Abstract
The initial virulence and invasiveness of a bacterial strain may play an important role in leading to a maximally efficacious attenuated live vaccine. Here we show that χ9909, derived from Salmonella Typhimurium UK-1 χ3761 (the most virulent S. Typhimurium strain known to us), is effective in protecting mice against lethal UK-1 and 14028S (less virulent S. Typhimurium strain) challenge. As opposed to this, 14028S-derived vaccine χ12359 induces suboptimal levels of protection, with survival percentages that are significantly lower when challenged with lethal UK-1 challenge doses. T-cell assays have revealed that significantly greater levels of Th1 cytokines IFN-γ and TNF-α were secreted by stimulated T-lymphocytes obtained from UK-1(ΔaroA) immunized mice than those from mice immunized with 14028S(ΔaroA). In addition, UK-1(ΔaroA) showed markedly higher colonizing ability in the spleen, liver, and cecum when compared to 14028S(ΔaroA). Enumeration of bacteria in fecal pellets has also revealed that UK-1(ΔaroA) can persist in the host for over 10 days whereas 14028S(ΔaroA) titers dropped significantly by day 10. Moreover, co-infection of parent strains UK-1 and 14028S resulted in considerably greater recovery of the former in multiple mucosal and gut associated lymphatic tissues. Mice immunized with UK-1(ΔaroA) were also able to clear UK-1 infection remarkably more efficiently from the target organs than 14028S(ΔaroA). Together, these results provide ample evidence to support the hypothesis that attenuated derivatives of parent strains with higher initial virulence make better vaccines.
Introduction
Salmonella enterica serovar Typhimurium is a Gram-negative, facultative anaerobic pathogen that causes substantial burden of disease globally [1, 2]. Unlike the typhoidal serovars Salmonella Typhi and Salmonella Paratyphi A that are human-restricted [3], S. Typhimurium is a non-typhoidal serovar (NTS) with a diverse host range [3, 4]. Due to its ability to invade and adapt to a wide range of agriculturally important hosts including pigs, poultry, calves, and sheep [4], S. Typhimurium-mediated food-borne illnesses are an international public health concern. Although vaccination is one of the best prophylactic measures to control Salmonella infection, a licensed vaccine against S. Typhimurium for human use does not exist [5]. A multitude of vaccine development endeavors utilizing attenuated live bacteria, whole-cell killed vaccines, and subunit vaccines aimed at inducing long-term, cross protective immunity against multiple Salmonella NTS serovars are underway [6–10]. The ability of live attenuated Salmonella strains to stimulate strong humoral and cellular immunity has rendered them as an attractive option for development as homologous vaccines and for use as carriers of heterologous antigens [11–14].
Our group has devised several modulating strategies to develop recombinant attenuated Salmonella vaccines (RASVs) with enhanced safety, efficacy and biological containment [15–18]. Delayed onset of attenuation strategy ensures that the vaccine strain maintains wild-type phenotype until achieving colonization in effector lymphoid tissues but becomes self-limiting upon delivering protective antigens using a balanced-lethal vector-host system [18–20]. S. Typhimurium strain UK-1 (ATCC 68169) is the wild-type parent strain used as the foundation for the majority of the attenuated recombinant and non-recombinant vaccine derivatives studied in our laboratory [21]. UK-1 strain χ3761 was the parent strain from which many commercial vaccines for poultry such as Megan®Egg and Megan®Vac were also derived [21–25]. Since UK-1 is highly pathogenic in several hosts including mice and chickens [21, 26–28], these vaccine derivatives, when orally administered, are presumed to be more immunogenic and hence trigger a greater level of protective immunity than vaccine derivatives from less-virulent S. Typhimurium strains 14028S and SL1344 [26, 29]. Previous work in our laboratory involving characterization of UK-1 and SL1344 mutants with deletions of the crp and cdt genes has demonstrated that UK-1 derived Δcrp and Δ(crp-cdt) strains conferred complete protection against challenge with its own parent as well as with the less-virulent S. Typhimurium strains [30]. We have also utilized comparative genome analysis in a different study [28] to examine the phenotypic impact of genomic differences between UK-1 and other S. Typhimurium strains. Significant differences in genomic features which might contribute to virulence were identified between UK-1 and four other strains, LT2, 14028S, D23580, and SL1344.
In the present study, we compared the immunogenicity and protective efficacy of a single-dose oral administration of two aroA-deficient strains χ9909 and χ12359, derived from UK-1 χ3761 and 14028S, respectively. UK-1 and 14028S are highly similar, virulent, non-host-adapted strains, although the former has 4-fold lower LD50 (2.5 x 104 CFU) than the latter (9.6x104 CFU) [28, 30]. AroA is part of the shikimate pathway, and is involved in the synthesis of aromatic amino acids and several vitamins. As these aromatic amino acids and vitamins are not readily available in the mammalian host, lack of aroA results in auxotrophy and attenuation of Salmonella [31–33]. Hence, such attenuated metabolic mutants of Salmonella can be used as potential vaccine candidates against lethal homologous and heterologous infections [34, 35]. In this capacity, the current study investigates the magnitude of protection offered by the two strains UK-1(ΔaroA) and 14028S(ΔaroA) against lethal challenge with both parent strains, in a dose-dependent fashion. In addition to survival rate, these aroA-deficient strains were also evaluated in terms of antibody responses, cell-mediated cytokine production, pathogen clearance, and colonization of intestinal mucosa and deeper tissues. Findings reported in this paper provide evidence to further corroborate the notion that attenuated vaccines derived from hypervirulent strain UK-1 induce protective immunity that is markedly superior to those derived from the less-virulent S. Typhimurium strains such as 14028S.
Results
Evaluation of protective immunity
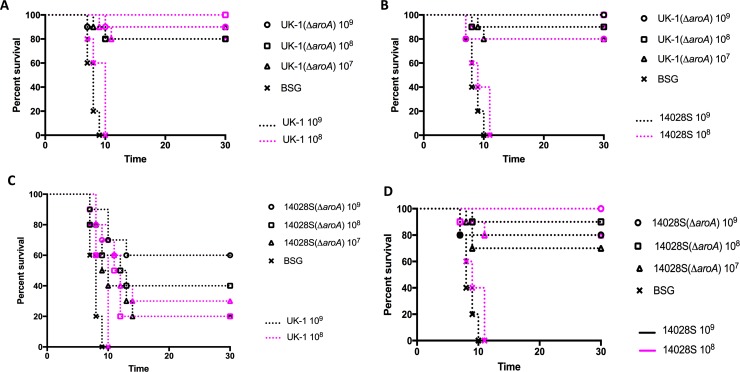
Given that UK-1 χ3761 was more highly virulent when compared to 14028S [28], we sought to determine whether immunization with its attenuated derivative UK-1(ΔaroA) induces a greater level of protective immunity against lethal oral challenge than strain 14028S(ΔaroA). To this end, BALB/c mice were orally immunized with 1x109, 1x108 or 1x107 CFU doses of UK-1(ΔaroA) or 14028S(ΔaroA). 35 days after immunization, mice were orally challenged with a 1x109 (~105 x LD50) or 1x108 (~104 x LD50) CFU dose of parent strains UK-1 or 14028S. A schematic of the vaccination schedule is presented in S1 Fig. We observed consistently higher survival percentages (80–100%) against both challenge strains and challenge doses in mice that were immunized with UK-1(ΔaroA) (Fig 1A and 1B). In contrast, varying levels of protection ranging from 20–100% were observed in mice immunized with 14028S(ΔaroA) (Fig 1C and 1D). The survival rate appeared to be dose dependent. Specifically, the 1x107 CFU immunizing dose of 14028S(ΔaroA) conferred only 20 and 30% protection against 1x109 (P < 0.01) and 1x108 (P < 0.05) CFU challenge doses of UK-1, respectively (Fig 1C). The 1x108 CFU dose of 14028S(ΔaroA) also yielded suboptimal levels of protection ranging between 20–40% against UK-1 challenge (P < 0.001). 60% survival was observed with 1x109 CFU of 14028S(ΔaroA) against both 1x109 and 1x108 CFU doses of UK-1, which is still lower, although not significantly so, than the 80–100% protection offered against 14028S challenge (Fig 1D). Evidently, 14028S(ΔaroA) provided reasonably good percentages of protection (70–100%) at all doses against 14028S challenge (Fig 1D) that were better than against UK-1 challenge. These results suggest that UK-1(ΔaroA) could effectively protect immunized mice at all doses against both UK-1 and 14028S challenge. Contrary to this, although 14028S(ΔaroA) performed well against challenge with its parent 14028S, it failed to elicit optimal protection against challenge with the more virulent UK-1, specifically at lower immunization doses.
