Overexpression or suppression of OsDER1 in rice leads to an unfolded protein response and ER stress hypersensitivity, and suppression results in floury, shrunken seeds.
Abstract
Endoplasmic reticulum-associated protein degradation (ERAD) plays an important role in endoplasmic reticulum (ER) quality control. To date, little is known about the retrotranslocation machinery in the plant ERAD pathway. We obtained a DERLIN-like protein (OsDER1) through a SWATH-based quantitative proteomic analysis of ER membrane proteins extracted from ER-stressed rice (Oryza sativa) seeds. OsDER1, a homolog of yeast and mammal DER1, is localized in the ER and accumulates significantly under ER stress. Overexpression or suppression of OsDER1 in rice leads to activation of the unfolded protein response and hypersensitivity to ER stress, and suppression results in floury, shrunken seeds. In addition, the expression levels of polyubiquitinated proteins increased markedly in OsDER1 overexpression or suppression transgenic rice. Coimmunoprecipitation experiments demonstrated that OsDER1 interacted with OsHRD1, OsHRD3, and OsCDC48, the essential components of the canonical ERAD pathway. Furthermore, OsDER1 associated with the signal peptide peptidase, a homolog of a component of the alternative ERAD pathway identified recently in yeast and mammals. Our data suggest that OsDER1 is linked to the ERAD pathway.
Secretory and membrane proteins are synthesized on the endoplasmic reticulum (ER) membrane and translocated into the ER lumen for folding and modification, which is under the control of the ER-mediated protein quality control (ERQC) system in eukaryotes. In the ER, the accumulation of unfolded, unassembled, or damaged proteins causes ER stress, which activates the unfolded protein response (UPR) signaling pathway to increase the expression of ER chaperones to facilitate protein folding. Another process, called endoplasmic reticulum-associated protein degradation (ERAD), also responds by stimulating the degradation of unfolded proteins; ERAD is an integral part of the ERQC and also can regulate the turnover of ER-resident proteins (Liu et al., 2011, 2015; van den Boomen et al., 2014).
To date, most ERAD studies have focused on yeast and mammalian systems. Currently, the commonly accepted view of ERAD is that CDC48 (an AAA-ATPase motor) mediates protein retrotranslocation across the ER membrane and ultimately is degraded by the ubiquitin-proteasome system in the cytosol (Ye et al., 2001, 2003; Avci and Lemberg, 2015). In this process, there are two distinct degradation routes according to the substrate in yeast. Membrane proteins with lesions in their cytosolic domains are recognized by the E3 ubiquitin ligase DOA10 complex (Habeck et al., 2015), whereas proteins with folding problems in their transmembrane or luminal domains are targeted to the E3 ubiquitin ligase HRD1 complex (Sato et al., 2009; Ruggiano et al., 2014). The yeast HRD1 was identified as a retrotranslocation channel for the movement of misfolded polypeptides through the ER membrane (Baldridge and Rapoport, 2016; Schoebel et al., 2017). The HRD1 E3 ligase complex includes multiple receptors for ERAD substrates, such as HRD3, YOS9, and USA1, and another subunit, DER1, is recruited to HRD1 through USA1 (Knop et al., 1996; Kim et al., 2005; Denic et al., 2006; Gauss et al., 2006a, 2006b; Horn et al., 2009). Notably, the DER1 subunit is linked to substrate recognition, ubiquitination, and extraction of ERAD substrates (Mehnert et al., 2014). A high activation barrier must be overcome to extract membrane integral proteins from the ER, and CDC48 can provide the pulling force needed for extraction (Cymer et al., 2015). However, an alternative CDC48-independent retrotranslocation pathway has been reported (Carlson et al., 2006; Hampton and Sommer, 2012; Avci et al., 2014). In this pathway, aberrant proteins are recognized by a complex composed of signal peptide peptidase (SPP); an E3 ubiquitin ligase, TRC8; and DER1 and released from the ER by intramembrane proteolysis (Stagg et al., 2009; Chen et al., 2014). SPPs cleave the aberrant proteins in the plane of the membrane (Lemberg et al., 2001) and release peptide fragments from the lipid bilayer by catalyzing the cleavage of tail-anchored proteins (Boname et al., 2014; Hsu et al., 2015), while DER1 acts as a substrate receptor, recognizing the misfolded proteins (Chen et al., 2014).
Multiple studies have demonstrated that plants have ERAD machinery to degrade aberrant proteins. A number of ERAD-related genes similar to those in yeast and mammals have been identified in plants. Arabidopsis (Arabidopsis thaliana) HRD3 is involved in the degradation of mutated brassinosteroid receptor BRI (Su et al., 2011). OsHRD3 plays an important role in ER quality control by interacting with OsOS-9 and OsHRD1 in rice (Oryza sativa; Ohta and Takaiwa, 2015). Plant CDC48 is involved in the retrotranslocation of the mutated mildew resistance O protein and ricin (Müller et al., 2005). The ubiquitin conjugase UBC32, an ERAD component, is involved in brassinosteroid-mediated salt stress tolerance in Arabidopsis (Cui et al., 2012). However, compared with the knowledge gained in yeast and mammalian studies, our understanding of the retrotranslocation machinery of the plant ERAD pathway is still limited, especially about its core component, DERLIN. Kirst et al. (2005) reported the discovery of four DERLIN genes in maize (Zea mays), and these ZmDERLINs functionally complemented a yeast DER1 deletion mutant. Kamauchi et al. (2005) found that AtDER1 was up-regulated under ER stress in a fluid microarray analysis of tunicamycin-treated plantlets. These studies suggested a potential role for a plant DER1 homolog in an ERAD pathway; however, no biochemical and genetic evidence exists to support this hypothesis. In fact, the precise functions of DERLINs in yeast and mammals remain unclear. It has been suggested they are a factor that links the recognition, ubiquitination, and extraction of ERAD substrates. Due to their functional role in retrotranslocation, DERLINs also have been suggested to form a protein-conducting channel (Lilley and Ploegh, 2004), but recent studies have demonstrated that DER1 is an inactive rhomboid intramembrane protease (Greenblatt et al., 2011) and acts in the SPP-TRC8-DER1 complex as a substrate receptor by recognizing misfolded substrates (Avci et al., 2014; Chen et al., 2014). The current lack of a crystal structure suitable for modeling the structure of DERLIN has hindered understanding the structure-function relationships underlying DERLIN-mediated biological processes.
Adverse environmental conditions during seed development induce ER stress. Since ERAD serves to remove unfolded proteins, it is crucial for maintaining functional and healthy proteostasis in the ER. Previously, we developed seed-specific ER stress transgenic plants by endosperm-specific knockdown of OsSAR1 (secretion-associated, Ras-related protein1; Tian et al., 2013). SAR1, a small GTPase, acts as a molecular switch to regulate the assembly of coat protein complex II, which exports secretory protein from the ER to the Golgi apparatus. Suppression of OsSAR1 induced ER stress by blocking the exit of secretory proteins from the ER (Tian et al., 2013). The differentially expressed proteins in the OsSAR1 RNA interference (RNAi) transgenic rice seed were analyzed via proteomic strategies (Qian et al., 2015). However, due to their low abundance in low-salt-soluble protein extraction, ER-localized membrane proteins were almost undetectable. In this study, we isolated the ER from developing seeds of OsSAR1 RNAi transgenic rice and performed proteomic analysis of the ER to identify differentially expressed membrane proteins. Focusing on the ERAD pathway, we found that 14 proteins that may be involved in protein processing in the ER pathway were differentially expressed. Among these differentially expressed proteins, OsDER1, a homolog of yeast DER1, was up-regulated dramatically in the ER of stressed seeds. Consequently, we analyzed the function of OsDER1 and illustrated its role in ERAD. This study sheds light on the ERAD machinery in plants.
