PHOSPHATE STARVATION RESPONSE1, encoding the key regulator of responses to Pi starvation, is the target gene of the transcription factors ARF7 and ARF19 and is positively regulated by auxin signaling in Arabidopsis roots.
Abstract
PHOSPHATE STARVATION RESPONSE1 (PHR1) is a key regulatory component of the response to phosphate (Pi) starvation. However, the regulation of PHR1 in this response remains poorly understood. Here, we report that PHR1 is a target of the transcription factors AUXIN RESPONSE FACTOR7 (ARF7) and ARF19 and is positively regulated by auxin signaling in Arabidopsis (Arabidopsis thaliana) roots. PHR1 expression was induced by exogenous auxin and suppressed by auxin transport inhibitors in Arabidopsis roots. In the PHR1 promoter, three auxin-response elements, which are bound directly by ARF7 and ARF19, were shown to be essential for PHR1 expression. The arf7, arf19, and arf7 arf19 mutants showed down-regulated expression of PHR1 and downstream Pi starvation-induced genes in roots; they also exhibited defective Pi uptake in roots and overaccumulation of anthocyanin in shoots. The induction of lateral root formation in response to low Pi and to exogenous auxin was decreased in the phr1 mutant, whereas the expression of LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16) and LBD29 was not changed significantly. PHR1 acted independently of LBD16 and LBD29 in the regulation of lateral root formation in response to low Pi. Under low-Pi conditions, lateral root impairment in the arf7 arf19 mutant was partially rescued by constitutive expression of PHR1, demonstrating that reduced PHR1 expression contributed to the arf7 arf19 phenotype. In addition to PHR1, other genes encoding MYB-CC members also were targets of ARF7 and ARF19. Our work thus reveals a mechanism coordinating auxin signaling and the PHR1 regulon in Arabidopsis responses to Pi deficiency.
Phosphorus (P) is one of the essential nutrients required for plant growth, development, and reproduction. Although P is abundant in many soils, very little of it is present in phosphate (Pi) forms available for plant usage (Holford, 1997). In order to improve Pi uptake from soil, plants have developed numerous adaptive responses to low Pi availability (Raghothama, 1999; Williamson et al., 2001; Vance et al., 2003). In these responses, the expression of a subset of genes is up- or down-regulated in plants (Raghothama, 1999; Wu et al., 2003; Misson et al., 2005). The Pi starvation-induced (PSI) genes are regulated by several transcription factors, with PHOSPHATE STARVATION RESPONSE1 (PHR1) being the key transcription factor in vascular plants (Rubio et al., 2001; Devaiah et al., 2007, 2009). The phr1 mutant displays decreased expression of a subset of PSI genes, reduced contents of Pi, and minimal induction of anthocyanin accumulation in responses to Pi starvation (Rubio et al., 2001; Misson et al., 2005). On the contrary, the overexpression of PHR1 in Arabidopsis (Arabidopsis thaliana) leads to an increased content of Pi in shoots and an elevated expression of a subset of PSI genes (Nilsson et al., 2007). PHR1 is considered a central positive regulator of most PSI genes (Rubio et al., 2001; Bari et al., 2006). PHR1 belongs to the MYB-CC family of transcription factors, which are encoded by 15 genes in Arabidopsis. Besides PHR1, the other two MYB-CC members, namely PHR1-LIKE1 (PHL1) and PHL2, also are regarded as the key components of the central regulatory system controlling Arabidopsis transcriptional responses to Pi starvation (Bustos et al., 2010; Sun et al., 2016). PHR1, PHL1, and PHL2, and perhaps additional MYB-CC members, act redundantly or function in different plant tissues to regulate plant transcriptional responses to Pi starvation. The orthologs of PHR1 have been found in other plant species, including Phaseolus vulgaris (Valdés-López et al., 2008), Oryza sativa (Zhou et al., 2008), Brassica napus (Ren et al., 2012), and Triticum aestivum (Wang et al., 2013), where they function similarly in response to Pi starvation. The transcription of the PHR1 gene seems independent of the Pi status of plants (Rubio et al., 2001; Zhou et al., 2008; Ren et al., 2012). A more detailed understanding of the transcriptional regulation of PHR1 in plant tissues and organs is so far unavailable.
Much progress has been made in research into Pi deficiency-induced root architecture remodeling (Williamson et al., 2001; López-Bucio et al., 2002). The early data suggest that the ability of the root system to respond to Pi availability is independent of auxin signaling, because the root system architectures (RSAs) of the auxin-resistant mutants axr1, aux1, and axr4 appear to respond normally to changes of Pi availability (Williamson et al., 2001). However, further research suggests that auxin plays an important role in mediating the Pi starvation effects on RSAs (López-Bucio et al., 2002; Al-Ghazi et al., 2003; Nacry et al., 2005; Jain et al., 2007; Pérez-Torres et al., 2008). The increase in lateral root formation in Pi-deprived Arabidopsis seedlings is mediated, at least in part, by an increase in the auxin sensitivity of root cells, and the Pi availability modulates the expression of the TIR1 gene encoding an auxin receptor (Pérez-Torres et al., 2008). AUXIN RESPONSE FACTOR (ARF) gene family products regulate auxin-mediated transcriptional activation and repression (Salehin et al., 2015). In Arabidopsis, ARF19 is reported to play an important role in lateral root formation in response to Pi starvation (Pérez-Torres et al., 2008).
Although no data directly support the cross talk between the PHR1 regulon and auxin signaling in Pi starvation responses so far, the PSI genes IPS1, At4, and Pht1;1/PT1 are down-regulated in the roots of slr-1 and arf7 arf19 mutants during Pi-starvation responses (Narise et al., 2010). IPS1, At4, and Pht1;1/PT1 are the target genes of PHR1 (Rubio et al., 2001; Nilsson et al., 2007). slr-1 is a gain-of-function mutant of IAA14 (a repressor of auxin signaling), and the arf7 arf19 double mutant is a loss-of-function mutant of ARF7 and ARF19. However, low Pi-induced anthocyanin accumulation, which also is controlled by PHR1, is increased rather than reduced in slr-1 and arf7 arf19 mutants (Rubio et al., 2001; Narise et al., 2010). Based on these conflicting results, previous researchers deduced that the reduction of low Pi-dependent gene expression in slr-1 and arf7 arf19 mutants was not caused by the inactivation of PHR1 (Narise et al., 2010).
To date, the relationship between auxin signaling and the PHR1 regulon in low-Pi responses is still unclear. This relationship is uncovered by our work, which indicates that auxin signaling is more intimately linked with the PHR1 regulon in low-Pi responses than was thought previously. Some questions regarding low-Pi responses can be answered by the regulating mechanism from our work.
RESULTS
Transcriptional Regulation of PHR1 in Arabidopsis
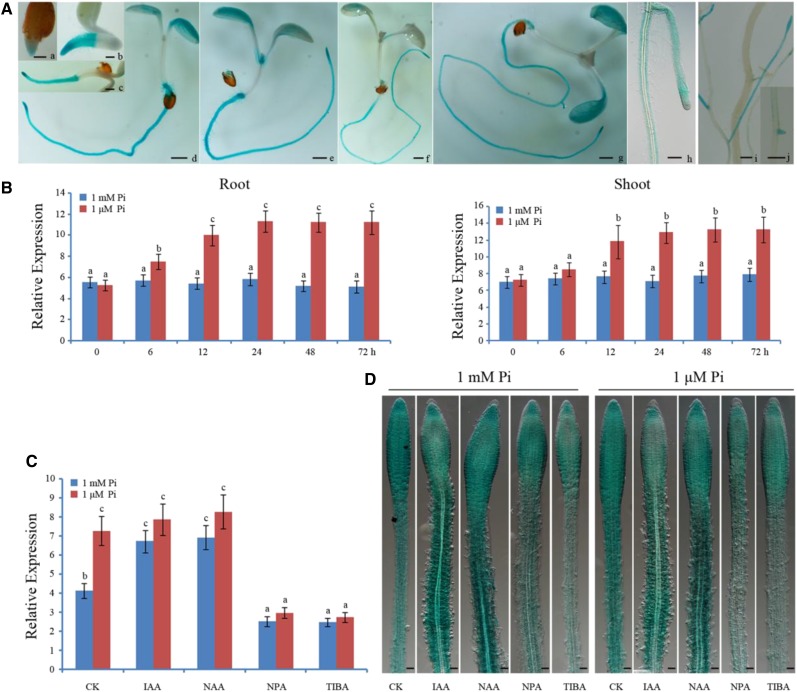
To investigate the transcriptional profiles of PHR1, we analyzed PHR1 promoter activity using transgenic Arabidopsis carrying a fusion of the PHR1 promoter to the reporter gene encoding GUS. Analysis of GUS activity revealed that the PHR1 promoter is active in roots (Fig. 1A). As early as 1 d after germination, GUS activity was detectable in roots of young seedlings. GUS activity in roots then increased continuously up to 5 d after germination. At the flowering stage, GUS activity was clearly detectable in young lateral roots, whereas it was weakly detected in lignified lateral roots and almost undetectable in lignified primary roots (Fig. 1A). Expression analysis showed that PHR1 expression is induced in both Arabidopsis roots and shoots by Pi deficiency (Fig. 1B). These results suggest that PHR1 expression is modulated by external or internal Pi levels in Arabidopsis.
