Dear Editor,
The PHYTOCHROME B (PHYB) gene, encoding the apoprotein of the major red/far-red photoreceptor phyB, has been one of the most extensively studied genes in Arabidopsis (Arabidopsis thaliana) for the past 2 decades (Casal, 2013). During these research efforts, the phyB-9 allele (formerly named hy3-EMS142) has played a key role in molecular genetic analyses as the favorite choice among phyB loss-of-function mutants. Its genetic background is Col-0, that it harbors an EMS-induced point mutation that facilitates facilitating further crosses or transgenic modifications, and that this mutation causes the complete disruption of phyB function by the presence of a premature stop codon in the chromophore-binding GAF domain(Reed et al., 1993).
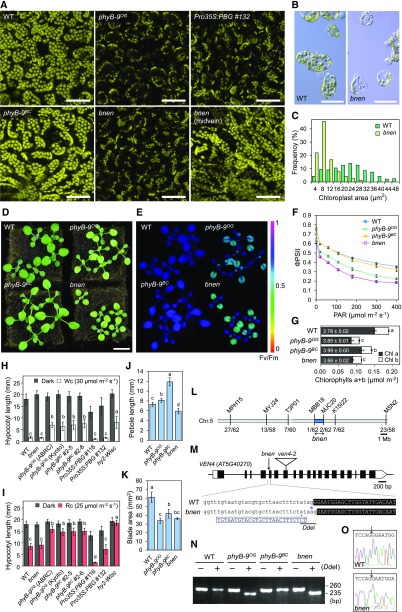
Leaves of phyB-9 plants are yellowish green, seemingly consistent with the positive regulatory role of phyB in chlorophyll biosynthesis (Reed et al., 1993). We found that the subepidermal layer of phyB-9 leaf palisade cells contains chloroplasts reduced in size, which did not fully undercoat the paradermal, apical surface of these cells (Fig. 1A). Such alteration of chloroplast size, however, was not rescued by constitutive expression of a functional PHYB-GFP fusion construct, suggesting that this phenotype was not caused by the loss of phyB function (Fig. 1A). We backcrossed phyB-9 to the wild type and found that the yellowish leaf color phenotype segregated independently from the long hypocotyl caused by phyB. Therefore, we concluded that the original phyB-9 stock carries a second-site mutation that enhances the leaf color phenotype, which we named phyB-nine-enhancer (bnen).
Figure 1.
The original stock of phyB-9 carries the bnen allele of the VEN4 gene. A, Chlorophyll autofluorescence of palisade layer cells. Adaxial side of the first-node foliage leaves at 14 d after sowing were observed with a confocal laser scanning microscope. Maximum intensity projection images were generated from optical sections of 1-μm intervals. Pro-35S:PBG, 35S promoter-driven PHYB-GFP in phyB-9OG background; WT, wild type. Scale bars = 50 μm. B, Light micrographs of macerated mesophyll cells isolated from wild-type and bnen leaves. Scale bars = 50 μm. C, Size distribution of chloroplasts in the palisade layer cells of the wild type and bnen. D, Seedlings (13 d after sowing) grown under long-day (16 h light/8 h dark) condition. Scale bar = 1 cm. E, Pseudocolor image showing the maximum quantum yield of photosystem II (Fv/Fm). F, Light response of the effective quantum yield of photosystem II (ΦPSII). Average ± sd (n = 5). G, Quantification of total chlorophyll extracted from first-node foliage leaves. White numbers indicate chlorophyll a/b ratio. Average ± sd (n = 3). Different letters indicate statistically significant difference (Tukey HSD multiple comparison test, P < 0.05). H and I, Hypocotyl length of seedlings (7 d after sowing) grown on agar plates under constant white light (Wc; H) or constant red light (Rc; I) in comparison to darkness control. Average ± sd (n = 32∼40). Different letters indicate statistically significant difference (Tukey HSD multiple comparison test, P < 0.05). J and K, Leaf size measurement. Fully expanded first-node foliage leaves were harvested at 30 d after sowing to measure petiole length (J) and blade area (K). Average ± sd (n = 4–10). Different letters indicate statistically significant difference (Tukey HSD multiple comparison test, P < 0.05). L, Mapping of bnen. SSLP markers on chromosome 5 and recombination ratio are shown. M, Structure of the VEN4 (AT5G40270) locus and the DNA sequence around the bnen mutation site. Black boxes and white boxes indicate coding sequences and untranslated regions, respectively. Triangle indicates the position of T-DNA insertion in the ven4-2 allele. A single nucleotide substitution found in bnen (asterisk) disrupts the splicing acceptor site of the intron 4, which shifts the first nucleotide of the exon 5. A 26-nt fragment below the bnen sequence is the dCAPS forward primer (YY1102) with a single mismatch (dot) to create a DdeI site only in the bnen allele. N, Detection of bnen mutation by the dCAPS marker. See also Supplemental Figure S2. O, Sequencing electropherograms of bulk RT-PCR products of VEN4 cDNA from the wild type and bnen. Arrows indicate the junction between exons 4 and 5.
