RHA2b promotes the degradation of MYB30 via the 26S proteasome, and sumoylation enhances the stability of MYB30 by occupying the same locus.
Abstract
The phytohormone abscisic acid (ABA) is critical for plants encountering abiotic stress. We reported previously that the Arabidopsis (Arabidopsis thaliana) transcription factor MYB30 participates in ABA responses via SUMO ligase SAP-MIZ Domain-Containing SIZ1-mediated sumoylation. Here, we show that the RING-type ubiquitin E3 ligase RHA2b, which positively regulates ABA signaling, interacted with and ubiquitinated MYB30 to modulate MYB30 stability through the 26S proteasome pathway. The degradation rate of MYB30 was repressed significantly in the rha2b-1 mutant. Phenotypic analyses showed that MYB30 acts genetically downstream of RHA2b in ABA signaling. Substitutions of lysine-283 (K283) and K165 blocked ubiquitination, suggesting that these residues are sites of ubiquitination. K283 residue substitution significantly inhibited the degradation of MYB30 induced by ABA. The K165 site functioned additively with K283 in ABA-induced MYB30 degradation and ABA responses. At the same time, sumoylation protected MYB30 from degradation under cycloheximide and ABA treatment. Compared with MYB30, overexpression of MYB30-SUMO1 partially recovered the ABA sensitivity of siz1-2. But MYB30-SUMO1 exhibited similar localization with MYB30 in nuclei. Overall, our results suggest that RHA2b targets MYB30 for degradation to modulate ABA signaling. Considering that the K283 residue also is the major site for sumoylation, we propose that sumoylation and ubiquitination act antagonistically in the ABA response to regulate the stability of MYB30 by occupying the same residue.
The phytohormone abscisic acid (ABA) plays important roles in plant growth and development processes, such as seed germination, cotyledon greening, and seedling growth. ABA also is involved in regulating plant adaptations to various environmental challenges, including drought, high salinity, extreme temperature, and other abiotic stresses (Leung and Giraudat, 1998; Finkelstein et al., 2002; Nakashima et al., 2009b). For example, during water deficit conditions, synthesized ABA induces stomatal closure via the efflux of K+ and anions from guard cells, leading to a decrease in the water loss rate through transpiration (Schroeder et al., 2001; Pandey et al., 2007). This ABA-controlled process is vital for plant survival, and ABA-deficient and ABA-responsive mutants are susceptible to water stress (Kang et al., 2002).
With the identification of ABA receptors, ABA signal transduction has become relatively well understood (Raghavendra et al., 2010; Umezawa et al., 2010; Guo et al., 2011). Under normal conditions, Protein Phosphatase 2Cs (PP2Cs) interact with SNF1-Related Protein Kinases (SnRKs), keeping them in an inactive state via dephosphorylation. Under stress conditions, ABA is perceived by Pyrabactin Resistance (PYR)/PYR1-Like (PYL) regulatory components of ABA receptors. ABA receptors undergo conformational changes after binding to ABA that facilitate their interaction with PP2Cs. This interaction leads to the inhibition of PP2C dephosphorylation activity and the removal of PP2C-mediated SnRK inhibition (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009). Subsequently, SnRKs phosphorylate downstream ion channels (Geiger et al., 2009; Lee et al., 2009) and transcription factors (Fujii et al., 2009; Sato et al., 2009), such as ABA-Responsive Elements-Binding Factors (AREBs/ABFs), to trigger the ABA response.
The transcriptional activity and protein stability of the basic leucine zipper (bZIP) transcription factor ABA Insensitive5 (ABI5; Finkelstein and Lynch, 2000), which binds to ABA-responsive elements, are regulated by the phosphorylation of SnRK2s (Nakashima et al., 2009a). ABI5 participates in ABA signaling by regulating the expression of downstream target genes, such as Early Methionine-Labeled1 (EM1) and EM6 and Polygalacturonase Inhibiting Protein1 (PGIP1) and PGIP2, which are associated with the germination process (Finkelstein and Lynch, 2000; Kanai et al., 2010). In addition to ABI5, MYB transcription factors that contain the MYB DNA-binding domain also play an important role in the ABA response. The MYB family is characterized by the MYB DNA-binding domain. Several R2R3-type MYB transcription factors, including MYB20, MYB30, and MYB96, are involved in ABA and a variety of stress responses, such as osmotic, drought, and salt tolerance responses (Seo et al., 2009; Zheng et al., 2012; Cui et al., 2013). For example, Arabidopsis (Arabidopsis thaliana) mutants lacking MYB30 are hypersensitive to ABA during germination and seedling growth, while the overexpression of MYB30 in wild-type seedlings results in an ABA-insensitive phenotype (Zheng et al., 2012).
The activity of transcription factors is regulated by many posttranslational modifications. The ubiquitin/26S proteasome pathway, as the dominant selective protein turnover system, plays a very important role in ABA signaling. Three enzymes, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3), act sequentially to catalyze the covalent addition of ubiquitin to the target protein, and E3 is the key factor that defines substrate specificity (Vierstra, 2009; Sadanandom et al., 2012). Among the more than 1,400 different E3s in Arabidopsis, ∼470 proteins belong to the Really Interesting New Gene (RING)-type E3 protein family (Stone et al., 2005; Lee and Kim, 2011). Remarkably, a significant number of RING-type E3 enzymes are responsible for ABA signal transduction. For example, the RING-type E3 ligase RING Finger of Seed Longevity1 (RSL1) interacts with the ABA receptors Pyrabactin Resistance1 (PYR1) and PYR1-Like4 (PYL4) at the plasma membrane to promote PYR1 and PYL4 degradation through ubiquitination (Bueso et al., 2014). The RING-type E3 ligase ABI3-Interacting Protein2 (AFP2) negatively regulates ABA signaling by targeting the B3 domain transcription factor ABI3 for protein degradation (Zhang et al., 2005). ABI5 and ABF1/3 also are proteolytically degraded by the RING finger protein Keep On Going (Stone et al., 2006; Liu and Stone, 2010). The RING finger ubiquitin E3 ligase Salt and Drought Induced RING Finger1 (SDIR1) serves as a positive regulator of ABA signaling in seed germination, stomatal closure, and drought tolerance by promoting the degradation of SDIR1-Interacting Protein1 (Zhang et al., 2007, 2015). The RING-H2 E3 ligases RING-H2 Finger Protein A2a (RHA2a) and RHA2b also regulate ABA-dependent seed germination, early seedling development, and drought tolerance (Bu et al., 2009; Li et al., 2011). The RING-type E3 ligase MYB30-Interacting E3 Ligase1 (MIEL1) regulates ABA sensitivity by promoting MYB96 turnover in Arabidopsis (Marino et al., 2013; Lee and Seo, 2016).
In addition to ubiquitination, sumoylation also participates in ABA signaling by affecting the subcellular localization, transcriptional activity, and stability of proteins. The mechanism of sumoylation is similar to that of ubiquitination; the covalent addition of small ubiquitin-like modifier (SUMO) proteins to the target protein is catalyzed primarily by four SUMO E3 ligases, SIZ1, Methyl Methanesulfonate-Sensitive21 (MMS21/HPY2), Protein Inhibitor of Activated Stat Like1 (PIAL1), and PIAL2, in Arabidopsis (Miura et al., 2007a; Tomanov et al., 2014; Han et al., 2016; Liu et al., 2016; Zhang et al., 2017). The siz1 and mms21 mutants are hypersensitive to ABA (Miura et al., 2009; Zhang et al., 2013). Previous studies have shown that SIZ1 participates in ABA signaling by protecting target proteins from degradation through sumoylation. For example, the sumoylation of ABI5 and MYB30 is important for their functions in the response to ABA. In siz1-2 mutant seedlings, ABI5 and MYB30 are unstable under ABA treatment. When the major sumoylation sites (i.e. Lys-391 [K391] in ABI5 and K283 in MYB30) are mutated, the expression of ABI5 or MYB30 cannot rescue the ABA phenotype of the abi5 or myb30 mutant, respectively (Miura et al., 2009; Zheng et al., 2012).
