Analysis of integrated prior knowledge and ensemble networks highlights a previously unidentified connection between ethylene and salicylic acid signaling modules in potato.
Abstract
To develop novel crop breeding strategies, it is crucial to understand the mechanisms underlying the interaction between plants and their pathogens. Network modeling represents a powerful tool that can unravel properties of complex biological systems. In this study, we aimed to use network modeling to better understand immune signaling in potato (Solanum tuberosum). For this, we first built on a reliable Arabidopsis (Arabidopsis thaliana) immune signaling model, extending it with the information from diverse publicly available resources. Next, we translated the resulting prior knowledge network (20,012 nodes and 70,091 connections) to potato and superimposed it with an ensemble network inferred from time-resolved transcriptomics data for potato. We used different network modeling approaches to generate specific hypotheses of potato immune signaling mechanisms. An interesting finding was the identification of a string of molecular events illuminating the ethylene pathway modulation of the salicylic acid pathway through Nonexpressor of PR Genes1 gene expression. Functional validations confirmed this modulation, thus supporting the potential of our integrative network modeling approach for unraveling molecular mechanisms in complex systems. In addition, this approach can ultimately result in improved breeding strategies for potato and other sensitive crops.
Plants have evolved a multilayered immune system to cope with the potential invasion of pathogens (Jones and Dangl, 2006). The recognition of invading organisms triggers a rapid induction of signaling cascades, leading to diverse defense responses (Pieterse et al., 2012). The effectiveness of these downstream events is crucially dependent on salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), but other hormones also were shown to play important roles in plant immunity (Verma et al., 2016). Hormonal signals differ considerably in timing, quantity, and composition, depending on the type of attacker (Blüthgen, 2015). Cross talk between hormonal pathways can have antagonistic or synergistic effects and is largely multidimensional (Tsuda and Somssich, 2015). This interconnected plant hormonal network provides an important regulatory mechanism, granting plants quick adaptation abilities via intruder-specific alterations (Pieterse et al., 2012). At the molecular level, cross talk between signaling pathways with several regulatory feedback loops adds robustness to the plant immune signaling network (Windram and Denby, 2015). Network analysis, the application of mathematical graph theory approaches, also continues to be paramount in systems biology investigations of complex systems (Barabási, 2009). This approach, which involves thorough analyses of critical system properties, facilitates the discovery of novel key players or interactions, making it suitable for providing new insights into plant defense specificities (McCormack et al., 2016).
While heterogenous technologies of high-content omics allow us to capture snapshots of the systems, the challenge now lies in the integration of knowledge into a coherent systems view (Hillmer and Katagiri, 2016). Network inference from omics data sets allows us to deduce the underlying structure of activated processes. However, due to the high noisiness, high dimensionality, and low sample sizes of data, this is a nontrivial task (Veiga et al., 2010). Thus, additional improvements are needed; for example, the incorporation of prior knowledge can greatly improve reconstructed network accuracy, simultaneously reducing noise and sparsity effects of the source data without inflating the computational cost (Ghanbari et al., 2015).
Despite extensive potato (Solanum tuberosum) breeding programs, average potato yields still do not reach their physiological potential (Singh, 2008). This is the result of the sensitivity of potato to a wide range of environmental factors. The aim of this study was to improve our understanding of potato immune signaling using network modeling and, thus, in the long term, to provide means for novel crop breeding strategies directed toward high and sustainable yields. We built on a manually curated plant immune signaling model (Miljkovic et al., 2012), complementing it with knowledge from various public resources, the majority of the available data coming from the model plant Arabidopsis (Arabidopsis thaliana). We also inferred networks using time-resolved transcriptomics data of both compatible and incompatible potato-virus interactions (Baebler et al., 2014; Stare et al., 2015) and superimposed them with our knowledge network. We tested the resulting network for its potential for generating novel hypotheses and show that network analysis revealed a previously unknown connection between ET and SA signaling, namely that activation of the ET signaling module, through Ethylene Insensitive3 (EIN3), induces the expression of Nonexpressor of PR Genes1 (NPR1), an important regulator of SA signaling. This newly identified cross talk was experimentally validated in potato.
RESULTS
Construction of the Comprehensive Knowledge Network
First, a previous plant immune signaling model (PIS-v1; Miljkovic et al., 2012) was expanded with manually curated knowledge from recently published literature. The addition of 64 Arabidopsis genes to the existing model resulted in an expanded plant immune signaling model, version 2 (PIS-v2), with 212 biological components (177 genes, 31 metabolites, and four small RNAs), categorized into 108 component families as defined by Miljkovic et al. (2012). Following the abstraction of the component families, we added 32 new reactions, with the total number of reactions reaching 111 (Supplemental Data Set S1).
We combined the graph of binary PIS-v2 interactions with three layers of publicly available information: protein-protein interactions (PPIs), transcriptional regulation (TR), and regulation through microRNA (miRNA). This resulted in an Arabidopsis thaliana comprehensive knowledge network (AtCKN; Fig. 1A) with 20,012 nodes (19,812 genes, 186 miRNA families, three metabolites, and 11 viral proteins) and 70,091 connections (Supplemental Table S1). Each data layer covers unique gene or miRNA subsets in the entire network, with only six nodes present in all four layers, which indicates that our layer selection was well suited for inclusion (Fig. 2).
Figure 1.
Schematic overview of network construction and analyses. A, Networks of four prior knowledge layers were merged into the AtCKN. B, Orthologous relationships were used to translate from Arabidopsis to potato, forming the Solanum tuberosum comprehensive knowledge network (StCKN). C, Starting with two time-resolved transcriptome data sets (tolerant or hypersensitive response, HR), gene coexpression networks (GCNs; relationships between coexpressed genes) and gene regulatory networks (GRN; transcription-factor-to-regulated-gene relationships) were inferred using three methods. For each inference method, two subnetworks were generated for mock-inoculated and virus-infected samples. Removing all connections present in all coexpression or gene regulatory networks resulted in differential networks. PVY, Potato virus Y. D, StCKN and differential networks were merged into the Solanum tuberosum integrated network (StIN). E, The created networks were analyzed using network analysis approaches.
Figure 2.
Contribution of the four layers to node coverage in AtCKN. The overlap display shows the four layers contributing to AtCKN.
