Abstract
Specialized DNA damage-bypass Y-family DNA polymerases contribute to cancer prevention by providing cellular tolerance to DNA damage that can lead to mutations and contribute to cancer progression by increasing genomic instability. Y-family polymerases can also bypass DNA adducts caused by chemotherapy agents. One of the four human Y-family DNA polymerases, DNA polymerase (pol) κ, has been shown to be specific for bypass of minor groove adducts and inhibited by major groove adducts. In addition, mutations in the gene encoding pol κ are associated with different types of cancers as well as with chemotherapy responses. We characterized nine variants of pol κ whose identity was inferred from cancer-associated single nucleotide polymorphisms for polymerization activity on undamaged and damaged DNA, their abilities to extend from mismatched or damaged base pairs at primer termini, and overall stability and dynamics. We find that these pol κ variants generally fall into three categories: similar activity to wild-type (WT) pol κ (L21F, I39T, P169T, F192C, and E292K), more active than WT pol κ (S423R), and less active than pol κ (R219I, R298H, and Y432S). Of these, only pol κ variants R298H and Y432S had markedly reduced thermal stability. Molecular dynamics (MD) simulations with undamaged DNA revealed that the active variant F192C and more active variant S423R with either correct or incorrect incoming nucleotide mimic WT pol κ with the correct incoming nucleotide, whereas the less active variants R219I, R298H, and Y432S with the correct incoming nucleotide mimic WT pol κ with the incorrect incoming nucleotide. Thus, the observations from MD simulations suggest a possible explanation for the observed experimental results that pol κ adopts specific active and inactive conformations that depend on both the protein variant and the identity of the DNA adduct.
INTRODUCTION
DNA damage is a constant threat, from both endogenous and exogenous sources. DNA adducts can lead to mutations and, in turn, to disease and cell death. Specialized Y-family DNA polymerases (pols) replicate past DNA adducts in a damage tolerance process called translesion synthesis.1 Y-family DNA pols, conserved in all domains of life, are characterized by low fidelity on undamaged DNA and the ability to bypass DNA adducts. Y-family DNA pols bypass specific lesions and have different accuracies and efficiencies of bypass depending on the adduct. Pol κ is one of four Y-family polymerases in humans. Pol κ is able to bypass N2-dG adducts such as N2-furfuryl-dG and N2-(1-carboxyethyl)-dG as well as larger, bulkier adducts like N2-benzo[a]pyrene diol epoxide-dG2–6 and has been shown to be inhibited by major groove adducts.7–10 Pol κ is also able to extend from mispaired primer termini on both undamaged DNA and DNA containing 8-oxo-7,8-dihydrogua-nine and O6-methyl-dG, as well as from a dG:dT mismatch at the 3′-end of a thymine–thymine dimer.11–16
Single nucleotide polymorphisms (SNPs) are differences in base pair positions in genomic DNA between individuals.17 The different base in the DNA could be translated to a mutated protein or cause a truncated protein to be translated. Mutated or truncated proteins could cause disease or cell death. Swett et al. reported several SNPs of pol κ based on a targeted search with the hypothesis-driven SNP search (HyDn-SNP-S) method. HyDn-SNP-S provides a way to search and statistically validate SNPs associated with a given disease phenotype on selected genes using data from genome-wide association studies. The initial search performed by Swett et al. found various SNPs that result in missense mutations on pol κ, however, none of these SNPs were found to be statistically significantly associated with the four cancer phenotypes (lung, breast, melanoma, and prostate) tested in that study.18 Some of these SNPs have been recently subsequently reported: Dai et al., found two noncoding POLK SNPs that were associated with breast cancer risk.19 Shao et al., found several noncoding POLK SNPs linked to platinum chemotherapy toxicity and response as well as progression-free survival in nonsmall cell lung cancer patients.20 Four missense mutations in pol κ are associated with early onset prostate cancer.21 Several POLK SNPs in the dbSNP have been analyzed with in silico prediction tools to determine those that would affect pol κ activity and were biochemically characterized.22,23 Variants L21F, I39T, D189G, R219I, E419G, S423R, and Y432S (in a construct containing pol κ residues 1–526) were biochemically characterized using undamaged DNA and DNA containing the minor groove adduct N2-CH2 (9-anthracenyl)G.23 On undamaged DNA, WT pol κ and the L21F, I39T, D189G, R219I, and S423R variants were all able to extend a primer to the end of the template, while E419G and Y432S variants had much less activity than WT pol κ. WT pol κ is able to perform full-length primer extension with DNA substrates containing N2-CH2(9-anthracenyl)G, as are R219I and S423R variants but not as well as with the undamaged template. L21F, I39T, R219I, D189G, E419G, and S423R variants were much less active on the damaged DNA template.23 In the second study, E29K, T44M, F192C, E292K, R298H, A471V, T473A, and R512W variants were characterized on DNA templates containing N2-CH2(9-anthracenyl)G, 8-oxo-7,8-dihydroguanine (8-oxoG), O6-methyl-G, or an abasic site.22 The SNP variants E29K, T44M, F192C, E292K, and A471V had similar activity as WT pol κ on undamaged DNA, while R298H, T473A, and R512W variants had much lower activity that WT. The same trend was observed with N2-CH2(9-anthracenyl)G, 8-oxoG, O6-methyl-G, or DNA containing an abasic site, except T44M had a decrease in activity opposite every lesion except 8-oxoG.22
In this work POLK SNP variants L21F, I39T, P169T, F192C, R219I, E292K, R298H, S423R, and Y432S (Table 1, Figure 1) were biochemically characterized using undamaged DNA, or DNA containing N2-furfuryl-dG (N2ffdG), N6-furfuryl-dA (N6ffdA), and etheno-dA, for both insertion and extension activities. We chose these lesions because of prior work showing that pol κ prefers N2ffdG and is inhibited by major groove lesions N6ffdA and etheno-dA4,10 and because structural characterization reveals that the presence of N2ffdG does not appreciably perturb the structure or stability of the DNA helix.24 The SNP variants were identified in prior work18 and found in dbSNP.25 It was found that the SNP variants could be grouped into three different categories: those with activity similar to WT (L21F, I39T, P169T, F192C, and E292K), more active than WT (S423R), and less active than WT (R219I, R298H, Y432S).
Table 1.
POLK Single Nucleotide Polymorphisms Studied
| ID | amino acid mutation | domain | tumor site |
|---|---|---|---|
| rs3104729 | L21F | N-clasp | prostate |
| rs3094258 | I39T | N-clasp | prostate, melanoma |
| rs148385845 | P169T | fingers | lunga |
| rs150515841 | F192C | palm | |
| rs3104717 | R219I | palm | prostate |
| rsl42203892 | E292K | palm | |
| rs151251843 | R298H | palm | large intestinea |
| rs35257416 | S423R | little finger (PAD) | melanoma |
| rs77612491 | Y432S | little finger (PAD) | melanoma |
From 1000 genomes.
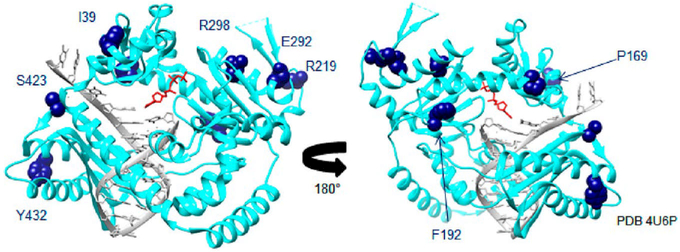
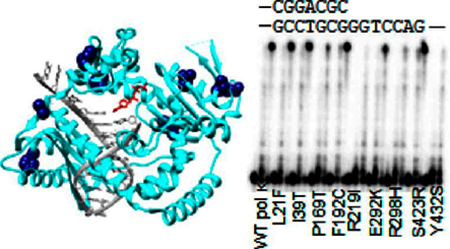
Figure 1.
SNP locations on pol κ crystal structure (PDB 4U6P)59 highlighted in dark blue. DNA is in gray, and the incoming nucleotide is in red. L21 is not shown, as it is not observed in the crystal structure.
EXPERIMENTAL PROCEDURES
Proteins and DNA.
Pol κ (residues 19–526) was expressed and purified as described previously.26 This construct has been shown to have very similar polymerase activity as full-length pol κ.27 Pol κ SNP variants, listed in Table 1, were created by using Quikchange site-directed mutagenesis kits (Agilent). Mutations were confirmed by DNA sequencing (Macrogen, Cambridge, MA; or Eton Bioscience, Charlestown, MA). The DNA template containing a single N2ffdG was prepared as previously described.4,28 The DNA template containing a single N6ffdA was prepared as previously described from O6-phenyl-deoxyinosine (Glen Research).29 The unmodified DNA and etheno-dA templates and primers (running start, standing start, MatchC, MatchG, and MatchT) were from Eurofins Operon. The DNA sequences for templates and primers are in Table 2. DNA was purified by denaturing polyacryalmide gel electrophoresis and the crush and soak method.30 DNA primers (running start, standing start, and MatchC/G/T) were end labeled with32P as previously described.30
Table 2.