RESULTS
Mass Spectrometric Analysis of the ER Enrichment Fraction in Rice Seeds
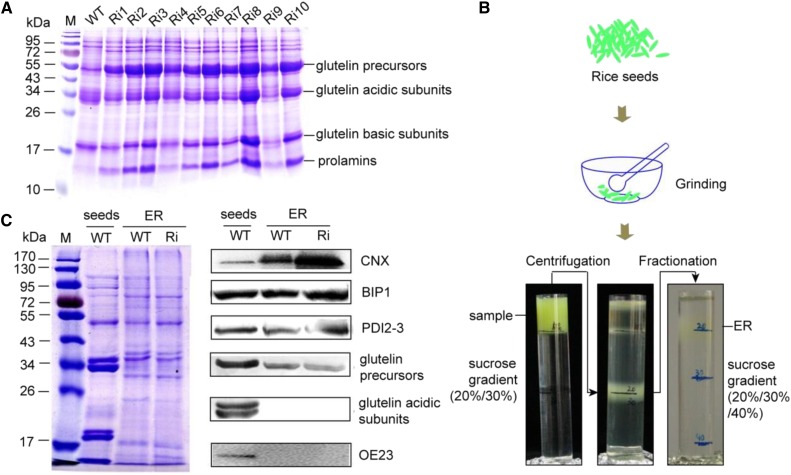
A previous study found that knockdown of OsSAR1 led to ER stress in rice seeds, and the resulting seeds had reduced levels of the glutelin acidic and basic subunits and increased levels of glutelin precursors (Qian et al., 2015). The ER-stressed rice seeds were generated as reported previously (Tian et al., 2013; Qian et al., 2015). SDS-PAGE analysis of the seeds showed that the glutelin precursors had accumulated (Fig. 1A), indicating that we had obtained ER-stressed rice seeds as reported (Tian et al., 2013). The ER was extracted from the developing seeds by grinding, differential centrifugation, and Suc density gradient centrifugation (Fig. 1B). To estimate the enrichment of the ER membrane fraction, some ER-localized proteins were detected by antibodies, such as calnexin (CNX), BIP, and PDI. Compared with the total protein (marked as seeds), the enriched ER also contained CNX, BIP, and PDI (marked as ER; Fig. 1C). Glutelins are initially synthesized on the ER as 57-kD precursors and then cleaved to 37- to 39-kD acidic and 19- to 20-kD basic subunits and stored in the protein storage vacuole (Yamagata et al., 1982; Yamagata and Tanaka, 1986). Glutelin acidic subunits or basic subunits can serve to evaluate the purity of the ER in ER-enriched fractions and to demonstrate the separation of the ER from protein storage vacuoles, which tend to sediment along with the ER. No glutelin acidic subunits were detected in our samples (Fig. 1C), suggesting that we obtained the ER enrichment fraction without vacuole contamination.
Figure 1.
Extraction of the ER enrichment fraction from rice seeds. A, Coomassie Blue-stained gel showing the increase in glutelin precursors in ER-stressed rice seeds. M, Marker; WT, wild-type rice seeds; Ri, OsSAR1 RNAi rice seeds (the numbers represent different transgenic rice lines). B, A brief workflow of the extraction of the ER enrichment fraction from rice seeds. C, CNX, BIP1, PDI2-3, and glutelin subunits were detected by Coomassie Blue staining and immunoblotting. OE23 (the 23-kD subunits of the oxygen-evolving complex) serves as a marker protein of chloroplasts. Seeds indicates total proteins from rice seeds, and ER indicates proteins of the ER enrichment fraction.
To identify the ER membrane proteins differentially expressed in the stressed condition, we performed a quantitative proteomics analysis of the ER fractions using a SWATH strategy, which combines the advantages of both shotgun and targeted proteomics (Tang et al., 2015). Each biological sample was composed of four individual extractions of ER enrichment fractions. After searching the NCBI O. sativa database using ProteinPilot 5.0 at a 1% critical false discovery rate, we identified 1,270 proteins (Supplemental Data Set S1). Of these, 1,129 proteins had quantitative information, of which only the peptides with confidence greater than 99% and without modification could be used for quantitative analysis.
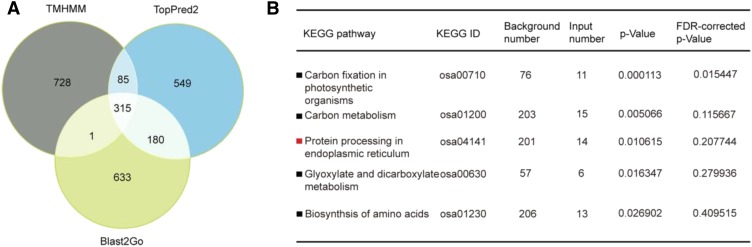
Among the 1,129 proteins, 403, 549, and 879 proteins were predicted to be membrane proteins after analysis with the Transmembrane Hidden Markov Model (TMHMM) Server version 2.0 algorithm, Blast2GO, and TopPred II (topology prediction of membrane proteins), respectively. Based on the consideration that the proteins present in at least two analyses were reliable, 581 proteins were predicted as membrane proteins (Fig. 2A). These predicated membrane proteins accounted for 51.5% of the total proteins identified, indicating that this method was more effective for identifying ER membrane proteins than the conventional proteomics strategies using total seed proteins (Qian et al., 2015).
Figure 2.
Membrane association and functional analysis of differentially expressed proteins. A, Venn diagram of the predicted numbers of membrane proteins using Blast2GO, TMHMM, and TopPred2. B, KEGG pathway analysis of differentially expressed proteins. All differentially expressed proteins were subjected to KEGG pathway analysis.
Membrane Proteins Differentially Expressed in OsSAR1 RNAi Rice Seeds
A total of 263 differentially expressed proteins were identified by a greater than 1.5-fold difference in quantity (P < 0.05), with 103 proteins up-regulated and 160 proteins down-regulated (Supplemental Data Set S2). Many of the up-regulated proteins were storage proteins, reflecting the fact that these proteins were blocked in the ER due to the repression of SAR1 (Tian et al., 2013).
These differentially expressed proteins were used subsequently for bioinformatics and pathway analyses. Based on a hypergeometric distribution, the 263 differentially expressed proteins were predicted to be involved mainly in five pathways (P < 0.05): carbon fixation in photosynthetic organisms (11 proteins), carbon metabolism (15 proteins), protein processing in the ER (14 proteins), glyoxylate and dicarboxylate metabolism (six proteins), and biosynthesis of amino acids (13 proteins; Fig. 2B). Fourteen differentially expressed proteins were involved in protein processing in the ER pathway, two of which were up-regulated: BIP1 (Os02g02410) and DER1-like protein (Os05g09550; OsDER1 hereafter). The others were down-regulated, including OsPDIL2-1 (Os05g06430), calreticulin (Os03g61670), HSP70 (Os03g16920, Os11g47760, and Os03g16860), ribophorin II precursor (Os01g68324), HSP20 (Os03g16030, Os03g14180, and Os03g16020), OsPDIL5-2 (Os04g35290), SAR1 (Os12g37360), and DNAK family protein (Os05g23740). Most proteins are chaperones that facilitate folding, sorting, or degradation, relieving ER stress. To investigate whether these genes responded to ER stress, their expression in DTT- and tunicamycin-induced ER-stressed seedlings was analyzed by reverse transcription quantitative PCR (RT-qPCR). The results showed that the expression of OsDER1 increased more than 20-fold in the ER-stressed seedlings, which was comparable to the change in BIP1 expression (Supplemental Fig. S1). Thus, we focused on the study of OsDER1.
