A feed-forward loop facilitates the expression of the sporopollenin biosynthesis pathway genes, thereby promoting rapid sexine formation.
Abstract
Sporopollenin is the major component of the outer pollen wall (sexine). It is synthesized using a pathway of approximately eight genes in Arabidopsis (Arabidopsis thaliana). MALE STERILITY188 (MS188) and its direct upstream regulator ABORTED MICROSPORES (AMS) are two transcription factors essential for tapetum development. Here, we show that all the sporopollenin biosynthesis proteins are specifically expressed in the tapetum and are secreted into anther locules. MS188, a MYB transcription factor expressed in the tapetum, directly regulates the expression of POLYKETIDE SYNTHASE A (PKSA), PKSB, MALE STERILE2 (MS2), and a CYTOCHROME P450 gene (CYP703A2). By contrast, the expression of CYP704B1, ACYL-COA SYNTHETASE5 (ACOS5), TETRAKETIDE a-PYRONE REDUCTASE1 (TKPR1) and TKPR2 are significantly reduced in ams mutants but not affected in ms188 mutants. However, MS188 but not AMS can activate the expression of CYP704B1, ACOS5, and TKPR1. In ms188, dominant suppression of MS188 homologs reduced the expression of these genes, suggesting that MS188 and other MYB family members play redundant roles in activating their expression. The expression of some sporopollenin synthesis genes (PKSA, PKSB, TKPR2, CYP704B1, and ACOS5) was rescued when MS188 was expressed in ams. Therefore, MS188 is a key regulator for activation of sporopollenin synthesis, and AMS and MS188 may form a feed-forward loop that activates the expression of the sporopollenin biosynthesis pathway for rapid pollen wall formation.
The cell wall is the fundamental support structure of a plant cell. The pollen wall is the most complicated wall of all plant cells. It is composed of the inner intine and the outer exine. The intine composition is similar to normal plant cell walls (Edlund et al., 2004). The exine is divided into the sexine and nexine. The nexine is a flat layer localized between the sculptured sexine and intine. Glycoproteins are essential components of the nexine layer (Lou et al., 2014; Jia et al., 2015). The sexine is mainly composed of complex chemical compounds of sporopollenin (Scott, 1994; Piffanelli et al., 1998; Blackmore et al., 2007). The morphological pattern of sexine varies among species, which is the basis of the discipline of palynology (Blackmore et al., 2007). As one of the most complex structures of the extracellular matrix, the pollen wall plays an important role in protecting pollen against microorganism invasion and mediating species-specific pollen-stigma recognition during the pollination process.
Sporopollenin is a highly resistant biopolymer, which is thought to be composed of aromatics, phenolics, and long-chain aliphatic acids (Piffanelli et al., 1998; Ariizumi and Toriyama, 2011). The components of sporopollenin precursors are produced by and stored in tapetum cells after meiosis. They are subsequently transported out of the tapetum cells and deposited on the surface of microspores to form the sexine layer (Heslop-Harrison, 1962; Dickinson and Heslop-Harrison, 1968; Zhou et al., 2015; Xu et al., 2016). In Arabidopsis (Arabidopsis thaliana), eight proteins are reported to participate in the synthesis of sporopollenin precursors. Among them, ACYL-CoA SYNTHETASE5 (ACOS5) catalyzes mid-/long-chain fatty acids into fatty acyl-CoA, which is hydroxylated by members of the cytochrome P450 superfamily, CYP703A2 and CYP704B1. The hydroxylated products are catalyzed by the chalcone synthase family POLYKETIDE SYNTHASE A (PKSA) and PKSB into triketide and tetraketide α-pyrones, which are the substrates of TETRAKETIDE α-PYRONE REDUCTASE1 (TKPR1) and TKPR2, respectively. As a fatty acyl reductase, MALE STERILE2 (MS2) catalyzes the palmitoyl acyl carrier protein into a fatty alcohol (Aarts et al., 1997; Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009, 2010; Grienenberger et al., 2010; Kim et al., 2010; Chen et al., 2011). The resultant sporopollenin precursors were predicted to be synthesized in the tapetum and transported to the anther locule by the member of ATP-binding cassette transporter superfamily (ABCG26; Quilichini et al., 2010; Choi et al., 2011; Dou et al., 2011).
In Arabidopsis, several transcription factors are essential for tapetum development and pollen formation including two basic helix-loop-helix (bHLH) transcription factors, DYSFUNCTIONAL TAPETUM1 (DYT1) and ABORTED MICROSPORES (AMS), two MYB transcription factors, DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (TDF1) and MALE STERILITY188 (MS188, also named MYB103 or MYB80), and a PLANT HOMEODOMAIN finger member MS1 (Wilson et al., 2001; Sorensen et al., 2003; Zhang et al., 2006, 2007; Zhu et al., 2008). DYT1 is the most upstream regulator in tapetum development. DYT1 affects many downstream genes related to tapetum development and pollen wall formation by directly regulating TDF1 (Gu et al., 2014). TDF1 directly regulates AMS, AMS regulates MS188, and MS188 regulates MS1 (Lou et al., 2014, 2018; J.Y. Lu, S.X. Xiong, W.Z. Yin, X.D. Teng, Y. Lou, J. Zhu, C. Zhang, J.N. Gu, Z.A. Wilson, and Z.N. Yang, unpublished data). Thus, these transcription factors form a genetic pathway (DYT1-TDF1-AMS-MS188-MS1) in controlling tapetum development. In this pathway, AMS can bind to the promoters of CYP703A2, PKSB, TKPR1, and CYP704B1 in vivo (Xu et al., 2014). The ams mutant displays aberrantly enlarged tapetal cells and aborted microspores (Xu et al., 2010; Zhu et al., 2011). The pollen sexine is absent in the ms188 mutant (Zhang et al., 2007). Recently, we found that MS188 interacts with AMS to regulate the expression of CYP703A2 (Xiong et al., 2016). The pollen wall is formed in a short time to protect the development of the microspore into mature pollen. It is uncertain how the tapetum regulates the sporopollenin synthesis pathway. In this work, we show that MS188 acts as the major regulator to activate the sporopollenin synthesis pathway. As an upstream regulator, AMS may interact with MS188 to form a feed-forward loop to facilitate the expression of sporopollenin synthesis genes for pollen wall formation.
