The acquisition of a single chromosome set by gametes generated during sexual reproduction depends on cohesion proteins that are protected by a phosphatase enzyme complex including the proteins PP2AB′α and β.
Abstract
The correct separation of homologous chromosomes during meiosis I, and sister chromatids during meiosis II, relies on the tight control of the cohesion complex. The phosphorylation and subsequent cleavage of the meiotic recombination protein REC8 (REC8-like family protein [SYN1] in Arabidopsis [Arabidopsis thaliana]), the α-kleisin subunit of the cohesion ring, along the chromosome arms at meiosis I allows crossovers and separation of homologous chromosomes without chromatid dissociation. REC8 continues to localize and function at the centromeres up to metaphase II and, in yeast and vertebrates, is protected from cleavage by means of protein phosphatase 2A (PP2A)-mediated dephosphorylation. Here, we show that, in plants, centromeric sister chromatid cohesion until meiosis II also requires the activity of a PP2A-type phosphatase complex. The combined absence of the regulatory subunits PP2AB′α and PP2AB′β leads to the premature loss of chromosome cohesion in meiosis I. Male meiocytes of the pp2ab′αβ double mutant display premature depletion of SYN1. The PP2AA1 structural and B′α regulatory subunit localize specifically to centromeres until metaphase II, supporting a role for the PP2A complex in the SYN1-mediated maintenance of centromeric cohesion in plant meiosis.
Meiosis is a specialized cell division program essential for sexual reproduction in which two rounds of chromosome segregations follow a single DNA replication step, leading to halving of the chromosome set. In meiosis I, homologous chromosomes are separated while sister chromatid association is maintained. In meiosis II, sister chromatids separate, which resembles a mitotic division (Zickler and Kleckner, 1999). One of the crucial processes ensuring accurate chromosome segregation during meiosis is the stepwise loss of cohesion, starting at the sister chromatid arms in meiosis I followed by the loss of cohesion at the centromeres in meiosis II (Bolaños-Villegas et al., 2017). The removal of chromosome arm cohesion promotes homologous chromosome segregation and the resolution of chiasmata in anaphase I. At this stage, sister chromatids stay together through centromeric cohesion and monopolar kinetochore orientation (Marston, 2014). The programmed loss of centromeric cohesion at the onset of anaphase II triggers the separation of sister chromatids to opposite poles, eventually generating four haploid nuclei out of a diploid mother nucleus (Marston and Amon, 2004).
In mitotic cells, sister chromatid cohesion is mediated by the multisubunit cohesin complex, which forms a ring-shaped structure that encompasses the two sister chromatids formed by DNA replication (Nasmyth and Haering, 2009). The ring-shaped cohesion complex is composed of four core subunits: Structural Maintenance of Chromosomes1 (SMC1) and SMC3 and Sister Chromatid Cohesion1 (SCC1) and SCC3 (Ishiguro and Watanabe, 2007). At the onset of anaphase, the kleisin protein SCC1 is targeted by separase (ESP), which cleaves the cohesin ring structure and locally eliminates sister chromatic cohesion (Uhlmann et al., 2000; Hauf et al., 2001). In the eukaryotic clade, the cohesin complex is highly conserved and shows close similarity to bacterial cohesion-like proteins, indicating the ancient nature of these proteins (Peters et al., 2008).
Sister chromatid cohesion in meiotically dividing cells also is established by a ring-shaped cohesion complex. Here, the SCC1/RAD21 subunit is replaced by the meiosis-specific variant α-kleisin cohesin REC8. During the first meiotic cell division, REC8 is cleaved along the chromosome arms through a separase-mediated mechanism. The REC8 proteins residing at the centromeric region are protected from cleavage throughout meiosis I to maintain chromatid cohesion until metaphase II (Kitajima et al., 2006). This regulation involves the phosphorylation of REC8, performed by the polo-like kinase, cell division cycle5-like protein (CDC5) in budding yeast, whereas in fission yeast this function is executed by casein kinase 1δ/ɛ (CK1δ/ɛ) and DBF4-dependent CDC7 kinase (Clyne et al., 2003). Phosphorylation marks REC8 for cleavage by ESP but is antagonized by protein phosphatase 2A (PP2A), which protects against cleavage (Ishiguro et al., 2010; Rumpf et al., 2010). The PP2A complex interacts with Shugoshin (SGO), and both are targeted cooperatively to the centromere (Kitajima et al., 2006). In plants, REC8-like family protein (SYN1) and SGO have been identified in several species, and their function as a protector of centromeric cohesion in meiosis is conserved (Hamant et al., 2005; Wang et al., 2011; Cromer et al., 2013; Zamariola et al., 2013). PP2A is a family of holoenzyme phosphatases that account for the majority of Ser/Thr phosphatase activities in many processes and cell types. It consists of the scaffolding A subunit, a catalytic C subunit, and a regulatory/targeting B subunit. The B subunit family consists of structurally different proteins separated into four groups, B, B′, B′′ and B′′′, that are responsible for the specificity or targeting of the PP2AA and PP2AC subunits (Janssens and Goris, 2001). In Arabidopsis (Arabidopsis thaliana), the B′ family comprises nine members (α, β, γ, η, θ, ζ, δ, κ, and ε; Farkas et al., 2007). The PP2A complexes in plants have functions in cell division, hormone signaling, development, and biotic stress resistance (Tang et al., 2011; Durian et al., 2016; Wang et al., 2016; Yue et al., 2016).
We previously reported that the double mutant of the PP2AB′ α- and β-subunits are semisterile, implying a function during sexual reproduction (Jonassen et al., 2011). Here, we report that the activity of PP2AB′α and PP2AB′β is required for maintaining SYN1 at centromeres from metaphase I to II, presumably by promoting SYN1 dephosphorylation. Since AtPP2AB′α and AtPP2AB′β are dispensable for mitotic cell division and both are expressed predominantly in flower buds undergoing meiosis, we propose that these B′ subunits exert a specific function in the regulation of chromosome cohesion during meiosis.
RESULTS
The PP2AB′ α- and β-Subunits Are Involved in Male and Female Sporogenesis
In line with a function in sexual reproduction, the mining of public expression data indicated that PP2AB′α and PP2AB′β are expressed predominantly in the anthers during meiosis (Supplemental Fig. S1). PP2AB′α and PP2AB′β contain putative nuclear targeting sequences (Latorre et al., 1997), suggesting that these isoforms may be involved in the recruitment of PP2A enzyme components into nuclei and, hence, to meiotic chromosomes.