OsDER1 Is an ER-Localized Membrane Protein, and the OsDER1 Gene Shows Significant Increase under ER Stress
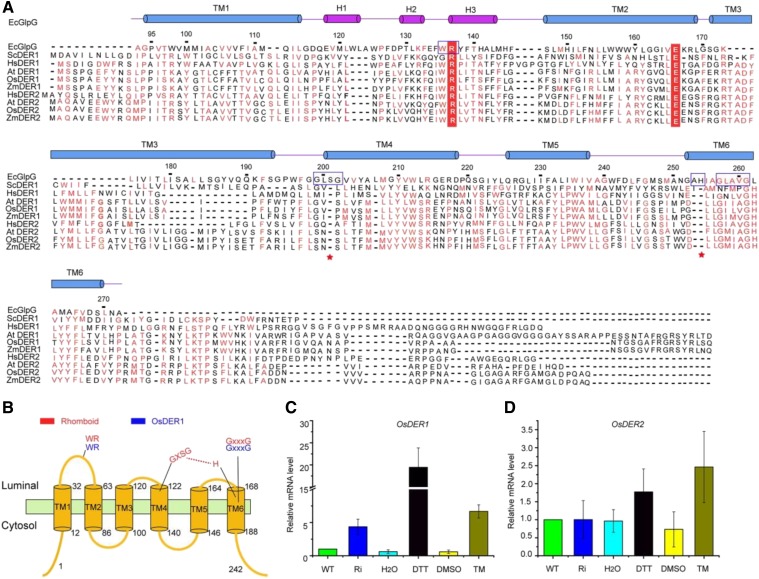
A survey of the rice genome revealed two DERLIN homologs, named OsDER1 and OsDER2. OsDER1 and OsDER2 encode polypeptides of 242 and 249 amino acids, respectively. The amino acid sequence of OsDER1 shares only 36% identity with the OsDER2 sequence, whereas it is nearly 64% and 81% identical to those of AtDER1 and ZmDER1, respectively. Based on a sequence alignment, OsDER1 and OsDER2 showed high similarity to the rhomboid intramembrane protease GlpG. Like mammalian DERLINs (Greenblatt et al., 2011), OsDER1 is a rhomboid pseudoprotease without the active site Ser-His dyad but with the conserved WR motif in loop L1, which connects transmembrane domain 1 (TM1) and TM2, and the conserved GxxxG transmembrane dimerization motif in TM6 (Fig. 3, A and B). The sequence similarity between the TM3 regions of OsDER1 and Escherichia coli GlpG is extremely low, but this region is highly conserved among other species (Fig. 3A). The hydropathy of TM3 of OsDER1 was further subjected to Kyte-Doolittle hydropathy analysis, and the results suggested that this region is a possible transmembrane region (Supplemental Fig. S2). In addition, it has been demonstrated that the human DER1 (HsDER1) spans the ER membrane six times (Greenblatt et al., 2011). TM3 of OsDER1 is highly similar to that of HsDER1; thus, OsDER1 also was proposed to have a six-pass transmembrane topology (Fig. 3B). There also is a possibility that it has four or five TMs instead of six due to their length; a 3D crystal structure is needed to clarify this issue.
Figure 3.
OsDER1 gene expression increased markedly under ER stress. A, Sequence alignment of OsDER1 with other homologs. Blue boxes indicate highly conserved motifs. Red stars indicate active sites in GlpG. The hydropathy of TM3 of OsDER1 was analyzed by Kyte-Doolittle software, and the results suggested that this region is a possible transmembrane region. There also is a possibility that this protein has four TMs instead of six TMs due to the lengths of some TMs. B, Transmembrane topology of OsDER1. Red indicates the highly conserved motif in GlpG; the active site Ser-His dyad is shown. Blue indicates the highly conserved motif in OsDER1. C and D, RT-qPCR of OsDER1 (C) and OsDER2 (D) mRNA levels under ER stress. The expression of the OsDERs was normalized to that of ACTIN. All data are averages of three independent experiments, and error bars represent sd. WT, Wild-type rice seeds; Ri, OsSAR1 RNAi rice seeds; DMSO, dimethyl sulfoxide; TM, tunicamycin treatment.
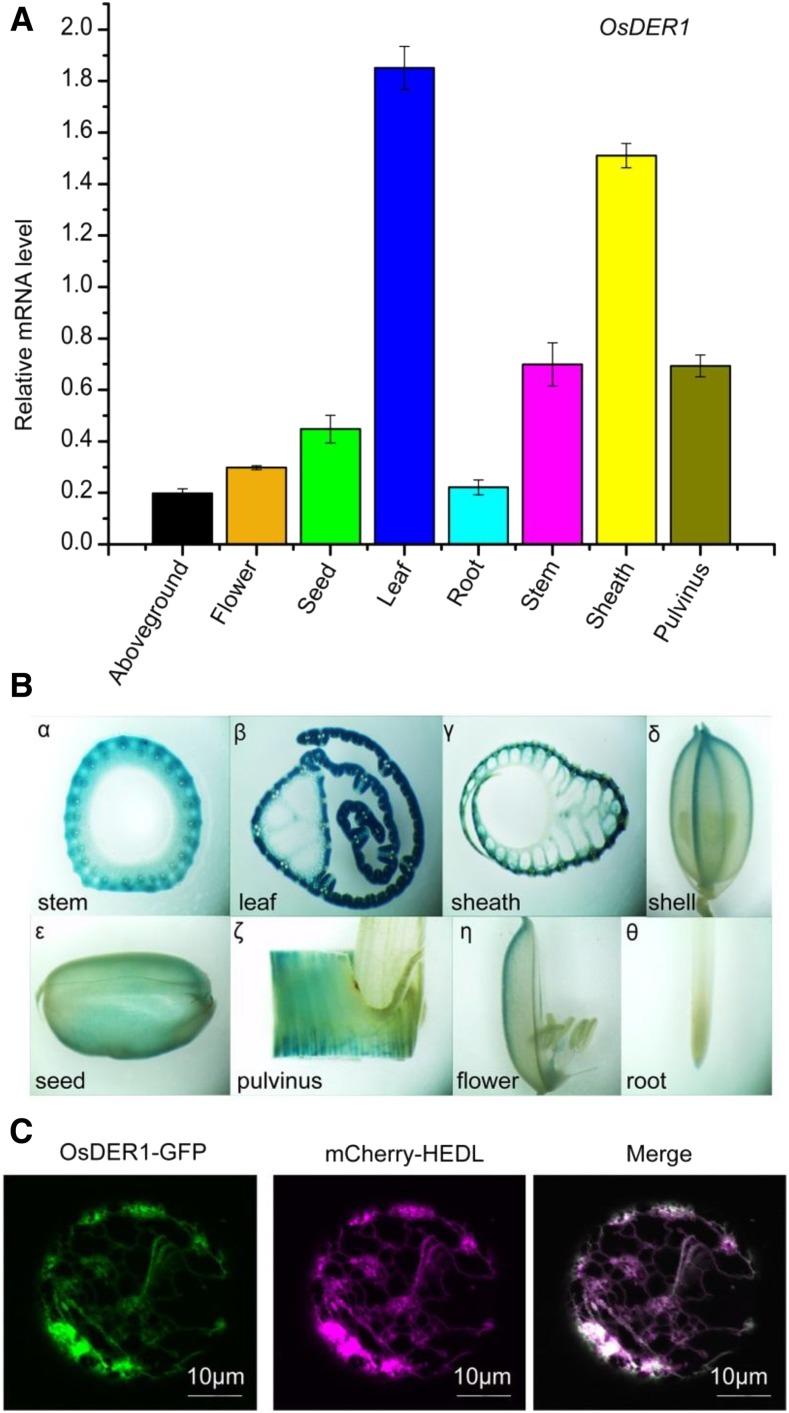
DERLIN genes in plants are grouped into two subfamilies (Supplemental Fig. S3). To test which gene in rice has the same function as the yeast and mammalian DERLINs, we analyzed the expression of the two genes under ER stress. The expression level of OsDER1 increased markedly under ER stress, while no significant change was observed in OsDER2, indicating that OsDER1 is part of the UPR in rice (Fig. 3, C and D). An RT-qPCR analysis revealed that OsDER1 was expressed in all tissues and has the highest expression level in leaves (Fig. 4A). In addition, the specific