Abstract
The LOINC (Logical Observation Identifier Names and Codes) vocabulary is a set of more than 10,000 names and codes developed for use as observation identifiers in standardized messages exchanged between clinical computer systems. The goal of the study was to create universal names and codes for clinical observations that could be used by all clinical information systems. The LOINC names are structured to facilitate rapid matching, either automated or manual, between local vocabularies and the universal LOINC codes. If LOINC codes are used in clinical messages, each system participating in data exchange needs to match its local vocabulary to the standard vocabulary only once. This will reduce both the time and cost of implementing standardized interfaces. The history of the development of the LOINC vocabulary and the methodology used in its creation are described.
We have recently recognized a strong dependency between the development of medical vocabularies (medical concept representation) and health care data exchange standards. These two endeavors have had rather independent histories, but it is apparent that they are closely related and mutually dependent activities. In this paper we briefly describe each endeavor and then describe in detail the process used to develop the LOINC (Logical Observation Identifier Names and Codes) vocabulary. We use creation of the LOINC vocabulary as one model for the synergistic development of health information exchange standards and coded medical vocabularies. A previous article1 has described some of the background, rationale, and content of the LOINC vocabulary for laboratory procedure codes.
Background
Medical concept representation is not an end in itself but is desirable for the capabilities it provides.2,3,4 There are at least three activities that would be made possible by a consistent representation of medical concepts: 1) the real-time exchange of medical data; 2) the exchange of decision-support programs including alerts, protocols, care pathways, and care plans; and 3) the pooling of data for outcomes research, quality assurance programs, and clinical research. One proposed model of medical concept representation, which we adopt here,5 breaks the representation model into three components: a medical vocabulary or lexicon, a semantic data model (an information model), and a knowledge base. We discuss only the first two of these components in the following paragraphs.
The first component, a structured medical vocabulary, is an organized set of terms or words with associated codes. Coded medical vocabularies are one of the foremost issues in the field of medical informatics. Indeed, Sittig6 noted that a unified controlled medical vocabulary was one of the grand challenges of medical informatics. There is a large and growing set of publications related to the development and use of structured medical vocabularies.7,8,9,10,11,12,13
The second component of medical concept representation, a semantic data model (SDM), is a description (template or data structure) of how vocabulary items can be combined to make a valid representation of medical information. Semantic data models have also generated a large number of publications.3, 14,15,16,17,18,19,20,21 Often unrecognized as SDMs are the health care data exchange standards, particularly CEN TC251/PT-00822 and CEN TC251/PT-02223 of Technical Committee 251 (Health Informatics) of the European Committee for Standardization; HL7 (Health Level 7)24; the DICOM standards25,26,27,28; and specifications E123829 and E139430 of the American Society for Testing and Materials (ASTM). These standards describe the interchange format and syntax for messages that can be exchanged between medical computing systems.
In talking about these standards, it is appropriate to establish a common definition of terms. In Europe, an important distinction is made between an interchange format and message syntax. An interchange format is a syntax-independent description of the structure and content of information within a message. As used in this article, the SDM for a message is the same thing as an interchange format. Message syntax, however, is the specific way a message is encoded for transmission. Examples of message syntaxes include: ED-IFACT, BER (Binary Encoding Rules) of ASN.1 (Abstract Syntax Notation One), and the delimited record format of HL7.
We illustrate how message standards represent SDMs by reference to HL7. As shown in ▶, an HL7 unsolicited observation result (ORU) message is divided into distinct parts or segments. The MSH segment contains information that identifies the kind of message that follows. The PID and PV1 segments identify a patient and a particular visit to which the message pertains. The ORC and OBR segments describe information about who ordered an observation and the common context in which the observation was made. The repeating OBX segments represent individual observations (results or measurements).
Figure 1.
An HL7 unsolicited observation result (ORU) message. The message is composed of segments (MSH, EVN, PID, PV1, ORC, OBR, OBX), and each segment contains fields that are separated by a vertical bar (|). The MSH segment contains information that identifies the kind of message that follows. The PID and PV1 segments identify a patient and a particular visit to which the message pertains. The ORC and OBR segments provide information about who ordered an observation and the common context in which the observation was made. The repeating OBX segments represent individual observations (results or measurements). The structure of HL7 messages represents a simple semantic data model.
It is not obvious at first that this kind of message syntax represents an SDM. However, on further inspection (and by reference to the HL7 specification) it is clear that an OBR segment has many associated OBX segments. This structure reflects a model of laboratory testing where an ordered procedure can result in a number of laboratory observations or none. Considering the substructure of an OBX segment, we can assert that each observation is measured in particular units and can be classified as normal or abnormal. Thus, HL7 message syntax defines a simple SDM.
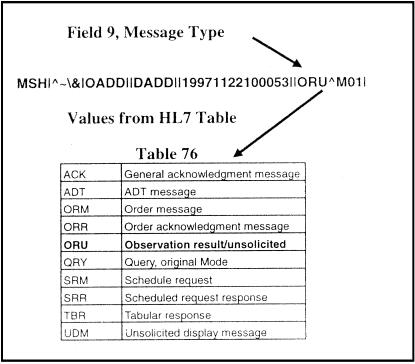
In the HL7 standard, vocabulary elements are linked to the message structure by the use of identifier (ID) and coded element (CE) data-type fields. Data fields of format type ID contain a reference to HL7 tables that define the set of allowed values for the given field. In the ORU example of ▶, the ninth field of the MSH segment is Message Type, which specifies the type of message that is to follow. As shown in ▶, all valid message types are defined in table 0076 of the HL7 specification. Many vocabulary tables exist in the HL7 specification, including definitions for patient type, specimen type, and order priorities.
Figure 2.
Use of a coded identifier (ID) field in the MSH segment of the HL7 message. Field 9 of the MSH segment is used to indicate the type of message that follows. Message Type is an ID field, meaning that its value must come from an HL7 table, in this case table 0076.
Coded element data fields allow the set of values for a field to be defined by reference to a coded vocabulary that is external to the HL7 standard. This allows reuse of the many existing medical vocabularies and spares the HL7 group the time and effort of recreating terms that have been painstakingly created by other organizations.
A CE field has six subparts. The first subpart is a code, the second subpart is a textual description of the meaning of the code, and the third subpart is the name of the external coding system that is being used. The fourth, fifth, and sixth subparts allow for sending an alternate code and text from a second coding scheme. In ▶, the fourth field of the OBX segment—the Observation Identifier field—is a CE field. The code is 2951-2, the description is Serum Sodium, and the coding system is LN (for LOINC). Coded element field types are used in messages to define diagnoses, procedure batteries, observation identifiers, and units of measure.
Figure 3.
Use of a coded element (CE) field in an OBX segment of the HL7 message. Field 4 of the OBX segment contains the Observation Identifier (what was observed) and is of type CE (Coded Element). Each CE type field consists of six parts, but only three parts are shown in this example. The first part of a CE field is the code, the second part is the description (meaning or text) of the code, and the third part is the coding scheme from which the code was selected. This example shows a LOINC coding (LN) being used as the observation identifier.
Besides the model implicit in the HL7 specification, work on version 3.0 of HL7 includes an explicit Reference Information Model. Standards other than HL7 also have underlying SDMs. The orders and results portions of the ASTM 1238 standard29 are technically aligned with HL7, and the implicit model is similar to the HL7 model. Several other ASTM standards are models of clinical processes or of the computer-based patient record.13, 17, 31, 32 In the development of the DICOM standard, an explicit SDM was declared early on and the standard was developed using the model.27, 33, 34 DICOM messages use coded vocabulary to describe procedures, image acquisition context, and observational findings. In the first versions of the standard, controlled terminology was embedded in the normative text as “enumerated values” and “defined terms” ad hoc. Recent versions of the standard have included references to externally controlled vocabularies (principally SNOMED7) via the SNOMED DICOM microglossary.35,36,37
The work within CEN TC251 in working groups PT3-008 (Messages for Exchange of Laboratory Information) and PT3-022 (Request and Report Messages for Diagnostic Services Departments) is also strongly based on SDMs, called Domain Information Models (DIMs) within the CEN TC251 context. The model is expressed using a formal literary and graphic notation.22 Objects modeled within the clinical laboratory DIM include subject of investigation, sample, laboratory investigation, and laboratory investigation result item. The DIM incorporates references to external vocabularies using the coded value and list of coded values data types. The coded value data type is nearly identical to the CE data type used in HL7 and consists of three parts: a health care coding scheme designator, a code value, and a code meaning.
