Sucrose nonfermenting 1-related protein kinase 1 targets the geminivirus Rep protein to interfere with viral infection.
Abstract
Geminiviruses are single-stranded DNA viruses that infect a wide variety of plants and cause severe crop losses worldwide. The geminivirus replication initiator protein (Rep) binds to the viral replication origin and catalyzes DNA cleavage and ligation to initiate rolling circle replication. In this study, we found that the Tomato golden mosaic virus (TGMV) Rep is phosphorylated at serine-97 by sucrose nonfermenting 1-related protein kinase 1 (SnRK1), a master regulator of plant energy homeostasis and metabolism. Phosphorylation of Rep or the phosphomimic S97D mutation impaired Rep binding to viral DNA. A TGMV DNA-A replicon containing the Rep S97D mutation replicated less efficiently in tobacco (Nicotiana tabacum) protoplasts than in wild-type or Rep phosphorylation-deficient replicons. The TGMV Rep-S97D mutant also was less infectious than the wild-type virus in Nicotiana benthamiana and was unable to infect tomato (Solanum lycopersicum). Nearly all geminivirus Rep proteins have a serine residue at the position equivalent to TGMV Rep serine-97. SnRK1 phosphorylated the equivalent serines in the Rep proteins of Tomato mottle virus and Tomato yellow leaf curl virus and reduced DNA binding, suggesting that SnRK1 plays a key role in combating geminivirus infection. These results established that SnRK1 phosphorylates Rep and interferes with geminivirus replication and infection, underscoring the emerging role for SnRK1 in the host defense response against plant pathogens.
The relationship between a pathogen and its host is determined by the history of their coevolution (Sacristán and García-Arenal, 2008). Biological organisms constantly face new challenges from rapidly evolving infectious agents that are more virulent and spread to new hosts and geographical locations. Global climate changes also alter interactions within ecosystems, providing opportunities for pathogens to exploit new hosts. These new encounters assert pressure on the host to evolve mechanisms to tolerate or resist pathogens. Conversely, pathogens also might need to adjust their virulence to conserve the pool of hosts necessary for their own survival. Geminiviruses are a family of plant single-stranded DNA (ssDNA) viruses that have high DNA mutation and recombination rates and are excellent models for studying the interplay between pathogens, their hosts, and the environment (Seal et al., 2006; Duffy and Holmes, 2008).
Geminiviruses infect many plant species, including both monocots and dicots, and cause substantial crop losses every year in developing and developed countries (Mansoor et al., 2003, 2006; Navas-Castillo et al., 2011). The family Geminiviridae is divided into nine genera based on their genome structures and their insect vectors (Zerbini et al., 2017). Begomoviruses constitute the largest genus, which includes more than 320 viral species, with new species being identified regularly (Brown et al., 2015; Zerbini et al., 2017). Some begomoviruses are associated with satellite DNAs that can increase virulence (Zhou, 2013).
Begomovirus genomes consist of one or two circular DNA molecules of approximately 2,700 nucleotides in size (Rojas et al., 2005). Each viral DNA molecule is packaged as an ssDNA moiety into a twinned icosahedral capsid. Begomovirus genomes encode six to eight proteins involved in viral DNA replication and transcription, movement in the host plant, encapsidation, and insect transmission (Rojas et al., 2005; Fondong, 2013; Hanley-Bowdoin et al., 2013). The viral proteins also reprogram plant cell cycle controls, alter host gene expression and DNA methylation, interfere with host cell signaling, and suppress host defense pathways (Hanley-Bowdoin et al., 2004, 2013; Rojas et al., 2005; Burgyán and Havelda, 2011; Fondong, 2013).
Geminiviruses replicate their ssDNA genomes through double-stranded DNA (dsDNA) intermediates using a combination of rolling circle and recombination-dependent replication (Jeske et al., 2001; Hanley-Bowdoin et al., 2004). They replicate in the nuclei of infected plant cells using host DNA polymerases (Nagar et al., 1995). The viral replication initiator protein (Rep; also known as AC1, C1, or AL1) acts as the origin recognition protein (Fontes et al., 1992, 2004), endonuclease/DNA ligase, and DNA helicase (Desbiez et al., 1995; Laufs et al., 1995; Clérot and Bernardi, 2006) during the initiation of rolling circle replication. Rep is a 40-kD basic protein with a modular structure (Fig. 1A). Its N terminus mediates binding to and cleavage/ligation of viral DNA, while the C terminus unwinds DNA (Orozco and Hanley-Bowdoin, 1998; Clérot and Bernardi, 2006). A central domain is involved in the formation of Rep oligomers and interactions with other viral proteins (Settlage et al., 2005) and host factors (Kong et al., 2000; Orozco et al., 2000; Castillo et al., 2003). The Rep DNA-binding site is located in the 5′-intergenic region of the viral genome that separates divergent transcription units (Fontes et al., 1994). The binding site, which is characterized by direct repeats (Argüello-Astorga and Ruiz-Medrano, 2001), is located upstream of a conserved hairpin structure that contains the cleavage/ligation site (Orozco and Hanley-Bowdoin, 1996). The Rep N terminus contains four conserved sequence motifs that have essential roles in DNA binding and cleavage (Koonin and Ilyina, 1992; Orozco and Hanley-Bowdoin, 1998; Nash et al., 2011). Rep also plays an essential role in reprogramming the plant cell cycle through its interaction with the host retinoblastoma-related protein, leading to the synthesis of the host replication machinery required for viral replication in infected cells (Kong et al., 2000).
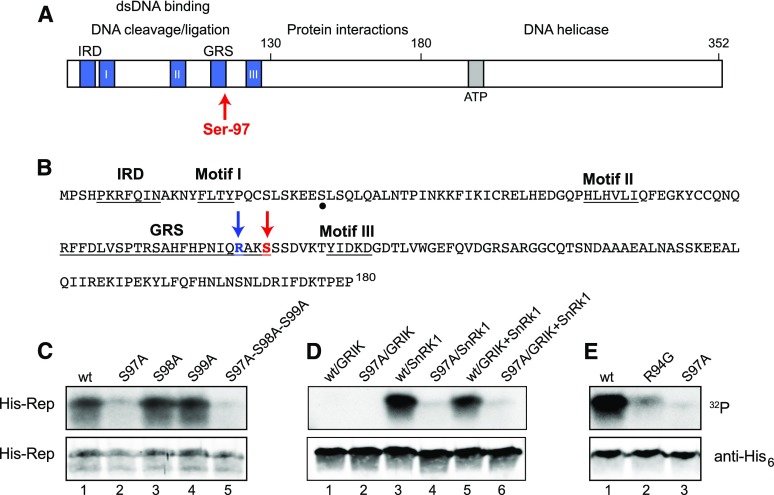
Figure 1.
TGMV Rep is phosphorylated by SnRK1 at Ser-97. A, Schematic diagram of the TGMV Rep showing functional domains, conserved motifs, and Ser-97. B, Amino acid sequence of the TGMV Rep N terminus (1–180). The SnRK1 phosphorylation site Ser-97 and its upstream Arg-94 are indicated by red and blue arrows, respectively. Conserved motifs are underlined. A potential secondary phosphorylation site is marked by a dot. C, Wild-type (wt) His6-Rep(1–131), the S97A, S98A, and S99A single mutants, and the S97A-S98A-S99A triple mutant phosphorylated by GRIK+SnRK1. D, Wild-type His6-Rep(1–180) and its S97A mutant phosphorylated by GRIK, preactivated SnRK1, or GRIK+SnRK1. E, Wild-type His6-Rep(1–180) and the R94G or S97A mutants phosphorylated by preactivated SnRK1. In C to E, recombinant His6-tagged TGMV Rep proteins were incubated in vitro in kinase reactions in the presence of [γ-32P]ATP. The proteins were resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Protein phosphorylation was detected by autoradiography (top gels), and the loading of His6-Rep was assessed by immunoblotting with anti-His6 antibodies (bottom gels).
Given its essential role in viral replication, the Rep protein is an excellent target for modulating virulence via host defense pathways. Posttranslational modifications, like phosphorylation, enable an organism to quickly alter protein activity, localization, and/or stability. The phosphorylation status of a protein is controlled by the opposite actions of protein kinases and phosphatases. Sucrose nonfermenting 1 (SNF1)-related protein kinase 1 (SnRK1) is the best characterized host protein kinase known to be involved in geminivirus infection (Shen et al., 2011, 2014; Hulsmans et al., 2016). It is a heterotrimeric complex composed of an α-catalytic subunit and β- and γ-regulator/adaptor subunits related to SNF1 in budding yeast and AMP-activated protein kinase (AMPK) in animals (Polge and Thomas, 2007; Ghillebert et al., 2011; Emanuelle et al., 2016; Roustan et al., 2016). SnRK1 plays a central role in regulating energy and metabolism in plants and has been implicated in responses to abiotic and biotic stresses (Baena-González et al., 2007; Polge and Thomas, 2007; Broeckx et al., 2016; Hulsmans et al., 2016). As a primary metabolic regulator, SnRK1 is crucial for the maintenance of energy balance during stress and is targeted by pathogens to interfere with host defenses.
The antiviral role of SnRK1 is supported by the observation that Nicotiana benthamiana SnRK1 RNA interference plants are more susceptible to geminivirus infection, while SnRK1-overexpressing plants are more resistant (Hao et al., 2003). SnRK1 phosphorylates some begomovirus AL2 proteins and the satellite-encoded βC1 protein to suppress infection (Shen et al., 2011, 2014; Zhong et al., 2017). Conversely, the viral AL2/C2 protein interacts with SnRK1 and inhibits its kinase activity to counter host defenses (Hao et al., 2003). AL2/C2 also inhibits host adenosine dikinase, which catalyzes the salvage synthesis of 5′-AMP that is required to sustain SnRK1 activity (Wang et al., 2003, 2005).
SnRK1 is activated by another set of plant protein kinases, the GRIKs (Geminivirus Rep-Interacting Kinases). The GRIKs phosphorylate a Thr residue in the SnRK1 T-loop, leading to a conformational change and activation (Sugden et al., 1999; Shen and Hanley-Bowdoin, 2006; Hey et al., 2007; Shen et al., 2009; Glab et al., 2017). As their names indicate, the GRIKs were first identified through their interactions with geminivirus Rep proteins, and their levels were shown to be elevated in infected leaves (Kong and Hanley-Bowdoin, 2002; Shen and Hanley-Bowdoin, 2006). In this study, we show that SnRK1 phosphorylates Rep at a Ser residue in the overlapping DNA-binding and cleavage domains. This phosphorylation event compromises the interaction of Rep with viral dsDNA, reduces viral DNA replication in infected plants, and delays symptom onset and development. Our observations identify a molecular mechanism for a role for the GRIK-SnRK1 protein kinase cascade in plant defense against geminiviruses.
RESULTS
SnRK1 Phosphorylates Tomato golden mosaic virus Rep on Ser-97
To investigate if Rep is phosphorylated by SnRK1, we examined the amino acid sequence of Tomato golden mosaic virus (TGMV) Rep for potential SnRK1 target motifs (Vlad et al., 2008). We found two Ser residues in contexts that resemble the SnRK1 consensus (φX[R/K]XX[S/T]XXφ), with a basic residue at the −3 position and flanked by hydrophobic residues (Fig. 1B). Both Ser residues are located in the Rep N terminus at residue positions 29 and 97 (Fig. 1B).
We next examined if TGMV Rep is phosphorylated by the GRIK-SnRK1 cascade using recombinant proteins in an in vitro kinase assay. His6-tagged TGMV Rep(1–131) produced in Escherichia coli was incubated with GST-tagged Arabidopsis (Arabidopsis thaliana) GRIK1 and the His6-tagged Arabidopsis SnRK1.1 kinase domain in the presence of [γ-32P]ATP as a phosphate donor. Labeled His6-Rep was resolved by SDS-PAGE and visualized by autoradiography, indicating that TGMV Rep is phosphorylated by the GRIK-SnRK1 cascade (Fig. 1C, lane 1).
Mass spectrometry was used to identify the phosphorylated residues in His6-TGMV Rep(1–131) after incubation with GST-GRIK, His6-SnRK1 kinase domain, and unlabeled ATP. After phosphorylation, His6-Rep was resolved by SDS-PAGE and in gel digested with chymotrypsin to generate peptides for liquid chromatography–mass spectrometry with electrospray ionization (LC/MSE) analysis. A single putative phosphopeptide, (p)HPNIQRAKSSSDVKTY (residues 89–104), was detected at a measured [M+H]+ monoisotopic mass of 1,910.9202 (Supplemental Fig. S1A). A reconstructed LC/MSE product ion spectrum revealed that one of the three consecutive Ser residues (Ser-97, Ser-98, or Ser-99) in this peptide contained a phosphate group, but the detected product ions could not be used to distinguish which Ser was phosphorylated (Supplemental Fig. S1B). Therefore, we generated three recombinant proteins corresponding to single Ala substitutions at each of the Ser residues, S97A, S98A, and S99A, and a recombinant protein containing all the three Ala mutations, S97A-S98A-S99A. In in vitro assays with GRIK, SnRK1, and [γ-32P]ATP, the S98A (Fig. 1C, lane 3) and S99A (lane 4) mutants were phosphorylated to levels similar to that of wild-type Rep (lane 1), whereas the S97A (lane 2) and S97A-S98A-S99A (lane 5) mutants were only weakly labeled. These results indicated that Ser-97 is the major phosphorylation site in TGMV Rep(1–131) in reactions containing both GRIK and SnRK1. The low level of phosphorylation of the S97A-S98A-S99A mutant may reflect another GRIK-SnRK1 target site in the Rep N terminus, potentially Ser-29. We decided not to pursue the site(s) of residual phosphorylation and, instead, focused on Ser-97.
