Consensus co-expression networks generate robust gene clusters and yield important insights into fertilization-induced iron transport and key floral regulators in wild strawberry.
Abstract
The diploid strawberry, Fragaria vesca, is a developing model system for the economically important Rosaceae family. Strawberry fleshy fruit develops from the floral receptacle and its ripening is nonclimacteric. The external seed configuration of strawberry fruit facilitates the study of seed-to-fruit cross tissue communication, particularly phytohormone biosynthesis and transport. To investigate strawberry fruit development, we previously generated spatial and temporal transcriptome data profiling F. vesca flower and fruit development pre- and postfertilization. In this study, we combined 46 of our existing RNA-seq libraries to generate coexpression networks using the Weighted Gene Co-Expression Network Analysis package in R. We then applied a post-hoc consensus clustering approach and used bootstrapping to demonstrate consensus clustering’s ability to produce robust and reproducible clusters. Further, we experimentally tested hypotheses based on the networks, including increased iron transport from the receptacle to the seed postfertilization and characterized a F. vesca floral mutant and its candidate gene. To increase their utility, the networks are presented in a web interface (www.fv.rosaceaefruits.org) for easy exploration and identification of coexpressed genes. Together, the work reported here illustrates ways to generate robust networks optimized for the mining of large transcriptome data sets, thereby providing a useful resource for hypothesis generation and experimental design in strawberry and related Rosaceae fruit crops.
Fragaria vesca, the alpine or woodland strawberry, has been cultivated since at least the fourteenth century (Darrow, 1966). More recently, F. vesca has been developed as a model for the commercial strawberry, the octoploid Fragaria ananassa, and other members of the economically important Rosaceae family due to its diploidy (2n = 14), small genome size (240 Mb), and amenability to transformation (Slovin et al., 2009; Shulaev et al., 2011). In particular, the external seed configuration of strawberry fruit is ideal for studying cross-tissue communication during fruit development. The strawberry botanical fruit, the achene, is derived from the ovary wall and houses a single seed. The fleshy fruit is derived from the receptacle, or stem tip. Previous work indicates that the hormones auxin and gibberellic acid (GA) are synthesized in the achene, particularly in the endosperm and seed coat, and are transported to the underlying receptacle where they initiate fleshy fruit development (Nitsch, 1950; Kang et al., 2013). The primary model systems for studying fruit development have historically been Arabidopsis (Arabidopsis thaliana; Roeder and Yanofsky, 2006) and tomato (Solanum lycopersicum; Kimura and Sinha, 2008), the fruits of which are dry and fleshy, respectively, and develop from the ovary wall (Gasser and Robinson-Beers, 1993; Ferrándiz et al., 1999). Studying the strawberry accessory fruit expands our knowledge of general developmental processes.
A number of research efforts have focused on the late stages of strawberry fruit development to facilitate studies of ripening, flavor, aroma, and nutritional content (Aharoni and O’Connell, 2002; García-Gago et al., 2009; Estrada-Johnson et al., 2017; Sánchez-Sevilla et al., 2017). However, knowledge of molecular events underlying fruit set and the early stages of development is equally critical and useful for ensuring consistent crop yield. Previously, we conducted a detailed developmental characterization of strawberry flowers and early-stage fruit to generate morphological markers corresponding to successive developmental stages (Hollender et al., 2012). Subsequently, we generated a large and comprehensive set of RNA sequencing (RNA-seq) data from F. vesca flower and fruit tissues at multiple early- and mid-developmental stages (Kang et al., 2013; Hollender et al., 2014; Hawkins et al., 2017). This wealth of spatial and temporal transcriptome data provides genome-scale insight into the biological processes and molecular events underlying early fruit development.
Recent technological advancements in transcriptome sequencing, coupled with decreasing costs, have created unprecedented opportunities to study nonmodel and developing model systems (Strickler et al., 2012). RNA-seq experiments can be designed to study a plethora of topics, including comparison of mutant and wild-type organisms, development, and abiotic and biotic stress response. Transcriptome data are also highly versatile and can be used to characterize gene expression across space and time (Rowland et al., 2012; Pattison et al., 2015), to identify alternative splicing events (Li et al., 2017), to identify novel transcripts (Chettoor et al., 2014), and to identify key biological processes. However, remaining challenges include how to best visualize large data sets and identify information of interest. Gene coexpression network analysis, first developed for microarray data analysis (Eisen et al., 1998; Ficklin and Feltus, 2011; Sato et al., 2011; De Bodt et al., 2012), can be used to identify sets of functionally related genes.
Coexpression network analysis is based on correlations between gene expression values. It describes correlation patterns between genes in a pairwise fashion across multiple microarray or RNA-seq samples. Coexpression networks are useful for exploring large, complex data sets and are powerful tools for predicting gene function. Exploration of neighborhoods of connected genes invokes the “guilt by association” principle in that genes that are highly connected and have very similar expression patterns are more likely to function in the same pathways or regulate the same biological processes (Ravasz et al., 2002; Spirin and Mirny, 2003; Singer et al., 2005; Wolfe et al., 2005).
Recent analyses have sought to test and optimize gene coexpression networks generated with RNA-seq data (Iancu et al., 2012; Sekhon et al., 2013; Huang et al., 2017). However, the accuracy of gene functions, relationships predicted by network approaches, and the overall utility of coexpression networks is still largely unknown. Computationally, one method to test the stability and robustness of clusters is to use a post-hoc consensus clustering approach (Monti et al., 2003). If clusters accurately represent subpopulations of a larger data set, the number and composition of clusters should not vary greatly if clustering is repeatedly conducted using different parameters or different subsets of the full data set. Clusters that are robust in response to sampling variability are more likely to represent true relationships between genes. Therefore, consensus clusters are potentially more reliable for predicting gene functions and interactions, though experimental data are still necessary to assess their accuracy.
In this study, we used the Weighted Gene Co-Expression Network Analysis (WGCNA) package available in R (Langfelder and Horvath, 2008) to generate three sets of coexpression networks. The first incorporates early stage F. vesca floral tissues dissected by laser capture microdissection (LCM). The second includes hand-dissected flower and fruit tissues spanning prefertilized flowers to fruit just prior to ripening. The third set concerns receptacle fruit tissues at the ripening stages. Our use of multiple, comprehensive RNA-seq libraries provides extensive gene expression information for network construction and therefore increases the likelihood of identifying genetic correlations (Lee et al., 2004; Wren, 2009; Ballouz et al., 2015). Additionally, we tested a consensus clustering add-on to the WGCNA algorithm and used bootstrapping to test the reliability of clusters generated with both WGCNA alone and with WGCNA plus consensus clustering. We demonstrate that coexpression network analyses can illuminate molecular processes underlying developmental events and are useful tools for hypothesis generation and experimental design. We further explore experimental validation for gene relationships predicted by our consensus networks, including evidence of increased iron transport to the seed immediately postfertilization and characterization of an F. vesca floral mutant.
RESULTS
WGCNA Network Analysis of Comprehensive Flower and Fruit RNA-Seq Data
RNA-seq data for 46 different tissues/stages (46 tissues × 2 biological replicates) were generated previously using the F. vesca accession Yellow Wonder 5AF7 (Supplemental Table S1; Kang et al., 2013; Hollender et al., 2014; Hawkins et al., 2017). Most tissues were harvested by hand-dissection (HD) under a stereomicroscope, and young floral tissues were isolated using LCM. Therefore, LCM and HD data were analyzed separately to avoid variation introduced by different techniques (Hollender et al., 2014; Supplemental Fig. S1). Coexpression networks were generated using the WGCNA package (Langfelder and Horvath, 2008), in which all coexpressed genes are connected to each other with varying correlation strengths (Supplemental Data Set S1). This is accomplished using soft thresholding, thereby preserving the continuous nature of the data set and eliminating the need to set an arbitrary correlation score cutoff.