Thus, the development of the health care data and image exchange standards has clarified and strengthened the connection between vocabularies and data models. The data model is an essential skeleton, without which the coded terms would be an ambiguous jumble. The named slots or data elements in the model provide semantic labels for the vocabulary items contained in the model, making the context and meaning of the terms more explicit and computable. It is the combination of the SDM and a structured medical vocabulary that makes the representation of a medical concept complete. The recognition of this dependency should guide vocabulary development efforts related to the electronic medical record.
The interdependence of the vocabulary and data model are especially evident in the context of data exchange standards. Let's assume, for example, that we are developing a message to send medication orders between two systems. One approach would be to define the message so that separate fields in the message convey the name of the drug, its dose, form, and manufacturer. Each coded field in the message would have a well-defined set of possible values, such as Drugs (e.g., digoxin, penicillin V potassium) and Forms (e.g., capsule, tablet, suppository). A second possibility would be to define the message so that the only field it contained was National Drug Classification (NDC) code. Each NDC code represents a complex aggregate name for a medication. The name of each NDC code includes the drug, its form, dose, manufacturer, and package size. So, if I choose to use NDC codes in my messages, I need only one field to express the medication information. However, if I use the multi-axial strategy, I will have four fields in my message, each field having a set of possible values specific to its purpose. I might also choose to include both the composite field and the four individual parts. The point is that decisions about what vocabulary to use in an interface message are not independent of the data structures that will be used to carry the information. The choice and use of vocabularies in message standards is inexorably tied to the structure and organization of fields in the messages, and vice versa.
Finally, the Canon Group2 has described the need for “a scientific methodology” for developing medical concept representations so that the work would be reproducible, extensible, testable, expressive, and understandable. A similar goal has been expressed by CEN TC251 Project Team 2.38 Recognizing this goal, we describe in the following section the process and rationale that lead to the development of the LOINC vocabulary. The LOINC vocabulary was developed with an understanding of the interdependency of structured medical vocabulary and an SDM. As such, the LOINC experience may provide insights into how this process can be made reproducible, extensible, testable, and understandable.
The LOINC Development Process
Problem Selection
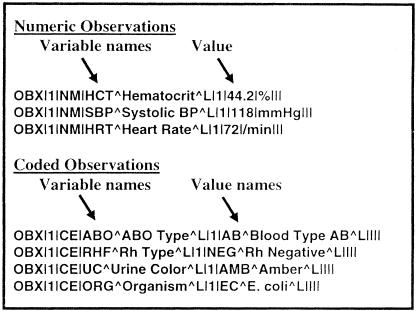
An obvious but important first step for the LOINC committee was to select a domain of interest. The initial need we chose to address was the set of codes that could be used as observation identifiers in the HL7, ASTM 1238, and CEN TC251/PT3-008 and PT3-022 result messages. The observation identifier is the part of the result message that expresses what kind of observation is being made. It is important to distinguish the names of observations (also called variable names) from the coded value of the observation. As shown in a series of OBX segments in ▶, observation names (variable names) include hematocrit, systolic blood pressure, heart rate, ABO blood type, Rh type, urine color, and organism identified by culture. Depending on the particular variable, the value associated with a variable in a message can be a numeric quantity, a titer, a range, or a coded value. For instance, hematocrit, heart rate, and blood pressure are names of numeric variables, whereas ABO blood type, Rh type, urine color, and organism identified by culture are the names of coded variables. The possible coded values for ABO blood type are A, B, O, and AB, while the values for Rh type are Positive, Du Positive, and Negative. Possible values for organism identified include E. coli, P. aeruginosa, P. mirabilis, and others. Variable names correspond exactly to the role played by the observation identifier in an HL7 OBX segment, whereas value names correspond to the value field in an OBX segment.
Figure 4.
Variable names and value names as used in a series of HL7 OBX segments. The kind of observation is indicated using a variable name. The value of the observation can be any of the allowed HL7 types, including NM (numeric) or CE (coded element). For numeric observations, the value of the observation is number, whereas for coded observations the actual value of the observation is indicated by a code. Possible value codes could come from other vocabularies, such as SNOMED, Read Codes, or the UMLS Metathesaurus. In this example, all codes are local, as indicated by the L following each code description.
There were at least four reasons why we chose the creation of observation identifiers as our initial task. First, creating a consistent set of observation identifiers appeared to have an immediate benefit. Even though the HL7 and ASTM 1238 standards were being used in a growing number of health care facilities worldwide, laboratories and clinical systems continued to report procedure results using internally defined names and codes that are idiosyncratic to the institution, as shown in ▶. Installing a new system in a health care network meant painstakingly matching codes sent by the laboratory to the codes used within the local clinical information system. We estimate that at least 90 percent of the effort of installing a new interface is spent in matching vocabulary between sending and receiving systems. It is not unusual for a complete matching of codes to require a year of earnest investigation. Our desire was to create names and codes that could be used by all laboratories and clinical systems where the names were structured to facilitate rapid matching, either automated or manual. If such a universal vocabulary existed, each system participating in data exchange would need to match their local vocabulary to the standard vocabulary once, but thereafter would need to maintain the mappings only as new results were added. By not having to map codes each time a new system was installed, a good deal of time and money could be saved.
Figure 5.
Institution codes used as variable names in an HL7 message. As shown by the L following the description of the code, each site is using its own local coding scheme for variable names (observation identifiers). Each site has chosen a different code to represent a serum sodium concentration measurement. The purpose of creating LOINC codes is so that all sites can use universal codes for the names of observations.
Second, when the LOINC committee began, no structured vocabulary existed that was appropriate for naming clinical, laboratory, or physiologic measurements in result messages and that also had the correct degree of granularity to represent the names of procedures as commonly defined in laboratory information systems and in clinical information systems. For example, SNOMED procedure codes are typically the names of small classes of measurements rather than specific measurements. For instance, P3-71260 Albumin measurement in SNOMED does not specify whether the measurement was on urine, or serum. The type of specimen could be expressed in an associated modifier field in the message, but this approach would be inconsistent with the common practice of using a single field to name an observation.
Many Current Procedural Terminology (CPT) codes39 also represent classes of analytes rather than specific measurements. This reflects the purpose for which CPT codes were created—billing for procedures and not reporting clinical results. In contrast, the multiaxial approach used in EUCLIDES40 and the Open-Labs system41 is very flexible but does not match up well with the data structures available in HL7/ASTM or with common usage in clinical applications. The strategy in EUCLIDES was to represent result names by combining several codes from a canonical set of axes to express the kind of observation that was made. The initial system used minimal precoordination. This provided a robust mechanism for representing observation identifiers, but was not compatible with the message structure available in HL7 or ASTM messages or with the corresponding data structures used in a typical laboratory information system. The final version of the CEN TC251/PT3-008 standard allows for the use of either single aggregate codes or the multi-axial codes. At the time that the LOINC work began, however, the pre-coordinated terms did not exist in EUCLIDES.
Third, good vocabularies, such as SNOMED, did exist for expressing many of the coded values of the observations, so the creation of sets of coded values was not a high priority. Also, creating coded value sets is a large task and more than we thought we could accomplish within the scope of the LOINC committee.
Fourth, while no standard set of observation identifiers existed, good starting lists were available. All laboratory information systems and most clinical information systems have internal lists of observation names in electronic form. The availability of good starter sets of observation identifiers made the task look doable.