Figure 1.
Expression profiles of PHR1 in Arabidopsis. A, Expression of GUS in PHR1p:GUS transgenic Arabidopsis. Histochemical assay of GUS activity in 1- (a), 2- (b), 3- (c), 4- (d), 5- (e), 6- (f), 7- (g), and 14-d-old seedlings (h) on 1 mm Pi medium and in roots of 25-d-old plant grown in soil (i and j). Bars = 100 μm. B, Expression analysis of PHR1 in roots and shoots. Five-day-old seedlings were transferred to 1 μm Pi for 0, 6, 12, 24, 48, and 72 h, and then total RNA was isolated from roots and shoots for reverse transcription quantitative PCR (RT-qPCR) analysis. The data are presented as means ± sd. The significance of differences was analyzed by Duncan’s test (P < 0.05; n = 9). Different lowercase letters indicate statistically significant differences. C, RT-qPCR analysis of PHR1 expression in roots treated with auxin (0.05 μm IAA or 0.05 μm NAA) and auxin transport inhibitors (10 μm NPA or 10 μm TIBA) for 24 h under 1 mm Pi or 1 μm Pi conditions, respectively. CK, Nontreated control. The data are presented as means ± sd. The significance of differences was analyzed by Duncan’s test (P < 0.05; n = 9). Different lowercase letters indicate statistically significant differences. D, Expression of GUS in roots of PHR1p:GUS transgenic Arabidopsis treated with auxin (0.05 μm IAA or 0.05 μm NAA) and auxin transport inhibitors (10 μm NPA or 10 μm TIBA) for 24 h under 1 mm Pi or 1 μm Pi conditions, respectively.
In Arabidopsis responses to Pi starvation, auxin is involved in the remodeling of Arabidopsis RSA (Pérez-Torres et al., 2008). Additionally, gain- or loss-of-function mutations of PHR1 result in the respective modification of Arabidopsis RSA (Nilsson et al., 2007; Ren et al., 2012). Thus, PHR1 may be a component of the auxin regulatory pathway involved in low-Pi responses. To investigate this possibility, we examined the effects of exogenous auxin and auxin transport inhibitors on PHR1 expression. RT-qPCR analysis showed that, on both 1 mm Pi and 1 μm Pi, the expression of PHR1 was obviously increased in Arabidopsis roots exposed to 0.05 μm indole-3-acetic acid (IAA; natural auxin) or 0.05 μm naphthylacetic acid (NAA; synthetic auxin; Fig. 1C). Promoter activity analysis also showed that, on both 1 mm Pi and 1 μm Pi, although no significant difference was found in root tips, the activity of the PHR1 promoter was obviously increased in the upper region of Arabidopsis roots exposed to 0.05 μm IAA or 0.05 μm NAA (Fig. 1D). On the contrary, the expression of PHR1 was decreased significantly in the upper region of Arabidopsis roots that were exposed to auxin transport inhibitors (Fig. 1, C and D).
Auxin-Response Elements in the PHR1 Promoter Essential for PHR1 Expression
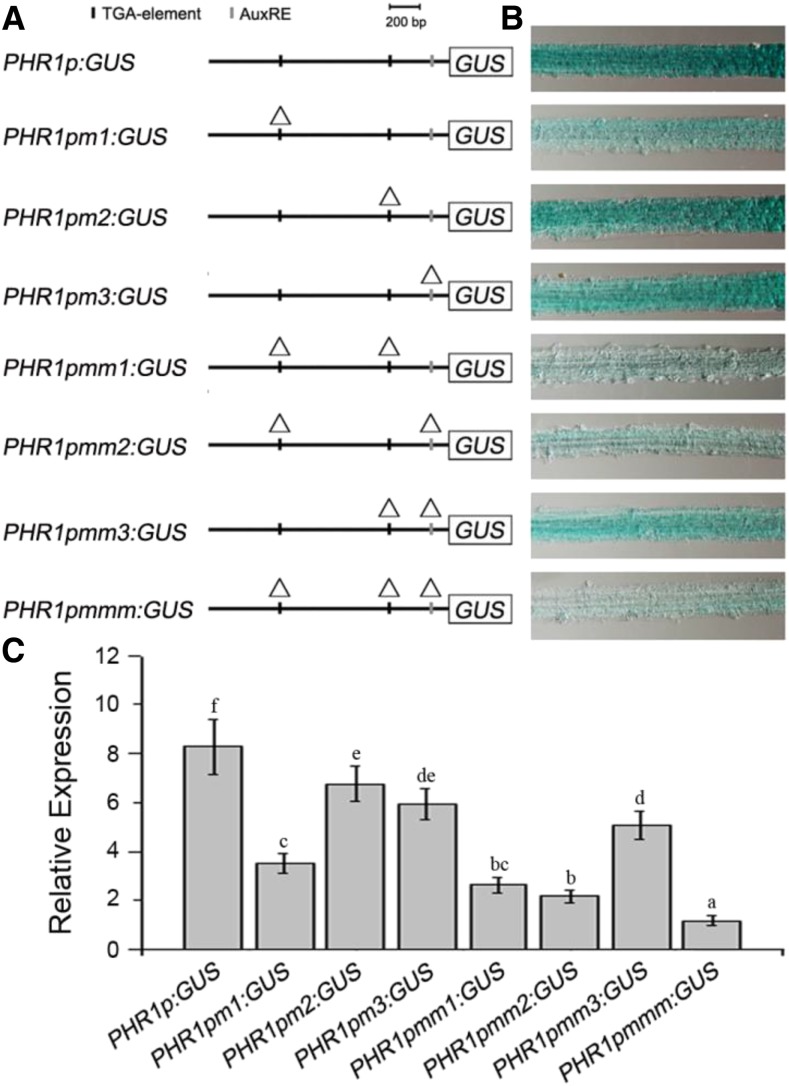
Using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), we found several cis-elements in the PHR1 promoter. Among these, there are three auxin-response elements, namely one copy of the AuxRE (GAGACA, −103 to −108) and two copies of the TGA (TGA-1 AACGAC, −1,037 to −1,042; and TGA-2 AACGAC, −353 to −358) elements (Supplemental Fig. S1).
To confirm that these elements are required for PHR1 transcription, we carried out a deletion-mutation analysis using transgenic Arabidopsis. A set of constructs were generated and introduced into Arabidopsis (Fig. 2A). Histochemical staining of GUS activity revealed that PHR1 promoter activity was decreased in the roots of transgenic Arabidopsis following the deletion mutation of auxin-response elements (Fig. 2B). RT-qPCR analysis also indicated that expression of the GUS reporter gene under the control of the mutated PHR1 promoter was lower compared with that under the control of the intact PHR1 promoter (Fig. 2C). In Arabidopsis carrying single point mutations, GUS activity was obviously decreased in the roots of PHR1pm1:GUS transgenic plants. In Arabidopsis carrying double point mutations, GUS activity was decreased significantly in the roots of PHR1pmm1:GUS and PHR1pmm2:GUS transgenic plants. In roots of PHR1pmmm:GUS transgenic plants, although the GUS activity was decreased dramatically, it was still weakly detectable. These results suggest that the three auxin-response elements are essential; however, the TGA-1 element is the most important of them for PHR1 transcription in Arabidopsis roots.
Figure 2.
Functional sequence analysis of auxin-response elements in the PHR1 promoter. A, Schematic representations of the constructs showing the deletion mutation of auxin-response elements in the PHR1 promoter. The triangles indicate the positions of deleted auxin-response elements. B, GUS staining showing the activity of the PHR1 promoter with deletion mutation of auxin-response elements. C, Expression analysis of the GUS gene in roots of PHR1p:GUS, PHR1pm1:GUS, PHR1pm2:GUS, PHR1pm3:GUS, PHR1p:GUSmm1, PHR1p:GUSmm2, PHR1p:GUSmm3, and PHR1pmmm:GUS transgenic plants. Total RNAs were isolated from roots of 5-d-old seedlings for RT-qPCR analysis. The data are presented as means ± sd. The significance of differences was analyzed by Duncan’s test (P < 0.05; n = 9). Different lowercase letters indicate statistically significant differences.
ARF7 and ARF19 Binding to Auxin-Response Elements of the PHR1 Promoter
Using ARF7p:GUS and ARF19p:GUS transgenic plants, ARF7 and ARF19 promoter activity in Arabidopsis roots was investigated (Supplemental Fig. S2A). Under 1 mm Pi conditions, GUS activity was weak in the roots of ARF7p:GUS transgenic plants and was almost undetectable in root tips. Under 1 μm Pi conditions, GUS activity was increased in roots (except for root tips) of ARF7p:GUS transgenic plants. GUS activity was easily detectable in the whole root system of ARF19p:GUS transgenic Arabidopsis under 1 mm Pi conditions. Under 1 μm Pi conditions, the GUS activity in roots of ARF19p:GUS transgenic plants was increased dramatically. RT-qPCR analysis also showed that ARF7 and ARF19 are induced by low Pi in Arabidopsis roots (Supplemental Fig. S2B). These results indicate that the expression patterns of ARF7 and ARF19 are similar to that of PHR1 in Arabidopsis roots.