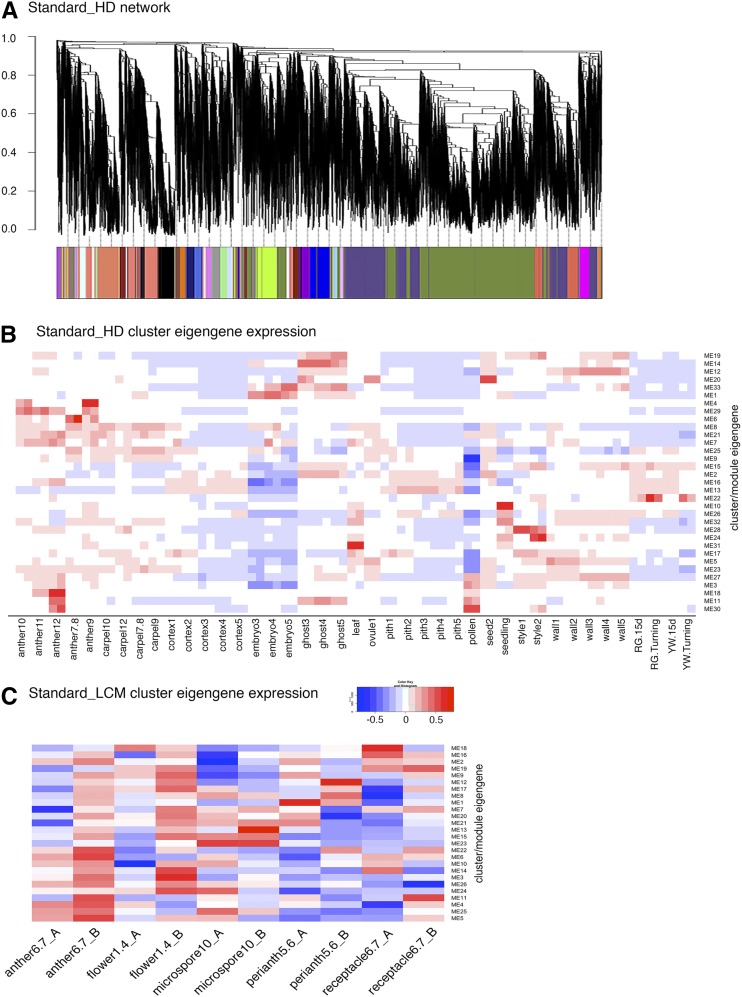
When standard parameters (see “Materials and Methods”) are used, the HD tissue network incorporates 33 clusters of coexpressed genes (Fig. 1A). Eigengenes, the first principal component of a cluster, can be thought of as a representative of a cluster’s expression profile. The expression value of each cluster’s eigengene in each of the HD tissues is plotted in a heat map (Fig. 1B; Supplemental Data Set S2), which allows for easy visualization of the cluster-tissue association. For example, genes in cluster 22 are expressed most highly in ripening fruits at the turning stage. Cluster 4 correlates with stage 9 to 10 anthers. Cluster 10 is more specific to seedlings and leaves. A similar network analysis was applied to the LCM samples and led to 26 clusters. The expression profile of each cluster eigengene is also shown as a heat map (Fig. 1C; Supplemental Data Set S2). The large number of tissues and stages enabled the development of robust coexpression networks, a significant improvement over our previous network analysis utilizing only 16 floral tissues/stages (Hollender et al., 2014).
Figure 1.
Standard (non_consensus) WGCNA network analyses of 82 hand-dissected flower, fruit, and vegetative samples and 10 LCM flower samples. A, Dendrogram showing coexpression modules (clusters) identified by standard WGCNA across HD flower and fruit tissues. Each leaf in the tree is one gene. The major tree branches constitute 33 modules labeled with different colors. B, Heat map showing cluster-tissue associations of the standard HD network. Each row corresponds to a cluster. Each column corresponds to a specific tissue/stage. The color of each cell at the row-column intersection indicates the eigengene expression value. Blue color indicates a negative association and red color indicates a positive association between the cluster and the tissue. Specific cluster eigengene values are provided in Supplemental Data Set S2. C, Heat map showing cluster-tissue associations of the standard LCM network. A total of 26 clusters were identified. Each cluster eigengene expression value may be found in Supplemental Data Set S2.
Ghost-Associated Modules Provide Insight into Iron Transport during Fruit Development
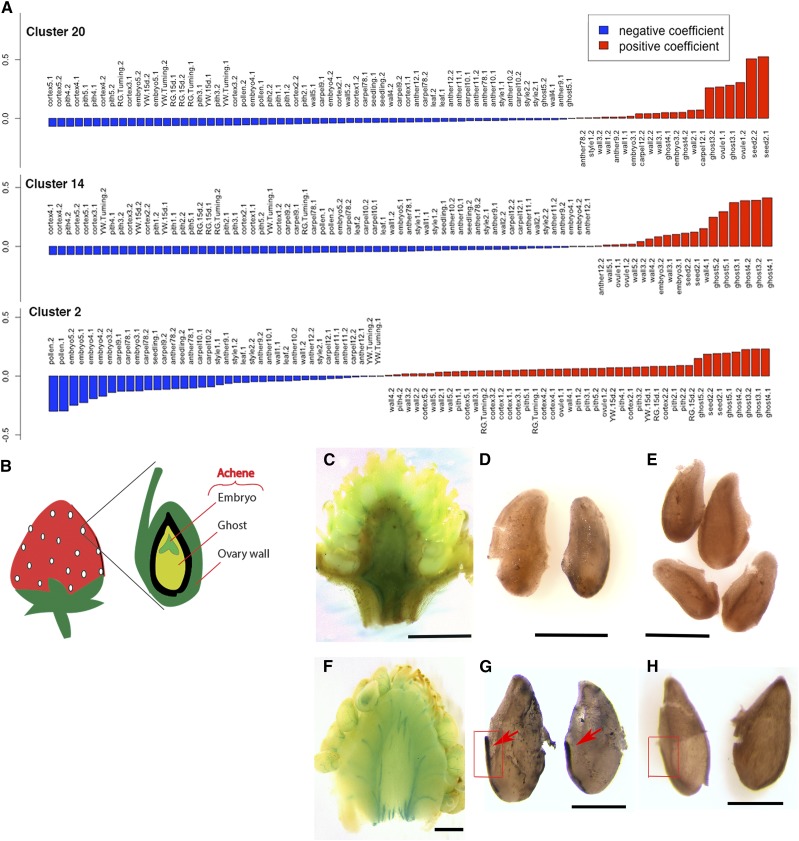
Fertilization initiates the biosynthesis of auxin and GA in seeds; these phytohormones subsequently stimulate fruit set in strawberry (Nitsch, 1950; Kang et al., 2013). When a fertilized seed is dissected to remove the embryo, the remaining seed tissue containing the endosperm and seed coat is referred to as the “ghost” (Fig. 2B). Previous transcriptome analysis revealed that auxin and GA biosynthesis genes were transcriptionally induced in the ghost upon fertilization, implicating the importance of the ghost in F. vesca fruit set (Kang et al., 2013). Clusters 2, 14, and 20 from the HD network show clear association with the ghost (Figs. 1B and 2A). Cluster 20 is correlated with the stage 1 ovule (prefertilization) and the stage 2 seed (immediately after fertilization). The top two gene ontology (GO) terms in cluster 20 are “Regulation of Fertilization” and “Regulation of Double Fertilization” (Supplemental Data Set S3). Abundant MADS box genes were found among genes in cluster 20 including three annotated as AGAMOUS-LIKE80-like (genes 04949, 15899, and 22916) and four AGAMOUS-LIKE62-like (genes 01789, 07364, 30567, and 07361). Cluster 14 is most strongly associated with ghost stages 3, 4, and 5, all of which are postfertilization stages. Interestingly, the enriched GO terms are completely distinct from those of cluster 20, suggesting very different molecular events in similar tissues postfertilization.
Figure 2.
Ghost-associated clusters indicate active iron transport postfertilization. A, Eigengene expression values of clusters 20, 14, and 2 in different tissues. Highest eigengene values are with seed (cluster 20) and ghost (cluster 2 and 14). B, Diagram of the strawberry receptacle in relation to the achene, which consists of the ovary wall and seed. Each seed houses the ghost (endosperm and seedcoat) and embryo. C, Perls staining of the prefertilization (stage 1) receptacle. D, DAB-enhanced Perls staining of iron in fixed ovules at prefertilization. No difference is seen from the DAB-only control in E. E, DAB-only staining of fixed ovules at prefertilization. F, Perls staining of the receptacle at stage 3 (postfertilization). Blue lines are stained vascular strands connecting the receptacle to individual achenes. G, DAB-enhanced Perls staining of iron in fixed stage 3 seeds (postfertilization). Strong vascular strand staining is seen along the side of the seeds (arrows). This positive staining contrasts with the negative control (rectangle) shown in H. H, DAB-only staining of fixed stage 3 seeds. Scale bars in C and F, 1 mm; in D, E, G, and H, 0.4 mm.