Use of Prior Knowledge to Improve the Plant Immune Signaling Model
To assess the potential of using prior knowledge for the improvement of the mechanistic model of plant immune signaling, we extracted a subnetwork of AtCKN with all components of PIS-v2. This subnetwork consisted of 391 connections between 212 nodes in the fully expanded version or 254 connections between 108 nodes at the level of component families. By comparing the PIS-v2 layer against the remaining layers of AtCKN (PPI, TR, and miRNA), we found that 45 connections were present in both subnetworks, 67 were present only in PIS-v2, and 142 novel reactions were found from the remaining AtCKN layers. These represent model upgrades, demonstrating the value of dispersed knowledge sources also for the construction of detailed mechanistic models. Inspecting these new connections showed that manual curation is more successful in knowledge extraction within a signaling module (Fig. 3B) than between signaling modules (Fig. 3A; Supplemental Data Set S2).
Figure 3.
Connections between gene families of three plant hormone immune signaling pathways (ET, JA, and SA) and other signaling modules. The contribution of the PIS-v2 layer in contrast to that of the remaining AtCKN layers (PPI, TR, and miRNA) is shown in terms of connections between signaling modules (i.e. cross talk connections; A) and connections within a signaling module (B). Solid blue lines represent novel connections present in PPI, TR, and miRNA layers of AtCKN; solid gray lines represent connections present only in the PIS-v2 layer; and dotted gray lines represent connections existing in both compared subsets.
Translation of Knowledge to Potato and Integration with Experimental Data
Based on predictions of orthologous relationships, we translated AtCKN from Arabidopsis to potato to form the StCKN. This resulted in an intermediary abstracted network with 9,679 nodes (9,497 ortholog groups, 168 miRNA families, three metabolites, and 11 viral proteins) and 43,393 connections. Next, we inferred all combinations between potato genes of the same ortholog group for each abstracted connection; this resulted in the StCKN (Fig. 1B) having 18,036 nodes (17,855 genes, 168 miRNA families, three metabolites, and 11 viral proteins) and 296,834 connections. This expansion is the result of a many-to-many relationship of orthologous genes between species. But it also includes cases where no homologous gene is found.
To identify transcriptional modules in potato that contribute to potato immune signaling in Potato virus Y infection, we selected data sets profiling temporal response dynamics in potato genotypes displaying either a tolerant or a hypersensitive response (108 samples). Out of 17,855 potato StCKN genes, 10,920 (61%) had a microarray probe assigned. We used the expression values of these genes to infer a targeted and nontargeted coexpression network and a gene regulatory network: the former, in order to propose genes controlled by the same transcriptional regulatory program; and the latter, to propose potential regulators (Fig. 1C).
Two types of subnetworks generated (mock inoculated and virus infected) allowed us to examine differences between gene connections. Subnetworks of mock-inoculated plants reflecting developmental cues were all of similar size (25,916, 25,910, and 30,570 connections for targeted, nontargeted coexpression, and gene regulatory network, respectively). The sizes the of subnetworks reflecting plant responses to viral infection were between 56% and 64% of the sizes of mock-inoculated subnetworks but, again, were similar to each other (16,716, 15,993, and 17,204 connections in the same order as above). The difference in connection count per method could be explained by a greater variability of the viral subset (tolerant, hypersensitive), which resulted in a smaller number of inferred connections. Conversely, as developmental profiles of all genotypes share greater similarities, the number of inferred connections was larger in the mock-inoculated subnetwork. Comparison of the predicted connections among all three approaches revealed that they are largely independent, as the three methods shared only 111 out of 116,391 total unique connections (Supplemental Fig. S1).
To elucidate perturbations in network topologies in plant immunity, we extracted differential networks by removing any connections shared between the mock-inoculated and virus-infected treatments. The differential networks were merged as new layers with StCKN into a StIN (Fig. 1D) with 402,277 connections between 19,801 nodes (19,619 genes, 168 miRNA families, three metabolites, and 11 viral proteins).
To validate our approach of StIN construction, we compared interactions covered by selected layers of biological information against a gold standard, a set of highly reliable reactions from the manually curated plant immune signaling model (Table 1). The PPI layer covered 50% of all reactions identified by manual literature curation in our PIS-v2 model. The TR layer had even greater concordance with the gold-standard reactions (80%). On the other hand, connections resulting from gene regulatory network inference covered only 20% of interactions in the gold standard. We must note, however, that some PIS-v2 model connections might be triggered in nonviral infections instead.
Table 1. StIN validation by comparing connection existence in selected data layers against the gold standard.
Connections are grouped by their reaction effect types (activation, binding, and inhibition) and their interaction types (B, protein binding; and T, transcriptional regulation). Compared layers include partial StCKN (PPI and TR) and two differential networks (merged targeted and nontargeted coexpression and gene regulatory). Coverage of a gold-standard reaction (on the component family level) is indicated by + (reaction present in the layer), +n (reaction present in the layer, but not in the gold standard), – (reaction not present in the layer), or n.a. (not a relevant comparison [e.g. transcriptional regulation cannot validate a protein-binding connection]). Coexpression results were not included in the validation, as they represent the coregulation of genes and not transcriptional regulation.