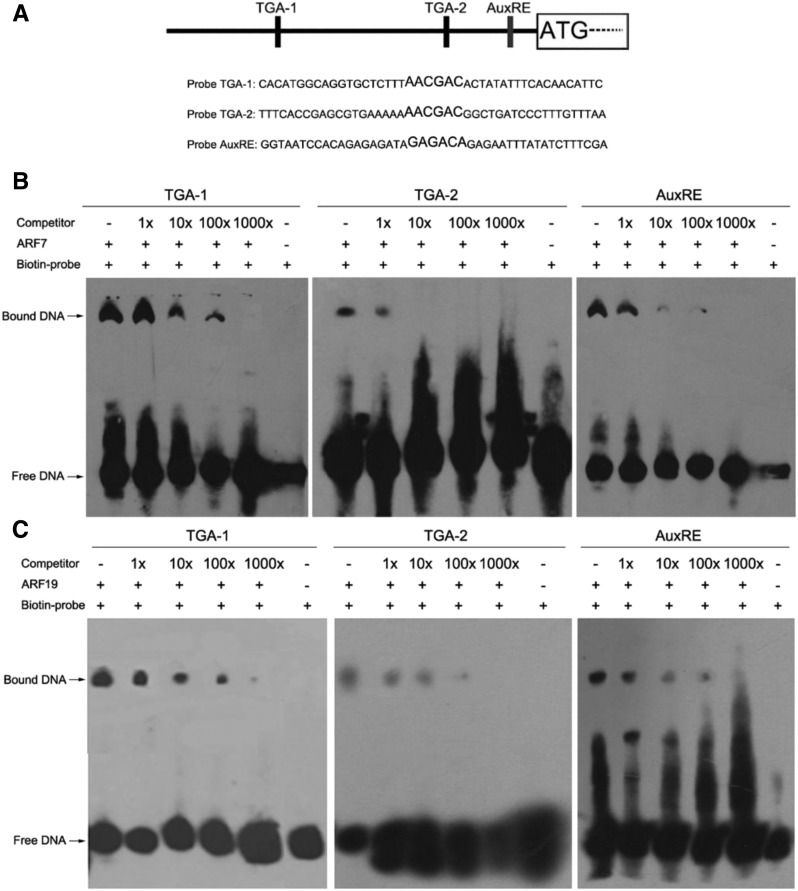
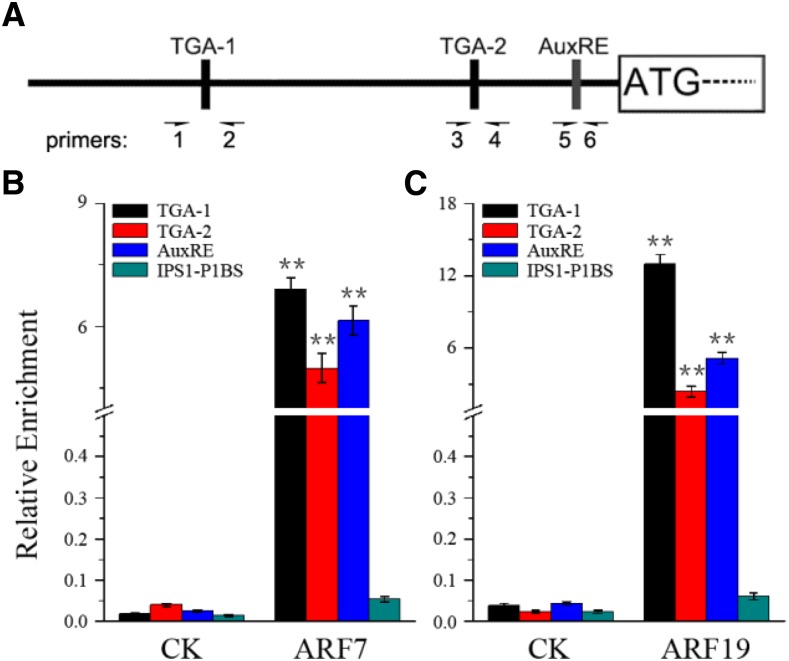
Possibly, the regulation of PHR1 by auxin is carried out directly by ARF7 and/or ARF19. To test this possibility, a yeast one-hybrid assay was performed. The yeast cells with the effector AD-ARF7 or AD-ARF19 and the reporter PHR1p:AUR1-C (pPHR1-AbAi) grew well on both SD/-Leu medium and SD/-Leu/AbA medium (Supplemental Fig. S3). These data indicated that both ARF7 and ARF19 are able to bind the PHR1 promoter in yeast cells. To further confirm that ARF7 and ARF19 can bind directly to auxin-response elements of the PHR1 promoter, we performed electrophoretic mobility shift assays (EMSAs; Fig. 3). The EMSA results indicated that ARF7 and ARF19 proteins caused a mobility shift of biotin-labeled TGA-1, TGA-2, and AuxRE sequences (Fig. 3, B and C). Furthermore, the unlabeled TGA-1, TGA-2, and AuxRE sequences specifically competed with labeled sequences in binding to ARF7 and ARF19, indicating that the binding is sequence specific (Fig. 3, B and C). These results suggest that ARF7 and ARF19 are capable of binding auxin-response elements of the PHR1 promoter in vitro. To confirm that PHR1 is a target gene of ARF7 and ARF19 in vivo, interaction between ARF7/ARF19 and the PHR1 promoter was detected by chromatin immunoprecipitation (ChIP) assay (Fig. 4). The ChIP assay indicated that ARF7 and ARF19 interact with the PHR1 promoter encompassing TGA-1, TGA-2, or AuxRE and that there is a similar pattern of interaction between both ARF7 and ARF19 and the PHR1 promoter (Fig. 4, B and C). Comparatively, the interaction with TGA-1 is strong and the interaction with TGA-2 is weak. The results demonstrate that ARF7 and ARF19 are able to bind to the auxin-response elements of the PHR1 promoter in vivo.
Figure 3.
EMSAs for ARF7 and ARF19 binding to the auxin-response elements of the PHR1 promoter in vitro. A, Diagram of the PHR1 promoter showing the relative positions of the auxin-response elements (TGA-1, TGA-2, and AuxRE) and the probes used in the EMSAs. B and C, EMSAs of the binding of ARF7 (B) and ARF19 (C) to auxin-response elements of the PHR1 promoter. Each biotin-labeled DNA probe was incubated with ARF7 or ARF19 protein. An excess of unlabeled probe was added to compete with labeled promoter sequence. Biotin-labeled probes incubated without recombinant ARF7 or ARF19 protein served as the negative control.
Figure 4.
ChIP assay of ARF7 and ARF19 binding to the auxin-response elements of the PHR1 promoter in vivo. A, Diagram of the PHR1 promoter showing the relative positions of two TGA elements and one AuxRE. The arrows show the positions of primers used in ChIP-qPCR. B and C, ChIP-qPCR analysis of the PHR1 promoter sequence. ChIP assays were performed with chromatin prepared from wild-type Arabidopsis. CK represents the ChIP signals without antibody, and ARF7 (B) and ARF19 (C) represent the ChIP signals with the addition of anti-ARF7 and anti-ARF19 serum, respectively. The data are presented as means ± sd. The significance of differences was analyzed by Student’s t test (**, P < 0.01; n = 6).
PHR1 Is Positively Regulated by ARF7 and ARF19
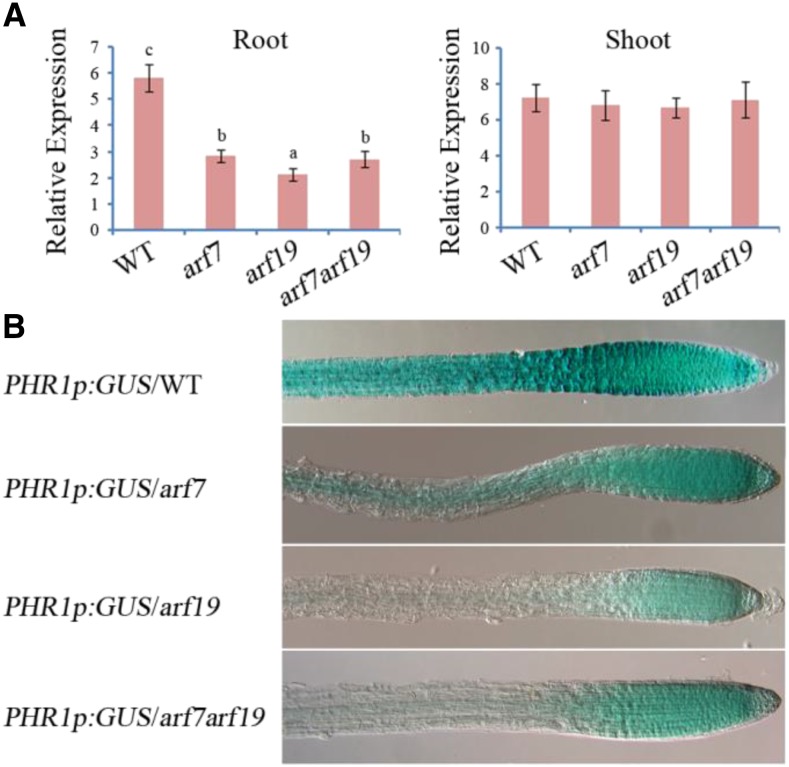
Based on the results described above, we further hypothesized that ARF7 and ARF19 are able to positively regulate PHR1 expression in Arabidopsis roots. To test this hypothesis, we first analyzed the expression of PHR1 in arf7, arf19, and arf7 arf19 mutants by RT-qPCR (Fig. 5A). PHR1 expression was obviously decreased in roots of arf7, arf19, and arf7 arf19 mutants compared with that of wild-type plants. However, in Arabidopsis shoots, PHR1 expression was not significantly different between arf7, arf19, and arf7 arf19 mutants and wild-type plants. These results indicate that ARF7 and ARF19 are positive upstream regulators of PHR1 expression in Arabidopsis roots.