RESULTS
Sporopollenin Synthesis Enzymes Are Localized in the Tapetum and Locules
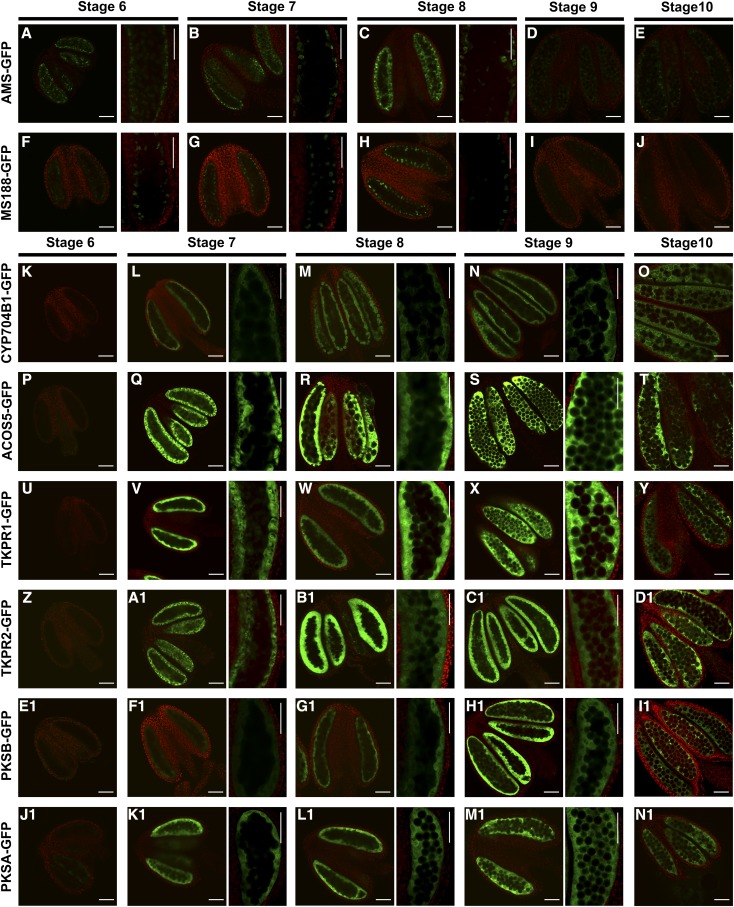
During Arabidopsis anther development, AMS and MS188 accumulate in tapetum cells at stages 6 to 9, in which the tapetal cells transfer into the secretory type and provide the sporopollenin materials for sexine formation (Xiong et al., 2016). We confirmed this finding by GFP observation in current study (Fig. 1, A–J). The sporopollenin synthesis enzyme, CYP703A2, is localized in both the tapetal cells and anther locule at stages 7 through 9 (Xiong et al., 2016). The MS2 protein accumulates in tapetal cells (Chen et al., 2011). To understand the expression pattern of other sporopollenin synthesis genes, we fused the GFP gene to the genomic sequences of PKSA, PKSB, ACOS5, CYP704B1, TKPR2, and TKPR1 driven by their native promoters. The TKPR1-GFP construct was transferred to wild-type Arabidopsis plants to obtain transgenic lines. All other constructs were transferred to their corresponding mutants with heterozygous backgrounds. Transgenic lines with homozygous backgrounds were identified. The transgenes successfully complemented the fertility of the mutants (Supplemental Fig. S1). Hence, the GFP tagging of these genes did not interfere with their biological function. The transgenic plants were designated as PKSA-GFP, PKSB-GFP, ACOS5-GFP, CYP704B1-GFP, TKPR1-GFP, and TKPR2-GFP (Fig. 1, K–N1). Observations using confocal microscopy indicated that these sporopollenin synthesis proteins first accumulated in anthers at stage 7 and then decreased significantly at stage 10 (Fig. 1, K–N1). The expression pattern of these proteins overlap with that of AMS and MS188 (Fig. 1, B, C, G, and H). The GFP fluorescent signals in transgenic plants appeared initially in the tapetal layer. When the microspores were released, the GFP signals were observed in both the tapetum and the locule and accumulated to high levels during the late stage of anther development. No GFP signals were detected inside the tetrads and free microspores (Supplemental Fig. S2). It has been reported that these genes are transcribed in tapetal cells (Aarts et al., 1997; Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2010; Chen et al., 2011), consistent with our results that indicate that the sporopollenin synthesis proteins are translated in the tapetum and gradually transported into the locules during anther development.
Figure 1.
Protein localization of AMS-GFP, MS188-GFP, and sporopollenin synthesis protein-GFPs. Confocal images of the fluorescence of the AMS-GFP, MS188-GFP, CYP704B1-GFP, ACOS5-GFP, TKPR1-GFP, TKPR2-GFP, PKSB-GFP, and PKSA-GFP fusion proteins from stages 6 through 10. GFP expression (530 nm) is shown in the green channel, while chlorophyll autofluorescence (>560 nm) is shown in the red channel. A to C and F to H, The fluorescence of AMS-GFP and MS188-GFP at stages 6 through 8. Right, high-magnification images of GFP localization. D, E, I, and J, The fluorescence of AMS-GFP and MS188-GFP at stages 9 and 10. K, P, U, Z, E1, and J1, The fluorescence of CYP704B1-GFP, ACOS5-GFP, TKPR1-GFP, TKPR2-GFP, PKSB-GFP, and PKSA-GFP at stage 6. L to N, Q to S, V to X, A1 to C1, F1 to H1, and K1 to M1, The fluorescence of CYP704B1-GFP, ACOS5-GFP, TKPR1-GFP, TKPR2-GFP, PKSB-GFP, and PKSA-GFP at stages 7 to 9. Right, high-magnification images. O, T, Y, D1, I1, and N1, The fluorescence of CYP704B1-GFP, ACOS5-GFP, TKPR1-GFP, TKPR2-GFP, PKSB-GFP, and PKSA-GFP at stage 10. Scale bars, 50 μm.