The in vitro kinase assay contained two protein kinases, GRIK and SnRK1. Therefore, it does not provide information about whether one or both kinases are necessary to phosphorylate Rep. To distinguish the activities of the two kinases, we developed a protocol using GRIK to preactivate SnRK1 before performing affinity purification to separate the two kinases prior to incubation with TGMV Rep. Phosphorylation assays using a specific peptide substrate for SnRK1 (Huang and Huber, 2001) showed that the purified, activated SnRK1 did not require the presence of GRIK for full kinase activity (Supplemental Fig. S2).
To determine if Rep Ser-97 is phosphorylated by GRIK, SnRK1, or both together, we incubated His6-tagged Rep(1–180) with GRIK alone, preactivated SnRK1, or GRIK+SnRK1 (Fig. 1D). GRIK did not phosphorylate Rep(1–180) (lane 1) or its S97A mutant (lane 2). In contrast, preactivated SnRK1 phosphorylated Rep(1–180) (lane 3) and very weakly labeled the S97A mutant (lane 4). There was no difference in the phosphorylation signals when both SnRK1 and GRIK were included in the kinase reactions with Rep(1–180) (lane 5) or the S97A mutant (lane 6) when compared with preactivated SnRK1. Based on these results, we concluded that TGMV Rep Ser-97 is phosphorylated by SnRK1 and that GRIK has no direct role in Rep phosphorylation. However, given that Rep(1–180) binds to GRIK (Kong and Hanley-Bowdoin, 2002), GRIK may serve as an adaptor for Rep phosphorylation in vivo.
The context of Ser-97 conforms to the common SnRK1 phosphorylation target sequence, with a basic Arg-94 at the −3 position and hydrophobic Ile-92 and Val-101 residues at the −5 and +4 positions, respectively (Fig. 1B; Vlad et al., 2008). Replacing Arg-94 with a Gly, which occurs in many other geminivirus Rep proteins at the equivalent position, greatly reduced SnRK1 phosphorylation of TGMV Rep(1–180) compared with the wild-type control (Fig. 1E, compare lanes 1 and 2). The R94G mutant incorporated more label than the S97A mutant (lane 3), indicating that the Gly replacement did not eliminate Ser-97 phosphorylation. However, Arg-94 is necessary for the maximal SnRK1 phosphorylation of Ser-97 in TGMV Rep.
SnRK1 Phosphorylation of Rep Ser-97 Inhibits DNA Binding But Not DNA Cleavage Activity
The DNA-binding and cleavage/ligation domains are located in the N terminus of the Rep protein and contain four conserved motifs necessary for these activities (Fig. 1A; Laufs et al., 1995; Orozco and Hanley-Bowdoin, 1998; Nash et al., 2011). Ser-97 is the last amino acid residue of the TGMV Rep Geminivirus Rep Sequence (GRS) domain, which is required for DNA cleavage but not for DNA binding (Fig. 1B; Nash et al., 2011). We first examined if phosphorylation of Ser-97 impacts the DNA cleavage activity of His6-tagged TGMV Rep(1–180) using a phosphomimic S97D mutation as a proxy for phospho-Ser. TGMV Rep(1–180), the phosphonull S97A mutant, or the phosphomimic S97D mutant was incubated with a fluorescently labeled, single-stranded oligonucleotide substrate that contains the loop and the 3′ stem sequences of the hairpin and includes the DNA cleavage site (Supplemental Table S1; Orozco and Hanley-Bowdoin, 1998). Cleavage was detected by resolving the 23-nucleotide fluorescent substrate and the 11-nucleotide fluorescent product on an acrylamide gel and visualization using an infrared fluorescence scanner. Similar levels of the cleavage product were detected for the three recombinant proteins over a 100-fold concentration range (Fig. 2A). Thus, we concluded that phosphonull and phosphomimic mutations at the Ser-97 site do not alter Rep DNA cleavage activity.
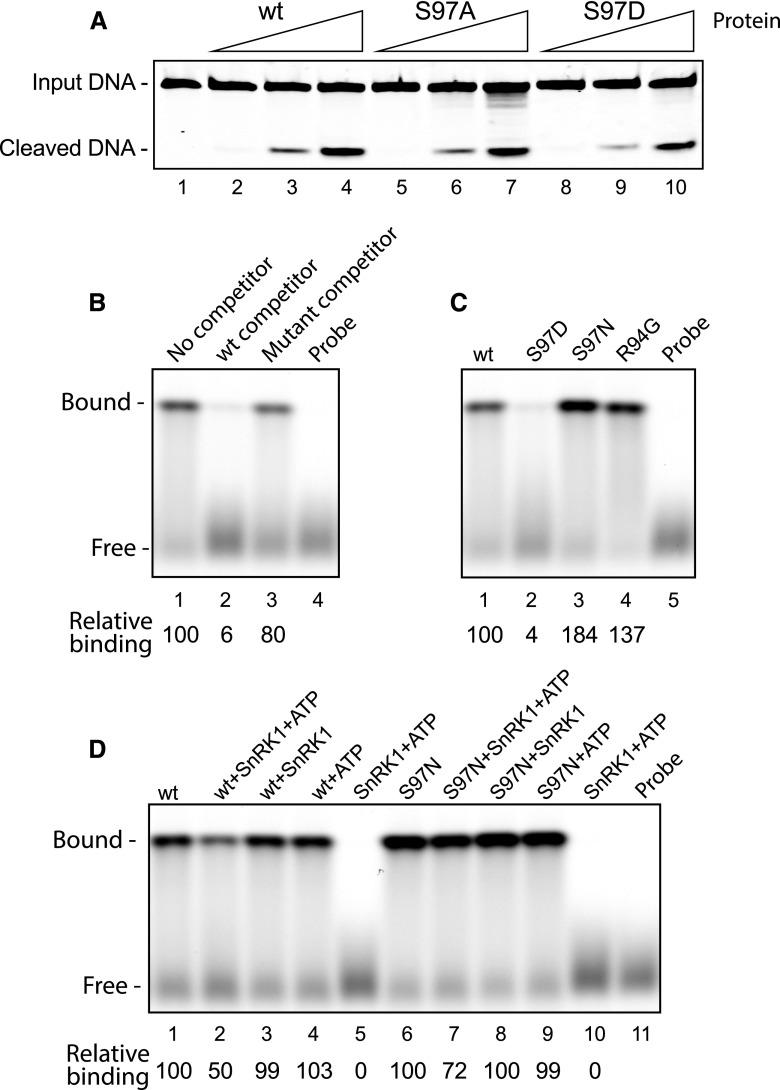
Figure 2.
Phosphorylation of Ser-97 in TGMV Rep impairs DNA binding but not cleavage activity. A, Increasing amounts of the wild-type (wt) His6-tagged TGMV Rep(1–180) or its S97A and S97D mutants were assayed for cleavage activity. The substrate was a fluorescently labeled, single-stranded oligonucleotide corresponding to the loop and left stem of the hairpin regions in the viral origin of replication. The Rep protein amounts were 0.5 pmol (lanes 2, 5, and 8), 5 pmol (lanes 3, 6, and 9), and 50 pmol (lanes 4, 7, and 10). There was no protein in lane 1. B, Sequence-specific DNA-binding activity of wild-type TGMV Rep(1–180) (50 pmol) in EMSAs using a fluorescently labeled, double-stranded oligonucleotide corresponding to the TGMV Rep-binding site in the origin of replication. The specificity of binding was verified in the presence of a 1,000-fold excess of an unlabeled wild-type dsDNA competitor (lane 2) or a mutant competitor (lane 3). Relative binding (shown below) is the ratio of the fluorescence of the shifted bands in lanes 2 and 3 compared with lane 1, which was set to 100. C, Binding assays with wild-type Rep (lane 1) or its S97D (lane 2), S97N (lane 3), or R94G (lane 4) mutant. Relative binding (shown below) is the ratio of the fluorescence of the shifted bands in lanes 2 to 4 compared with lane 1, which was set to 100. D, The effect of SnRK1 phosphorylation of Rep Ser-97 on DNA binding was assayed by preincubation of the wild-type or the S97N mutant Rep with activated SnRK1 and ATP (lanes 2 and 7), activated SnRK1 alone (lanes 3 and 8), or ATP alone (lanes 4 and 9). Relative binding is shown for wild-type Rep (ratio of lanes 2–5 to lane 1) or Rep S97N (ratio of lanes 7–10 to lane 6).
We also used Rep mutants to examine if SnRK1 phosphorylation of Ser-97 impacts the sequence-specific, dsDNA-binding activity of TGMV Rep(1–180) in electrophoretic mobility shift assays (EMSAs) using a fluorescent double-stranded oligonucleotide probe corresponding to the Rep DNA-binding site upstream of the conserved hairpin (Supplemental Table S1; Fontes et al., 1994; Nash et al., 2011). A single shifted band was observed when His6-tagged TGMV Rep(1–180) was incubated with probe DNA (Fig. 2B, lane 1). The addition of a 1,000-fold excess of an unlabeled, wild-type oligonucleotide with an intact binding site nearly abolished Rep binding (lane 2), while inclusion of an oligonucleotide with a mutant binding site (Supplemental Table S1) reduced binding only slightly (lane 3), thereby establishing the specificity of the Rep DNA-binding assays. We then compared the binding activity of TGMV Rep(1–180) with three Rep mutants: the phosphomimic S97D mutant, the phosphonull S97N mutant, and the near-phosphonull R94G mutant (Fig. 1E). A bulkier Asn that cannot be phosphorylated was used as a phosphonull mutation because it is more similar to phospho-Ser in side chain volume. Strikingly, the S97D mutant was severely impaired for DNA binding (Fig. 2C, compare lanes 1 and 2), while the S97N (lane 3) and R94G (lane 4) mutants were both capable of DNA binding. These results suggested that the negative charge associated with phosphorylation of Ser-97 interferes with Rep binding to viral DNA.
We then directly tested whether SnRK1 phosphorylation of Ser-97 in TGMV Rep interferes with DNA binding. TGMV Rep(1–180) was preincubated with activated SnRK1 and ATP to generate the Rep phosphoform and then used in EMSAs with the fluorescent dsDNA probe. The amount of probe bound by phosphorylated Rep was only 50% of the amount bound by untreated Rep (Fig. 2D, compare lanes 1 and 2). Exclusion of either ATP (lane 3) or SnRK1 (lane 4) in the kinase reaction abolished the reduction in DNA-binding activity, and no shifted probe was observed in the presence of SnRK1+ATP only (lane 5). Hence, the reduction cannot be attributed to the inclusion of ATP and/or SnRK1 in the binding assays and, instead, reflects SnRK1 phosphorylation of Rep. A Rep S97N mutant, which cannot be phosphorylated on Ser-97, showed a smaller reduction in DNA-binding activity than wild-type Rep after preincubation with activated SnRK1 and ATP (Fig. 2D, compare lanes 2 and 7). Together, these results show that SnRK1 phosphorylation of TGMV Rep Ser-97 compromises its interaction with viral DNA. The 50% residual binding activity of wild-type Rep preincubated with SnRK1 and ATP most likely is attributable to Rep protein that was not phosphorylated during the kinase reaction and, thus, was able to bind to DNA.
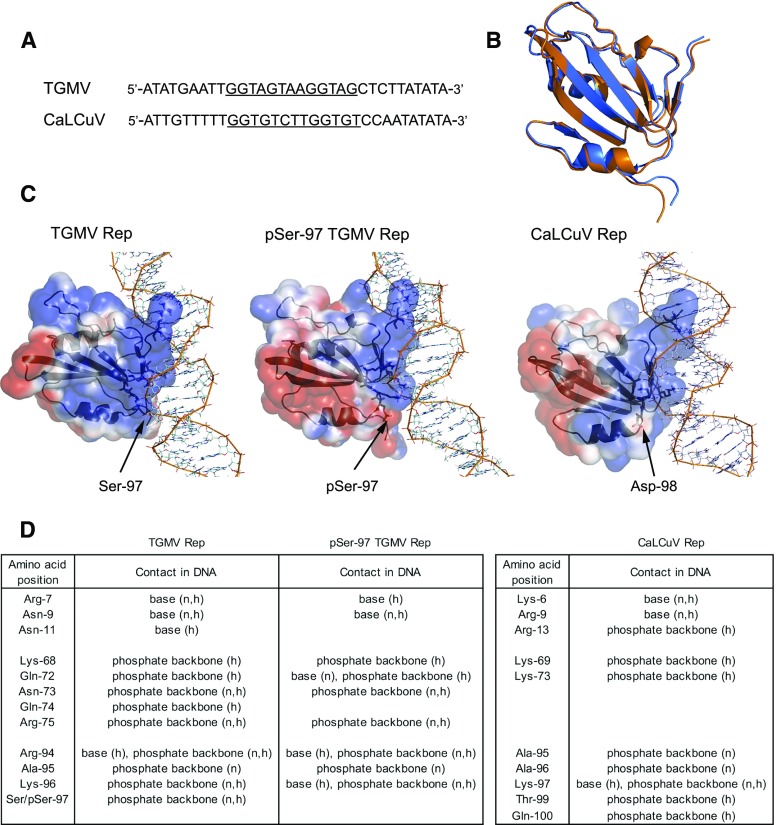
To determine if phosphorylation of Ser-97 interferes physically with the interaction between Rep and viral DNA, we simulated the structures of TGMV Rep(7–122) and its phopho-Ser-97 (pSer-97) modification based on the published NMR structure of Tomato yellow leaf curl virus (TYLCV) Rep(4–121) (Campos-Olivas et al., 2002). Docking the models of wild-type or pSer-97 Rep onto B-form dsDNA derived from the 5′ intergenic region of TGMV DNA containing the Rep-binding sequence (Fig. 3A; Fontes et al., 1994) revealed that phosphorylation does not significantly alter the overall structure of the protein, as shown by the overlay of the two dsDNA-bound median structures (Fig. 3B). For both forms of the protein, the Rep-dsDNA interaction occurs across both DNA strands (Fig. 3C; Supplemental Fig. S3). The Rep amino acid residues that directly contact dsDNA cluster in three regions: (1) Arg-7, Gln-9, and Asn-11; (2) Lys-68 and Gln-72 through Arg-75; and (3) Arg-94 through Ser-97 (Fig. 3D). Rep interacts with the DNA bases via Arg-7, Gln-9, Asn-11, and Arg-94 and with the DNA phosphate backbone via Lys-68, Gln-72 through Arg-75, and Arg-94 through Ser-97. Both types of contacts occur through a combination of nonbonded interactions and hydrogen bonds (Supplemental Fig. S3). When Ser-97 is phosphorylated, the distribution of negative charges on the surface of Rep increases substantially at the Rep-DNA interface, resulting in diminished contacts with DNA involving Arg-7, Asn-11, Gln-74, and pSer-97 (Fig. 3, C and D). The number of total contacts by pSer-97 Rep decreases by 60%, leading to weaker Rep-DNA binding, consistent with the EMSA results.