| Gold-Standard Connections | Validation of StIN | ||||||
|---|---|---|---|---|---|---|---|
| Group | Node 1 | Node 2 | Type | PPI | TR | Coexpression | Gene Regulatory |
| Activation | MPK | LOX | B | – | n.a. | n.a. | n.a. |
| MPK3 | EDS1/PAD4 | B | – | n.a. | + | n.a. | |
| MPK6 | EDS1/PAD4 | B | – | n.a. | n.a. | n.a. | |
| EIN2 | EIN2 | B | + | n.a. | n.a. | n.a. | |
| EIN2 | EIN3(like) | B | – | n.a. | n.a. | n.a. | |
| RTE1 | CTR | B | – | n.a. | n.a. | n.a. | |
| EDS1/PAD4 | EDS5 | B | – | n.a. | n.a. | n.a. | |
| EDS5 | ICS | B | – | n.a. | + | n.a. | |
| NPR | MOS | B | – | n.a. | n.a. | n.a. | |
| MYC | PR3 | T | n.a. | + | – | – | |
| MYC | PR4 | T | n.a. | + | – | – | |
| EIN3(like) | EBF | T | n.a. | + | + | + | |
| EIN3(like) | ERF/EDF | T | n.a. | – | – | – | |
| EIN3(like) | PR3 | T | n.a. | + | – | – | |
| EIN3(like) | PR4 | T | n.a. | + | – | – | |
| ERF/EDF | PDF1.2 | T | n.a. | – | – | – | |
| MYC | JAZ | T | n.a. | + | – | – | |
| MYC | LOX | T | n.a. | + | + | + | |
| TGA | PR1 | T | n.a. | + | – | – | |
| Protein complex formation | CTR | ETR | B | + | n.a. | n.a. | +n |
| COI1 | SCF | B | + | n.a. | n.a. | n.a. | |
| EBF | SCF | B | + | n.a. | n.a. | n.a. | |
| RBX | CUL | B | + | n.a. | n.a. | n.a. | |
| EDS1 | PAD4 | B | + | n.a. | n.a. | n.a. | |
| GSNO | NPR | B | – | n.a. | n.a. | n.a. | |
| NPR | TGA | B | + | n.a. | +n | +n | |
| Inhibition | JAZ | EIN3(like) | B | – | n.a. | n.a. | n.a. |
| EIN5 | EBF | B | – | n.a. | + | n.a. | |
| ETR/CTR | EIN2 | B | + | n.a. | n.a. | n.a. | |
| JAM | MYC | B | – | n.a. | n.a. | n.a. | |
| JAZ | MYC | B | + | n.a. | n.a. | n.a. | |
| COI1 | JAZ | B | + | n.a. | n.a. | n.a. | |
| EBF | EIN3(like) | B | + | n.a. | + | n.a. | |
| NIMIN | NPR | B | + | n.a. | n.a. | n.a. | |
| Confirmed reactions (%) | 50 | 80 | 20 | ||||
| Novel reactions (no.) | 0 | 2 | 2 | ||||
Integrated Network-Driven Hypotheses: Ethylene Modulates NPR1 Gene Expression
First, we analyzed the topologies of the generated networks, namely AtCKN, StCKN, and StIN. AtCKN showed some bias toward high-degree nodes, a direct result of two included data sets from chromatin immunoprecipitation sequencing experimental data (Supplemental Table S1). On the other hand, the expansion to all potato genes performed for StCKN and StIN distorted the network topological indices (many-to-many phylogenetic relationships). Further network analyses (Fig. 1E) aimed at targeted identification of novel cross talk connections between receptors and transmitters of seven plant hormonal pathways (Supplemental Table S2). Due to topology distortion in translated potato networks, we performed the initial search in AtCKN, afterward analyzing the connections in StCKN.
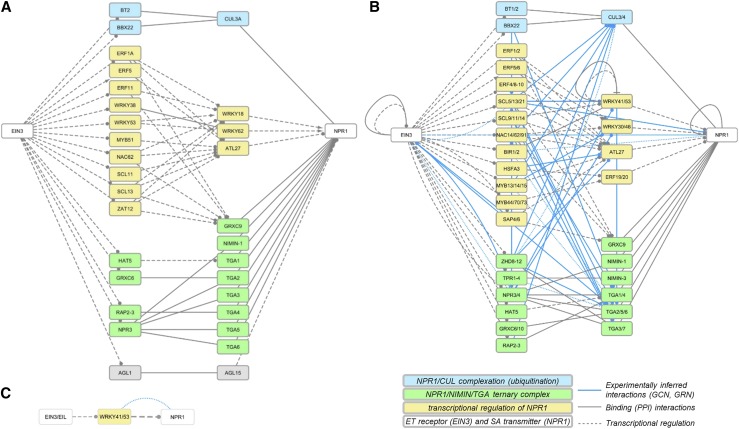
One of the most interesting findings was the shortest path from the ET pathway transmitter EIN3 to the SA receptor NPR1. In AtCKN, we identified several shortest paths of three-step length, involving 32 genes and 61 connections (Fig. 4A; Supplemental Data Set S3). One-third of these were binding (PPI) connections, and the remainder were TRs. In StCKN, we searched for walks (length of three steps) from EIN3 to NPR1 and then superimposed coexpression and gene regulatory network connections from StIN (Fig. 4B; Supplemental Data Set S3). The potato EIN3-to-NPR1 walk subnetwork included 32 genes, 57 StCKN, and 48 experimentally inferred connections. Searching for walks of a specific length was required to ease comparisons, as the shortest path between EIN3 and NPR1 after translation was of two-step length (Fig. 4C).
Figure 4.
Results of shortest paths or walks from EIN3 (ET) toward NPR1 (SA). A, Shortest path (three steps) in AtCKN. B, Walk of length three in StCKN. C, Shortest path (two steps) in StCKN. Line type and color indicate the interaction type: binding (solid gray), regulation by transcription factors (dashed gray), coexpression (dotted blue), and gene regulatory (solid blue). Target arrows indicate the action of the connection: activatory (arrowhead), inhibitory (T), unknown (circle), or undirected in the case of binding or coexpression (no arrow). Gene group identifiers corresponding to these images are given in Supplemental Data Set S3.
The majority of the binding-type connections in the potato EIN3-NPR1 walk can be attributed to the formation of a ternary complex between NPR1, NIMIN1, and TGA factors (Fig. 4, green), which, in turn, modulates PR1 gene expression (Weigel et al., 2005). The other set of binding-type connections relates to the complexation of NPR1 with cullin (Fig. 4, blue), which is important for plant immunity regulation (Spoel et al., 2009). The remaining shortest paths indicate potential TR of NPR1 through ET signaling (Fig. 4, yellow). As expected, superimposed connections from the transcriptomics data for potato also were denser in this area (Fig. 4B, blue connections). The first step of all shortest paths describing TR includes several ET-responsive factors (ERFs) and specific members of the C2C2, GRAS, NAC, MYB, and HSF families. These transcription factors then target two WRKY transcription factors, in particular WRKY18 and WRKY62, where the former was shown to have an important role in plant responses to bacterial and fungal pathogens (Xu et al., 2006; Chen et al., 2010), besides another ERF and an ATL.