Additional Vocabulary Principles
There were characteristics of some of the existing vocabularies, other than content, that made them undesirable for use in messages. Many of the desirable characteristics of coded vocabularies have been described previously.5,13,42,43 We wanted to adhere to some additional principles related to aspects of a good vocabulary:
There should be version control associated with the coding system. The version identifier should be a prominent component in each distribution.
The code associated with a term should have no intrinsic or embedded meaning.
The term component should stand on its own without additinal components being required to convey the meaning of the concept.
There should be an organization capable of extending, correcting, and maintaining the coding system. This should include evidence of organizational stability and adequate funding for the coding activity.
There should be easy access to the vocabulary. It should be available by both paper and electronic distribution.
There should be no limitation on who can acquire copies of the coding system.
There should be no cost or minimal cost associated with access to and use of the coding system. Likewise, maintenance fees are undesirable.
The coding system should have training materials, such as tutorials, training syllabi, and printed user manuals. People with in-depth understanding of the coding system should be available to provide help and consultation.
The coding system should be acceptable for use internationally.
The coding system should be extensible. There should be no inherent limits on the number or types of codes that can be created.
The coding system should be compositional. The creation of new codes should be guided by a data model.
Organizational Structure
There were at least three possible ways in which a vocabulary committee could have been organized: 1) as part of an existing standards development organization like ASTM, the DICOM Standards Committee, or HL7; 2) as a commercial venture to produce a forprofit product; or 3) as a group of interested individuals with support from governmental or private sources. There were two major factors that guided how the LOINC committee was organized. First, we wanted the product (a set of names and codes) to be freely available to the public, in accordance with the principles stated earlier. This could not be guaranteed if the work were done as a part of an existing standards organization, where the organization would typically hold the copyright to any materials developed within the organization. Second, we wanted to be able to provide a vocabulary quickly and provide fast changes and revisions. The structure of most standards development groups, with formal ballots and procedures, means that they typically have a change and revision cycle of two to five years, but we wanted to be able to make revisions and additions within weeks or months. As a result of these factors, the LOINC committee was initially created by invitation to a small group of interested individuals. We have accepted a number of additional volunteer experts as the process has proceeded. The complete list of past and current participants is available at the LOINC Web site, in the Introduction, at http://www.mcis.duke.edu/standards/termcode/loinclab/LOININTR.TXT.
Financial support has come from a number of sources. The Regenstrief Institute was largely responsible for supporting the first two meetings. Subsequently, there has been support from the Agency for Health Care Policy and Research, the Hartford Foundation, and the National Library of Medicine.
Selection of Participants
The danger of a small-group approach is that all perspectives on a problem may not be adequately represented. In the case of LOINC, care was taken to have representation from several areas related to laboratory data representation. The committee ultimately included individuals with the following backgrounds: physicians in active clinical practice, clinical chemists, pathologists, laboratory technologists, medical informaticists, managers from large reference laboratories, and specialists in the naming of laboratory analytes. Many participants also had previous experience in developing or implementing the CEN TC251/PT3-008, ASTM 1238, 1633, and 1712, DICOM, and HL7 standards. This mixture of individuals proved to be essential to ensuring that the codes and names produced by the group would be appropriate for use in private hospitals, Veterans Health Administration hospitals, and commercial reference laboratories.
Focusing the Scope
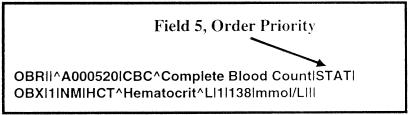
Once the participants had been selected, the initial meetings commenced. The first meetings were used to focus the scope of the activity and to mutually educate one another. Again, the general goal of the LOINC committee was to produce observation identifiers for the reporting of clinical observations in HL7, ASTM, and CEN messages. The initial scope was specifically focused on these standards, although it was later recognized that any other standard, such as DICOM, that had a similar logical model could also make use of the vocabulary. Restricting the focus of LOINC to a message context where there was a known, fixed data structure was essential in making decisions about what information should be carried in the code used in the observation identifier field of the observation segment and what information could appropriately be sent in another part of the message. For instance, because order priority (STAT, ASAP, Routine) is carried in the observation header of the message, there was no need to include priority as a part of the observation name itself. In other words, it would be inappropriate in HL7 messages to make names like “STAT Hematocrit,” because this information can be easily represented in such messages by sending the order priority in the OBR (observation header) segment while sending “Hematocrit” in the OBX (observation) segment, as shown in ▶.
Figure 6.
Using other parts of an HL7 message to carry important information. Since order priority is sent as part of the OBR segment (in this case STAT), there is no need to make a name for “STAT Hematocrit” within the LOINC vocabulary.
Another key decision was to limit the initial scope to the representation of names and codes for clinical laboratory result observations. The global scope was the creation of names and codes for all clinical observations, including direct patient observations as well as clinical laboratory observations. Laboratory observations were chosen as the initial focus because the committee had ready access to a number of good starter sets from clinical laboratories. Later, when substantial progress had been made in the area of laboratory identifiers, attention was turned back to clinical observations such as vital signs, 12-lead ECG measurements, fluid intake and output measurements, and measurements taken during imaging studies. Since some clinical measurements might be sent using either the HL7 or DICOM standards, it was important that the same vocabulary be used by both standards to ensure that they could intercommunicate.
The names of orderable laboratory batteries were excluded from the initial focus because they are names for a group of two or more laboratory observations. For example, serum glucose is an appropriate observation name, whereas Liver Panel, Chem 7, or Urine Electrolytes are names of laboratory batteries. Accurate names for the laboratory observations must exist before well-defined battery names can be created. Later, after we had created many of the laboratory result names, we returned to create the names of common laboratory procedure batteries, and this work is currently in progress.
It was determined that the names created by the LOINC committee would be “fully specified” names, which would need no further definition when used for matching to local terms and codes. As will be shown below, the fully specified names are created by a combination of five (and sometimes six) independent axes. It was postulated that the fully specified names would typically be too long and complex to be used as common names on clinical printouts or reports. Instead, we decided to maintain common names in a “related names” column in the LOINC database. It was assumed that local sites would supply their own list of preferred terms (which would correspond one-to-one with LOINC terms) for use in clinical reports and displays. We recognized that it would be desirable to have a standardized short name (suitable as a column or field label) for each observation, but this task was left for a later date.
Developing the Model and the Vocabulary Content
General Approach
The process of developing LOINC content was, and continues to be, an iterative combination of both bottom-up (empirical) and top-down (conceptual modeling) approaches. In the case of the top-down approach, we did not start from scratch in developing the initial conceptual model of the vocabulary. We were influenced indirectly by principles and techniques reported by several groups that are working on concept representation in medicine,12,22,38 but we were also directly influenced by four ongoing medical vocabulary initiatives.
First, the multi-axial representational approach as used in SNOMED7 was an important basic tenet. We recognized very early that we were making aggregate or pre-coordinated expressions and that it was important to know the set of atomic terms that could be used to construct the molecular expressions. The multi-axial approach in medical vocabularies has become so common that it is almost overlooked as a basic principle.
Second, we were influenced by the work of IUPAC (International Union of Pure and Applied Chemistry) as published in the Compendium of Terminology and Nomenclature of Properties in Clinical Laboratory Sciences (The Silver Book).44 The model presented by IUPAC is an outgrowth of metrology (the theory of measurement) and establishes a framework for naming clinical chemistry measurements. There are three main concepts, or axes, in the IUPAC model. First, there is a system, which establishes a context in which a measurement is made. The system in clinical laboratory measurements is usually the specimen type, like urine, serum, cerebral spinal fluid, or stool. Second, there is some component of the system that is being evaluated, observed, or measured. The component is usually a particular chemical compound, ion, or cell type, like sodium ion, potassium ion, white blood cell, or coagulation factor VIII. Third, there is a particular property of the component that is being measured in the system. Examples of kinds-of-property are mass concentration (mass per unit volume), catalytic activity, number concentration (number of items per unit volume), and color. This three-axis model is a very useful way of characterizing clinical chemistry measurements. We extended the model and found that the principles underlying it were useful for many measurements like blood pressure, heart rate, and deep tendon reflexes. IUPAC has carefully established naming conventions for systems and components and maintains an extensive list of kinds-of-property. More information about the coding of properties in the clinical laboratory sciences is available at http://inet.unic.dk/home/ifcc_iupac_cnpu/.