DNA Primer and Template Sequences
| DNA | length | sequence |
|---|---|---|
| Running start | 30 mer | 5’GCATATGATAGTACAGCTGCAGCCGGACGC3’ |
| Standing start | 31 mer | 5’GCATATGATAGTACAGCTGCAGCCGGACGCC3’ |
| MatchC | 32 mer | 5’GCATATGATAGTACAGCTGCAGCCGGACGCCC3’ |
| MatchT | 32 mer | 5’GCATATGATAGTACAGCTGCAGCCGGACGCCT3’ |
| MatchG | 32 mer | 5’GCATATGATAGTACAGCTGCAGCCGGACGCCG3’ |
| ddP13 | 13 mer | 5’GCTTGCCGGACGC3’ |
| T18 | 18 mer | 5’CCTAXGCGTCCGGCAAGC3’ |
| Template DNAa | 61 mer | 5’CGTTACTCAGATCAGGCCTGCGAAGACCtXGGCGTCCGGCTGCAGCTGTACTATCATATGC3’ |
Where X = A, G, N2ffdG, N6ffdA, EthenodA; in the case of the N6ffdA template, the T following the modification is replaced with G (indicated with a lower-case t here and in the figures).
Primer Extension Assays.
DNA template (unmodified, N2ffdG, N6ffdA, or etheno-dA) was combined with32P-labeled primer (running start, standing start, MatchC, MatchG, or MatchT) in a 1:1 ratio (500 nM) and annealed with annealing buffer (20 mM HEPES (pH 7.5) and 5 mM Mg(OAc)2) by heating for 2 min at 95 °C, incubating at 50 °C for 60 min, and cooling to 37 °C. Reactions were carried out in 1 × reaction buffer (30 mM HEPES (pH 7.5), 20 mM NaCl, 7.5 mM MgSO4, 2 mM β-mercaptoethanol, and 1% bovine serum albumin), with 100 nM32P-labeled primer/template, 5 nM protein, and 500 μM dNTPs. For misincorporation assays, the final concentrations in 1 × reaction buffer were 100 nM32P-labeled primer/template, 5 nM protein, and 1 mM dATP, dCTP, dGTP, or dTTP. Gels shown are representative of at least three replicates.
Steady-state kinetics assays were carried out using the method of initial rates31 with standing start primer and N2ffdG-containing DNA template, 1 nM (pol κ F192C or pol κ S423R) or 2.5 nM (WT pol κ) enzyme, varying the concentration of dCTP as the incoming nucleotide. The template base is N2ffdG, with the next template base T, and thus only a single C is incorporated into the primer. The dCTP concentration was varied from 1 μM to 100 μM or 200 μM, depending on the variant assayed. Six time points, including the zero-time point, were taken for each [dCTP] concentration. Representative gels are shown in Figure S1 of the Supporting Information. Kinetics parameters represent the average, and errors indicate the standard deviation from three trials analyzed by fits to the Michaelis–Menten equation using GraphPad Prism (Figure S1). DNA was detected using a Storm 860 phosphorimager and quantified using ImageQuant.
Thermal Shift Assays.
Thermal shift assays were conducted using dideoxy-terminated Primer13 (IDT) and undamaged Template18 as well as N2ffdG, N6ffdA, and etheno-dA Template18 DNA (sequences in Table 2). DNA was annealed in the same manner as stated for the primer extension assays except at a final concentration of 75 μM. Protein, at a final concentration of 5 μM, was combined in the presence or absence of DNA (at a final concentration of 10 μM) and dNTP (dCTP or dTTP at 1 mM) in 15 μL of 1 × assay buffer (30 mM HEPES (pH 7.5), 20 mM NaCl, 7.5 mM MgSO4, 2 mM β-mercaptoethanol). The reactions were incubated for 20 min at room temperature before Sypro Orange (20X, Life Technologies) was added. The fluorescence was detected as the temperature was increased from 10 to 90 °C in a Bio-Rad CFX real-time PCR instrument. Values represent the average of at least three trials, and error bars represent the standard deviation.
Molecular Dynamics Simulations.
Molecular dynamics (MD) simulations were performed to investigate the effect of five of the mutations on the structure/function/dynamics of pol κ. All systems, including WT pol κ and five variant structures (F192C, R219I, R298H, S423R, and Y432S), were modeled based on the ternary structure for pol κ (PDB ID: 2OH2) with both thymidine and cytidine triphosphate ligands. The DNA sequence was adjusted to be the same as that used in the experiments but one nucleotide shorter (P12:T17). A large section of 60 residues in the palm domain (224–282) that is unresolved in crystal structures27 was replaced with a three-residue linker of Gln225, Ser226, and Gly281 using Modeller,32 since it was too large to create a reasonable model for this region from sparse structural information. Previous MD simulations of pol κ show that this section of WT pol κ is not present in its homologue, Dpo4, but that the palm structure is still essentially the same even without this region,27,33 and so we conclude that this is a reasonable approximation. The crystal structure was checked and protonated using MolProbity prior to simulation.34 Parameters for the triphosphate ligands were downloaded as fragments from the RED database.35 After running simulations of WT pol κ for the C and T incoming nucleotides, their relaxed structures were mutated with the primary Dunbrack rotamers in Chimera.36,37 Additionally, the R47 residue was identified in Chimera to be very close to other residues and was manually changed to its primary Dunbrack rotamer to alleviate clashes.
Each system was neutralized to a net charge of 0 with K+ ions using AMBER’s tleap program.38 The systems were then solvated using a TIP3P water box extending at least 12 Å from the surface of the protein–DNA complex.39 All MD simulations were performed with pmemd.cuda from the AMBER16 software suite using the ff14SB force field for the protein and OL15 force field for the DNA.40–43 The Berendsen thermostat was used for production in the NVT44 SHAKE was applied to bonds involving hydrogen, and long-range Coulomb interactions were treated with the smooth particle mesh Ewald method, with an 8 Å cutoff for nonbonded interactions.45 Individual simulations were run in triplicate for 100 ns (~3 μs in total) with a 2 fs time-step.
AMBER’s cpptraj program was used to calculate RMSD, RMSF, hydrogen bonding, and correlational analyses. VMD was used for principal component (PCA) and normal mode (NMA) analyses by means of the ProDy interface using 5000 snapshots from each trajectory to calculate 100 PCA modes.46,47 NCIPlot was used to display the noncovalent interactions for the Y432S mutant.48
RESULTS
Strategy.
Our goal in this work was to characterize the primer extension of pol κ variants whose amino acid changes were inferred from cancer-linked SNPs. We began with a set of nine cancer-associated SNP variants from a prior analysis where any SNP found in four specific cancer phenotype databases associated with DNA polymerase genes was determined and tested for statistical significance (Table 1).18 However, in our initial study, none of the SNPs obtained for POLK were found to be statistically significantly associated with disease status for the tested databases (lung, breast, prostate, and melanoma, dbGaP access request #1961).18,49–56During the course of this work, several of these SNPs were added to cancer databases, and characterization of some of the variants mainly with DNA containing dG adducts was reported.22,23 Here, we characterized the nine cancer-associated SNP variants of pol κ for their primer extension and mismatch extension activities on both preferred minor-groove DNA damage and nonpreferred major-groove DNA damage, determined overall stability, and characterized their dynamics with simulations.
Some SNP Variants Are as Active as or More Active than WT Pol κ.