Superimposing inferred connections based on transcriptional profiles of the potato response to viral infection, although incomplete, confirmed the potential regulation of NPR1 gene activity through several WRKY (WRKY30/46 and WRKY41/53), zinc-finger (SAP4/6), and MYB (MYB13/14/15 and MYB44/70/73) transcription factors (Fig. 4B). Interestingly, when searching in a reduced StIN with only transcriptional layer connections, we predicted the regulation of NPR1 through ERF (ERF2a), WRKY (WRKY34 and WRKY41/53), MYB (MYB18/19 and MYB52/54), and bHLH (bHLH84/84) transcription factors (Supplemental Fig. S2).
For further evaluation of these findings, we scanned Arabidopsis and potato promoters of the NPR1 gene for known cis-regulatory elements. Apart from containing known conserved motifs for light and development responses, both promoters contain motifs specific for several hormones (abscisic acid, gibberellic acid, JA, and SA; Supplemental Data Set S4). In terms of general stress responses, both promoter regions contain binding sites for responses to heat, drought, and defense. In addition, we detected a wounding motif, MYB, and WRKY-binding motifs in the potato promoter only.
Experimental Validation of TR of NPR1 by ET
To validate our network-generated hypothesis, we tested the TR of NPR1 following the induction of the ET pathway in potato to show the potential of such translation of knowledge to a crop. We additionally checked for the potential of the SA signaling module to participate in this process. Thus, we induced the SA signaling module by 2,6-dichloroisonicotinic acid (INA; a functional analog of SA that is not accessible to degradation by salicylate hydroxylase [NahG]) while either leaving the ET module active or blocking its activity (treatment with 1-methylcyclopropene [1-MCP]). Alternatively, we tested the regulatory potential of the ET module, while SA signaling was blocked by using transgenic plants expressing NahG, which degrade any internally produced SA (Fig. 5A). The induction of Aminocyclopropanecarboxylate Oxidase4 (ACO4) gene expression was used as a marker for the efficient activation of the ET signaling module and Pathogenesis-Related Protein1b (PR1b) as a marker of SA signaling module activation.
Figure 5.
Validation of the direct transcriptional regulation of NPR1 by the ET signaling module in potato leaves. A, Scheme of the underlying biological pathways of ET (yellow) and SA (blue), with interactions on the protein or transcriptional level (solid or dashed lines, respectively). Interactions can activate (arrowhead) or inhibit (circle) downstream signaling events. Blue lines denote treatments of the functional validation (ET, INA, and 1-MCP) or NahG plants (deficient in SA signaling). The red dashed arrow denotes our tested hypothesis. B and C, Plants belonging to potato cv. Rywal (B) and its transgenic line NahG-Rywal (C) were treated with ETs (yellow), INA (SA analog; blue), or a combination of INA with 1-MCP (ET inhibitor [INA+MCP]; gray). Log2 fold changes (logFC) in gene expression in treated and control plants are shown (*, P < 0.05 [n = 3], Student’s t test) for ACO4 (ET signaling marker), PR1b (SA signaling marker), and NPR1 (SA signal transmitter). D, Arabidopsis ecotype Columbia wild-type (wt) and ein3-1 mutant plants were treated with INA or INA+MCP, and log2 fold change in gene expression in treated and control plants for NPR1 (*, P < 0.05 [n = 6], Student’s t test) is shown. Error bars denote se of biological replicates. Note the different y axis scales for different genes. Results of the second independent potato experiment are provided in Supplemental Figure S3.
The significant up-regulation (P < 0.05, Student’s t test) of NPR1 gene expression after ET treatment substantiated our network-generated hypothesis in potato plants (Fig. 5B, ET treatment). Strong induction of the PR1b gene after ET treatment additionally confirmed the regulation of the SA signaling module by ET. Induction of PR1b by ET was even stronger than its expected induction by SA signaling (Fig. 5B, INA treatment). Tight interaction between both modules also was confirmed by ACO4 induction by both ET and INA treatment. When SA signaling was blocked (using NahG transgenic plants; Fig. 5C), the induction of NPR1 gene expression by ET was similar to that in nontransgenic plants, confirming that the string of events leading to activation is not dependent on SA. All other module cross talk observed in nontransgenic plants also was confirmed in the SA-depleted plants.
The direct role of EIN3 in this cross talk was evaluated using the Arabidopsis ET-insensitive mutant ein3-1. The results show that SA triggers the expression of the NPR1 gene but that this induction is blocked in the ein3-1 mutant and diminished if the activation of the ETR receptor is blocked by 1-MCP (Fig. 5D).
DISCUSSION
Plants have evolved a complex immune system to defend themselves against diverse pathogens and herbivores. This plant immune signaling network with its tightly interconnected signaling modules ensures a timely, precise, and effective response to attackers (Coolen et al., 2016). Many regulatory mechanisms are buffered by the network, rendering them undetectable by traditional genetic approaches of single-gene null-mutant analyses (Hillmer et al., 2017), thus making network modeling algorithms invaluable tools to expand our understanding of plant immunity (Windram and Denby, 2015).
An ideal network model would encompass all components of the biological system and all interactions between them. However, due to limited knowledge and data availability, this is not possible at present. To circumvent this problem, researchers studying plant immune signaling have adopted various approaches. Bottom-up approaches based on manual literature curation led to detailed and accurate models (Miljkovic et al., 2012; Naseem et al., 2012), but their extensiveness is limited. However, the majority of published research builds on networks inferred entirely from experimental data (Vermeirssen et al., 2014; Ebrahim et al., 2016) or a combination of network inference with prior knowledge (Sabaghian et al., 2015; Jiang et al., 2016), which can substantially simplify the computational burden of network inference (Windram and Denby, 2015).