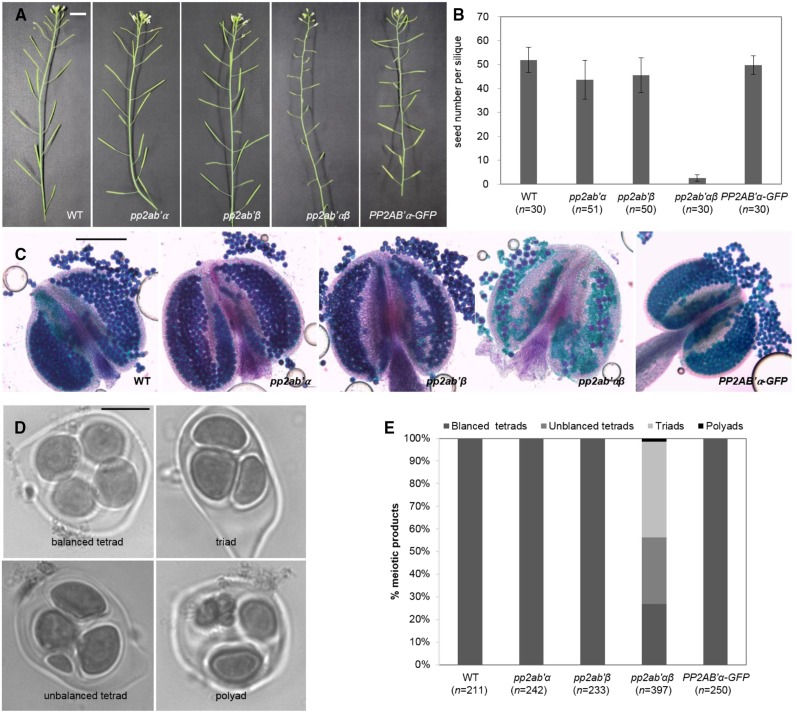
A possible role for PP2AB′α and PP2AB′β subunits in meiosis was investigated by analyzing the male reproductive system of T-DNA mutants carrying insertions in pp2ab′α (SALK_077700) and pp2ab′β (SALK_103167). The pp2ab′α and pp2ab′β single mutants produced anthers that were filled with pollen and produced regular amounts of seeds per silique (Fig. 1, A–C), indicating that meiosis, gametogenesis, and fertilization were not compromised. Considering the overlapping expression profiles of PP2AB′α and PP2AB′β and the potential functional redundancy, we generated pp2ab′αβ double mutants carrying an insertion in both isoforms. pp2ab′αβ double mutants did not show any defects in vegetative development or growth, but they showed a substantial decrease in silique length, suggesting reduced seed set (Fig. 1A). To quantify seed set, we counted the seeds of at least 30 siliques per genotype and found that the number of seeds per silique in pp2ab′αβ (2.6 ± 1.4) was decreased compared with that in the wild type (51.9 ± 5.3), pp2ab′α (43.6 ± 8.1), and pp2ab′β (45.5 ± 7.4; Fig. 1B). The anthers from the double mutant contained a severely reduced number of viable pollen grains, and these varied in size compared with the uniformly sized pollen from wild-type anthers (Fig. 1C; Supplemental Fig. S2). These observations support a function of PP2AB′α and PP2AB′β in male sporogenesis.
Figure 1.
The pp2ab′αβ double mutant shows reduced fertility. Phenotypic analysis is shown for the wild type, pp2ab′α, pp2ab′β, pp2ab′αβ (double mutant), and PP2AB′α-GFP (pp2ab′αβ double mutant expressing PP2AB′α-GFP). A, Phenotypes of mature plants producing siliques. Bar = 1 cm. B, Quantification of seeds per silique (means ± sd). Compared with the wild type, pp2ab′α, pp2ab′β, and pp2ab′αβ show significant differences (Tukey’s test in SPSS, P < 0.001), but not PP2AB′α-GFP (Tukey’s test in SPSS, P < 0.05). C, Analysis of pollen grain viability. Viable pollen grains are stained in purple, whereas dead pollen grains appear green. Bar = 200 μm. D, Analysis of the meiotic products: polyads, triads, unbalanced tetrads, and balanced tetrads. Bar = 10 µm. E, Quantification of the meiotic products shown in D.
Arabidopsis male meiosis yields four haploid cells organized as a balanced tetrad (Fig. 1D). The pp2ab′αβ double mutant produced balanced tetrads (27%), unbalanced tetrads (29.2%), triads (42.3%), and polyads (1.5%; Fig. 1E), indicating defects in chromosome segregation and/or meiotic cytokinesis. Female reproduction also was affected in the pp2ab′αβ double mutant, as a reciprocal cross between the wild type and pp2ab′αβ yielded reduced seed set. Crosses using the pp2ab′αβ double mutant as the pollen donor yielded only 1.1 seeds per pod (n = 23; Supplemental Fig. S3). When pp2ab′αβ was used as the female parent, the resulting seed pods contained on average 2.3 seeds (n = 44; Supplemental Fig. S3). These data indicate that both PP2AB′α and PP2AB′β are redundantly required for male as well as female reproduction.
The pp2ab′αβ Double Mutant Generates Aneuploid Offspring
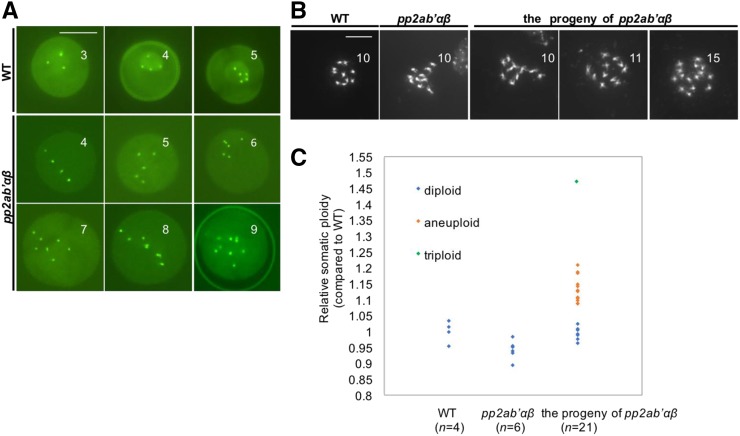
In contrast to the correct distribution of chromosomes in somatic cells, we found that pp2ab′αβ plants produced a substantial proportion of unbalanced tetrads, pointing toward a meiosis-specific chromosome segregation defect. Next, the chromosome number was determined in male gametophytes using the CENCOUNTER marker (De Storme et al., 2016). Uninuclear microspores of wild-type plants typically showed four or five centromeric dots corresponding to the haploid chromosome number in Arabidopsis (2x = 10; Fig. 2A). In contrast to the wild type, microspores of the pp2ab′αβ double mutant had six to nine centromeric signals, indicative of aneuploidy (Fig. 2A). Because of the abundance of aneuploid pollen, we assessed whether these were fertile and gave rise to aneuploid offspring. The double mutant pp2ab′αβ had severely reduced seed set (see above) yet allowed the collection of some seeds for ploidy analysis. Flow cytometry identified seven diploids, 13 aneuploids, and one triploid (Fig. 2, B and C). The aneuploid and triploid plants showed reduced stature and exhibited differences in rosette size and shape, flowering time, and fertility (Supplemental Fig. S4B). The original pp2ab′αβ plants derived from a segregating pp2ab′α−/−β+/− line, on the other hand, all were diploids (n = 15) and developed similarly to wild-type plants, in support of the view that the phenotypes observed in the progeny of pp2ab′αβ are to be attributed to the deviant ploidy status rather than a direct defect in PP2A enzyme activity.