In this study, we found that the RING-type E3 ligase RHA2b, which was reported previously as a positive regulator of the ABA response, interacts with MYB30 and promotes its degradation through ubiquitination. MYB30 acts genetically downstream of RHA2b in ABA signaling. The K283 and K165 residues were identified as the critical sites for protein stability and the function of MYB30 in the ABA response. Moreover, sumoylation protected MYB30 from degradation after cycloheximide (CHX) and ABA treatment, and the stability conferred by sumoylation plays an important role in MYB30-mediated ABA signaling. Overall, our results suggest that RHA2b-mediated ubiquitination and sumoylation act antagonistically in the regulation of MYB30 protein stability to function in the ABA response.
RESULTS
The Ubiquitin E3 Ligase RHA2b Interacts with MYB30 in Vivo
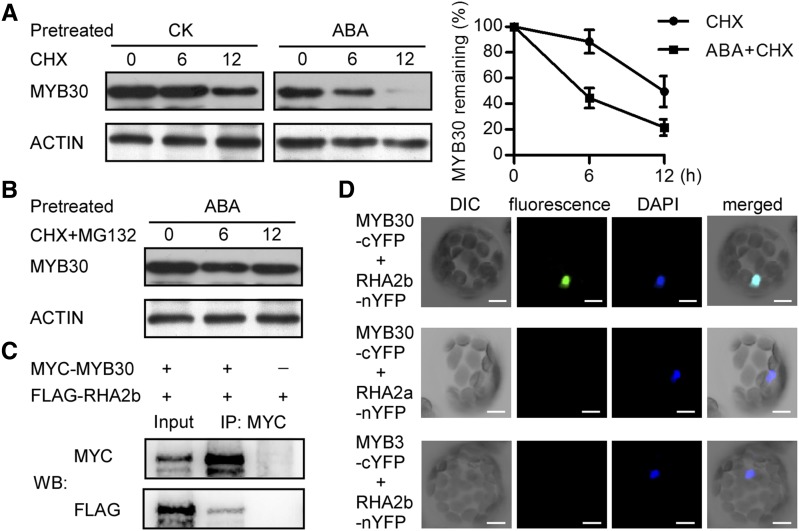
As a transcription factor, MYB30 plays a very important role in ABA signaling by regulating the expression of downstream genes (Zheng et al., 2012). Although we have known that sumoylation is important for the stability of MYB30 during ABA treatment (Zheng et al., 2012), it is still unclear how the stability of MYB30 is regulated precisely in ABA signaling. We first checked the stability of MYB30 with or without ABA treatment in the presence of CHX, which can block de novo protein synthesis. Figure 1A shows that ∼88% and ∼50% MYB30 remained at 6 and 12 h under the CHX treatment, and ABA accelerated the rate approximately 2-fold (∼55% loss at 6 h and ∼78% loss at 12 h). The ABA-induced degradation of MYB30 was repressed by the 26S proteasome inhibitor MG132 (Fig. 1B). These results suggest that the function of MYB30 in ABA signaling can be regulated at the protein level via the 26S proteasome.
Figure 1.
The ubiquitin E3 ligase RHA2b interacts with MYB30 in vivo. A, ABA induced the degradation of MYB30. Eight-day-old Col-0/35S:FLAG-MYB30 transgenic seedlings were treated without or with 100 μm ABA for 24 h and then incubated in the same medium with 100 μm CHX. Samples were collected 0, 6, and 12 h after treatment. MYB30 protein was detected with anti-FLAG antibody, and ACTIN was used as a loading control. The protein level compared with ACTIN at the initial time 0 was identified as 100%. Data represent mean values of three independent experiments ± sd. B, ABA-induced degradation of MYB30 was mediated by the 26S proteasome. Eight-day-old Col-0/35S:FLAG-MYB30 transgenic seedlings were treated with 100 μm ABA for 24 h and then incubated in the same medium with 100 μm CHX and 75 μm MG132. C, Coimmunoprecipitation (Co-IP) assays between MYB30 and RHA2b. MYC-MYB30 and FLAG-RHA2b were transiently expressed in Arabidopsis protoplasts. Soluble extracts from the protoplasts were detected directly (Input) or immunoprecipitated with an anti-MYC antibody (IP: MYC) and detected with anti-MYC and anti-FLAG antibodies (WB: MYC or FLAG). D, Split-YFP complementation imaging assays in Arabidopsis protoplasts. MYB30-cYFP and RHA2b-nYFP or RHA2a-nYFP were coexpressed in Arabidopsis protoplasts. MYB3 was used as a negative control. 4′,6-Diamino-phenylindole (DAPI) was used to stain the nuclei. DIC, Differential interference contrast. Bars = 50 μm.
As MYB30 negatively regulates ABA signaling, we inferred that a ubiquitin E3 ligase may serve as a positive regulator of the ABA response by mediating the degradation of MYB30. RHA2b, which is a RING-H2 ubiquitin E3 ligase that exhibits E3 ligase activity and positively regulates ABA signaling during seed germination and postgerminative growth (Li et al., 2011), was identified to interact with MYB30. When MYC-MYB30 and FLAG-RHA2b were transiently expressed in Arabidopsis protoplasts, FLAG-RHA2b was detected in protein extracts purified by an anti-MYC antibody (Fig. 1C). Next, a bimolecular fluorescence complementation assay was performed to verify this interaction. When MYB30-cYFP and RHA2b-nYFP were coexpressed in Arabidopsis protoplasts, fluorescence was detected exclusively in the nuclei. These results indicate that RHA2b interacts with MYB30 in vivo (Fig. 1D). We tested for interaction between MYB30 and RHA2a, which is the closest homolog of RHA2b and acts additively with RHA2b in the regulation of ABA-mediated inhibition of seedling growth. Co-IP and bimolecular fluorescence complementation assays revealed that MYB30 did not interact with RHA2a (Fig. 1D; Supplemental Fig. S1), indicating that MYB30 may be ubiquitinated specifically by RHA2b.
MYB30 Is a Substrate of RHA2b
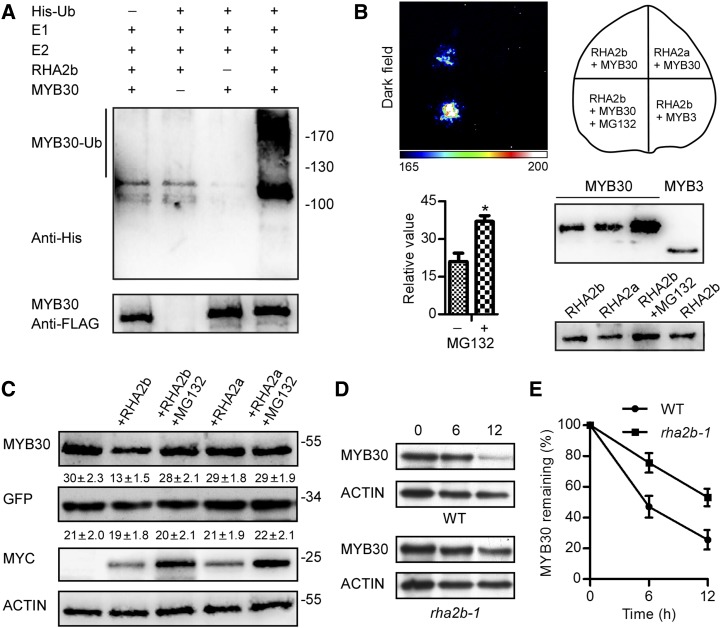
Because RHA2b is an active RING finger E3 ligase (Li et al., 2011) and MYB30 degradation is dependent on the 26S proteasome pathway, we investigated whether MYB30 is a substrate of RHA2b. In vitro ubiquitination assays were performed using transiently expressed MYC-RHA2b and FLAG-MYB30 purified from Arabidopsis protoplasts. In the presence of human His-ubiquitin, UBE1, and UbcH5b (ubiquitin E2), MYC-RHA2b catalyzed the ubiquitination of FLAG-MYB30, as evidenced by the higher molecular mass forms of MYB30 detected using anti-His antibodies (Fig. 2A). When any of these proteins in the reaction was omitted, the ubiquitinated form of MYB30 was not found. These results demonstrate directly that MYB30 is a substrate of RHA2b in vitro.