Fig 1. Protective efficacy of ΔaroA mutants UK-1(ΔaroA) and 14028S(ΔaroA) against UK-1 and 14028S challenge.
BALB/c mice (n = 10 per immunization dose) were orally immunized with UK-1(ΔaroA) or 14028S(ΔaroA) at doses 1x109, 1x108 or 1x107 CFU in 20 μl BSG. Mice were also mock-vaccinated with 20 μl BSG (n = 5). Oral challenge at doses 1x109 or 1x108 CFU were performed with UK-1 or 14028S 35 days after immunization. Mice were monitored for mortality and signs of morbidity for 30 days after challenge. Survival curve of (A) UK-1(ΔaroA)-vaccinated UK-1 challenged mice, (B) UK-1(ΔaroA)-vaccinated 14028S challenged mice, (C) 14028S(ΔaroA)-vaccinated UK-1 challenged mice, and (D) 14028S(ΔaroA)-vaccinated 14028S challenged mice. Log-rank (Mantel-Cox) test was used to plot and analyze survival curves.
Antibody responses to SOMPs and LPS
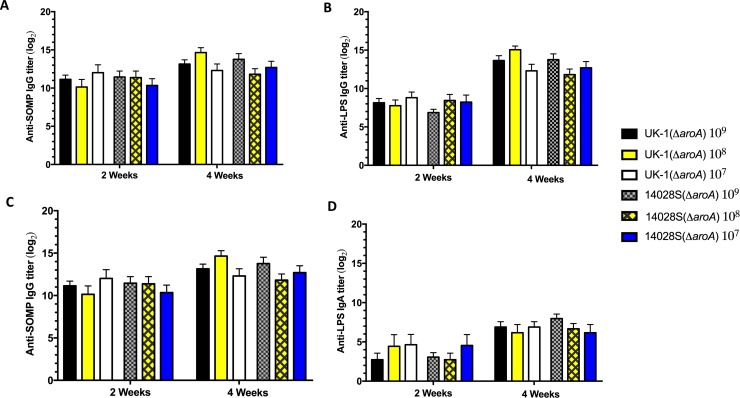
Serum immunoglobulin G (IgG) and mucosal IgA responses to Salmonella outer membrane proteins (SOMPs) and lipopolysaccharide (LPS) were measured by ELISA (Fig 2A–2D). Both aroA mutants UK-1(ΔaroA) and 14028S(ΔaroA) induced equivalent anti-SOMP and anti-LPS serum IgG responses (Fig 2A & 2B). At 4 weeks, the IgG titers in mice immunized with either UK-1(ΔaroA) or 14028S(ΔaroA) were slightly higher than those at 2 weeks, with no significant differences between the groups. Additionally, the secretory IgA endpoint titers induced by both vaccine strains were suboptimal at both 2 and 4 weeks (Fig 2C & 2D). No serum IgG or secretory IgA titers were detected in mice that were mock-vaccinated with BSG, as expected (data not shown). The type of immune response to SOMPs and LPS was determined by measuring the levels of IgG isotype subclasses IgG1 and IgG2a (S2A–S2D Fig). A Th1- and Th2-type mixed response was observed against LPS antigen since both strains induced high IgG1 and IgG2a responses. The IgG2a titers to SOMP at 4 weeks in both UK-1(ΔaroA) (P < 0.001) and 14028S(ΔaroA) (P < 0.0001) groups were significantly higher than IgG1 titers. Collectively, these data suggest that there is no difference in antibody responses between UK-1(ΔaroA) and 14028S(ΔaroA)-immunized mice, suggesting that antibodies might play a role in vaccine-mediated protection against challenge with wild-type parent strains, but are not the sole determining factor.
Fig 2. Anti-SOMP and anti-LPS antibody responses in mice.
Serum IgG responses to (A) SOMP, and (B) LPS as well mucosal IgA responses to (C) SOMP, and (D) LPS were determined by ELISA at 2 and 4 weeks after oral immunization in sera and vaginal washes from BALB/c mice (n = 6) orally immunized with UK-1(ΔaroA) or 14028S(ΔaroA) at doses 1x109, 1x108 or 1x107 CFU. No statistically significant difference was observed between vaccine groups.
Antigen presentation and cytokine production
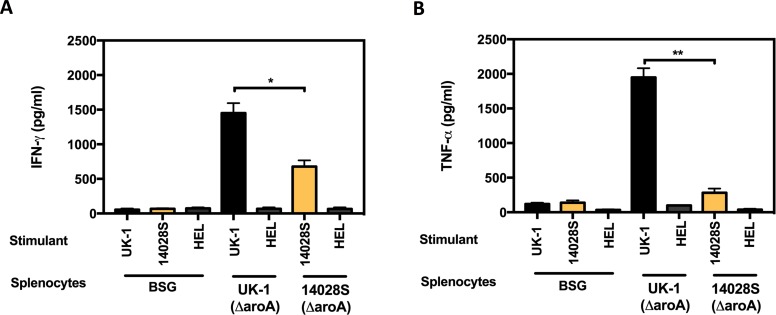
Since both UK-1(ΔaroA) and 14028S(ΔaroA) elicited similar levels of antibody responses despite differential protective efficacies, cytokine concentrations were assayed to compare with protection data. Antigen-presenting cells (APCs) were generated by treating the spleen cells obtained from naïve BALB/c mice with mitomycin C, followed by removal of non-adherent cells. APCs (2 x 105 cells/well) were then infected with UV-inactivated UK-1 or 14028S (MOI of 10) for 2 h. Hen egg lysozyme (HEL) was included as an unrelated antigen for comparison. These antigen-loaded APCs were co-cultured for 72 h with T-lymphocytes obtained from spleens of orally immunized mice (UK-1(ΔaroA) or 14028S(ΔaroA)). T-cells obtained from the BSG (mock-vaccinated) group were also co-cultured with Salmonella or HEL treated APCs as control. IFN-γ, TNF-α, IL-4 and IL-12 levels in the culture supernatants were assayed following incubation. As shown in Fig 3A and 3B, T-lymphocytes obtained from BSG group when co-cultured with APCs previously infected with Salmonella or HEL, had minimal cytokine induction. In contrast, T-lymphocytes from mice primed by oral vaccination with UK-1(ΔaroA) exhibited markedly elevated levels of IFN-γ and TNF-α upon incubation with APCs infected with UV-inactivated UK-1. However, T-lymphocytes from mice similarly immunized with 14028S(ΔaroA) and co-cultured with APCs previously infected with UV-inactivated 14028S showed significantly lower levels of IFN-γ (P < 0.05) and TNF-α (P < 0.01) in the supernatants compared with UK-1(ΔaroA). In addition, incubation of UK-1 or 14028S infected APCs with T-cells from mice immunized with respective mutant strains also did not cause significant induction of IL-4 or IL-12 (S3A and S3B Fig). Thus, proinflammatory responses such as increased secretion of IFN-γ and TNF-α appear to contribute positively to the observed superior protective efficacy of UK-1(ΔaroA).