expression pattern of the promoter of OsDER1 was analyzed using GUS as a reporter gene. GUS expression was detected in the root, stem, leaf, flower, sheath, pulvinus, and seed, and the leaf showed the highest expression (Fig. 4B), which was consistent with the RT-qPCR result. The subcellular localization of OsDER1 was examined by confocal microscopy using a transient expression system in rice protoplasts. The result indicated that OsDER1-GFP showed typical ER localization. The ER localization of OsDER1-GFP was confirmed further by coexpression with the ER marker mCherry-HDEL; the green fluorescence of OsOsDER1-GFP merged well with the ER marker (Fig. 4C). These results confirmed that OsDER1 is localized to the ER.
Figure 4.
Tissue-specific expression of OsDER1. A, RT-qPCR of OsDER1 mRNA levels in different tissues. The expression of OsDER1 was normalized to that of ACTIN. All data are averages of three independent experiments, and error bars represent sd. B, Histochemical GUS staining of different tissues. α, Stem; β, leaf; γ, sheath; δ, shell; ε, seed; ζ, pulvinus; η, flower; θ, root. C, Subcellular localization of OsDER1 analyzed by confocal microscopy. OsDER1-GFP colocalized with mCherry-HEDL.
Endosperm-Specific Knockdown of OsDER1 Induced UPR and Resulted in an Abnormal Phenotype
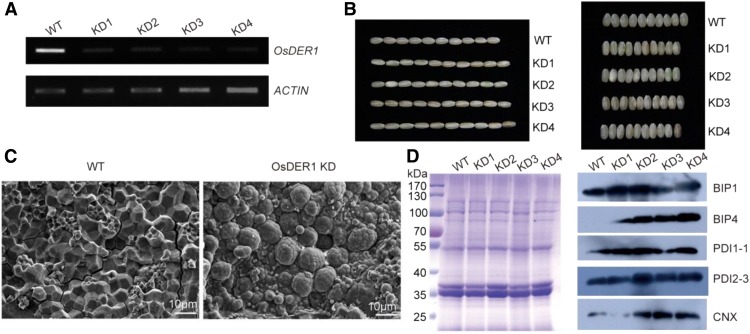
To further elucidate the function of OsDER1, transgenic rice was produced in which OsDER1 was knocked down (KD) by RNAi under the control of the endosperm-specific promoter GluA-2. The OsDER1 mRNA levels in OsDER1 KD were lower than those in the wild type (Fig. 5A). The seeds of the OsDER1 KD lines were longer and thinner than those of the wild-type control (Fig. 5B; Supplemental Fig. S4). Furthermore, opaque, floury, and shrunken seeds were found in OsDER1 KD. The starch granules were observed by scanning electron microscopy. In the wild type, the starch granules were densely packed and similar in size and had irregular polyhedral shapes with sharp edges. In the OsDER1 KD lines, the starch granules were loosely packed, round, and heterogeneous in size (Fig. 5C). To verify whether OsDER1 KD induced UPR, the levels of ER-resident proteins in mature seeds were investigated by western-blot analysis. The results showed that the expression levels of BIP1, PDI1-1, PDI2-3, and CNX increased obviously in OsDER1 KD compared with those in the wild type (Fig. 5D). It is noteworthy that OsBIP4, which is expressed only under ER stress conditions (Wakasa et al., 2012), was detected in OsDER1 KD. These results showed that knockdown of OsDER1 induced UPR and resulted in an abnormal phenotype, indicating that OsDER1 plays an essential role in maintaining protein homeostasis in the ER.
Figure 5.
Analysis of the changes in OsDER1 KD transgenic rice. A, RT-PCR of OsDER1 mRNA levels in transgenic rice plants with knocked down expression of OsDER1 by RNAi, under the control of the endosperm-specific promoter GluA-2. B, Phenotypic analysis of OsDER1 KD transgenic rice seeds. C, The starch granules of transgenic rice seeds were observed by scanning electron microscopy. D, Changes in the expression of BIP1, BIP4, PDI1-1, PDI2-3, and CNX in transgenic rice seeds detected by SDS-PAGE and immunoblotting. WT, Wild type.