Figure 2.
RHA2b is involved in the degradation of MYB30. A, MYC-RHA2b was capable of ubiquitinating FLAG-MYB30 in vitro in the presence of His-ubiquitin (His-Ub), UBE1, and UbcH5b (E2). MYB30-Ub was analyzed using an anti-His antibody. The loading of MYB30 protein was detected by anti-FLAG antibody. B, Split-luciferase complementation imaging assays in N. benthamiana. The luminescence intensity was analyzed quantitatively. Relative values are presented as means ± sd (n = 3). Higher luminescence intensity was observed after MG132 treatment compared with the control plants (*, P < 0.05, Student’s t test). The expression of protein was detected by anti-LUC antibody. C, RHA2b promoted MYB30 degradation. FLAG-tagged MYB30 and GFP were coexpressed in Arabidopsis protoplasts alone or with MYC-tagged RHA2b or RHA2a and treated with or without 75 μm MG132 as indicated. Densitometry values (means ± se) for FLAG-MYB30 and GFP were calculated from three independent experiments and are shown at the bottom of each immunoblot. ACTIN was used as a loading control of protoplasts. D, ABA-induced MYB30 degradation was delayed in the rha2b-1 mutant. Col-0/35S:FLAG-MYB30 and rha2b-1/35S:FLAG-MYB30 seedlings were pretreated with 100 μm ABA for 24 h and then incubated in the same medium with 100 μm CHX. Samples were collected 0, 6, and 12 h after treatment. WT, Wild-type Col-0. E, MYB30 protein levels were quantified and normalized. Three independent biological repetitions were performed and analyzed. Data presented are means ± sd.
RHA2b Promotes MYB30 Degradation
Next, we investigated whether RHA2b could mediate the degradation of MYB30 in vivo. When transiently coexpressed in Nicotiana benthamiana, MYB30-nLUC and cLUC-RHA2b yielded weak fluorescence signals, suggesting that the two proteins can interact with each other. If the combination of RHA2b and MYB30 led to the degradation of MYB30 by the 26S proteasome, this interaction could be enhanced by the addition of the 26S proteasome inhibitor MG132. As expected, the fluorescence intensity increased almost twice after MG132 treatment (Fig. 2B). The MYB30 protein level was stable when FLAG-MYB30 was transiently expressed in Arabidopsis protoplasts. When FLAG-MYB30 was coexpressed with RHA2b, MYB30 protein accumulation was reduced by 55.2%, while coexpression with RHA2a did not affect the stability of MYB30 (Fig. 2C), suggesting that the degradation of MYB30 is affected by the coexpression of RHA2b. Additionally, MG132 treatment protected MYB30 from degradation, which confirmed that the RHA2b-mediated degradation of MYB30 was proteasome dependent.
Moreover, the degradation rate of MYB30 in the rha2b-1 mutant, in which the expression of RHA2b was reduced by approximately 50% (Li et al., 2011), also was studied. Columbia-0 (Col-0)/35S:FLAG-MYB30 and rha2b-1/35S:FLAG-MYB30 transgenic lines with comparable FLAG-MYB30 protein levels were used to study the stability of MYB30 in the presence of ABA. Figure 2, D and E, showed that, under ABA treatment, the RHA2b knockdown mutation efficiently delayed MYB30 degradation, with ∼74% and ∼56% MYB30 protein remaining at 6 and 12 h under the treatment, suggesting that RHA2b negatively regulated the stability of MYB30. Based on these results, we conclude that RHA2b can promote the degradation of MYB30 via the 26S proteasome in Arabidopsis.
RHA2b Negatively Regulates MYB30 Transcription Activity
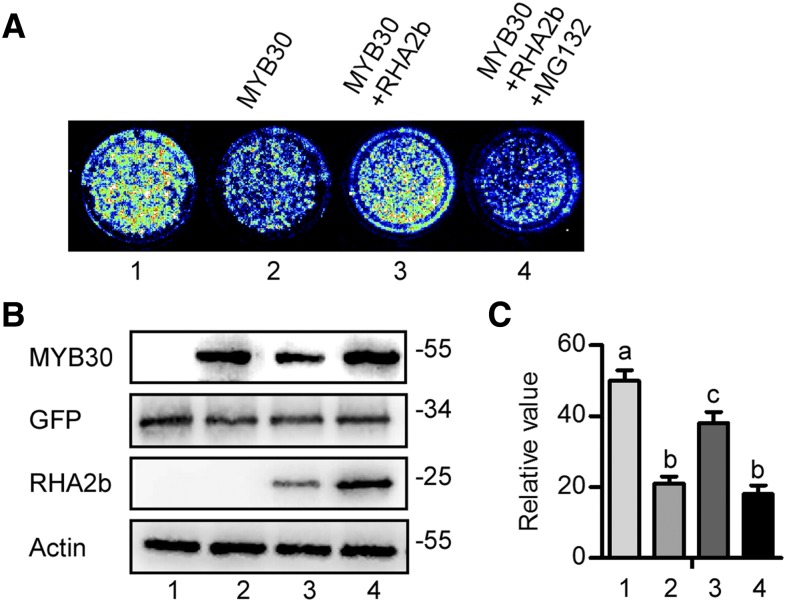
Previous studies have shown that MYB30 binds to the promoter of AtANNEXIN4 (ANN4), which encodes a Ca2+-regulated membrane-binding protein that modulates cytosolic calcium signatures, to repress its expression in oxidative and heat stress responses (Liao et al., 2017). ANN4 also participates in the ABA response (Lee et al., 2004), and its expression can be induced by ABA (Supplemental Fig. S2). To elucidate the functional role of the interaction between MYB30 and RHA2b, the effect of RHA2b on MYB30 transcription activity was studied by transiently expressing a Luciferase (LUC) reporter gene fused with the ANN4 promoter (ProANN4) that is bound by MYB30 directly in Arabidopsis protoplasts (Liao et al., 2017). Consistent with previous results, the overexpression of MYB30 repressed ProANN4:LUC transcription (Fig. 3). In agreement with our observation that RHA2b reduced MYB30 protein accumulation, MYB30-mediated transcriptional repression of ProANN4:LUC was reduced efficiently when MYB30 was coexpressed with RHA2b (Fig. 3). When the degradation of MYB30 was inhibited by MG132 treatment, the transcriptional repression of ProANN4:LUC also was recovered (Fig. 3). These results indicate that RHA2b negatively regulates the transcription activity of MYB30.
Figure 3.
RHA2b negatively regulates MYB30 transcription activity. A, A transcription activation assay was performed in Arabidopsis protoplasts with the following constructs: ProANN4:LUC reporter alone (1) or with FLAG-MYB30 (2); FLAG-MYB30 + MYC-RHA2b (3); or FLAG-MYB30 + MYC-RHA2b + MG132 (4). Representative bioluminescence images are shown. B, Immunoblot showing the expression of the FLAG-MYB30 and MYC-RHA2b proteins. The protein levels of GFP and ACTIN were used to confirm equal transformation rates and loading. C, Relative luminescence signal intensity in A. The data represent means ± sd of at least three replicate experiments. Significant differences (P < 0.05, Student’s t test) are indicated by different lowercase letters.