Fig 3. APC-mediated T-cell activation and cytokine secretion.
T-lymphocytes obtained from BALB/c mice (n = 5) orally immunized with UK-1(ΔaroA), 14028S(ΔaroA) or BSG (mock) were co-incubated with respective UV-inactivated UK-1 or 14028S-infected and mitomycin C—treated APCs. APCs treated with an unrelated antigen, HEL, were also included for comparison. The co-cultures were incubated for 72 h and supernatants were collected for determination of (A) IFN-γ, (B) TNF-α. *P ≤ 0.05, **P ≤ 0.01.
Co-infection with UK-1 and 14028S
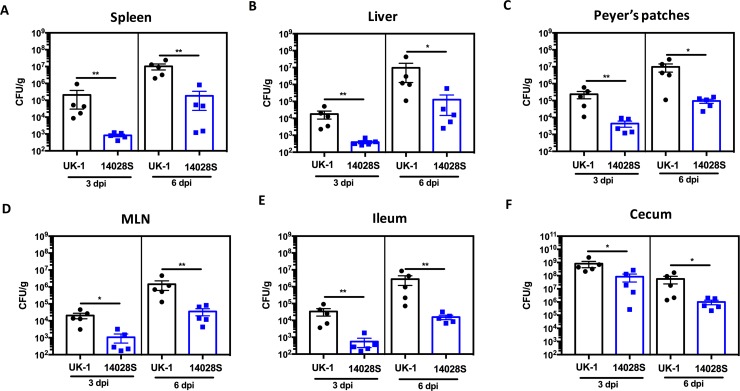
UK-1(Nalr) and 14028S(Cmr Tcr) were mixed equivalently and inoculated orally into mice (1 x 109 CFU/mouse) to test the fitness of the two strains directly against each other. Bacteria were enumerated in spleen, liver, Peyer’s patches, MLN, cecum, and ileum on days 3 and 6 post infection. Strains were distinguished phenotypically by using antibiotic markers. UK-1(Nalr) outcompeted 14028S(Cmr Tcr) and hence, a significantly higher UK-1 burden was observed (P < 0.05) in gastrointestinal tissues as well as systemic sites on both 3 and 6-day time points post infection when compared to 14028S (Fig 4A–4F). Nevertheless, when colonization was compared by infecting the mice with single strains, 14028S colonized as efficiently as UK-1 in all the tissues (S4A–S4F Fig). Conversely, when colonization was compared by inoculating the mice with UK-1(ΔaroA) or 14028S(ΔaroA), UK-1(ΔaroA) showed higher burdens in systemic tissues compared to 14028S(ΔaroA) (spleen P < 0.01, liver P < 0.01). Higher UK-1(ΔaroA) titers were also found in cecum at 6 days post infection (P < 0.01) (S5A–S5F Fig). Interestingly, 14028S(ΔaroA) was shed at significantly reduced numbers in feces by day 7 (P < 0.01) when compared to UK-1(ΔaroA) (Fig 5).
Fig 4. Co-infection with UK-1 and 14028S.
BALB/c mice were orally challenged with a 1:1 mixture of 1 x 109 CFU of UK-1(Nalr) and 14028S(Cmr Tcr). Groups of mice (n = 5 per group) were euthanized on days 3 and 6 post challenge. (A) Spleen, (B) liver, (C) Peyer’s patches, (D) MLN, (E) ileum, and (F) cecum were collected to determine the bacterial burdens. Significantly higher UK-1 burdens were observed in all tested organs on both day 3 and 6 compared to 14028S burdens. *P ≤ 0.05, **P ≤ 0.01.
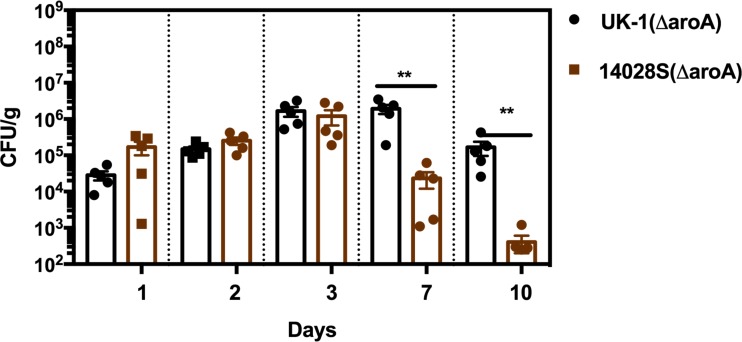
Fig 5. Vaccine strain detection in fecal pellets following oral immunization.
BALB/c mice (n = 5) were orally immunized with UK-1(ΔaroA), 14028S(ΔaroA) or BSG (mock). Fecal pellets were collected on days 0, 1, 2, 3, 7, and 10 post immunization for enumeration of bacterial shedding post vaccination. 14028S(ΔaroA) titers in the fecal pellets declined significantly by day 7 and continued to decline at day 10. Mice vaccinated with UK-1(ΔaroA) maintained significantly higher bacterial titers (**P ≤ 0.01) by day 10 post immunization (last time-point analyzed) when compared to 14028S(ΔaroA)-vaccinated mice.