Cluster 2 is not only positively correlated with the ghost but is also negatively correlated with embryos, suggesting endosperm-/seed coat-specific molecular events. This cluster is also positively correlated with the cortex and pith tissues of the receptacle. Twenty-eight out of a total of forty enriched GO terms (Biological Process) in cluster 2 are related to iron transport and iron sequestration (Supplemental Data Set S3). Cluster 2 contains gene 19831, which is annotated as a homolog of the phloem-specific iron transporter OLIGOPEPTIDE TRANSPORTER3 (Stacey et al., 2002, 2008; Zhai et al., 2014), three metal binding proteins (genes 08918, 10308, and 18489), and two members of the VACUOLAR IRON TRANSPORTER1 family (genes 32625 and 17575; Kim et al., 2006). The GO terms enriched in cluster 2 suggest that the ghost and receptacle, but not the embryo, carry out active iron transport during the earliest stages of strawberry fruit development.
To test the above hypothesis, we stained free iron in the receptacle and seed using the iron-specific Perls stain (Green and Rogers, 2004; Stacey et al., 2008; Roschzttardtz et al., 2009; Brumbarova and Ivanov, 2014). Potassium ferrocyanide, a component of the Perls reagent, reacts with iron to form an insoluble pigment known as Prussian blue. Increased free iron in the vascular tissue of the receptacle was observed postfertilization as abundant blue strands connecting the receptacle to individual achenes (Fig. 2, C and F). A subsequent intensification reaction with 3,3′-diaminobenzidine (DAB) was previously shown to enhance Perls staining and produce a dark brown pigment (Nguyen-Legros et al., 1980; Roschzttardtz et al., 2009). Achenes were dissected to isolate seeds, which were stained with Perls and then DAB. The strongest staining was observed in the strands of vasculature connecting the seed to the subtending receptacle (Fig. 2G, arrows). The ovules (precursors of seeds) did not show vascular strand staining (Fig. 2D). Seeds fixed and treated with only DAB served as negative controls (Fig. 2, E and H) since DAB alone is unable to directly stain iron (Roschzttardtz et al., 2009; see “Materials and Methods”). The significant increase in iron transport from the receptacle to the seed postfertilization is consistent with increased iron transporter expression in the cluster 2 network. The requirement of an iron cofactor for the GA biosynthetic enzymes GA20ox and GA3ox (Huang et al., 2015; White and Flashman, 2016) exemplifies one of the many postfertilization molecular events that require iron.
Consensus Networks Provide Robust and Reproducible Clusters
Because of the potential for noise and instability in standard coexpression network analysis, a consensus-clustering approach (Wu et al., 2002; Monti et al., 2003) was applied as an extension to WGCNA. This strategy is independent of parameter selection and ensures module reproducibility. Spurious coclusterings are reduced by testing the stability of clusters in response to sampling variability. Sampling variability, or perturbations of the data set, can be simulated with a resampling approach. Therefore, 1,000 runs of the WGCNA clustering algorithm were performed with each run resampling 80% of the genes and using randomly generated parameter selections (detailed in “Materials and Methods”).
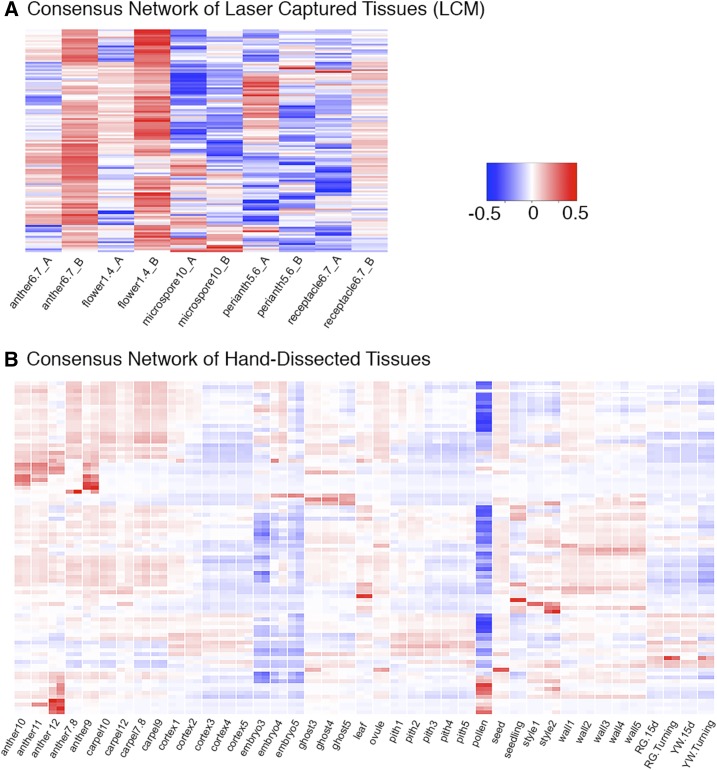
Consensus clustering of HD and LCM samples yielded 86 and 123 clusters, respectively (Fig. 3, A and B; Supplemental Data Set S1). The consensus HD clusters were evaluated against the standard (WGCNA) HD clusters (Fig. 4). Although both approaches yielded a similar number of clusters of comparable size (Fig. 4A), the clusters had little overlap as shown by the Jaccard index (Fig. 4B). The Jaccard index is a statistic used for comparing the similarity and diversity of sample sets ranging from 0% to 100% (Fuxman Bass et al., 2013). The higher the percentage, the more similar the two clusters. While many clusters had a significant overlap (hypergeometric P value < 0.05) across methods, the Jaccard index was never higher than 50%.
Figure 3.
Consensus network analyses of LCM and HD flower, fruit, and vegetative tissues. A, Heat map showing cluster-tissue associations of the consensus LCM network. A total of 123 clusters were identified. Each row corresponds to a cluster. Each column corresponds to a specific tissue/stage. The color of each cell at the row-column intersection indicates the eigengene value. Blue color indicates a negative association and red color indicates a positive association between the cluster and the tissue. Specific eigengene values are provided in Supplemental Data Set S2. B, Heat map showing cluster-tissue association of the consensus_HD network. Eighty-six clusters were identified. Two replicates of each tissue are labeled with one name at the bottom.
Figure 4.
Comparison of standard WGCNA to the consensus clustering method. A, Statistics describing the clustering behaviors of both methods. B, Similarity between standard and consensus clusters with significant overlap. C, Bootstrap confidence intervals of gene pairs within the same cluster.
Since the standard WGCNA and consensus algorithms define clusters that are not known a priori, the quality of clustering was measured by internal evaluation criterion. Typical objective functions in clustering aim to attain high intracluster similarity and low intercluster similarity. The RS value, sometimes referred to as the pseudo-F statistic, is a ratio of the variance between clusters to the variance within clusters, thereby defining the proportion of variation explained by a particular clustering of genes (Sharma, 1995; He et al., 2015; details in “Materials and Methods”). Both the standard WGCNA and consensus methods performed similarly with regard to the RS statistic and intracluster correlation (Fig. 4A). However, bootstrap confidence intervals are significantly higher for consensus clustering (Wilcoxon P value < 0.05; Fig. 4C). Bootstrapping allows the assignment of measures of accuracy; a high confidence interval implies that if the entire study were repeated ad infinitum, the resulting gene pairs would be the same. This result demonstrates that the consensus clustering method, while preserving the same level of correlation between clustered gene pairs as the standard WGCNA method, also produces clusters with higher reliability.
To maximize the potential of identifying pairs of genes with functional relationships, consensus90 and consensus100 networks were also generated. These networks apply stringent cutoffs of 90% and 100% to the consensus matrix; only genes that cluster together 90% or 100% of the time in the consensus network appear in the consensus90 and consensus100 networks, respectively. The HD and LCM consensus90 networks contain 2,870 and 6,332 clusters, respectively (Supplemental Table S2), a significant increase over the number of clusters in the consensus network. Accordingly, each cluster in the consensus90 HD and LCM networks has fewer genes. On average, there are six genes per cluster in the HD network and five genes per cluster in the LCM network (Supplemental Table S2). The HD and LCM consensus100 networks contain 962 and 3,814 clusters, respectively. This decrease in cluster number as compared to the consensus90 networks is due to the decreased total number of genes included in each network as the majority of genes do not cluster with any partners 100% of the time in the consensus matrix. However, genes paired in the consensus100 networks are reliable candidates for functional relationships.