We established the purified phyB-9 and bnen single mutant lines after two rounds of backcrossing. Hereafter, phyB-9OG designates the original strain containing the bnen mutation, and phyB-9BC designates the genuine phyB-9 single mutant. Leaf palisade cells of phyB-9BC had chloroplasts comparable to those of the wild type in size and distribution, while those of bnen were smaller and scattered as in phyB-9OG (Fig. 1, A and C). This phenotype also was confirmed by observing macerated leaf mesophyll cells under a light microscope (Fig. 1B). Interestingly, not all mesophyll cells of bnen showed the chloroplast size defect, exceptions being the perivascular cells adjacent to the midvein (Fig. 1A), similar to a previously characterized reticulate mutant, dov1 (Kinsman and Pyke, 1998). In addition, immature leaves were reticulate and pale, but a dark green color was progressively restored in expanded leaves, similar to that described for other reticulate mutants (for review, see Lundquist et al., 2014). Although the main scope of this letter is to validate the integrity of phyB-9, the molecular function of the gene mutated in bnen (VEN4; see below) is of significant interest for studies on chloroplast size regulation.
Leaf color of the phyB-9BC single mutant was darker than that of phyB-9OG, which was consistent with the restoration of chloroplast size and distribution in palisade cells (Fig. 1D). We tested whether such difference in leaf color had any consequence in photosynthetic efficiency, using an Imaging-PAM fluorometer (Walz). The phyB-9OG line and the bnen mutant showed significantly lower Fv/Fm value than the wild type or the phyB-9BC single mutant (Fig. 1E), indicating the occurrence of PSII photoinhibition. In addition, the quantum yield of PSII (ΦPSII) in phyB-9OG and the bnen mutant was lower than that of the wild type and the phyB-9BC single mutant at any light intensity (Fig. 1F). Because phyB is known to promote light-dependent chlorophyll biosynthesis, we also measured the total chlorophyll content in foliage leaves. This confirmed reduced chlorophyll levels in phyB-9BC compared with the wild type, but phyB-9OG showed further reduction in chlorophyll content, as expected from its rudimentary chloroplasts (Fig. 1G). The presence or absence of PHYB did not further affect low chlorophyll levels in the bnen mutation background. Taken together, these results indicate that the photosynthetic defects of phyB-9OG are largely attributable to the bnen mutation.
Loss of phyB results in longer hypocotyl than the wild type under white or red light, and this visible phenotype has been considered useful to assess phyB activity (Chory et al., 1989). No significant differences in hypocotyl length were observed between phyB-9OG and phyB-9BC under either white light or red monochromatic light, indicating that the bnen mutation does not affect this well-studied trait of phyB, at least in the fluence rate examined (Fig. 1, H and I).
In Arabidopsis, leaf blade and petiole are known to show opposite growth responses to light intensity in a phyB-dependent manner. Namely, light-activated phyB promotes leaf blade expansion but represses petiole elongation in wild-type plants. Loss of phyB, in turn, represses leaf blade expansion and promotes petiole elongation, causing a constitutive shade avoidance response (Tsukaya et al., 2002). We measured the size of foliage leaves in each genotype and confirmed that the bnen mutation represses both leaf blade expansion and petiole elongation, regardless of the PHYB genotype (Fig. 1, D, J, and K). Therefore, the leaf growth phenotypes observed in earlier studies using the original phyB-9OG line were in fact altered by the bnen mutation.
The bnen mutation was mapped to a 1.38-Mb candidate interval at the bottom arm of chromosome 5 (Fig. 1L). In an independent screen (Berná et al., 1999), we isolated several venosa4 (ven4) alleles that cause a reticulate phenotype similar to that caused by bnen, and we identified AT5G40270 as the VEN4 gene. VEN4 mapped within the bnen interval and, hence, was considered a good candidate gene (R. Sarmiento-Mañús, M.R. Ponce, and J.L. Micol, unpublished data). F1 plants from a bnen × ven4-2 cross showed noncomplementation, revealing that bnen is a novel allele of VEN4 (Supplemental Fig. S1). VEN4 gene sequence in bnen had a G-to-A transition disrupting the splicing acceptor site of its intron 4 (Fig. 1M). We designed a dCAPS marker to detect the bnen mutation by PCR amplification followed by DdeI digestion (Supplemental Methods S1). We confirmed that phyB-9BC lacks the bnen mutation (Fig. 1N). To determine the effect of bnen mutation on VEN4 pre-mRNA splicing, we sequenced the corresponding RT-PCR products and confirmed the presence of more than one type of transcript. The major mRNA variant was misspliced by moving the splicing acceptor site one nucleotide, resulting in a frameshift whose translation generates a truncated protein of 111 amino acids, the 11 last residues being different from those of the wild-type VEN4 protein (473 aa; Fig. 1O). A detailed characterization of VEN4 gene function will be described elsewhere (R. Sarmiento-Mañús, unpublished data).