UK-1 burden following immunization with UK-1(ΔaroA) and 14028S(ΔaroA)
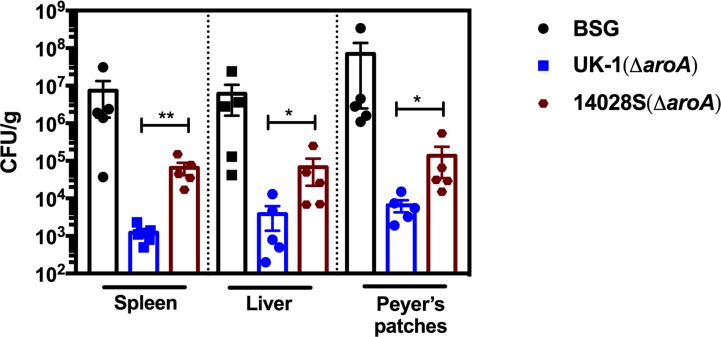
Since UK-1(ΔaroA) and 14028S(ΔaroA) showed differential levels of protection to UK-1 challenge, the ability of these strains in clearing UK-1 from various organs was tested. When mice were immunized with a 1x109 CFU dose of UK-1(ΔaroA) or 14028S(ΔaroA) and challenged 35 days after immunization, significantly higher bacterial burdens were observed after 6 days in all 14028S(ΔaroA) immunized mouse tissues that were enumerated (spleen (P < 0.01), liver (P < 0.05), and Peyer’s patches (P < 0.05)) compared to in UK-1(ΔaroA) immunized mouse tissues (Fig 6). Consistently high UK-1 burdens were also observed in all organs of the BSG control group mice. Thus, UK-1(ΔaroA)-immunized mice showed significant restriction of UK-1 spread in all target organs compared to 14028S(ΔaroA) as well as BSG-vaccinated groups.
Fig 6. UK-1 burdens are controlled efficiently by UK-1(ΔaroA), but not 14028S(ΔaroA).
BALB/c mice (n = 5) were orally immunized with UK-1(ΔaroA), 14028S(ΔaroA) or BSG (mock) on day 0 and orally challenged with UK-1 or 14028S on day 35. Spleen, liver, and Peyer’s patches were collected on day 6 post challenge to determine the bacterial burdens. UK-1 burdens were significantly reduced in all target organs when mice were immunized with UK-1(ΔaroA), but not when immunized with 14028S(ΔaroA), as compared to mock-vaccinated mice. *P ≤ 0.05, **P ≤ 0.01.
Discussion
While much research effort has focused on utilizing various S. Typhimurium strains to develop vaccines against salmonellosis and a variety of other infectious diseases, the initial virulence of the parent strain is seldom considered important in predetermining a vaccine’s efficacy. Two previous studies from our laboratory [28, 30] have suggested that the hypervirulent nature of UK-1 might impart its attenuated derivatives an advantage over similarly attenuated mutants derived from relatively less-virulent S. Typhimurium strains. The current study is the first report that provides conclusive evidence towards this proposition. We have demonstrated here that a single oral immunization with aroA-deficient UK-1 mutant, UK-1(ΔaroA), presents a greater level of protection against infection with UK-1 as well as 14028S than afforded by immunization with the less-virulent 14028S(ΔaroA) strain. Consistent with this, UK-1(ΔaroA)-vaccinated mice displayed significantly reduced UK-1 burden in Peyer’s patches, spleen and liver 6 days after lethal challenge, indicating efficient pathogen clearance. In contrast, the 14028S derivative, 14028S(ΔaroA), was effective in protecting against a challenge with its parent, and somewhat effective against a UK-1 challenge. The difference became more prominent at lower doses of immunization. At 107 CFU (low dose) of 14028S(ΔaroA), a majority of the mice succumbed to UK-1 infection by day 10, which is likely due to the relatively high UK-1 burden in the target organs.
A similar study conducted by Zhang et al. [30] describes deletions of the crp (cyclic AMP receptor protein) and cdt (colonization of deep tissues) genes in UK-1 and SL1344. These Δcrp and Δ(crp-cdt) mutants conferred complete protection against challenge with SL1344. However, neither UK-1 nor SL1344 based attenuated strains could fully protect mice against a UK-1 challenge even though the UK-1 attenuated strains performed significantly better than the latter. Furthermore, these Δcrp and Δ(crp-cdt) mutants were also unable to efficiently colonize internal lymphoid organs, suggesting poor induction of systemic or cellular immune responses. On the other hand, the current study illustrates that the UK-1 derived ΔaroA mutant UK-1(ΔaroA) not only provides complete protection against challenge with the less-virulent 14028S, but also fully protects against UK-1 challenge with a single immunization dose. Moreover, UK-1(ΔaroA) efficiently colonizes both gut-associated lymphoid tissue (GALT) as well as deeper tissues such as spleen and liver. While both UK-1(ΔaroA) and 14028S(ΔaroA) induce strong IgG responses, T-cell assays have shown that only the former could induce high-level secretion of IFN-γ and TNF-α. This trend parallels the poor colonizing ability of 14028S(ΔaroA) in internal lymphoid organs as opposed to that of UK-1(ΔaroA). Accordingly, colonization of deeper tissues and secretion of proinflammatory cytokines appear to be the underlying mechanisms mediating the induction of seemingly superior protection in UK-1(ΔaroA)-immunized mice. Even though both UK-1(ΔaroA) and 14028S(ΔaroA) could effectively colonize GALT, neither were able to elicit considerable secretory IgA responses. Overall, UK-1(ΔaroA) could afford excellent protection despite low mucosal antibody responses, and 14028S(ΔaroA) only suboptimal levels of protection even with high levels of IgG, thus antibodies seem not to be the principal basis for protective immunity to Salmonella infection.
Cytokine induction in response to Salmonella is crucial for the control and resolution of an infection [36–38]. Previous studies have reported that IFN-γ, TNF-α, IL-12, IL-15, IL-18, and IL-1β play a vital role in anti-Salmonella defense whereas IL-4 and IL-10 inhibit these protective host defenses [37, 39]. We examined the secretion of IFN-γ, TNF-α, IL-12, and IL-4 in vitro 35 days after immunization. T-lymphocytes obtained from UK-1(ΔaroA) immunized mice, upon incubation with APCs that were infected with UV-inactivated UK-1 released significant amounts of IFN-γ and TNF-α into the supernatants. Since IL-12 is known to be essential for promoting type 1 cytokine responses and cell-mediated immunity against Salmonella [37], we expected to find significant levels of this cytokine in the supernatants following 72 h incubation. However, we detected only suboptimal amounts of IL-12 upon stimulation with UV-inactivated UK-1 that may be due to the time of assessment of the supernatants. As IL-12 is produced early in the immune response to Salmonella infection, maximal levels might have been detected if analyzed at earlier time points. Degree and duration of IL-12 response may have significant consequences in terms of induction of cellular immunity and protection via production of Th1 cytokines. Interestingly, consistent with the mixed Th1/Th2 response exhibited by serum antibodies against LPS and SOMPs, a noticeable amount of IL-4 was also detected in both UK-1(ΔaroA) and 14028S(ΔaroA) groups upon stimulation. Nevertheless, it is also possible that the cytokine profile could have been considerably different if the mice were euthanized at an earlier time following immunization. However, since the mice were challenged with UK-1 or 14028S 35 days after immunization during survival and burden studies, we chose the day 35 time point for assessing cytokine production as well.
Besides differential Th1 cytokine production and colonization of internal lymphoid organs, another striking difference observed between UK-1(ΔaroA) and 14028S(ΔaroA) was the cecal and fecal titers following immunization. UK-1(ΔaroA) was detected in the fecal pellets for a significantly longer duration than 14028S(ΔaroA). This greater level of fecal shedding also correlated with the relatively higher level of cecal colonization as compared to 14028S(ΔaroA). However, these observations do not seem to accurately reflect intestinal colonization; as 14028S(ΔaroA) also colonizes GALT efficiently although it is unable to disseminate to systemic sites. UK-1(ΔaroA) has a significantly greater presence in the ceca and fecal pellets as well as in the deeper tissues, which likely contributes to its protective efficacy. We rule out the possibility that this greater persistence of UK-1(ΔaroA) could prevent subsequent colonization of the wildtype challenge strains since UK-1(ΔaroA) was not detected in the fecal pellets collected at 2 and 4 weeks post immunization. The hypervirulent nature of UK-1 likely drives these protective phenomena. In this regard, co-infection of UK-1 with 14028S clearly portrayed the ability of UK-1 to outcompete 14028S and aggressively colonize both gastrointestinal and systemic tissues.