A User-Friendly Interface for Exploring Coexpression Networks
To facilitate utilization, exploration, and visualization of the coexpression networks, we generated the F. vesca gene coexpression network explorer, a user-friendly web interface, using the Shiny application from R Studio (http://shiny.rstudio.com). The site (www.fv.rosaceaefruits.org) hosts data from the standard, consensus, consensus100, and consensus90 networks for both the HD and LCM data sets as well as the ripening fruit tissue-only data set (4 network types × 3 data sets = 12 networks). Users can first choose a specific network to explore (Supplemental Fig. S2A) and subsequently retrieve general information such as the number of clusters in the network or search for a specific strawberry gene to determine in which cluster the gene resides (Supplemental Fig. S2B). Users may also identify a cluster with a specific tissue expression profile by selecting the “Clusters” tab (Supplemental Fig. S2C). This generates a list of the top five positively and negatively associated correlated tissues for the cluster, based on the eigengene expression value. This information is also visually displayed on a heat map under the “Tissue-Eigengene Expression” tab (such as Fig. 1, B and C, and Fig. 3, A and B). Users can further obtain detailed information for a specific cluster by choosing a cluster number from the drop-down menu (Supplemental Fig. S2D). Cluster-specific information includes a list of genes in the cluster with annotations and Arabidopsis homologs, a bar graph plotting the cluster’s eigengene expression in each of the profiled tissues in the network (such as Fig. 2A), enriched GO terms, and a plot indicating correlation between clusters (Supplemental Fig. S2E). A downloadable connectivity score file available for each cluster under the “Downloads” tab can easily be imported into Cytoscape for network visualization as shown in Figure 5.
Figure 5.
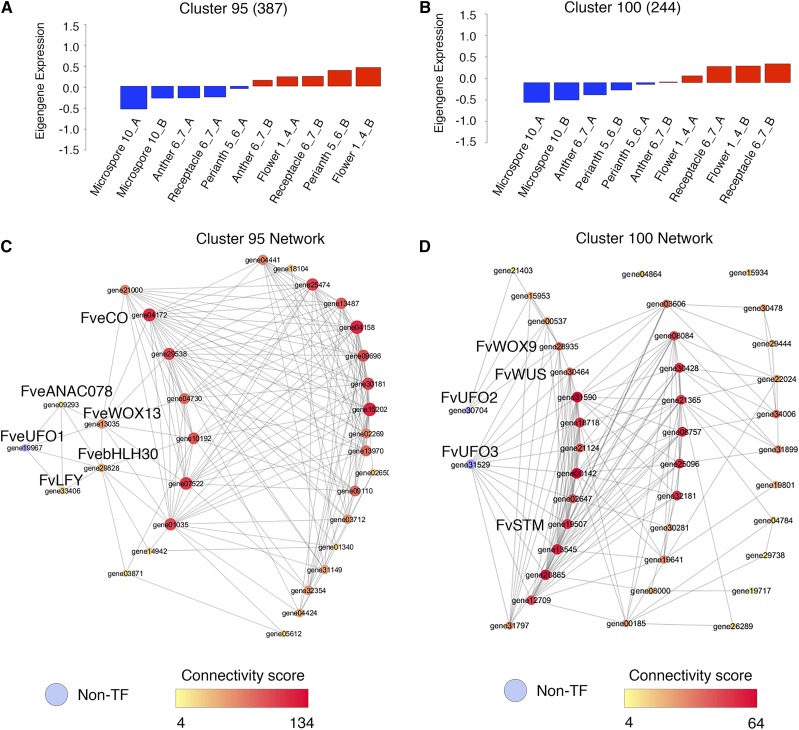
Young floral meristem and developing receptacle-associated clusters and transcription factor networks. A, Consensus_LCM cluster 95 eigengene expression value. B, Consensus_LCM cluster 100 eigengene expression value. C, Network showing connections between FveUFO1 and transcription factors in cluster 95. Edge cutoff is 0.8. Each colored circle (node) represents one gene. Larger node size and darker red node color indicate greater connectivity within the network. D, Network showing connections between FveUFO2, FveUFO3, and transcription factors in cluster 100. Edge cutoff is 0.8. Larger node size and darker red node color indicate greater connectivity within the network.
Consensus Networks Identify Potential Floral Meristem and Receptacle Meristem Regulators
We sought to test the ability of the consensus clusters to predict functional relationships between genes. Based on the consensus_LCM network, cluster 95 (387 genes) appears to correlate more strongly with the young floral meristem (floral stages 1–4), while cluster 100 (244 genes) correlates with floral stages 6 and 7, the stage at which the receptacle enlarges (Supplemental Data Set S2). Transcription factors with the strongest connections (cutoff 0.8 on a scale of 0 to 1) from clusters 95 and 100 were visualized using Cytoscape (Fig. 5, A and B). Cluster 100 is particularly rich in transcription factors involved in meristem regulation; 15 of the 38 transcription factors encode meristem regulators, including FveWUSCHEL (gene30464), FveSHOOT MERISTEMLESS (gene19507), and FveWUSCHEL-RELATED HOMEOBOX9 (gene28935). Interestingly, seven TALE homeodomain proteins are in cluster 100, while cluster 95 has only one. Together, the abundance of meristem regulators in cluster 100 supports previous anatomical and network analyses (Hollender et al., 2012, 2014), suggesting that the receptacle is a floral organ with meristematic activity.
Cluster 95 is more closely correlated with the young floral bud (floral stages 1–4). Among the 31 transcription factors in this cluster, eight are meristem regulators. Interestingly, FveLEAFY (FveLFY; gene33406) is found in this cluster but not cluster 100, implicating FveLFY in promoting the early stage floral meristem development. Gene04172, annotated as a CONSTANS-like transcription factor, is also in this cluster, suggesting a role for a CONSTANS family member in regulating the floral meristem.
Although UNUSUAL FLORAL ORGAN (UFO) does not encode a transcription factor, it was shown in Arabidopsis to be an important regulator of LFY. The interaction between UFO and LFY is required to activate the B class gene APETALA3 (AP3) for petal and stamen identity specification (Lee et al., 1997; Chae et al., 2008). Three UFO homologs in F. vesca (Supplemental Fig. S3) are found in clusters 95 and 100; FveUFO1 (gene19967) is in cluster 95 (Fig. 5C; Supplemental Data Set S4) and is strongly correlated with FveLFY (gene33406; edge score of 0.86). However, FveUFO2 (gene30704) and FveUFO3 (gene31529) are in cluster100 (Fig. 5D; Supplemental Data Set S4), suggesting that perhaps FveUFO2 and FveUFO3 act later during flower development and may not be involved in the regulation of FveLFY, which is absent from cluster 100. This fine separation between FveUFO1 and FveUFO2/3 indicates the possibility that FveUFO1 is not functionally redundant with FveUFO2 and FveUFO3. Altered or abolished FveUFO1 function would hence be likely to affect FveLFY and floral homeotic genes regulated by FveLFY.
We searched for F. vesca floral ABCE genes (Hollender et al., 2014) in the clusters containing FveUFO1/FveLFY in both the standard_LCM and consensus_LCM networks. In the standard_LCM network, FveUFO1 and FveLFY are clustered together with class A genes FveAP1 (gene04562) and FveAP2 (gene23876) and class E genes FveSEPALLATA1 (FveSEP1; gene04229), FveSEP4 (gene26118), and FveSEP1-like (gene04563; cluster 9; Supplemental Fig. S4). In contrast, no ABCE genes are clustered together with FveUFO1/FveLFY in the consensus_LCM network (Supplemental Fig. S4). However, consensus_LCM cluster 95, which contains FveUFO1 and FveLFY, has the highest Pearson correlation score (0.92009 on a scale of −1 to 1; Supplemental Data Set S5) with consensus_LCM cluster 119 containing FveAP1 and FveSEP1. This result suggests that, while the consensus network may predict close and immediate partnerships between FveUFO and FveLFY and between FveAP1 and FveSEP1, the standard network may predict genes acting in the same pathway. Further, the coclustering between FveUFO/FveLFY and class A and E genes predicts that FveUFO/FveLFY may regulate class A and E genes. In contrast, the three F. vesca class B genes, FveAP3 (gene14896), FvePISTILLATA-a (gene11267) and FvePISTILLATA-b (gene11268), which themselves cluster together in both the standard_LCM and consensus_LCM networks, do not cluster with FveUFO/FveLFY (Supplemental Fig. S4). Hence, strawberry class B genes may not be regulated by FveUFO/FveLFY as they are in Arabidopsis.