In contrast to other efforts of prior knowledge integration (Dai et al., 2016), in our study, we based our knowledge network on the manually built, highly reliable model of plant immune signaling (Miljkovic et al., 2012) and complemented it with data from various publicly available databases or data sets for Arabidopsis that were published as supplements to articles. Therefore, our results represent the most current and comprehensive knowledge network of immune signaling and related processes in Arabidopsis (see “Data Availability” below). Compared with data for immune signaling in this model species, those for immune signaling in crop species are much sparser; therefore, the translation of knowledge is essential for crop resistance breeding. It has been shown that knowledge can be transferred across species based on orthology, with higher reliability transfer between related species, whereas translation from dicotyledons to monocotyledons is less predictive (Lee et al., 2015b). Molecular network rewiring leads to functional divergence and, thus, plays a central role in speciation (Chae et al., 2012). The speed of network rewiring depends on several factors, including the type of interaction, with transcriptional regulatory networks having one of the fastest rewire speeds (Shou et al., 2011). However, functional modules often experience evolutionary cohesiveness (Chae et al., 2012), which can be a basis for network translation, as in our case. Because of the sparsity of network data for potato, any new information from these evolutionarily conserved modules alone is extremely valuable. We translated the AtCKN to StCKN and subsequently inferred networks from time-resolved experimental data on the potato-virus interaction. To alleviate unknowns arising from speciation and/or evolution-related events and to include the dynamics of the potato response to Potato virus Y, we applied different methods to construct both coexpression and gene regulatory networks (Fig. 1). Such ensemble solutions have been shown to match or outperform single methods, particularly in revealing the true underlying network structure (Vermeirssen et al., 2014).
To assess the validity of our approach, we compared interactions covered by different layers of biological information against a gold standard (i.e. a set of reactions from the manually curated plant immune signaling model; Table 1). We show a coverage of 58% of known interactions in the newly built network. Biologically more relevant, our integrated network approach predicted 142 additional connections between components of the manually built PIS model, which shows the potential of our integrated network approach in generating new testable hypotheses about biological systems (Fig. 3).
As the comparison of the PIS model with other knowledge sources revealed low coverage of connections between signaling modules in the literature (Fig. 3; Amar and Shamir, 2014), we tested the power of our newly built model to identify cross talk connections. Intuitively, a regulatory pathway is unlikely to repeatedly pass a node. Given that shortest paths are a subset of simple paths in graph theory, they do not contain any repeated nodes and, hence, could represent the most optimal explanation for interdependence between two analyzed nodes (Shih and Parthasarathy, 2012). Walks of specific length become of use in the case of between-species translations, which also was the case in our potato network searches. We have indeed discovered an interesting novel TR connection between ET and SA signaling modules, where EIN3 regulates the transcription of the NPR1 gene (Fig. 4). Furthermore, we confirmed this predicted mechanism of cross talk by performing both in silico promoter analysis (Supplemental Data Set S4) and a set of experiments in potato and Arabidopsis (Fig. 5; Supplemental Fig. S3), showing that short paths and walks from network analysis allow for the discovery of the underlying signaling pathways.
Evidence on the importance of ET in plant immune signaling is emerging from several perspectives (Broekgaarden et al., 2015). For example, ET biosynthesis was found to be crucial for the induction of programmed cell death during the interaction of Nicotiana umbratica with Alternaria alternata (Mase et al., 2012) and Pseudomonas syringae-triggered Arabidopsis susceptibility to herbivory (Groen et al., 2013). Several experiments have shown that SA can modulate ET signaling (Van der Does et al., 2013; Zander et al., 2014; Guan et al., 2015; Caarls et al., 2017). This regulation, in most cases, is implicated in the context of JA/SA antagonism (Robert-Seilaniantz et al., 2011; Derksen et al., 2013; Caarls et al., 2015). Only a few studies indicate that ET might be an important regulator of SA signaling. It was shown that EIN3 transcription factors directly target the promoter of ICS2, negatively regulating SA biosynthesis (Chen et al., 2009). On the other hand, Frye et al. (2001), Mikkelsen et al. (2003), and Leon-Reyes et al. (2009) have shown ET potentiation of PR1 gene expression in Arabidopsis. We observed the same effects in our potato experiments (Fig. 5). Chromatin immunoprecipitation sequencing of EIN3 targets revealed NPR3 promoter as its direct target (Chang et al., 2013), providing further evidence that the two modules are connected.
Most studies involving NPR1 as a master regulator of SA signaling have focused on its posttranslational modifications or loss-of-function effects. The ET modulation of NPR1’s role in JA/SA antagonism was studied by performing a series of experiments with the npr1 mutant, which did not allow identification of the effects of transcriptional, translational, and posttranslational regulation (Leon-Reyes et al., 2009). Further studies showed the importance of the proteasome-mediated degradation of NPR1 (Fu et al., 2012; Saleh et al., 2015; Ding et al., 2016) and nuclear import (Fu et al., 2012; Kovacs et al., 2015; Lee et al., 2015a) for effective SA perception. To our knowledge, no study performed so far has focused on the regulation of NPR1 gene expression.
Detailed inspection of our integrated network shows several potential transcription regulation paths from EIN3 to NPR1 (Fig. 4B), which involve a cascade of one or two transcription factors. Some of the transcription factors are well characterized (ERF and WRKY), while some were not investigated in detail (ATL27 or bHLH) or at least not in relation to immune signaling (MYB). As our in silico analyses of the NPR1 promoter identified WRKY and MYB transcription factor-binding motifs, these are the most likely candidates for signal transduction. Considering different experiments, including ours, we reason that, apart from the regulation of NPR1 activity on the protein level, transcriptional regulation of the NPR1 gene also contributes to immune signaling in plants.
CONCLUSION
We conclude that the integration of prior knowledge and experimental data sets followed by network modeling is useful for hypothesis generation, as suggested previously (Medeiros et al., 2015). Network analysis results thus help us to understand the complex interactions and the information flow between a causal and affected gene within a system of interest. However, one must note that, while network analysis is useful in helping us understand the organization and information-processing capabilities of the system, its results are still a static view of the system (Chae et al., 2012). Additionally, connectivity between components does not automatically imply that signals are propagated through them. In order to understand how this organization enables differential responses based on particular triggers, changes in time and space after receiving the stimuli must be observed. Thus, to understand all emerging properties of immune signaling, network analysis should be combined with dynamic modeling.