A recent study [13] testing the efficacy of UK-1 and 14028S based LPS core deletion (ΔrfaG) mutant strains in cancer therapy has demonstrated that the UK-1 background carried a significantly higher therapeutic capacity than the 14028S background. Unlike UK-1(ΔaroA) and 14028S(ΔaroA) from our study, ΔrfaG mutants from both UK-1 and 14028S backgrounds showed similar organ colonization and TNF-α induction. Another study [40] showed that ΔaroA strains exhibited increased in vivo immunogenicity and pathogenicity compared to its highly immunogenic parent. However, we did not observe increased virulence or tissue burdens during our studies with ΔaroA mutants. One explanation could be the intravenous route of inoculation used in the abovementioned studies. To resolve this discrepancy, further testing is required to investigate whether parenteral administration of ΔaroA mutants may lead to increased immunostimulatory capacity. Future studies should also focus on comparing UK-1 and other S. Typhimurium based ΔaroA vaccine strains containing various other mutations that enable delivery of homologous and heterologous antigens. Additional attenuating mutations will also be introduced to shut off the virulence phenotypes in vivo, in order to enhance the safety of these strains prior to their use as antigen-delivering vectors.
To conclude, results of our study strongly indicate that vaccines derived from highly pathogenic UK-1 achieve maximal immunogenicity and protection when compared to less-virulent S. Typhimurium strains. These findings can be applied towards rational vaccine design to select the best vaccine candidates, especially among isogenic strains exhibiting distinct pathogenic properties, for further optimization and clinical testing.
Materials and methods
Ethics statement
All procedures involving the use of mice were thoroughly reviewed and approved by the University of Florida, Gainesville IACUC, the Institutional Animal Care and Use Committee (Study #201509049). All protocols conform to the federal regulations and policies outlined by the United States Department of Agriculture, Animal and Plant Health Inspection Service (USDA-APHIS) and Office of Laboratory Animal Welfare (OLAW).
For assessment of survival, death was used as the endpoint for evaluating protective efficacy of the vaccine strains. Specific criteria for establishing early endpoints were discussed and approved by IACUC and animal care representatives to achieve optimal scientific results without compromising conscientious ethical standards. To this end, mice exhibiting signs of distress such as labored breathing, acute dehydration, severe hunching and impaired motility were considered moribund and humanely euthanized within 4 h after onset of symptoms via carbon dioxide asphyxiation followed by cervical dislocation. Professional advice from veterinary technicians was also sought by research staff for mice in question.
Bacterial strains, plasmids, growth conditions, and reagents
Bacterial strains and plasmids used in this study are listed in Table 1. aroA mutants were generated from S. Typhimurium using suicide vector pYA3600 [41]. For routine use, S. Typhimurium vaccine strains were grown with aeration in LB medium supplemented with 0.1% glucose at 37°C [42]. Strains with the ΔaroA21419 deletion were grown with the addition of PABA (Para-aminobenzoic acid) and DHB (2,3-dihydroxybenzoic acid) at concentrations of 2 μg/ml. MacConkey plates with 1% lactose, necessary supplements, and antibiotics were used for colonization and coinfection experiments. All media were purchased from BD Difco (Franklin Lakes, NJ) unless otherwise indicated. Antibiotics were added as needed at the following concentrations: chloramphenicol, 20 μg/ml; tetracycline, 50 μg/ml; nalidixic acid, 30 μg/ml. All antibiotics and chemicals were purchased from Sigma (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA).
Table 1. Characteristics of the strains and plasmids used in this study.
| Strain or plasmid | Relevant Characteristics | Designation | Source |
|---|---|---|---|
| χ3761 | UK-1 wild-type Salmonella Typhimurium | UK-1 | [21] |
| ATCC 14028 | 14028S wild-type Salmonella Typhimurium | 14028S | [43] |
| χ9909 | ΔaroA21419, derived from χ3761 | UK-1(ΔaroA) | [41] |
| χ12359 | ΔaroA21419, derived from 14028S | 14028S(ΔaroA) | Curtiss lab chi collection |
| χ4138 | gyrA1816, derived from χ3761 | UK-1(Nalr) | [43] |
| pYA3600 | Suicide vector for generation of ΔaroA21419 | - | [41] |
| pACYC184 | E. coli plasmid cloning vector (p15A ori) with chloramphenicol and tetracycline resistance genes | Cmr Tcr | [44] |
Mice
Female BALB/c mice were obtained from Charles River Laboratories (Wilmington, MA) and acclimated for 7 days after arrival in the general housing facility. Mice were seven-weeks-old at the time of commencement of the experiments.
Immunization and challenge
Mice (5 to 10 per group) were deprived of food and water for 6 h before and 30 min after oral immunization. Salmonella strains UK-1(ΔaroA) and χ14028S(ΔaroA) were cultured in LB and supplements with aeration to an OD600 of 0.85. Cultures were centrifuged at 4,000 rpm at room temperature and resuspended in buffered saline containing 0.01% gelatin (BSG) [45]. Vaccine strains were orally administered to groups of mice at doses 1x109, 1x108 or 1x107 CFU in 20 μl BSG. 20 μl of BSG was orally administered to control groups (mock immunized). Challenge strains UK-1 and 14028S were grown and administered similarly on day 35 post immunization at 1x109 or 1x108 CFU in 20 μl BSG. Animals were monitored closely twice a day for 30 days for signs of mortality and morbidity following lethal challenge. The actual vaccination and challenge doses administered were determined by dilution plating on LB agar with supplements.
Assessment of antibody responses
Blood and vaginal washes were collected individually from immunized and control groups on the day of immunization, as well as week 1 and week 2 post immunization. 100 μl blood was collected by submandibular bleeding, and samples were incubated at 37°C for 1 h before separating the serum fractions by centrifugation at 13,000 rpm for 10 min in an Eppendorf 5415D digital tabletop microfuge (Marshall Scientific, NH). Vaginal secretions were collected as 50 μl BSG washes and stored at -20°C. Antibody responses were assayed by enzyme-linked immunosorbent assay (ELISA) [46]. Salmonella serovar Typhimurium outer membrane proteins (SOMPs) were purified from χ9761 as described previously [47]. Purified lyophilized lipopolysaccharide (LPS) isolated from S. enterica serovar Typhimurium (O-4,5,12) was obtained from Sigma. LPS and SOMPs were used at 1 μg/ml and 2 μg/ml, respectively for sensitization of 96-well flat-bottom microtiter plates (Nalge Nunc. Rochester, NY, USA). They were then incubated overnight at 4°C. Sera were diluted starting from 1:200 for detection of total IgG, and 1:100 for IgG1 and IgG2a titers, before adding to individual wells in triplicate. Vaginal secretions were pooled and diluted 1:10 before adding 100 μl of sample to wells in triplicate. Biotinylated goat anti-mouse IgA (vaginal washes), IgG, IgG1 or IgG2a were diluted 1: 10,000 for detection of anti-LPS and anti-SOMP specific antibody titers.