Overexpression or Suppression of OsDER1 Affects Steady-State Polyubiquitylation and Leads to Hypersensitivity to ER Stress
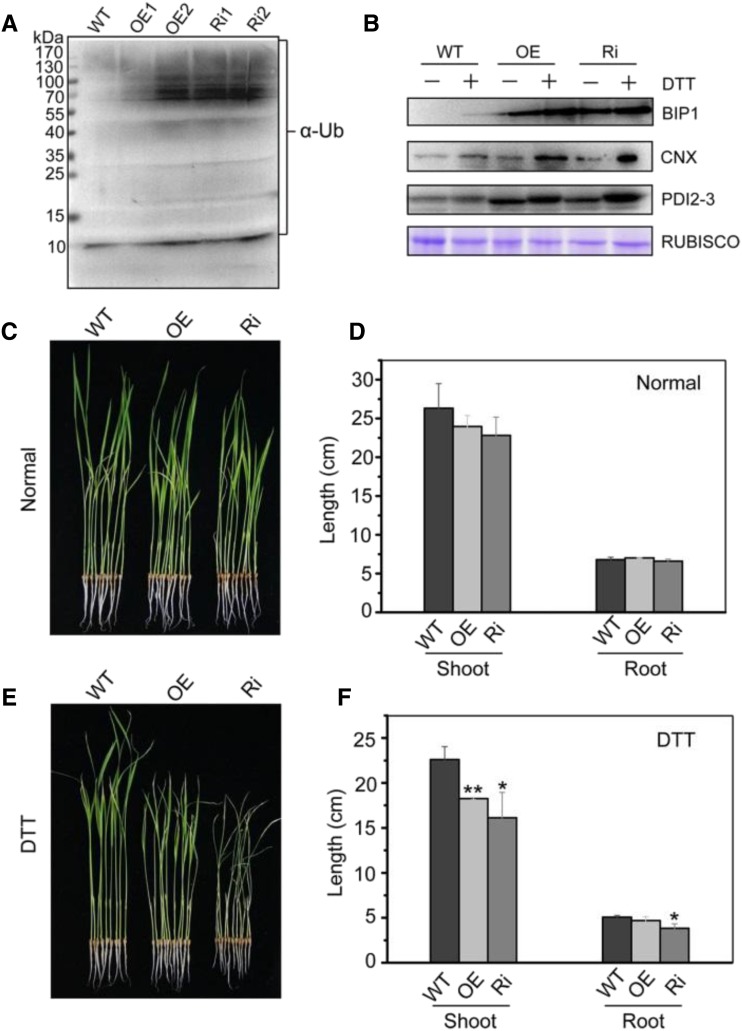
Transgenic rice was generated with overexpression (OsDER1 OE) and suppression (OsDER1 Ri) of OsDER1 under the control of the maize Ubiquitin promoter. The expression of OsDER1 was measured by RT-qPCR. The mRNA levels of OsDER1 were lower in OsDER1 Ri leaves and higher in OsDER1 OE leaves than in the leaves of the wild type (Supplemental Fig. S5, A and B), indicating that OsDER1 was successfully overexpressed or suppressed, respectively. To investigate whether OsDER1 is involved in the ubiquitination of unfolded proteins in rice, the levels of polyubiquitinated proteins in the transgenic plants were analyzed. Total proteins were extracted from OsDER1 OE and OsDER1 Ri seeds and subjected to immunoblot analysis using anti-α-ubiquitin as an antibody. The results showed that the levels of ubiquitinated proteins in the OsDER1 OE and OsDER1 Ri lines were both higher than those in the wild type (Fig. 6A). These results suggested that OsDER1 affects steady-state polyubiquitination, hinting that OsDER1 is related to the ubiquitination of unfolded proteins.
Figure 6.
Effects of overexpression and repression of OsDER1. A, Changes in the levels of ubiquitinated proteins in transgenic rice seeds with increased and repressed expression of OsDER1. Immunoblot analysis of the ubiquitinated proteins was performed with anti-α-ubiquitin (α-Ub). The loading control is shown in Supplemental Figure S5C. B, Changes in the expression of BIP1, PDI2-3, and CNX in seedlings (wild type [WT], OE, and Ri) treated with 2 mm DTT for 4 h. Equal volumes of water were added as negative controls. C to F, The growth of OsDER1-overexpressing and OsDER1-repressed rice (OsDER1 OE and OsDER1 Ri) in liquid Murashige and Skoog (MS) medium with and without 1 mm DTT refreshed daily was observed over a period of 2 weeks. Wild-type rice served as a control. C and E, Representative images of wild-type, OsDER1 OE, and OsDER1 Ri rice in the presence and absence of 1 mm DTT after 2 weeks. D and F, Quantitative measurements of the lengths of shoots and roots of wild-type, OsDER1 OE, and OsDER1 Ri seedlings in three trials. Error bars represent sd. *, P < 0.05 and **, P < 0.01 (Student’s t test).
To further investigate the effect of overexpressed or suppressed OsDER1 on the UPR signaling pathway, seedlings of OsDER1 OE, OsDER1 Ri, and the wild type were treated with 2 mm DTT for 4 h, and the expression levels of BIP1, CNX, and PDI2-3 were detected. Their expression was higher in OsDER1 OE and OsDER1 Ri seedlings than in wild-type seedlings (Fig. 6B). These results demonstrated that the alteration of the expression of OsDER1 enhanced the UPR signaling in response to ER stress. In addition, we also observed the growth of OsDER1 OE and OsDER1 Ri rice under ER stress. Under normal conditions, the lengths of the shoot and root were not significantly different among the seedlings of the wild type, OsDER1 OE, and OsDER1 Ri (Fig. 6, C and D). After treatment with 1 mm DTT for 14 d, the shoot lengths of OsDER1 OE and OsDER1 Ri were significantly (OE, P = 0.0065, n = 30; Ri, P = 0.02, n = 30) shorter than those of the wild type. The root length of OsDER1 Ri also was shorter than that of the wild type (Fig. 6, E and F). These results indicated that alteration of the expression of OsDER1 led to hypersensitivity to ER stress and affected the growth of rice. Overall, the expression level of OsDER1 should be controlled precisely in rice.
OsDER1 Is Linked to the ERAD Pathway
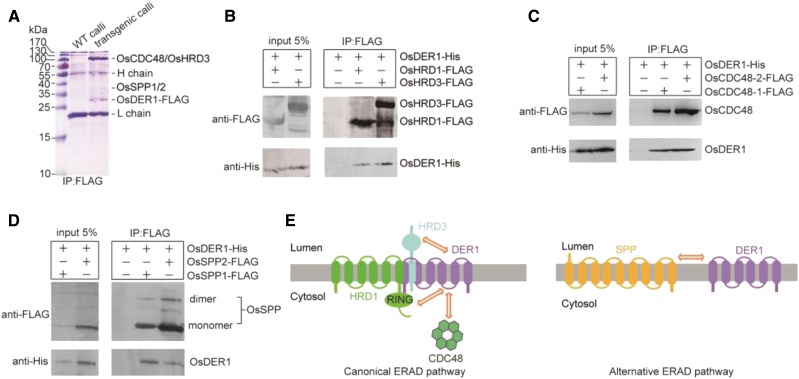
To search for proteins interacting with OsDER1 in rice, a series of coimmunoprecipitation assays was performed to uncover OsDER1-interacting proteins. Transgenic calli were generated with OsDER1-3×FLAG tag under the control of the Ubiquitin promoter. After detergent extraction, the supernatants from the transgenic calli were incubated with anti-FLAG resin; the wild type calli acted as a negative control. After SDS-PAGE and staining, an intense band and a faint band were detected, with no band observed at the equivalent positions in the control lane. The different bands were identified by mass spectrometry. The results showed that the tryptic peptides of the intense band aligned well with the primary sequences deduced from OsCDC48-1 (Os03g05730), OsCDC48-2 (Os10g30580), and OsHRD3 (Os03g15350), while that from the faint band aligned with OsSPP1 (Os02g02530) and OsSPP2 (Os05g36070; Fig. 7A).
Figure 7.
Interactions of OsDER1 with OsHRD1, OsHRD3, OsCDC48s, and OsSPPs. A, Coimmunoprecipitation assay of transgenic OsDER1-FLAG and endogenous interacting proteins. WT, Wild type. B, Overexpression-based coimmunoprecipitation assay of Sf9 cells coexpressing FLAG-epitope-tagged OsHRD1 or OsHRD3 and His-tagged OsDER1. C, Overexpression-based coimmunoprecipitation assay in Sf9 cells coexpressing FLAG-epitope-tagged OsCDC48-1 or OsCDC48-2 and His-tagged OsDER1. D, Overexpression-based coimmunoprecipitation assay of Sf9 cells coexpressing FLAG-epitope-tagged OsSPP1 or OsSPP2 and His-tagged OsDER1. E, Schematic diagram of the interactions found in this study. Two-way arrows indicate interactions.