Figure 2.
The pp2ab′αβ double mutant generates aneuploid offspring. A, Analysis of the chromosome number in male gametophytes. Microspores expressing CENH3-GFP typically show four or five centromeres in the wild type, whereas those of the pp2ab′αβ double mutant frequently show more than six centromeres. Bar = 10 µm. B, Chromosome spreads stained by 4′,6-diamidino-2-phenylindole (DAPI) of mitotic cells containing condensed chromosomes of the wild type, the pp2ab′αβ double mutant, and the progeny derived from the pp2ab′αβ double mutant. Bar = 10 µm. C, Relative somatic ploidy of the wild type, the pp2ab′αβ double mutant, and its offspring by DNA flow cytometry.
PP2AB′α and PP2AB′β Are Required for Sister Chromatid Cohesion in Male Meiosis I
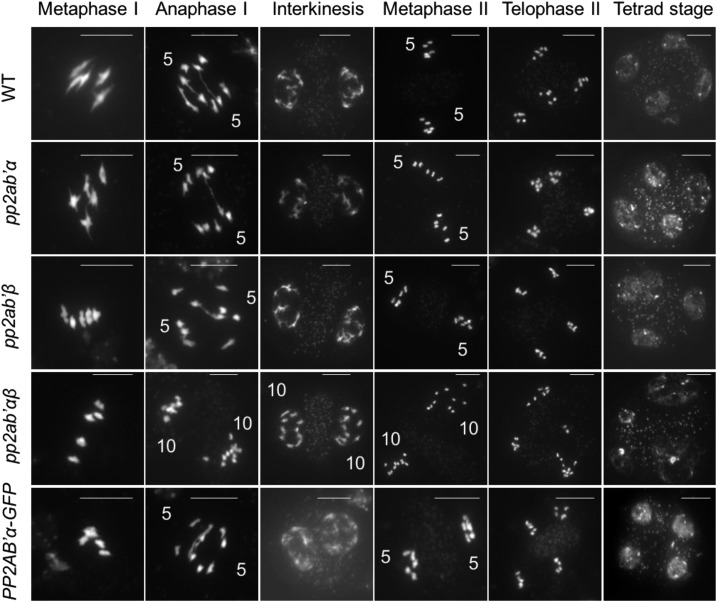
To determine the chromosome behavior underlying the formation of aneuploid microspores, we performed meiotic spread analyses. Key stages of wild-type male meiosis are shown in Figure 3 (top row). At metaphase I, five bivalents align, each composed of two homologous chromosomes connected by one or more crossovers (Fig. 3). Cohesion release along the chromosome arms together with monopolar spindle attachment at metaphase I allows the balanced segregation of homologous chromosomes at anaphase I (Fig. 3). Next, at telophase I, chromosomes decondense and, at interkinesis, form two distinct nuclei (Fig. 3) containing X-shaped chromosomes that reflect the residual cohesion at the centromeric region (Fig. 3). At the start of meiosis II, the two groups of five condensed chromosomes align on two perpendicularly oriented metaphase II plates (Fig. 3). Then, the programmed release of centromeric cohesion at the onset of anaphase II enables the equal segregation of sister chromatids to the opposing poles, leading to the formation of four haploid nuclei (Fig. 3). Meiotic chromosome behavior in pp2ab′αβ is highly similar to that in the wild type until early anaphase I. The formation of chiasmata, bivalent alignment at metaphase I (Fig. 3), and segregation of homologous chromosomes at early anaphase I (Supplemental Fig. S5) are indistinguishable from those in the wild type. Starting from late anaphase I, differences were observed in pp2ab′αβ showing 10 single chromosome units (likely reflecting chromatids) instead of five at each pole, indicating premature loss of centromeric sister chromatid cohesion (Fig. 3). Next, at metaphase II, both sets of single chromatids aligned at the two sides of the organelle band (Fig. 3). As five chromosomes at each pole were not observed at interkinesis or metaphase II, the loss of cohesion was fully penetrant. Finally, during anaphase II, both groups of single chromatids exhibited an erratic segregation pattern leading to microspores with unbalanced (aneuploid) chromosome number (Fig. 3). The chromosome behavior in both pp2ab′α and pp2ab′β single mutants appeared highly similar to that of control lines, in support of the functional redundancy of PP2AB′α and PP2AB′β (Fig. 3).
Figure 3.
Male meiosis in the wild type, pp2ab′α, pp2ab′β, the pp2ab′αβ double mutant, and PP2AB′α-GFP (pp2ab′αβ double mutant expressing PP2AB′α-GFP). Chromosome spreads were stained by DAPI. Numbers at anaphase I indicate the chromosomes counted at each side of the equatorial plate, and numbers at interkinesis and metaphase II indicate the number of chromosomes counted at each side of the organelle band. Bars = 10 µm.