RHA2b Interacts Genetically with MYB30 in the ABA Response
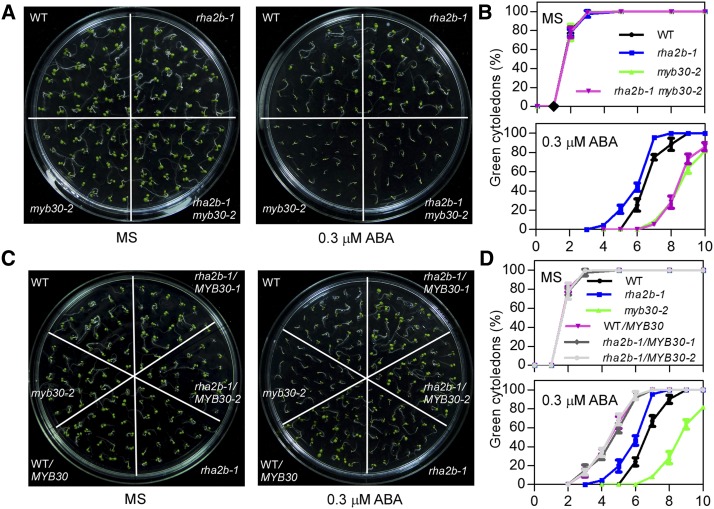
To analyze the genetic relationship between MYB30 and RHA2b in the ABA response, we generated a rha2b-1 myb30-2 double mutant. Wild-type, rha2b-1, myb30-2, and rha2b-1 myb30-2 plants were germinated on Murashige and Skoog (MS) medium containing 0, 0.3, or 0.5 μm ABA. In the presence of 0.3 μm ABA, 75.4% of the wild-type seedlings grew green cotyledons, as measured by the cotyledon greening percentage, but 100% of the rha2b-1, 5.5% of the myb30-2, and 6.2% of the rha2b-1 myb30-2 mutant seedlings grew green cotyledons after 7 d of growing (Fig. 4, A and B; Supplemental Fig. S3). In the presence of 0.5 μm ABA, after 8 d, 21.2% of the wild-type seedlings grew green cotyledons, but 46.4% of the rha2b-1, 2.9% of the myb30-2, and 3.3% of the rha2b-1 myb30-2 mutant seedlings grew green cotyledons (Supplemental Fig. S3, C and D). These results showed that rha2b-1 is hyposensitive and myb30-2 is hypersensitive to ABA relative to the wild type during postgerminative growth, as reported previously. The rha2b-1 myb30-2 double mutant exhibited the same ABA sensitivity as the myb30-2 mutant. These results suggest that the ABA-insensitive phenotype displayed by the rha2b-1 mutant could be suppressed in the myb30 mutant background.
Figure 4.
RHA2b interacts genetically with MYB30 in the ABA response. A, ABA phenotypes of the wild type (WT) and the myb30-2 rha2b-1 double mutant. Seeds were sown on MS medium without (left) or with 0.3 μm ABA (right), and photographs were taken after 7 d. B, Green cotyledons were scored at the indicated times and are represented as averages of 100 seeds from at least three independent experiments ± sd. C and D, ABA phenotypes of the rha2b-1/MYB30 transgenic plants. FLAG-MYB30 was expressed in wild-type and rha2b-1 mutant plants. T4 transgenic lines were used for phenotypic analyses, and photographs were taken after 6 d. ABA sensitivity was monitored as described in A and B.
Additionally, the overexpression of MYB30 in wild-type plants resulted in transgenic plants that were insensitive to ABA. After 6 d of growth, 91.1% of the wild type/MYB30 in the presence of 0.3 μm ABA and 69.6% of the wild type/MYB30 in the presence of 0.5 μm ABA grew green cotyledons, and these percentages were much higher than that of the rha2b-1 mutant seedlings, while the rha2b-1 plants containing a similar level of MYB30 protein (rha2b-1/MYB30) exhibited the same ABA phenotype as the wild-type transgenic plants (Fig. 4, C and D; Supplemental Fig. S3). At the same time, the fresh weights of the different lines in response to ABA also were consistent with our conclusion (Supplemental Fig. S3). Together, these results suggest that MYB30 acts genetically downstream of RHA2b in the ABA response.
Residues K283 and K165 Are Critical for MYB30 Degradation by the 26S Proteasome
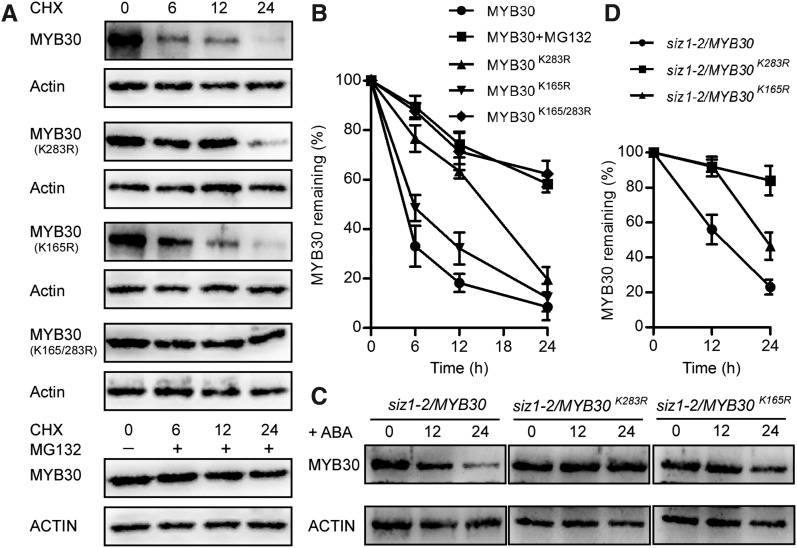
Next, the ubiquitination sites in MYB30 were detected. Since MYB30 was slightly unstable under CHX and this degradation was via the 26S proteasome, CHX treatment was used to screen for ubiquitination sites in MYB30. To identify the ubiquitination sites, Lys (K) residues of MYB30 were mutated to Arg (R). The mutated proteins were then transiently expressed in Arabidopsis protoplasts, and the stability of MYB30 was studied under the CHX treatment. We first split MYB30 into two parts, MYB30-N (amino acids 1–159) and MYB30-C (amino acids 160–323). MYB30-N was stable under the CHX treatment (Supplemental Fig. S4). In MYB30-C, we found that residues K283 and K165 were critical for MYB30 stability. After 6 h of CHX treatment, the protein level of MYB30 was reduced by 70% compared with its original level, but almost 80% of the MYB30K283R and 50% of the MYB30K165R proteins still remained, while the other mutation sites did not elicit significant differences (P < 0.05) from the wild-type protein (Supplemental Fig. S5). We also investigated the degradation rates of MYB30K283R and MYB30K165R proteins. After 12 h of CHX treatment, 63.4% of the MYB30K283R and 32.3% of the MYB30K165R proteins remained, compared with 18.2% of the MYB30 protein. After 24 h, most MYB30 protein was degraded, leaving only 8.5% of the MYB30 protein, compared with 32.3% of the MYB30K283R and 19.6% of the MYB30K165R proteins (Fig. 5, A and B). When treated with MG132, the degradation of MYB30 by the 26S proteasome was inhibited and degradation of MYB30 was efficiently delayed. The degradation rate under MG132 treatment was slower than that of the MYB30K283R protein and similar to that of the MYB30K165/283R double mutant protein (Fig. 5, A and B), which indicated that the K165 site functions additively with K283 in regulating the degradation of MYB30.
Figure 5.
The K283 and K165 residues in MYB30 are important for its degradation by the 26S proteasome. A, Degradation of MYB30, MYB30K165R, MYB30K283R, and MYB30K165/283R protein in Arabidopsis protoplasts. FLAG-MYB30, FLAG-MYB30K165R, FLAG-MYB30K283R, and FLAG-MYB30K165/283R plasmids were transiently expressed overnight in Arabidopsis protoplasts, which were then treated with 100 μm CHX and 75 μm MG132 for the indicated periods of time. ACTIN was used as a loading control. B, The FLAG-MYB30 protein levels in A were quantified. All protein levels were normalized to ACTIN and their initial value. Three independent biological replicates were analyzed. Data presented are means ± sd. C, ABA-induced MYB30, MYB30K165R, and MYB30K283R degradation in the siz1-2 mutant. Seedlings were treated with 100 μm ABA for 12 or 24 h. The MYB30 levels were detected by immunoblotting with an anti-FLAG antibody. ACTIN was used as a loading control. D, The protein levels in C were normalized to that of FLAG-MYB30 normalized to ACTIN at time 0. Three independent biological replicates were analyzed. Data presented are means ± sd.