Assessment of cell-mediated responses
Antigen-presenting cells (APCs) were generated from spleens removed aseptically from seven-week-old naïve mice. Single-cell suspensions were counted by trypan blue exclusion [48], and treated with mitomycin C (5 μg/106 cells) for 30 min in a 75-cm2 cell culture flask (Fisher Scientific, PA), incubated at 37°C in 5% CO2, followed by 2 h of incubation to obtain adherent APCs. 2 × 105 APCs were seeded in each well (Corning 96-well TC-treated flat bottom microplates, Sigma), and infected with UV-inactivated UK-1 or 14028S (10 MOI) for 2 h. These infected APCs were cocultured for 72 h with T-lymphocytes obtained from vaccinated mice.
To obtain primed T-lymphocytes, 5 mice per group were orally immunized with 1x109 CFU dose of UK-1(ΔaroA), 14028S(ΔaroA) or BSG (control). The mice were euthanized on day 35. Spleens were disrupted to create single-cell suspensions. These suspensions were treated with RBC lysing buffer (Sigma, MO) at 37°C for 5 min to lyse the erythrocytes. Cells were washed, and viability was assessed by trypan blue exclusion. These single cell suspensions were enriched using an EasySep mouse T-cell enrichment kit (Stemcell Technologies, Vancouver, Canada). 1 x 106 cells per well were cocultured with UK-1 or 14028S-infected APCs. Following the coculture with APCs for 72 h, cytokine levels in the culture supernatants were assayed to examine APC-mediated T-cell activation.
Colonization, coinfection and bacterial shedding
Groups of mice (5 mice per time point) were immunized orally with 1x109 CFU doses of strains UK-1(ΔaroA) and 14028S(ΔaroA) or one of the two wild-type strains: χ4138 (UK-1 with nalidixic acid resistance) [43] and 14028S (with pACYC184 plasmid–for chloramphenicol and tetracycline resistance [44]). Spleen, liver, Peyer’s patches, mesenteric lymph nodes (MLN), cecum, and ileum were removed on days 3 and 6 following immunization or challenge. Bacterial counts in the homogenized tissues were determined by dilution plating on MacConkey plates containing 1% lactose and appropriate antibiotics when required. 2 μg/ml each of PABA and DHB were also added to the MacConkey plates when determining UK-1(ΔaroA) and 14028S(ΔaroA) counts. Colonies obtained were screened for dependence on PABA and DHB by patching on LB agar ± PABA and DHB. For burden studies, mice were immunized with UK-1(ΔaroA) or 14028S(ΔaroA) and challenged with UK-1 on day 35 post immunization. Bacteria were enumerated in spleen, liver, and Peyer’s patches on day 6 post challenge.
Wild-type strains were also administered in pairs to evaluate the ability of UK-1 to colonize in the presence of the less-virulent 14028S. Bacteria were enumerated in spleen, liver, Peyer’s patches, (MLN), cecum, and ileum on days 3 and 6 after challenge.
Fecal pellets were collected from mice immunized with strains UK-1(ΔaroA) or 14028S(ΔaroA) on days 1, 2, 3, 7 and 10 after immunization for enumeration of bacterial shedding post vaccination. Colonies obtained on MacConkey plates with 1% lactose, PABA, and DHB (2 μg/ml) were screened for dependence on PABA and DHB by patching on LB agar ± PABA and DHB.
Statistics
Statistical analyses were performed by using the GraphPad Prism 5 software package (Graph Software, San Diego, CA). Log-rank (Mantel-Cox) test was applied to compare survival curves following oral challenge. Antibody responses as well as bacterial titers in organs and fecal pellets were expressed as means ± standard deviations. One-way ANOVA test was used to evaluate differences in antibody titers between various groups of immunized mice. Mann-Whitney U test was used to compare bacterial titers in corresponding organs. Differences were considered significant at a P value of ≤0.05. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. When required, data were transformed to log10 or log2 prior to these calculations.
Supporting information
BALB/c mice (n = 10 per immunization dose) were orally immunized with UK-1(ΔaroA) or 14028S(ΔaroA) at doses 1x109, 1x108 or 1x107 CFU in 20 μl BSG. Mice were also mock-vaccinated with 20 μl BSG. Blood and vaginal washes were collected on day 14 and day 28 post immunization for evaluating antibody responses against LPS and SOMP. Oral challenge at doses 1x109 or 1x108 CFU were performed with UK-1 or 14028S 35 days after immunization. Mice were monitored for mortality and signs of morbidity for 30 days after challenge.
(TIFF)
Serum IgG1 and IgG2a responses to (A) LPS in UK-1(ΔaroA)-immunized mice, (B) LPS in 14028S(ΔaroA)-immunized mice, (C) SOMP in UK-1(ΔaroA)-immunized mice, and (D) SOMP in 14028S(ΔaroA)-immunized mice were determined by ELISA at 2 and 4 weeks after oral immunization in sera and vaginal washes from BALB/c mice (n = 6).
(TIFF)
T-lymphocytes obtained from BALB/c mice (n = 5) orally immunized with UK-1(ΔaroA), 14028S(ΔaroA) or BSG (mock) were co-incubated with respective UV-inactivated UK-1 or 14028S-infected and mitomycin C—treated APCs. APCs treated with an unrelated antigen, HEL, were also included for comparison. The co-cultures were incubated for 72 h and supernatants were collected for determination of (A) IL-4, and (B) IL-12.
(TIFF)
BALB/c mice were orally challenged with 1 x 109 CFU of UK-1(Nalr) or 14028S(Cmr Tcr). Groups of mice (n = 5 per group) were euthanized on days 3 and 6 post challenge. (A) Spleen, (B) liver, (C) Peyer’s patches, (D) MLN, (E) ileum, and (F) cecum were collected to determine the bacterial burdens. No statistically significant difference was observed between challenge groups.
(TIFF)
BALB/c mice were orally immunized with 1 x 109 CFU UK-1(ΔaroA) or 14028S(ΔaroA). Groups of mice (n = 5 per group) were euthanized on days 3 and 6 post challenge. (A) Spleen, (B) liver, (C) Peyer’s patches, (D) MLN, (E) ileum, and (F) cecum were collected for enumeration of bacteria.