PP2AA1 and PP2AB′α Localize to Chromosomes from Pachytene up to Metaphase II
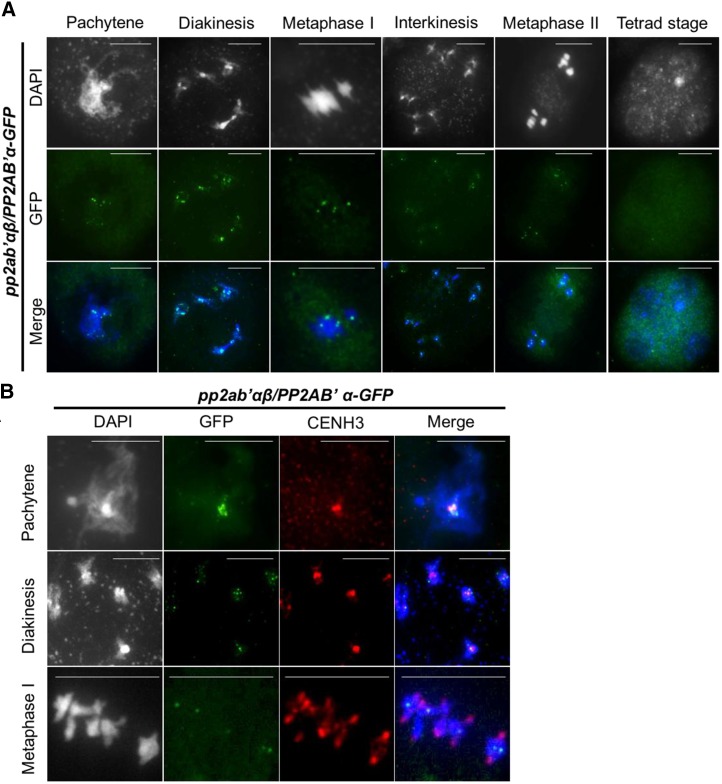
The putative protective role of the PP2A complex for cohesion would require appropriate targeting to chromosomes, which we assessed using GFP-tagged PP2A subunits PP2AA1 and PP2AB′α. Expression of pPP2AB′α:PP2AB′α-GFP restored the fertility of the pp2ab′αβ double mutant (Fig. 1, A–C), as it yielded balanced meiotic products (Fig. 1E) and caused regular chromosome segregation in male meiosis (Fig. 3). The GFP fluorescence in pPP2AB′α:PP2AB′α-GFP-expressing meiocytes was weak. Therefore, we monitored pPP2AB′α:PP2AB′α-GFP by immunostaining using a GFP antibody. A punctate pattern of immunofluorescent spots colocalized with DAPI-stained centromeres of fully paired chromosomes at pachytene and of bivalents at diakinesis (Fig. 4A). At metaphase I, PP2AB′α-GFP was detected at the centers of the five bivalents (Fig. 4A). During later stages (i.e. interkinesis and metaphase II), centromeres also were immunostained, albeit with reduced intensity. PP2AB′α-GFP was no longer detected beyond metaphase II, in line with a role for PP2AB′α in sister chromatid cohesion (Fig. 4A). To test whether PP2AB′α-GFP indeed localized to the centromere, meiocytes were costained with centromeric histone H3 (CENH3) antibody. PP2AB′α-GFP was first observed at the heterochromatin of pachytene chromosomes overlapping with CENH3 immunofluorescence signals (Fig. 4B). At diakinesis, PP2AB′α-GFP concentrated at foci overlapping with the centromeres and also associated with pericentromeric regions (Fig. 4B). During metaphase, PP2AB′α-GFP was concentrated at a single location of each bivalent chromosome between the pulled-apart centromeres of the sister chromosomes. This shows that, when chromosomes are under tension in metaphase I, PP2AB′α-GFP localization is no longer defined by the CENH3 loaded centromeres. We were unable to detect a CENH3 signal above the background after metaphase I in pPP2AB′α:PP2AB′α-GFP-expressing meiocytes. Given that PP2A subunits form a complex to exert phosphatase activity, we immunolocalized the GFP-tagged PP2AA1 structural subunit. PP2AA1-GFP displayed a punctate pattern in male meiocytes from pachytene stage up to metaphase II and no longer was detected in tetrad stage meiocytes (Supplemental Fig. S6). The timing and pattern are almost identical to those of PP2AB′α-GFP, and PP2AA1-GFP also colocalized with CENH3, with some protein detected in regions close to the centromeres (Supplemental Fig. S6). These observations are in support of a function for PP2A-type phosphatase during spindle-mediated dynamics and the segregation of meiotic chromosomes.
Figure 4.
Immunolocalization of PP2AB′α-GFP and centromeres in male meiocytes at different meiotic stages. A, Row 1, DNA stained by DAPI (grayscale); row 2, localization of PP2AB′α-GFP using GFP antibody (green); row 3, merge of DAPI (blue) and GFP (green) immunodetection. Bars = 10 µm. B, Column 1, DNA stained by DAPI (grayscale); column 2, localization of PP2AB′α-GFP using GFP antibody (green); column 3, localization of centromeres using CENH3 antibody (red); column 4, merge of DAPI (blue), GFP (green), and CENH3 immunodetection (red). Bars = 10 µm.
PP2AB′α and PP2AB′β Maintain SYN1 at the Centromeres in Male Meiosis I and II
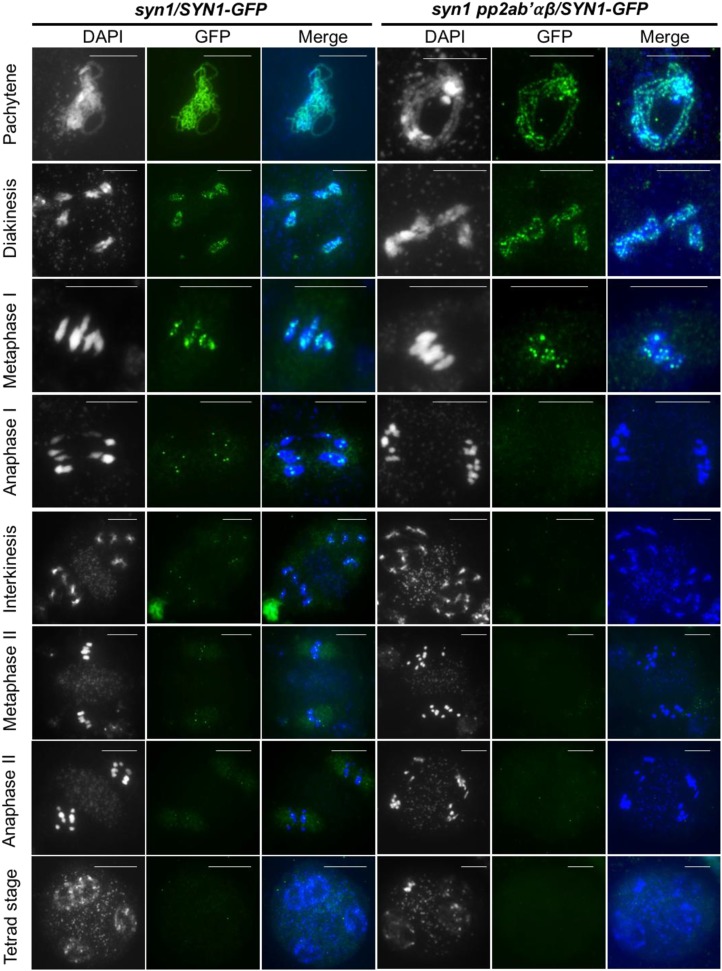
A key question is whether the PP2A complex protects centromeric SYN1 from proteolytic cleavage at the end of meiosis I. To assess the stability of the SYN1 protein throughout meiosis, we performed immunolocalization experiments monitoring SYN1-GFP localization in meiocytes from wild-type and pp2ab′αβ plants (Fig. 5). Heretofore, we used a syn1 mutant complemented with a construct delivering a SYN1-GFP fusion protein. SYN1 was then visualized with an antibody against GFP and the concomitant staining of DNA by DAPI in spreads of male meiotic chromosomes. Introgression of the pSYN1:SYN1-GFP construct in the pp2ab′αβ background was achieved by intercrossing the syn1 pSYN1:SYN1-GFP line with the pp2ab′α−/−β+/− line and recovering triple homozygous syn1 pp2ab′αβ plants that additionally contained the pSYN1:SYN1-GFP construct.