AMS and MS188 Are Required for the Expression of Sporopollenin Synthesis Genes
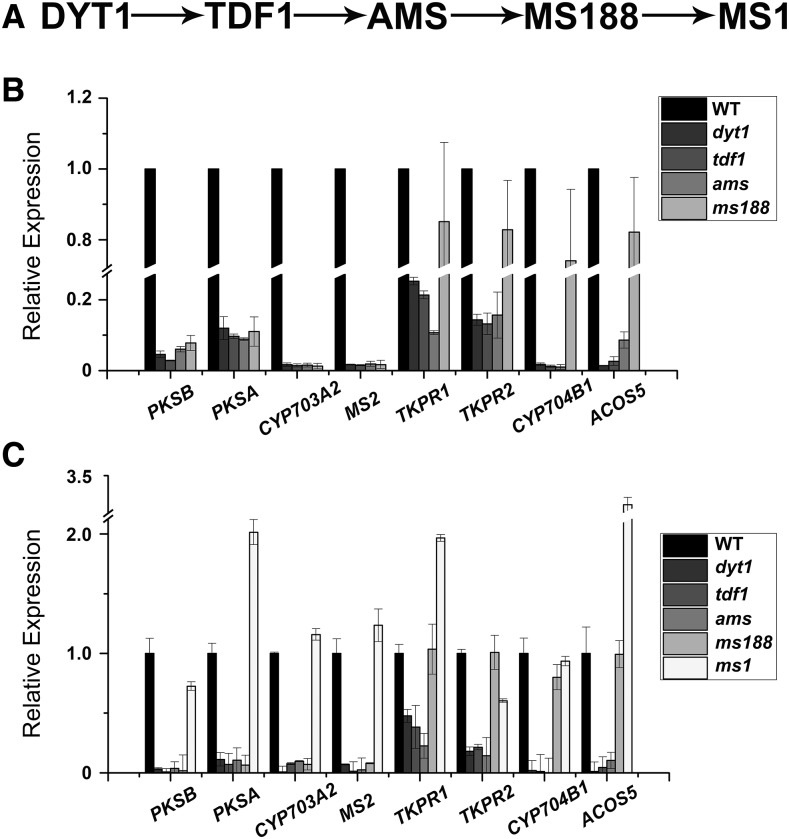
DYT1, TDF1, AMS, MS188, and MS1 form a genetic pathway that is important for tapetal development and pollen wall formation (Zhu et al., 2011; Gu et al., 2014; Lou et al., 2014, 2018; Fig. 2A). We analyzed the expression of sporopollenin synthesis genes in the mutants of these transcription factors. Results from microarray analysis showed that expression of PKSA, PKSB, MS2, and CYP703A2 was suppressed in dyt1, tdf1, ams, and ms188 mutants (Fig. 2B; Zhu et al., 2010; Li et al., 2017). Since DYT1, TDF1, and AMS are upstream regulators of MS188, it is likely that these sporopollenin synthesis genes are downstream of MS188. The expression of CYP704B1, ACOS5, TKPR1, and TKPR2 was reduced in dyt1, tdf1, and ams mutants, but not in ms188 (Fig. 2B). This indicates that these four genes are downstream of AMS. We utilized reverse transcription quantitative PCR (RT-qPCR) analysis to further verify these data. The results of RT-qPCR were consistent with the microarray data (Fig. 2C). MS1 is thought to be the last transcription factor of the genetic pathway. RT-qPCR analysis showed that the expression of these genes was not affected in the ms1 mutant (Fig. 2C). In summary, these results suggest that AMS and MS188 are two regulators closely related with the expression of sporopollenin synthesis genes.
Figure 2.
Expression of the sporopollenin synthesis genes in tapetal transcription factor mutants. A, The proposed hierarchical relationship between the tapetal transcription factors required for exine development. B, Results from microarray analysis of PKSA, PKSB, CYP703A2, MS2, TKPR1, TKPR2, CYP704B1, and ACOS5 expression in the inflorescences of wild type (WT), dyt1, tdf1, ams, and ms188 mutants. C, RT-qPCR analysis of the above sporopollenin synthesis genes in the inflorescences of wild-type, dyt1, tdf1, ams, ms188, and ms1 plants. x axis represents the genes, and y axis is shown as relative expression. sd is indicated as error bar.
MS188 Directly Activates the Expression of PKSB, PKSA, and MS2
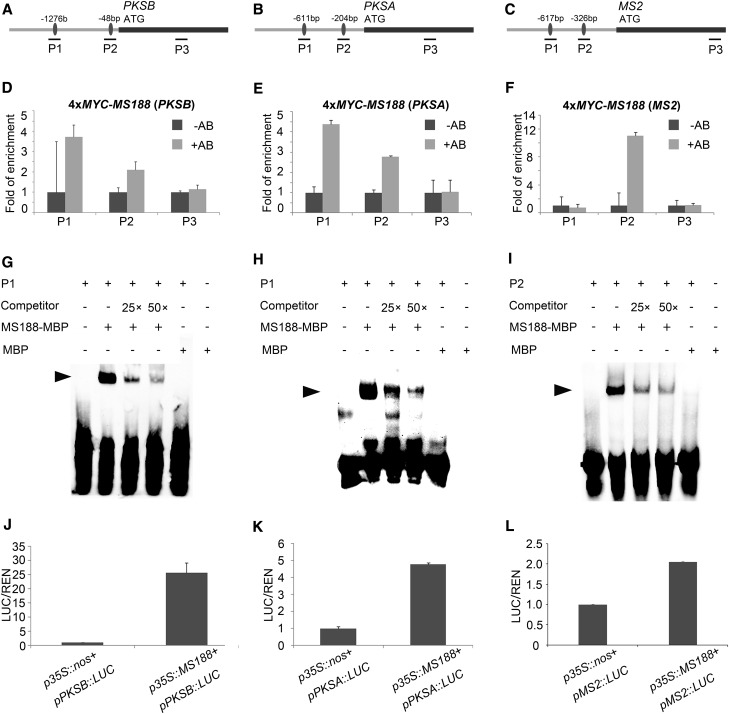
MS188 encodes a member of the MYB family (Zhang et al., 2007). AACC is the core MYB binding motif (Phan et al., 2011; Xiong et al., 2016). The promoter regions of PKSA, PKSB, and MS2 genes all contain several MYB core binding motifs (Phan et al., 2011; Xiong et al., 2016). In a previous study, we obtained transgenic lines in which Myc-tag fused with MS188 fully complemented the ms188 phenotype (Xiong et al., 2016). We used this transgenic line to perform chromatin immunoprecipitation (ChIP) to understand whether MS188 was directly associated with these promoters. For each promoter, two probes spanning the MYB binding motifs (probe 1 and 2) and one probe in its coding region as a control were synthesized for analysis (Fig. 3, A–C). Our results showed that MYC-MS188 binding was stronger in the promoter regions of these genes than in the control coding regions (Fig. 3, D–F; Supplemental Fig. S3). Results from electrophoretic mobility shift assays (EMSA) showed that Maltose binding protein (MBP)-MS188 recombinant protein (Supplemental Fig. S4) bound to these cis-elements containing “AACC” core motifs. A 25-fold and 50-fold excess of unlabeled AACCT-containing probes was able to compete for the binding of MS188 to labeled probes (Fig. 3, G–I). Hence, MS188 is able to directly bind to the promoter sequences of PKSA, PKSB, and MS2. To analyze whether MS188 could activate the expression of these genes, a transient overexpression assay using Arabidopsis protoplasts was performed. The firefly luciferase (LUC) reporter gene individually driven by PKSA, PKSB, and MS2 promoters and the Renilla gene driven by the 35S promoter were coconstructed as reporters, while MS188 driven by the 35S promoter was used as an effector. When each reporter paired with the p35S::MS188 effector were cotransformed into protoplasts, the amount of LUC luminescence driven by PKSB, PKSA, and MS2 promoters increased, compared with the background level in the negative control (Fig. 3, J–L). These results indicate that MS188 directly binds to the promoters of PKSA, PKSB, and MS2 to activate their expression.