Third, we took advantage of the conceptual frame-work described as part of the EUCLIDES,40 Open-Labs,41 and CEN TC251/PT3-008 efforts.22 OpenLabs describes 42 canonic classes of information that can be used to describe a clinical laboratory measurement. The set of classes is quite comprehensive and describes not only the component, system, and kind-of-property of the measurement, but also things like the urgency of the measurement, sample collection conditions, preservative used in specimen collection, reagent used in testing, equipment type, method of analysis, and reason for investigation. Many of these dimensions were the basis for the parts of the fully specified names in LOINC, whereas other classes represent information that is sent in other fields of an HL7 or ASTM message. As a direct collaboration with the OpenLabs team, we used many of their concepts to build our initial LOINC database. In some sense, part of the task of the LOINC committee was to decide (recognizing that we were creating a single name and code to be used as the variable name within the message standard) which OpenLabs dimensions should be part of the aggregate LOINC term and which represented information that should be sent in other fields in the message. The LOINC committee defined the agreed-on sets of pre-coordinated concepts that were to be used as observation identifiers. However, in the process we found some gaps and discovered important distinctions that led us to many new (more refined) concepts within what would have been a single axis. We created additional axes to distinguish measures that needed to be distinguished and created small syntaxes to deal with special complexities. We also added more formality to some of the component names, which we describe in the following section.
Fourth, as initial vocabulary lists we used a number of sources. One was a table of chemistry tests submitted by Arden Forrey. He has been working on conventions for naming orderable laboratory procedures, orderable batteries, and results. He has identified at least three important axes in creating result names: the type of the specimen, the substance being measured, and the precision of the measurement. The items submitted by Arden were translated into more formal name structures by Tom Fiers. The translation was accomplished using software developed by the OpenLabs group. We also used laboratory files submitted by seven participating laboratories. In the initial stages, individual committee members also developed starting sets of LOINC names for assigned subject matter.
The Initial Model
Given the conceptual background as defined in preceding paragraphs, we began with a simple four-axis model. The four axes were the component being measured, the specimen type (system), the precision of the measurement, and the method by which the measurement or observation was made. Precision indicated whether a given observation was quantitative, semi-quantative (ordinal, i.e., selected from a ranked set of possible values), or qualitative (nominal, i.e., selected from an unranked set of possible values). We decided that parts of the fully specified name would be separated by a colon. The choice for the delimiter was arbitrary and only for the convenience of the LOINC committee, since each independent part is actually stored as a separate column in the LOINC database table. Represented in an informal notation, our first model looked like this:
Model 1
<component>:<specimen>:<precision>:<method>
Example: HEPATITIS B VIRUS SURFACE AG:SER:SQ:EIA
Rapid Evolution of the Model
In the preliminary stages of LOINC development, the model evolved rapidly as the committee gained understanding and experience. The first model was just a starting point, and it was substantially enhanced even before the first LOINC committee meeting was held. Members of the committee compared the model with result names in their own systems and found further complexity that needed to be expressed. Even at this early stage, we recognized special circumstances in which we needed to specify the timing of specimen collection (12- or 24-hour collection), and conditions or challenges at the time of specimen collection (peak, trough, or random). In the first model, these details were just appended to the component part of the name. As the work of the committee progressed, we became more systematic in the evolution of the model.
An underlying requirement for the LOINC committee was that we would be able to create fully specified names for all results that are commonly found in laboratory and clinical information systems. To this end, we collected laboratory result names from several clinical systems and began creating result names according to our simple model. Seven sites contributed result names: Corning Medical Laboratories, Rutherford, New Jersey; Associated Regional and University Pathologists, Salt Lake City, Utah; Intermountain Health Care, Salt Lake City; Mayo Medical Laboratories, Rochester, Minnesota; Indiana University Laboratories, Indianapolis; Department of Veteran's Affairs, Dallas, Texas; and University of Washington Laboratories, Seattle. Using the names submitted by these institutions, we used word processors and manual editing to create LOINC names using the simple model.
Using the pooled set of result names, the need for enhancements to the model became more evident and other issues surfaced. The issues were of two types: the information to populate the model was not readily available or the model was insufficient to distinguish results that existed in typical systems. We describe issues with the lack of information first.
One problem was the lack of specificity in the names commonly used in clinical systems. Most of the seven systems did not include precision as a formal part of the name. The precision could often be inferred by the unit of measure of the item, or the result name would contain the word “screen,” implying a semi-quantitative result. However, since technologists in the laboratory know what the precision is, it usually is not stated explicitly anywhere in the result definition or configuration file. Less commonly, the specimen type (system) was not specified. When this occurred, it was typically because a given compound is routinely measured in only one kind of specimen, and laboratory technologists know this implicitly. Thus, we could not make fully specified names for many results without asking for further information from laboratory specialists. Asking for more information from laboratory experts became a common activity as name creation progressed.
A second problem was the use of abbreviations. Abbreviations are not typically standardized, so SE might denote serum or semen, and BM might mean bone marrow or bowel movement. Besides ambiguity, there was also inconsistency. Serum might be abbreviated (or truncated) as S, SE, SER, or SM for different results in the same procedure file. Other acronyms for the component part of a name, like ANA for antinuclear antibody, are commonly understood by medical personnel but are not understood by computer programmers. A medical expert can disambiguate abbreviations or acronyms given the appropriate context of use, but in the general case abbreviations are an obstruction to computerized matching techniques. Again, we often found it necessary to ask laboratory technologists to explain the meaning of an abbreviation. In some cases the abbreviations are used so frequently that even the lab technologists have forgotten what they represent. With only a few exceptions, all abbreviations in the component name are prohibited. Examples of common abbreviations that may be used in the component name include RNA (ribonucleic acid), DNA (deoxyribonucleic acid), HLA (human histocompatibility-complex-derived antigen), AB (antibody), and AG (antigen). All allowed abbreviations are expanded in the LOINC manual. There has been an attempt to keep these items aligned with those found in ASTM 1712, but the two lists are not always identical.
A third problem was with capitalization, punctuation, word order, word form, and other conventions. We wanted the LOINC vocabulary to easily allow case-insensitive comparisons, so LOINC names are always stated using uppercase characters. This strategy has problems when capitalization is used to distinguish subtypes in a given domain, as with red cell antigens. For example, r and R represent different rhesus blood group antigens. We chose to represent the lowercase characters using the key word LITTLE, so r became LITTLE R. In naming components, it was decided that the substance name should come first, like “Hepatitis A antibody,” not “Antibody, Hepatitis A.” Another convention related to the use of anti-: “Anti-hepatitis A antibody,” became just “Hepatitis A antibody,” since the anti- prefix is redundant with the meaning of antibody itself. Similarly, conventions were adopted to deal with anion and acid forms of compounds, the naming of alcohols, noun and adjectival forms of tissues, Greek letters in names, hyphens in names, and others. Where possible, we adopted conventions recommended by IUPAC or other national or international standards groups. More than 20 conventions related to capitalization, word order, punctuation, and other strategies for normalizing LOINC names are specified in the LOINC manual.