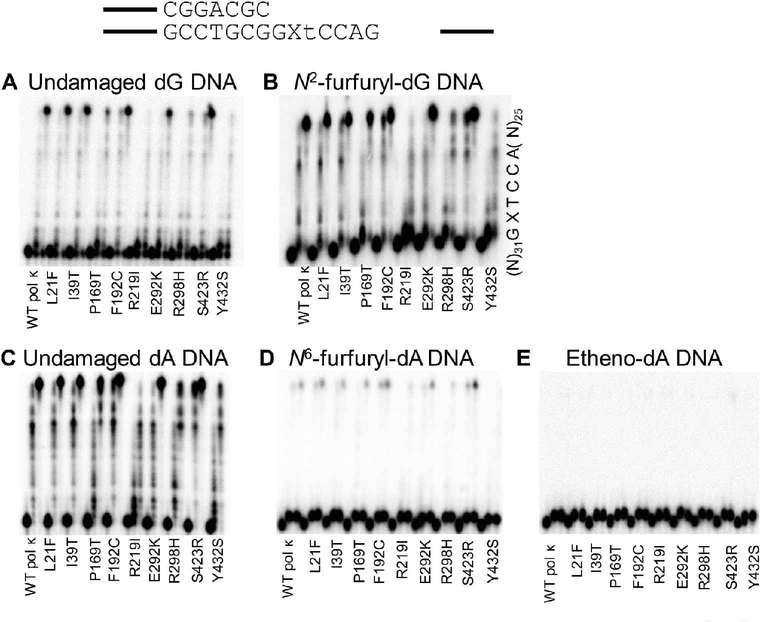
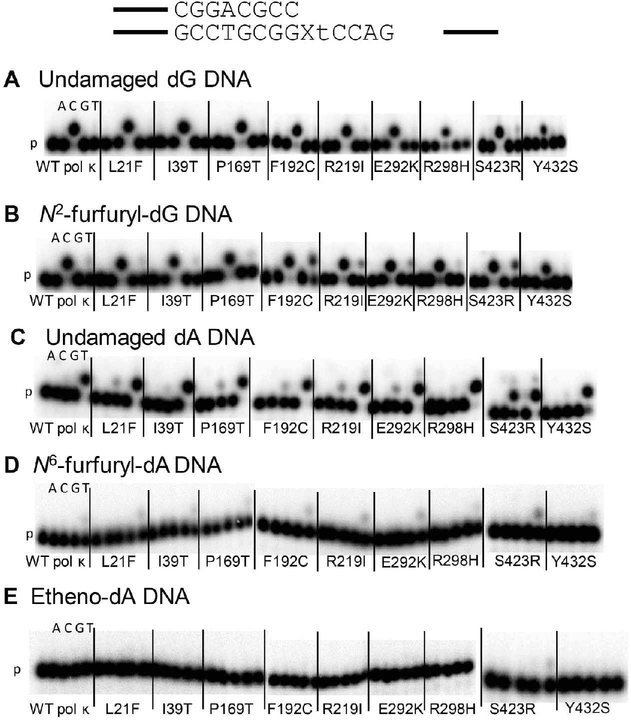
Using running start primer, we assayed the insertion and extension ability of WT pol κ and SNP variants on undamaged DNA (dG and dA) and N2ffdG, N6ffdA, and etheno-dA damaged DNA templates. We have previously shown that WT pol κ is not as active on DNA templates harboring N6ffdA as it is on an undamaged DNA template.10,57 SNPs encoding pol κ variants L21F, I39T, P169T, F192C, E292K, and S423R have similar activity as WT on the undamaged template (Figure 2, Table S1). The remaining variants, R219I, R298H, and Y432S, have markedly lower activity than WT pol κ. A similar trend is observed for both of the furfuryl-damaged templates, those containing N2ffdG or N6ffdA. With both major groove adducts, N6ffdA and etheno dA, WT pol κ and the variants are able to insert efficiently one nucleotide before the lesion but very poorly extend the newly generated primer to the end of the template, unlike the activity with minor groove N2ffdG or undamaged templates (Figure 2).
Figure 2.
Pol κ SNPs retain similar running start primer extension activity as WT pol κ on undamaged DNA templates (A, C) as well as both minor groove (B) and major groove (D, E) DNA adducts with the exception of R219I, R298H, and Y432S, which are markedly less active than WT on all templates. Time points are 0, 20, and 60 min for each variant; quantitation is reported in Table S1.
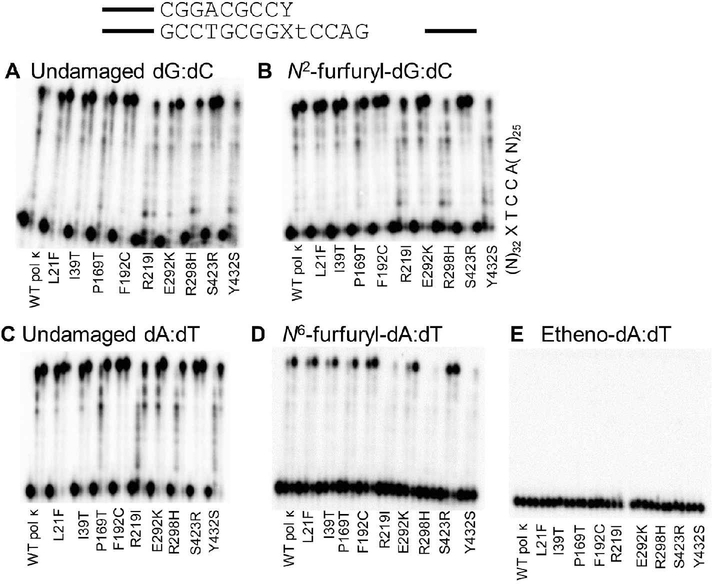
SNP Variants Can Extend Primers Annealed to Damaged Templates with the Correct Base Pairing.
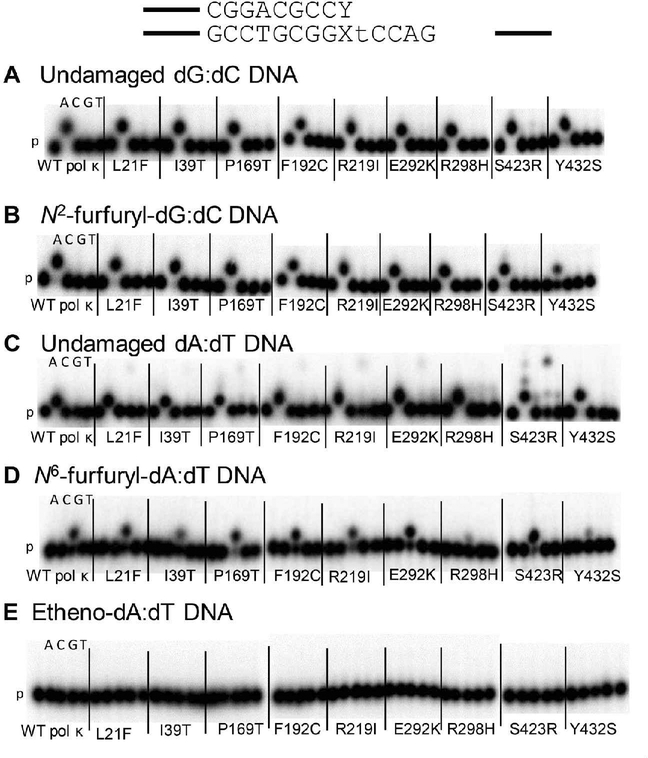
We assayed the extension ability of WT pol κ and SNP variants on damaged DNA templates with a primer that creates the correct Watson–Crick base pair with the damaged base (dC opposite N2ffdG and dT opposite N6ffdA and etheno-dA). There is an increase in extension of primers that form terminal base pairs containing a damaged template base (Figure 3, Table S2) compared to insertion and extension using running start primer (Figure 2). Pol κ F192C and S423R variants show an increase in primer extension ability on templates with either N2ffdG or N6ffdA compared to WT pol κ. Steady-state kinetics analysis with these two variants and standing-start primer revealed an increase in the effciency of insertion of dCTP opposite N2ffdG for S423R but not F192C relative to WT pol κ (Table S3 and Figure S1). Although WT pol κ is known to act as an extender in TLS,11–13,15 etheno-dA remains a blocking lesion for pol κ as an extender since there is no primer extension from the correct etheno-dA:dT base pair.
Figure 3.
Pol κ SNPs retain similar extension activity from the correct base pair as WT pol κ on undamaged DNA templates (A, C) as well as both minor groove (B) and major groove (D, E) DNA adducts with the exception of R219I, R298H, and Y432S, which are less active than WT on all templates. Time points are 0, 20, and 60 min for each variant; quantitation is reported in Table S2.
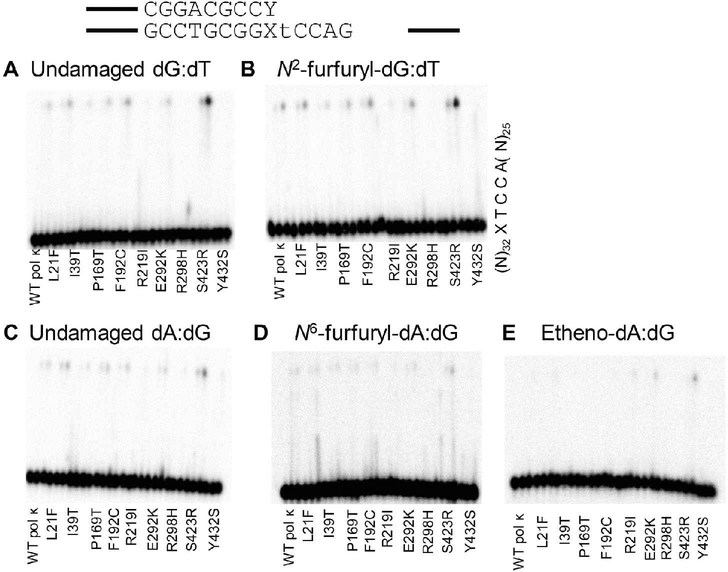
SNP Variants Can Extend from Mismatched Termini on Damaged Templates.