Figure 3.
Transcription factor MS188 directly regulates the expression of PKSA, PKSB, and MS2. A to C, The designed probes of PKSB, PKSA, and MS2, respectively. Probe 1 (P1) and P2 contain the binding sites (black dots) of MS188, and P3 was designed in the coding region as a control. The black lines show the fragments of the probes for the ChIP-qPCR assay. D to F, The fragments of the above probes in PKSB, PKSA, and MS2 promoters specifically amplified by ChIP-qPCR using 4xMYC-MS188 transgenic plant samples. The fold of enrichment is the average of three replicates, and sd is indicated as error bar. −AB, absence of antibody; +AB, presence of antibody. G to I, EMSA assay showing MS188 binding to probes in vitro. The first panel represents free probe, and the last panel indicates the mixture of free probe and MBP tag; both are used as negative controls. The shift band is indicated by the arrowhead, which is highlighted by the positive control of mixture of biotin-tagged probe and non-biotin-tagged probe and MBP-MS188. J to L, p35s::MS188 was cotransformed with pPKSB::LUC, pPKSA::LUC, and pMS2::LUC, respectively, and a transient dual-luciferase assay was conducted in the Arabidopsis protoplasts. Three replicates were performed, and the y axis is shown as the ratio of Luciferase/Renilla. sd is indicated as error bar.
MS188, Not AMS, Activates the Expression of ACOS5, CYP704B1, and TKPR1
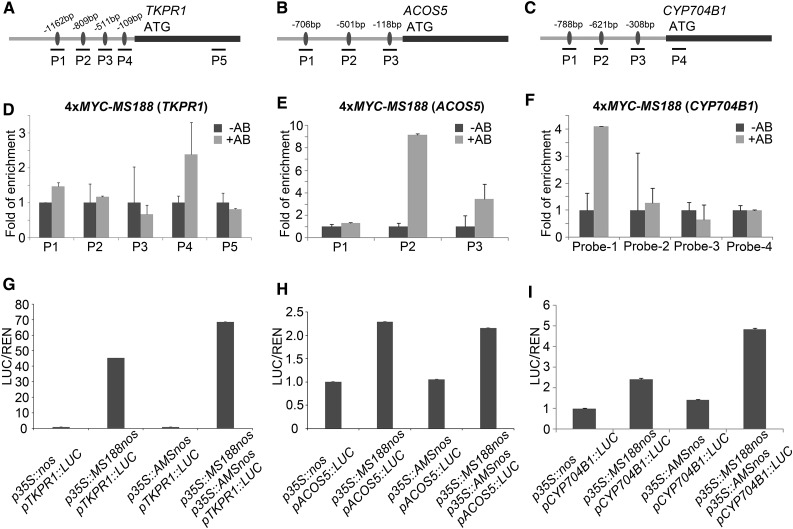
Through cytological analysis, MS188 was shown to be the key regulator for sexine formation (Zhang et al., 2007). However, results from microarray data and RT-qPCR analysis suggest that ACOS5, CYP704B1, and TKPR1 are downstream of AMS (Fig. 2, B and C). AMS can also bind to the promoters of ACOS5, CYP704B1, and TKPR1 (Xu et al., 2014; Supplemental Fig. S5). We further looked into this issue using a ChIP assay. The promoter regions of ACOS5, CYP704B1, and TKPR1 contain several MYB binding sites (Fig. 4, A–C). ChIP assay and EMSA showed that MS188 bound the promoters of ACOS5, CYP704B1, and TKPR1 (Fig. 4, D–F; Supplemental Figs. S3 and S6). Since both AMS and MS188 can bind to the promoters of these genes, a dual-luciferase assay was performed to understand whether they could activate the expression of these genes. When the constructs of pTKPR1::LUC, pACOS5::LUC, and p35S::AMS were cotransformed into Arabidopsis leaf protoplasts, LUC activity was hardly induced (Fig. 4, G and H). The pCYP704B1::LUC could be slightly induced with p35S::AMS in the protoplasts (Fig. 4I). This indicates that AMS alone could hardly activate the expression of ACOS5, CYP704B1, and TKPR1. However, the LUC reporter driven by the promoters of the three genes above was strongly induced by MS188 protein (Fig. 4, G–I). Once pCYP704B1::LUC and pTKPR1::LUC were cotransformed with p35S::AMS and p35S::MS188, respectively, the luminescence signals increased (Fig. 4, G and I). For the cotransformation of pACOS5::LUC with p35S::AMS and p35S::MS188, LUC activity did not obviously increase (Fig. 4H). These results show that MS188 directly regulates the expression of ACOS5, and AMS and MS188 synergistically activate the expression of TKPR1 and CYP704B1.
Figure 4.
MS188 but not AMS activates the expression of ACOS5, CYP704B1, and TKPR1. A to C, The designed probes in the promoter and genomic regions of TKPR1, ACOS5, and CYP704B1, respectively. D to F, The fragments of the above probes in TKPR1, ACOS5, and CYP704B1 promoters were specifically amplified by ChIP-qPCR using 4xMYC-MS188 transgenic plant samples. The fold enrichment is the average of three replicates (error bar is shown by sd). −AB, absence of antibody; +AB, presence of antibody. G to I, Transient dual-luciferase assays were performed in Arabidopsis leaf protoplasts. The p35S::NOS vector was used as negative control. The p35S::MS188 and p35S::AMS were transformed into the protoplasts with reporter plasmids separately. The last column of three panels was cotransformation of p35S::MS188, p35S::AMS, and each reporter plasmid. sd is indicated as error bar.