Figure 5.
Immunolocalization of SYN1-GFP in male meiocytes at different meiotic stages. The construct pSYN1:SYN1-GFP was introduced into the triple mutant pp2ab′αβ syn1, and SYN1-GFP protein was detected using GFP antibody. Columns 1 and 4, DNA stained by DAPI (grayscale); columns 2 and 5, localization of SYN1-GFP using GFP antibody (green); columns 3 and 6, merge of DAPI (blue) and GFP immunodetection (green). Bars = 10 µm.
In male meiocytes of syn1 plants, SYN1-GFP localized at dots spread along the whole pachytene and diakinesis chromosomes (Fig. 5). Immunostaining of metaphase I meiocytes showed fewer dots along the bivalents. Later, during anaphase I, the chromosome arms were no longer labeled, and 10 distinct SYN1-GFP foci corresponding to centromeres were observed. These observations are in agreement with a gradual loss of cohesion along the chromosome arms and the maintenance of cohesion at the centromeric region of metaphase I and anaphase I chromosomes (Fig. 5). Until metaphase II and during anaphase II, SYN1-GFP localized specifically at the centromeric domains of all chromosomes (Fig. 5). The intensity of SYN1-GFP foci decreased strongly at telophase II and was no longer observed at the tetrad stage (Fig. 5). The SYN1-GFP immunolocalization pattern in the syn1 pp2ab′αβ background was highly similar to that of the wild type up to metaphase I. The SYN1-GFP foci were no longer present in anaphase I chromosomes and also were not observed in any of the later meiotic stages. The loading of SYN1-GFP to the chromosomes and selective maintenance at the centromeric region appeared not to be affected in the pp2ab′αβ mutant. These observations support a role for the PP2A complex in preventing the precocious release of SYN1-mediated cohesins during meiosis I and II, thereby ensuring the cohesion of the sister chromatids up to metaphase II.
Plant Growth and Mitotic Chromosome Segregation Appear Normal in the pp2ab′αβ Double Mutant
Because pp2ab′αβ double mutants produced mainly aborted seeds (Fig. 1, A and B), we performed phenotypic analysis using seeds harvested from a mutant line that was genotyped as homozygous for the T-DNA insertion in the pp2ab′α gene and heterozygous for the T-DNA in the pp2ab′β gene. Progeny genotyped as pp2ab′αβ double mutant developed normally and started to flower at about the same time as wild-type plants (Supplemental Fig. S4A). Upon extended cultivation of the pp2ab′αβ double mutant plants, however, many more axillary branches were observed than in wild-type plants (Supplemental Fig. S4C). This excessive branching is typical for sterile and partially sterile plants (Hensel et al., 1994; Noodén and Penney, 2001; Wuest et al., 2016) and also occurred in the meiotic recombination mutant spo11 and the aborted microspores mutant cultivated alongside the pp2ab′αβ double mutant (Supplemental Fig. S4C).
To further investigate putative somatic phenotypes of the pp2ab′αβ double mutant, we examined the primary root growth and hypocotyl elongation of progeny derived from a pp2ab′α−/−β+/− plant. The average primary root length of 1-week-old pp2ab′α single and pp2ab′αβ double mutants was indistinguishable from that of the wild type (Supplemental Fig. S7A). Hypocotyl lengths were determined from plants grown in the dark. Seeds from wild-type, pp2ab′α, pp2ab′β, pp2ab′αβ, and pp2ab′α−/−β+/− mother plants were incubated in the dark, and the hypocotyls were measured after 6 d. The hypocotyl lengths were distributed nonuniformly, with more plants having short hypocotyls (less than 1 cm) found in the progeny of pp2ab′αβ (23.5%) than in the wild type (5.6%), pp2ab′α (2.6%), pp2ab′β (0%), and pp2ab′α−/−β+/− (4.8%). For simplicity, we genotyped 83 plants from a total of 189 plants derived from pp2ab′α−/−β+/−. In the end, 21 plants (about 25%) were identified as pp2ab′αβ (Supplemental Fig. S7D). The elongated hypocotyls of these plants were, on average, the same as those of wild-type plants, indicating that the short-hypocotyl growth phenotype did not correlate with the pp2ab′αβ genotype.
To investigate whether the pp2ab′αβ double mutant causes subtle defects in somatic divisions that would not necessarily cause an obvious alteration of organ size, we analyzed chromosome segregation in somatic cells of pp2ab′α−/−β+/− descendants using the centromeric marker CENCOUNTER (Supplemental Fig. S7C; De Storme et al., 2016). In wild-type petal cells, we typically observed 10 signals corresponding to the diploid number of chromosomes in somatic cells (40.9%; n = 308). In the other 59.1% of the cells, we detected fewer chromosomes (e.g. between seven and nine), most likely due to centromere colocalization or signal overlap. Cells with more than 10 centromeric signals were not observed. In pp2ab′αβ petal cells, we counted predominantly 10 centromeres (42.7%; n = 213) or fewer (47.4% between seven and nine). Cells containing more than 10 centromeric signals were not observed (Supplemental Fig. S7B). These findings indicate that PP2AB′α and PP2AB′β are not required for chromosome segregation in mitotic cells and the maintenance of ploidy in somatic cell types.
DISCUSSION
In this study, a specific role of PP2AB′ cofactors α and β in Arabidopsis chromosome cohesion during meiosis is described. The inactivity of either AtPP2AB′α or AtPP2AB′β alone does not have a noticeable effect on meiotic chromosome segregation, indicating that these proteins are redundant and fully complement each other’s function during meiosis. Moreover, we found no evidence for a role of these subunits in mitotic cell divisions and organ growth.