In the siz1-2 mutant, ABA induced the degradation of the MYB30 protein due to the absence of sumoylation mediated by the SUMO E3 ligase SIZ1. We then examined the functions of the K283 and K165 residues in ABA-induced MYB30 degradation. After 12 h of ABA treatment, 56% of the MYB30 protein remained, but 92% of the MYB30K283R and 90% of the MYB30K165R proteins remained. In the presence of ABA for 24 h, 23% of the MYB30 proteins remained, while 84% of the MYB30K283R and 46% of the MYB30K165R proteins remained (Fig. 5, C and D). Considering the degradation of MYB30 under ABA and MG132 treatment (Fig. 1A), these results also indicated that the K165 site functions additively with K283 in ABA-induced MYB30 degradation. At the same time, when residue K283 was mutated, the polyubiquitination of MYB30 was repressed (Supplemental Fig. S6), indicating that K283 is required for ubiquitination. Together with the K165 residue that functioned in addition to K283 in the ABA-induced MYB30 degradation, we conclude that the K283 and K165 residues are critical sites for the ubiquitination of MYB30 in ABA-induced degradation.
Residues K283 and K165 Also Are Important for the Function of MYB30 in the ABA Response
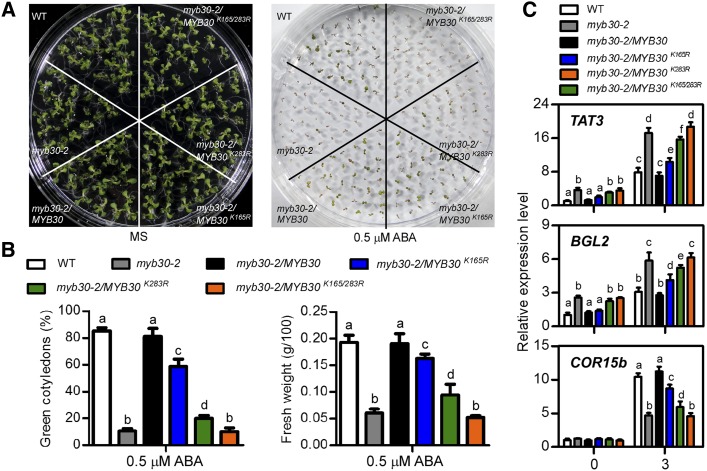
The expression of MYB30K283R only partially reduced the ABA sensitivity of the myb30-2 mutant (Zheng et al., 2012), suggesting that the sumoylation or ubiquitination of K283 is critical for MYB30 to function during ABA signaling. As K165 also is a key site for ubiquitination, we examined its function in the ABA response by expressing FLAG-MYB30K165R, FLAG-MYB30K283R, or FLAG-MYB30K165/283R protein in myb30-2 plants. Transgenic lines with equivalent protein expression of MYB30 (Supplemental Fig. S7) were tested for ABA sensitivity. In the presence of 0.5 μm ABA, after 10 d, 86.1% of the wild-type seedlings and 82.5% of myb30-2/MYB seedlings grew green cotyledons, but 6.9% of the myb30-2, 58.4% of the myb30-2/MYB30K165R, 19.5% of the myb30-2/MYB30K283R, and 5.6% the myb30-2/MYB30K165/283R mutant seedlings grew green cotyledons. The expression of the MYB30K165R protein did not completely rescue the ABA phenotype of myb30-2, while the plants expressing MYB30K165/283R exhibited nearly the same ABA sensitivity as the myb30-2 mutants, suggesting that K165 ubiquitination also is important for the function of MYB30 in the ABA response (Fig. 6, A and B).
Figure 6.
Residues K283 and K165 are important for the function of MYB30 in the ABA response. A, ABA phenotypes of myb30/MYB30K165R. Photographs were taken 10 d after the seeds were sown on MS medium without or with 0.5 μm ABA. B, Germination frequencies were scored after 10 d of growth and are represented as averages of 100 seeds from three independent experiments ± se. Genotypes with different letters are significantly different according to Student’s t test (P < 0.05). C, The transcript abundances of TAT3, LOX3, and COR15b in the different transgenic lines were determined by reverse transcription quantitative PCR using 1-week-old seedlings without or with 100 μm ABA treatment for 3 h. The mRNA levels are expressed relative to the value of the wild-type (WT) seedlings before ABA treatment. The data are represented as means ± sd for three independent experiments. Significant differences (P < 0.05, Student’s t test) are indicated by different lowercase letters.
Tyrosine Aminotransferase3 (TAT3), Lipoxygenase3 (LOX3), and Cold Regulated15b (COR15b) are genes whose expression is regulated by MYB30 in the ABA response (Zheng et al., 2012). Because the K165 and K283 residues are important for maintaining the stability of MYB30 in the regulation of ABA signaling, we investigated whether they functioned at the transcriptional level. In the myb30-2 mutant, the expression of TAT3 and LOX3 was up-regulated, and that of COR15b was down-regulated, after ABA treatment, and these changes were rescued by the expression of MYB30. In contrast to this result, the expression of MYB30K165/283R in the myb30-2 mutant did not change the expression of these genes, while MYB30K165R and MYB30K283R partially recovered the expression of these genes, which is consistent with their ABA phenotypes (Fig. 6C). These results indicate that ubiquitination also plays an important role in regulating the expression of MYB30-associated ABA-responsive genes.
The Stability of MYB30 Is Important for the Function of MYB30 in ABA Signaling
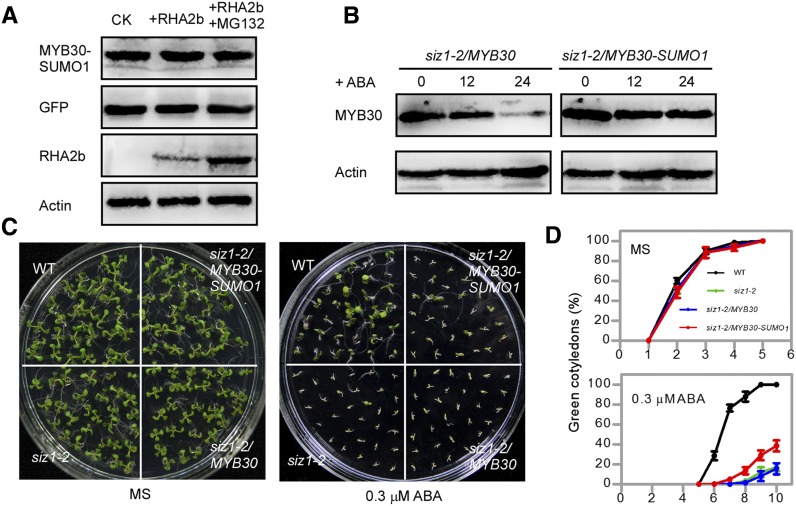
Next, we investigated the function of maintaining MYB30 stability in ABA signaling. We fused SUMO1 to the C terminus of MYB30 to mimic the sumoylation of MYB30 and detected the function of MYB30-SUMO1 in ABA signaling (Jiang et al., 2011; Wu et al., 2011). Compared with that of wild-type MYB30, the degradation rate of MYB30-SUMO1 was efficiently delayed (P < 0.01) upon CHX treatment (Supplemental Fig. S8A). When coexpressed with RHA2b, the protein level of MYB30-SUMO1 was almost not affected (Fig. 7A). In the siz1-2 mutant seedlings, the MYB30-SUMO1 protein also was more stable than MYB30 after ABA treatment (Fig. 7B). Considering that ABA induces the degradation of MYB30 in the siz1-2 mutant seedlings (Fig. 7B), overexpression of MYB30 in siz1-2 (siz1-2/MYB30) does not change the ABA sensitivity of the siz1-2 mutant (Fig. 7, C and D; Zheng et al., 2012). Since sumoylation can protect MYB30 from degradation under ABA treatment, we investigated the function of MYB30-SUMO1 in the siz1-2 mutant. When MYB30-SUMO1 was overexpressed in siz1-2 (siz1-2/MYB30-SUMO1), the ABA phenotype of siz1-2 was partially recovered (Fig. 7, C and D). These results show that maintaining MYB30 stability plays an important role in the function of MYB30 in ABA signaling.
Figure 7.