Other MYBs, Together with MS188, Regulate the Expression of CYP704B1, ACOS5, TKPR1, and TKPR2
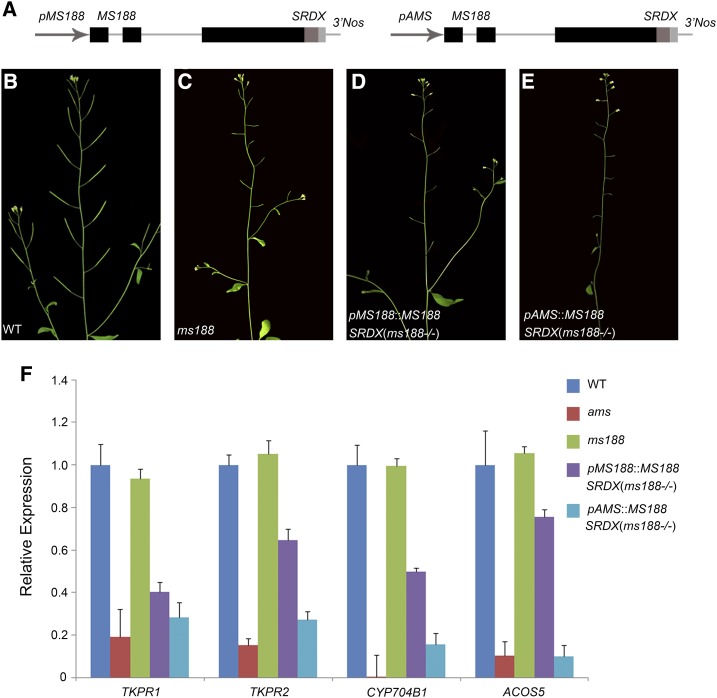
MS188 was shown to activate the expression of ACOS5 and TKPR1 in Arabidopsis protoplasts; however, their expression was not affected in the ms188 mutant (Fig. 2, B and C). This may be caused by functional redundancy with other gene family members. To investigate this further, we made constructs of MS188 fused with a 12-amino acid SRDX motif (LDLDLELRLGFA) driven by either the promoter of MS188 or AMS (Fig. 5A). The chimeric protein with this motif was reported to act in a dominant manner to inhibit the activation of redundant transcription factors (Hiratsu et al., 2003). The two constructs were named pMS188::MS188-SRDX and pAMS::MS188-SRDX and were individually transformed into MS188+/− heterozygous plants. Transgenic lines with the ms188 homozygous background were identified (Fig. 5, B–E). In the pMS188::MS188-SRDX transgenic line, the expression of CYP704B1, ACOS5, TKPR1, and TKPR2 was decreased compared with that in ms188 (Fig. 5F). In the transgenic line of pAMS::MS188-SRDX, the transcript levels of AMS compared to wild-type plants were not affected (Supplemental Fig. S7A), but the expression of all these genes was further reduced compared with that in the pMS188::MS188-SRDX transgenic line (Fig. 5F). The expression level of these genes in the pAMS::MS188-SRDX line was close to that of the ams mutant (Fig. 5F). AMS acts upstream of MS188, and its expression is earlier than MS188 (Zhu et al., 2011). The earlier expression of MS188-SRDX conferred by the AMS promoter may lead to a higher level of the chimeric MS188-SRDX protein thereby enhancing the repression of its targets. These results indicate that other MYB transcription factors are also involved in regulating the expression of CYP704B1, ACOS5, TKPR1, and TKPR2.
Figure 5.
Redundant genes of the MYB family are involved in regulating the expression of sporopollenin synthesis genes. A, pMS188::MS188-SRDX and pAMS::MS188-SRDX constructs. B to E, Fertility comparison between wild-type (WT), ms188, pMS188::MS188-SRDX (ms188−/−), and pAMS::MS188-SRDX (ms188−/−) plants, respectively. F, Reverse transcription quantitative PCR (RT-qPCR) analysis of TKPR1, TKPR2, CYP704B1, and ACOS5 expression in the inflorescences of WT, ams, ms188, pMS188::MS188-SRDX (ms188−/−), and pAMS::MS188-SRDX (ms188−/−) plants. y axis is shown as relative expression, and error bar is indicated as sd.
The Expression of MS188 in ams Rescues the Expression of Most Sporopollenin Synthesis Genes
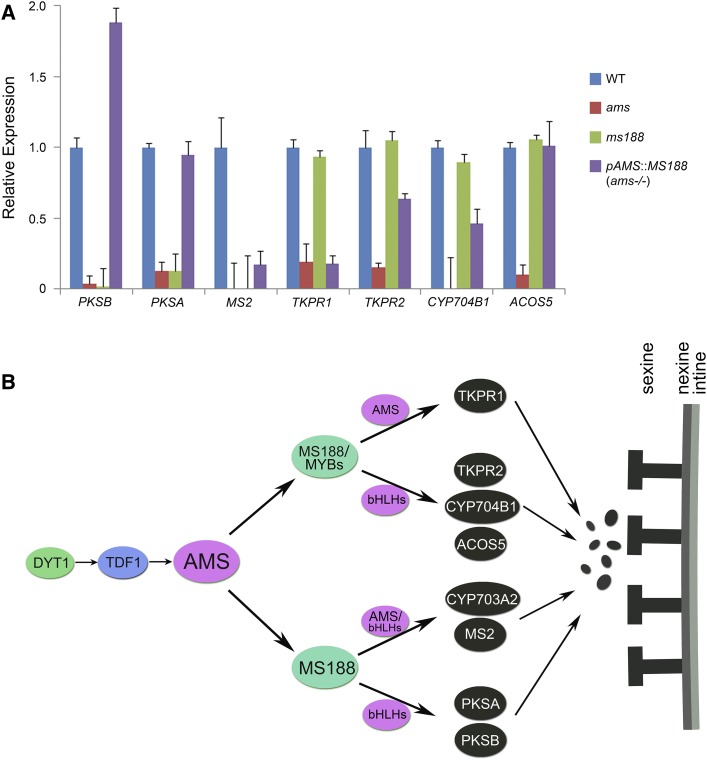
In previous work, we obtained pAMS::MS188 transgenic plants where MS188 is expressed normally in the ams mutant background (Xiong et al., 2016). We used this transgenic line to analyze if MS188 without AMS could activate the expression of sporopollenin synthesis genes. Of PKSA, PKSB, MS2, and CYP703A2, which were severely suppressed in both ams and ms188 mutants (Fig. 2, B and C), the expression of PKSA and PKSB was fully restored in this transgenic line. The expression of MS2 was partially restored (Fig. 6A), as was CYP703A2 (Xiong et al., 2016). In ams, the expression of CYP704B1, ACOS5, and TKPR2 decreased. The expression of these genes was largely restored in the transgenic line (Fig. 6A). However, the expression of TKPR1 did not show any recovery (Fig. 6A). These results demonstrate that MS188 acts as a major regulator to activate the expression of most genes for sporopollenin synthesis.
Figure 6.