Lack of Evidence for a Role of PP2AB′α and PP2AB′β in Somatic Growth and Development
The role of PP2A phosphatases has been associated with physiological and developmental processes, including auxin transport, stress response, phototropism, and lateral root initiation (Janssens and Goris, 2001; Rashotte et al., 2001; Michniewicz et al., 2007; Tseng and Briggs, 2010). Since the structural core subunit PP2AA1 is essential for activity and expressed ubiquitously, the diversification of PP2A phosphatase activity requires other subunits. The B, B′, and B′′ subunits mediate target specificity by cell type-specific expression and subcellular targeting (Janssens and Goris, 2001). The Arabidopsis genome encodes nine PP2AB′ isoforms, yet only the α and β variants appear to be essential for regulating meiotic chromosome segregation, supporting the tissue-specific functional divergence of PP2AB′ regulatory subunits (Wang et al., 2016). PP2AB′α and PP2AB′β are expressed predominantly in anther tissue and, albeit to a lesser degree, in sporophytic tissues. Immunolocalization of pPP2AB′α:PP2AB′α-GFP corroborates expression in male meiocytes as well as in sporophytic tissues, pointing to a role in vegetative as well as reproductive development. Based on the expression profile and earlier functional studies, we expected defects in growth and development during the vegetative growth phase of young pp2ab′αβ double mutants. Surprisingly, plants derived from seeds from a pPP2AB′α−/− parent segregating for pPP2AB′β-/+ were morphologically and developmentally not different from wild-type plants apart from excessive branching and flowering, which we attribute to semisterility and the subsequent poor seed filling and reduced sink formation (Hensel et al., 1994; Noodén and Penney, 2001; Wuest et al., 2016). Whereas dwarfism and other developmental defects were reported for pp2ab′αβ double mutants (Tang et al., 2011), we did not observe these phenotypes in the original mutants. The descendants from the original mutants, however, did show a range of morphological and developmental defects, including dwarfism. The phenotypes of the progeny of the double mutant were similar to those of the progeny of triploids and of sexual reproduction mutants that are known to generate aneuploid offspring (Huettel et al., 2008; Henry et al., 2010; Jonassen et al., 2011; Tang et al., 2011). The cytological analysis of centromere-labeled cells corroborated the production of aneuploid offspring from self-fertilized pp2ab′αβ double mutants. Therefore, the various defects of the offspring of pp2ab′αβ double mutants were likely the result of aneuploidy rather than a direct consequence of the loss of PP2A functionality in sporophytic tissue.
PP2A Protects the Cohesin SYN1 Subunit at Centromeres during Meiosis I and Meiosis II
Meiosis-specific orthologs of REC8 have been identified in Arabidopsis (SYN1/DIF1/AtREC8), rice (Oryza sativa; OsRAD21-4), and maize (Zea mays; ZmREC8/AFD1), and their function as a meiosis-specific α-kleisin cohesin subunit was confirmed (Bai et al., 1999; Bhatt et al., 1999; Chelysheva et al., 2005; Golubovskaya et al., 2006; Zhang et al., 2006). Immunolocalization of SYN1 protein using commercial and custom-made antibodies has revealed some differences in localization in different studies and is not fully consistent with what has been reported in animal and yeast systems. Here, we used GFP antibody to detect a SYN1-GFP chimeric protein, which has been shown to restore the fertility of syn1 mutants. Immunostaining assays revealed strong and specific SYN1-GFP signals along the whole length of chromosomes in prophase I and metaphase I, whereas SYN1-GFP localized specifically at the centromeres during anaphase I. In male meiosis of rice, OsREC8 also was found to localize across the chromosome arms from interphase until metaphase I, whereas no fluorescence signals were retrieved in later meiotic stages (Shao et al., 2011). This may result from either different spatiotemporal patterns of cohesin targeting during the meiosis I-meiosis II progression or because of structural modification changing the antibody-binding capacity. The cohesin subunit REC8 indeed undergoes multiple phosphorylations that are geared to promoting its degradation by the protease ESP (Ishiguro et al., 2010). The phosphorylation is countered by PP2A phosphatase to ensure residual cohesion at the centromere (Kitajima et al., 2006; Riedel et al., 2006; Ishiguro et al., 2010). Similar to other eukaryotes, PP2A may protect centromeric cohesion by the dephosphorylation of SYN1 and possibly other cohesion components. REC8 cleavage at anaphase I depends on its phosphorylation by a highly conserved casein kinase1 Hrr25 (CK1δ) or Dbf4-dependent Cdc7 kinase (Ishiguro et al., 2010; Katis et al., 2010). The orthologous function in budding yeast is executed by polo kinase CDC5 (Attner et al., 2013). In plants, evidence for SYN1 phosphorylation is missing, and further studies are required to put forward a candidate kinase responsible for sensitizing SYN1 for degradation.
PP2A Is Not Required for Kinetochore Monoorientation
In addition to its role in the cohesion of sister chromatids, SYN1 plays a role in establishing monopolar attachment at meiosis I (Chelysheva et al., 2005). This raises the question of whether PP2A also would be required for controlling this function of SYN1. In the pp2ab′αβ double mutant, bivalents nicely align at metaphase I in a single plane and chromosomes are pulled to the opposite poles, creating two equivalent euploid chromosome sets. Because in the pp2ab′αβ double mutant the chromosomes dissociate prematurely at anaphase I, sets of 10 chromatids appear at the poles, indicating a monopolar orientation of the sister kinetochores in the pp2ab′αβ double mutant in meiosis I. Thus, PP2A is not required for monoorientation and becomes active after anaphase onset. Likewise, SGO is not essential for monoorientation but is required for sister chromatid cohesion during anaphase I (Zamariola et al., 2013). In both sgo1 and the pp2ab′αβ double mutant, the pairing of homologous chromosomes, chromosome condensation, and alignment along the metaphase plate are the same as in the wild type. SGO1 and PP2AB′αβ, therefore, seem to be specifically critical for the transition from metaphase I to anaphase I. Unfortunately, attempts to localize SGO1 with an antibody targeted against mammalian SGO1 or by expressing N- and C-terminal fusions of Arabidopsis SGO1 with GFP were not successful, leaving us to speculate that SGO1 and AtPP2AB′αβ colocalize at centromeres. The SGO ortholog OsSGO1 in rice has been localized to the pericentromeric region, which does not fully overlap with the functional kinetochore labeled with CENH3 (Wang et al., 2011). During metaphase, early anaphase of the first meiotic division, we observed that PP2AB′α-GFP also does not colocalize with the CENH3-labeled kinetochore, suggesting that the putative Arabidopsis SGO1-PP2A complex is targeted to the pericentromeric region of the chromosomes.
GFP-tagged PP2AB′α in Arabidopsis, ZmSGO1, and OsSGO1 are detected in early prophase (i.e. on pachytene and diakinesis chromosomes). Thus, these proteins appear to be targeted to the (peri)centromeres prior to the moment when they are required (i.e. at the beginning of anaphase I; Hamant et al., 2005; Wang et al., 2011; Zamariola et al., 2013). The GFP-tagged A1 structural subunit was found to associate with the metaphase I chromosomes, suggesting that formation of the functional PP2A complex occurs during metaphase I at the moment that its activity matters.