Figure 3.
Structural modeling shows that TGMV pSer-97 Rep has fewer contacts with its DNA-binding site. A, Viral DNA sequences used for modeling. The TGMV and CaLCuV Rep-binding sites are underlined. B, Overlay of dsDNA-bound median structures of wild-type TGMV Rep (blue) and pSer-97 Rep (orange). The Cα root mean square deviation between the two structures is 0.75 Å. C, Electrostatic plot of TGMV Rep(7–122) (left), its pSer-97 form (middle), and CaLCuV Rep(5–116) (right) in association with their cognate B-form viral dsDNAs. The electrostatic surface of the protein was created with Adaptive Poisson-Boltzmann Solver and PyMOL. The blue color denotes positive electrostatics, while the red color denotes negative electrostatics. Residues labeled as active in the molecular docking are shown in stick format, and residues defined as passive are shown in line format. D, List of contacts of amino acid residues of TGMV Rep (from a total of 28 structures), pSer-97 Rep (39 structures), and CaLCuV Rep (31 structures) with viral dsDNA via nucleotide base or phosphate backbone. h, Hydrogen bond; n, nonbonded contact.
The TGMV Rep DNA-binding domain (amino acid residues 1–120) is only functional for DNA binding when it is fused to the oligomerization domain (residues 134–180; Orozco et al., 1997; Orozco and Hanley-Bowdoin, 1998). Hence, the Rep-DNA interface could differ slightly from the model generated using Rep amino acid residues 5 to 122.
Phosphorylation of TGMV Rep Ser-97 Compromises Viral DNA Replication
The binding of Rep to the origin of replication is an essential step during the initiation of viral replication (Fontes et al., 1994). Thus, we examined if SnRK1 phosphorylation of TGMV Rep Ser-97 impairs viral DNA replication. In many geminivirus genomes, including that of TGMV, the Rep coding sequence containing the Ser-97 codon overlaps with the AL4 viral open reading frame, and it is not possible to mutate the codon without also altering the AL4 coding sequence. Earlier studies indicated that the AL4 gene is not required for TGMV replication or infection (Pooma and Petty, 1996). Therefore, we generated an AL4 knockout in TGMV DNA-A for testing Rep mutants. The ATG start codon for AL4 and an upstream in-frame ATG codon were altered to ACG codons to block AL4 translation (Supplemental Fig. S4A). These T-to-C transitions occur in wobble positions in the Rep coding sequence and do not introduce amino acid residue changes into the Rep gene (Supplemental Fig. S4A). We transfected a wild-type TGMV DNA-A replicon and the AL4-knockout replicon into protoplasts prepared from tobacco (Nicotiana tabacum) suspension cells and monitored viral DNA accumulation 3 d posttransfection on DNA gel blots. Slightly lower levels of double-stranded TGMV DNA-A were detected for the AL4-knockout-transfected protoplasts than for the wild-type TGMV DNA-A replicons (Supplemental Fig. S4B, compare lanes 2–4 with 5–7), indicating that knocking out AL4 has a minimal impact on TGMV accumulation.
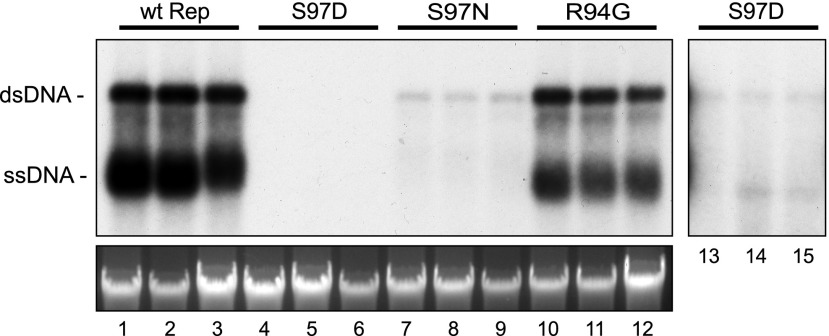
We then used the TGMV DNA-A AL4 knockout to generate replicons carrying the Rep S97D phosphomimic, the S97N phosphonull, or the R94G phosphodeficient mutations. Replication of the mutant replicons was examined in the tobacco protoplast assay as described above. The TGMV DNA-A replicon with the AL4 knockout had high levels of accumulation of both double- and single-stranded viral DNA forms (Fig. 4, lanes 1–3). In contrast, replication of the Rep S97D phosphomimic mutant could be detected only after overexposure of the blot (lanes 13–15). The phosphonull S97N mutant also showed a major reduction in viral DNA replication (lanes 7–9) but still accumulated to a higher level than the S97D mutant (lanes 4–6), while the phosphodeficient R94G mutant was reduced only slightly for replication (lanes 10–12). These data indicate that viral DNA replication is sensitive to the amino acid residue composition at TGMV Rep position 97. However, the negatively charged S97D mutation that mimics phosphorylation of Ser-97 is more detrimental than the uncharged phosphonull S97N replacement. Moreover, the R94G mutation, which greatly reduces SnRK1 phosphorylation of Ser-97, has a minimal effect on viral replication. Together, these results are consistent with a model in which SnRK1 phosphorylation of Rep Ser-97 interferes with Rep DNA binding to reduce viral DNA replication.
Figure 4.
Mutation of TGMV Rep Ser-97 reduces viral replication. Protoplasts prepared from tobacco suspension cells (NT1) were transfected with DNA (10 µg) corresponding to an AL4-knockout TGMV-A replicon (lanes 1–3) or AL4-knockout replicons containing the Rep S97D (lanes 4–6), S97N (lanes 7–9), or R94G (lanes 10–12) mutation, all of which impair SnRK1 phosphorylation of Ser-97. Total DNA was extracted at 3 d post transfection, cut with XhoI, and digested with DpnI to cut Dam-methylated input plasmid DNA but not nascent viral DNA. The DNA was analyzed on a DNA gel blot using a radiolabeled TGMV-A probe (top left gel). A longer exposure (10-fold; lanes 13–15) shows weak signals from the Rep S97D mutant. The bottom gel shows ethidium bromide-stained, digested DNA before transfer. wt, Wild type.
Phosphorylation of TGMV Rep Ser-97 Delays Symptom Development and Viral DNA Accumulation in Planta
Wild-type TGMV infects N. benthamiana, causing leaf curling and chlorosis (Elmer et al., 1988). We cobombarded N. benthamiana plants with a TGMV DNA-B replicon and replicons corresponding to wild-type TGMV DNA-A or the AL4 knockout. Similar symptoms appeared at the same time for both treatments, indicating that the AL4 mutations do not impair TGMV infection of N. benthamiana (Supplemental Fig. S4C). TGMV DNA-A accumulation was slightly lower in N. benthamiana plants infected with the AL4-knockout mutant than those infected with wild-type virus at 10 d post inoculation (dpi; Supplemental Fig. S4D). Thus, we could use the TGMV AL4-knockout background to examine the infectivity of replicons carrying mutations in the Rep N terminus.
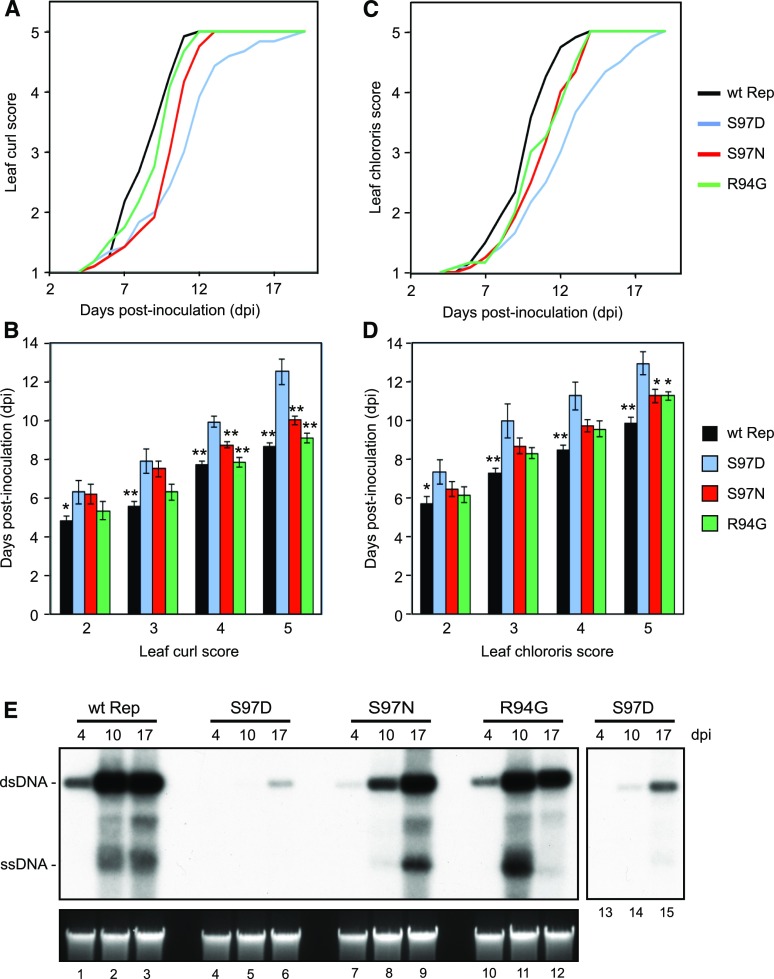
We generated TGMV DNA-A replicons containing the Rep S97D phosphomimic, the S97N phosphonull, or the R94G phosphodeficient mutations. The mutations were cloned into both the full and partial copies of the tandemly repeated TGMV DNA-A replicon. N. benthamiana plants were cobombarded with a TGMV DNA-B replicon and TGMV DNA-A replicons corresponding to the AL4 knockout (our wild-type Rep control) or the S97D, S97N, and R94G mutants in the AL4-knockout background. Leaf curl and chlorosis symptoms were scored daily up to 17 dpi on a scale of 1 (no symptoms) to 5 (severe symptoms), and the average score at each day was plotted over time (Fig. 5, A and C). The phosphomimic S97D mutant TGMV was able to infect N. benthamiana, but the leaf curl and chlorosis symptoms appeared 2 to 4 d after they appeared on plants infected with the AL4 knockout. The S97N-phosphonull mutant also showed slower symptom development, but the delay was only 1 to 2 d. The timing of leaf curl symptom development was similar for the R94G mutant and the AL4-knockout control, but the R94G mutant showed a delay in leaf chlorosis development, similar to the S97N mutant. We also determined the average time it took infected plants to reach a given leaf curl or chlorosis symptom score (Fig. 5, B and D). The S97D mutant took the longest to reach each symptom score, with the difference being more apparent for high scores. The difference was statistically significant (P < 0.05, Student’s t test) at all symptom scores compared with the AL4 knockout and at the higher scores relative to the phosphonull and phosphodeficient mutants.
Figure 5.
The TGMV Rep phosphomimic mutation S97D delays symptoms and viral DNA accumulation in infected plants. N. benthamiana (12 plants per treatment) was inoculated by cobombardment of a TGMV-B replicon with either an AL4-knockout TGMV-A replicon carrying wild-type (wt) Rep or the S97D, S97N, or R94G Rep mutant. A to D, Symptoms corresponding to leaf curl (A and B) and chlorosis (C and D) were scored on a scale of 1 (none) to 5 (severe). The average symptom scores (A and C) and average time of appearance for a given score (B and D) were calculated. The asterisks indicate significant symptom delays in the phosphomimic S97D mutant compared with the wild-type Rep or the other mutants as determined using Student’s t test (*, P < 0.05; **, P < 0.01). Error bars represent sE. E, Total DNA was extracted from leaves at 4, 10, and 17 dpi, linearized with XhoI, and analyzed on a DNA gel blot using a radiolabeled TGMV-A probe (top left gel). A longer exposure (3-fold; lanes 13–15) shows weak signals from the Rep S97D mutant. The bottom gel shows ethidium bromide-stained, digested DNA before transfer.
We also examined the accumulation of TGMV DNA-A in infected N. benthamiana plants at 4, 10, and 17 dpi using DNA gel blots. The AL4 knockout (Fig. 5E, lanes 1–3) and the Rep R94G mutant (lanes 10–12) showed similar viral dsDNA accumulation patterns, with viral DNA readily detectable at 4 dpi and at maximal levels by 10 dpi. Viral ssDNA accumulation was detected at 10 dpi and showed similar patterns for the AL4 knockout and the R94G mutant. The reduced level of ssDNA in plants at 17 dpi (lane 12) suggested that position 94 could modulate viral ssDNA accumulation. Viral DNA accumulation was delayed for the S97N-phosphonull mutant but reached a high level similar to that in the AL4 knockout by 17 dpi (lanes 7–9). In contrast, viral DNA corresponding to the S97D phosphomimic mutant was not detected at 4 dpi and was observed only at 10 dpi upon overexposure of the blot (lanes 4–6 and 13–15). Strikingly, the level of S97D was lower at 17 dpi than the levels detected for the AL4 knockout and the R94G mutant at 4 dpi (compare lanes 6 and 1 or 10). The patterns of viral DNA accumulation of the TGMV Rep mutants in N. benthamiana compared with the AL4 knockout correlate with the patterns of symptom development (Fig. 5) and the viral DNA replication results in transfected tobacco protoplasts (Fig. 4). Taken together, our results show that SnRK1 phosphorylation of TGMV Rep Ser-97 impairs viral DNA replication and causes a delay and reduction in viral DNA accumulation, leading to delayed symptom development during infection.