The stability of MYB30 is important for the function of MYB30 in ABA signaling. A, The degradation of MYB30 promoted by RHA2b was repressed by sumoylation. FLAG-tagged MYB30-SUMO1 or GFP was expressed alone or with MYC-tagged RHA2b in Arabidopsis protoplasts, which were treated with or without the proteasome inhibitor MG132 as indicated. ACTIN was used as a loading control. B, ABA-induced MYB30 degradation in the siz1-2 mutant was repressed by sumoylation. Eight-day-old seedlings were treated with 100 μm ABA for 12 or 24 h. ACTIN was used as a loading control. C, The expression of MYB30-SUMO1 can partially recover the ABA phenotype of siz1-2. ABA phenotypes of wild-type (WT), siz1-2, siz1-2/MYB30, and siz1-2/MYB30-SUMO1 seedlings are shown. Photographs were taken 9 d after the seeds were sown on MS medium without or with 0.3 μm ABA. D, Green cotyledons were scored at the indicated times and represent averages of 100 seeds from at least three independent experiments ± se.
Ubiquitination and Sumoylation Act Antagonistically in the MYB30-Associated ABA Response
Since sumoylation also regulates the stability of MYB30 in the ABA response, we investigated the relationship between sumoylation and ubiquitination. When MYC-SUMO1 was cotransformed with FLAG-MYB30 into wild-type protoplasts, the MYB30 protein levels decreased to a low level after 6 h of treatment with CHX, while the sumoylated MYB30 protein was not affected, indicating that sumoylation protects MYB30 from degradation (Supplemental Fig. S8B). When transiently expressed in Arabidopsis protoplasts, MYB30-SUMO1-GFP exhibited a similar localization in nuclei to MYB30-GFP (Supplemental Fig. S8C). Moreover, the K165R mutation of MYB30 did not affect MYB30 sumoylation, indicating that there may be no feedback control of sumoylation by ubiquitination (Supplemental Fig. S8D). Because the K283 residue is the critical site for both sumoylation and ubiquitination, and sumoylation does not change the localization of MYB30 in nuclei, we hypothesized that ubiquitination and sumoylation may act antagonistically in the regulation of MYB30 stability by occupying the same site.
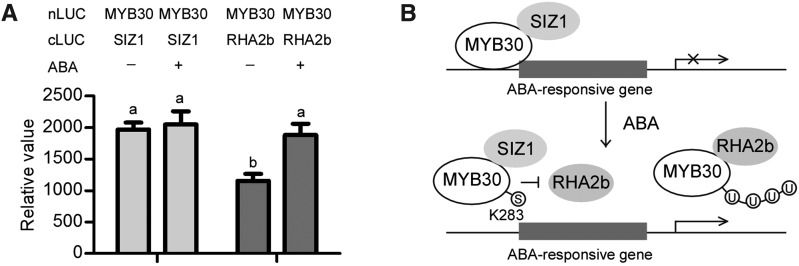
Finally, we attempted to reveal how the function of MYB30 was regulated in the ABA response by analyzing the interactions of MYB30 with RHA2b and SIZ1 after ABA treatment. nLUC-MYB30 was transiently expressed with RHA2b-cLUC or SIZ1-cLUC in N. benthamiana. After 48 h of incubation, the luminescence intensity induced by ABA treatment was detected. When MYB30 was coexpressed with RHA2b, the luminescence intensity increased upon ABA treatment, while the luminescence intensity in leaves coexpressing MYB30 and SIZ1 was much higher before ABA treatment but showed no difference after ABA treatment (Fig. 8A). Based on these results, we propose that ABA may promote the degradation of MYB30 by enhancing the interaction between RHA2b and MYB30. Moreover, the interaction between SIZ1 and MYB30 did not change after ABA treatment, suggesting that SIZ1 stabilizes MYB30 by sumoylation to protect MYB30 from degradation.
Figure 8.
Potential antagonistic relationship between sumoylation and ubiquitination in the regulation of the stability of MYB30 during the ABA response. A, Interaction between MYB30 and SIZ1 or RHA2b after ABA treatment. nLUC-MYB30 was transiently expressed with RHA2b-cLUC or SIZ1-cLUC in N. benthamiana, and the luminescence intensities were detected before or after treatment with 100 μm ABA. The significance of the luminescence intensity difference was determined by Student’s t test. Significant differences (P < 0.05) are indicated by different lowercase letters. B, Working model for the cooperation of sumoylation and ubiquitination in the regulation of MYB30 stability during the ABA response. In the absence of ABA, little MYB30 is degraded by RHA2b to maintain its turnover, and MYB30 negatively regulates the expression of ABA-responsive genes. When the ABA signal is present, RHA2b promotes the degradation of MYB30 to release the expression of ABA-responsive genes. Meanwhile, SIZ1 protects MYB30 from degradation to maintain its function in other responses, such as the hypersensitive response (HR) and the oxidative response.
DISCUSSION
We previously reported that sumoylation of the transcription factor MYB30, which is mediated by the SUMO E3 ligase SIZ1, is important for its stability and role in the ABA response (Zheng et al., 2012). In this study, we identified that the stability and function of MYB30 also are regulated by ubiquitination. The ubiquitin E3 ligase RHA2b ubiquitinates MYB30 directly, and the degradation rate of MBY30 is repressed in rha2b-1 under ABA treatment. The ABA phenotype of the rha2b-1 myb30-2 double mutant was the same as that of the myb30-2 mutant, and overexpression of MYB30 in the rha2b-1 mutant caused similar ABA insensitivity to MYB30-overexpressing plants, indicating that MYB30 acts genetically downstream of RHA2b in ABA signaling. When the ubiquitination sites are mutated, the function of MYB30 also is disrupted. Overexpression of MYB30-SUMO1, but not MYB30 protein, can partially recover the ABA sensitivity of siz1-2. All these data suggest that the stability of MYB30 is critical for its role in ABA signaling. Together with the results that ABA can enhance the interaction between RHA2b and MYB30, we speculate that RHA2b may function in ABA signaling by mediating the degradation of MYB30.
Cross talk between ubiquitin and SUMO in signal transduction has been reported in yeast (Saccharomyces cerevisiae) and humans (Homo sapiens). Sumoylation and ubiquitination can interact competitively or cooperatively to regulate protein stability and function (Ulrich, 2005; Praefcke et al., 2012). For example, phosphorylation of the human Flap Endonuclease1 induces SUMO conjugation, which, in turn, leads to ubiquitination and subsequent proteasomal degradation (Guo et al., 2012). In contrast, sumoylation of IκBα, an inhibitor of nuclear factor-κB, stabilizes the protein and protects it from proteasomal degradation. Interestingly, sumoylation and ubiquitination occur at the same residue (i.e. K21), which has led to the concept of ubiquitin and SUMO as biological antagonists (Desterro et al., 1998). In plants, many proteins, such as ABI5 and Inducer of CBF Expression1, are modified by both sumoylation and ubiquitination (Miura et al., 2007b, 2009). For example, ABI5, which is a bZIP transcription factor that functions as a positive regulator in ABA signaling, is sumoylated at K339 and ubiquitinated at K344. In the siz1-2 mutant, ABI5 was less abundant than it was in wild-type seedlings, suggesting that the sumoylation of ABI5 may increase its stability. However, how sumoylation and ubiquitination function together remains unclear.
The fact that MYB30 is regulated by both modifications provides material for studying the relationship between sumoylation and ubiquitination. The RHA2b-mediated degradation of MYB30 is repressed by sumoylation, and ABA induces the degradation of MYB30 but not MYB30-SUMO1 in the siz1-2 mutant. Considering that the K283 residue is the major site for both sumoylation and ubiquitination, and MYB30-SUMO1 protein exhibits a similar localization with MYB30 in nuclei, these results suggest that sumoylation can protect, at least partially, MYB30 from proteasomal degradation by occupying the same locus. ABA treatment enhances the interaction between RHA2b and MYB30, which may promote the degradation of MYB30. At the same time, a high luminescence intensity demonstrates the strong interaction between MYB30 and SIZ1, and the fact that the intensity does not change after ABA treatment provides an explanation for the stability of the MYB30-SUMO1 protein. These results provide a model of how sumoylation functions together with ubiquitination in the regulation of protein stability during the ABA response in plants (Fig. 8B). In the absence of ABA, RHA2b interacts with a portion of MYB30 in the cell to maintain its turnover, and MYB30 negatively regulates the expression of ABA-responsive genes. When the ABA signal arrives, MYB30 degradation is induced by RHA2b to release the suppression of ABA-responsive genes. Moreover, the interaction between SIZ1 and MYB30 remains unchanged to protect MYB30 from degradation and allow it to continue to function in other responses, such as the HR and the oxidative and heat stress responses (Vailleau et al., 2002; Liao et al., 2017).