What is the origin of bnen? We obtained the original phyB-9 seeds from two sources, both of which independently maintained the stock for many years (Supplemental Methods S1), and we confirmed the presence of the bnen mutation in both lineages (Supplemental Fig. S2). The G-to-A substitution found in bnen is consistent with the mutations usually induced by EMS, the mutagen that was used to obtain phyB-9 (Reed et al., 1993). Hence, we infer that the bnen mutation was induced at an early phase of phyB-9 isolation, and that it has been widely distributed throughout the Arabidopsis research community by hitchhiking phyB-9. To date, the original seed stock of phyB-9 (CS6217) has been distributed to more than 200 research groups, according to the order history of the Arabidopsis Biological Resource Center (ABRC). Therefore, we encourage many colleagues who are using phyB-9 to check the VEN4 genotype of their seed stocks. Previous analyses of phyB-9, especially those focusing on photosynthesis and/or leaf growth (biomass), require critical reevaluation to separate the effects of the phyB-9 and bnen mutations. It should also be noted that the PHYB and VEN4 genes are not linked. Hence, crosses between phyB-9OG and other mutants to make double mutants can readily result in unaware assortment of bnen in independent F2 plants, which will further complicate the reproducibility and interpretation of the genetic interaction. For instance, it was reported that the reduced chlorophyll content of phyB-9 was largely restored in the phyA-211 phyB-9 double mutant (Rusaczonek et al., 2015), which is in contradiction with the well-established synergism of phyA and phyB in chlorophyll biosynthesis. We found that phyA-211 phyB-9 has a wild-type VEN4 allele (Supplemental Fig. S2), raising the possibility that the chlorophyll phenotype was determined by the presence or absence of the bnen mutation and not due to the phyA mutation.
An increasing number of cases have been reported of Arabidopsis lines that had been considered to be single mutants but were later found to carry second-site, extragenic mutations that alter the original phenotype or even confer a new phenotype (Bergelson et al., 2016). It thus is likely that many more cases of second-site mutations are slightly or largely affecting phenotypes of other mutant strains without recognition. Here, we emphasize again the importance of backcrossing, and the risk of using only a single favored allele, even if the line has been studied for a long time and considered a de facto standard. The seed stocks used in this report have been donated to the ABRC (https://www.arabidopsis.org/abrc/index.jsp) and are publicly available at the time of publication of this letter under the stock numbers CS71624 (phyB-9OG), CS71625 (phyB-9BC), and CS71626 (bnen). As alternative choices, T-DNA insertional alleles of phyB in the Col-0 genetic background are available (Seo et al., 2006). In addition, phyB and other phytochrome mutant alleles isolated from Ler and Ws-2 accessions have provided important insights into the common and distinct functions of phytochromes in a genetic background different from Col-0 (Hu et al., 2013).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Allelism test between bnen and ven4-2.
Supplemental Figure S2. Genotyping of VEN4 in phyB-9-derived lines.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We are grateful to Akira Nagatani, Toshiaki Kozuka, ABRC, and Nottingham Arabidopsis Stock Centre for providing seeds. Daisuke Sugiura and Ichiro Terashima helped with photosynthetic analysis. Akihiko Nakano and Tomohiro Uemura granted use of a confocal laser scanning microscope.
Footnotes
This research was supported by a Grant-in-Aid for Scientific Research on Innovation Areas (#25113002) from Japan Society for Promotion of Science (JSPS) to H.T., KAKENHI Grant Number 16H06552 from JSPS to W.Y., and grants from the Ministerio de Economía y Competitividad of Spain (BIO2014-53063-P to J.L.M., and BIO2014-56889-R to M.R.P.).
References
- Bergelson J, Buckler ES, Ecker JR, Nordborg M, Weigel D (2016) A proposal regarding best practices for validating the identity of genetic stocks and the effects of genetic variants. Plant Cell 28: 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná G, Robles P, Micol JL (1999) A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152: 729–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC (2013) Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci USA 110: 1542–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsman EA, Pyke KA (1998) Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125: 1815–1822 [DOI] [PubMed] [Google Scholar]
- Lundquist PK, Rosar C, Bräutigam A, Weber AP (2014) Plastid signals and the bundle sheath: mesophyll development in reticulate mutants. Mol Plant 7: 14–29 [DOI] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusaczonek A, Czarnocka W, Kacprzak S, Witoń D, Ślesak I, Szechyńska-Hebda M, Gawroński P, Karpiński S (2015) Role of phytochromes A and B in the regulation of cell death and acclimatory responses to UV stress in Arabidopsis thaliana. J Exp Bot 66: 6679–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, et al. (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48: 354–366 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Kozuka T, Kim GT (2002) Genetic control of petiole length in Arabidopsis thaliana. Plant Cell Physiol 43: 1221–1228 [DOI] [PubMed] [Google Scholar]