An earlier study showed that a TGMV Rep mutant severely impaired for replication is delayed for symptom development and reverts at a high frequency during infection (Arguello-Astorga et al., 2007). In this study, we altered all three nucleotides, TCG for Ser, to GAT for Asp, and it would be difficult for reversion to happen. To confirm that the S97D mutation is stable during infection, we isolated DNA from symptomatic leaves of 12 plants when infection was fully established at 28 dpi. We amplified the Rep coding region and sequenced PCR products corresponding to the entire Rep coding region. Of the six plants from which sequences could be amplified successfully by PCR, all contained the original S97D mutation and no additional mutations were detected in Rep, indicating that the S97D mutation is stable during infection.
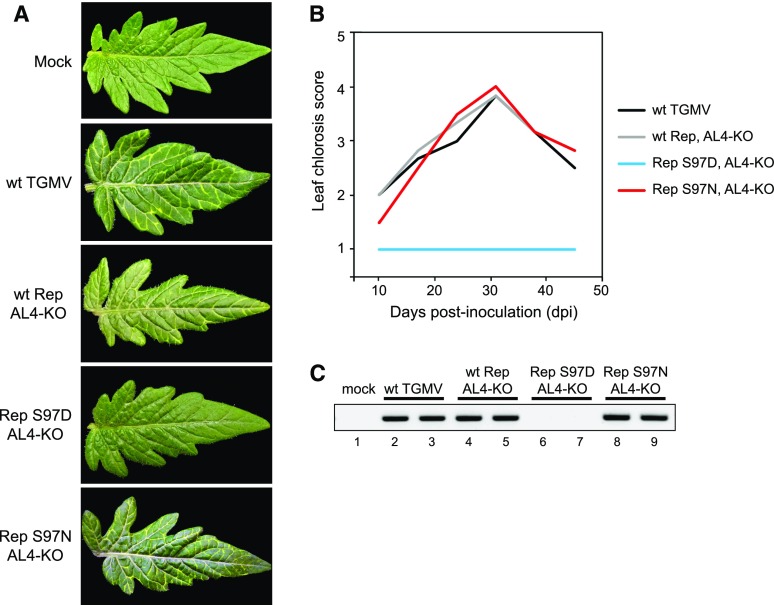
N. benthamiana carries a mutation that impairs silencing and, as such, is a permissive host for viral infection (Yang et al., 2004). To gain further insight into the role of SnRK1 phosphorylation of TGMV Rep, we examined the Ser-97-phosphomimic and Ser-97-phosphonull mutants in tomato (Solanum lycopersicum), which was the original source of the TGMV infectious clones. The tomato variety Florida Lanai reliably develops leaf chlorosis symptoms after being inoculated with TGMV DNA-A and DNA-B replicons (Rajabu et al., 2018). In this study, we observed that both wild-type TGMV and the AL4-knockout mutant caused bright yellow coloration along the veins of tomato leaves, starting at 10 dpi, peaking around 30 dpi, and then decreasing with new leaves showing a recovery phenotype (Fig. 6, A and B). The similarity between wild-type and AL4 mutant virus indicates that TGMV AL4 is not essential for establishing and maintaining infection in tomato plants under the conditions used for this study. In the AL4-knockout background, the Rep S97D-phosphomimic mutant did not cause any symptoms in infected plants, while the Rep S97N-phosphonull mutant resembled the wild-type and AL4-knockout viruses, indicating that the reduced interaction of Rep with viral DNA also compromises infection in tomato (Fig. 6, A and B). The lack of symptoms in tomato plants infected with the phosphomimic mutant compared with the delayed development of leaf curl and chlorosis symptoms in N. benthamiana plants (Fig. 5) suggested that the threshold for the establishment of TGMV infection is higher in tomato than in N. benthamiana. Consistent with this idea, viral DNA was detected readily by PCR in tomato plants infected with wild-type TGMV (Fig. 6C, lanes 2 and 3), the AL4-knockout mutant (lanes 4 and 5), and the Rep S97N-phosphonull mutant (lanes 8 and 9) but not in plants infected with the Rep S97D-phosphomimic mutant (lanes 6 and 7). Hence, we concluded that phosphorylation of TGMV Rep Ser-97 by SnRK1 reduces virulence in a species like N. benthamiana, which has an impaired host defense system, and prevents infection in tomato, which has a robust defense system.
Figure 6.
The TGMV Rep phosphomimic S97D mutant does not infect tomato. Tomato Florida Lanai plants were inoculated by cobombardment of a wild-type TGMV-B replicon with a wild-type (wt) TGMV-A or an AL4-knockout (KO) mutant with wild-type Rep or the S97D or S97N Rep mutant. The mock controls were inoculated with the TGMV-B alone. A, Leaflets from plants at 24 dpi. B, Leaf chlorosis symptoms were scored on a scale of 1 to 5 at 10, 17, 24, 31, 38, and 45 dpi. The average symptom scores from 10 plants for each treatment are shown. C, The presence of TGMV DNA-A was assessed by PCR at 28 dpi using specific primers that yielded a 403-bp product. Results from two independent plants are shown for each treatment, except for mock.
SnRK1 Phosphorylation of Rep May Be a Common Mechanism for Regulating Geminivirus Infection by the Host Plant
TGMV Rep Ser-97 is the last amino acid residue in the highly conserved GRS motif (Nash et al., 2011), raising the possibility that SnRK1 phosphorylation of Rep is a common geminivirus-host interaction during infection. We constructed a phylogenetic tree based on the amino acid sequences of the Rep proteins of 337 geminivirus species representing seven of the genera (Muhire et al., 2013; Varsani et al., 2014a, 2014b; Brown et al., 2015) and examined them for the occurrence of a Ser or a Thr at a position equivalent to Ser-97 (Supplemental Fig. S5; Supplemental Data Set S1). The vast majority of the begomovirus Rep proteins have a Ser at this position (271 out of 300 species; one has a Thr). The only exceptions are members of the Squash leaf curl (SLC) clade (26 species), which predominantly have an acidic amino acid (24 Asp, one Glu, and one Asn) that might mimic SnRK1 phosphorylation. Very few begomovirus Reps have a residue other than Ser, Thr, Asp, or Glu at this position. The Rep proteins from the topocuvirus, turncurtovirus, eragrovirus genera and Beet curly top virus of the curtovirus genus have a Ser, while those from becurtoviruses and the other curtoviruses do not. About half of the mastrevirus Rep proteins contain a Ser or an Asp at the corresponding position. However, only eight begomovirus Rep proteins (3%) sporadically distributed in the phylogenetic tree have an Arg (or Lys for one species) at the −3 position.
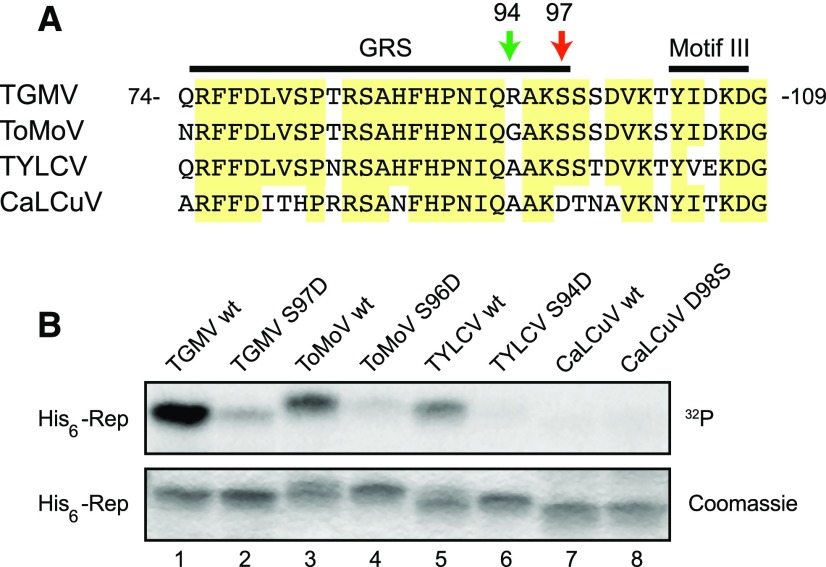
The SnRK1 phosphorylation consensus sequence includes a basic amino acid at the −3 position relative to the target Ser/Thr that is thought to greatly enhance phosphorylation (Vlad et al., 2008). The vast majority of begomovirus Reps have a Gly (82%) or an Ala (10%) at the −3 position (Supplemental Data Set S1). Hence, we were interested to know if Rep proteins without an Arg or Lys also are phosphorylated by SnRK1. Tomato mottle virus (ToMoV) and TYLCV, a bipartite New World begomovirus and a monopartite Old World begomovirus, respectively, were selected for further investigation. ToMoV and TYLCV Reps have a Gly or an Ala, respectively, at the −3 position of the Ser equivalent to Ser-97 of TGMV Rep (Fig. 7A). We first examined if recombinant proteins corresponding to His6-tagged ToMoV Rep(1–179) and TYLCV Rep(1–177) are phosphorylated by SnRK1 in vitro. Both ToMoV Rep(1–179) (Fig. 7B, lane 3) and TYLCV Rep(1–177) (lane 5) were phosphorylated by SnRK1, albeit at lower levels than TGMV Rep(1–180) (lane 1). We also tested Asp substitution mutations at ToMoV Rep Ser-96 and TYLCV Rep Ser-94, which correspond to TGMV Ser-97 (Fig. 7A), in the kinase assay. Both the ToMoV S96D (lane 4) and TYLCV S94D (lane 6) mutants showed significantly decreased phosphorylation, indicating that ToMoV Rep Ser-96 and TYLCV Rep Ser-94 are the SnRK1 phosphorylation targets.
Figure 7.
SnRK1 phosphorylation of begomovirus Rep proteins. A, Alignment of the amino acid sequences of the begomovirus TGMV, ToMoV, TYLCV, and CaLCuV Rep protein regions containing the TGMV Rep Ser-97 (red arrow) that is phosphorylated by SnRK1. The amino acid positions for the TGMV Rep are indicated. The Arg-94 of TGMV Rep is indicated with the green arrow. Conserved amino acids in three or more of the viruses are highlighted in yellow. GRS and motif III also are indicated. B, Phosphorylation assay of His6-tagged recombinant proteins corresponding to the N-terminal halves of TGMV (lane 1), ToMoV (lane 3), TYLCV (lane 5), and CaLCuV (lane 7) Rep proteins and their S97D (lane 2), S96D (lane 4), S94D (lane 6), and D98S (lane 8) mutants, respectively. These Ser or Asp residues correspond to the TGMV Rep Ser-97. The phosphorylation reactions were resolved by SDS-PAGE and visualized by autoradiography (top gel), and the gels were stained with Coomassie Brilliant Blue (bottom gel).
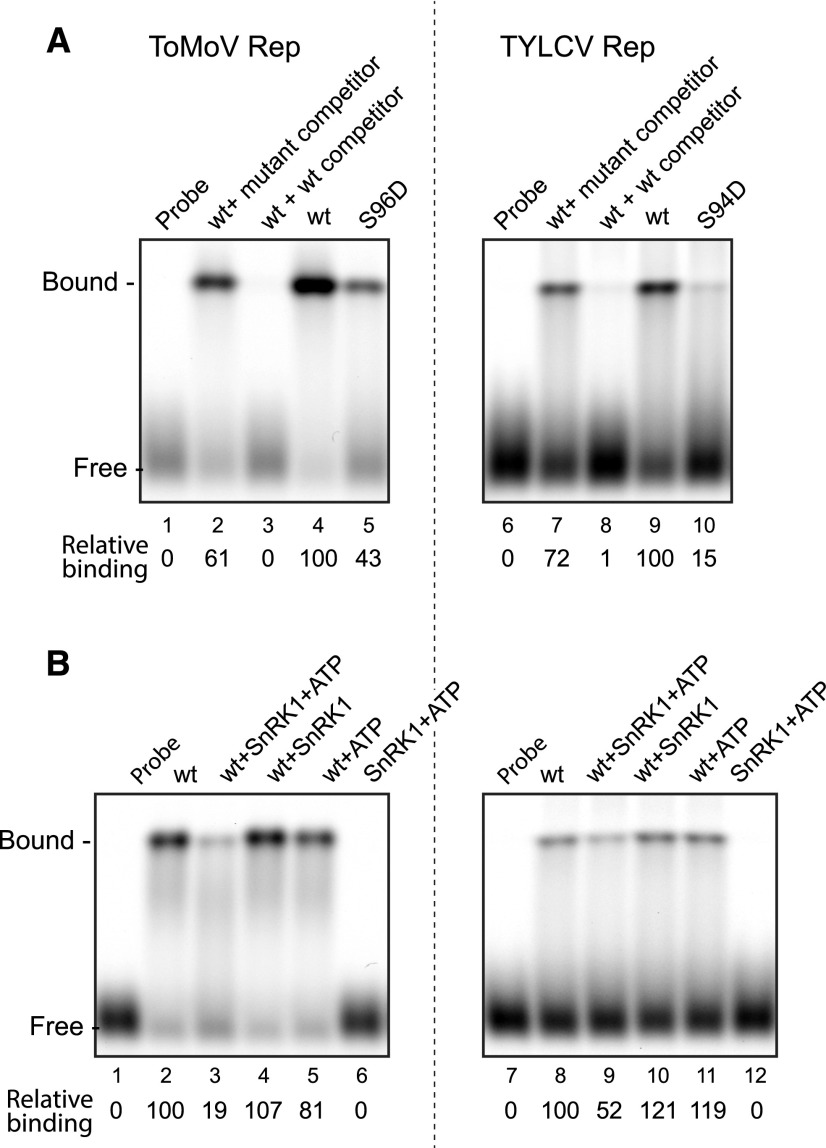
We next examined if SnRK1 phosphorylation interferes with DNA binding by ToMoV and TYLCV Rep proteins, analogous to TGMV Rep. We first used the ToMoV S96D and TYLCV S94D phosphomimics as proxies for the phosphorylated form in DNA-binding assays. We predicted the binding site sequences for both Reps based on iteron analyses (Fontes et al., 1994; Argüello-Astorga and Ruiz-Medrano, 2001) and generated the corresponding fluorescently labeled dsDNA probes for EMSAs (Supplemental Table S1). A single shifted band was observed when His6-tagged ToMoV Rep(1–179) (Fig. 8A, lane 4) and TYLCV Rep(1–177) (lane 9) were incubated with their predicted probe DNAs. Competition assays using unlabeled wild-type and mutant probes confirmed that both Reps bind specifically to their predicted binding sites (Fig. 8A, lanes 2, 3, 7, and 8). The ToMoV Rep S96D (lane 5) and TYLCV Rep S94D (lane 10) mutants had only 43% and 15% of the wild-type binding activity, respectively, indicating that the phosphomimic mutations impair the DNA binding of both Rep proteins. We then preincubated the wild-type ToMoV and TYLCV Rep proteins with preactivated SnRK1 and ATP prior to performing the DNA-binding assays. SnRK1 phosphorylation reduced the DNA-binding activities of ToMoV (Fig. 8B, lane 3) and TYLCV (lane 9) Reps to 19% and 52%, respectively, of their unphosphorylated Rep controls (lanes 2 and 8). Together, these results suggested that, like TGMV Rep, the Rep proteins of ToMoV, TYLCV, and probably many other begomoviruses are phosphorylated by SnRK1, leading to reduced interaction with viral DNA.