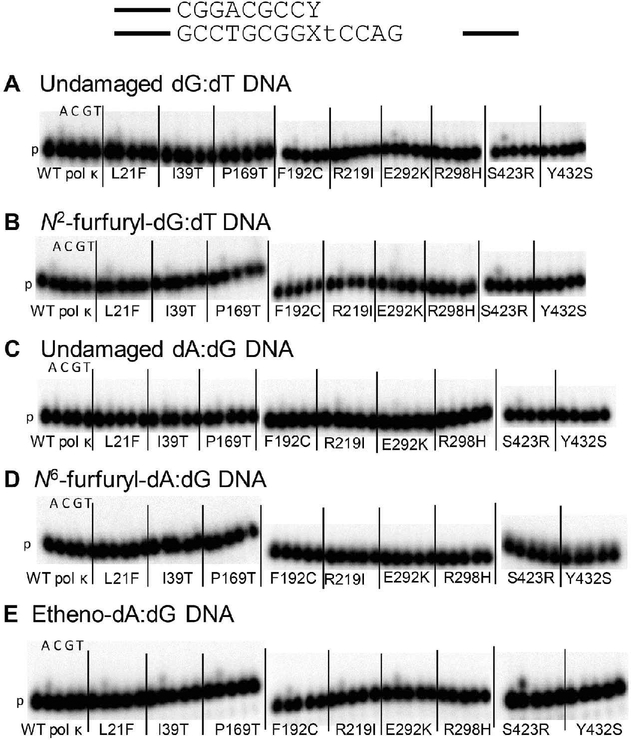
We next assayed the ability of the WT pol κ and the variants to extend primers containing mismatches at the damaged template base (N2ffdG:dT, N6ffdA:dG, or etheno-dA:dG and undamaged controls) (Figure 4, Table S4). It has been shown previously that WT pol κ can extend from a G:T primer terminus mismatch.11–13,15 In general, slightly more primer extension is observed from the N2ffdG:dT mismatch than the undamaged dG:dT mismatch (Figure 4). Pol κ F192C and S423R show more primer extension than WT pol κ from the N2ffdG:dT mismatch. The two proteins that have the weakest primer extension activity with running start primer, pol κ R298H and Y432S (Figure 2), have very little visible primer extension activity from the dG:dT mismatch on either template, those containing undamaged dG or N2ffdG (Figure 4). As with undamaged dG:dT, there is little extension activity from the A:G mismatch. WT pol κ and variants exhibit even less primer extension activity on DNA templates containing the N6ffdA major groove adduct (Figure 4D) relative to dA:dG, as compared to the slight increase in activity observed with the minor groove N2ffdG adduct relative to dG:dT (Figure 4B). WT pol κ and most variants show similar primer extension activity with an etheno-dA:dG mismatch at the primer terminus as compared to the undamaged dA:dG mismatch. The pol κ S423R variant has more extension activity in general compared to WT pol κ, whereas pol κ L21F, R219I, and R298H variants have poor primer extension activity on DNA containing etheno-dA:dG at the primer terminus compared to WT pol κ.
Figure 4.
Pol κ SNPs retain similar extension activity from mismatched base pairs as WT pol κ on undamaged DNA templates (A, C) as well as both minor groove (B) and major groove (D, E) DNA adducts with the exception of R219I, R298H, and Y432S which are less active than WT on all templates. Pol κ S423R, on the other hand, shows more primer extension from both the dG:dT mismatches and the ethenodA:dG mismatch as compared to WT. Time points are 0, 20, and 60 min for each variant; quantitation is reported in Table S4.
SNP Variants Have Similar Nucleotide Incorporation Pattern as WT Pol κ Opposite Lesions.
On an undamaged template G, WT pol κ incorporates dC, and the only variant to show dT misincorporation is S423R (Figure 5, Table S5). However, when inserting opposite N2ffdG, WT pol κ and the pol κ SNP variants correctly incorporate dC and also misincorporate dT. With an undamaged A template base, WT pol κ correctly inserts dT and misinserts dC, as do the SNP variants. In contrast, with the major groove adducts N6ffdA and etheno-dA, WT pol κ and the variants correctly incorporate dT opposite only N6ffdA. With etheno-dA as the template base, only S423R shows correct dT incorporation, while WT pol κ and the remaining variants exhibit no incorporation after 30 min.
Figure 5.
Pol κ SNPs have similar nucleotide incorporation patterns as WT pol κ. (A) Opposite undamaged dG, only correct dC incorporation is observed except for the S423R variant which also exhibits dT misincorporation. (B) Opposite N2ffdG, there is an increase in dT misincorporation by all variants. (C) For the undamaged dA control, there is dC misincorporation for the most active variants, which is higher for the S423R variant. (D) Opposite N6ffdA, no misincorporation is observed for WT or any variant (only dT incorporation). (E) With ethenodA as the template, there is correct dT incorporation by the most active variant S423R; other variants show no activity. P = primer, 30 min time points; quantitation is reported in Table S5.
Insertion from a Correct Base Pair Containing a Lesion Is Mostly Accurate.
We determined the pattern of nucleotide incorporation from a correct base pair containing a lesion as well as the respective undamaged controls. The DNA template used in all experiments carried out here harbors a T as the template base after the lesion, with the exception of the template containing N6ffdA which is followed by a G, as the template containing T induced sequence-specific misincorpo-ration by WT pol κ. WT pol κ and the SNP variants insert dA correctly after the undamaged dG:dC base pair (Figure 6, Table S6). WT pol κ and the SNP variants also insert mainly the correct dA after a base pair containing the minor groove lesion N2ffdG (Figure 6). When extending from the correct undamaged dA:dT base pair, WT pol κ and most variants incorporate A but also weakly misincorporate what appears to be three guanines, except the two least active variants pol κ R298H and Y432S, which incorporate almost exclusively dA. The second and third dG residues would be correctly incorporated opposite the two dC residues in the template following T (Table 2). A second dA and possibly a third dA are also misincorporated by S423R (Figure 6). Surprisingly, extension from the major groove adduct N6ffdA:dT base pair results in predominately correct incorporation of dC by all proteins (Figure 6). Extension from the major groove adduct etheno-dA:dT base pair yielded no visible incorporation of any nucleotide after 30 min (Figure 6).
Figure 6.
Pol κ SNPs have similar nucleotide incorporation patterns extending from the correct base pair as WT pol κ (the templating base is dT). From both the (A) dG:dC and (B) N2ffdG:dC base pairs, there is only correct dA incorporation. (C) For the undamaged dA:dT control, dA incorporation is observed for all the variants, and the S423R variant incorporates multiple dAs and misincorporates every nucleotide, including multiple dGs. (D) In assays of incorporation after the N6ffdA:dT base pair, in which the template base is dG, no misincorporation by WT or any variant is observed; only correct dC incorporation is observed. With ethenodA:dT base pair, very little activity is observed. P = primer, 30 min time points; quantitation is reported in Table S6.
Insertion from a Mismatched Base Pair Containing a Lesion Is Much Weaker but Accurate.
We next determined the pattern of nucleotide incorporation after a mismatched base pair containing a lesion compared to undamaged DNA controls. In general, all of the proteins studied here exhibit low activity extending from a dG:dT mismatch (Figure 4). With undamaged dG:dT mismatch, mainly the correct dA (the templating base is T) incorporation is observed with WT pol κ, and the variants exhibit primer extension activity similar to or greater than WT pol κ. The less active R298H and Y432S variants did not exhibit observable nucleotide insertion under these conditions (Figure 7, Table S7). A similar incorporation pattern is observed with the N2ffdG:dT mismatch, as mainly correct dA incorporation is observed with WT pol κ and most variants; no incorporation of any nucleotide is observed with either R298H or Y432S even after 30 min, which correlates with the low primer extension activity by both variants from the dG:dT and N2ffdG:dT mismatches (Figure 7).
Figure 7.
Pol κ SNPs have similar nucleotide incorporation patterns extending from the mismatched base pair as WT pol κ (the templating base is dT). From both the (A) dG:dT and (B) N2ffdG:dT base pairs, there is little correct dA incorporation. (C) For the undamaged dA:dG control, dA incorporation is observed for the most active variants. (D) From the N6ffdA:dG base pair, there is no incorporation by WT or any variant after 30 min, which corresponds to the low primer extension activity (Figure 4). (E) With ethenodA:dG base pair, there is correct dA incorporation by all variants. P = primer, 30 min time points; quantitation is reported in Table S7.
Extension from the undamaged dA:dG mismatch also shows a similar pattern of very low incorporation levels, with only dA incorporation (T is the template base) observed with R219I, E292K, and S423R variants (Figure 7). WT pol κ and variants show no incorporation after the N6ffdA:dG mismatch (Figure 7), in agreement with the primer extension experiments as this activity is quite low with templates containing either dA:dG or N6ffdA:dG mismatches (Figure 4). Insertion after the ethenodA:dG mismatch has the greatest amount of incorporation. There is correct dA incorporation by all variants, except the two variants with the weakest primer extension activity (R298H and Y432S), for which little activity is observed (Figure 7).
Thermal Shift Assays Show Stabilization of Proteins with Preferred Substrates.