The second set of problems related to the insufficiency of the simple model. Although a large number of laboratory results are adequately distinguished by the four-component model, many results are not. For instance, most laboratory systems distinguish creatinine concentrations measured on a 24-hour sample from those measured on a one-time (spot) sample. To handle the set of measurements in which this distinction is important, timing was moved from being a part of the component to being an independent axis. Hence, our second model looked like this:
Model 2
<component>:<timing>:<specimen>:<precision>:<method>
Example: HEPATITIS B VIRUS SURFACE AG:PT:SER:SQ:EIA
The timing axis has possible values of :PT (at a point or moment in time), 24 hours, 12 hours, 4 hours, etc. Timed is a special value for the timing axis where the exact duration of collection is sent as a separate part of the result message.
Chemical Subspecies and Kind-of-Property
Given the new model, the process of examining data continued, and other problems were encountered. It was important to distinguish a substance that was “free” in solution from the same substance that was protein-bound, so a notation was adopted for the component part of the name that allowed subspecies to be specified. For example, free ionic calcium is named “calcium.free.” Recognizing that the context in which a measurement was made might have a broader scope than just the laboratory (and following the lead of IUPAC) we renamed “specimen” as “system.”
It also became important to distinguish the kind-of-property of a given component that was being measured. For example, in urinalysis it is important to distinguish the measurement of the concentration of sodium ion in a 24-hour urine sample (typically reported in the United States as grams per liter) from the measurement of the total amount of sodium excreted in urine by the patient (typically reported in grams per 24 hours). In the first case, the property of sodium ion being measured is the mass of sodium ion per unit volume of urine (a mass concentration, MCNC), whereas in the second case the property being measured is the rate of sodium ion excretion in urine (a mass rate, MRAT). In order to make these kinds of distinctions in the LOINC fully specified name, the kind-of-property being measured was moved from being a subpart of the component to being an independent axis in the model. Given these additions, a third version of the model could be represented as:
Model 3
<component>.<subspecies>:<property>:<timing>:<system>:<precision>:<method>
Example: HEPATITIS B VIRUS CORE AB.IGM:ACNC:PT:SER:QN:EIA
Challenge Tests and Observation Methods
As we continued to create result names using the model, the inherent complexity of challenge tests was better understood. Examples of challenge tests are glucose tolerance tests, insulin tolerance tests, and dexamethasone suppression tests. In these kinds of procedures, the common feature is that some intervention takes place (often the administration of a drug, hormone, or nutrient) and at some point in time after the intervention one or more measurements are taken to evaluate the response of the patient to the intervention. The important consideration for the fully specified LOINC name is that we need to indicate in the name, as precisely as possible, what the intervention was and the temporal relationship of the measurement to the intervention. We decided to express this as a separate part of the component name. The carat (^) was chosen to delimit the challenge information as a subpart of the component name. Again, the choice of delimiters is for convenience only and has no semantic importance for the standard. With this addition, the component portion of the name for one measurement in a glucose tolerance test is:
Partial Model
<component>^<chall>
Example: GLUCOSE^30 MN POST 100 GM ORAL GLUCOSE
One of the most problematic and subjective areas addressed by the LOINC model is the question of observation methods. Generally, the method by which a measurement is made is not important, given that the property measured and the precision of the measurement are truly the same. For instance, with serum sodium measurements most laboratories do not report a different result name for sodium measured by ion sensitive electrodes versus sodium measured by flame ionization. However, in other situations, typically those dealing with antigen-antibody reactions, the sensitivity and specificity of the measurement may be dramatically different depending on whether the method is a latex agglutination method or a flocculation method. Addressing this need, we modified the rules related to the specification of methods in the LOINC name. The method axis is used only when information specified in the other five axes is insufficient for distinguishing clinical measurements that have very different reference ranges, sensitivities, or specificities. Whether the method should be stated or not can usually be decided by answering the question, “Would clinicians caring for a patient want to see observations made by this method in the same column on a clinical report as those made by a different methodology, or would they want it displayed in a separate column?” If the clinicians think of it as the same thing and if it can be compared with similar measurements made by other methods to track a physiologic variable over time, then a distinct name incorporating the method should not be made. If clinicians think about it as different, because of a very different reference range, sensitivity, or specificity, then the measurement should have a name that distinguishes the method.
With the addition of the challenge information (and a better understanding when to use the method field), the final informal representation of the model is:
Model 4
<component>.<subspecies>^<chall>:<property>:<timing>:<system>:<prec>:<method>
Example: GLUCOSE^30 MN POST 100 GM ORAL GLUCOSE:MCNC:PT:SER:QN
This iterative process of examining data, matching to the model, and modifying the model when necessary is the core process in creating the LOINC vocabulary. Over the course of 18 months, the LOINC model became progressively more sophisticated (complex), and it continues to evolve to the present day. The complexity is not an artifact of poor design; it is a direct result of trying to systematically distinguish different results that exist in real-world laboratory and clinical information systems. The complexity comes from trying to closely model the real-world measurements that we are describing. The complete model is more complex than we have presented here. In a later section, we describe the representation of the LOINC SDM using a formal notation. The complete model is available at the LOINC Web site at http://www.mcis.duke.edu/standards/termcode/loinc.htm/.
Policies for Creating LOINC Names Using the Model
Once a reasonably robust model was in place, policies and procedures were instituted that made creation of names using the model consistent. One such policy was that no parts of the name would be implied. For example, because most laboratory procedures are measurements at a point in time, it is common to mention the timing aspect of a measurement only if it involves a 12-hour or 24-hour sample collection. In other words, the timing aspect is implied to be “point in time” if not otherwise stated explicitly. In LOINC naming, we chose to always be explicit and say “point in time” in every name where it applies. This kind of consistency facilitates automated matching and comparison of LOINC to other vocabularies.
A second convention was the use of the virgule (/) and the plus sign (+) in names. The virgule is used to represent a Boolean OR. For example, if a given measurement can be made on serum or plasma and has the same clinical significance regardless of which type of specimen is used, then the system part of the name would be SER/PLAS. The plus sign is used to represent a Boolean AND. It is commonly used in the component part of the name when a given procedure measures more than one chemical species. For example, if a procedure measures both doxepin and desdoxepin, the component part of the name is represented as DOXEPINE+DESDOXEPINE.
A third policy or practice was that we would create only names that exist in real systems. We would not create names based on allowable permutations of the six axes within the LOINC name. This policy is necessary to avoid combinatorial explosion, since the potential name space based on arbitrary combinations would be huge. Other policies related to name creation are described in detail in the LOINC manual.
The final step in establishing a LOINC term is creation of a unique code for each fully specified name. The LOINC code is a sequentially assigned unique number that has no embedded meaning. The number of digits in the code is flexible and will increase as the number of items in the LOINC vocabulary increases. A MOD10 check digit is included as a hyphen-separated suffix as part of the LOINC code. The check digit can be used to check for common typographic errors when LOINC codes are entered manually. Details of the MOD10 check digit calculation are available in the LOINC manual.
The LOINC Model in a Formal Notation
One of the strengths of the LOINC terminology is its underlying SDM. A previous publication describes an earlier version of the SDM.21 A current version using Abstract Syntax Notation One (ASN.1) is available at http://www.mcis.duke.edu/standards/termcode/loinc.htm. The document also includes a brief description of ASN.1 syntax. ASN.1* is an international standard for describing abstract syntax (data models).
The main objective of having a formal model is to make explicit all the discrete domains that are used by the different axes of the LOINC fully specified names. The identification of the discrete domains helps categorize the concepts used by the LOINC names and will enable the LOINC names to be expressed using other natural languages and coding schemes.21 This functionality does not affect the operational aspect of information systems adopting LOINC, because only the LOINC codes are necessary to exchange data. The ASN.1 specification does provide a computable model that can be used to support automated processes that map local vocabularies to the LOINC database.
According to Rossi Mori et al.,45 LOINC has the characteristics of a “second-generation” terminology system. These include a categoric structure (identified as the SDM) and a well-defined set of descriptors (collections of terms for the various LOINC name axes). The ASN.1 version of the LOINC SDM helps refine the categoric structure of the laboratory result names and facilitates the utilization of a more elaborate vocabulary model. Both aspects are crucial for the transformation of the LOINC vocabulary into an extensible and compositional terminology system. The envisioned terminology system should ensure the canonic representation of the fully specified LOINC names and make LOINC compatible with other clinical models.