To confirm the interaction between OsDER1 and these proteins, we performed a coimmunoprecipitation assay in Spodoptera frugiperda (Sf9) insect cells. His-tagged OsDER1 was coexpressed separately with FLAG-epitope-tagged OsHRD1, OsHRD3, OsCDC48-1, and OsCDC48-2. The results showed that OsDER1 was immunoprecipitated with each of these proteins (Fig. 7, B and C). OsHRD1, OsHRD3, OsCDC48-1, and OsCDC48-2 are essential components of the HRD1 complex; their interaction with OsDER1 demonstrated that the HRD1-mediated ERAD pathway is conserved in rice and involves OsDER1.
SPP1 and SPP2 are components of the DER1-SPP-TRC8 complex involved in the alternative ERAD pathway reported in yeast and mammalian systems (Avci et al., 2014; Chen et al., 2014). Our data suggested that OsDER1 was coimmunoprecipitated with OsSPP1 and OsSPP2 (Fig. 7D), indicating that the alternative DER1-SPP-TRC8-mediated proteolysis pathway also might be conserved in rice.
DISCUSSION
The ERQC system recognizes and eliminates misfolded, unassembled, or damaged proteins. To date, most insights into ERQC have been obtained through the analysis of plants and plantlets in which ER stress was induced by treating them with the chemical agent tunicamycin or DTT, which interferes with N-glycosylation or disulfide bond formation, respectively (Kamauchi et al., 2005). However, the use of these chemical agents to induce ER stress is not possible in seeds. Previously, we reported that endosperm-specific knockdown of OsSAR1 blocked the seed storage proteins glutelin and α-globulin in the ER and accumulated in the ER lumen, eliciting UPR (Tian et al., 2013; Qian et al., 2015). The UPR mitigates ER stress by up-regulating the expression of genes encoding components of the protein-folding machinery or the ER-associated degradation system. BIP1, BIP4, PDIL1-1, PDIL2-3, PDIL5-1, calreticulin, and ERO1 were up-regulated in OsSAR1 RNAi seeds based on iTRAQ analysis (Qian et al., 2015). However, many factors involved in protein processing and modification identified previously were not detected, or the expression levels of these proteins were down-regulated in this study (Supplemental Data Set S2). These results might be due to the fact that chaperones reside mainly in the ER lumen but are not ER membrane proteins, which might leak out during the extraction of ER membranes. It is also possible that this phenomenon is caused by an attenuation in bulk protein synthesis, which happens in mammalian cells to alleviate ER stress (Rutkowski and Kaufman, 2004) but has not been reported in plant cells.
In this study, we isolated the ER from ER-stressed developing rice seeds and performed a SWATH-based proteomic analysis aiming to identify global differentially expressed ER membrane proteins. Through this strategy, we identified 581 putative ER membrane proteins (Fig. 2). Many more membrane proteins were identified in this study compared with those obtained in a previously reported proteomics study of total soluble proteins extracted from the same OsSAR1 RNAi seed samples (Qian et al., 2015), indicating that it is effective to identify organelle-resident proteins by proteomic analysis of the organelle. Among the differentially expressed proteins, an obviously up-regulated protein, OsDER1, was observed. In contrast to the diverse biological functions of DER1 in yeast or mammalian systems, relatively little is known about DER1 in plants. In this study, we found that endosperm-specific knockdown of OsDER1 induced UPR, which is similar to a result in OsHRD3 knockdown rice seeds (Ohta and Takaiwa, 2015), and resulted in opaque and floury rice seeds (Fig. 5, A and B). In addition, the overexpression or repression of OsDER1 led to higher expression of ubiquitinated proteins. Proteins that fail to fold correctly are labeled with ubiquitin and escorted by a series of chaperones to the proteasome. Ubiquitination in the Ri lines was stronger than in the OE lines (Fig. 6A). Ubiquitination in the OE2 line was stronger than that in the OE1 line (Fig. 6A), corresponding to the higher expression level of OsDER1 mRNA in the OE2 line (Supplemental Fig. S5B). These results suggest that OsDER1 plays a key role in the ER stress response in rice and that the stoichiometry of OsDER1 relative to other components is important to the HRD complex. Loss of OsDER1 promotes ubiquitination, while the increase of ubiquitination in the OE line could be caused by some nonspecific effect on the UPR signaling pathway. Similar effects have been reported for BIP1 in rice, in which either underexpression or overexpression of BIP1 activates an ER stress response (Wakasa et al., 2011).
HRD3 is a component of the HRD1 ubiquitin ligase complex in yeast. OsHRD3 is involved in the ubiquitination of RM1 in rice endosperm, and the aberrant distribution of RM1 leads to the deformation of PB-I in OsHRD3 KD seeds (Ohta and Takaiwa, 2015). To investigate whether protein body formation was altered in OsDER1 KD seeds, we performed an immunohistochemical analysis using transmission electron microscopy. Rice endosperm has two types of PBs: the spherical protein body PB-I is formed by prolamin, and the irregularly shaped, electron-dense PB-II contains glutelin and α-globulin (Tian et al., 2013). Therefore, the anti-prolamin antibody and anti-glutelin antibody were used to label PB-I and PB-II, respectively. In OsDER1 KD endosperm, PB-II was identical to that in the wild type. In addition to the normal PB-I in OsDER1 KD endosperm, some abnormal PB-Is with venation or cracks were observed (Supplemental Fig. S6). However, the storage proteins of OsDER1 KD were not significantly different from those in the wild type, and the proportion of abnormal PB-I was not large. The regulatory mechanism of OsDER1 on PB-I formation remains to be investigated.
Recent studies revealed that, in addition to the canonical ERAD pathway mediated by the HRD1 complex, an alternative ERAD pathway mediated by the DER1-SPP-TRC8 complex exists in yeast and mammals. However, the existence of this alternative ERAD pathway in plants was unconfirmed. In this study, we found that OsDER1 interacted with OsSPP1 and OsSPP2 (Fig. 7E). These results indicated that the alternative ERAD pathway may be conserved in plants. It is noteworthy that no ortholog of TRC8 was found in rice. Whether an E3 ubiquitin ligase in rice has functional roles akin to those of TRC8, and whether the entire alternative pathway is present in plants, remain to be demonstrated.
MATERIALS AND METHODS
Generation of Transgenic Plants
ER-stressed transgenic rice (Oryza sativa) was generated by repressing OsSAR1a/b/c using RNAi, as described previously. Developing seeds at 10 to 15 d after fertilization were used for the SWATH analysis. Endosperm-specific knockdown transformants for OsDER1 (OsDER1 KD) were generated by RNAi. The inserted gene fragment was 309 bp (bp 229–538). This fragment was inserted as positive and negative into a modified GluA2-pTCK303 vector with the endosperm-specific GluA2 promoter replacing the ubiquitin promoter. Transgenic rice plants (cv Zhonghua 11) were generated by Agrobacterium tumefaciens-mediated transformation (Qu et al., 2005).
To produce the gene overexpression transformants (OsDER1 OE), the coding region of OsDER1 was amplified using rice cDNA as a template and connected with a 3×FLAG sequence by PCR. The DNA fragment was inserted into the pCAMBIA1301 vector with the ubiquitin promoter. To produce the RNAi transformants (OsDER1 Ri), the same fragment was used for insertion into the pTCK303 vector containing the ubiquitin promoter. Transgenic rice plants (cv Kitaake) were generated by A. tumefaciens-mediated transformation (Qu et al., 2005). The calli of OsDER1 OE and OsDER1 Ri were analyzed on selective medium. To construct the OsDER1 pro-GUS vector, the promoter of OsDER1 was amplified using rice genomic DNA as a template. The DNA fragment was introduced into the pCAMBIA1301 vector, which contains the gusA gene encoding GUS. All primer sequences are listed in Supplemental Table S1. The constructs are listed in Supplemental Table S2.