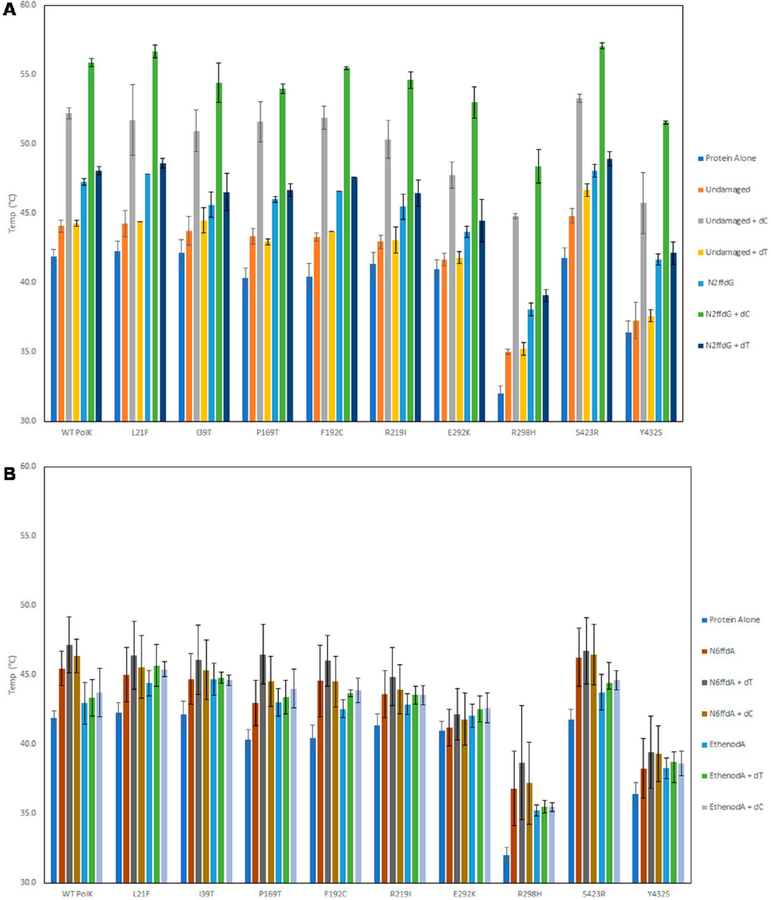
We next assessed the overall stability of WT pol κ and variants alone and with their substrates using a thermal shift assay. Many of the pol κ SNP variants have similar melting temperatures as WT pol κ (41.9 ± 0.5 °C), except R298H (32.0 ± 0.6 °C) and Y432S (36.4 ± 0.8 °C), which are lower (Figure 8). Pol κ R298H and Y432S are also the least active of the all the variants. As shown previously with WT pol κ and other variants, there is a small increase in the melting temperature in the presence of any primer:template DNA;7,57,58 however, with the addition of DNA and the correct nucleotide, the melting temperature increases by 7–12 °C for all the proteins. A large stabilization is not observed with the incorrect nucleotide or the nonpreferred DNA lesions, which for pol κ are the major groove lesions, as we have shown previously.7,57,58
Figure 8.
Pol κ SNP variants, like WT, show an increase in melting temperature in the presence of preferred DNA substrates and the correct nucleotide. (A) Melting temperatures of pol κ variants in the presence of undamaged DNA, N2ffdG, and incoming nucleotides dC (correct nucleotide) or dT (incorrect nucleotide). Stabilization of the variants is observed in the presence of both undamaged and N2ffdG DNA templates with the correct nucleotide present but not with the incorrect nucleotide or with DNA alone. (B) The same increase is not seen with the major groove adducts, N6ffdA and ethenodA. There is no increase in melting temperature in the presence of either major groove adduct or in the presence of the correct nucleotide (dT for the major groove adducts) and DNA.
Computational Analysis Reveals Differences in Active and Inactive Variants.
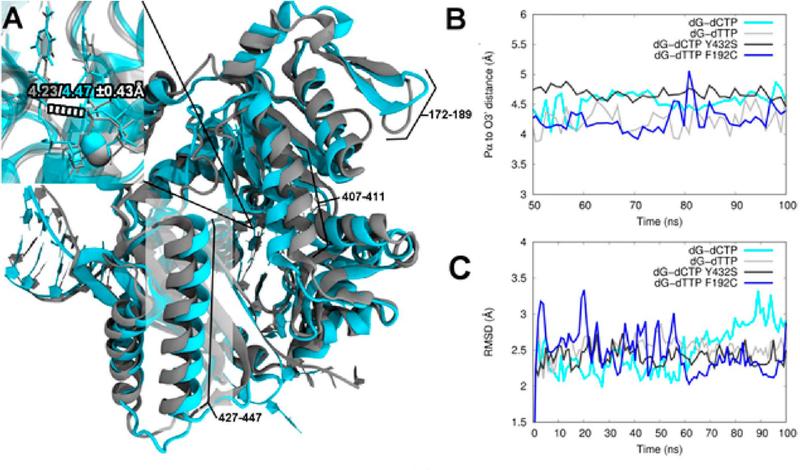
In order to gain structural and dynamical insight into the differences in activity observed with the pol κ SNP variants, we carried out MD simulations. Several significant structural differences of WT pol κ between the dG:dC and dG:dT structures are observed in four areas, three of which can be seen in Figure 9A. The largest area of deviation occurs from residues ~427–447 (highlighted), with smaller areas from 407 to 411 (top center), 172 to 189 (top right), and 325 to 334 (not pictured; on the back center). Thus, pol κ has a noticeable overall structural change based on the incoming nucleotide. These results are consistent with the experimental results showing that WT pol κ successfully continues synthesis from a base pair mismatch, albeit less efficiently than with correct base pairs. In addition, the simulation results suggest there is a slight decrease in distance between the Pα of the incoming nucleotide and the O3′ on the primer strand (−0.24 Å on average, Figure 9A inset).
Figure 9.
Structural comparison between WT pol κ and selected variants. (A) Overlay of the WT structure with the correct incoming nucleotide (cyan) compared with the WT structure with an incorrect incoming nucleotide (gray). Distances in the active site close-up denote the average O3′ and Pα distance (in Å) for correct (cyan) and mismatched (gray) incoming dNTP. (B) O3′ to Pα distance as a function of time for WT and selected variants. (C) Average RMSD for WT and selected variants.
The Pα–O3′ distance fluctuates between 4.17 and 4.58 Å for the variant systems that display higher activity, essentially within the standard deviation for the WT systems (Table S8). Interestingly, the less active R219I and R298H variants display higher overall Pα–O3′ distances for all simulations, and the Y432S variant shows a particularly large increase for the simulations with incorrect incoming nucleotides. The R298H dG:dT system resulted in unstable simulations because of the close proximity of the triphosphate moiety of the incoming nucleotide to various charged residues in the active site and thus was not considered further. The variation in the distances suggests some likelihood that mutations that decrease activity disturb the active site. All of the simulations show an overall backbone RMSD of around 3 Å or less (Figure 9C and Figure S2). WT pol κ with incoming dG:dC and the active F192C variant with incoming dG:dT show similar levels of RMSD fluctuations over time (Figure 9C). Further details of the dynamic analyses are in the Supporting Information, including correlation and principal component normal-mode analyses.
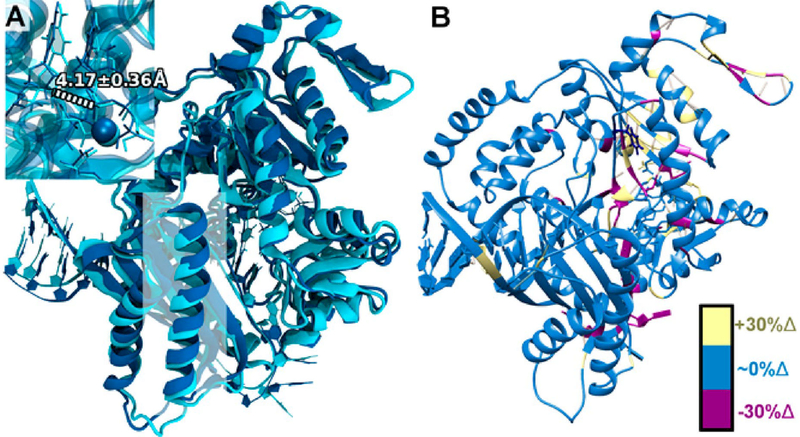
As indicated above, the more active variants are structurally more similar to WT pol κ with correct incoming dNTP. For example, the F192C variant with dG:dT shows the most similarity to the correctly paired WT pol κ with dG:dC, despite having a mispaired base itself. The F192C variant dG:dT shows a resemblance to WT pol κ in most regions of the protein, including the highlighted ~427–447 region (Figure 10A), and in the close-up of the active site. This is a marked difference compared with WT pol κ seen in Figure 9A, which showed numerous structural differences as previously discussed. The F192C variant with dG:dT also shows the smallest average difference in backbone RMSD from WT pol κ with dG:dC as compared to the other systems, averaging ~1.8 Å. WT pol κ with incoming dG:dT shows a larger average difference compared with the WT dG:dC of ~2.2 Å. From this, we hypothesize that the modestly increased activity of pol κ F192C with dG:dT or N2ffdG:dT mismatches involves a structural shift in this variant that induces a favorable geometrical arrangement of the incoming mispair in the active site, which could lead to improved proficiency.