Figure 5.
Expression of PHR1 in arf7, arf19, and arf7 arf19 mutants. A, Expression analysis of the PHR1 gene in roots and shoots of the wild type (WT), arf7 and arf19 single mutants, and the arf7 arf19 double mutant. Total RNAs were isolated from roots and shoots of 5-d-old Arabidopsis seedlings for RT-qPCR analysis. The data are presented as means ± sd. The significance of differences was analyzed by Duncan’s test (P < 0.05; n = 9). Different lowercase letters indicate statistically significant differences. B, GUS staining showing PHR1 expression patterns in transgenic plants carrying the PHR1p:GUS construct in the wild type and arf7, arf19, and arf7 arf19 mutants. The histochemical assay of GUS activity was performed in roots of 5-d-old Arabidopsis germinated on 1 mm Pi medium.
To obtain a more detailed understanding of the transcriptional regulation of PHR1 expression by ARF7 and ARF19, the construct PHR1p:GUS was introduced into arf7, arf19, and arf7 arf19 mutants. In 5-d-old seedlings, GUS signal was detectable in roots of each transgenic line (Fig. 5B). In wild-type plants, GUS signal was strong in the entire root system. In the arf7 single mutant, the GUS signal was reduced in most regions of the root system, whereas GUS signal was obviously detectable at root steles and root tips. In the arf19 single mutant, the GUS signal was weak in most regions of the root system, whereas GUS signal was still detectable at the root tips. In the arf7 arf19 double mutant, which shows an impaired lateral root formation phenotype, the GUS signal also was weak in most of the root system and detectable only at the tip of the primary root. The results suggest that the transcription of PHR1 is positively regulated by ARF7 and ARF19 in Arabidopsis roots.
Expression of PSI Genes in arf7, arf19, and arf7 arf19 Mutants
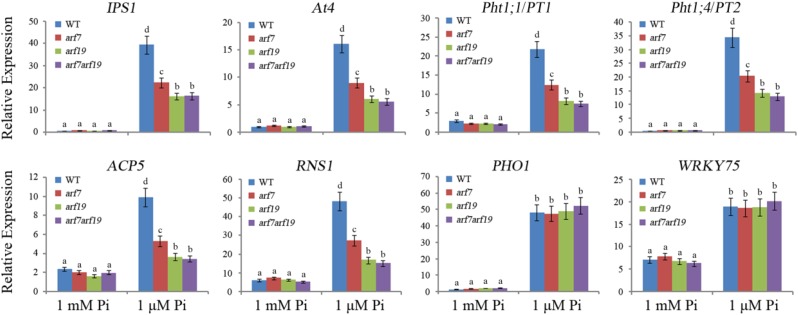
Theoretically, if ARF7 and ARF19 positively modulate PHR1 expression in Arabidopsis roots, the PSI genes, whose expression regulation is downstream of PHR1, could be down-regulated in arf7, arf19, and arf7 arf19 mutants. To confirm this possibility, the expression of eight PSI genes in roots of arf7, arf19, and arf7 arf19 mutant and wild-type plants was measured (Fig. 6). Under 1 mm Pi conditions, no remarkable difference in expression was found in the roots of each plant line for all PSI genes. Under 1 μm Pi conditions, the expression of PSI genes aside from PHO1 and WRKY75 was decreased in roots of the arf7 single mutant and was decreased significantly in roots of the arf19 single mutant and the arf7 arf19 double mutant. These results indicate that the loss of function of ARF7 and/or ARF19 has a vital effect on the expression of PSI genes under low-Pi conditions.
Figure 6.
RT-qPCR analysis of PSI genes in roots of arf7, arf19, and arf7 arf19 mutants. Five-day-old Arabidopsis seedlings were transferred to 1 mm Pi or 1 μm Pi medium for 24 h, and then total RNAs were isolated from roots for RT-qPCR analysis. The data are presented as means ± sd. The significance of differences was analyzed by Duncan’s test (P < 0.05; n = 9). Different lowercase letters indicate statistically significant differences. WT, Wild type.
arf7, arf19, and arf7 arf19 Phenotypes under Low-Pi Conditions
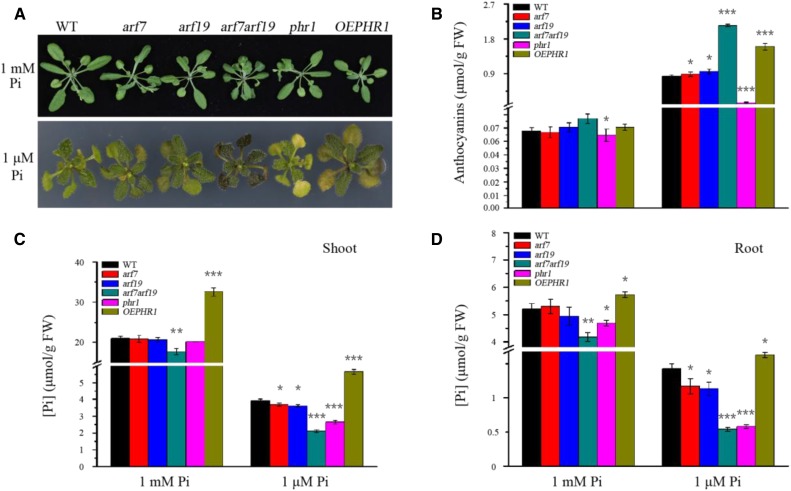
As upstream regulators of PHR1, the loss-of-function mutation of ARF7 or ARF19 could affect Arabidopsis responses to low Pi. As described previously, there was no obvious difference among arf7 and arf19 single mutants and wild-type plants on 1 mm Pi medium, whereas the arf7 arf19 double mutant displayed an obvious mutant phenotype with impaired formation of lateral roots (Supplemental Fig. S4). On 1 μm Pi medium, arf7 arf19 double mutants exhibited this same phenotype as well as dark-purple leaves that show an overaccumulation of anthocyanin (Fig. 7A). Under low-Pi conditions, the content of anthocyanin in shoots of wild-type plants and the arf7, arf19, and arf7 arf19 mutants was 0.85, 0.89, 0.97, and 2.18 μmol g−1, respectively (Fig. 7B), meaning that there was higher anthocyanin content in shoots of the three mutant lines compared with that in wild-type plants, especially for the arf7 arf19 double mutant (Fig. 7B). Under low-Pi conditions, the Pi content in arf7, arf19, and arf7 arf19 mutants is lower than that in wild-type plants (Fig. 7, C and D). Whether in roots or in shoots, the Pi content of the arf7 arf19 double mutant was remarkably lower than that in the other plants (Fig. 7, C and D). The Pi content analysis indicates that, under low-Pi conditions, the loss-of-function mutation of ARF7 and/or ARF19 results in reduced Pi uptake in Arabidopsis.
Figure 7.
Phenotypes of arf7, arf19, and arf7 arf19 mutants under low-Pi conditions. A, Twenty-day-old Arabidopsis seedlings of the wild type (WT), arf7 and arf19 single mutants, the arf7 arf19 double mutant, the phr1 mutant, and the OEPHR1-5 transgenic line grown in hydroponics with 1 mm Pi and 1 μm Pi. B, Content of anthocyanin in shoots of the wild type, arf7 and arf19 single mutants, the arf7 arf19 double mutant, the phr1 mutant, and the OEPHR1-5 transgenic line grown in hydroponics with 1 mm Pi and 1 μm Pi. C, Content of free Pi in shoots of the wild type, arf7 and arf19 single mutants, the arf7 arf19 double mutant, the phr1 mutant, and the OEPHR1-5 transgenic line grown in hydroponics with 1 mm Pi and 1 μm Pi. D, Content of free Pi in roots of the wild type, arf7 and arf19 single mutants, the arf7 arf19 double mutant, the phr1 mutant, and the OEPHR1-5 transgenic line grown in hydroponics with 1 mm Pi and 1 μm Pi. The data are presented as means ± sd. The significance of differences was analyzed by Student’s t test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001; n = 5). FW, Fresh weight.