Other Cohesion Components Putatively Dephosphorylated by PP2A
Arabidopsis harbors two redundant wings apart-like protein (WAPL) helicases that are required for opening the cohesion ring during meiosis I along the chromosome arms (De et al., 2014). In a WAPL mutant background, SYN1 is no longer removed from the prophase I chromosomes, indicating that WAPL activates SYN1 removal and instigates the onset of cohesion loss. This meiosis-specific function of WAPL in Arabidopsis contrasts with the regulation of REC8 in other eukaryotes, where WAPL is responsible for activating the selective degradation of the mitotic kleisin subunit RAD21 and not REC8. An additional specialization of plant meiosis is suggested by the discovery of the plant-specific cohesion factor PATRONUS (PANS). PANS plays a role in the close association of chromatids during meiosis I and is critically required for cohesion during the interphase between meiosis I and II up to anaphase II (Cromer et al., 2013; Zamariola et al., 2014; Singh et al., 2015). PANS and SGO are required to maintain SYN1 localization at metaphase II chromatids and, hence, secure cohesion, particularly during metaphase II (Cromer et al., 2013; Zamariola et al., 2014; Singh et al., 2015). PANS operates as an inhibitor of the anaphase-promoting complex or cyclosome (APC/C), linking the maintenance of centromeric cohesion with meiotic cell cycle progression (Singh et al., 2015). A current hypothesis is that either the stability or centromeric loading of SYN1-protecting PP2A-SGO complexes at the onset of anaphase II is reduced by the activation of the APC/C and that PANS inhibits APC/C activation from metaphase I to metaphase II. Alternatively, PANS-directed control of the meiotic cell cycle may include the proper regulation of centromeric properties and, hence, indirectly determine spatiotemporal patterns of centromeric cohesion. Although evidence in yeast supports the former hypothesis (Miller et al., 2012), more experiments are needed to fully elucidate the molecular mechanism(s) controlling the loading, stability, and removal of centromeric cohesion during plant meiosis I and II.
Taken together, our analyses reveal that PP2AB′α and PP2AB′β are redundantly required for the protection of centromeric sister chromatid cohesion in Arabidopsis male meiosis I and II, likely by protecting the cohesin REC8 α-kleisin subunit from cleavage. Based on the similarity with other eukaryotic organisms, it is further presumed that Arabidopsis PP2AB′α and PP2AB′β localize specifically to the (peri)centromere from meiosis I to meiosis II through its binding with SGO1, where it reverses the phosphorylated status of SYN1. In conclusion, PP2AB′α and PP2AB′β constitute an essential part of the molecular machinery that controls reductional cell division in meiosis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used in the study. The plant material in all experiments was in the Columbia-0. Arabidopsis seeds were germinated on K1 medium and grown for 6 to 8 days under 100 to 150 µmol m−2 s−1 fluorescent warm white light, 12 h of light/12 h of dark, 20°C, and 70% humidity. Then, seedlings were transferred to soil and cultivated in controlled-climate chambers with similar conditions. Metric analyses of root and hypocotyl were conducted with seed batches from all lines harvested at the same time. Plants were grown on vertical agar plates (12 × 12 cm) containing K1 medium. Images were analyzed in ImageJ.
Isolation of PP2A Mutants
pp2ab′α corresponds to MIPS AT5G03470 and pp2ab′β to MIPS AT3G09880. T-DNA insertions in AT5G03470 (SALK_077700) and AT3G09880 (SALK_103167) were from the Nottingham Arabidopsis Stock Centre. Plants were genotyped using primers SALK_077700LP (5′-TCGTGTCTTTGGTTCTGATTTG-3′), SALK_077700RP (5′-AAGGGCCTGTGAACCATAAAC-3′), LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′), SALK_103167LP (5′-TTGCGGATACAGTAATCAGGG-3′), and SALK_103167RP (5′-TTCTTCTCCTCCTCCTCCATC-3′). Confirmed homozygous single-insertion mutants were crossed with each other to generate pp2ab′αβ double mutants. Transcriptional levels of PP2AB′α and PP2AB′β have been analyzed before (Jonassen et al., 2011; Tang et al., 2011).
Molecular Cloning and Generation of Transgenic Arabidopsis
The full-length open reading frame of PP2AB′α without a stop codon was cloned into pDONR221 vector (Invitrogen) and subcloned into the Gateway-compatible pH7m34GW to generate C-terminal fusions with GFP. The full-length open reading frame of SYN1 without a stop codon was cloned into pENTR vector and subcloned into the Gateway-compatible pGWB501 to generate C-terminal fusions with GFP. The constructs were introduced into Agrobacterium tumefaciens strain GV3101 and used for the transformation of Arabidopsis by the floral dip method (Clough and Bent, 1998).
The CENH3-GFP-expressing lines were described previously (De Storme et al., 2016). The pPP2AA1:PP2AA1-GFP-expressing line was supplied by M. Michniewicz (Michniewicz et al., 2007).
Phenotypic Analysis
Immature flower buds were squashed in a solution of 45% (v/v) lactopropionic orcein solution, and tetrads were observed under bright-field conditions in an Olympus IX81 inverted fluorescence microscope equipped with an X-Cite Series 120Q UV lamp and an Olympus XM10 camera.
DNA Flow Cytometry
The somatic ploidy level was determined using DNA flow cytometry (Epics Altra; Beckman) using the Galbraith extraction method (Galbraith et al., 1983). The sample preparation was performed as described previously (De Storme and Geelen, 2011).
Meiotic Chromosome Spreads and Immunofluorescence
Chromosome spreads were essentially as described (Armstrong et al., 2009). Slides were stained with 10 μL of a 2 μg mL−1 solution of DAPI in Vectashield antifade mounting medium (Vector Laboratories). Immunofluorescence was performed using the microwave technique (Chelysheva et al., 2010). The anti-CENH3 antibody was used as described diluted 1:400 (Talbert et al., 2002). The anti-GFP antibody was from Vector Laboratories and was used at 1:100.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NC_003076.8 (SYN1), NC_003076.8 (PP2AB′α), NC_003074.8 (PP2AB′β), and NC_003070.9 (PP2AA1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. B′α and B′β are expressed predominantly in flower anthers.
Supplemental Figure S2. Microscopy of complete anthers.
Supplemental Figure S3. Quantification of seeds per silique.
Supplemental Figure S4. Morphology at the flowering stage.
Supplemental Figure S5. Male meiosis in the pp2ab′αβ double mutant.
Supplemental Figure S6. Immunolocalization of PP2AA1-GFP in male meiocytes.
Supplemental Figure S7. pp2ab′αβ double mutant plants show regular root growth and ploidy.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Footnotes
G.Y. was awarded China Scholarship Council Grant 201406320206, and N.D.S. was supported by the Fund for Scientific Research (FWO-Flanders, FWO16/PDOH1/049).
Articles can be viewed without a subscription.