Figure 8.
SnRK1 phosphorylation interferes with ToMoV and TYLCV Rep binding to DNA. A, The sequence-specific DNA-binding activities of wild-type (wt) ToMoV Rep(1–179) (lanes 2–4) and TYLCV Rep(1–177) (lanes 7–9) proteins and their respective S96D (lane 5) and S94D (lane 10) mutants were examined in EMSAs. The assays contained 5 pmol of ToMoV or 100 pmol of TYLCV recombinant His6-tagged Rep protein and fluorescently labeled, double-stranded oligonucleotides corresponding to their predicted binding sites. The specificity of binding was verified in the presence of a 1,000-fold excess of unlabeled wild-type dsDNA competitors (lanes 3 and 8) or mutant competitors (lanes 2 and 7). Relative binding (shown below) is the ratio of the fluorescence of the shifted bands compared with lanes 4 and 9, which were set to 100, respectively, for ToMoV Rep and TYLCV Rep. B, The effects of SnRK1 phosphorylation on DNA binding by wild-type ToMoV or TYLCV Rep were assayed by preincubation of the Rep proteins with activated SnRK1 and ATP (lanes 3 and 9), activated SnRK1 alone (lanes 4 and 10), or ATP alone (lanes 5 and 11). Relative binding is shown for ToMoV Rep (ratio of lanes 3–6 to lane 2) or TYLCV Rep (ratio of lanes 9–12 to lane 8).
We also examined if Cabbage leaf curl virus (CaLCuV) Rep, which has an Asp instead of Ser at the corresponding residue position (Fig. 7A), is phosphorylated by SnRK1. No phosphorylation was detected when His6-tagged CaLCuV Rep(1–177) was incubated with activated SnRK1 and [γ-32P]ATP (Fig. 7B, lane 7). Moreover, the CaLCuV Rep D98S mutant was not phosphorylated by SnRK1 (lane 8), even though it contained a Ser residue at the equivalent position of the TGMV, ToMoV, and TYLCV Rep proteins. Thus, unlike most begomovirus Rep proteins, members of the SLC clade, which includes CaLCuV, may not be regulated by SnRK1 phosphorylation in the N-terminal region that mediates viral DNA binding. Modeling of CaLCuV Rep binding to its cognate viral dsDNA revealed that, like the TGMV Rep–DNA interaction, the contacts occur predominantly in three regions in Rep, but Asp-98 is not involved directly in the interaction (Fig. 3, C and D; Supplemental Fig. S3). In addition, the lowest energy Rep–DNA binding structures are very different between CaLCuV and TGMV, with a significant shift of interaction in CaLCuV toward the 5′ end of the iteron region (Supplemental Fig. S3), reflecting the differences in both the iteron sequences and the Rep sequences between the two viruses.
DISCUSSION
Posttranslational modifications, including phosphorylation of host and pathogen proteins, have been implicated in disease processes and defense responses in plants (Waigmann et al., 2000; Hulsmans et al., 2016). Host protein kinases that are present at functional levels in a plant can respond quickly to infection and have the potential to be among the earliest defenders against disease. The protein kinase SnRK1, a master regulator of plant energy homeostasis and primary metabolism, has been implicated in the host defense response by phosphorylating both pathogen and plant proteins (Hulsmans et al., 2016). We report here that SnRK1 phosphorylates the essential geminivirus replication protein, Rep, and interferes with its ability to bind to the viral origin of replication and to support efficient viral replication and infection. These results, in combination with earlier studies showing that SnRK1 phosphorylation of two other geminivirus proteins also impairs infection (Shen et al., 2011, 2014), underscore the importance of protein phosphorylation in the host defense response against geminiviruses.
We used a combination of mass spectrometry (Supplemental Fig. S1) and site-directed mutagenesis (Fig. 1) to identify Ser-97 as a major SnRK1 phosphorylation site in the TGMV Rep protein. The amino acid sequence surrounding Ser-97 conforms to the SnRK1 consensus target site, with a basic Arg at position −3 and hydrophobic Ile and Val at positions −5 and +4, respectively (Vlad et al., 2008). SnRK1 weakly phosphorylates an S97A mutant of both Rep(1–131) and Rep(1–180) (Fig. 1, C and E). The residual phosphorylation of the S97A mutant may be an artifact of the in vitro kinase assay, or the Rep N terminus may contain another SnRK1 phosphorylation site. One candidate is Ser-29, which is located in a SnRK1 consensus sequence (Fig. 1B), but structural modeling suggested that Ser-29 is not on the surface of Rep and not accessible to SnRK1. We also cannot rule out that SnRK1 phosphorylates another Ser or Thr in a context that does not conform to its consensus (Nukarinen et al., 2016). It is important to point out that we did not detect any other phospho-Sers or phospho-Thrs by mass spectrometry analysis, and if alternative SnRK1 target sites exist in TGMV Rep(1–180), they are minor targets of unknown functional significance. Because of the poor expression and solubility of full-length Rep and its C terminus in E. coli, we did not examine if the Rep C terminus is phosphorylated by SnRK1.
Amino acid residues 1 to 180 of TGMV Rep contain several conserved motifs and include the DNA-binding, DNA cleavage/ligation, and protein interaction domains (Fig. 1A). Ser-97 is the last amino acid residue of the highly conserved GRS motif (Nash et al., 2011). Functional in vitro assays showed that phosphorylation of TGMV Rep Ser-97 has no impact on DNA cleavage activity (Fig. 2A) but compromises dsDNA-binding activity (Fig. 2, C and D). Earlier studies indicated that the DNA-binding and cleavage activities of Rep are distinct and have different requirements. Rep binds and cleaves different sequences in the viral origin of replication (Orozco and Hanley-Bowdoin, 1996). Rep DNA-binding activity requires the formation of oligomers, while cleavage does not depend on oligomerization (Orozco et al., 2000). Other mutations in the GRS, which includes Ser-97, differentially impact binding and cleavage (Nash et al., 2011).
The ability of Rep to bind specifically to its recognition sequence in the origin is essential for viral DNA replication and infection (Fontes et al., 1992, 1994). Hence, it was not surprising that a TGMV DNA-A component with a Rep S97D mutation as a proxy for phosphorylation replicated poorly in tobacco protoplast assays (Fig. 4). Symptom development was delayed in N. benthamiana plants inoculated with the Rep S97D mutant, but eventually the plants infected with the mutant virus developed wild-type symptoms even though viral DNA levels were very low (Fig. 5), suggesting that the development of severe symptoms does not require viral DNA to reach wild-type levels. In contrast, the S97D mutant was unable to infect tomato (Fig. 6). This difference may indicate that N. benthamiana carries a mutation in host silencing pathways (Yang et al., 2004) and, as such, is a permissive host for many plant viruses.
Although the S97N Rep phosphonull mutation did not impair Rep dsDNA-binding activity (Fig. 2C), it was associated with a significant reduction in viral DNA accumulation, albeit to a lesser extent than the S97D phosphomimic mutant (Figs. 4 and 5E). This reduction suggested that the Rep S97N mutant is impaired for some other activity necessary for efficient replication. For example, the S97N mutation might impact the ability of Rep to interact with a viral or host protein necessary for efficient viral replication (Kong et al., 2000; Castillo et al., 2003; Settlage et al., 2005). The R94G phosphodeficient mutation did not impact Rep dsDNA-binding activity or viral replication (Figs. 4 and 5E). However, even though R94G was not a good SnRK1 substrate (Fig. 1E), it was not more infectious than wild-type virus (Fig. 5), suggesting that Arg-94 is involved in some other Rep function or that the Rep Ser-97 phosphoform plays a role in infection. It is also possible that only a small fraction of wild-type Rep Ser-97 is phosphorylated in infected plants, thereby minimizing any enhancement of infection by the Rep R94G mutation.
A structural model of the N terminus of TGMV Rep in association with its cognate dsDNA recognition site (Fig. 3) provided insight into how phosphorylation of Rep Ser-97 impairs DNA binding. The model identified three regions in TGMV Rep that contact dsDNA directly (Fig. 3D). The first region (residues 7, 9, and 11) is located immediately upstream of motif I and overlaps the iteron-related domain (Orozco and Hanley-Bowdoin, 1998; Chatterji et al., 1999; Argüello-Astorga and Ruiz-Medrano, 2001). The second interacting region (residues 68 and 72–75) is located between motif II and the GRS, while the third region (residues 94–97) overlaps the C-terminal end of the GRS (Orozco and Hanley-Bowdoin, 1998; Nash et al., 2011). The model shows amino acids Arg-7, Asn-9, Asn-11, and Arg-94 contacting DNA bases directly (Fig. 3D; Supplemental Fig. S3), suggesting that they mediate sequence specificity, consistent with earlier studies proposing that residues in the iteron-related domain confer binding specificity (Chatterji et al., 1999; Argüello-Astorga and Ruiz-Medrano, 2001). The model also shows that Arg-94 contacts the DNA phosphate backbone along with Lys-68, Gln-72 through Arg-75, Ala-95, and Lys-96. These interactions likely contribute to the overall strength of the Rep-DNA interaction. With the exception of Arg-75 in motif II, the model did not detect any direct contacts between DNA and motifs I, II, and III. This result was surprising given that Ala substitutions in the three motifs impair DNA-binding activity (Orozco and Hanley-Bowdoin, 1998). All of the motif mutations also impair DNA cleavage activity (Orozco and Hanley-Bowdoin, 1998), indicating that they may impose allosteric restrictions on Rep structure that interfere with its overall activity.
The model revealed that Ser-97 is located in a loop between the eighth β-strand and the second α-helix of the Rep structure and is exposed on the protein surface when it is not in association with DNA (Fig. 3B). Hence, Ser-97 is readily accessible to SnRK1 for phosphorylation. Phosphorylation of Ser-97 increases the negative electric potential at Ser-97, substantially increasing the overall negative electrostatic distribution at the Rep interface with dsDNA. Phosphorylation also introduces repelling forces between Ser-97 and the DNA phosphate backbone, both of which are negatively charged. Consequently, neighboring amino acid residues undergo allosteric reorientation and reorganization, changing the Rep-DNA interface. Interactions of Ser-97 and the surrounding residues with dsDNA are greatly reduced, and the overall strength of Rep-DNA binding decreases.
Over 90% of begomovirus Rep proteins have a Ser or a Thr at the equivalent position to TGMV Rep Ser-97 (Supplemental Fig. S5; Supplemental Data Set S1). The Ala and Lys residues at positions 95 and 96, respectively, also are highly conserved in begomovirus Rep proteins. In contrast, position 94, which most often is a Gly, is more variable and also can be an Ala, Ser, Arg, Thr, Lys, Asp, or Asn. This variability is striking because position −3 in the SnRK1 consensus sequence (equivalent to Rep position 94) is typically a basic amino acid (Vlad et al., 2008). We showed that SnRK1 can phosphorylate Rep proteins with a Gly (ToMoV) or an Ala (TYLCV) at the −3 position, but the level of phosphorylation is less than that of TGMV Rep with an Arg at −3 (Fig. 7). Moreover, phosphorylation of ToMoV and TYLCV Rep proteins reduced their binding to their cognate dsDNA recognition sites (Fig. 8). Hence, variation at the −3 position provides a mechanism for minimizing or modulating the impact of SnRK1 phosphorylation on begomovirus infection. The prevalence of Gly (80%; Supplemental Data Set S1) suggests that it could be the ancestral amino acid residue at this position. Mutations that rendered a virus with an Arg or Lys would make it more susceptible to SnRK1 interference and, hence, less virulent. This could offer an evolutionary benefit by increasing the survival rate of infected plants that can serve as sources for viral inoculum. Alternatively, the same virus could accumulate to lower levels and eventually disappear, consistent with the low frequency (3%) of Arg or Lys at the −3 position (Supplemental Data Set S1).