Arabidopsis Sensitivity to Auxin Is Changed by PHR1 Mutation
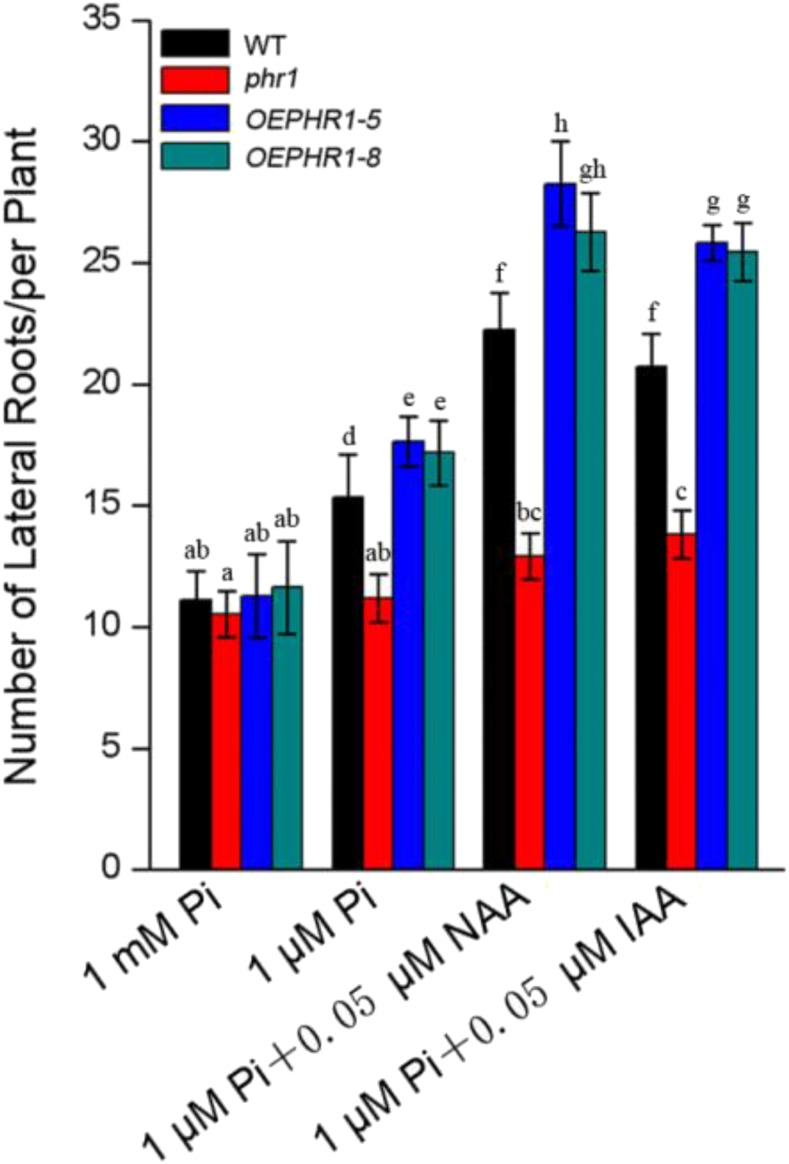
Under low-Pi conditions, the increase in lateral root formation in Arabidopsis is mediated by an increase in auxin sensitivity of root cells, a process in which ARF7 and ARF19 function as the crucial mediators (Pérez-Torres et al., 2008). As PHR1 is a target gene of ARF7 and ARF19, PHR1 could be involved in the regulation of RSA. To confirm this hypothesis, a phr1 T-DNA insertion mutant (SALK_067629) was identified and a PHR1 overexpression transgenic Arabidopsis line (OEPHR1-5/-8) was generated (Supplemental Fig. S5). On 1 mm Pi medium, no significant difference was found among the RSAs of wild-type, phr1, and OEPHR1-5/-8 seedlings (Fig. 8; Supplemental Fig. S5). On 1 μm Pi medium, the number of lateral roots of phr1 mutant and OEPHR1-5/-8 transgenic plants changed compared with that of wild-type plants, whereby they decreased in phr1 mutants and increased in OEPHR1-5/-8 transgenic plants (Fig. 8; Supplemental Fig. S5). On 1 μm Pi medium with auxin (0.05 μm NAA or 0.05 μm IAA), the number of lateral roots in wild-type and OEPHR1-5/-8 transgenic plants increased dramatically, whereas they did not increase significantly in the phr1 mutant (Fig. 8; Supplemental Fig. S5). These results show that the loss- or gain-of-function mutation of PHR1 changes Arabidopsis sensitivity to auxin.
Figure 8.
Number of lateral roots of the phr1 mutant and PHR1 overexpression transgenic plants after exogenous auxin treatment. Five-day-old seedlings germinated on Murashige and Skoog (MS) medium were transferred to 1 mm Pi, 1 μm Pi, 1 μm Pi + 0.05 μm NAA, or 1 μm Pi + 0.05 μm IAA medium and cultured vertically for 3 d. The data are presented as means ± sd. The significance of differences was analyzed by Duncan’s test (P < 0.05; n = 5). Different lowercase letters indicate statistically significant differences. WT, Wild type.
Overexpression of PHR1 Partially Rescues Lateral Root Impairment of the arf7 arf19 Mutant
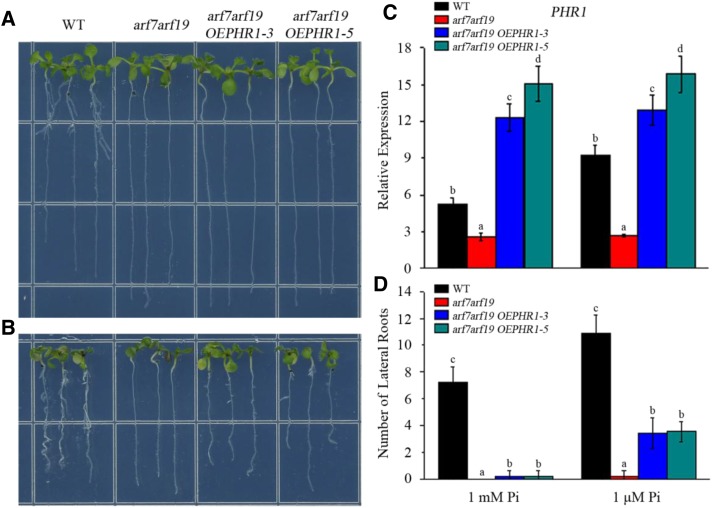
From the above data, PHR1, as a target gene of ARF7 and ARF19, is involved in the initiation and development of lateral roots under low-Pi conditions. The representative phenotype of arf7 arf19 mutants is lateral root impairment. To investigate the contribution of down-regulated PHR1 expression to the lateral root impairment of the arf7 arf19 mutant, PHR1 overexpression transgenic Arabidopsis in the arf7 arf19 mutant background (arf7 arf19 OEPHR1-3/-5) was generated (Fig. 9). On 1 mm Pi medium, almost no lateral roots were observed in both arf7 arf19 double mutant and arf7 arf19 OEPHR1-3/-5 transgenic plants (Fig. 9A). On 1 μm Pi medium, lateral roots were still not observed in the arf7 arf19 mutant, whereas lateral roots were obviously observed in arf7 arf19 OEPHR1-3/-5 transgenic plants (Fig. 9B). However, the lateral root number of arf7 arf19 OEPHR1-3/-5 transgenic plants was significantly less than that of wild-type plants (Fig. 9, B and D). These results indicate that, under low-Pi conditions, the lateral root impairment of the arf7 arf19 mutant could be partially rescued by the constitutive expression of PHR1.
Figure 9.
Overexpression of PHR1 partially rescues the lateral root impairment of the arf7 arf19 mutant. A, Five-day-old seedlings of wild-type (WT), arf7 arf19 mutant, and arf7 arf19 OEPHR1-3/-5 transgenic plants transferred to 1 mm Pi medium and cultured vertically for 3 d. B, Five-day-old seedlings of wild-type, arf7 arf19 mutant, and arf7 arf19 OEPHR1-3/-5 transgenic plants transferred to 1 μm Pi medium and cultured vertically for 3 d. C, Expression analysis of PHR1 in wild-type, arf7 arf19 mutant, and arf7 arf19 OEPHR1-3/-5 transgenic plants. Total RNAs were isolated from roots of Arabidopsis seedlings for RT-qPCR analysis. The data are presented as means ± sd. The significance of differences was analyzed by Duncan’s test (P < 0.05; n = 9). Different lowercase letters indicate statistically significant differences. D, Number of lateral roots of wild-type, arf7 arf19 mutant, and arf7 arf19 OEPHR1-3/-5 transgenic plants. The data are presented as means ± sd. The significance of differences was analyzed by Student’s t test (P < 0.05; n = 9). Different lowercase letters indicate statistically significant differences.