Only a few proteins are quantitatively sumoylated to constitutively perform their different functions. Instead, only a small percentage of most targets appear to be modified during steady-state conditions (Geiss-Friedlander and Melchior, 2007). In our study, sumoylated MYB30 accounted for only a fraction of total MYB30 protein. However, when coexpressing cLUC-SIZ1 and MYB30-nLUC, a high luminescence intensity indicated strong interaction between MYB30 and SIZ1. These results suggest that, when SIZ1 combines with MYB30, its function may be not limited to the sumoylation of MYB30. SIZ1 has been found to localize to the nuclear foci, and the sumoylation of transcription factors facilitates their recruitment into nuclear bodies (Miura et al., 2005; Geiss-Friedlander and Melchior, 2007). However, the sumoylation of MYB30 does not affect its localization in nuclei. Based on the above, it is plausible that the combination of SIZ1 may directly inhibit the transcriptional activity of MYB30.
In addition to ABA signaling, MYB30 also is involved in the HR, hypoxia response, brassinosteroids, calcium signaling, and oxidative and heat stress responses (Vailleau et al., 2002; Raffaele et al., 2008; Li et al., 2009; Xie et al., 2015; Liao et al., 2017). As a transcription factor, MYB30 participates in these signaling pathways by regulating the expression of downstream target genes, such as very-long-chain fatty acid-related genes in the HR and hypoxia response and ANNEXIN genes in oxidative and heat stress responses. However, how MYB30 is regulated in different responses remains poorly understood. In the HR, another RING-type E3 ubiquitin ligase, MIEL1, interacts with MYB30 and participates in the HR by mediating the ubiquitination and proteasomal degradation of MYB30 (Marino et al., 2013). Furthermore, MIEL1 also is involved in the ABA response via its mediation of the ubiquitination of MYB96, which is one of the closest homologs of MYB30 among the R2R3-type MYB transcription factors. The miel1 mutant is hypersensitive to ABA. In miel1 mutant seedlings, the turnover of MYB30 is affected, and the expression of the transcription factor ABI4 is up-regulated, but the function of MYB30 seems to be most likely independent of MIEL1 because both of their mutants are hypersensitive to ABA. Combined with our results, these findings suggest that the stability of MYB30 can be regulated by MIEL1 and RHA2b to participate in the HR and ABA response, respectively. In the rha2b-1 mutant, although the degradation rate was delayed, the MYB30 protein continued to decrease under the CHX treatment, suggesting that other E3 ligases also may be involved in the ABA response and may cooperate with RHA2b by regulating the stability of MYB30.
Here, we demonstrate that ABA can induce the degradation of MYB30 protein and that this degradation is mediated by the ubiquitin E3 ligase RHA2b via the 26S proteasome. The stability of MYB30 plays an important role in the regulation of ABA signaling, and RHA2b may participate in ABA responses by promoting the degradation of MYB30. Sumoylation can protect MYB30 from degradation, and the K283 residue is identified as the major site for both sumoylation and ubiquitination, providing a model for sumoylation and ubiquitination acting antagonistically in the ABA response to regulate the stability of MYB30 by occupying the same residue.
MATERIALS AND METHODS
Plant Growth and Treatment
Arabidopsis (Arabidopsis thaliana, Col-0 ecotype) was used as the wild type. Seeds of each genotype were sown on MS medium with 0.3% (w/v) agar, stratified at 4°C in darkness for 3 d, and then grown in a growth chamber at 23°C in constant light. To monitor ABA sensitivity, seeds were sown on MS medium or MS medium containing the indicated concentrations of ABA (Sigma). Germination frequencies were obtained by scoring green cotyledons of approximately 100 seeds from three independent experiments ± se.
For ABA and CHX treatments, 8-d-old Col-0/35S:FLAG-MYB30 or rha2b-1/35S:FLAG-MYB30 transgenic seedlings were treated with 100 μm ABA for 24 h and then treated with the same medium with 100 μm CHX for the indicated times with or without 75 μm MG132. Protoplasts that were transiently expressed with various MYB30 proteins were incubated with 100 μm CHX at 23°C for the indicated times after a 20-h incubation. Eight-day-old siz1-2/MYB30 seedlings were treated with water (0 h) or ABA for 12 or 24 h.
Plasmid Construction and Generation of Transgenic Plants
Full-length coding sequences of MYB30, MYB30K165R, MYB30K283R, MYB30K165/283R, MYB3, RHA2b, RHA2a, and SUMO1 were cloned into pCAMBIA1307-3×FLAG or pCAMBIA1307-6×Myc binary vector with the 35S promoter between the BamHI and KpnI sites. MYB30 and MYB3 coding sequences without stop codons were cloned into pCAMBIA1300-nLUC or pSPYCE, and RHA2b and RHA2a coding sequences were cloned into pCAMBIA1300-cLUC pSPYNE between the KpnI/SalI or BamHI/SalI sites.
The resulting constructs were introduced into Arabidopsis using Agrobacterium tumefaciens-mediated floral transformation and verified by immunoblot analysis. T3 transgenic plants were used for analysis.
Complementation Assays
DNA 2,234 bp upstream of the MYB30 translation start codon and full-length coding sequences of MYB30 without stop codons were cloned into pCAMBIA1300 with a 3×FLAG tag at the C-terminal end. The resulting construct was introduced into the myb30-2 mutant using A. tumefaciens-mediated floral transformation and verified by immunoblot analysis. T3 transgenic plants were used for analysis.
Co-IP and Immunoblot Analysis
For Co-IP assays, MYC-MYB30 and FLAG-RHA2b were coexpressed in protoplasts by polyethylene glycol-mediated transformation with 75 μm MG132. After a 20-h incubation at 23°C, protoplasts were harvested with 1 mL of extraction buffer (10 mm Tris [pH 7.5], 0.5% Nonidet P-40, 2 mm EDTA, 150 mm NaCl, 1 mm PMSF, and 1% [v/v] protease inhibitor mixture [Sigma]). The extracts were centrifuged at 12,000g for 15 min at 4°C, and 20 μL of the resulting supernatant was used as the input. The rest was incubated with 15 μL of an anti-MYC agarose conjugate (Sigma) for 2 h at 4°C to purify the MYB30 protein. After five washes in 1 mL of extraction buffer, the RHA2b proteins were detected by immunoblot analysis using an anti-FLAG antibody.
For the quantitation of protein expression, protein levels of MYB30 and ACTIN were quantified by the FusionCapt Advance system. Then, protein levels compared with ACTIN at the initial time were identified as 100%. Protein levels compared with ACTIN at the indicated times were normalized to the initial value.
In Vitro Ubiquitination Assay
For the in vitro ubiquitination assay, ubiquitination reaction mixtures (30 μL) contained 30 ng of UBE1 (E1; Boston Biochem), 30 ng of UbcH5b (E2; Boston Biochem), 5 μg of His-tagged ubiquitin (Sigma), 50 ng of MYC-RHA2b, and 200 ng of FLAG-MYB30 or FLAG-MYB30K283R in a reaction buffer containing 50 mm Tris, pH 7.5, 10 mm MgCl2, 2 mm ATP, and 0.5 mm DTT. MYC-RHA2b and FLAG-MYB30 or FLAG-MYB30K283R were transiently expressed in Arabidopsis protoplasts and purified. After 2 h of incubation at 30°C, the reactions were stopped by adding 4× sample buffer. Samples were then separated onto 8% SDS-PAGE gels. Ubiquitinated FLAG-MYB30 was detected using anti-His antibody (Xu et al., 2014). The loading of MYB30 protein (without reaction) was detected using anti-FLAG antibody.