Figure 10.
Structural comparisons between WT pol κ with the correct incoming nucleotide (cyan) and the active F192C variant with the incorrect incoming nucleotide (dark blue). (A) Structural overlay between WT pol κ with incoming dG:dC (cyan) and pol κ F192C with incoming dG:dT (dark blue) with α helix residues ~427–447 highlighted in a white square. Distances in the active site close-up denote the average O3′ and Pα distance (in Å) for the F192C variant with dG:dT. (B) Changes in hydrogen bonding for the F192C variant with incoming dG:dT compared with WT pol κ with incoming dG:dC. Residues with an increase of 30% or more in hydrogen-bonding character compared with WT pol κ are highlighted in yellow; residues with similar hydrogen-bonding character are in blue; and residues with a decrease of 30% or more in hydrogen-bonding character compared with WT pol κ are highlighted in magenta.
Further evidence for this can be seen in the hydrogen-bond changes shown in Figure 10B. In the pol κ F192C simulation with dG:dT, there are a large number of hydrogen bonds that are present in the palm domain and specifically in the active site, highlighted in yellow (Figure 10B), that are present far less or not at all in WT pol κ with incoming dG:dC. In particular, D198, a member of the catalytic triad, forms a hydrogen bond with S196 for ~63% of the F192C dG:dT simulation. This interaction is present only 8% of the time in the simulation of WT pol κ with dG:dC. Further details on the hydrogen-bonding analyses can be found in Table S9.
The dynamics of WT pol κ with incoming dG:dC and pol κ F192C with incoming dG:dT are also quite similar. Difference correlation analysis and normal-mode analysis indicate a high level of dynamic movement in the two systems with similar features (Figures S3–S7). The S423R variant is also more active in primer extension experiments than WT pol κ (Figures 2–4) and shows results analogous to F192C, which has similar activity to WT pol κ (Figures S8–S10).
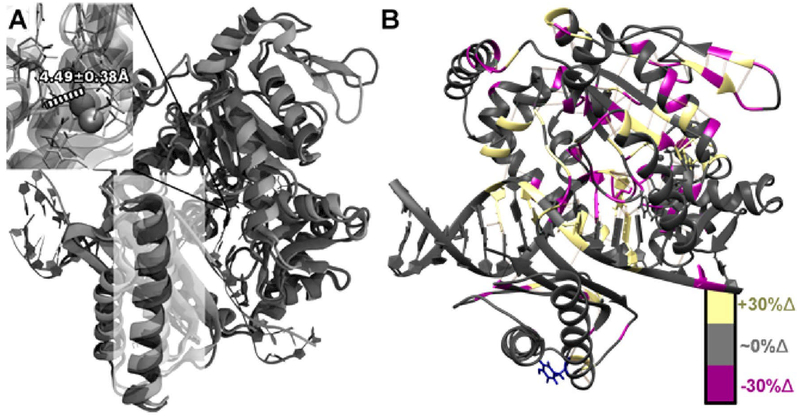
In contrast to the active variants, the less active variants are more comparable to WT pol κ with a mismatched incoming dNTP. For example, the pol κ Y432S variant with incoming dG:dC shows a larger overall level of structural similarity to WT with a mismatched incoming dNTP, as can again be seen in the region comprising residues ~427–447 (Figure 9A). In addition, the Y432S variant shows a large shift in the active site and the metal location (Figure 11A), although the Pα–O3′ distance increases only slightly (~0.02 Å on average up to ~0.21 Å at various points in the simulation). The average RMSD for the Y432S variant with incoming dG:dC compared with WT with incoming dG:dT is below 1 Å and is much less dynamic than the more active variants and WT pol κ with the correct base pair (Figure 9C and Figure S2). This suggests that the active site shift is likely due to a shift in the binding orientation of the DNA.
Figure 11.
Structural comparison between WT pol κ with incorrect incoming nucleotide (light gray) and the less active Y432S variant with the correct incoming nucleotide (dark gray). (A) Structural overlay between WT pol κ with incoming dG:dC (light gray) and Y432S variant with incoming dG:dC (dark gray) with α helix residues ~427–447 highlighted in a white square. Distances in the active site close-up denote the average O3′ and Pα distance (in Å) for the Y432S variant with incoming dG:dC indicated in white text. (B) Changes in hydrogen bonding for the Y432S variant with incoming dG:dC compared to the WT pol κ with dG:dT. Residues with an increase of 30% or more in hydrogen-bonding character compared with WT pol κ are highlighted in yellow; residues with similar hydrogen-bonding character are in dark gray; and residues with a decrease of 30% or more in hydrogen-bonding character compared with WT pol κ are highlighted in magenta.
The structural changes can be particularly seen from the hydrogen bonding analysis, where there are a large number of hydrogen bonds broken from the backbone of the primer strand and in the little finger domain, particularly in the β sheet strand closest to the DNA (Figure 11B). There are also new hydrogen bonds formed with the primer strand backbone, though not enough to compensate for those broken (~4 broken vs ~2 formed). A corresponding hydrogen bond is formed between the template strand backbone and the thumb domain on the opposite side, which could indicate that the DNA strands are pulling away from each other internally. There are additional changes in noncovalent interactions throughout the protein in key areas close to the DNA, including the elimination of a key stabilizing bond between the 3′-end of the primer strand and S160. This is consistent with the overall indication that, while the protein structure is not overly distorted, the location and structure of the DNA within the polymerase are disturbed by the mutation. It is also interesting to note that the structure for the correctly paired base more closely resembles the WT structure with a mispair, indicating that the pol κ structure is sensitive to multiple types of changes in DNA. The normal-mode analysis also shows that K328, which is hydrogen bonded to the incoming dNTP, and its corresponding α helix show a large increase in contribution for the first normal mode for both the Y432S variant, the other less active variants, and the WT dG:dT mismatch. This contribution is essentially not present in the active variants and the WT dG:dC system, providing support for a shift in dynamics as well as structural impact from the less active variants. Further details of the dynamic analysis and the other less active variants can be found in Figures S11–S16.
DISCUSSION
Pol κ has been shown to bypass minor groove adducts efficiently and is inhibited by major groove adducts.2–6,8,10 It has also been shown to be an extender from mismatched primer termini.11–16 In this work, we investigated the in vitro activity of nine variants encoded by single nucleotide polymorphisms of human pol κ. The nine variants can be separated into three categories: enhanced insertion and extension activity compared to WT, similar activity to WT, and diminished insertion and extension activity compared to WT. We found that the most active variants, F192C and S423R, efficiently insert nucleotides opposite minor groove and major groove lesions as well as extend from correct and mismatched damaged base pairs. Pol κ R298H and Y432S variants are the least active variants and most blocked by the major groove adducts. R298H and Y432S variants cannot extend from mismatched damaged bases but can extend from the minor groove lesion with the correct base pair. The variants L21F, I39T, and P169T have similar insertion and extension activity as WT pol κ. R219I and E292K variants have decreased extension activity from the lesions with both correct and mismatched base pairs, but their insertion activity is similar to that of WT.