Genes Encoding MYB-CC Family Members Are Regulated by ARF7 and ARF19
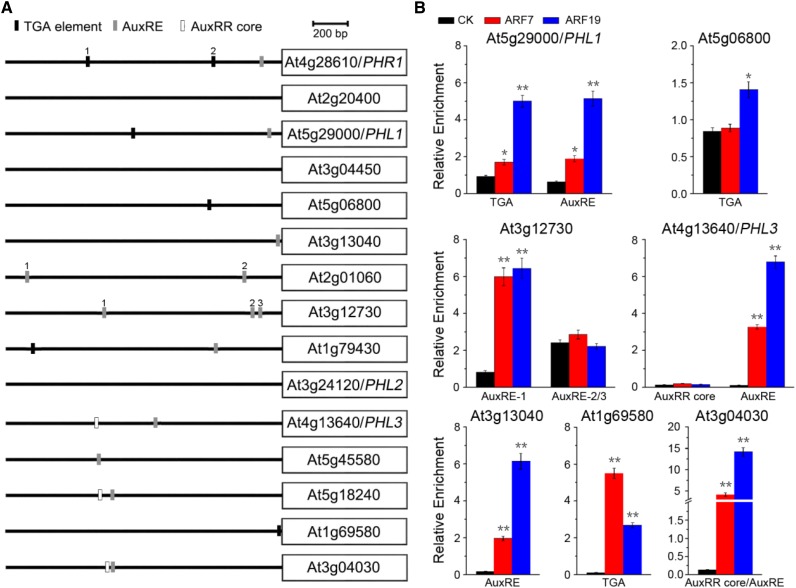
PHR1 is part of the MYB-CC transcription factor family, which includes 15 members in Arabidopsis (Rubio et al., 2001). The functional redundancy between PHR1 and other MYB-CC members has been shown previously (Bustos et al., 2010; Sun et al., 2016). It is possible that genes encoding MYB-CC transcription factors other than PHR1 also are modulated by ARF7 and ARF19. To test this possibility, auxin-response elements were identified within the 1.5-kb sequence upstream of each gene start codon. Interestingly, auxin-response elements were found in the promoters of most MYB-CC family genes (Fig. 10A). Specifically, the TGA element was found in the promoters of At5g29000/PHL1, At5g06800, At1g79430, and At1g69580; AuxREs were found in the promoters of At5g29000/PHL1, At3g13040, At2g01060, At3g12730, At1g79430, At4g13640/PHL3, At5g45580, At5g18240, and At3g04030; and the core auxin-responsive region (AuxRR core) was found in the promoters of At4g13640/PHL3, At5g18240, and At3g04030. The auxin-response elements were not found in the promoters of only three genes, namely At2g20400, At3g04450, and At3g24120/PHL2.
Figure 10.
ChIP assay of ARF7 and ARF19 binding to the auxin-response elements of the promoters of MYB-CC genes in vivo. A, Diagrams of the promoters of MYB-CC genes showing the relative positions of auxin-response elements. B, ChIP-qPCR analysis of the MYB-CC promoters. ChIP assays were performed with chromatin prepared from wild-type Arabidopsis. CK represents ChIP signals without antibody, and ARF7 and ARF19 represent ChIP signals with the addition of anti-ARF7 and anti-ARF19 serum, respectively. The experiments were repeated three times. The data are presented as means ± sd. The significance of differences was analyzed by Student’s t test (*, P < 0.05 [n = 9] and **, P < 0.01 [n = 6]).
Furthermore, the interaction of ARF7 and ARF19 with these auxin-response elements was detected by ChIP assay (Fig. 10B). This showed that, in addition to the PHR1 promoter, ARF7 and ARF19 were able to interact with the auxin-response elements of the promoters of At5g29000/PHL1, At5g06800, At3g13040, At3g12730, At4g13640/PHL3, At1g69580, and At3g04030. These results demonstrate that ARF7 and ARF19, functioning as transcription factors, are the upstream regulators of the genes encoding MYB-CC family members.
DISCUSSION
PHR1 Is Regulated by Auxin on a Transcriptional Level
In Arabidopsis, the PHR1 gene is not itself particularly Pi responsive, and its transcript is only induced 2-fold by low-Pi stress (Rubio et al., 2001). In contrast, PHOSPHORUS STARVATION RESPONSE1, the counterpart of PHR1 in Chlamydomonas reinhardtii, is induced 13-fold by Pi deprivation (Wykoff et al., 1999). As in Arabidopsis, the expression of PHR1 homologs in other plant species is not very responsive to Pi deprivation (Valdés-López et al., 2008; Zhou et al., 2008). It seems that the transcription of PHR1 is independent of the plant Pi status. C. reinhardtii is a unicellular alga and, thus, does not often suffer the stress of Pi deficiency. By contrast, terrestrial plants require adequate Pi for their growth and development, yet they often suffer Pi deficiency from the environment. So, under high-Pi conditions, the transcription of PHR1 is maintained at a high level in preparation for the low-Pi stress that can occur at any time. Our data support that a complicated regulation network exists in Arabidopsis for controlling PHR1 expression, especially under low-Pi conditions.
What is the upstream regulator of PHR1 during Arabidopsis development or under low-Pi stress? To answer this question, we investigated potential candidates, and auxin, one of the pivotal phytohormones, was found to play a positive regulatory role in PHR1 expression. In Arabidopsis, several studies have examined the role of auxin as a mediator in the responses to Pi starvation, and increased auxin sensitivity, rather than increased auxin transport or synthesis, is considered a mediator of proliferative lateral root growth in low-Pi responses (Williamson et al., 2001; López-Bucio et al., 2002; Jain et al., 2007; Pérez-Torres et al., 2008). The induction of PHR1 by Pi starvation is consistent with the increase of auxin sensitivity by Pi starvation in Arabidopsis roots. Additionally, the up- or down-regulation of PHR1 by the application of exogenous auxin or auxin inhibitor, respectively, also supports that PHR1 is regulated by auxin.
PHR1 Is the Target Gene of ARF7 and ARF19 in Roots
In plants, auxin plays a crucial role in various aspects of physiological and developmental processes (Vanneste and Friml, 2009). In these processes, auxin coordinates the transcription of numerous auxin-mediated genes through the actions of ARFs and Aux/IAA proteins (Salehin et al., 2015). The ARFs bind specific auxin-response elements in their target genes (Ulmasov et al., 1997). The Aux/IAAs function as repressors of ARF-mediated transcription by forming multimers with ARFs and recruiting the Groucho/Tup family corepressor TOPLESS and its family proteins (TPRs; Long et al., 2006; Szemenyei et al., 2008; Korasick et al., 2014; Nanao et al., 2014). To date, some kinds of auxin-response elements, which include the AuxRE (TGTCTC), TGA element (AACGAC), and AuxRR core (GGTCCAT), have been identified in the promoters of auxin-responsive genes (Nagao et al., 1993; Ballas et al., 1995; Pastuglia et al., 1997). In the PHR1 promoter, three auxin-response elements (one copy of AuxRE and two copies of TGA elements) are certainly found, and each of them contributes to the transcription of PHR1.
In Arabidopsis, ARF7 and ARF19 play essential roles in auxin-mediated plant development by regulating both unique and partially overlapping sets of target genes (Okushima et al., 2005). ARF7 and ARF19 regulate lateral root formation by interacting directly with AuxREs in promoters of LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16) and LBD29 (Okushima et al., 2007). ARF19 also plays an important role in lateral root formation in response to Pi starvation (Pérez-Torres et al., 2008). PHR1 has a similar expression pattern to ARF7 and ARF19 in Arabidopsis roots under both high- and low-Pi conditions. The almost parallel expression patterns imply that PHR1 and ARF7/ARF19 could be involved in a mutual biological process. In the PHR1 promoter, in addition to the AuxRE, two TGA elements also are binding sites of ARF7 and ARF19. Although several cis-elements have been identified as auxin-response elements in plants, only AuxRE is confirmed as a binding site of ARFs by protein-DNA interaction assay (Ulmasov et al., 1997; Okushima et al., 2007; Cole et al., 2009). We suggest that the TGA element, as an auxin-response element, can be bound by ARFs in plants.
ARFs bind to the promoters of auxin-responsive genes and activate or repress their transcription (Guilfoyle and Hagen, 2007). As transcription activators, ARF7 and ARF19 positively regulate LBD16 and LBD29 to regulate lateral root formation (Okushima et al., 2007). In roots of arf7, arf19, and arf7 arf19 mutants, the activity of the PHR1 promoter is decreased significantly, as is the transcription of PHR1. In Arabidopsis roots, the expression of ARF7 is weak in most regions of roots and impaired in root tips, whereas the expression of ARF19 is comparatively high in the whole root system. The activity of the PHR1 promoter in the arf7 and arf19 mutants is consistent with the expression patterns of ARF7 and ARF19, respectively, namely weakly reduced in roots of the arf7 mutant and significantly reduced in roots of the arf19 mutant. In the arf7 arf19 mutant, although the activity of the PHR1 promoter is low in most regions of the roots, it is still detectable in root tips. This suggests that other regulators (other ARFs) could be involved in controlling PHR1 expression in root tips. Considering lateral root impairment in the arf7 arf19 double mutant, auxin could be converged in roots, which results in expression pattern changes of other ARF genes. Accordingly, the expression pattern of PHR1 in the arf7 arf19 double mutant is not as expected. The data prove that PHR1 is positively regulated by ARF7 and ARF19 in roots. Our data also indicate that ARF7 cooperates with ARF19 to control PHR1 expression in roots, whereas ARF7 and ARF19 act redundantly to control lateral root development (Okushima et al., 2005, 2007).