Identification of a Nonsense Mutation in FveUFO1
In an effort to provide experimental validation of the network analyses described above, an ethyl methanesulfonate (EMS) mutagenesis screen produced a floral mutant, hereafter called extra floral organs (efo), with defects in both floral meristem determinacy and floral organ development (Fig. 6). Specifically, the floral meristem has shoot meristem characteristics; a single flower can give rise to secondary and tertiary flowers (Fig. 6, B, C, and E). The secondary and tertiary floral buds arise from the axials of sepals or leaf-like organs and resemble the Arabidopsis ap1 mutants, where new flowers are formed in the axials of sepals (Irish and Sussex, 1990). The efo mutant also exhibits a repeated sepal-petal-stamen pattern before terminating in an enlarged receptacle topped with supernumerary carpels (Fig. 6H), which resembles the weak Arabidopsis agamous-4 mutant flower (Sieburth et al., 1995). In addition, the sepals, petals, and stamens of the efo mutant often exhibit mosaic organ identity (Fig. 6I); sepals contain white petal-like patches and petals develop out of the anthers. This mosaic organ phenotype bears resemblance to the Arabidopsis ufo mutants (Levin and Meyerowitz, 1995). Hence, the efo mutant appears to exhibit defects similar to those of A, B, and C classes of floral homeotic mutants.
Figure 6.
Phenotype characterization of efo/FveUFO1. A, A wild-type shoot showing the primary flower (bending) and the secondary and tertiary flowers. B, An efo mutant flower showing many more flowers originating from what would be a single flower in wild type. C, An efo flower showing elongated internodes between whorls of sepals/leaves and axillary flower buds. D, The back of a wild-type flower showing five bracts in the outermost whorl and five sepals in alternating positions. E, The back of a mutant flower showing many whorls of leaves (L) or leaf-like organs, in the axils of which many young flower buds reside (arrows). F, Wild-type flower. G, A wild-type petal (top) and three wild-type stamens (bottom). H, An efo flower showing a larger central receptacle giving rise to many more carpels/ovaries than in the wild-type in F. The central receptacle is flanked by a whorl of stamens, then a whorl of petals, then a whorl of sepal-like organs. This central flower is on top of additional whorls of stamens, petals, and sepals. I, Mosaic stamen/petal organs are often seen in the mutant flowers. J, Mosaic sepal/petal organs are often seen in the mutant flowers.
Using a bulk segregant mapping-by-sequencing approach (Schneeberger et al., 2009; Cuperus et al., 2010; Hartwig et al., 2012), we identified a candidate mutation in the efo mutant. Specifically, genomic DNA from 3 mutant and 22 wild type plants, all of which were derived from an M2 phenotypically normal parent plant heterozygous for the mutation, was pooled and submitted for whole-genome sequencing. An analysis pipeline with a series of filtering steps yielded 97 variants (Supplemental Data Set S6), only one of which was in a gene previously known to regulate flower development. Gene19967 encodes FveUFO1; a highly conserved W residue in the C terminus was mutated to a STOP codon in the mutant (Supplemental Fig. S5). Reverse transcription quantitative PCR (RT-qPCR) indicates that the transcript level of FveUFO1 is increased by 13-fold in stage 1 to 4 flowers of the mutant versus the wild type (Supplemental Fig. S6). This suggests that the mutant phenotype is not a result of the nonsense-mediated decay pathway targeting UFO1 transcripts for degradation, perhaps due to the nonsense mutation occurring near the C-terminal end. The increased expression of FveUFO1 in the efo mutant may result from a significantly increased number of young floral organs in the mutant flower.
DISCUSSION
We have shown how standard coexpression network and consensus network analyses can be used to highlight phenomena like increased iron transport to developing fruit and seeds immediately postfertilization and the meristem-like nature of the receptacle. Our work demonstrates the power of coexpression network analyses in hypothesis building and testing, especially in a developing model system. An intuitive and freely available web interface makes it possible for biologists to explore and mine these networks.
Consensus versus Standard Networks
In this study, we generated two types of networks. First, standard networks were generated by following the published WGCNA analysis pipeline (Langfelder and Horvath, 2008). Second, robust consensus networks were generated by varying parameters and simulating sampling variability over 1,000 runs of the WGCNA clustering algorithm. In total, twelve independent networks were generated by combining four network types (standard [nonconsensus], consensus, consensus90, and consensus100) with three data sets (HD tissues, LCM tissues, and ripening fruit tissues; Supplemental Data Sets S1 and S2; Supplemental Tables S1 and S2).
Recent applications of WGCNA have included resampling as a means of quantifying cluster stability. Shannon et al. (2016) estimated the cluster stability of WGCNA output by comparing coclustering of resampled WGCNA bootstrap iterations. An et al. (2016) started their analysis with a Self-Organizing Map clustering and used bootstrapped WGCNA to identify hub genes in the Self-Organizing Map clusters based on average node connectivity. Instead of starting with an initial clustering, our consensus-clustering implementation uses the subsampled coclustering matrix as the input to a final run of WGCNA.
For the standard networks, we chose parameters with the goal of generating a smaller number of clusters. However, correlation scores between pairs of genes are sensitive to the user-selected parameters, including minimum module size, power transformation, and merging on eigengenes. The consensus networks (Wu et al., 2002; Monti et al., 2003) are less influenced by parameter selection due to 1,000 iterations of the WGCNA clustering algorithm with each run using randomly generated algorithm parameters and resampling 80% of the genes. This simulates sampling variability. For most statistics, the standard and consensus methods perform similarly; however, the consensus method greatly increases bootstrapping confidence. As a result, gene pairs that reliably cluster together in a consensus network are more likely to represent true relationships. The consensus clustering approach produces a more robust network without sacrificing the high intracluster similarity or low intercluster similarity produced by the WGCNA algorithm. Further, our interrogation of standard and consensus clusters containing FveUFO1 and FveLFY suggests that the consensus network may identify direct interactions and close functional partnerships among genes while the standard network may identify genes acting in the same or related pathways or biological processes.
Further, we applied 90% and 100% cutoffs to the consensus matrix and used the remaining genes to generate consensus90 and consensus100 networks, respectively. These networks contain fewer total genes than the consensus networks but many more clusters with fewer genes per cluster. By adding stringent 90% and 100% cutoffs, we restricted clusters to genes that are the most likely to have close or direct relationships. Six iron transporter genes (gene17575, gene10308, gene19831, gene08918, gene18489, gene32625) are found in cluster 2 of the standard HD network, which prompted us to investigate iron transport in the developing receptacle and seeds (Fig. 2). However, these six iron transporter genes reside in five different consensus HD clusters due to the stringent cut off used in consensus network construction. Hence, the trade-off between sensitivity and confidence allows the user to explore both clusters of small numbers of closely related genes (consensus network) and larger numbers of less closely related genes (standard network).
The Role of FveUFO1 in Strawberry Flower Development
UFO, an F box protein, associates with an Skp1-Cul1-F-box protein complex and targets proteins for degradation via ubiquitination (Samach et al., 1999; Ni et al., 2004). In Arabidopsis, UFO was shown to promote LFY (Weigel et al., 1992) transcription factor activity in a positive feedback manner by targeting LFY for degradation (Chae et al., 2008). The primary floral defects reported in Arabidopsis ufo mutants include reduced B class gene expression and mosaic organs with unclear boundaries between petals and stamens, though an important hallmark is also a high degree of phenotypic variation across individual mutants (Levin and Meyerowitz, 1995).
While we have not yet validated with a transgenic approach that the efo phenotype is indeed caused by the nonsense mutation in FveUFO1, we noticed there is a striking similarity in the mutant phenotype between the F. vesca efo mutant and mutants of UFO orthologs in pea (Pisum sativum; stp), Lotus japonicus (pfo), tomato (an), and Torenia fournieri (Tfufo), including loss of floral meristem determinacy, proliferating sepals, and in particular the production of ectopic flowers within the primary flowers of pea stp mutants (Taylor et al., 2001; Zhang et al., 2003; Lippman et al., 2008; Sasaki et al., 2012). Further, the wide range of phenotypes exhibited by the efo mutant is also consistent with the high degree of phenotypic variation reported in Arabidopsis ufo mutants compared to other Arabidopsis mutants (Levin and Meyerowitz, 1995). These similarities provide initial support for FveUFO1 as the gene that underlies the efo mutant phenotype in F. vesca and suggests a different role for UFO/LFY in the Solanaceae and Rosaceae families compared to their counterparts in Brassicaceae.
Our network analyses also support FveUFO1 as the candidate gene for efo. First, the network analysis predicts a nonredundant function between FveUFO1 and FveUFO2/3 as they reside in two temporally separated consensus clusters. Second, a tight partnership between FveUFO1 and FveLFY in early-stage flower development is predicted due to coclustering in both consensus and standard networks, thereby suggesting a conserved relationship between FveUFO1 and FveLFY in multiple plant species. Third, the coclustering between FveUFO/FveLFY and FveAP1/FveSEP1 genes in the standard_LCM network as well as the very high correlation score (Pearson correlation = 0.92009; Supplemental Data Set S5) between their respective consensus_LCM clusters (95 and 119) predicts that FveUFO/FveLFY may positively regulate FveAP1/FveSEP1 expression during early-stage flower development. This could explain why the efo mutant develops secondary and tertiary flowers in the axils of sepals (Fig. 6, B and E), which does not resemble the Arabidopsis ufo mutant but does resemble the Arabidopsis ap1 mutant (Irish and Sussex, 1990). Further, the additional floral phenotypes of the efo mutant could be explained by a reduction of class E genes such as FveSEP1, which may lead to defective A, B, and C complexes. Taken together, FveUFO1 may differ from its Arabidopsis homolog in mutant phenotypes due to a primary role in regulating class A and E genes instead of class B genes. It will be interesting to knock out FveLFY in F. vesca and compare phenotypes to determine if all aspects of the fveufo1 phenotype are mediated through FveLFY.