Structurally these variants are found in the N-clasp, fingers, palm, and little finger domains of pol κ (Table 1). The activity of pol κ F192C is similar to WT pol κ and modestly more active than WT pol κ in extension from mismatches. In the structure of pol κ (Figure 1), F192 is in the palm domain adjacent to the linker between the polymerase domain and little finger or PAD. F192 interacts with the adjacent α helix, and thus changing this phenylalanine to a cysteine results in greater flexibility and movement as observed in our MD simulations. This increased flexibility may be partly responsible for the experimentally observed increase in activity. Pol κ F192C has a melting temperature of 40.4 °C; in the presence of undamaged or N2ffdG-containing DNA, there is a modest increase in Tm of 43.3 and 46.6 °C, respectively. In the presence of DNA and the correct nucleotide, there is a larger increase with undamaged DNA of approximately 7 °C, while with N2ffdG, there is an increase of approximately 9 °C. Pol κ S423R is overall more active than WT pol κ, particularly when extending from major groove adducts, whether paired with correct or incorrect bases at the primer termini. S423 is in the little finger, or PAD, and is solvent accessible. Replacing this hydrophilic residue with a larger charged arginine residue apparently does not affect the overall stability given that the melting temperature of this variant is similar to that of WT pol κ (Figure 8). MD simulations show that the overall structure and hydrogen-bonding network remain largely unchanged from the WT, with some shifts in hydrogen bonding between the DNA template backbone and the PAD. Hydrogen-bond analysis reveals that the R at position 423 forms a salt bridge with the DNA backbone around 100% of the simulation time for the correctly paired base system, suggesting increased noncovalent interactions compared with the WT. Both F192C and S423R variants have similar patterns of incorporation and misincorporation as WT pol κ. Previous work with F192C and S423R variants (in a construct including pol κ amino acids 1–526) resulted in activity similar to WT on undamaged DNA and N2-AnthG-containing DNA, with similar kinetics parameters despite the differences in the DNA substrates in those studies relative to the DNA used here (Table S3).22,23
The L21F and I39T mutations are both in the N-clasp, and both variants retain activity similar to WT pol κ with both the minor groove N2ffdG adduct and major groove adducts N6ffdA and etheno-dA. It has been shown previously that both L21F and I39T variants were markedly less active in the bypass of the bulkier N2-AnthG, especially in the extension step.23 The bulkier adduct in the base pair at the primer terminus could cause more perturbation in the N-clasp compared to the smaller furfuryl adduct, as it has been proposed that the N-clasp can tolerate distortions at the primer terminus.16,33 The N2ffdG adduct is overall nondistorting to the DNA structure.24 Additionally, our construct, unlike the construct used by Song et al., does not contain the first 18 residues of pol κ, which have been shown to increase DNA binding as well as positively alter the ability of the N-clasp to bind DNA and extend from mismatches.16 This difference could account for the different activities by the same variants in different constructs. Both the L21F and I39T variants have a similar melting temperature compared to WT pol κ with and without the different DNA constructs studied here. In the presence of both undamaged DNA and N2ffdG DNA with the correct nucleotide dCTP, the melting temperatures of the L21F and I39T variants increase by approximately 10 °C, similar to that of WT pol κ.
Pol κ R219I has decreased insertion and extension activity compared to WT, while the E292K variant activity is similar to WT. R219 and E292 are both near a region of pol κ that has been unresolved in crystal structures, which means it is likely disordered, and it is unclear whether that region of approximately 60 amino acids interacts with the rest of the palm domain. This could explain why the R219I mutation has a greater effect on the activity of pol κ than E292K; there could be different interactions with the residues not observed in the crystal structure that are important in catalysis and especially the translocation step. However, the melting temperatures of pol κ R219I are similar to WT in the presence of all templates and the correct or incorrect nucleotides, indicating the R219I mutation likely does not have a large effect on protein folding or DNA or nucleotide binding. This is consistent with our MD simulations, which indicate that the structure of R219I is similar to WT with the incorrect nucleotide, but that the dNTP location is altered and that its Pα–O3′ distance increases substantially for both correct and incorrect base pairings. Therefore, the R219I mutation likely disturbs the location of the dNTP more than E292K and has a stronger effect on the overall activity. However, we are not able to determine the effect of the unstructured region from our simulations since this region is not present in the systems studied by simulation (see Experimental Procedures section).
The R298H and Y432S variants have much lower activity than WT pol κ on all primer:template combinations. R298 has strong hydrogen-bonding character with Q332 in the crystal structure and in WT pol κ simulations. Replacing arginine with histidine results in a loss of interaction with Q332. In the thermal shift assays, the melting temperature of the R298H variant is only 32.0 ± 0.6 °C, almost 10 °C lower than WT. When undamaged or N2ffdG DNA is present, there is a small increase (3–5 °C) in melting temperature, but when the correct dNTP is present, there is a 9–10 °C increase (to 44.8 ± 0.2 °C), similar to WT pol κ. The decrease in activity could be due to the low melting temperature of the variant. In the MD simulations, Y432 is essentially contained in a hydro-phobic pocket and shows evidence of a large number of weak interactions with the surrounding residues (Figure S16). Like R298H, the melting temperature of the Y432S variant is low, 36.4 ± 0.8 °C, and there is no or a relatively small increase in the presence of DNA, but there is a 10 °C increase in Tm with undamaged DNA or N2ffdG-containing DNA and dCTP. Of the variants characterized by Song et al., only Y432S showed a defect in DNA binding.23 The change from the larger phenol ring of tyrosine to the much smaller serine appears to cause a substantial change in crucial intermolecular interactions between the little finger (PAD) and the DNA in the MD simulations as previously discussed, leading to a large distortion and potential instability of the DNA binding orientation. Additionally, many hydrogen bonds are broken in the palm domain that could denote a strong overall structural shift in the protein and a noticeable shift in dynamics as seen in the normal-mode analysis, providing a potential explanation for the change in melting temperature. The low activity of these two proteins could be correlated with the low melting temperatures, although it appears that both variants are stabilized in the presence of DNA and the correct nucleotide based on the corresponding thermal shifts.
Overall, most of the SNP variants, like WT pol κ, have a greater increase in melting temperature in the presence of N2ffdG and dCTP compared to undamaged DNA and dCTP, although many have similar in vitro activity as WT. There is no clear increase in activity for the SNP variants on undamaged DNA versus DNA containing N2ffdG. The SNP variants are all slightly blocked by major groove adducts, like WT pol κ, but can extend better from the N6ffdA:dT base pair than from the etheno-dA:dT or mismatched dA:dG primer termini. These observations together with the MD simulations provide further support for the model that pol κ adopts specific conformations that correlate with activity.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely acknowledge Ke Zhang and Xueguang Lu of Northeastern University for assistance with the synthesis of the modified DNA substrates. We thank F. Peter Guengerich of Vanderbilt University for the gift of the pol κ expression plasmid.
Funding
This work was supported by American Cancer Society grant RSG-12–161-01-DMC to P.J.B., NSF MCB-1615946 to P.J.B., R01GM108583 to G.A.C., and NSF CHE-1531468. Research support to collect data and develop an application to support this project was provided by 3P50CA093459, 5P50CA097007, 5R01ES011740, and 5R01CA133996.
ABBREVIATIONS
- N2ffdG
N2-furfuryl-dG
- N6ffdA
N6-furfuryl-dA
- SNP
single nucleotide polymorphism
- MD
molecular dynamics
- PCA
principal component analysis
- NMA
normal-mode analysis
- NCI
noncovalent interaction
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.8b00055.
Quantitation of primer extension activity and kinetics parameters and analysis of molecular dynamics simulations (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Friedberg EC, Walker G, Siede W, Wood R, Schultz R, and Ellenberger T (2006) DNA Repair and Mutagenesis, 2nd ed, ASM Press, Washington, DC. [Google Scholar]
- (2).Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, and Geacintov NE (2002) trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J. Biol. Chem. 277, 30488–30494. [DOI] [PubMed] [Google Scholar]
- (3).Choi JY, Angel KC, and Guengerich FP (2006) Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 281, 21062–21072. [DOI] [PubMed] [Google Scholar]
- (4).Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, and Walker GC (2006) A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439, 225–228. [DOI] [PubMed] [Google Scholar]
- (5).Yuan B, Cao H, Jiang Y, Hong H, and Wang Y (2008) Efficient and accurate bypass of N2-(1-carboxyethyl)-2’-deoxyguano-sine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 105, 8679–8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhang Y, Wu X, Guo D, Rechkoblit O, and Wang Z (2002) Activities of human DNA polymerase κ in response to the major benzo[a]pyrene DNA adduct: error-free lesion bypass and extension synthesis from opposite the lesion. DNA Repair 1, 559–569. [DOI] [PubMed] [Google Scholar]
- (7).Nevin P, Lu X, Zhang K, Engen JR, and Beuning PJ (2015) Noncognate DNA damage prevents the formation of the active conformation of the Y-family DNA polymerases DinB and DNA polymerase κ. FEBS J. 282, 2646–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, and Guengerich FP (2006) Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 281, 38244–38256. [DOI] [PubMed] [Google Scholar]