MATERIALS AND METHODS
Network-Based Knowledge Integration
First, a previously inferred plant immune signaling model (Miljkovic et al., 2012) was upgraded by adding manually curated reactions from the recent literature (forming PIS-v2). Furthermore, we transformed the model reactions to a graph of binary interactions, forming the first knowledge layer. Next, we retrieved additional binary connections from different public resources representing additional knowledge layers (for full description, see Supplemental Table S1): PPIs from databases AtPIN and STRING-v10, two yeast two-hybrid experiments, and three experiments on plant-pathogen interactions; TR from atRegNet and ATRM, two chromatin immunoprecipitation sequencing experiments, and one predicted data set; and regulation through miRNA from miRTarBase, PMRD, and PNRD.
These prior knowledge layers were integrated into the Arabidopsis (Arabidopsis thaliana) AtCKN (Fig. 1A). Reliability ranks were assigned to each connection between two nodes in AtCKN (Supplemental Table S1). To translate the network from Arabidopsis to potato (Solanum tuberosum), a union of three ortholog clusterings was used (available in the GoMapMan database as OCD_all [www.gomapman.org/exports/]; Ramšak et al., 2014). Only connections where both Arabidopsis nodes had a defined ortholog in potato were kept in the StCKN (Fig. 1B).
Network Inference from Experimental Data Sets
Two microarray data sets profiling temporal response dynamics in potato genotypes with a tolerant (GEO:GSE58593; Stare et al., 2015) or hypersensitive (GEO:GSE46180; Baebler et al., 2014) response to viral infection were used. Microarray features (microarray probes) were translated to potato gene models (Ramšak et al., 2014). For potato genes covered by several microarray features, one was selected as a representative microarray feature based on the maximum number of differentially expressed time points per feature between mock-inoculated and virus-infected samples (false discovery rate-corrected P < 0.05). When several such features were present, those with the highest log fold change and the highest average expression across time points were prioritized.
Three network inference methods were applied to the gene expression data. Nontargeted coexpression networks were inferred with BioLayout (Theocharidis et al., 2009), afterward running the Markov clustering algorithm (Van Dongen, 2008) to divide the graph into discrete subsets. Pearson correlation coefficients (PCCs) were calculated on the gene expression of representative microarray features for 156 PIS-v2 potato genes and their 2,548 first neighbors in StCKN (PCC ≥ 0.98, top 1 percentile). Targeted coexpression networks were inferred with CoExpNetViz (Tzfadia et al., 2016), calculating coexpression values using mutual information and PCCs (percentiles set between 1 and 99). As bait, 156 potato genes of PIS-v2 were used against all 17,171 representative microarray features. Inference with GENIE3 (Huynh-Thu et al., 2010) was performed on the same subset of microarray features as for nontargeted coexpression network inference (weight ≤ 6 × 10−3, top 1 percentile). Thresholds for BioLayout and GENIE3 were determined empirically, so that the resulting networks followed the scale-free and small-world properties of complex networks. Each method was used to generate a mock-inoculated and a virus-infected subnetwork from data for 16 biological samples each (Fig. 1C). By removing any connections shared between networks from both treatments (mock inoculated and virus infected), a differential network was created for each inference type. Finally, binary interactions in StCKN and both differential networks were merged to create the StIN.
Validation of the Network Construction Approach
To assess and estimate the importance and contributions of various knowledge layers in StIN, a gold standard (set of reliable connections) was constructed from manually curated PIS-v2. Genes were grouped into so-called component families (for representation levels, see Miljkovic et al., 2012), and connections were compared at this level of abstraction. We kept 37 reactions, where all components had both an Arabidopsis and a potato ortholog. StCKN prior knowledge layers (PPI, TR, and miRNA) and differential networks were then compared against the gold standard.
Network Analyses
NetworkAnalyzer (Doncheva et al., 2012) was applied to calculate graph indices, and MCODE (Bader and Hogue, 2003) was used to search for highly interconnected subgraphs in the constructed networks. Pajek (Batagelj and Mrvar, 1998) was applied to search for shortest paths and walks between all combinations of 14 manually selected genes known to be involved in plant signaling (specifically, receptors and transmitters for seven plant hormones; Supplemental Table S2). For all network visualizations, Cytoscape (Shannon et al., 2003) was used.
In Silico Promoter Analyses
Sequences 1,500 nucleotides upstream of the NPR1 gene translation start site were extracted for Arabidopsis (Araport 11) and potato (SpudDB) and scanned for known cis-regulatory elements with TRANSFAC (Matys et al., 2003) and PlantCARE (Lescot et al., 2002).
Plant Growth and Treatments
The potato cv Rywal and its transgenic line NahG-Rywal, which is deficient in SA accumulation (by expressing SA hydroxylase; Baebler et al., 2014), were cultivated as described previously (Baebler et al., 2009). Arabidopsis ecotype Columbia wild-type and mutant ein3-1 plants, with reduced responsiveness to ET (Chao et al., 1997; TAIR germplasm identifier CS8052), were grown in soil under long-day conditions as described for potato. Treatments were performed on 4-week-old plants. For SA treatments, plants were sprayed with 300 µm INA (Aldrich) dissolved in ethanol; control plants were sprayed with 1% (v/v) ethanol solution. For ET treatments, plants, sealed in air-tight clear plastic containers, were treated with 50 µL L−1 ET (Messer); control plants were sealed in identical containers without ET. To inhibit the ET signaling pathway, plants were first treated with SmartFresh (containing 0.14% [w/w] 1-MCP [AgroFresh]) according to the manufacturer’s protocol; this was followed by INA and 1-MCP treatment after 2 h. Plant leaves were sampled 24 h after treatment and immediately frozen in liquid nitrogen (three to six plants per treatment and genotype). The experiment was performed twice for potato and once for Arabidopsis.