In its current form, the LOINC ASN.1 SDM is still an incomplete model, since it is not sufficiently detailed to allow only reasonable and sensible names to be created. The effort to create a more detailed model will require a more “atomic” vocabulary, similar to EUCLIDES,40 and the stronger semantics available in GRAIL.46 The level of detail present in any model is determined by the purpose for which the model was created. Our goal is to use the LOINC ASN.1 SDM to facilitate discussions of the LOINC approach, to automate semantically based matching processes, and to assist users in the creation of new names.
Distribution of and Additions to the LOINC Vocabulary
The LOINC vocabulary is maintained as a single table, with the formal parts of the LOINC name and the LOINC code being separate columns in the table. Other columns in the table are used for maintaining cross-references to other vocabularies and for managing maintenance information (who, when, and why) about additions and deletions. The complete specification of the LOINC table structure is available in the LOINC manual.
The first release of the LOINC vocabulary was ready for external comment and review in March 1995. ▶ shows dates and the number of terms in each LOINC release over the last two years. The two largest growth periods for the LOINC vocabulary (after the initial load) are represented by version 1.0h, which followed the first widespread implementations of LOINC in working systems, and version 1.0i, which was the first version to include terms used outside the clinical laboratory. In accordance with initial goals, the LOINC vocabulary is distributed free of charge on diskette, or it can be downloaded via anonymous ftp over the Internet. It is available on the standards server at Duke University Medical Center:
Table 1.
Growth of the LOINC Vocabulary
| Version | Date | No. of Terms |
|---|---|---|
| 1.0 | 04/24/95 | 5,905 |
| 1.0a | 05/24/95 | 5,906 |
| 1.0b | 06/23/95 | 5,927 |
| 1.0c | 06/28/95 | 5,929 |
| 1.0d | 07/14/95 | 6,295 |
| 1.0e | 09/15/95 | 6,296 |
| 1.0f | 12/21/95 | 6,490 |
| 1.0g | 04/28/96 | 6,711 |
| 1.0h* | 08/21/96 | 8,458 |
| 1.0i† | 01/08/97 | 10,773 |
Version 1.0h had increased content based on input from laboratories as the vocabulary was put into clinical service.
Version 1.0i was the first version to include terms outside clinical laboratory use.
The portion of the LOINC vocabulary that pertains to clinical laboratory measurements is maturing rapidly. We have recently adopted policies and procedures for submitting requests for new laboratory terms. The strategy is to reduce the work of maintaining the vocabulary by requiring that submitters provide accurate information about the items to be added. A summary of the rules is found in the LOINC manual. If these rules are followed, the goal of the LOINC committee is to add new codes within two weeks after receipt of a request.
Also available from the Web site is RELMA, a program that helps users map their local vocabulary to LOINC terms. The RELMA program accepts a file of local result names as input and then interactively assists the user in locating LOINC terms that are possible matches.
Implementing and Evaluating the LOINC Vocabulary
The LOINC codes have been adopted as standard identifiers by several vendors of laboratory information system software and by several large commercial laboratories, including Quest Diagnostics (formerly Corning Clinical Laboratories), LabCorp, LifeChem, and ARUP (Associated Regional and University Pathologists). LOINC is also used by the U.S. Veteran's Administration, the U.S. Navy, Kaiser Permanente, Clarion of Indianapolis, and Partners of Boston. It has been adopted as a national standard in New Zealand, and the Province of Ontario is using it for a pilot study. Additionally, LOINC is the basis for the clinical identifier used in the 3M Health Information System Lifetime Data Repository. It has also been endorsed by the American Clinical Laboratory Association and the Andover Working Group for OHI (Open Healthcare Interoperability). While use provides some strong evidence of merit, formal evaluations of the LOINC vocabulary are just beginning, and published reports are not currently available.
For purposes of evaluation, the potential utility of LOINC centers on two propositions: the use of LOINC as a universal coding scheme, and the use of LOINC to improve mapping between systems. Concerning the first, LOINC is designed to provide a universal coding system to which users can map their existing master files. Without a universal vocabulary, mapping can only occur point-to-point between each communicating system. This means that if there are N systems that are interconnected, N × (N - 1) mappings will need to be done, as noted by Simborg et al.47 more than ten years ago. However, if each institution can map to a universal vocabulary, each system would need to map only once, resulting in a tremendous reduction in the total number of mappings that need to be done. Of course, the same benefit would accrue to the use of any universal vocabulary. Tests of the first proposition, then, should focus on how well LOINC serves as a universal vocabulary within its scope of coverage.
The question of whether LOINC is a suitable universal vocabulary relates directly to adequate converage. Despite the initial consideration of various clinical laboratory vocabularies as sources of terms and the actual size of the LOINC vocabulary (more than 10,000 terms), there may be subdomains not completely represented in LOINC. We recognize that the LOINC vocabulary will be ever growing, but we expect to reach a stage at which the only maintenance of the corpus is due to new laboratory procedures and refinements on the existing names. Evaluations of LOINC coverage would be useful in focusing future work on areas where greater coverage is needed.
The second proposition is that the use of LOINC can improve the process of mapping. Improvements could be in the form of greater accuracy in mapping or decreased time and costs. These benefits could be realized whether the mapping were done manually or by an automated process. Evaluations of the second proposition then would focus on the speed, efficiency, and accuracy of mapping between systems using LOINC.
Evaluations of the second proposition could be rephrased by the question, “Is the fully specified LOINC name really fully specified?”—that is, is there sufficient information in the LOINC name to match laboratory result names across heterogeneous systems. The LOINC data model was designed to include all the attributes of a laboratory result name needed to create an explicit and unambiguous meaning for each term. There are at least two possible error sources in manual mapping: human errors related to simple typographic or selection errors, and errors caused by inadequacy of the LOINC names. In the second case, errors might occur because the fully specified LOINC name may not be sufficiently explicit to unambiguously distinguish different procedures, or there could be systematic or random errors in creating the LOINC names that make the names difficult to understand or use. One approach to answering this question may be to wait until LOINC is in use in production systems. At that stage, data in production databases can be pooled and statistical comparisons can be made to determine whether a LOINC code in one system has the same meaning as a LOINC code in another system. If the meanings of LOINC codes are found to be different in the different systems, it will be important to analyze the sources of the error. Analysis of errors that are attributable to LOINC itself should lead to improvements in the LOINC structure and would be directed at removing any remaining ambiguity in the fully specified name.
A closely related issue is whether the LOINC approach is clear and implementable. At the present time it takes approximately four person-months to completely map a complex laboratory. It may be that the process is too complex and is not reproducible when used by people outside of the LOINC committee. The existence of the SDM, standard naming conventions, and standard abbreviations as developed by the LOINC committee should facilitate the elaboration of new names. The main issue may be the understanding and interpretation of the properties and methods used by LOINC. Despite the fact that the properties and methods were obtained from “standard” sources, their interpretation and use may not be clear to the end user. The LOINC committee may have to educate end users on how to interpret and apply the various properties and methods adopted, and perhaps even publish a set of rules and examples that will guide them.
Another issue related to the creation of new result names is the submission of these terms to the LOINC committee. The centralized control adopted by LOINC is certainly necessary, but the potential overhead to review and advise end users may require a more elaborate people and database infrastructure than is currently in place. The collaboration of the users in this process will be very important, and the various interpretations that can be made regarding the specimens, kinds-of-property, and methods will have to be progressively standardized and disseminated to all users. The process of adding new LOINC terms will need to be evaluated frequently to ensure that the needs of end users are being met in a timely fashion.