Another mechanism to counter SnRK1 interference with Rep would be to mutate the Ser/Thr target residue to an amino acid that cannot be phosphorylated. The TGMV S97N mutant provides insight into why this has not been the preferred mechanism. We showed that this mutation reduces the ability of TGMV Rep to support efficient viral replication even though it cannot be phosphorylated and has wild-type dsDNA-binding activity (Figs. 2C and 4). Similar results were seen with a TGMV S97A mutant (W. Shen and L. Hanley-Bowdoin, unpublished data). These results, in combination with the strong evolutionary conservation of Ser-97, indicated that this residue is involved in an unknown but important Rep function in addition to dsDNA binding. Interestingly, the SLC clade provides an example of a group of begomoviruses that have replaced the Ser with an Asp (Supplemental Fig. S5; Supplemental Data Set S1). This is striking given that Asp is the phosphomimic residue that impairs TGMV Rep DNA-binding and replication activities (Figs. 2C and 4). CaLCuV Rep and other members of the SLC clade also contain a number of other amino acid substitutions and small insertions/deletions in and around the GRS (Nash et al., 2011). Many of these changes target highly conserved amino acids, suggesting that the emergence of the SLC clade was a complex evolutionary process and/or involved an uncharacterized recombination event. The SLC clade originated after begomoviruses migrated to the New World, and its emergence is a recent event in begomovirus history (Lefeuvre et al., 2011), which may explain why most begomoviruses have Rep proteins that can be phosphorylated by SnRK1.
Geminiviruses are typically associated with vascular tissue, and virus-positive cells constitute only 1% to 10% of the total cells in an infected leaf (Nagar et al., 1995; Morra and Petty, 2000). Therefore, total protein extracts from infected leaves contain very low levels of the Rep protein, and it is not technically feasible to detect phosphorylated Rep in vivo. However, both the Rep and SnRK1 proteins have been shown to localize to nuclei in leaves, providing an opportunity for phosphorylation (Nagar et al., 1995; Kong and Hanley-Bowdoin, 2002; Ascencio-Ibáñez et al., 2008; Bitrián et al., 2011; Williams et al., 2014). In addition, SnRK1 is widely distributed in leaves, including vascular tissue where geminiviruses occur (Hannappel et al., 1995; Bitrián et al., 2011; Williams et al., 2014). Moreover, GRIK, which interacts with Rep and activates SnRK1, localizes specifically to nuclei of infected cells (Kong and Hanley-Bowdoin, 2002; Shen et al., 2009). Together, these observations strongly suggest that SnRK1 phosphorylates Rep in planta to limit viral replication and attenuate infection.
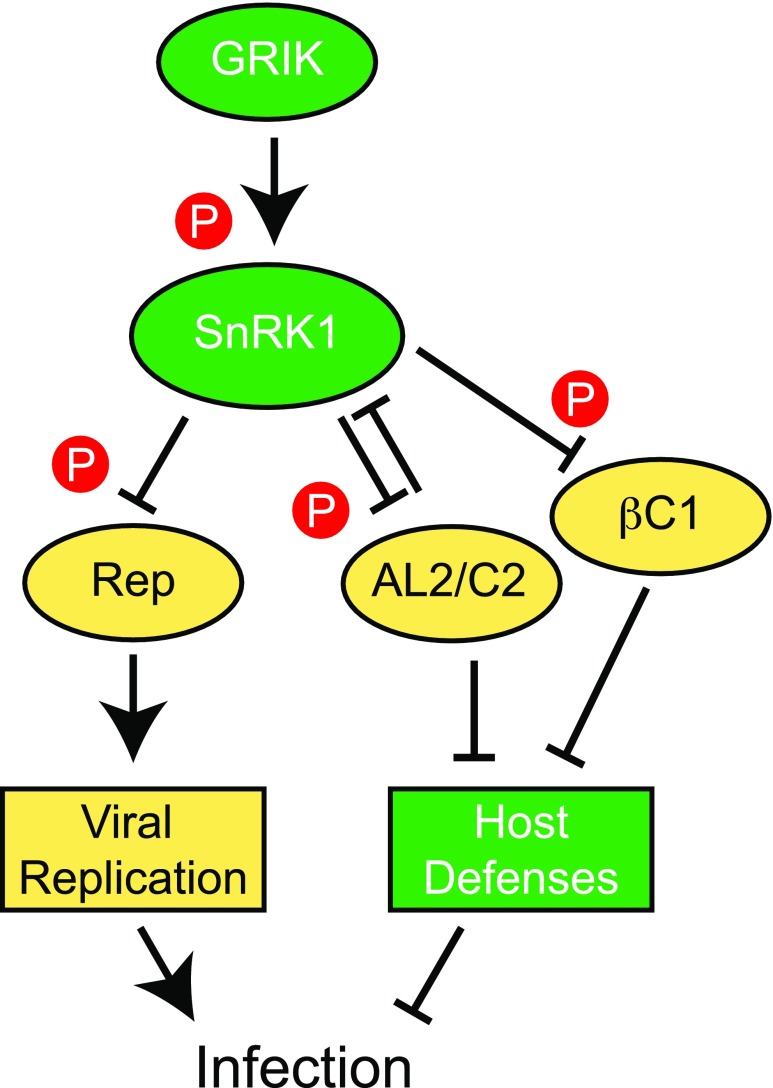
The replication of viral and host DNA and other processes necessary for geminivirus infection require a significant expenditure of metabolites and energy that can pose a major stress on the host. Given the central role of SnRK1 in sensing decreasing levels of cellular energy or nutrients in plants (Tomé et al., 2014), it is not surprising that SnRK1 phosphorylates multiple geminivirus proteins to suppress their activities necessary for the establishment and progression of infection (Fig. 9). The viral proteins targeted by SnRK1 have diverse functions and different consequences. We showed here that SnRK1 phosphorylation of Rep interferes with viral replication. In contrast, the host defense response is enhanced by SnRK1 phosphorylation of AL2/C2 (Shen et al., 2014) and the β-satellite-encoded βC1 protein (Shen et al., 2011; Zhong et al., 2017), both of which suppress transcriptional and posttranscriptional gene silencing (Cui et al., 2005; Wang et al., 2005; Buchmann et al., 2009; Yang et al., 2011; Zhang et al., 2011; Castillo-González et al., 2015).
Figure 9.
Model showing that SnRK1 phosphorylates multiple viral proteins to interfere with geminivirus infection. The host protein kinase SnRK1 is activated by the upstream kinase GRIK, which is up-regulated by geminivirus infection. SnRK1, in turn, phosphorylates several viral proteins, including Rep, AL2/C2, and the β-satellite-encoded βC1 protein. Phosphorylation of Rep inhibits its dsDNA-binding activity to interfere with viral DNA replication and infection. Phosphorylation of AL2/C2 and βC1, which are viral suppressors of gene silencing, also makes the virus less infectious by enhancing the host defense response.
Among the three viral proteins known to be phosphorylated by SnRK1, Rep and AL2/C2 are encoded by all begomoviruses. It is noteworthy that the prevalence of the phosphorylation target sites differs between Rep and AL2/C2. The AL2/C2 Ser residue phosphorylated by SnRK1 occurs only in a subset of New World begomoviruses and may have arisen during the adaptation of begomoviruses to the Americas (Shen et al., 2014). In contrast, the SnRK1 target site in Rep is found in the vast majority of begomoviruses in both the Old and New Worlds as well as in many geminivirus species in other genera (Supplemental Fig. S5; Supplemental Data Set S1). Interestingly, members of the SLC clade contain the AL2 target site but lack the Rep target site. The impact of SnRK1 phosphorylation also differs for the two viral proteins. Phosphorylation of AL2/C2 is associated with a minor but significant delay in symptom development (Shen et al., 2014), while the effect of SnRK1 phosphorylation of Rep is more profound, as demonstrated by the complete loss of infectivity of a TGMV Rep S97D phosphomimic mutant in tomato (Figs. 5 and 6).
Although SnRK1 is generally viewed as part of the host defense response, it also may play a positive role in the infection process. Rep is a multifunctional protein that is involved in a variety of host protein interactions as well as viral DNA replication (Fondong, 2013; Hanley-Bowdoin et al., 2013). It is not known how the various Rep functions are modulated. During infection, it is likely that only a fraction of the Rep protein is phosphorylated by SnRK1, and the unphosphorylated form is competent for dsDNA binding and supports viral replication. In contrast, phosphorylated Rep may have a specific function distinct from replication (i.e. interacting with a host protein or the viral REn protein). According to this model, SnRK1 phosphorylation of Rep would be regulatory, and the impact of phosphorylation on infection would depend on the stoichiometric ratio of unphosphorylated and phosphorylated Rep, which could be modulated by the selection of the amino acid residue at the −3 position in the SnRK1 recognition sequence of Rep. This scenario also provides a rationale for the maintenance of the SnRK1 target site in Rep over time and underscores the complex relationship between SnRK1 and geminiviruses.
MATERIALS AND METHODS
Plasmid Construction
Oligonucleotides and plasmids used in this study are listed in Supplemental Tables S1 and S2, respectively. The Escherichia coli expression plasmid for His6-tagged TGMV Rep(1–131) was generated by inserting an NdeI-KasI(filled) fragment containing the Rep coding sequence from pNSB828 (Kong et al., 2000) into pET16b, which had been digested with BamHI, filled, and digested again with NdeI. His6-tagged TGMV Rep(1–180) has been published previously (Nash et al., 2011). To construct expression plasmids for His6-tagged ToMoV Rep(1–179) and TYLCV Rep(1–177), the Rep coding regions were amplified from FS577pGEMX (provided by J. Polston of Florida State University) and pTYLC2 (Settlage et al., 2005), respectively, using the Phusion DNA polymerase (New England Biolabs). The PCR products were digested partially with NdeI and completely with BamHI and ligated to pET16b cut with the same enzymes. The expression plasmid pNSB1044 for His6-tagged full-length CaLCuV Rep was constructed by cloning an NdeI-XhoI fragment from pNSB981 (Kong and Hanley-Bowdoin, 2002) into pET16b digested with the same enzymes. The expression plasmid for His6-tagged CaLCuV Rep(1–177) was generated by amplifying the coding sequence from pNSB1044 followed by digestion with NdeI and XhoI and cloning into pET16b cut with the same enzymes. Rep mutants were generated using pairs of complementary primers containing the desired mutations and Pfu Ultra DNA polymerase (Agilent Technologies) to copy plasmids containing wild-type Rep coding sequences. The resulting DNAs were treated with DpnI and transformed into E. coli DH5α cells. Transformants were screened for the correct mutations by DNA sequencing.
To construct viral replicons for tobacco (Nicotiana tabacum) protoplast studies, a TGMV AL4-knockout mutant (M1T-M3T) was generated by site-directed mutagenesis of pMON344, which has a single copy of TGMV DNA-A in pUC18 (Elmer et al., 1988). Next, the Rep S97D, S97N, or R94G mutation was introduced into the AL4-knockout mutant background. Mutant viral replicons were generated by replacing a 1.1-kb HindIII-SalI fragment in pMON1565, which has a partial tandem copy of TGMV DNA-A with duplicated 5′ intergenic regions (Elmer et al., 1988), with the corresponding mutant fragments. For plant infection assays, the AL4-knockout and the Rep phosphorylation mutations also were introduced into the partial copy of TGMV DNA-A. The plasmids were partially digested with EcoRI and XhoI to generate 5.6-kb plasmid backbones that either lost a 750-bp fragment containing the mutations in the full repeat or a fragment of the same size in the partial repeat without the mutation. These fragments were then replaced by the corresponding 750-bp EcoRI-XhoI fragment derived from the plasmid that has a single copy of the DNA-A replicon containing the same mutations. Plasmids with two copies of each mutation were selected by DNA sequencing.
Recombinant Protein Production and Purification
The production and purification of recombinant Arabidopsis (Arabidopsis thaliana) SnRK1.1 and GRIK1 proteins in E. coli have been reported previously (Shen et al., 2009). Plasmids generated in strain DH5α were introduced into the strain BL21(DE3) or BL21(DE3)pLysE for protein production. Late log-phase cultures grown at 37°C in 250 mL of Luria-Bertani medium were induced by 0.5 mm IPTG at 16°C for 18 h. Cells were pelleted by centrifugation at 4,000g for 15 min and washed in TBS (25 mm Tris-HCl, pH 7.5, 140 mm NaCl, and 2.7 mm KCl). Bacteria were disrupted by 12 rounds of 10-s sonication pulses in 10 mL of binding buffer (20 mm Tris-HCl, pH 8, 500 mm NaCl, 10 mm imidazole, and 0.1% Triton X-100). Lysates were cleared by centrifugation at 32,500g for 30 min and applied to a 0.4-mL bed volume of Ni-NTA agarose (Qiagen) for batch-wise purification. After two 10-mL washes with the binding buffer and one 10-mL wash with binding buffer adjusted to 30 mm imidazole, the His6-tagged proteins were eluted in three 400-μL fractions of elution buffer (20 mm Tris-HCl, pH 8, 500 mm NaCl, and 500 mm imidazole). Purified proteins were concentrated, equilibrated to TBS using Amicon Ultra centrifugal filter devices, and stored at −20°C in 0.5× TBS, 0.5 mm DTT, and 50% (v/v) glycerol.
Protein Quantification, SDS-PAGE, and Immunoblotting
Protein concentration was measured by the Bradford method with the Protein Assay Reagent (Bio-Rad). BSA was used as a standard protein. Standard procedures were followed for SDS-PAGE with a stacking gel for protein electrophoresis. Coomassie Brilliant Blue-stained protein bands were quantified by scanning with a 700-nm laser in a LI-COR Odyssey Infrared Imager. For immunoblotting, proteins were transferred to nitrocellulose membranes and probed with mouse anti-His6 monoclonal antibodies (Takara Bio; 1:20,000 dilution). Fluorescently labeled secondary antibodies (1:20,000 dilution; Alexa Fluor 680 [Thermo Fisher Scientific] or IRdye 680 [LI-COR]) were used to detect the primary antibodies in the Odyssey Imager.