Gene Expression Analysis
Leaf samples (∼100 mg) were homogenized with the FastPrep Instrument (MP Biomedicals). Total RNA extraction, DNase treatment, RNA quality control, and reverse transcription were performed as described previously (Baebler et al., 2009). For potato, expression was measured using high-throughput quantitative PCR for NPR1, PR1b, and ACO4 genes. The Cytochrome Oxidase (COX) and Elongation Factor1 genes were used as endogenous controls. TATAA PreAmp GrandMaster Mix (TATAA Biocenter) was used for cDNA preamplification (two dilutions per sample) according to the manufacturer’s specifications. Gene expression analysis of the samples was conducted in Fluidigm BioMark HD System Real-Time PCR (Fluidigm) using 48.48 Dynamic Arrays IFC. The sample reaction mix contained preamplified sample DNA (10-fold diluted), DNA Sample Loading Reagent (Fluidigm), and FastStart Universal Probe Master (Rox; Roche). The assay reaction mix included the Assay Loading Reagent (Fluidigm) and a mix of 2.5 µm TaqMan probe and 9 µm forward and reverse primers. IFC Controller (Fluidigm) was used to prime and load the IFC according to the manufacturer’s protocol and under standard PCR conditions. A second independent potato experiment was performed, and the expression of the same genes was analyzed using the QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific). Reactions were set as described before (Baebler et al., 2014). For Arabidopsis samples, the expression of NPR1 was analyzed and normalized to the expression of COX as described above for the second potato experiment. Detailed information on all quantitative PCR assays performed is presented in Supplemental Table S3. For relative gene expression quantification using a standard curve, quantGenius (Baebler et al., 2017; http://quantgenius.nib.si) was used. To determine differences in gene expression between treated and control sample groups, Student’s t test was performed.
Data Availability
Microarray transcriptomics data are available from the Gene Expression Omnibus (GSE58593 and GSE46180). The AtCKN network is available from NDEx (Pratt et al., 2015; http://www.ndexbio.org/#/) with the uuid 67507c30-995f-11e7-a10d-0ac135e8bacf.
Accession Numbers
Sequence data from this article can be found in the National Center for Biotechnology Information gene data library under gene identifiers 842733 (AT1G64280, AtNPR1), XM_006357647 (Sotub07g011600.1.1, StNPR1), and NM_001288166 (Sotub09g006090.1.1, StPR1b).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of predicted connections for three selected network inference algorithms.
Supplemental Figure S2. Shortest path search from EIN3 to NPR1 in StIN.
Supplemental Figure S3. Results of the replicated experiment for the validation of direct TR of NPR1 by the ET signaling module in potato leaves.
Supplemental Table S1. Contribution of four knowledge layers to the built AtCKN.
Supplemental Table S2. List of selected plant hormone pathway receptors and transmitters.
Supplemental Table S3. Primers and probes used for functional validation in Arabidopsis and potato and their properties according to MIQE guidelines.
Supplemental Data Set S1. PIS-v2.
Supplemental Data Set S2. Comparison of contributions in the PIS model and AtCKN subnetwork.
Supplemental Data Set S3. Gene connections for network analysis results between EIN3 and NPR1.
Supplemental Data Set S4. Results of in silico regulatory element search for AtNPR1 and StNPR1.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Katja Stare and Lidija Matičič for excellent technical assistance.
Footnotes
This work was supported by grants from the Slovenian Research Agency (P4-0165, J4-7636, J7-7303, and N4-0026).
Articles can be viewed without a subscription.
References
- Amar D, Shamir R (2014) Constructing module maps for integrated analysis of heterogeneous biological networks. Nucleic Acids Res 42: 4208–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CWV (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler Š, Krečič-Stres H, Rotter A, Kogovšek P, Cankar K, Kok EJ, Gruden K, Kovač M, Žel J, Pompe-Novak M, et al. (2009) PVY(NTN) elicits a diverse gene expression response in different potato genotypes in the first 12 h after inoculation. Mol Plant Pathol 10: 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler Š, Witek K, Petek M, Stare K, Tušek-Žnidarič M, Pompe-Novak M, Renaut J, Szajko K, Strzelczyk-Żyta D, Marczewski W, et al. (2014) Salicylic acid is an indispensable component of the Ny-1 resistance-gene-mediated response against potato virus Y infection in potato. J Exp Bot 65: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baebler Š, Svalina M, Petek M, Stare K, Rotter A, Pompe-Novak M, Gruden K (2017) quantGenius: implementation of a decision support system for qPCR-based gene quantification. BMC Bioinformatics 18: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási AL. (2009) Scale-free networks: a decade and beyond. Science 325: 412–413 [DOI] [PubMed] [Google Scholar]
- Batagelj V, Mrvar A (1998) Pajek: program for large network analysis. Connections 21: 47–57 [Google Scholar]
- Blüthgen N. (2015) Signaling output: it’s all about timing and feedbacks. Mol Syst Biol 11: 843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekgaarden C, Caarls L, Vos IA, Pieterse CMJ, Van Wees SCM (2015) Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol 169: 2371–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls L, Pieterse CMJ, Van Wees SCM (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci 6: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls L, Van der Does D, Hickman R, Jansen W, Verk MC, Proietti S, Lorenzo O, Solano R, Pieterse CMJ, Van Wees SCM (2017) Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol 58: 266–278 [DOI] [PubMed] [Google Scholar]
- Chae L, Lee I, Shin J, Rhee SY (2012) Towards understanding how molecular networks evolve in plants. Curr Opin Plant Biol 15: 177–184 [DOI] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen S, Proietti S, Hickman R, Davila Olivas NH, Huang PP, Van Verk MC, Van Pelt JA, Wittenberg AH, De Vos M, Prins M, et al. (2016) Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant J 86: 249–267 [DOI] [PubMed] [Google Scholar]
- Dai X, Li J, Liu T, Zhao PX (2016) HRGRN: a graph search-empowered integrative database of Arabidopsis signaling transduction, metabolism and gene regulation networks. Plant Cell Physiol 57: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen H, Rampitsch C, Daayf F (2013) Signaling cross-talk in plant disease resistance. Plant Sci 207: 79–87 [DOI] [PubMed] [Google Scholar]
- Ding Y, Dommel M, Mou Z (2016) Abscisic acid promotes proteasome-mediated degradation of the transcription coactivator NPR1 in Arabidopsis thaliana. Plant J 86: 20–34 [DOI] [PubMed] [Google Scholar]
- Doncheva NT, Assenov Y, Domingues FS, Albrecht M (2012) Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc 7: 670–685 [DOI] [PubMed] [Google Scholar]