Expression of MS188 in ams rescues the expression of sporopollenin synthesis-related genes. A, Reverse transcription quantitative PCR (RT-qPCR) analysis of PKSB, PKSA, MS2, TKPR1, TKPR2, CYP704B1, and ACOS5 in the inflorescences of wild-type (WT), ams, ms188, and pAMS::MS188 (ams−/−) plants. y axis is shown as relative expression, and error bar is indicated as sd. B, A proposed model of MS188 acting as the main regulator together with AMS and other MYB family and bHLH family members to modulate sexine formation. For the genetic pathway (DYT1-TDF1-AMS-MS188-MS1) in the tapetal layer, MS188 is directly regulated by AMS and MS188 interacts with AMS to form a feed-forward loop to regulate the expression of sporopollenin synthesis genes. Some other MYB family and bHLH family members participate in the regulation process. The colorful ellipses indicate the transcription factors, and the black ones indicate the sporopollenin enzymes. The black circles represent the sporopollenin precursors.
DISCUSSION
The Localization of Sporopollenin Synthesis Proteins Suggests the Synthesis of Sporopollenin Precursors in the Locule
In Arabidopsis, sporopollenin precursor synthesis involves at least eight genes that are expressed in the tapetal layer (Morant et al., 2007; de Azevedo Souza et al., 2009; Dobritsa et al., 2009, 2010; Grienenberger et al., 2010; Kim et al., 2010; Chen et al., 2011). Immunochemical localization under transmission electron microscope reveals that ACOS5, PKSA/B, TKPR1/2 are localized in tapetal cells (Lallemand et al., 2013). In the transgenic line with CYP703A2-GFP fully rescuing the cyp703a2 phenotype, CYP703A2-GFP is localized in both the tapetum and anther locule (Xiong et al., 2016). In this study, we used the same strategy to understand the expression pattern of sporopollenin synthesis genes during anther development. The protein products of these genes were initially detected in tapetal cells, then accumulated in the tapetum and anther locules (Fig. 1, K–N1). The expression pattern of these proteins is quite similar to that of CYP703A2. The similarity of their expression patterns supports that they belong to the same pathway of metabolic synthesis. Tapetal degeneration is first detected at stage 10, and is completed at stage 12 during anther development (Sanders et al., 1999). These proteins were observed in the locule at stage 8 (Fig. 1, M, R, W, B1, G1, and L1) indicating that they were secreted from the tapetum into the anther locule. It was proposed that the sporopollenin precursors are synthesized in the tapetal cells and transported onto the surface of microspores (Ariizumi and Toriyama, 2011; Quilichini et al., 2015). The high accumulation of sporopollenin synthesis proteins in locule suggests that sporopollenin precursors could be synthesized not only in the tapetum but also in the locule. The synthesis of sporopollenin precursors in the locule may facilitate sporopollenin deposition on the microspore surface to form the sexine layer.
MS188 Plays a Key Role in Activating the Expression of Sporopollenin Synthesis Genes
In the genetic pathway (DYT1-TDF1-AMS-MS188-MS1), AMS and MS188 are two regulators important for the expression of sporopollenin synthesis genes. All eight genes were downregulated in ams, and four of them were suppressed in the ms188 mutant (Fig. 2, B and C). In this work, we demonstrate that MS188 acts as a key regulator for sporopollenin synthesis gene expression based on the following evidence: (1) MS188 bound to the promoters of these genes (Figs. 3, D–I, and 4, D–F; Supplemental Fig. S6);( 2) MS188, but not AMS, activated their transcription (Fig. 3, J–L, and 4, G and H); (3) without AMS, MS188 could activate the expression of most sporopollenin synthesis genes in vivo (Fig. 6A). The key role of MS188 for regulating sporopollenin synthesis is consistent with the phenotype of an absent sexine layer in the ms188 mutant (Zhang et al., 2007). Among these sporopollenin synthesis genes, the expression of ACOS5, CYP704B1, and TKPR1/2 were not affected in the ms188 mutant (Fig. 2, B and C). A dominant suppression assay suggests that other MYB members play a redundant role to activate the expression of these genes (Fig. 5F). In Arabidopsis, MYB33 and MYB65 redundantly facilitate proper tapetal development. The tapetal cells of the myb33 myb65 double mutant undergo hypertrophy during meiosis resulting in abortion of pollen development (Millar and Gubler, 2005). Additionally, MYB32 is reportedly involved in the regulation of phenylpropanoid pathways, affecting the composition of the pollen wall in Arabidopsis (Preston et al., 2004). These MYB family transcription factors might play a redundant role with MS188 to regulate sporopollenin synthesis for sexine formation.
AMS Interacting with MS188 May Form a Feed-Forward Loop to Highly Regulate Sporopollenin Synthesis
MYB family members frequently interact with bHLH transcription factors to regulate downstream genes during plant growth and development. In Arabidopsis, bHLH family members GLABROUS3 (GL3) and TRANSPARENT TESTA8 interact with MYB family members TRANSPARENT TESTA2, GL1, and MYB61 to regulate the expression of DIHYDROFLAVONOL REDUCTASE and LEUCOANTHOCYANIDIN OXIDASE for anthocyanin synthesis (Koes et al., 2005; Appelhagen et al., 2011). The GL3-GL1 complex is also required for root epidermal and trichome development (Schiefelbein, 2003; Zhao et al., 2008). bHLH-MYB complexes regulate the jasmonate-mediated stamen and seed maturation (Qi et al., 2015). The bHLH family member MYC2 interacts with MYB2 to regulate the gene expression of the abscisic acid-inducible pathway (Abe et al., 2003; Kazan and Manners, 2013). MS188, a MYB gene family member, is the key regulator in activating the expression of sporopollenin synthesis genes for pollen wall formation. In the tapetum, AMS (bHLH family) is reported to interact with MS188 (MYB family; Xiong et al., 2016; Lou et al., 2018). AMS can bind to the promoters of several sporopollenin synthesis genes (Xu et al., 2014). It is likely that the activation of MS188 for sporopollenin synthesis requires its interaction with bHLH transcription factor AMS. During anther development, three other bHLH family members (bHLH010, bHLH089, and bHLH091) are also involved in tapetum development and male fertility (Zhu et al., 2015; Cui et al., 2016). Yeast two-hybrid assays showed that MS188 also interacted with both AMS and bHLH010 (Supplemental Fig. S8). Therefore, MS188 or other MYBs may form different combinations with AMS or other bHLHs to regulate the expression of relative sporopollenin synthesis genes as shown in the model (Fig. 6B). In this model, MS188 and other MYBs play a redundant role in activating the expression of CYP704B1, ACOS5, TKPR1, and TKPR2. Among them, AMS is critical for TKPR1 expression because it could not be restored by MS188 in ams mutants. AMS and other bHLHs may also participate in activating the expression of CYP704B1, ACOS5, and TKPR2. MS188 was essential to activate the expression of CYP703A2, PKSB, PKSA, and MS2. As the expression of CYP703A2 and MS2 was partially restored by MS188 in ams mutants, AMS is important and other bHLHs play a partial redundant role to regulate their expression. The expression of PKSB and PKSA was fully restored, indicating that other bHLHs can replace AMS to regulate their expression (Fig. 6B).