Iron Transport during Fruit Set
Fruit set is the process of fruit initiation triggered by fertilization-induced auxin and GA production. Previously, we showed increased abundance of auxin and GA biosynthesis gene transcripts in the ghost (endosperm and seed coat) immediately after fertilization, suggesting the ghost as a possible site of auxin and GA biosynthesis postfertilization (Kang et al., 2013). Since GA biosynthetic enzymes GA20ox and GA3ox are both 2-oxoglutarate/Fe (II)-dependent dioxygenases (Yamaguchi, 2008; Huang et al., 2015; White and Flashman, 2016), the increased iron transport to the ghost may ensure iron cofactor availability for GA20ox and GA3ox. In addition to GA metabolism, iron may serve as a nutrient for later seedling growth as well as a cofactor for other 2-oxoglutarate/Fe (II)-dependent dioxygenases during seed development (Farrow and Facchini, 2014).
Previously, with a radiotracer technique, it was shown that iron is successfully phloem uploaded and transported to legume seeds (Grusak, 1994). Here, using the iron-specific Perls stain, we show that iron is uploaded to the vasculature and travels from the stem/receptacle to the achenes/seeds in F. vesca fruit. Further, the iron staining intensity in vasculature tissues is significantly increased postfertilization (Fig. 2, C, D, F, and G). How iron transport activities respond to fertilization remains an interesting and important question. Since the embryo is oppositely correlated with cluster 2, our result supports the hypothesis that it is the ghost (endosperm), and not the embryo, that is the major site of iron transport.
CONCLUSION
Our network analyses provide new insights into the biological processes underlying flower development and fruit set. This work demonstrates how coexpression networks can lead to new hypotheses and guide subsequent experiments. Further, the networks can be more broadly appreciated and utilized by the research community through our freely available web interface. Anyone with an interest in a specific biological process can easily explore and mine all twelve networks. Therefore, the work reported here sets an example of how coexpression network analyses generated with large-scale RNA-seq data can facilitate research in emerging model systems.
MATERIALS AND METHODS
WGCNA Network Analysis
Coexpression networks for Fragaria vesca were created using gene-level TPM (transcripts per million; Wagner et al., 2012) expression measurements from 92 RNA-seq libraries spanning the early developmental stages of plant tissues to ripening fruit. Genes with variance <0.05 were filtered out, and the results were used as input to the signed WGCNA network construction (WGCNA v1.60 package in R; Langfelder and Horvath, 2008). In standard WGCNA networks, power was set to 6, minModuleSize was set to 100, and initial clusters were merged on eigengenes. The mergeCutHeight value was set to 0.25 across all networks. Total connectivity was calculated for all genes in each network.
Consensus Network Construction
To construct the consensus network, 80% of genes were subsampled 1,000 times; paired with each subsampling was a set of randomized parameters standard to the WGCNA. These parameters consisted of power transformation [1, 2, 4, 8, 12, 16], minModuleSize [40, 60, 90, 120, 150, 180, 210], and merge on eigengenes [true/false]. After 1,000 runs of WGCNA were performed, a weighted adjacency matrix was computed to represent the connection strength between every gene pair. Letting p be the number of genes with count variance >0.5, the adjacency matrix (A) was calculated by
The adjacency matrix was then used as a basis for the consensus network, consensus90 network, and consensus100 network. The consensus network was constructed by clustering the adjacency matrix using WGCNA with power 6, minModuleSize 100, and no merging on eigengenes. Consensus90 was constructed by translating the weighted adjacency matrix to a graph by thresholding at a value of 0.90. The following clusters were then established by using the connected components function in graph. Consensus100 was performed similarly with a threshold of 1.
Description of stepwise construction of the consensus network and the associated python and R scripts used for consensus clustering are provided in the Supplemental Materials and Methods. These scripts are server specific, which likely limits their direct application. In brief, make_subsamp_wgcna.py creates a subsample and generates a bash script that runs subsamp_wgcna.R, which produces clusters of the subsamples using random parameters. seq_indicator_mat.R is then used to generate an indicator matrix for the clusters produced by subsamp_wgcna.R. Then, seq_adding_mat.R is run to output the number of times every pair of genes clusters together, and merge_mats.R is used to combine the matrices into a single indicator and a single coclustering matrix. Finally, the coclustering matrix is divided by the indicator matrix to make a consensus matrix, which is used as input to WGCNA (consensus_cluster.R), which produced the final clusters.
Network Visualization
Module eigengenes were calculated subsequent to network construction using the module Eigengenes function in WGCNA. This function calculates the first principal component of the genes’ TPM in a given cluster. As shown in previous work with WGCNA, this can be used as a summary statistic to relate clusters to sample-specific expression levels as detailed previously (Langfelder and Horvath, 2007). This is then visualized using R’s boxplot function. The networks were visualized using Cytoscape _v.3.5.1.
Comparisons between Standard WGCNA and Consensus Networks
Consensus clusters were compared to a standard WGCNA clustering approach. This comparison was based on gene expression values from the hand-dissected, log-transformed TPM values. The standard WGCNA approach was constructed using Pearson’s Correlation distance, power set to 6, and minModuleSize set to 100. The Jaccard index and hypergeometric P value were determined using the R package GeneOverlap (Shen and Sinai, 2013). The comparative statistics were computed using RS statistic, which is a measure of the variance between clusters to the variance within clusters, calculated by (TSS-SSE)/TSS, where TSS = SSE+SSB (TSS, total sum of squares; SSE, sum of square error; SSB, between group sum of squares). By definition, this statistic was based on the Euclidian distance between two genes (Liu et al., 2013). Bootstrap confidence intervals were determined by the frequency of coclustering between gene pairs in 1,000 runs of WGCNA with varied parameters and sampling.
GO Enrichment
GO enrichment tests were performed to understand potential functional relationships between coclustered genes. GO annotations were created using Blast2GO (Conesa and Götz, 2008). GO term enrichment P values were calculated using the Fisher's exact test in the TopGO R package (Alexa and Rahnenfuhrer, 2016).
Web-Based Application to Visualize and Download Network Data
All coexpression network data are available at www.fv.rosaceaefruits.org. The web application was generated with Shiny from RStudio (http://shiny.rstudio.com)
Free Iron Staining: Perls followed by DAB Enhancement
The following protocol is adapted from Roschzttardtz et al. (2009) and Brumbarova and Ivanov (2014). All solutions were made fresh on the day of treatment.
For Perls staining, tissues were collected from the wild type F. vesca accession Yellow Wonder 5AF7. Plants were grown in chambers set to 16 h of light at 25°C and 8 h of darkness at 20°C. Ovules and seeds were hand-dissected under a stereomicroscope. Tissues were first fixed for 1.5 h in a solution containing methanol:chloroform:glacial acetic acid (6:3:1). Then, tissues were vacuum infiltrated for 45 min in a solution containing equal volumes of 6% Perls (potassium ferrocyanide) and 4% HCl. Following infiltration, samples were incubated at room temperature under a fume hood for 15 min. Next, samples were washed three times with deionized water. At this point, receptacle samples were photographed under a stereomicroscope equipped with a Zeiss Axiocam 105 color camera.
Taking advantage of the redox activity of Prussian blue, 3,3′-Diaminobenzidine (DAB) was previously used to intensify Perls staining (Nguyen-Legros et al., 1980; Roschzttardtz et al., 2009). A 0.5% DAB stock solution was prepared by adding 0.05 g of DAB to 10 mL DI water and allowed to sit for 5 min with occasional vortexing. Next, 300 µL of 37% HCl was added, and the solution was left at room temperature for 5 min with occasional vortexing. Finally, the DAB solution was filtered through a 0.22 µm filter. To stain the tissue, plant material previously stained with Perls was incubated for 1 h in the preparation solution (0.01 m NaN3 and 0.3% H2O2 in methanol). Following this, samples were washed three times with 0.1 m PBS (pH 7.4). DAB intensification was carried out in a 0.1 m phosphate-buffered saline solution containing 0.0125% DAB, 0.005% H2O2, and 0.005% CoCl2. Intensification was carried out for 20 min at room temperature. Samples were washed three times in DI water to stop the DAB intensification reaction.