ER Membrane Protein Isolation
Developing seeds were dehusked and then milled in a mortar for 30 min with homogenization buffer (500 mm Suc, 10 mm KCl, 1 mm EDTA, 1 mm MgCl2, 2 mm DTT, and 150 mm Tricine-KOH, pH 7.5). The crude homogenate was filtered through a 100-μm mesh, and the filtrate was centrifuged at 1,000g and 4°C for 15 min. The supernatant was then centrifuged at 10,000g and 4°C for 15 min. Discontinuous Suc density gradients were prepared with 30% and 20% Suc and the supernatant of differential centrifugation with a ratio of 7:4:2 and then centrifuged at 100,000g for 2 h at 4°C. The membrane at the 20% to 30% Suc interface was removed with a homemade pipette and mixed with an equal volume of 60% Suc. Then, discontinuous Suc density gradients were prepared with the diluted membrane fractions using 40%, 30%, and 20% Suc with the ratio 3.5:4:3:2 and centrifuged at 250,000g for 22 h at 4°C. The purified ER was carefully removed from the 20% to 30% Suc interface and diluted with ice-cold water. Then, the membranes were pelleted by centrifugation, followed by snap freezing in liquid nitrogen and storage at −80°C. All operations were performed on ice, and all Suc solutions contained a protease inhibitor cocktail (Roche).
Membrane Proteomics Analysis
The ER membrane proteins were extracted with buffer (0.125 m Tris-HCl, 4 m urea, 4% SDS, and 2% β-mercaptoethanol, pH 6.8) from the wild type and OsSAR1 RNAi and quantified using a 2D Quant Kit (GE Healthcare); trypsin digestion was performed as described previously (Qian et al., 2015). Because the yield of extraction of the ER was very low, the digestion peptides from four independent experiments were pooled together as one sample and then subjected to SWATH analysis performed on a TripleTOF 5600 system (AB Sciex; Gillet et al., 2012). A total of three technical replicates were designed and conducted in the study. To analyze the membrane proteins identified, the TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and TopPred II (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::toppred) were used to predict the number of transmembrane helices, and the Blast2GO software was used to categorize their subcellular categories. A protein pathway analysis was performed using KOBAS 2.0.
DTT Treatment, RNA Isolation, and RT-qPCR
The seedlings of cv Kitaake were incubated in liquid MS medium that contained 2 mm DTT and 5 μg mL−1 tunicamycin for 4 h. Equal volumes of water and DMSO were added as negative controls. The treated materials with developing seeds at 10 d after fertilization acted as samples and were used to extract total RNA using TRIpure Reagent (Bioteke). RT-qPCR was performed on a LightCycler system (Roche Diagnostics) as described previously (Qian et al., 2015). The primers were designed by Beacon Designer 8.0 (Supplemental Table S3).
Seeds of OsDER1 OE, OsDER1 Ri, and wild-type rice were germinated in water for 5 d and then transferred to one-half-strength liquid MS medium with or without 1 mm DTT and cultured for 14 d. The lengths of the shoots and roots of the rice plants were calculated by ImageJ.
SDS-PAGE and Immunoblot Analysis
Total protein was extracted with buffer (0.125 m Tris-Cl, 4% SDS, 4 m urea, and 2% β-mercaptoethanol, pH 6.8) from seeds and leaves. After centrifugation at 12,000g for 5 min, the protein samples were subjected to 13.6% SDS-PAGE and then immunoblot analysis using a PVDF membrane (Millipore) and immunodetection with HRP-conjugated secondary antibodies. Antibodies against α-tubulin and FLAG were generated by Sigma, and antibodies against α-ubiquitin were generated by the Beijing Genomics Institute.
Transient Expression in Rice Protoplasts
GFP was fused to the C terminus of OsDER1. The signal peptide of AtWAK2 and the ER retention signal His-Asp-Glu-Leu were combined to create the ER marker (mCherry-HDEL). Then, the chimeric genes were subcloned into vector pBI221 containing the cauliflower mosaic virus 35S promoter and cotransformed into rice protoplasts as described by Chen et al. (2006). The transformed cells were observed through a confocal microscope (Olympus FV1000 MPE).
Analysis of GUS Expression
Different tissues of OsDER1 pro-GUS transgenic rice were incubated with GUS staining buffer (Real-Times; RTU4032) at 37°C for 12 h and then destained in (v/v) 70% ethanol until the chlorophyll was removed.
Transmission Electron Microscopy
Transverse sections of mature seeds were used for scanning electron microscopy analyses as described (Fujita et al., 2003) and then observed by transmission electron microscopy (Hitachi S-4800 scanning electron microscope).
Coimmunoprecipitation Assays
Transgenic calli containing the OsDER1-3×FLAG tag under the control of the ubiquitin promoter were generated (Wakasa et al., 2011). Successful transgenic calli were screened by anti-FLAG antibodies. In the coimmunoprecipitation assay, the wild type served as a negative control; the transgenic calli and wild-type calli were ground and extracted by lysis buffer (25 mm Tris-HCl, pH 8, 150 mm NaCl, 2.0% [w/v] n-dodecyl-β-d-maltopyranoside [Anatrace], and a protease inhibitor cocktail [Roche]). Briefly, after centrifugation, the supernatant was incubated with anti-FLAG resin for 2 h at 4°C, the immunoprecipitation complexes were washed three times using wash buffer (25 mm Tris-HCl, pH 8, 150 mm NaCl, and 0.02% [w/v] n-dodecyl-β-d-maltopyranoside), and the protein complexes were eluted by 2× SDS-PAGE loading buffer and subjected to immunoblot analysis. To assess coexpression with OsHRD1, OsHRD3, OsSPPs, or OsCDC48s, the recombinant proteins were expressed using the pFastBac baculovirus system (Invitrogen; Sokolenko et al., 2012). Briefly, recombinant bacmid DNAs were generated in DH10Bac cells and then transfected and amplified in Sf9 insect cells (Invitrogen). At 72 h after viral infection, the cells were harvested and extracted by lysis buffer, and the subsequent procedure was the same as that mentioned above.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project Database under accession numbers Os03g15350 (OsHRD3), Os03g05730 (OsCDC48-1), Os10g30580 (OsCDC48-2), Os05g09550 (OsDER1), Os03g63520 (OsDER2), Os02g02530 (OsSPP1), and Os05g36070 (OsSPP2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RT-qPCR analysis of the mRNA expression of nine differentially expressed genes.
Supplemental Figure S2. Kyte-Doolittle hydropathy analysis of the OsDER1 TM3.
Supplemental Figure S3. Phylogenetic tree of OsDERs with homologs in other species.
Supplemental Figure S4. Statistical analysis of the length and width of OsDER1 knockdown transgenic rice seeds.
Supplemental Figure S5. Analysis of the mRNA levels of OsDER1 in overexpressed and suppressed OsDER1 transgenic rice seeds.
Supplemental Figure S6. Immunohistochemical analysis of the alteration of protein body formation in OsDER1 KD seeds using transmission electron microscopy.
Supplemental Table S1. Primers used for vector construction.