The Responses to Low Pi Are Down-Regulated in ARF7 and ARF19 Loss-of-Function Mutants
As ARF7 and ARF19 are upstream positive regulators of PHR1, the loss-of-function mutation of ARF7 and/or ARF19 should affect the induction of PSI genes by low Pi. The PSI genes (IPS1, At4, Pht1;1/PT1, Pht1;4/PT2, ACP5, and RNS1) are the target genes of PHR1 during the response to low Pi (del Pozo et al., 1999; Rubio et al., 2001; Shin et al., 2004). Consistent with this, the induction of the six genes by Pi starvation is decreased significantly in roots of arf7, arf19, and arf7 arf19 mutants. In contrast, PHO1 and WRKY75 (not targets of PHR1) are induced normally. Logically, the down-regulation of Pht1;1/PT1 and Pht1;4/PT2 (even more genes encoding high-affinity Pi transporters) could inactivate the Pi uptake system triggered by Pi starvation. Indeed, Pi content was lower in arf7, arf19, and arf7 arf19 mutants compared with wild-type plants. Unlike the Pi content, the anthocyanin accumulation is enhanced in shoots of arf7, arf19, and arf7 arf19 mutants under low-Pi conditions. In Arabidopsis shoots, anthocyanin accumulation induced by low Pi is dependent on PHR1 (Rubio et al., 2001). It seems contradictory, now that ARF7 and ARF19 positively regulate PHR1, that the anthocyanin accumulation should be down-regulated in arf7, arf19, and arf7 arf19 mutants. Previously, it was discussed that PHR1 activity could not be affected in the arf7 arf19 double mutant (Narise et al., 2010). Considering the tissue specificity of PHR1 regulated by ARF7 and ARF19, it is logical that ARF7 and ARF19 specifically or dominantly control PHR1 in roots, whereas, in shoots, there are likely other ARF(s) controlling PHR1 expression. In roots of the mutants, the down-regulated PHR1 results in the decreased expression of PSI genes and Pi uptake. However, in shoots of the mutants, PHR1 is not affected and anthocyanin biosynthesis is intact. Moreover, the Pi deficiency in the arf7 arf19 mutant is more severe than that in arf7 and arf19 mutants and wild-type plants, so much more anthocyanin is accumulated in the double mutant under low-Pi conditions. Consistently, previous data also have shown that the induction of At4 and Pht1;1/PT1 by low Pi is decreased dramatically in arf7 arf19 roots, whereas it is not obviously affected in arf7 arf19 shoots (Narise et al., 2010).
Loss- and Gain-of-Function Mutation of PHR1 Affects Arabidopsis Sensitivity to Auxin
Under low-Pi conditions, the RSA of the phr1 mutant is changed (Rubio et al., 2001; Nilsson et al., 2007). Statistically, the number of lateral roots of phr1 mutant and PHR1 overexpression transgenic Arabidopsis is significantly decreased and weakly increased, respectively, under low-Pi conditions (Ren et al., 2012). By exogenous auxin treatment, the sensitivity of the phr1 mutant to auxin for inducing lateral root formation is reduced. PHR1 is at least partially involved in the modulation of Arabidopsis RSAs under low-Pi conditions. ARF7 and ARF19 act as key components in a developmental pathway regulating lateral root formation by directly regulating LBD16 and LBD29 (Okushima et al., 2005; Wilmoth et al., 2005). In roots of the phr1 mutant, the expression of LBD16 and LBD29 is not changed obviously (Supplemental Fig. S6). On the other hand, constitutive expression of PHR1 could partially rescue the lateral root impairment in the arf7 arf19 double mutant, showing the contribution of down-regulated PHR1 to the phenotype of the arf7 arf19 mutant. It seems that there is a bypass independent of LBD16 and LBD29 in which PHR1 as a component is involved in the regulation of lateral root formation under low-Pi conditions. In Arabidopsis, besides the ARF7/19-mediated pathway, a WOX11-mediated pathway also contributes to root system formation (Sheng et al., 2017). Potentially, PHR1 is the mutual component of ARF7/19- and WOX11-mediated pathways for regulating lateral root initiation and development.
Arabidopsis MYB-CC Family Members Are Regulated by ARF7 and ARF19
Arabidopsis PHR1 belongs to a 15-member family of MYB-CC transcription factors (Rubio et al., 2001). Furthermore, PHL1 and PHL2, the other two MYB-CC family members, are proven to be functionally redundant with PHR1 in Arabidopsis responses to Pi starvation (Bustos et al., 2010; Sun et al., 2016). It is highly possible that the genes encoding the other MYB-CC transcription factors also could be the targets of ARF7 and ARF19. Among 15 genes encoding MYB-CC family transcription factors, the auxin-response elements were found in the promoters of 13 genes, and the majority of them are bound by ARF7 and/or ARF19. Here, the TGA element, AuxRE, and AuxRR core element are tested in the promoters. It cannot be entirely excluded that the other cis-elements, which are unidentified so far, could be bound by ARF7 or ARF19. Moreover, it cannot be ruled out that the other ARFs, besides ARF7 and ARF19, could modulate the genes encoding MYB-CC family transcription factors.
CONCLUSION
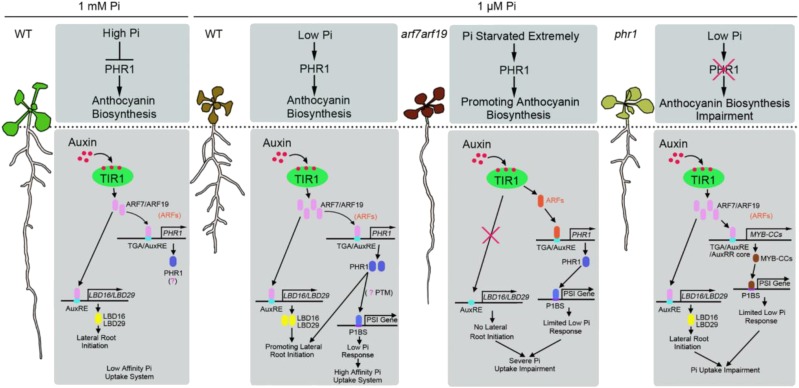
Based on the results obtained, we propose a model for how auxin signaling and the PHR1 regulon in Arabidopsis is coordinated during responses to Pi deficiency (Fig. 11). (1) Under high-Pi conditions, auxin signaling regulates PHR1 by ARF7 and ARF19 in roots, yet the necessity of PHR1 expression is unclear. In shoots, the high level of Pi suppresses the biosynthesis of anthocyanin. (2) When plants suffer Pi deficiency, the increased sensitivity of an auxin receptor, TIR1, results in up-regulated ARF7 and ARF19, which promote the expression of PHR1 in roots. PHR1 protein is activated by an unknown mechanism (probably posttranslational modification), then its downstream PSI genes are up-regulated. Simultaneously, PHR1 is involved in the modulation of lateral root initiation, which results in increased proliferation of lateral roots. In shoots, the biosynthesis of anthocyanin dependent on PHR1 is triggered by Pi deficiency. (3) Consistently, in roots of the arf7 arf19 double mutant, LBD16/LBD29 and PHR1 expression is inactivated, causing the suppression of lateral root initiation and molecular responses to Pi starvation, resulting in more severe impairment of Pi uptake. In shoots of the arf7 arf19 double mutant, PHR1 expression is intact and the severe deficiency of Pi results in increased biosynthesis of anthocyanin. (4) When phr1 mutants suffer from Pi deficiency, the basic lateral root initiation dependent on LBD16/LBD29 is active and the lateral root initiation dependent on PHR1 is interrupted, resulting in decreased proliferation of lateral roots compared with that of wild-type plants. In shoots of the phr1 mutant, although Pi is severely deficient, anthocyanin biosynthesis dependent on PHR1 is interrupted and anthocyanin accumulation is impaired.
Figure 11.
Model for the integration of auxin signaling and the PHR1 regulon in the Arabidopsis Pi-starvation response. Under both high- and low-Pi conditions, ARF7 and ARF19 regulate PHR1 in roots. When plants suffer Pi deficiency, PHR1 protein is activated by an unknown mechanism, then its downstream PSI genes are up-regulated. In roots of the arf7 arf19 mutant, PHR1 is inactivated, causing the suppression of molecular responses to Pi starvation. In shoots of the arf7 arf19 mutant, PHR1 expression is intact, and the severe deficiency of Pi results in increased biosynthesis of anthocyanin. In roots of the phr1 mutant, the lateral root initiation dependent on PHR1 is interrupted, resulting in decreased proliferation of lateral roots. In shoots of the phr1 mutant, anthocyanin accumulation dependent on PHR1 is impaired. WT, Wild type.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study were of the Columbia-0 ecotype background. The phr1 mutant line (SALK_067629) was obtained from the SALK collection. The homozygous phr1 mutant, which was selected by PCR-based genomic identification, was confirmed by RT-PCR. The ARF7p:GUS (CS24633) and ARF19p:GUS (CS24634) transgenic lines were obtained from the Arabidopsis Biological Resource Center. The seeds of arf7, arf19, and arf7 arf19 mutants were provided by Luis Herrera-Estrella and confirmed by RT-PCR using gene-specific primers (Supplemental Table S1). Arabidopsis seeds were surface sterilized and germinated on one-half-strength MS medium (pH 5.8) containing 0.8% (w/v) agar at 22°C to 25°C. After 4 d, the germinated seedlings were transferred onto high-Pi or low-Pi medium at 22°C to 25°C. MS medium with 1 mm KH2PO4 was used as high-Pi medium and that with 1 μm KH2PO4 was used as low-Pi medium. For hydroponics, 5-d-old seedlings were transferred in one-quarter-strength Hoagland nutrient solution containing 1 mm or 1 μm KH2PO4, respectively. Plants were grown under a long-day light cycle (16 h of light/8 h of dark), and the humidity was maintained at 65%.