Plant Growth Conditions and efo Mutant Isolation
Wild-type Yellow Wonder 5AF7 (YW5AF7), the efo mutant in the YW5AF7 background, and a segregating efo mutant population of M3 sister plants were all grown in a growth chamber with 16 h light at 25°C followed by 8 h dark at 20°C. The efo mutant was isolated from an EMS mutagenesis screen of YW5AF7 (Hollender, 2012).
Bulk Segregant Mapping by Sequencing of the F. vesca efo Mutant
The M3 mapping population consisted of three efo mutant plants and 22 sister plants with a wild-type phenotype. Genomic DNA was extracted from young leaves using the NucleoSpin Plant II kit (Machery-Nagel). Equal quantities of genomic DNA (gDNA) from each mutant plant were combined into one pool (mutant pool). Equal quantities of gDNA from each of the 22 sister plants were combined into a second pool (wild-type pool). A total of 2 μg gDNA from each pool was sequenced at the Genomics Resources Core Facility at Weill Cornell Medical College on an Illumina HiSeq 2000. The two libraries were bar coded and each were sequenced on one-half of one lane. A total of 92,632,150 and 89,565,704, 51 bp, single-end reads were generated for the mutant and wild-type pools, respectively.
Fifty-one base pair reads were mapped to the F. vesca reference genome v1.1 using Bowtie2 with default settings. Variants were called across the two samples using SAMtools. Of the 199,622 total variants called, about 87% were homozygous across both samples. Some of these variants represent differences between YW5AF7 and Hawaii-4, the accession used for the reference genome, and were discarded. Of the remaining variants, 11,242 are G/C to A/T single-nucleotide changes, which is the most common mutation induced by EMS treatment. We further filtered these 11,242 variants to select for those that (1) are homozygous in the mutant pool; (2) are heterozygous in the wild-type pool; (3) are located in exons; (4) cause amino acid changes; (5) are nonsense or nonsynonymous mutations; and (6) are present at 13% to 53% frequency in the wild-type pool. After filtering, variants in 97 genes remained (Supplemental Data Set S6). Forty-three of these genes had expression of RPKM (reads per kilobase million) 10 or higher in stage 1 to 4 flower buds and only one, gene19967 (FveUFO1), is a homolog of an Arabidopsis (Arabidopsis thaliana) gene known to control floral development (AT1G30950; UNUSUAL FLORAL ORGANS; Levin and Meyerowitz, 1995).
RT-qPCR to Test Transcript Levels of FveUFO1
Total RNA was isolated from stage 1 to 4 flower buds using an RNAeasy Plant Mini Kit (Qiagen). Three flower buds isolated from each of three individual plants were pooled into a single biological replicate (nine buds total/replicate). Four biological replicates were analyzed for both the wild type and efo mutant. RNA samples were treated with DNase I (Thermo Fisher) to remove contaminating genomic DNA and subsequently repurified with the NucleoSpin RNA XS kit (Machery-Nagel). cDNA was synthesized from 1 µg of total RNA in a 20 µL solution using the RevertAid First Strand cDNA synthesis kit (Thermo Fisher). 1:10 cDNA was used as the template in qPCR. SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was used to set up the PCR reactions, which were run and analyzed on a CFX96 Real-Time System. Forty cycles of two-step PCR were run as follows: 95 degrees, 30 s; 95 degrees, 15 s; 58 degrees, 30 s, followed by melt curve (65 to 95 degrees, 0.5 degree increments). Primer sequences to target FveUFO1, gene 19967, are listed in Supplemental Table S3. Gene03773, which is stably expressed across all profiled stages of receptacle development, was used as a control for normalization (Lin-Wang et al., 2014; primers in Supplemental Table S3). Data were analyzed with the 2∆Ct method and statistical significance was calculated with a Student’s t test.
Accession Numbers
All RNA-seq data used in this article can be found in Sequence Read Archive at NCBI. The accession numbers are SRA065786, SRP035308, and SRR5155708 to SRR515515.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. PCA plot showing separation of LCM and HD RNA-seq samples.
Supplemental Figure S2. Screenshots to illustrate navigation of web interface.
Supplemental Figure S3. Protein sequence alignment for three F. vesca UFOs and Arabidopsis UFO.
Supplemental Figure S4. F. vesca ABCE gene homologs and the eigengene expression profiles of clusters to which they belong in standard_LCM and consensus_LCM networks.
Supplemental Figure S5. Protein sequence alignment for UFO-like genes in multiple species.
Supplemental Figure S6. FveUFO1 RT-qPCR bar graph.
Supplemental Table S1. Descriptions of all RNA-Seq samples.
Supplemental Table S2. Information for each network: total number of genes, number of clusters, average number of genes per cluster, and notes on creation of each cluster.
Supplemental Table S3. Primers used in this study.
Supplemental Materials and Methods. Consensus_network instruction and scripts.pdf.
Supplemental Data Set S1. Excel file with each network, the numbers of clusters in each network, and the number of genes per cluster.
Supplemental Data Set S2. Excel file with connectivity scores for eigengene of each cluster with all tissues (colored based on connectivity).
Supplemental Data Set S3. Standard_hand dissected (Nonconsensus_HD) Clusters 2, 14, and 20 GO and full lists of genes.
Supplemental Data Set S4. Genes and lists of transcription factors in consensus_LCM clusters 95 and 100.
Supplemental Data Set S5. Pearson correlation scores of cluster 95 with each of the clusters in consensus_LCM.
Supplemental Data Set S6. List of efo candidate genes based on filtering.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Footnotes
R.S. is a recipient of a USDA predoctoral fellowship (NIFA 2016-67011-24629). H.W. is supported by the NSF Computation and Mathematics for Biological Networks Research Traineeship (1632976). W.W. was supported by the China Scholarship Council. The work is supported by National Science Foundation grants (MCB0923913 and IOS1444987) to Z.L.
Articles can be viewed without a subscription.