Split-YFP and Split-LUC Assays
For the split-YFP assay, YNE-RHA2b and MYB30-YCE were coexpressed in Arabidopsis protoplasts in the presence of 75 μm MG132. Fluorescence was measured using a Zeiss 510 confocal microscope (excitation at 488 nm and emission at 520 nm).
For the split-LUC assay, MYB30-nLUC or MYB3-nLUC was cotransformed with cLUC-RHA2b, cLUC-RHA2a, or cLUC-SIZ1 into Nicotiana benthamiana via A. tumefaciens and incubated for 2 d. MG132 was injected into the leaf tissues 12 h before observation, and 100 μm d-luciferin was sprayed on the leaves before image collection by a CCD camera. For the ABA treatment, leaves were divided into small discs and then incubated in water (control) or 100 μm ABA, and luminescence intensities were detected after the treatment.
Transactivation Activity Assays in Arabidopsis Protoplasts
A plasmid containing the ANN4 promoter upstream of the LUC gene was used as the reporter. The LUC reporter and GFP (internal control) were coexpressed in protoplasts with or without FLAG-MYB30, MYC-RHA2b, and MG132 as indicated. After an overnight incubation at 23°C, the protoplasts were transferred to a 12-well plate, and 50 μL of 1 mm d-luciferin (Promega) was added to each sample. The plate was kept in the dark for 5 min, and a CCD camera (Roper Scientific) was used to visualize the LUC signal. The primer pairs used are shown in Supplemental Table S1.
Sumoylation Assay
For the SUMO conjugation assay, FLAG-MYB30 and MYC-SUMO1 were coexpressed in protoplasts. FLAG-MYB30 was purified by an anti-FLAG agarose conjugate, and an anti-MYC antibody was used to detect the FLAG-MYB30-MYC-SUMO1 interaction. An anti-FLAG antibody was used to detect the abundance of FLAG-MYB30 as a control. All primer pairs used are shown in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MYB30, At3g28910; SIZ1, At5g60140; RHA2b, At2g01150; RHA2a, At1g15100; MYB3, At1g22640; ANN4, At2g38750; TAT3, At2g24850; LOX3, At1g17420; and COR15b, At2g42530.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Co-IP assays between MYB30 and RHA2a.
Supplemental Figure S2. ABA enhanced the expression of ANN4 in wild-type and myb30-2 seedlings.
Supplemental Figure S3. ABA responses of rha2b-1 myb30-2 double mutant plants and rha2b-1/MYB30 transgenic plants in early seedling growth.
Supplemental Figure S4. The ubiquitination sites of MYB30 localize at the C-terminal end.
Supplemental Figure S5. Screen for ubiquitination sites of MYB30.
Supplemental Figure S6. The K283 residue is required for ubiquitination.
Supplemental Figure S7. Protein levels of different types of MYB30 in myb30-2 mutant seedlings.
Supplemental Figure S8. Sumoylation protects MYB30 from degradation.
Supplemental Table S1. Primer sequences used in plasmid constructs and reverse transcription quantitative PCR.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yan Guo (State Key Laboratory of Plant Physiology and Biochemistry, China Agricultural University) for providing guidance and giving valuable advice. We thank Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for giving valuable advice on the in vitro ubiquitination assay. We thank Chuanyou Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the rha2b-1 (SALK_014943) mutant.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31400264) and The Opening Scientific Program of the State Key Laboratory of Plant Physiology and Biochemistry, China Agricultural University (grant no. SKLPPBKF1703 to Y.Z.).
References
- Bu Q, Li H, Zhao Q, Jiang H, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Wang D, et al. (2009) The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol 150: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Rodriguez L, Lorenzo-Orts L, Gonzalez-Guzman M, Sayas E, Muñoz-Bertomeu J, Ibañez C, Serrano R, Rodriguez PL (2014) The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J 80: 1057–1071 [DOI] [PubMed] [Google Scholar]
- Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, Ok SH, Yoo SD, Shin JS (2013) An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett 587: 1773–1778 [DOI] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT (1998) SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell 2: 233–239 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell (Suppl) 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Guo J, Yang X, Weston DJ, Chen JG (2011) Abscisic acid receptors: past, present and future. J Integr Plant Biol 53: 469–479 [DOI] [PubMed] [Google Scholar]
- Guo Z, Kanjanapangka J, Liu N, Liu S, Liu C, Wu Z, Wang Y, Loh T, Kowolik C, Jamsen J, et al. (2012) Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol Cell 47: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YF, Zhao QY, Dang LL, Luo YX, Chen SS, Shao CR, Huang HW, Li YQ, Li L, Cai T, et al. (2016) The SUMO E3 ligase-like proteins PIAL1 and PIAL2 interact with MOM1 and form a novel complex required for transcriptional silencing. Plant Cell 28: 1215–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Chiu SY, Hsu W (2011) SUMO-specific protease 2 in Mdm2-mediated regulation of p53. Cell Death Differ 18: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, Nishimura M, Hayashi M (2010) A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J 62: 936–947 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Seo PJ (2016) The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat Commun 7: 12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim WT (2011) Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK (2004) Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. Plant Cell 16: 1378–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49: 199–222 [DOI] [PubMed] [Google Scholar]
- Li H, Jiang H, Bu Q, Zhao Q, Sun J, Xie Q, Li C (2011) The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol 156: 550–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Zheng Y, Guo Y (2017) MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol 216: 163–177 [DOI] [PubMed] [Google Scholar]
- Liu H, Stone SL (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lai J, Yu M, Wang F, Zhang J, Jiang J, Hu H, Wu Q, Lu G, Xu P, et al. (2016) The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex in cell cycle regulation. Plant Cell 28: 2225–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, Pouzet C, Jauneau A, Roby D, Rivas S (2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat Commun 4: 1476. [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Hasegawa PM (2007a) Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol 10: 495–502 [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007b) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009a) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009b) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Zhang W, Assmann SM (2007) Roles of ion channels and transporters in guard cell signal transduction. FEBS Lett 581: 2325–2336 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, Hofmann K, Dohmen RJ (2012) SUMO playing tag with ubiquitin. Trends Biochem Sci 37: 23–31 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S (2012) The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol 196: 13–28 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ (2001) Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanov K, Zeschmann A, Hermkes R, Eifler K, Ziba I, Grieco M, Novatchkova M, Hofmann K, Hesse H, Bachmair A (2014) Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26: 4547–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. (2005) Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol 15: 525–532 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol 51: 1821–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang L, Zhou P, Wang G, Zeng Y, Wang Y, Liu J, Zhang B, Liu S, Luo H, et al. (2011) Regulation of REGγ cellular distribution and function by SUMO modification. Cell Res 21: 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie LJ, Yu LJ, Chen QF, Wang FZ, Huang L, Xia FN, Zhu TR, Wu JX, Yin J, Liao B, et al. (2015) Arabidopsis acyl-CoA-binding protein ACBP3 participates in plant response to hypoxia by modulating very-long-chain fatty acid metabolism. Plant J 81: 53–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Lin F, Jiang Y, Huang X, Li J, Ling J, Hettiarachchi C, Tellgren-Roth C, Holm M, Deng XW (2014) The RING-finger E3 ubiquitin ligase COP1 SUPPRESSOR1 negatively regulates COP1 abundance in maintaining COP1 homeostasis in dark-grown Arabidopsis seedlings. Plant Cell 26: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cui F, Wu Y, Lou L, Liu L, Tian M, Ning Y, Shu K, Tang S, Xie Q (2015) The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27: 214–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lai J, Wang F, Yang S, He Z, Jiang J, Li Q, Wu Q, Liu Y, Yu M, et al. (2017) A SUMO ligase AtMMS21 regulates the stability of the chromatin remodeler BRAHMA in root development. Plant Physiol 173: 1574–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Qi Y, Liu M, Yang C (2013) SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana. J Integr Plant Biol 55: 83–95 [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y (2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]