Sporopollenin synthesis genes are highly and specifically expressed from stage 7 (late tetrad stage) to stage 10 during anther development (Fig. 1, C–H), when sporopollenin synthesis and deposition occur. The activation of the sporopollenin synthesis pathway by the key activator MS188 may facilitate the quick initiation of sporopollenin synthesis to protect developing microspores. AMS directly regulates MS188 and interacts with MS188 (Xiong et al., 2016; Lou et al., 2018). This appears analogous to the regulatory relationship of TDF1 and AMS, in which TDF1 directly regulates AMS and interacts with AMS to form a feed-forward loop to facilitate downstream gene expression (Lou et al., 2018). In the tapetum, the expression of TDF1 is relatively low (Lou et al., 2018). These two feed-forward loops may act together to achieve the high levels of expression of sporopollenin synthesis genes required for rapid formation of the pollen wall.
MATERIALS AND METHODS
Plants Growth Conditions
The Arabidopsis (Arabidopsis thaliana) Colombia ecotype was used for wild-type plants. The seeds were placed in the dark at 4°C for 2 d before germination and then planted in a 22°C incubator with 16 h light and 8 h dark. Plant flowers were transformed with Agrobacterium tumefaciens containing the constructs in a binary vector, and the seeds obtained were screened on selective growth medium to obtain the transgenic plants. All T-DNA insertion mutant lines were provided by the Arabidopsis Biological Resource Center.
Expression Analysis
The RNA used for microarray and RT-qPCR analyses was derived from the inflorescences of wild-type, dyt1, tdf1, ams, ms188, and ms1 plants. Three replicates of independently grown materials were used. The RNA was extracted using TRIzol (Invitrogen), purified using a RNeasy Mini kit (Qiagen) and used as a template for cDNA synthesis. Cy3- and Cy5-labeled cRNA molecules were synthesized (Agilent; 5184-3523) and hybridized onto a 44 K Arabidopsis oligo microarray (Agilent; 5188-5242). Signals were detected according to the manufacturer’s protocol. The spots with 2-fold or more differential expression between the wild type and mutants (pValueLogRatio < 0.05; Flag = 0; S/n > 2.6) in three independent experiments were identified. RT-qPCR was carried out as previously described (Xiong et al., 2016). The primers were synthesized by Shanghai Generay Biotech. The primer sequences are presented in Supplemental Table S1.
Protein Localization
The transgenic plants of ACOS5-GFP, CYP704B1-GFP, TKPR1-GFP, TKPR2-GFP, PKSA-GFP, and PKSB-GFP were constructed by fusing GFP with ACOS5, CYP704B1, TKPR1, TKPR2, PKSA, and PKSB driven by their native promoters, and the binary vector pCAMBIA1300 was used for the construction. The constructed plasmids were transferred into A. tumefaciens to infect the flower buds. The flower buds of the plants were harvested, and the anthers were isolated. A Zeiss laser confocal microscope with a 40× oil objective was used to observe GFP green fluorescence (Xiong et al., 2016).
ChIP
The MS188-MYC transgenic plants were planted and their buds were collected as ChIP experimental materials. The MS188-MYC-tagged proteins and chromatin forming complexes in buds were cross linked in formaldehyde containing buffer. The nuclei were isolated, and the chromatin was sheared with an ultrasonic apparatus. The majority of the DNA fragments with a size between 300 and 800 bp were incubated with salmon sperm DNA/protein agarose mix (Millipore) for 1 h, and the supernatants were incubated with polyclonal antibody against MYC (Millipore) at 4°C overnight. The antibody-protein/DNA complexes were precipitated with magnetic beads protein G (Invitrogen). After isolation and purification, the DNA samples were subjected to real-time PCR analysis. The primer information is shown in Supplemental Table S1.
EMSAs
Induction and purification of MBP-MS188 fusion protein was based on a published protocol (Xiong et al., 2016). The biotin-labeled and unlabeled DNA probes were designed based on the MYB family binding sites on the cis-elements of PKSA, PKSB, MS2, and ACOS5 promoters (probe information in Supplemental Table S1). The EMSA was performed using the Light Shift Chemiluminescent EMSA Kit (Thermo Scientific). Reactions were performed in binding buffer (10 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm DTT) at room temperature for 30 min. Images were taken with a Tanon-5500 Chemiluminescent Imaging System (Tanon, China).
Protoplast Transformation
The promoters of sporopollenin synthesis genes were inserted into the pGreenII 0800-LUC vector. Arabidopsis (Col-0) leaves grown for 28 d were harvested and protoplasts were prepared using cellulase (0.015 g/mL) and pectinase (0.0035 g/mL) to lyse the cell wall. The above constructs and the p35S::MS188 construct were cotransfected into protoplasts in poly ethylene glycol solution (0.4g/mL PEG4000), and the cytoplasm was cultivated for 12 to 16 h of light treatment. After adding Passive Lysis buffer to lyse the protoplasts, the LUC substrate (Luciferase Assay Reagent II) was added to detect the firefly luminescence. The reaction was quenched, and then the Stop & Glo Reagent was added to test the Renilla luciferase. The above experiment was performed using luciferase assay kit obtained from Promega (catalog no. E1910) according to the manufacturer’s instructions.
Yeast Two-Hybrid Analysis
The yeast two-hybrid assay was achieved following the protocol of the Clontech two-hybrid system. The MS188 proteins without the self-activation fragment were obtained from previous work (Xiong et al., 2016), and the bHLH010 coding region was inserted into the pGADT7 plasmids. The Saccharomyces cerevisiae (yeast) strain AH109 was cotransformed with these constructs. The transformants were screened on the corresponding selective media of supplemented synthetic dextrose medium lacking Leu and Trp or supplemented synthetic dextrose medium lacking Leu, Trp, His, and adenine hemisulfate salt with X-α-Gal.
Accession Numbers
Microarray data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: GSE102528. DYT1, AT4G21330; TDF1, AT3G28470; AMS, AT2G16910; MS188/MYB80/MYB103, AT5G56110; MS1, AT5G22260; ACOS5, AT1G62940; MS2, AT3G11980; CYP703A2, AT1G01280; CYP704B1, AT1G69500; PKSA, AT1G02050; PKSB, AT4G34850; TKPR1, AT4G35420; TKPR2, AT1G68540; and bHLH010, AT2G31220.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The mutants of sporopollenin synthesis genes and their transgenic complementary plants.