Discussion
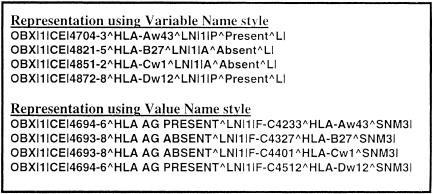
As we created the LOINC vocabulary we recognized problems that we were not solving. The process of trying to apply a rigorous methodology to name creation exposes issues that were previously hidden. One key issue was the recognition of alternative styles of representing data in messages. An example of the two alternative styles—which we have called the “variable” and “value” styles—is shown in ▶. The “variable” representation treats the concept as a variable (or field) that has a binary value, e.g., HLA B27 Antigen = Present. This style is usually used when there is a panel of such variables, each of which will be tested for and reported as present or absent. In the case of “value” representation, the variable is a more open-ended one and the value of the field is the antigen that was found, e.g., Antigen Found = HLA B27. Value representation is often used when testing to establish the HLA type of a person and when only the types of the antigens found will be reported. The use of the variable- and value-type styles of results-reporting is common in blood bank testing and microbiology cultures as well as in HLA typing.
Figure 7.
Variable-style names versus value-style names in HL7 OBX segments. The “variable” representation treats the concept as a variable (or field) that has a binary value, e.g., HLA B27 Antigen = Present. This style is usually used when there is a panel of such variables, each of which will be tested for and reported as present or absent. In the case of “value” representation, the variable is a more open-ended one and the value of the field is the antigen that was found, e.g., Antigen Found = HLA B27. Value representation is often used when testing is being done to establish the HLA type of a person, and only the types of the antigens found will be reported. The use of the variable- and value-type styles of results reporting is common in blood bank testing and microbiology cultures as well as HLA typing.
We have intentionally included both variable and value style names in the LOINC vocabulary, since both styles are in common use in systems today. Allowing both styles, however, points to the need for further standardization of the information model associated with medical data. Having both styles means that the same kind of result data coming from two different systems could have different representations, even though they are both using HL7 messages. This leads to either inconsistent representation of the data in patient databases or to increased complexity in computer-to-computer interfaces. These problems could be eliminated by adopting a single style of representation for HL7 interfaces or by making an explicit map between the synonymous representations. The goal would be to make user interfaces and HL7 interfaces smart enough that they would recognize the data in either form and be able to convert it to the most useful representation based on the clinical circumstances.
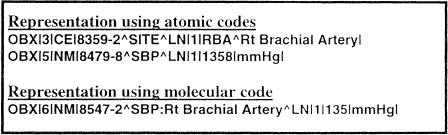
A second issue became apparent as we began to examine patient data outside the clinical laboratory. It became clear that different systems used different levels of aggregation (pre-coordination) in the names used to describe clinical measurements. As shown in ▶, some systems send the location of a given measurement as a separate observation in the message, whereas other systems express this information as part of a single name. For example, a systolic blood pressure of 135 mmHg measured in the right brachial artery could be represented as two separate observations: a body location observation with a value of right brachial artery and a second observation of systolic blood pressure with a value of 135. Alternatively, this same information could be represented as a single observation of systolic blood pressure in the right brachial artery with a value of 135. Again, these two styles of representation could lead to inconsistent representation of data in a patient database or to added complexity in computer-to-computer interfaces. The process of creating LOINC names has exposed these issues of alternative representation, enabling discussions that we hope can lead to further standardization of clinical data representations in medicine.
Figure 8.
Use of atomic (post-coordinated) codes versus a molecular code (pre-coordinated) in HL7 messages. Some systems send the location of a given measurement as a separate observation in the message, whereas other systems express this information as part of a single name.
The development paradigm of the LOINC vocabulary presents some interesting contrasts to the development paradigm of EUCLIDES and IUPAC. Systems like EUCLIDES and IUPAC were designed from a rather theoretic and wholesome viewpoint, proceeding where possible from well-established first principles and definitions. The systems were then tested in common implementations with limited real-world success because of the inherent complexity of implementing a multi-axial coding scheme in today's clinical information systems. LOINC came the other way around. It grew from a practical implementer's point of view, creating terms at the granularity usually found in working systems and gaining complexity as it grew without losing touch with common implementations. The danger of the LOINC approach is that it walks along the cliff of “combinatorial explosion.” If LOINC codes were created for all possible (or even sensible) combinations of the 42 EUCLIDES axes, there would be an unmaintainable number of terms. Combinatorial explosion is prevented by creating LOINC codes that only correspond to codes in real systems and by putting the other essential information in other fields of a message. Thus, the LOINC strategy is to create messages that are a careful balance between a pre-coordination approach (represented by using LOINC codes as observation identifiers) and a post-coordination approach (represented by the other ancillary and modifier fields present in the message).
Current/Future Activities
The content of the LOINC vocabulary continues to grow in the area of clinical laboratory measurements and especially in the area of direct patient measurements and observations. The LOINC vocabulary has recently been included in the UMLS Metathesaurus, and discussions are underway that should allow LOINC to be included in a future version of SNOMED International. Several clinical information systems are now using the LOINC vocabulary, and many other implementations are in progress. Suggestions from end users should lead to enhancements in both the structure and content of the LOINC vocabulary.
Conclusion
The accurate exchange of clinical data between computer systems requires the combination of an SDM and a vocabulary. The SDM provides structure and context that are essential for the correct interpretation and understanding of the terms that the structure contains. The creation of the LOINC vocabulary has lead to a greater understanding of the interdependency of the SDM and the vocabulary. It is an example of one of the first vocabularies specifically targeted for use in a data exchange standard. The experience of the LOINC committee in creating fully specified names for observations can serve as a useful foundation for other groups that might undertake the development of vocabulary for use in data exchange standards. Having the specific goal of creating names for observation identifiers for use in health care data exchange standards has been essential for focusing the work of the committee. Because the LOINC committee is relatively small, it has been able to make rapid progress, creating more than 10,000 terms in less than three years. The vocabulary has gained wide acceptance because it has useful content that is not found in other vocabularies and because it is freely available on the Internet. Formal evaluations of the content and structure of the LOINC vocabulary have not yet been published but would be very useful in improving the LOINC data model and contents.
Acknowledgments
The authors thank Henrik Olesen, Chairman of IUPAC, Commission on Quantities and Units in Clinical Chemistry, for his helpful comments and insights about laboratory test coding.
This work was supported in part by the John A. Hartford Foundation of New York, by contract NO-1-LM-3-3410 from the National Library of Medicine, and by grants HS 05626 and HS 07719-013 from the Agency for Health Care Policy and Research. Much of the work was performed under the auspices of the Regenstrief Institute.
Footnotes
International Organization for Standardization, 1990 #28; International Organization for Standardization, 1990 #222.
References
- 1.Forrey AW, McDonald CJ, DeMoor G, et al. Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem. 1996;42: 81-90. [PubMed] [Google Scholar]
- 2.Evans DA, Cimino JJ, Hersh WR, Huff SM, Bell DS, for the Canon Group. Toward a medical-concept representation language. J Am Med Inform Assoc. 1994;1: 207-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman C, Huff SM, Hersh WR, Pattison-Gordon E, Cimino JJ. The Canon Group's effort: working toward a merged model. J Am Med Inform Assoc. 1995;2: 4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dick RS, Steen EB. The Computer-based Patient Record: An Essential Technology for Health Care. Washington, DC: National Academy Press, 1991. [PubMed]
- 5.Huff SM, Cimino JJ. Medical data dictionaries and their use in medical information system development. In: Prokosh HU, Dudek J (eds): Hospital Information Systems: Design and Development Characteristics; Impact and Future Architecture. Amsterdam, The Netherlands: Elsevier, 1995: 53-75.
- 6.Sitting DF. Grand challenges in medical informatics? J Am Med Inform Assoc. 1994;1: 412-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Côté RA, Rothwell DJ, Palotay JL, Beckett RS, Brochu L. The Systematized Nomenclature of Human and Veterinary Medicine: SNOMED International. Northfield, Ill: College of American Pathologists, 1993.