(TIFF)
Acknowledgments
The aroA allele (aroA21419) is designated in honor of the memory of the pioneer Salmonella research scientist Bruce A. D. Stocker. Special thanks to Soo-Young Wanda, Katherine Gonzalez and Matt Bellefleur for technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institutes of Health Grants R01AI056289 (to RCIII) and R21AI126172 (to SFW) (https://www.niaid.nih.gov/) as well as the Florida Veterinary Scholars Program (http://research.vetmed.ufl.edu/research-programs/student-scholars-program/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rabsch W, Tschäpe H, Bäumler AJ. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 2001;3(3):237–47. . [DOI] [PubMed] [Google Scholar]
- 2.Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32(2):263–9. Epub 2001/01/15. 10.1086/318457 . [DOI] [PubMed] [Google Scholar]
- 3.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379(9835):2489–99. 10.1016/S0140-6736(11)61752-2 ; PubMed Central PMCID: PMCPMC3402672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winfield MD, Groisman EA. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl Environ Microbiol. 2003;69(7):3687–94. 10.1128/AEM.69.7.3687-3694.2003 ; PubMed Central PMCID: PMCPMC165204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortiz D, Siegal EM, Kramer C, Khandheria BK, Brauer E. Nontyphoidal cardiac salmonellosis: two case reports and a review of the literature. Tex Heart Inst J. 2014;41(4):401–6. Epub 2014/08/01. doi: 10.14503/THIJ-13-3722 ; PubMed Central PMCID: PMCPMC4120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desin TS, Koster W, Potter AA. Salmonella vaccines in poultry: past, present and future. Expert review of vaccines. 2013;12(1):87–96. 10.1586/erv.12.138 . [DOI] [PubMed] [Google Scholar]
- 7.MacLennan CA, Martin LB, Micoli F. Vaccines against invasive Salmonella disease: current status and future directions. Hum Vaccin Immunother. 2014;10(6):1478–93. 10.4161/hv.29054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erova TE, Kirtley ML, Fitts EC, Ponnusamy D, Baze WB, Andersson JA, et al. Protective Immunity Elicited by Oral Immunization of Mice with Salmonella enterica Serovar Typhimurium Braun Lipoprotein (Lpp) and Acetyltransferase (MsbB) Mutants. Front Cell Infect Microbiol. 2016;6:148 Epub 2016/11/10. 10.3389/fcimb.2016.00148 ; PubMed Central PMCID: PMCPMC5103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q, Zhao X, Liu T, Yi J, Liang K, Kong Q. Immunogenicity and Cross-Protective Efficacy Induced by Outer Membrane Proteins from Salmonella Typhimurium Mutants with Truncated LPS in Mice. Int J Mol Sci. 2016;17(3):416 Epub 2016/03/22. 10.3390/ijms17030416 ; PubMed Central PMCID: PMCPMC4813267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark-Curtiss JE, Curtiss R III. Salmonella Vaccines: Conduits for Protective Antigens. J Immunol. 2018;200(1):39–48. 10.4049/jimmunol.1600608 . [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, Mo H, Willingham C, Wang S, Park JY, Kong W, et al. Protection Against Necrotic Enteritis in Broiler Chickens by Regulated Delayed Lysis Salmonella Vaccines. Avian diseases. 2015;59(4):475–85. 10.1637/11094-041715-Reg . [DOI] [PubMed] [Google Scholar]
- 12.Sanapala S, Rahav H, Patel H, Sun W, Curtiss R, III. Multiple antigens of Yersinia pestis delivered by live recombinant attenuated Salmonella vaccine strains elicit protective immunity against plague. Vaccine. 2016;34(21):2410–6. 10.1016/j.vaccine.2016.03.094 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felgner S, Kocijancic D, Frahm M, Curtiss R III, Erhardt M, Weiss S. Optimizing Salmonella enterica serovar Typhimurium for bacteria-mediated tumor therapy. Gut Microbes. 2016;7(2):171–7. 10.1080/19490976.2016.1155021 ; PubMed Central PMCID: PMCPMC4856459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinod N, Noh HB, Oh S, Ji S, Park HJ, Lee KS, et al. A Salmonella typhimurium ghost vaccine induces cytokine expression in vitro and immune responses in vivo and protects rats against homologous and heterologous challenges. PLoS One. 2017;12(9):e0185488 Epub 2017/09/29. 10.1371/journal.pone.0185488 ; PubMed Central PMCID: PMCPMC5621678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtiss R III, Nakayama K, Kelly SM. Recombinant avirulent Salmonella vaccine strains with stable maintenance and high level expression of cloned genes in vivo. Immunol Invest. 1989;18(1–4):583–96. . [DOI] [PubMed] [Google Scholar]
- 16.Galán JE, Nakayama K, Curtiss R III. Cloning and characterization of the asd gene of Salmonella typhimurium: use in stable maintenance of recombinant plasmids in Salmonella vaccine strains. Gene. 1990;94(1):29–35. Epub 1990/09/28. doi: 0378-1119(90)90464-3 [pii]. . [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Li Y, Scarpellini G, Kong W, Shi H, Baek CH, et al. Salmonella vaccine vectors displaying delayed antigen synthesis in vivo to enhance immunogenicity. Infect Immun. 2010;78(9):3969–80. 10.1128/IAI.00444-10 ; PubMed Central PMCID: PMC2937466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galen JE, Wang JY, Chinchilla M, Vindurampulle C, Vogel JE, Levy H, et al. A new generation of stable, nonantibiotic, low-copy-number plasmids improves immune responses to foreign antigens in Salmonella enterica serovar Typhi live vectors. Infect Immun. 2010;78(1):337–47. Epub 2009/11/02. 10.1128/IAI.00916-09 ; PubMed Central PMCID: PMCPMC2798174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177(14):4121–30. ; PubMed Central PMCID: PMCPMC177145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtiss R III, Wanda SY, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, et al. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect Immun. 2009;77(3):1071–82. Epub 2008/12/22. 10.1128/IAI.00693-08 ; PubMed Central PMCID: PMCPMC2643627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtiss R III, Hassan JO. Nonrecombinant and recombinant avirulent Salmonella vaccines for poultry. Vet Immunol Immunopathol. 1996;54(1–4):365–72. . [DOI] [PubMed] [Google Scholar]
- 22.Hassan JO, Curtiss R III. Control of colonization by virulent Salmonella typhimurium by oral immunization of chickens with avirulent delta cya delta crp S. typhimurium. Res Microbiol. 1990;141(7–8):839–50. . [DOI] [PubMed] [Google Scholar]
- 23.Hassan JO, Porter SB, Curtiss R III. Effect of infective dose on humoral immune responses and colonization in chickens experimentally infected with Salmonella typhimurium. Avian Dis. 1993;37(1):19–26. . [PubMed] [Google Scholar]
- 24.Hassan JO, Curtiss R III. Development and evaluation of an experimental vaccination program using a live avirulent Salmonella typhimurium strain to protect immunized chickens against challenge with homologous and heterologous Salmonella serotypes. Infect Immun. 1994;62(12):5519–27. ; PubMed Central PMCID: PMC303297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aehle S. III RC. Current and Future Perspectives on Development of Salmonella Vaccine Technologies In: Ricke S, Gast R, editors. Producing Safe Eggs. San Diego, CA: Elsevier; 2016. p. 281–99. [Google Scholar]