- Ebrahim A, Brunk E, Tan J, O’Brien EJ, Kim D, Szubin R, Lerman JA, Lechner A, Sastry A, Bordbar A, et al. (2016) Multi-omic data integration enables discovery of hidden biological regularities. Nat Commun 7: 13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye CA, Tang D, Innes RW (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc Natl Acad Sci USA 98: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari M, Lasserre J, Vingron M (2015) Reconstruction of gene networks using prior knowledge. BMC Syst Biol 9: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen SC, Whiteman NK, Bahrami AK, Wilczek AM, Cui J, Russell JA, Cibrian-Jaramillo A, Butler IA, Rana JD, Huang GH, et al. (2013) Pathogen-triggered ethylene signaling mediates systemic-induced susceptibility to herbivory in Arabidopsis. Plant Cell 25: 4755–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Su J, Meng X, Li S, Liu Y, Xu J, Zhang S (2015) Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiol 169: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer RA, Katagiri F (2016) Toward predictive modeling of large and complex biological signaling networks. Physiol Mol Plant Pathol 95: 77–83 [Google Scholar]
- Hillmer RA, Tsuda K, Rallapalli G, Asai S, Truman W, Papke MD, Sakakibara H, Jones JDG, Myers CL, Katagiri F (2017) The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet 13: e1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P (2010) Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Dong X, Zhang Z (2016) Network-based comparative analysis of Arabidopsis immune responses to Golovinomyces orontii and Botrytis cinerea infections. Sci Rep 6: 19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kovacs I, Durner J, Lindermayr C (2015) Crosstalk between nitric oxide and glutathione is required for NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1)-dependent defense signaling in Arabidopsis thaliana. New Phytol 208: 860–872 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Park YJ, Seo PJ, Kim JH, Sim HJ, Kim SG, Park CM (2015a) Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell 27: 3425–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Kim H, Lee I (2015b) Network-assisted crop systems genetics: network inference and integrative analysis. Curr Opin Plant Biol 24: 61–70 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A, Spoel SH, De Lange ES, Abe H, Kobayashi M, Tsuda S, Millenaar FF, Welschen RAM, Ritsema T, Pieterse CMJ (2009) Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mase K, Mizuno T, Ishihama N, Fujii T, Mori H, Kodama M, Yoshioka H (2012) Ethylene signaling pathway and MAPK cascades are required for AAL toxin-induced programmed cell death. Mol Plant Microbe Interact 25: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack ME, Lopez JA, Crocker TH, Mukhtar MS (2016) Making the right connections: network biology and plant immune system dynamics. Curr Plant Biol 5: 2–12 [Google Scholar]
- Medeiros DB, Daloso DM, Fernie AR, Nikoloski Z, Araújo WL (2015) Utilizing systems biology to unravel stomatal function and the hierarchies underpinning its control. Plant Cell Environ 38: 1457–1470 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miljkovic D, Stare T, Mozetič I, Podpečan V, Petek M, Witek K, Dermastia M, Lavrač N, Gruden K (2012) Signalling network construction for modelling plant defence response. PLoS ONE 7: e51822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem M, Philippi N, Hussain A, Wangorsch G, Ahmed N, Dandekar T (2012) Integrated systems view on networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. Plant Cell 24: 1793–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pratt D, Chen J, Welker D, Rivas R, Pillich R, Rynkov V, Ono K, Miello C, Hicks L, Szalma S, et al. (2015) NDEx, the Network Data Exchange. Cell Syst 1: 302–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramšak Ž, Baebler Š, Rotter A, Korbar M, Mozetič I, Usadel B, Gruden K (2014) GoMapMan: integration, consolidation and visualization of plant gene annotations within the MapMan ontology. Nucleic Acids Res 42: D1167–D1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Sabaghian E, Drebert Z, Inzé D, Saeys Y (2015) An integrated network of Arabidopsis growth regulators and its use for gene prioritization. Sci Rep 5: 17617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Withers J, Mohan R, Marqués J, Gu Y, Yan S, Zavaliev R, Nomoto M, Tada Y, Dong X (2015) Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih YK, Parthasarathy S (2012) A single source k-shortest paths algorithm to infer regulatory pathways in a gene network. Bioinformatics 28: i49–i58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou C, Bhardwaj N, Lam HYK, Yan KK, Kim PM, Snyder M, Gerstein MB (2011) Measuring the evolutionary rewiring of biological networks. PLOS Comput Biol 7: e1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh HP. (2008) Policies and strategies conducive to potato development in Asia and the Pacific region. In Papademetriou MK, ed, Workshop to Commemorate the International Year of Potato-2008. Food and Agriculture Organization of the United Nations, Rome, pp 11–17 [Google Scholar]
- Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stare T, Ramšak Ž, Blejec A, Stare K, Turnšek N, Weckwerth W, Wienkoop S, Vodnik D, Gruden K (2015) Bimodal dynamics of primary metabolism-related responses in tolerant potato-potato virus Y interaction. BMC Genomics 16: 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharidis A, van Dongen S, Enright AJ, Freeman TC (2009) Network visualization and analysis of gene expression data using BioLayout Express(3D). Nat Protoc 4: 1535–1550 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Somssich IE (2015) Transcriptional networks in plant immunity. New Phytol 206: 932–947 [DOI] [PubMed] [Google Scholar]
- Tzfadia O, Diels T, De Meyer S, Vandepoele K, Aharoni A, Van de Peer Y (2016) CoExpNetViz: comparative co-expression networks construction and visualization tool. Front Plant Sci 6: 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T, et al. (2013) Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25: 744–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen S. (2008) Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl 30: 121–141 [Google Scholar]
- Veiga DFT, Dutta B, Balázsi G (2010) Network inference and network response identification: moving genome-scale data to the next level of biological discovery. Mol Biosyst 6: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V, Ravindran P, Kumar PP (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, De Clercq I, Van Parys T, Van Breusegem F, Van de Peer Y (2014) Arabidopsis ensemble reverse-engineered gene regulatory network discloses interconnected transcription factors in oxidative stress. Plant Cell 26: 4656–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel RR, Pfitzner UM, Gatz C (2005) Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram O, Denby KJ (2015) Modelling signaling networks underlying plant defence. Curr Opin Plant Biol 27: 165–171 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, Thurow C, Gatz C (2014) TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol 165: 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Microarray transcriptomics data are available from the Gene Expression Omnibus (GSE58593 and GSE46180). The AtCKN network is available from NDEx (Pratt et al., 2015; http://www.ndexbio.org/#/) with the uuid 67507c30-995f-11e7-a10d-0ac135e8bacf.