- (9).Choi JY, and Guengerich FP (2008) Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J. Biol. Chem. 283, 23645–23655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Walsh JM, Ippoliti PJ, Ronayne EA, Rozners E, and Beuning PJ (2013) Discrimination against major groove adducts by Y-family polymerases of the DinB subfamily. DNA Repair 12, 713–722. [DOI] [PubMed] [Google Scholar]
- (11).Wolfle WT, Washington MT, Prakash L, and Prakash S (2003) Human DNA polymerase κ uses template-primer misalignment as a novel means for extending mispaired termini and for generating single-base deletions. Genes Dev. 17, 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Washington MT, Johnson RE, Prakash L, and Prakash S (2002) Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc. Natl. Acad. Sci. U. S. A. 99, 1910–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Haracska L, Prakash L, and Prakash S (2002) Role of human DNA polymerase κ as an extender in translesion synthesis. Proc. Natl. Acad. Sci. U. S. A. 99, 16000–16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Niimi N, Sassa A, Katafuchi A, Gruz P, Fujimoto H, Bonala RR, Johnson F, Ohta T, and Nohmi T (2009) The steric gate amino acid tyrosine 112 is required for efficient mismatched-primer extension by human DNA polymerase κ. Biochemistry 48, 4239–4246. [DOI] [PubMed] [Google Scholar]
- (15).Vasquez-Del Carpio R, Silverstein TD, Lone S, Johnson RE, Prakash L, Prakash S, and Aggarwal AK (2011) Role of human DNA polymerase κ in extension opposite from a cis-syn thymine dimer. J. Mol. Biol. 408, 252–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, and Aggarwal AK (2007) Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell 25, 601–614. [DOI] [PubMed] [Google Scholar]
- (17).Brookes AJ (1999) The essence of SNPs. Gene 234, 177–186. [DOI] [PubMed] [Google Scholar]
- (18).Swett RJ, Elias A, Miller JA, Dyson GE, and Cisneros GA (2013) Hypothesis driven single nucleotide polymorphism search (HyDn-SNP-S). DNA Repair 12, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dai ZJ, Liu XH, Ma YF, Kang HF, Jin TB, Dai ZM, Guan HT, Wang M, Liu K, Dai C, Yang XW, and Wang XJ (2016) Association Between Single Nucleotide Polymorphisms in DNA Polymerase Kappa Gene and Breast Cancer Risk in Chinese Han Population: A STROBE-Compliant Observational Study. Medicine (Philadelphia, PA, U. S.) 95, e2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Shao M, Jin B, Niu Y, Ye J, Lu D, and Han B (2014) Association of POLK polymorphisms with platinum-based chemo-therapy response and severe toxicity in non-small cell lung cancer patients. Cell Biochem. Biophys. 70, 1227–1237. [DOI] [PubMed] [Google Scholar]
- (21).Yadav S, Mukhopadhyay S, Anbalagan M, and Makridakis N (2015) Somatic Mutations in Catalytic Core of POLK Reported in Prostate Cancer Alter Translesion DNA Synthesis. Hum. Mutat. 36, 873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kim JK, Yeom M, Hong JK, Song I, Lee YS, Guengerich FP, and Choi JY (2016) Six Germline Genetic Variations Impair the Translesion Synthesis Activity of Human DNA Polymerase κ. Chem. Res. Toxicol. 29, 1741–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Song I, Kim EJ, Kim IH, Park EM, Lee KE, Shin JH, Guengerich FP, and Choi JY (2014) Biochemical characterization of eight genetic variants of human DNA polymerase κ involved in error-free bypass across bulky N2-guanyl DNA adducts. Chem. Res. Toxicol. 27, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ghodke PP, Gore KR, Harikrishna S, Samanta B, Kottur J, Nair DT, and Pradeepkumar PI (2016) The N(2)-Furfuryldeoxyguanosine Adduct Does Not Alter the Structure of B-DNA. J. Org. Chem. 81, 502–511. [DOI] [PubMed] [Google Scholar]
- (25).Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, and Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Irimia A, Eoff RL, Guengerich FP, and Egli M (2009) Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase κ opposite the 7,8-dihydro-8-oxo-2’-deoxyguanosine adduct. J. Biol. Chem. 284, 22467–22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, and Aggarwal AK (2004) Crystal structure of the catalytic core of human DNA polymerase κ. Structure 12, 1395–1404. [DOI] [PubMed] [Google Scholar]
- (28).DeCorte BL, Tsarouhtsis D, Kuchimanchi S, Cooper MD, Horton P, Harris CM, and Harris TM (1996) Improved strategies for postoligomerization synthesis of oligodeoxynucleotides bearing structurally defined adducts at the N2 position of deoxyguanosine. Chem. Res. Toxicol. 9, 630–637. [DOI] [PubMed] [Google Scholar]
- (29).Larson CJ, and Verdine GL (1992) A high-capacity column for affinity purification of sequence-specific DNA-binding proteins. Nucleic Acids Res. 20, 3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sambrook J, Fritsch EF, and Maniatis T (1989) Molecular Cloning–A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- (31).Sheriff A, Motea E, Lee I, and Berdis AJ (2008) Mechanism and dynamics of translesion DNA synthesis catalyzed by the Escherichia coli Klenow fragment. Biochemistry 47, 8527–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sali A, and Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- (33).Jia L, Geacintov NE, and Broyde S (2008) The N-clasp of human DNA polymerase κ promotes blockage or error-free bypass of adenine- or guanine-benzo[a]pyrenyl lesions. Nucleic Acids Res. 36, 6571–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, and Dupradeau FY (2011) R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 39, W511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Dunbrack RL Jr (2002) Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 12, 431–440. [DOI] [PubMed] [Google Scholar]
- (37).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (38).Schafmeister CEAF, Ross WS, and Romanovski V (1995) LEAP; University of California, San Francisco. [Google Scholar]
- (39).Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. [Google Scholar]
- (40).Case DA, Cerutti SD, Cheatham TE III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Greene D, Homeyer N, Izadi S, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein D, Merz KM, Monard G, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith MW, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L, York MD, and Kollman PA (2017) AMBER 2017, University of California, San Francisco. [Google Scholar]
- (41).Cheatham TE 3rd, and Case DA (2013) Twenty-five years of nucleic acid simulations. Biopolymers 99, 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C (2015) ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, and Walker RC (2013) Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- (44).Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, and Haak JR (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. [Google Scholar]
- (45).Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, and Pedersen LG (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593. [Google Scholar]
- (46).Bakan A, Meireles LM, and Bahar I (2011) ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27, 1575–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Humphrey W, Dalke A, and Schulten K (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (48).Contreras-Garcia J, Johnson ER, Keinan S, Chaudret R, Piquemal JP, Beratan DN, and Yang W (2011) NCIPLOT: a program for plotting non-covalent interaction regions. J. Chem. Theory Comput. 7, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Waters KM, Le Marchand L, Kolonel LN, Monroe KR, Stram DO, Henderson BE, and Haiman CA (2009) Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol., Biomarkers Prev. 18, 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba II, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, and Meyerson M (2007) Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, and Houlston RS (2008) Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet. 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang SY, Kosoy R, Zhang MF, Qureshi AA, Vattathil S, Schacherer CW, Gardner JM, Wang YL, Bishop DT, Barrett JH, MacGregor S, Hayward NK, Martin NG, Duffy DL, Mann GJ, Cust A, Hopper J, Brown KM, Grimm EA, Xu YJ, Han YH, Jing KY, McHugh C, Laurie CC, Doheny KF, Pugh EW, Seldin MF, Han JL, and Wei QY (2011) Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet. 20, 5012–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF Jr., Hoover RN, Thomas G, and Chanock SJ (2007) A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF Jr., Hoover R, Hunter DJ, Chanock SJ, and Thomas G (2007) Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39, 645–649. [DOI] [PubMed] [Google Scholar]
- (55).Wang X, Pankratz VS, Fredericksen Z, Tarrell R, Karaus M, McGuffog L, Pharaoh PD, Ponder BA, Dunning AM, Peock S, Cook M, Oliver C, Frost D, EMBRACE, Sinilnikova OM, Stoppa-Lyonnet D, Mazoyer S, Houdayer C, GEMO, Hogervorst FB, Hooning MJ, Ligtenberg MJ, HEBON, Spurdle A, Chenevix-Trench G, kConFab, Schmutzler RK, Wappenschmidt B, Engel C, Meindl A, Domchek SM, Nathanson KL, Rebbeck TR, Singer CF, Gschwantler-Kaulich D, Dressler C, Fink A, Szabo CI, Zikan M, Foretova L, Claes K, Thomas G, Hoover RN, Hunter DJ, Chanock SJ, Easton DF, Antoniou AC, and Couch FJ (2010) Common variants associated with breast cancer in genome-wide association studies are modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. Hum. Mol. Genet. 19, 2886–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril MF, Azizi E, Bakker B, Bianchi-Scarra G, Bressac-de Paillerets B, Calista D, Cannon-Albright LA, Chin AWT, Debniak T, Galore-Haskel G, Ghiorzo P, Gut I, Hansson J, Hocevar M, Hoiom V, Hopper JL, Ingvar C, Kanetsky PA, Kefford RF, Landi MT, Lang J, Lubinski J, Mackie R, Malvehy J, Mann GJ, Martin NG, Montgomery GW, van Nieuwpoort FA, Novakovic S, Olsson H, Puig S, Weiss M, van Workum W, Zelenika D, Brown KM, Goldstein AM, Gillanders EM, Boland A, Galan P, Elder DE, Gruis NA, Hayward NK, Lathrop GM, Barrett JH, and Bishop JA (2009) Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 41, 920–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Antczak NM, Packer MR, Lu X, Zhang K, and Beuning PJ (2017) Human Y-Family DNA Polymerase κ Is More Tolerant to Changes in Its Active Site Loop than Its Ortholog Escherichia coli DinB. Chem. Res. Toxicol. 30, 2002–2012. [DOI] [PubMed] [Google Scholar]
- (58).Nevin P, Engen JR, and Beuning PJ (2015) Steric gate residues of Y-family DNA polymerases DinB and pol κ are crucial for dNTP-induced conformational change. DNA Repair 29, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Jha V, Bian C, Xing G, and Ling H (2016) Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human DNA polymerase κ. Nucleic Acids Res. 44, 4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.