Protein Phosphorylation Assay
Protein phosphorylation reactions were performed in 50 µL of 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 1 mm DTT, 1 mm EDTA, 0.1 mm ATP, 1 µCi of [γ-32P]ATP, and 25 pmol of each kinase and substrate protein. After incubation at 30°C for 30 min, the reactions were stopped by adding an equal volume of 2× SDS-PAGE sample buffer and boiled for 5 min before SDS-PAGE. Gels were stained with Coomassie Brilliant Blue and dried, and the labeled proteins were visualized by autoradiography. Alternatively, the separated proteins were transferred to nitrocellulose membrane for autoradiography and later quantified by immunoblotting with His6 antibodies. For the preparation of activated and purified His6-SnRK1.1 kinase domain, 2.5 nmol of each GST-GRIK1 and His6-SnRK1.1 kinase domain was incubated in a 300-µL kinase reaction mixture with unlabeled ATP at 30°C for 1 h. GST-GRIK1 was removed by two passes through a 50-µL bed volume of glutathione-Sepharose (GE Healthcare). The supernatant was then incubated with a 50-µL bed volume of Ni-NTA agarose, and His6-SnRK1.1 was eluted with four fractions of 50 µL of elution buffer. Pooled protein was concentrated to greater than 1 µg µL−1 and stored in 0.5 mm DTT and 50% (v/v) glycerol at −20°C. Peptide kinase assays using a peptide, acetyl-KGRMRRISSVEMMK (GenScript), containing a SnRK1 phosphorylation site from spinach (Spinacia oleracea) Suc-phosphate synthase was described previously (Huang and Huber, 2001; Shen et al., 2009).
Mass Spectrometry
A 300-pmol aliquot of the TGMV His6-Rep(1–131) recombinant protein was incubated with 100 nmol each of His6-GRIK1 and His6-SnRK1.1 kinase domain in a 120-μL phosphorylation reaction mixture with unlabeled ATP at 30°C for 1 h. The proteins were separated on a NuPAGE 4–12% Bis-Tris gradient gel (Invitrogen) and stained with NOVEX Colloidal Blue (Invitrogen), and the band for His6-Rep was excised and minced. The excised band was subjected to in-gel proteolysis according to a published protocol (Wilm et al., 1996), except that chymotrypsin was used as the protease. Following digestion, the resulting proteolytic peptides were analyzed by LC/MSE as described previously (Cheng et al., 2009). LC/MSE data files were processed and searched against the TAIR database, to which the His6-Rep sequence had been added, using ProteinLynx Global Server 2.4 (Waters). Reconstructed LC/MSE product ion spectra corresponding to putative phosphopeptides were inspected manually for phosphosite assignment.
Modeling of Rep-DNA Interaction
A structural model for TGMV Rep was created as described previously (Nash et al., 2011). The PyMOL Molecular Graphics System version 1.8 (Schrodinger) was used for generation of the posttranslational modification of pSer-97, structural analysis, and image creation. 3D-DART was used to generate a B-form dsDNA pdb (van Dijk and Bonvin, 2009) of the 5′-intergenic region of TGMV DNA (Fig. 3A), containing the sequence specifically recognized by TGMV Rep (Fontes et al., 2004; van Dijk and Bonvin, 2009).
Molecular dynamics (MD) simulations of TGMV Rep(7–122) and the viral DNA sequences were performed with the GROMACS 5.1 software package using the OPLS/AA force field and the flexible SPC water model individually (Abraham et al., 2015). The initial structures were immersed in a periodic dodecahedron water box of dodecahedron shape (1 nm thickness) and neutralized with counterions. Electrostatic energy was calculated using the particle mesh Ewald method with 1-nm cutoff distances for the Coulomb and van der Waals interactions. After energy minimization, the system was equilibrated to 300 K and normal pressure for 100 ps with position restraints for heavy atoms and LINCS constraints for all bonds. The system was coupled to the external bath by the Parrinello-Rahman pressure and temperature coupling. The final MD calculations were performed under the same conditions except that the position restraints were removed and the simulation was run for 100 ns. Every tenth frame from 10 to 100 ns of the simulation was extracted and combined into a conformational accurate description of the B-form dsDNA and TGMV Rep(7–122) in solution.
Default High Ambiguity Drive Docking (HADDOCK) parameters were used throughout the docking procedure on the Web server (van Dijk et al., 2012; van Zundert et al., 2016) with the following exceptions: auto_passive_radius = 6.5, create_narestraints = True, rotate180_0 = False, crossdock = True, and calcdesolv = True. Active residues for Rep(7-122) were defined as Arg-7, Lys-68, Arg-94, Ala-95, Lys-96, and Ser-97, and active bp corresponding to the Rep binding site (Fig. 3A) for the dsDNA ensemble pdb. One thousand structures were generated for the first iteration (rigid docking), and 200 were generated for each subsequent iteration (semiflexible docking and water refinement). The Cα/P root mean square deviation values of the complexes were calculated using ProFit version 3.1 (SciTech Software).
A cluster analysis was performed on the final docking solutions using a minimum cluster size of 4 and a root mean square deviation of 7.5 Å. The structures in the resulting clusters were scored using the FireDock scoring program/method (Andrusier et al., 2007; Mashiach et al., 2008) and plotted in a box plot to determine statistical outliers and median scores. The structures that represented the median score for the highest populated cluster were then visualized with PyMOL. The electrostatic surface of the protein was created by using the Adaptive Poisson-Boltzmann Solver in PyMOL (Baker et al., 2001; Dolinsky et al., 2004; Lerner and Carlson, 2006). The PDB2PQR Web server (Dolinsky et al., 2004) was used with the CHARMM force field and output naming schemes, which include parameters for phosphorylated Ser.
A model of CaLCuV Rep(5–116), its cognate dsDNA (Fig. 3A), MD simulations, and HADDOCK docking of the protein and DNA were generated using the same approaches described for TGMV Rep(7–122).
DNA Cleavage and Binding Assays
An IRDye-700-labeled single-stranded oligonucleotide (IDT) was used as a substrate in DNA cleavage assays for TGMV Rep. The reaction included 50, 5, or 0.5 pmol of recombinant His6-Rep(1–180) and 75 fmol of the probe in a 20-μL reaction mixture containing 25 mm Tris-HCl, pH 7.5, 75 mm NaCl, 5 mm MgCl2, 2 mm EDTA, 2.5 mm DTT, and 12.5 μg μL−1 poly(dI-dC). After incubation for 30 min at 22°C, the reaction was stopped by the addition of 10 μL of a gel electrophoresis loading buffer containing 50 mm EDTA, pH 8, and boiled for 2 min. DNA fragments were separated by electrophoresis on a 15% denaturing polyacrylamide gel in 0.5× TBE buffer and visualized by scanning the gel in a LI-COR Odyssey Imager.
For DNA-binding assays, double-stranded oligonucleotide probes were generated by annealing two complementary single-stranded oligonucleotides, one of which was 5′ labeled with IRDye-700 (IDT; Nash et al., 2011). Binding of 50 pmol of recombinant His6-tagged TGMV Rep(1–180), 5 pmol of ToMoV Rep(1–179), or 100 pmol of TYLCV Rep(1–177) with 50 fmol of their respective probe was carried out in 20 μL of 20 mm Tris-HCl, pH 8, 40 mm KCl, 5 mm MgCl2, 0.5 mm EDTA, 1 mm DTT, 0.5 μg μL−1 sheared salmon sperm DNA, 0.1 μg μL−1 BSA, and 5% (v/v) glycerol. After incubation at 22°C for 30 min, the protein-DNA mixture was applied to a 0.7% agarose gel in 0.5× TBE buffer for electrophoresis, and the gel was scanned in a LI-COR Odyssey Imager to visualize fluorescent signals and quantify band intensity. In some cases, 50 pmol of unlabeled double-stranded oligonucleotides was included in the 20-µL binding reaction for competition with the fluorescent dsDNA probe. To test directly the effect of SnRK1 phosphorylation of Rep on DNA binding, 100 pmol of TGMV Rep, 10 pmol of ToMoV Rep, or 200 pmol of TYLCV Rep was incubated with 10 pmol of purified, activated SnRK1.1 kinase domain in a 25-μL phosphorylation reaction mixture containing cold ATP and 0.16 μg μL−1 BSA at 30°C for 30 min. Half of the phosphorylation reaction volume was used in the 20-μL DNA-binding assay.
Viral DNA Replication in Tobacco Protoplasts
Protoplasts were prepared from tobacco NT1 cells and electroporated with 10 µg of pUC19-based plasmids containing TGMV DNA-A replicons (Fontes et al., 1994). The transfected protoplasts were incubated at 25°C for 3 d. The cells were then collected in a 2-mL screw-capped tube (Sarstedt) and ground in a Retsch MM301 grinder for 2 min at a frequency of 30 rounds per second prior to extraction, purification, and RNase A treatment of total DNA (Shen et al., 2014). The DNA (20 μg) was digested with XhoI and DpnI for DNA gel-blot analysis using a γ-32P-labeled 0.75-kb EcoRI-XhoI TGMV DNA-A fragment that spans the 5′ intergenic region as a probe. Hybridized DNA was visualized by autoradiography.
Virus Infection Assays in Nicotiana benthamiana and Tomato Plants
N. benthamiana and tomato (Solanum lycopersicum) Florida Lanai plants were grown at 25°C under a 16-h/8-h light/dark cycle. For TGMV infection, the apical tips of 4-week-old N. benthamiana plants (approximately eight leaves) or 3-week-old tomato plants were bombarded with 1-μm gold particles coated with viral replicon DNA using a hand-held microsprayer (Venganza) at pressures of 200 and 270 kPa, respectively (Cabrera-Ponce et al., 1997). Each plant was inoculated with a mixture of pUC19-based plasmids (750 ng each) containing partial tandem copies of TGMV DNA-A or DNA-B. Leaf curl and chlorosis symptoms were scored separately over time using five-point scales. For leaf curl, the scale was as follows: 1, no symptom; 2, leaf curl/wrinkles starting to appear; 3, small areas of curl/wrinkles on two leaves or large area on one leaf; 4, more than three leaves with curl/wrinkles; and 5, fully established leaf curl/wrinkles showing stunted growth. For leaf chlorosis, the scale was as follows: 1, no symptom; 2, small chlorotic spots on one leaf; 3, chlorosis spread to two leaves or a large area on one leaf; 4, three or more leaves showing chlorosis and some with yellow veins; and 5, fully established leaf chlorosis in large areas and/or yellow veins.
For infection in N. benthamiana, the first expanded leaf (1–2 cm long) and the shoot tip were collected and flash frozen in liquid nitrogen from plants at 4, 10, or 17 dpi. The frozen tissues were ground in a Retsch MM301 grinder for 2 min at a frequency of 30 rounds per second, and total DNA was extracted, purified, and treated with RNase A as published previously (Shen et al., 2014). DNA (12 μg) was digested with XhoI and DpnI and subjected to DNA gel-blot analysis as described above. For infected tomato plants, a bottom leaflet of the forth compound leaf counted from the top was collected for DNA extraction at 28 dpi. Total DNA (50 ng) was amplified in a 50-μL reaction mixture with Taq DNA polymerase (New England Biolabs) and a pair of specific primers for TGMV DNA-A (Supplemental Table S1).
Phylogenetic Analysis
The amino acid sequence of a Rep protein from one isolate of each geminivirus species assembled by several recent publications (Muhire et al., 2013; Varsani et al., 2014a, 2014b; Brown et al., 2015) was selected for alignment by Clustal and used to construct a phylogenetic tree by the neighbor-joining method with the software package MEGA7 (Kumar et al., 2016). A few of the begomovirus Rep proteins were not included due to poor DNA sequence quality or putative mutations that did not allow proper sequence alignment (Supplemental Data Set S1).
Accession Numbers
The gene and viral DNA accession numbers that were used in this study are as follows: AT3G01090 (SnRK1.1), AT3G45240 (GRIK1), NC_001507.1 (TGMV DNA A), NC_001508.1 (TGMV DNA B), NC_001938.1 (ToMoV DNA A), EF110890.1 (TYLCV), and NC_003866.1 (CaLCuV DNA A).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Mass spectrometry identification of phosphopeptides from His6-tagged TGMV Rep(1–131) treated with SnRK1.
Supplemental Figure S2. Affinity purification of GRIK-activated SnRK1 retains its kinase activity.
Supplemental Figure S3. Cartoon depictions of Rep-DNA contacts.
Supplemental Figure S4. AL4 knockout has a minimal impact on TGMV replication and symptom development.
Supplemental Figure S5. Phylogenetic tree of geminivirus Rep proteins highlighting putative SnRK1 phosphorylation sites.
Supplemental Table S1. List of oligonucleotides.
Supplemental Table S2. List of plasmids.
Supplemental Data Set S1. List of species and isolates used for the Rep phylogenetic tree.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Rajrani Ruhel (North Carolina State University) for comments on this article and Dr. Beverly M. Orozco (Roivant Sciences) for making plasmid pNSB873.
Footnotes
This work was supported by National Science Foundation Grant MCB 1052218 to L.H.-B. and M.B.G.
Articles can be viewed without a subscription.