- 26.Zhang X, Kelly SM, Bollen W, Curtiss R, III. Protection and immune responses induced by attenuated Salmonella typhimurium UK-1 strains. Microb Pathog. 1999;26(3):121–30. . [DOI] [PubMed] [Google Scholar]
- 27.Barrow PA, Page K, Lovell MA. The virulence for gnotobiotic pigs of live attenuated vaccine strains of Salmonella enterica serovars Typhimurium and Enteritidis. Vaccine. 2001;19(25–26):3432–6. . [DOI] [PubMed] [Google Scholar]
- 28.Luo Y, Kong Q, Yang J, Mitra A, Golden G, Wanda SY, et al. Comparative genome analysis of the high pathogenicity Salmonella Typhimurium strain UK-1. PLoS One. 2012;7(7):e40645 10.1371/journal.pone.0040645 ; PubMed Central PMCID: PMCPMC3391293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Covone MG, Brocchi M, Palla E, Dias da Silveira W, Rappuoli R, Galeotti CL. Levels of expression and immunogenicity of attenuated Salmonella enterica serovar Typhimurium strains expressing Escherichia coli mutant heat-labile enterotoxin. Infect Immun. 1998;66(1):224–31. ; PubMed Central PMCID: PMCPMC107881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Kelly SM, Bollen WS, Curtiss R, III. Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 delta crp and delta cdt deletion mutants. Infect Immun. 1997;65(12):5381–7. ; PubMed Central PMCID: PMCPMC175779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291(5812):238–9. . [DOI] [PubMed] [Google Scholar]
- 32.Bentley R. The shikimate pathway—a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25(5):307–84. 10.3109/10409239009090615 . [DOI] [PubMed] [Google Scholar]
- 33.Ruby T, McLaughlin L, Gopinath S, Monack D. Salmonella's long-term relationship with its host. FEMS Microbiol Rev. 2012;36(3):600–15. Epub 2012/03/09. 10.1111/j.1574-6976.2012.00332.x . [DOI] [PubMed] [Google Scholar]
- 34.Harrison JA, Villarreal-Ramos B, Mastroeni P, Demarco de Hormaeche R, Hormaeche CE. Correlates of protection induced by live Aro- Salmonella typhimurium vaccines in the murine typhoid model. Immunology. 1997;90(4):618–25. ; PubMed Central PMCID: PMCPMC1456680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villarreal-Ramos B, Manser J, Collins RA, Dougan G, Chatfield SN, Howard CJ. Immune responses in calves immunised orally or subcutaneously with a live Salmonella typhimurium aro vaccine. Vaccine. 1998;16(1):45–54. . [DOI] [PubMed] [Google Scholar]
- 36.Altare F, Lammas D, Revy P, Jouanguy E, Döffinger R, Lamhamedi S, et al. Inherited interleukin 12 deficiency in a child with bacille Calmette-Guérin and Salmonella enteritidis disseminated infection. J Clin Invest. 1998;102(12):2035–40. 10.1172/JCI4950 ; PubMed Central PMCID: PMCPMC509157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalmanach AC, Lantier F. Host cytokine response and resistance to Salmonella infection. Microbes Infect. 1999;1(9):719–26. . [DOI] [PubMed] [Google Scholar]
- 38.Jouanguy E, Döffinger R, Dupuis S, Pallier A, Altare F, Casanova JL. IL-12 and IFN-gamma in host defense against mycobacteria and salmonella in mice and men. Curr Opin Immunol. 1999;11(3):346–51. . [DOI] [PubMed] [Google Scholar]
- 39.Mizuno Y, Takada H, Nomura A, Jin CH, Hattori H, Ihara K, et al. Th1 and Th1-inducing cytokines in Salmonella infection. Clin Exp Immunol. 2003;131(1):111–7. 10.1046/j.1365-2249.2003.02060.x ; PubMed Central PMCID: PMCPMC1808588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felgner S, Frahm M, Kocijancic D, Rohde M, Eckweiler D, Bielecka A, et al. aroA-Deficient Salmonella enterica Serovar Typhimurium Is More Than a Metabolically Attenuated Mutant. MBio. 2016;7(5). Epub 2016/09/06. 10.1128/mBio.01220-16 ; PubMed Central PMCID: PMCPMC5013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Kong W, Wanda SY, Xin W, Alamuri P, Curtiss R, III. Generation of influenza virus from avian cells infected by Salmonella carrying the viral genome. PLoS One. 2015;10(3):e0119041 10.1371/journal.pone.0119041 ; PubMed Central PMCID: PMC4351096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62(3):293–300. Epub 1951/09/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gulig PA, Curtiss R III. Plasmid-associated virulence of Salmonella typhimurium. Infect Immun. 1987;55(12):2891–901. ; PubMed Central PMCID: PMCPMC260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134(3):1141–56. ; PubMed Central PMCID: PMCPMC222365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtiss R III, Charamella LJ, Berg CM, Harris PE. Kinetic and genetic analyses of D-cycloserine inhibition and resistance in Escherichia coli. J Bacteriol. 1965;90(5):1238–50. ; PubMed Central PMCID: PMCPMC315808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang HY, Srinivasan J, Curtiss R III. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar typhimurium vaccine. Infect Immun. 2002;70(4):1739–49. 10.1128/IAI.70.4.1739-1749.2002 ; PubMed Central PMCID: PMCPMC127874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24(3):330–2. ; PubMed Central PMCID: PMCPMC268907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strober W. Trypan blue exclusion test of cell viability. Curr Protoc Immunol. 2001;Appendix 3:Appendix 3B. 10.1002/0471142735.ima03bs21 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BALB/c mice (n = 10 per immunization dose) were orally immunized with UK-1(ΔaroA) or 14028S(ΔaroA) at doses 1x109, 1x108 or 1x107 CFU in 20 μl BSG. Mice were also mock-vaccinated with 20 μl BSG. Blood and vaginal washes were collected on day 14 and day 28 post immunization for evaluating antibody responses against LPS and SOMP. Oral challenge at doses 1x109 or 1x108 CFU were performed with UK-1 or 14028S 35 days after immunization. Mice were monitored for mortality and signs of morbidity for 30 days after challenge.
(TIFF)
Serum IgG1 and IgG2a responses to (A) LPS in UK-1(ΔaroA)-immunized mice, (B) LPS in 14028S(ΔaroA)-immunized mice, (C) SOMP in UK-1(ΔaroA)-immunized mice, and (D) SOMP in 14028S(ΔaroA)-immunized mice were determined by ELISA at 2 and 4 weeks after oral immunization in sera and vaginal washes from BALB/c mice (n = 6).
(TIFF)
T-lymphocytes obtained from BALB/c mice (n = 5) orally immunized with UK-1(ΔaroA), 14028S(ΔaroA) or BSG (mock) were co-incubated with respective UV-inactivated UK-1 or 14028S-infected and mitomycin C—treated APCs. APCs treated with an unrelated antigen, HEL, were also included for comparison. The co-cultures were incubated for 72 h and supernatants were collected for determination of (A) IL-4, and (B) IL-12.
(TIFF)
BALB/c mice were orally challenged with 1 x 109 CFU of UK-1(Nalr) or 14028S(Cmr Tcr). Groups of mice (n = 5 per group) were euthanized on days 3 and 6 post challenge. (A) Spleen, (B) liver, (C) Peyer’s patches, (D) MLN, (E) ileum, and (F) cecum were collected to determine the bacterial burdens. No statistically significant difference was observed between challenge groups.
(TIFF)
BALB/c mice were orally immunized with 1 x 109 CFU UK-1(ΔaroA) or 14028S(ΔaroA). Groups of mice (n = 5 per group) were euthanized on days 3 and 6 post challenge. (A) Spleen, (B) liver, (C) Peyer’s patches, (D) MLN, (E) ileum, and (F) cecum were collected for enumeration of bacteria.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.