 [DOI] [PubMed] [Google Scholar]
- Pittet M, Conzelmann A (2007) Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1771: 405–420 [DOI] [PubMed] [Google Scholar]
- Rosa MF, Correia ISA (1991). In vivo activation by ethanol of plasma membrane ATPase of Saccharomyces cerevisiae. Appl Environ Microbiol 57: 830–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC, Rothman JE (2009) Membrane fusion: grappling with SNARE and SM proteins. Science 323: 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C (2012) Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics 11: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Wallmeroth N, Berendzen KW, Grefen C (2016) Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol 171: 727–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Taniguchi N, Tanaka T, Ogino C, Fukuda H, Kondo A (2011) Direct ethanol production from cellulosic materials using a diploid strain of Saccharomyces cerevisiae with optimized cellulase expression. Biotechnol Biofuels 4: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Karnik R, Wang Y, Wallmeroth N, Blatt MR, Grefen C (2015) The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27: 1697–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Karnik R, Waghmare S, Donald N, Blatt MR (2017) VAMP721 conformations unmask an extended motif for K+ channel binding and gating control. Plant Physiol 173: 536–551 [DOI] [PMC free article] [PubMed] [Google Scholar]