References
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2: 19–25 [Google Scholar]
- Andrusier N, Nussinov R, Wolfson HJ (2007) FireDock: fast interaction refinement in molecular docking. Proteins 69: 139–159 [DOI] [PubMed] [Google Scholar]
- Arguello-Astorga G, Ascencio-Ibáñez JT, Dallas MB, Orozco BM, Hanley-Bowdoin L (2007) High-frequency reversion of geminivirus replication protein mutants during infection. J Virol 81: 11005–11015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello-Astorga GR, Ruiz-Medrano R (2001) An iteron-related domain is associated to motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Arch Virol 146: 1465–1485 [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibáñez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitrián M, Roodbarkelari F, Horváth M, Koncz C (2011) BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. Plant J 65: 829–842 [DOI] [PubMed] [Google Scholar]
- Broeckx T, Hulsmans S, Rolland F (2016) The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. J Exp Bot 67: 6215–6252 [DOI] [PubMed] [Google Scholar]
- Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos-Sobrinho R, Silva JC, Fiallo-Olivé E, Briddon RW, Hernández-Zepeda C, Idris A, et al. (2015) Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol 160: 1593–1619 [DOI] [PubMed] [Google Scholar]
- Buchmann RC, Asad S, Wolf JN, Mohannath G, Bisaro DM (2009) Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol 83: 5005–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgyán J, Havelda Z (2011) Viral suppressors of RNA silencing. Trends Plant Sci 16: 265–272 [DOI] [PubMed] [Google Scholar]
- Cabrera-Ponce JL, Lopez L, Assad-Garcia N, Medina-Arevalo C, Bailey AM, Herrera-Estrella L (1997) An efficient particle bombardment system for the genetic transformation of asparagus (Asparagus officinalis L.). Plant Cell Rep 16: 255–260 [DOI] [PubMed] [Google Scholar]
- Campos-Olivas R, Louis JM, Clerot D, Gronenborn B, Gronenborn AM (2002) The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc Natl Acad Sci USA 99: 10310–10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo AG, Collinet D, Deret S, Kashoggi A, Bejarano ER (2003) Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312: 381–394 [DOI] [PubMed] [Google Scholar]
- Castillo-González C, Liu X, Huang C, Zhao C, Ma Z, Hu T, Sun F, Zhou Y, Zhou X, Wang XJ, et al. (2015) Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. eLife 4: e06671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji A, Padidam M, Beachy RN, Fauquet CM (1999) Identification of replication specificity determinants in two strains of tomato leaf curl virus from New Delhi. J Virol 73: 5481–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng FY, Blackburn K, Lin YM, Goshe MB, Williamson JD (2009) Absolute protein quantification by LC/MS(E) for global analysis of salicylic acid-induced plant protein secretion responses. J Proteome Res 8: 82–93 [DOI] [PubMed] [Google Scholar]
- Clérot D, Bernardi F (2006) DNA helicase activity is associated with the replication initiator protein Rep of tomato yellow leaf curl geminivirus. J Virol 80: 11322–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Li G, Wang D, Hu D, Zhou X (2005) A Begomovirus DNAbeta-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J Virol 79: 10764–10775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbiez C, David C, Mettouchi A, Laufs J, Gronenborn B (1995) Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc Natl Acad Sci USA 92: 5640–5644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32: W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Holmes EC (2008) Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus tomato yellow leaf curl virus. J Virol 82: 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer JS, Sunter G, Gardiner WE, Brand L, Browning CK, Bisaro DM, Rogers SG (1988) Agrobacterium-mediated inoculation of plants with tomato golden mosaic virus DNAs. Plant Mol Biol 10: 225–234 [DOI] [PubMed] [Google Scholar]
- Emanuelle S, Doblin MS, Stapleton DI, Bacic A, Gooley PR (2016) Molecular insights into the enigmatic metabolic regulator, SnRK1. Trends Plant Sci 21: 341–353 [DOI] [PubMed] [Google Scholar]
- Fondong VN. (2013) Geminivirus protein structure and function. Mol Plant Pathol 14: 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EP, Luckow VA, Hanley-Bowdoin L (1992) A geminivirus replication protein is a sequence-specific DNA binding protein. Plant Cell 4: 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EP, Eagle PA, Sipe PS, Luckow VA, Hanley-Bowdoin L (1994) Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J Biol Chem 269: 8459–8465 [PubMed] [Google Scholar]
- Fontes EP, Santos AA, Luz DF, Waclawovsky AJ, Chory J (2004) The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev 18: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghillebert R, Swinnen E, Wen J, Vandesteene L, Ramon M, Norga K, Rolland F, Winderickx J (2011) The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: structure, function and regulation. FEBS J 278: 3978–3990 [DOI] [PubMed] [Google Scholar]
- Glab N, Oury C, Guérinier T, Domenichini S, Crozet P, Thomas M, Vidal J, Hodges M (2017) The impact of Arabidopsis thaliana SNF1-related-kinase 1 (SnRK1)-activating kinase 1 (SnAK1) and SnAK2 on SnRK1 phosphorylation status: characterization of a SnAK double mutant. Plant J 89: 1031–1041 [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Settlage SB, Robertson D (2004) Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol Plant Pathol 5: 149–156 [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S (2013) Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Hannappel U, Vicente-Carbajosa J, Barker JH, Shewry PR, Halford NG (1995) Differential expression of two barley SNF1-related protein kinase genes. Plant Mol Biol 27: 1235–1240 [DOI] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15: 1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey S, Mayerhofer H, Halford NG, Dickinson JR (2007) DNA sequences from Arabidopsis, which encode protein kinases and function as upstream regulators of Snf1 in yeast. J Biol Chem 282: 10472–10479 [DOI] [PubMed] [Google Scholar]
- Huang JZ, Huber SC (2001) Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase: influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol 42: 1079–1087 [DOI] [PubMed] [Google Scholar]
- Hulsmans S, Rodriguez M, De Coninck B, Rolland F (2016) The SnRK1 energy sensor in plant biotic interactions. Trends Plant Sci 21: 648–661 [DOI] [PubMed] [Google Scholar]
- Jeske H, Lütgemeier M, Preiss W (2001) DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J 20: 6158–6167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LJ, Hanley-Bowdoin L (2002) A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14: 1817–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong LJ, Orozco BM, Roe JL, Nagar S, Ou S, Feiler HS, Durfee T, Miller AB, Gruissem W, Robertson D, et al. (2000) A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J 19: 3485–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Ilyina TV (1992) Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J Gen Virol 73: 2763–2766 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs J, Traut W, Heyraud F, Matzeit V, Rogers SG, Schell J, Gronenborn B (1995) In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc Natl Acad Sci USA 92: 3879–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P, Harkins GW, Lett JM, Briddon RW, Chase MW, Moury B, Martin DP (2011) Evolutionary time-scale of the begomoviruses: evidence from integrated sequences in the Nicotiana genome. PLoS ONE 6: e19193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MG, Carlson HA (2006) APBS Plugin for PyMOL. University of Michigan, Ann Arbor [Google Scholar]
- Mansoor S, Briddon RW, Zafar Y, Stanley J (2003) Geminivirus disease complexes: an emerging threat. Trends Plant Sci 8: 128–134 [DOI] [PubMed] [Google Scholar]
- Mansoor S, Zafar Y, Briddon RW (2006) Geminivirus disease complexes: the threat is spreading. Trends Plant Sci 11: 209–212 [DOI] [PubMed] [Google Scholar]
- Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ (2008) FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res 36: W229–W232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra MR, Petty IT (2000) Tissue specificity of geminivirus infection is genetically determined. Plant Cell 12: 2259–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhire B, Martin DP, Brown JK, Navas-Castillo J, Moriones E, Zerbini FM, Rivera-Bustamante R, Malathi VG, Briddon RW, Varsani A (2013) A genome-wide pairwise-identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae). Arch Virol 158: 1411–1424 [DOI] [PubMed] [Google Scholar]
- Nagar S, Pedersen TJ, Carrick KM, Hanley-Bowdoin L, Robertson D (1995) A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash TE, Dallas MB, Reyes MI, Buhrman GK, Ascencio-Ibañez JT, Hanley-Bowdoin L (2011) Functional analysis of a novel motif conserved across geminivirus Rep proteins. J Virol 85: 1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Castillo J, Fiallo-Olivé E, Sánchez-Campos S (2011) Emerging virus diseases transmitted by whiteflies. Annu Rev Phytopathol 49: 219–248 [DOI] [PubMed] [Google Scholar]
- Nukarinen E, Nägele T, Pedrotti L, Wurzinger B, Mair A, Landgraf R, Börnke F, Hanson J, Teige M, Baena-Gonzalez E, et al. (2016) Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci Rep 6: 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Hanley-Bowdoin L (1996) A DNA structure is required for geminivirus replication origin function. J Virol 70: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco BM, Hanley-Bowdoin L (1998) Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J Biol Chem 273: 24448–24456 [DOI] [PubMed] [Google Scholar]
- Orozco BM, Miller AB, Settlage SB, Hanley-Bowdoin L (1997) Functional domains of a geminivirus replication protein. J Biol Chem 272: 9840–9846 [DOI] [PubMed] [Google Scholar]
- Orozco BM, Kong LJ, Batts LA, Elledge S, Hanley-Bowdoin L (2000) The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J Biol Chem 275: 6114–6122 [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M (2007) SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci 12: 20–28 [DOI] [PubMed] [Google Scholar]
- Pooma W, Petty IT (1996) Tomato golden mosaic virus open reading frame AL4 is genetically distinct from its C4 analogue in monopartite geminiviruses. J Gen Virol 77: 1947–1951 [DOI] [PubMed] [Google Scholar]
- Rajabu CA, Kennedy GG, Ndunguru J, Ateka EM, Tairo F, Hanley-Bowdoin L, Ascencio-Ibáñez JT (2018) Lanai: a small, fast growing tomato variety is an excellent model system for studying geminiviruses. J Virol Methods 256: 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas MR, Hagen C, Lucas WJ, Gilbertson RL (2005) Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Annu Rev Phytopathol 43: 361–394 [DOI] [PubMed] [Google Scholar]
- Roustan V, Jain A, Teige M, Ebersberger I, Weckwerth W (2016) An evolutionary perspective of AMPK-TOR signaling in the three domains of life. J Exp Bot 67: 3897–3907 [DOI] [PubMed] [Google Scholar]
- Sacristán S, García-Arenal F (2008) The evolution of virulence and pathogenicity in plant pathogen populations. Mol Plant Pathol 9: 369–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal SE, vandenBosch F, Jeger MJ (2006) Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit Rev Plant Sci 25: 23–46 [Google Scholar]
- Settlage SB, See RG, Hanley-Bowdoin L (2005) Geminivirus C3 protein: replication enhancement and protein interactions. J Virol 79: 9885–9895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Hanley-Bowdoin L (2006) Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol 142: 1642–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Liu Z, Song F, Xie Q, Hanley-Bowdoin L, Zhou X (2011) Tomato SlSnRK1 protein interacts with and phosphorylates βC1, a pathogenesis protein encoded by a geminivirus β-satellite. Plant Physiol 157: 1394–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Reyes MI, Hanley-Bowdoin L (2009) Arabidopsis protein kinases GRIK1 and GRIK2 specifically activate SnRK1 by phosphorylating its activation loop. Plant Physiol 150: 996–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Dallas MB, Goshe MB, Hanley-Bowdoin L (2014) SnRK1 phosphorylation of AL2 delays cabbage leaf curl virus infection in Arabidopsis. J Virol 88: 10598–10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG (1999) Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J 19: 433–439 [DOI] [PubMed] [Google Scholar]
- Tomé F, Nägele T, Adamo M, Garg A, Marco-Llorca C, Nukarinen E, Pedrotti L, Peviani A, Simeunovic A, Tatkiewicz A, et al. (2014) The low energy signaling network. Front Plant Sci 5: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk M, Bonvin AM (2009) 3D-DART: a DNA structure modelling server. Nucleic Acids Res 37: W235–W239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk M, Wassenaar TA, Bonvin AM (2012) A flexible, grid-enabled web portal for GROMACS molecular dynamics simulations. J Chem Theory Comput 8: 3463–3472 [DOI] [PubMed] [Google Scholar]
- van Zundert GCP, Rodrigues JPGLM, Trellet M, Schmitz C, Kastritis PL, Karaca E, Melquiond ASJ, van Dijk M, de Vries SJ, Bonvin AMJJ (2016) The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol 428: 720–725 [DOI] [PubMed] [Google Scholar]
- Varsani A, Martin DP, Navas-Castillo J, Moriones E, Hernández-Zepeda C, Idris A, Murilo Zerbini F, Brown JK (2014a) Revisiting the classification of curtoviruses based on genome-wide pairwise identity. Arch Virol 159: 1873–1882 [DOI] [PubMed] [Google Scholar]
- Varsani A, Navas-Castillo J, Moriones E, Hernández-Zepeda C, Idris A, Brown JK, Murilo Zerbini F, Martin DP (2014b) Establishment of three new genera in the family Geminiviridae: Becurtovirus, Eragrovirus and Turncurtovirus. Arch Virol 159: 2193–2203 [DOI] [PubMed] [Google Scholar]
- Vlad F, Turk BE, Peynot P, Leung J, Merlot S (2008) A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J 55: 104–117 [DOI] [PubMed] [Google Scholar]
- Waigmann E, Chen MH, Bachmaier R, Ghoshroy S, Citovsky V (2000) Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J 19: 4875–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hao L, Shung CY, Sunter G, Bisaro DM (2003) Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15: 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Buckley KJ, Yang X, Buchmann RC, Bisaro DM (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79: 7410–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SP, Rangarajan P, Donahue JL, Hess JE, Gillaspy GE (2014) Regulation of Sucrose non-Fermenting Related Kinase 1 genes in Arabidopsis thaliana. Front Plant Sci 5: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M (1996) Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379: 466–469 [DOI] [PubMed] [Google Scholar]
- Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS (2004) A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc Natl Acad Sci USA 101: 6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xie Y, Raja P, Li S, Wolf JN, Shen Q, Bisaro DM, Zhou X (2011) Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog 7: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A (2017) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98: 131–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, et al. (2011) BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23: 273–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Wang ZQ, Xiao R, Cao L, Wang Y, Xie Y, Zhou X (2017) Mimic phosphorylation of a βC1 protein encoded by TYLCCNB impairs its functions as a viral suppressor of RNA silencing and a symptom determinant. J Virol 91: e0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. (2013) Advances in understanding begomovirus satellites. Annu Rev Phytopathol 51: 357–381 [DOI] [PubMed] [Google Scholar]