References
- Armstrong SJ, Sanchez-Moran E, Chris F, Franklin H (2009) Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol Biol 558: 131–145 [DOI] [PubMed] [Google Scholar]
- Attner MA, Miller MP, Ee LS, Elkin SK, Amon A (2013) Polo kinase Cdc5 is a central regulator of meiosis I. Proc Natl Acad Sci USA 110: 14278–14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Peirson BN, Dong F, Xue C, Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19: 463–472 [DOI] [PubMed] [Google Scholar]
- Bolaños-Villegas P, De K, Pradillo M, Liu D, Makaroff CA (2017) In favor of establishment: regulation of chromatid cohesion in plants. Front Plant Sci 8: 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C, et al. (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118: 4621–4632 [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Grandont L, Vrielynck N, le Guin S, Mercier R, Grelon M (2010) An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res 129: 143–153 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K (2003) Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol 5: 480–485 [DOI] [PubMed] [Google Scholar]
- Cromer L, Jolivet S, Horlow C, Chelysheva L, Heyman J, De Jaeger G, Koncz C, De Veylder L, Mercier R (2013) Centromeric cohesion is protected twice at meiosis, by SHUGOSHINs at anaphase I and by PATRONUS at interkinesis. Curr Biol 23: 2090–2099 [DOI] [PubMed] [Google Scholar]
- De K, Sterle L, Krueger L, Yang X, Makaroff CA (2014) Arabidopsis thaliana WAPL is essential for the prophase removal of cohesin during meiosis. PLoS Genet 10: e1004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Geelen D (2011) The Arabidopsis mutant jason produces unreduced first division restitution male gametes through a parallel/fused spindle mechanism in meiosis II. Plant Physiol 155: 1403–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Keçeli BN, Zamariola L, Angenon G, Geelen D (2016) CENH3-GFP: a visual marker for gametophytic and somatic ploidy determination in Arabidopsis thaliana. BMC Plant Biol 16: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durian G, Rahikainen M, Alegre S, Brosché M, Kangasjärvi S (2016) Protein phosphatase 2A in the regulatory network underlying biotic stress resistance in plants. Front Plant Sci 7: 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049. [DOI] [PubMed] [Google Scholar]
- Golubovskaya IN, Hamant O, Timofejeva L, Wang CJR, Braun D, Meeley R, Cande WZ (2006) Alleles of afd1 dissect REC8 functions during meiotic prophase I. J Cell Sci 119: 3306–3315 [DOI] [PubMed] [Google Scholar]
- Hamant O, Golubovskaya I, Meeley R, Fiume E, Timofejeva L, Schleiffer A, Nasmyth K, Cande WZ (2005) A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr Biol 15: 948–954 [DOI] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM (2001) Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293: 1320–1323 [DOI] [PubMed] [Google Scholar]
- Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L (2010) Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics 186: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB (1994) The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol 106: 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kreil DP, Matzke M, Matzke AJM (2008) Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet 4: e1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K, Watanabe Y (2007) Chromosome cohesion in mitosis and meiosis. J Cell Sci 120: 367–369 [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Tanaka K, Sakuno T, Watanabe Y (2010) Shugoshin-PP2A counteracts casein-kinase-1-dependent cleavage of Rec8 by separase. Nat Cell Biol 12: 500–506 [DOI] [PubMed] [Google Scholar]
- Janssens V, Goris J (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353: 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen EM, Heidari B, Nemie-Feyissa D, Matre P, Lillo C (2011) Protein phosphatase 2A regulatory subunits are starting to reveal their functions in plant metabolism and development. Plant Signal Behav 6: 1216–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Lipp JJ, Imre R, Bogdanova A, Okaz E, Habermann B, Mechtler K, Nasmyth K, Zachariae W (2010) Rec8 phosphorylation by casein kinase 1 and Cdc7-Dbf4 kinase regulates cohesin cleavage by separase during meiosis. Dev Cell 18: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y (2006) Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52 [DOI] [PubMed] [Google Scholar]
- Latorre KA, Harris DM, Rundle SJ (1997) Differential expression of three Arabidopsis genes encoding the B′ regulatory subunit of protein phosphatase 2A. Eur J Biochem 245: 156–163 [DOI] [PubMed] [Google Scholar]
- Marston AL. (2014) Chromosome segregation in budding yeast: sister chromatid cohesion and related mechanisms. Genetics 196: 31–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Amon A (2004) Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol 5: 983–997 [DOI] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Miller MP, Ünal E, Brar GA, Amon A (2012) Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. eLife 1: e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Noodén LD, Penney JP (2001) Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae). J Exp Bot 52: 2151–2159 [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J (2008) The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, DeLong A, Muday GK (2001) Genetic and chemical reductions in protein phosphatase activity alter auxin transport, gravity response, and lateral root growth. Plant Cell 13: 1683–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Gálová M, Petronczki M, Gregan J, Cetin B, et al. (2006) Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441: 53–61 [DOI] [PubMed] [Google Scholar]
- Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, Ammerer G, Mechtler K, Gregan J (2010) Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle 9: 2657–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao T, Tang D, Wang K, Wang M, Che L, Qin B, Yu H, Li M, Gu M, Cheng Z (2011) OsREC8 is essential for chromatid cohesion and metaphase I monopolar orientation in rice meiosis. Plant Physiol 156: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, Spillane C, Siddiqi I (2015) PATRONUS1 is expressed in meiotic prophase I to regulate centromeric cohesion in Arabidopsis and shows synthetic lethality with OSD1. BMC Plant Biol 15: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS, et al. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TS, Briggs WR (2010) The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell 22: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Wang M, Tang D, Wang K, Shen Y, Qin B, Miao C, Li M, Cheng Z (2011) OsSGO1 maintains synaptonemal complex stabilization in addition to protecting centromeric cohesion during rice meiosis. Plant J 67: 583–594 [DOI] [PubMed] [Google Scholar]
- Wang R, Liu M, Yuan M, Oses-Prieto JA, Cai X, Sun Y, Burlingame AL, Wang ZY, Tang W (2016) The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B′ subunits. Mol Plant 9: 148–157 [DOI] [PubMed] [Google Scholar]
- Wuest SE, Philipp MA, Guthörl D, Schmid B, Grossniklaus U (2016) Seed production affects maternal growth and senescence in Arabidopsis. Plant Physiol 171: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue K, Sandal P, Williams EL, Murphy E, Stes E, Nikonorova N, Ramakrishna P, Czyzewicz N, Montero-Morales L, Kumpf R, et al. (2016) PP2A-3 interacts with ACR4 and regulates formative cell division in the Arabidopsis root. Proc Natl Acad Sci USA 113: 1447–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamariola L, De Storme N, Tiang CL, Armstrong SJ, Franklin FCH, Geelen D (2013) SGO1 but not SGO2 is required for maintenance of centromere cohesion in Arabidopsis thaliana meiosis. Plant Reprod 26: 197–208 [DOI] [PubMed] [Google Scholar]
- Zamariola L, De Storme N, Vannerum K, Vandepoele K, Armstrong SJ, Franklin FCH, Geelen D (2014) SHUGOSHINs and PATRONUS protect meiotic centromere cohesion in Arabidopsis thaliana. Plant J 77: 782–794 [DOI] [PubMed] [Google Scholar]
- Zhang L, Tao J, Wang S, Chong K, Wang T (2006) The rice OsRad21-4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol Biol 60: 533–554 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N (1999) Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754 [DOI] [PubMed] [Google Scholar]