- 8.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993;32: 281-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Neil MJ, Payne C, Read JD. Read codes, version 3: a userled terminology. Methods Inf Med. 1995;34: 187-92. [PubMed] [Google Scholar]
- 10.Kuperman GJ, Gardner RM, Pryor TA. HELP: A Dynamic Hospital Information System. New York: Springer-Verlag, 1991.
- 11.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J Am Med Inform Assoc. 1994; 1: 35-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rector AL, Nowlan WA, for the GALEN Consortium. The Galen project. Comput Methods Programs Biomed. 1994;45: 75-8. [DOI] [PubMed] [Google Scholar]
- 13.American Society for Testing and Materials. Guideline for Construction of a Clinical Nomenclature for the Support of Electronic Health Records. West Conshohocken, Pa: ASTM, 1996. Publication ASTM E1284-96.
- 14.Rector AL, Nowlan WA, Kay S. Foundations for an electronic medical record. Methods Inf Med. 1991;30: 179-86. [PubMed] [Google Scholar]
- 15.Rector AL, Nowlan WA, Kay S, Goble CA, Howkins TJ. A framework for modelling the electronic medical record. Methods Inf Med. 1993;32: 109-19. [PubMed] [Google Scholar]
- 16.Rector AL, Glowinski WA, Nowlan WA, Rossi-Mori A. Medical-concept models and medical records: an approach based on GALEN and PEN&PAD. J Am Med Inform Assoc. 1995;2: 19-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.American Society for Testing and Materials. Standard Practice for an Object-oriented Model for Registration, Admitting, Discharge, and Transfer (RADT) Functions in Computer-based Patient Record Systems. West Conshohocken, Pa: ASTM, 1995. Publication ASTM E1715-95.
- 18.Bell DS, Pattison-Gordon E, Greenes RA. Experiments in concept modeling for radiographic image reports. J Am Med Inform Assoc. 1994;1: 249-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huff SM, Rocha RA, Bray BE, Warner HR, Haug PJ. An event model of medical information representation. J Am Med Inform Assoc. 1995;2: 116-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pattison-Gordon E, Greenes RA. An empirical investigation into the conceptual structure of chest radiograph findings. J Am Med Inform Assoc. 1994;1: 257-61. [PMC free article] [PubMed] [Google Scholar]
- 21.Rocha RA. Development and evaluation of a semantic data model for chest radiology. Department of Medical Informatics. Salt Lake City, Utah: University of Utah, 1996: 268.
- 22.European Committee for Standardization (CEN), Technical Committee 251 (Health Informatics). Messages for Exchange of Clinical Laboratory Information. Brussels, Belgium: CEN TC251, 1994. Publication CEN ENV 1631.
- 23.European Committee for Standardization (CEN), Technical Committee 251 (Health Informatics). Request and Report Messages for Diagnostic Service Departments. Brussels, Belgium: CEN TC251, 1994. Publication CEN TC 251/PT022.
- 24.Health Level Seven, Inc. Health Level Seven, Standard Version 2.3: An Application Protocol for Electronic Data Exchange in Healthcare Environments. Ann Arbor, Mich: Health Level Seven, 1994.
- 25.National Electrical Manufacturers Association. Digital Imaging and Communications in Medicine (DICOM), Supplement 15: Visible Light Image for Endoscopy, Microscopy, and Photography. Rosslyn, Va: NEMA, 1997. Publication NEMA PS 3 Suppl 15.
- 26.National Electrical Manufacturers Association. Digital Imaging and Communications in Medicine (DICOM). Rosslyn, Va: NEMA, 1997. Publication NEMA PS 3.1-PS 3.12.
- 27.National Electrical Manufacturers Association. Digital Imaging and Communications in Medicine (DICOM), Supplement 23: Structured Reporting. Rosslyn, Va: NEMA, 1997. Publication NEMA PS 3 Suppl 23.
- 28.Bidgood WD Jr, Horri SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc. 1997; 4: 199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.American Society for Testing and Materials. Standard Specification for Transferring Clinical Observation Between Independent Computer Systems. West Conshohocken, Pa: ASTM, 1994. Publication ASTM E1238-94.
- 30.American Society for Testing and Materials. Specification for Transferring Information Between Clinical Instruments and Computer Systems. West Conshohocken, Pa: ASTM. Publication ASTM E1394.
- 31.American Society for Testing and Materials. Guide for Description of Reservation/Registration Admission, Discharge, Transfer (R-ADT) Systems for Automated Patient Care Information Systems. West Conshohocken, Pa: ASTM, 1994. Publication ASTM E1239-94.
- 32.American Society for Testing and Materials. Guide for the Functional Requirements of Clinical Laboratory Information Management Systems. West Conshohocken, Pa: ASTM. Publication ASTM E1639.
- 33.Bidgood WD Jr, Horii SC. Introduction to the ACR-NEMA DICOM standard. Radiographics. 1992;12: 345-55. [DOI] [PubMed] [Google Scholar]
- 34.Bidgood WD Jr. Documenting the information content of images. Proc AMIA Annu Fall Symp. 1997: 424-8. [PMC free article] [PubMed]
- 35.Bidgood WD Jr. The SNOMED DICOM microglossary: a controlled terminology resource for DICOM coded entry data elements. In: Chute CG (ed). IMIA Working Group 6. Jacksonville, Fla, 1997.
- 36.Korman LY, Bidgood WD Jr. Representation of the Gastrointestinal Endoscopy Minimal Standard Terminology in the SNOMED DICOM microglossary. Proc AMIA Annu Fall Symp. 1997: 434-8. [PMC free article] [PubMed]
- 37.Rossi Mori A, Galeazzi E, Consorti F, Bidgood WD Jr. Conceptual schemata for terminology: a continuum from headings to values in patient records and messages. Proc AMIA Annu Fall Symp. 1997: 650-4. [PMC free article] [PubMed]
- 38.European Committee for Standardization (CEN), Technical Committee 251 (Health Informatics), Project Team 2. Medical Informatics—Categorical Structure of Systems of Concepts—Model for Representation of Semantics. Brussels, Belgium: CEN TC251, 1995. Publication CEN ENV 12264.
- 39.American Medical Association. Physician's Current Procedural Terminology. Chicago, Ill: American Medical Association, 1991.
- 40.De Moor GJE. Towards a standard for electronic data interchange in laboratory medicine. Ghent, Belgium: University of Ghent, 1994.
- 41.De Moor GJE, Fiers T, Wieme R, Scott R. The research in semantics behind the OpenLabs coding system. Comput Methods Programs Biomed. 1996;50: 169-85. [DOI] [PubMed] [Google Scholar]
- 42.Cimino JJ, Hripcsak G, Johnson SB, Clayton PD. Designing an introspective, multipurpose, controlled medical vocabulary. Proc 13th Symp Comput Appl Med Care. 1989: 513-8.
- 43.Cimino JJ. Desiderata for controlled medical vocabularies in the 21st century. In: Chute CG (ed): IMIA Working Group 6. Jacksonville, Fla, 1997.
- 44.Rigg JC, Brown SS, Dybkaer R, Olesen H. Compendium of Terminology and Nomenclature of Properties in Clinical Laboratory Sciences. Cambridge, Mass: Blackwell Science, 1995.
- 45.Rossi Mori A, Consorti F, Galeazzi E. Standards to support development of terminological systems for healthcare telematics. In: Chute CG (ed): IMIA Working Group 6. Jacksonville, Fla, 1997. [PubMed]
- 46.Rector AL, Nowlan WA. The GALEN representation and integration language (GRAIL) kernel, version 1. In: The GALEN Consortium for the EC. Manchester, England: University of Manchester, 1993.
- 47.Simborg DW. Local area networks: Why? What? What if? MD Comput. 1984;1: 10-20. [PubMed] [Google Scholar]