Vector Construction and Plant Transformation
The PHR1 promoter, which is a 1.5-kb sequence upstream of the PHR1 start codon, was isolated from the Arabidopsis genome by PCR using a pair of primers and then cloned into pBluescript SK for sequence verification. The PHR1 promoter fragment was subcloned into pBI101 vector to generate the chimeric PHR1p:GUS construct. To investigate the necessity of auxin-response elements for PHR1 expression, one AuxRE and two TGA elements of the PHR1 promoter were deleted by primer-based mutagenesis, resulting in PHR1pm1:GUS, PHR1pm2:GUS, PHR1pm3:GUS, PHR1pmm1:GUS, PHR1pmm2:GUS, PHR1pmm3:GUS, and PHR1pmmm:GUS mutation constructs. With the gene-specific primers, the full-length cDNA of PHR1 was amplified and then cloned into pBluescript SK vector for sequence verification. The coding sequence of PHR1 was then cloned into pBI121 vector, replacing the GUS gene (35S:PHR1). The construct 35S:PHR1 was used to transform the wild type and the arf7 arf19 double mutant. The above-mentioned constructs were introduced into Arabidopsis by the floral dip method. Homozygous lines of the T3 and T4 generations were used in this study.
Histochemical Assay of GUS Activity
Histochemical assays of GUS activity in transgenic Arabidopsis were conducted according to a protocol described previously (Ren et al., 2014). The samples were incubated at 37°C in a GUS reaction buffer including 5-bromo-4-chloro-3-indolyl-β-d-GlcA. Chlorophyll was cleared from plant tissues by immersing them in 70% ethanol. GUS staining patterns were confirmed by observing at least five different transgenic lines. Representative stained seedlings or tissues were imaged using a Leica MZ16f stereomicroscope (Leica).
Pi and Anthocyanin Content Assay
Pi content and anthocyanin content were measured by the methods described previously (Ren et al., 2012).
RT-qPCR Analysis
The expression of the genes was analyzed by RT-qPCR using the fluorescent intercalating dye SYBR Green in a detection system (MJ Research; Opticon 2). The ACTIN2 gene was used as a standard control for RT-qPCR. A two-step RT-PCR procedure was performed using a method described earlier (Ren et al., 2014). All primers used in the RT-qPCR analyses are listed in Supplemental Table S1.
Yeast One-Hybrid Analysis
Yeast one-hybrid analysis was carried out using the Matchmaker Gold yeast one-hybrid system (Biosciences Clontech). PHR1 promoter was inserted upstream of the AUR1-C gene in the pAbAi vector and then efficiently integrated into the genome of the Y1HGold yeast strain by homologous recombination.
The truncated coding sequences of ARF7 and ARF19 (encoding amino acids 1–367 and 1–359, respectively, which include the DNA-binding domain and transcriptional factor domain) were cloned into pGADT7. This contains the GAL4 activation domain to create the fusion constructs GAL4-ARF7 and GAL4-ARF19, which were transformed into the Y1HGold/PHR1p reporter yeast strain. The resistance to AbA is conferred by the AbAr gene (AUR-1C), which is the reporter on the bait vector pAbAi. When ARF7 or ARF19 binds to the PHR1 promoter sequence, the GAL4 activation domain activates the expression of AbAr, which allows the cells to grow on medium containing the AbA antibiotic.
EMSA
The truncated coding sequences of ARF7 and ARF19 were cloned into pET32a. The ARF7 and ARF19 fusion proteins were purified from Escherichia coli strain BL21 (DE3) using an Ni-NTA resin column. Protein concentrations were determined using the Bio-Rad DC-assay kit. Three pairs of 45-bp oligonucleotides each containing one of the auxin-response elements were synthesized and labeled with biotin. Each pair of oligonucleotides is annealed to double-chain DNA as DNA probes. The labeled probes were incubated with the recombinant proteins in a buffer solution (750 mm KCl, 0.5 mm DTT, 0.5 mm EDTA, and 50 mm Tris, pH 7.4) and incubated at room temperature for 20 min. The DNA-protein reaction mixtures were separated by nondenaturing PAGE. The free and protein-bound DNAs were detected using the Light Shift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer’s instructions.
Antibody Generation and ChIP Assay
The polyclonal antibodies against ARF7 or ARF19 were generated by inoculation of rabbits with ARF7 and ARF19 fusion proteins. Seven-day-old seedlings were harvested for ChIP experiments. The chromatin samples for ChIP experiments were obtained according to a previously described protocol (Saleh et al., 2008). The plants were first cross-linked by formaldehyde, and the purified cross-linked nuclei were then sonicated to shear the chromatin into suitably sized fragments. The antibody that specifically recognizes ARF7 or ARF19 was used to immunoprecipitate DNA/protein complexes from the chromatin preparation. The DNA in the precipitated complexes was recovered and analyzed by RT-qPCR as described above. The chosen primer sets can amplify fragments of 150 to 200 bp within the promoters of PHR1 and the genes coding other MYB-CC family members. To ensure the reliability of ChIP data, the input sample and nonantibody control sample were analyzed with each primer set. On the other hand, the fragment including the P1BS element of the IPS1 promoter was amplified as a negative control.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PHR1 (AT4G28610), ARF7 (AT5G20730), ARF19 (AT1G19220), ARF5 (AT1G19850), ARF10 (AT2G28350), IPS1 (AT3G09922), At4 (AT5G03545), Pht1;1 (AT5G43350), Pht1;4 (AT2G38940), ACP5 (AT3G17790), RNS1 (AT2G02990), PHO1 (AT3G23430), WRKY75 (AT5G13080), LBD16 (AT2G42430), and LBD29 (AT3G58190).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Schematic representation of the cis-elements in a 1.5-kb sequence upstream of the start codon of the PHR1 gene.
Supplemental Figure S2. Expression analysis of ARF7 and ARF19 in Arabidopsis roots.
Supplemental Figure S3. Analysis of ARF7 and ARF19 binding to the PHR1 promoter via a yeast one-hybrid assay.
Supplemental Figure S4. Phenotypes of arf7 and arf19 single mutants and the arf7 arf19 double mutant.
Supplemental Figure S5. Phenotypes of the wild type, phr1 single mutants, and PHR1 overexpression lines.
Supplemental Figure S6. Expression analysis of LBD16 and LBD29 in Arabidopsis roots.
Supplemental Table S1. Primers or oligonucleotides used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Luis Herrera-Estrella for providing seeds of arf7, arf19, and arf7 arf19 mutants.
Footnotes
This work was supported by The National Natural Sciences Foundation of China (grant nos. 31271637 and 31571572) and The Project of National Research and Development of China (grant no. 2016YFD0100202).
References
- Al-Ghazi Y, Muller B, Pinloche S, Tranbarger TJ, Nacry P, Rossignol M, Tardieu F, Doumas P (2003) Temporal responses of Arabidopsis root architecture to phosphate starvation: evidence for the involvement of auxin signaling. Plant Cell Environ 26: 1053–1066 [Google Scholar]
- Ballas N, Wong LM, Ke M, Theologis A (1995) Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid-inducible gene PS-IAA4/5. Proc Natl Acad Sci USA 92: 3483–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W (2009) DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136: 1643–1651 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143: 1789–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Madhuvanthi R, Karthikeyan AS, Raghothama KG (2009) Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol Plant 2: 43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Holford ICR. (1997) Soil phosphorus: its measurement, and its uptake by plants. Aust J Soil Res 35: 227–239 [Google Scholar]
- Jain A, Poling MD, Karthikeyan AS, Blakeslee JJ, Peer WA, Titapiwatanakun B, Murphy AS, Raghothama KG (2007) Differential effects of sucrose and auxin on localized phosphate deficiency-induced modulation of different traits of root system architecture in Arabidopsis. Plant Physiol 144: 232–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC (2014) Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci USA 111: 5427–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P (2005) A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol 138: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao RT, Goekjian VH, Hong JC, Key JL (1993) Identification of protein-binding DNA sequences in an auxin-regulated gene of soybean. Plant Mol Biol 21: 1147–1162 [DOI] [PubMed] [Google Scholar]
- Nanao MH, Vinos-Poyo T, Brunoud G, Thévenon E, Mazzoleni M, Mast D, Lainé S, Wang S, Hagen G, Li H, et al. (2014) Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun 5: 3617. [DOI] [PubMed] [Google Scholar]
- Narise T, Kobayashi K, Baba S, Shimojima M, Masuda S, Fukaki H, Ohta H (2010) Involvement of auxin signaling mediated by IAA14 and ARF7/19 in membrane lipid remodeling during phosphate starvation. Plant Mol Biol 72: 533–544 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M, Roby D, Dumas C, Cock JM (1997) Rapid induction by wounding and bacterial infection of an S gene family receptor-like kinase gene in Brassica oleracea. Plant Cell 9: 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L (2008) Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20: 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB (2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE 7: e44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Zhao CZ, Liu CS, Huang KL, Guo QQ, Chang LL, Xiong H, Li XB (2014) A Brassica napus PHT1 phosphate transporter, BnPht1;4, promotes phosphate uptake and affects roots architecture of transgenic Arabidopsis. Plant Mol Biol 86: 595–607 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L, Hu X, Du Y, Zhang G, Huang H, Scheres B, Xu L (2017) Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture. Development 144: 3126–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D (2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G (2008) Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ 31: 1834–1843 [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, et al. (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot 111: 1139–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43: 118–130 [DOI] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA 96: 15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]