Supplemental Figure S2. Bright-field images of the tetrads of AMS-GFP, MS188-GFP, CYP704B1-GFP, ACOS5-GFP, TKPR1-GFP, TKPR2-GFP, PKSB-GFP, and PKSA-GFP at stage 7.
Supplemental Figure S3. Percentage input of the binding probes of PKSA, PKSB, MS2, ACOS5, TKPR1, and CYP704B1 in ChIP assays.
Supplemental Figure S4. The expression and purification of MS188 and AMS recombinant proteins.
Supplemental Figure S5. AMS directly binds to the promoters of CYP704B1 and ACOS5 in vivo.
Supplemental Figure S6. MS188 directly binds to the promoters of TKPR1 and ACOS5 in vitro.
Supplemental Figure S7. RT-qPCR analysis of the expression of AMS, MYB32, MYB33, and MYB65.
Supplemental Figure S8. MS188 interacts with bHLH010 in yeast two-hybrid assays.
Supplemental Table S1. List of primers used in the study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Xin Zhou for providing the pGreenII-0800-LUC vector for protoplast transformation. We thank Professor Jeremy Murray for critical reading and revising of the manuscript.
Footnotes
This work was supported by the grants from the National Key R&D Program of China (2016YFD0100902) and the National Science Foundation of China (31670314).
Articles can be viewed without a subscription.
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhagen I, Jahns O, Bartelniewoehner L, Sagasser M, Weisshaar B, Stracke R (2011) Leucoanthocyanidin Dioxygenase in Arabidopsis thaliana: characterization of mutant alleles and regulation by MYB-BHLH-TTG1 transcription factor complexes. Gene 484: 61–68 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Blackmore S, Wortley AH, Skvarla JJ, Rowley JR (2007) Pollen wall development in flowering plants. New Phytol 174: 483–498 [DOI] [PubMed] [Google Scholar]
- Chen W, Yu XH, Zhang K, Shi J, De Oliveira S, Schreiber L, Shanklin J, Zhang D (2011) Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol 157: 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Jin JY, Choi S, Hwang JU, Kim YY, Suh MC, Lee Y (2011) An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J 65: 181–193 [DOI] [PubMed] [Google Scholar]
- Cui J, You C, Zhu E, Huang Q, Ma H, Chang F (2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28: 1078–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ (2009) A novel fatty Acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson HG, Heslop-Harrison J (1968) Common mode of deposition for the sporopollenin of sexine and nexine. Nature 220: 926–927 [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D (2009) CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou XY, Yang KZ, Zhang Y, Wang W, Liu XL, Chen LQ, Zhang XQ, Ye D (2011) WBC27, an adenosine tri-phosphate-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. J Integr Plant Biol 53: 74–88 [DOI] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16 (Suppl): S84–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza CdeA, Heitz T, Douglas CJ, Legrand M (2010) Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JN, Zhu J, Yu Y, Teng XD, Lou Y, Xu XF, Liu JL, Yang ZN (2014) DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J 80: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1962) Origin of exine. Nature 195: 1069–1071 [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Jia QS, Zhu J, Xu XF, Lou Y, Zhang ZL, Zhang ZP, Yang ZN (2015) Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for Nexine formation. Mol Plant 8: 251–260 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM (2013) MYC2: the master in action. Mol Plant 6: 686–703 [DOI] [PubMed] [Google Scholar]
- Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, Souza CdeA, Geoffroy P, Heintz D, Krahn D, Kaiser M, et al. (2010) LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Lallemand B, Erhardt M, Heitz T, Legrand M (2013) Sporopollenin biosynthetic enzymes interact and constitute a metabolon localized to the endoplasmic reticulum of tapetum cells. Plant Physiol 162: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DD, Xue JS, Zhu J, Yang ZN (2017) Gene regulatory network for tapetum development in Arabidopsis thaliana. Front Plant Sci 8: 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Xu XF, Zhu J, Gu JN, Blackmore S, Yang ZN (2014) The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat Commun 5: 3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Zhou HS, Han Y, Zeng QY, Zhu J, Yang ZN (2018) Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol 217: 378–391 [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17: 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Ross JHE, Murphy DJ (1998) Biogenesis and function of the lipidic structures of pollen grains. Sex Plant Reprod 11: 65–80 [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40: 979–995 [DOI] [PubMed] [Google Scholar]
- Qi T, Huang H, Song S, Xie D (2015) Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27: 1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ (2010) ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol 154: 678–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini TD, Grienenberger E, Douglas CJ (2015) The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 113: 170–182 [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11: 297–322 [Google Scholar]
- Schiefelbein J. (2003) Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr Opin Plant Biol 6: 74–78 [DOI] [PubMed] [Google Scholar]
- Scott RJ. (1994) Molecular and Cellular Aspects of Plant Reproduction. University Press, Cambridge, UK [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Xiong SX, Lu JY, Lou Y, Teng XD, Gu JN, Zhang C, Shi QS, Yang ZN, Zhu J (2016) The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J 88: 936–946 [DOI] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Ding Z, Vizcay-Barrena G, Shi J, Liang W, Yuan Z, Werck-Reichhart D, Schreiber L, Wilson ZA, Zhang D (2014) ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26: 1544–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Zhang C, Zhou Q, Yang ZN (2016) Pollen wall pattern in Arabidopsis. Sci Bull (Beijing) 61: 832–837 [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, et al. (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999 [DOI] [PubMed] [Google Scholar]
- Zhou Q, Zhu J, Cui YL, Yang ZN (2015) Ultrastructure analysis reveals sporopollenin deposition and nexine formation at early stage of pollen wall development in Arabidopsis. Sci Bull (Beijing) 60: 273–276 [Google Scholar]
- Zhu E, You C, Wang S, Cui J, Niu B, Wang Y, Qi J, Ma H, Chang F (2015) The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J 83: 976–990 [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55: 266–277 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang G, Chang Y, Li X, Yang J, Huang X, Yu Q, Chen H, Wu T, Yang Z (2010) AtMYB103 is a crucial regulator of several pathways affecting Arabidopsis anther development. Sci China Life Sci 53: 1112–1122 [DOI] [PubMed] [Google Scholar]
- Zhu J, Lou Y, Xu X, Yang ZN (2011) A genetic pathway for tapetum development and function in Arabidopsis. J Integr Plant Biol 53: 892–900 [DOI] [PubMed] [Google Scholar]