Supplemental Table S2. Details for construction.
Supplemental Table S3. Primers used for RT-qPCR.
Supplemental Data Set S1. List of the total proteins identified by mass spectra.
Supplemental Data Set S2. List of the differentially expressed proteins identified by mass spectra.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Xiangbai Dong for valuable suggestions throughout this project.
Footnotes
This work was supported by the Ministry of Science and Technology of China (grant no. 2016YFD0100500) and the National Natural Science Foundation of China (nos. 31770271, 31171368, and 31300212).
References
- Avci D, Lemberg MK (2015) Clipping or extracting: two ways to membrane protein degradation. Trends Cell Biol 25: 611–622 [DOI] [PubMed] [Google Scholar]
- Avci D, Fuchs S, Schrul B, Fukumori A, Breker M, Frumkin I, Chen CY, Biniossek ML, Kremmer E, Schilling O, et al. (2014) The yeast ER-intramembrane protease Ypf1 refines nutrient sensing by regulating transporter abundance. Mol Cell 56: 630–640 [DOI] [PubMed] [Google Scholar]
- Baldridge RD, Rapoport TA (2016) Autoubiquitination of the HRD1 ligase triggers protein retrotranslocation in ERAD. Cell 166: 394–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boname JM, Bloor S, Wandel MP, Nathan JA, Antrobus R, Dingwell KS, Thurston TL, Smith DL, Smith JC, Randow F, et al. (2014) Cleavage by signal peptide peptidase is required for the degradation of selected tail-anchored proteins. J Cell Biol 205: 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson EJ, Pitonzo D, Skach WR (2006) p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation. EMBO J 25: 4557–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, Lemberg MK (2014) Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J 33: 2492–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Cui F, Liu L, Zhao Q, Zhang Z, Li Q, Lin B, Wu Y, Tang S, Xie Q (2012) Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24: 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymer F, von Heijne G, White SH (2015) Mechanisms of integral membrane protein insertion and folding. J Mol Biol 427: 999–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS (2006) A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126: 349–359 [DOI] [PubMed] [Google Scholar]
- Fujita N, Kubo A, Suh DS, Wong KS, Jane JL, Ozawa K, Takaiwa F, Inaba Y, Nakamura Y (2003) Antisense inhibition of isoamylase alters the structure of amylopectin and the physicochemical properties of starch in rice endosperm. Plant Cell Physiol 44: 607–618 [DOI] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C (2006a) A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol 8: 849–854 [DOI] [PubMed] [Google Scholar]
- Gauss R, Sommer T, Jarosch E (2006b) The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J 25: 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 11: 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR (2011) Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol 18: 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habeck G, Ebner FA, Shimada-Kreft H, Kreft SG (2015) The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J Cell Biol 209: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Sommer T (2012) Finding the will and the way of ERAD substrate retrotranslocation. Curr Opin Cell Biol 24: 460–466 [DOI] [PubMed] [Google Scholar]
- Horn SC, Hanna J, Hirsch C, Volkwein C, Schütz A, Heinemann U, Sommer T, Jarosch E (2009) Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell 36: 782–793 [DOI] [PubMed] [Google Scholar]
- Hsu FF, Yeh CT, Sun YJ, Chiang MT, Lan WM, Li FA, Lee WH, Chau LY (2015) Signal peptide peptidase-mediated nuclear localization of heme oxygenase-1 promotes cancer cell proliferation and invasion independent of its enzymatic activity. Oncogene 34: 2410–2411 [DOI] [PubMed] [Google Scholar]
- Kamauchi S, Nakatani H, Nakano C, Urade R (2005) Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J 272: 3461–3476 [DOI] [PubMed] [Google Scholar]
- Kim W, Spear ED, Ng DTW (2005) Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell 19: 753–764 [DOI] [PubMed] [Google Scholar]
- Kirst ME, Meyer DJ, Gibbon BC, Jung R, Boston RS (2005) Identification and characterization of endoplasmic reticulum-associated degradation proteins differentially affected by endoplasmic reticulum stress. Plant Physiol 138: 218–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Finger A, Braun T, Hellmuth K, Wolf DH (1996) Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J 15: 753–763 [PMC free article] [PubMed] [Google Scholar]
- Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B (2001) Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol 167: 6441–6446 [DOI] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL (2004) A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429: 834–840 [DOI] [PubMed] [Google Scholar]
- Liu L, Cui F, Li Q, Yin B, Zhang H, Lin B, Wu Y, Xia R, Tang S, Xie Q (2011) The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res 21: 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang C, Wang D, Su W, Liu L, Wang M, Li J (2015) EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc Natl Acad Sci USA 112: 12205–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert M, Sommer T, Jarosch E (2014) Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol 16: 77–86 [DOI] [PubMed] [Google Scholar]
- Müller J, Piffanelli P, Devoto A, Miklis M, Elliott C, Ortmann B, Schulze-Lefert P, Panstruga R (2005) Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell 17: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Takaiwa F (2015) OsHrd3 is necessary for maintaining the quality of endoplasmic reticulum-derived protein bodies in rice endosperm. J Exp Bot 66: 4585–4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian D, Tian L, Qu L (2015) Proteomic analysis of endoplasmic reticulum stress responses in rice seeds. Sci Rep 5: 14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q, Yoshihara T, Ooyama A, Goto F, Takaiwa F (2005) Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta 222: 225–233 [DOI] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P (2014) ER-associated degradation: protein quality control and beyond. J Cell Biol 204: 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Kaufman RJ (2004) A trip to the ER: coping with stress. Trends Cell Biol 14: 20–28 [DOI] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY (2009) Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell 34: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, Baker D, Chambers MG, Su H, Li D, et al. (2017) Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 548: 352–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko S, George S, Wagner A, Tuladhar A, Andrich JMS, Aucoin MG (2012) Co-expression vs. co-infection using baculovirus expression vectors in insect cell culture: benefits and drawbacks. Biotechnol Adv 30: 766–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg HR, Thomas M, van den Boomen D, Wiertz EJHJ, Drabkin HA, Gemmill RM, Lehner PJ (2009) The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol 186: 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Liu Y, Xia Y, Hong Z, Li J (2011) Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc Natl Acad Sci USA 108: 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Meng Q, Gao J, Zhang S, Zhang H, Zhang M (2015) Label-free quantitative analysis of changes in broiler liver proteins under heat stress using SWATH-MS technology. Sci Rep 5: 15119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Dai LL, Yin ZJ, Fukuda M, Kumamaru T, Dong XB, Xu XP, Qu Q (2013) Small GTPase Sar1 is crucial for proglutelin and α-globulin export from the endoplasmic reticulum in rice endosperm. J Exp Bot 64: 2831–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boomen DJH, Timms RT, Grice GL, Stagg HR, Skødt K, Dougan G, Nathan JA, Lehner PJ (2014) TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc Natl Acad Sci USA 111: 11425–11430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, Hayashi S, Yang L, Takaiwa F (2011) Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J 65: 675–689 [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Hayashi S, Takaiwa F (2012) Expression of OsBiP4 and OsBiP5 is highly correlated with the endoplasmic reticulum stress response in rice. Planta 236: 1519–1527 [DOI] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K (1986) The site of synthesis and accumulation of rice storage proteins. Plant Cell Physiol 27: 135–145 [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z (1982) Biosynthesis of storage proteins in developing rice seeds. Plant Physiol 70: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol 162: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]