References
- Aharoni A, O’Connell AP (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J (2016) topGO: enrichment analysis for gene ontology. R package version 2.26.0 https://bioconductor.org/packages/release/bioc/html/topGO.html
- An CI, Ichihashi Y, Peng J, Sinha NR, Hagiwara N (2016) Transcriptome dynamics and potential roles of Sox6 in the postnatal heart. PLoS One 11: e0166574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballouz S, Verleyen W, Gillis J (2015) Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics 31: 2123–2130 [DOI] [PubMed] [Google Scholar]
- Brumbarova T, Ivanov R (2014) Perls staining for histochemical detection of iron in plant samples. Bio Protoc 4: e1245 [Google Scholar]
- Chae E, Tan QK-G, Hill TA, Irish VF (2008) An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Chettoor AM, Givan SA, Cole RA, Coker CT, Unger-Wallace E, Vejlupkova Z, Vollbrecht E, Fowler JE, Evans MM (2014) Discovery of novel transcripts and gametophytic functions via RNA-seq analysis of maize gametophytic transcriptomes. Genome Biol 15: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008: 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus JT, Montgomery TA, Fahlgren N, Burke RT, Townsend T, Sullivan CM, Carrington JC (2010) Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc Natl Acad Sci USA 107: 466–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow GM. (1966) The Strawberry: History, Breeding, and Physiology. Holt, Rinehart and Winston, New York [Google Scholar]
- De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inzé D (2012) CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol 195: 707–720 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Johnson E, Csukasi F, Pizarro CM, Vallarino JG, Kiryakova Y, Vioque A, Brumos J, Medina-Escobar N, Botella MA, Alonso JM, et al. (2017) Transcriptomic analysis in strawberry fruits reveals active auxin biosynthesis and signaling in the ripe receptacle. Front Plant Sci 8: 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow SC, Facchini PJ (2014) Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front Plant Sci 5: 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C, Pelaz S, Yanofsky MF (1999) Control of carpel and fruit development in Arabidopsis. Annu Rev Biochem 68: 321–354 [DOI] [PubMed] [Google Scholar]
- Ficklin SP, Feltus FA (2011) Gene coexpression network alignment and conservation of gene modules between two grass species: maize and rice. Plant Physiol 156: 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxman Bass JI, Diallo A, Nelson J, Soto JM, Myers CL, Walhout AJM (2013) Using networks to measure similarity between genes: association index selection. Nat Methods 10: 1169–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gago JA, Posé S, Muñoz-Blanco J, Quesada MA, Mercado JA (2009) The polygalacturonase FaPG1 gene plays a key role in strawberry fruit softening. Plant Signal Behav 4: 766–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser CS, Robinson-Beers K (1993) Pistil development. Plant Cell 5: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Rogers EE (2004) FRD3 controls iron localization in Arabidopsis. Plant Physiol 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak MA. (1994) Iron transport to developing ovules of Pisum sativum (I. seed import characteristics and phloem iron-loading capacity of source regions). Plant Physiol 104: 649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig B, James GV, Konrad K, Schneeberger K, Turck F (2012) Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol 160: 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C, Caruana J, Li J, Zawora C, Darwish O, Wu J, Alkharouf N, Liu Z (2017) An eFP browser for visualizing strawberry fruit and flower transcriptomes. Hortic Res 4: 17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Gao X, Zhang Y, Zhou ZH, Liu ZY, Fu B, Hu F, Zhang Z, editors (2015) Intelligence Science and Big Data Engineering. Big Data and Machine Learning Techniques: 5th International Conference, IScIDE 2015, Suzhou, China, June 14–16, 2015, Revised Selected Papers. Springer, Basel, Switzerland [Google Scholar]
- Hollender CA. (2012) Molecular and genetic analysis of flower development in Arabidopsis thaliana and the diploid strawberry, Fragaria vesca. PhD dissertation. University of Maryland, College Park, MD [Google Scholar]
- Hollender CA, Geretz AC, Slovin JP, Liu Z (2012) Flower and early fruit development in a diploid strawberry, Fragaria vesca. Planta 235: 1123–1139 [DOI] [PubMed] [Google Scholar]
- Hollender CA, Kang C, Darwish O, Geretz A, Matthews BF, Slovin J, Alkharouf N, Liu Z (2014) Floral transcriptomes in woodland strawberry uncover developing receptacle and anther gene networks. Plant Physiol 165: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Vendramin S, Shi L, McGinnis KM (2017) Construction and optimization of a large gene coexpression network in maize using RNA-Seq data. Plant Physiol 175: 568–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang X, Ge S, Rao GY (2015) Divergence and adaptive evolution of the gibberellin oxidase genes in plants. BMC Evol Biol 15: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu OD, Kawane S, Bottomly D, Searles R, Hitzemann R, McWeeney S (2012) Utilizing RNA-Seq data for de novo coexpression network inference. Bioinformatics 28: 1592–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish VF, Sussex IM (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C, Darwish O, Geretz A, Shahan R, Alkharouf N, Liu Z (2013) Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML (2006) Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- Kimura S, Sinha N (2008) Tomato (Solanum lycopersicum): a model fruit-bearing crop. CSH Protoc 2008: pdb.emo105. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 1: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P (2004) Coexpression analysis of human genes across many microarray data sets. Genome Res 14: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Wolfe DS, Nilsson O, Weigel D (1997) A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol 7: 95–104 [DOI] [PubMed] [Google Scholar]
- Levin JZ, Meyerowitz EM (1995) UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7: 529–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dai C, Hu C, Liu Z, Kang C (2017) Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J 90: 164–176 [DOI] [PubMed] [Google Scholar]
- Lin-Wang K, McGhie TK, Wang M, Liu Y, Warren B, Storey R, Espley RV, Allan AC (2014) Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front Plant Sci 5: 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, Abu-Abied M, Pekker I, Paran I, Eshed Y, Zamir D (2008) The making of a compound inflorescence in tomato and related nightshades. PLoS Biol 6: e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li Z, Xiong H, Gao X, Wu J, Wu S (2013) Understanding and enhancement of internal clustering validation measures. IEEE Trans Cybern 43: 982–994 [DOI] [PubMed] [Google Scholar]
- Monti S, Tamayo P, Mesirov J, Golub T (2003) Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn 52: 91–118 [Google Scholar]
- Nguyen-Legros J, Bizot J, Bolesse M, Pulicani JP (1980) [“Diaminobenzidine black” as a new histochemical demonstration of exogenous iron (author’s transl)]. Histochemistry 66: 239–244 [DOI] [PubMed] [Google Scholar]
- Ni W, Xie D, Hobbie L, Feng B, Zhao D, Akkara J, Ma H (2004) Regulation of flower development in Arabidopsis by SCF complexes. Plant Physiol 134: 1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch JP. (1950) Growth and morphogenesis of the strawberry as related to auxin. Am J Bot 37: 211–215 [Google Scholar]
- Pattison RJ, Csukasi F, Zheng Y, Fei Z, van der Knaap E, Catalá C (2015) Comprehensive tissue-specific transcriptome analysis reveals distinct regulatory programs during early tomato fruit development. Plant Physiol 168: 1684–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL (2002) Hierarchical organization of modularity in metabolic networks. Science 297: 1551–1555 [DOI] [PubMed] [Google Scholar]
- Roeder AHK, Yanofsky MF (2006) Fruit development in Arabidopsis. Arabidopsis Book 4: e0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Conéjéro G, Curie C, Mari S (2009) Identification of the endodermal vacuole as the iron storage compartment in the Arabidopsis embryo. Plant Physiol 151: 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland LJ, Alkharouf N, Darwish O, Ogden EL, Polashock JJ, Bassil NV, Main D (2012) Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol 12: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J 20: 433–445 [DOI] [PubMed] [Google Scholar]
- Sánchez-Sevilla JF, Vallarino JG, Osorio S, Bombarely A, Posé D, Merchante C, Botella MA, Amaya I, Valpuesta V (2017) Gene expression atlas of fruit ripening and transcriptome assembly from RNA-seq data in octoploid strawberry (Fragaria × ananassa). Sci Rep 7: 13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yamaguchi H, Aida R, Shikata M, Abe T, Ohtsubo N (2012) Mutation in Torenia fournieri Lind. UFO homolog confers loss of TfLFY interaction and results in a petal to sepal transformation. Plant J 71: 1002–1014 [DOI] [PubMed] [Google Scholar]
- Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y (2011) RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res 39: D1141–D1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU (2009) SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM (2013) Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CP, Chen V, Takhar M, Hollander Z, Balshaw R, McManus BM, Tebbutt SJ, Sin DD, Ng RT (2016) SABRE: a method for assessing the stability of gene modules in complex tissues and subject populations. BMC Bioinformatics 17: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. (1995) Applied Multivariate Techniques. Wiley, New York [Google Scholar]
- Shen L, Sinai M (2013) GeneOverlap: test and visualize gene overlaps. R package version 1.14.0.http://shenlab-sinai.github.io/shenlab-sinai/.
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Running MP, Meyerowitz EM (1995) Genetic separation of third and fourth whorl functions of AGAMOUS. Plant Cell 7: 1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer GAC, Lloyd AT, Huminiecki LB, Wolfe KH (2005) Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol Biol Evol 22: 767–775 [DOI] [PubMed] [Google Scholar]
- Slovin JP, Schmitt K, Folta KM (2009) An inbred line of the diploid strawberry Fragaria vesca f. semperflorens for genomic and molecular genetic studies in the Rosaceae. Plant Methods 5: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin V, Mirny LA (2003) Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci USA 100: 12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Koh S, Becker J, Stacey G (2002) AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell 14: 2799–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickler SR, Bombarely A, Mueller LA (2012) Designing a transcriptome next-generation sequencing project for a nonmodel plant species. Am J Bot 99: 257–266 [DOI] [PubMed] [Google Scholar]
- Taylor S, Hofer J, Murfet I (2001) Stamina pistilloida, the pea ortholog of Fim and UFO, is required for normal development of flowers, inflorescences, and leaves. Plant Cell 13: 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131: 281–285 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- White MD, Flashman E (2016) Catalytic strategies of the non-heme iron dependent oxygenases and their roles in plant biology. Curr Opin Chem Biol 31: 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe CJ, Kohane IS, Butte AJ (2005) Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren JD. (2009) A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics 25: 1694–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LF, Hughes TR, Davierwala AP, Robinson MD, Stoughton R, Altschuler SJ (2002) Large-scale prediction of Saccharomyces cerevisiae gene function using overlapping transcriptional clusters. Nat Genet 31: 255–265 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Zhai Z, Gayomba SR, Jung HI, Vimalakumari NK, Piñeros M, Craft E, Rutzke MA, Danku J, Lahner B, Punshon T, et al. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26: 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sandal N, Polowick PL, Stiller J, Stougaard J, Fobert PR (2003) Proliferating Floral Organs (Pfo), a Lotus japonicus gene required for specifying floral meristem determinacy and organ identity, encodes an F-box protein. Plant J 33: 607–619 [DOI] [PubMed] [Google Scholar]