Abstract
Post-translational modifications, such as methylation, acetylation and phosphorylation, of histone proteins play important roles in regulating dynamic chromatin structure. Histone demethylation has become one of the most active research areas of epigenetics in the past decade. To date, with the exception of histone H3 lysine 79 methylation, the demethylases for all major lysine methylation sites have been discovered. These enzymes have been shown to be involved in various biological processes, with embryonic development being an exciting emerging area. This review will primarily discuss the involvement of these demethylases in the regulation of mammalian embryonic development, including their roles in embryonic stem cell pluripotency, primordial germ cell (PGC) formation and maternal-to-zygotic transition.
Introduction
In eukaryotic cells, DNA is packed together with histone proteins in a highly organized form called chromatin. Post-translational modifications of histones, such as acetylation, methylation, phosphorylation and ubiquitination, have been shown to influence chromatin structure and thus play critical roles in regulating gene transcription and genome integrity.1 Of the different histone modifications, methyl groups have been found to have the lowest turn-over rate; therefore, histone methylation was originally thought to be a permanent mark.2 The discovery of the first histone demethylase, KDM1A/LSD1, remarkably changed our view on the dynamic nature of lysine methylation. So far, ~20 histone lysine demethylases have been discovered, and these cover most of the major lysine methylation sites, including H3K4, H3K9, H3K27, H3K36 and H4K20; however, the demethylase for H3K79 has not yet been identified (Box 1).
Box 1 Histone demethylases and their substrate specificities.
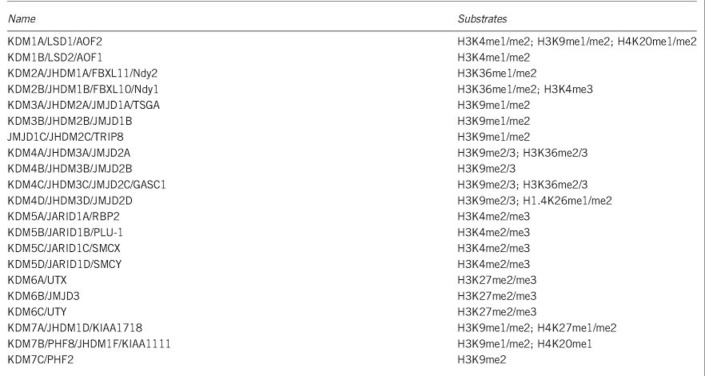
One of the major features of histone lysine methylation is that its regulation and function is highly site dependent. For example, H3K4me3 and H3K36me3 positively correlate with transcription and are usually restricted to the promoters and gene bodies of actively transcribed genes, respectively. H3K4me1 alone is the hallmark of primed enhancers, when presents with H3K27me3 or H3K27Ac, the combinatorial modifications are regarded as the marks of poised (H3K4me1/H3K27me3) and active (H3K4me1/H3K27Ac) enhancers, respectively.3 Several genome-wide studies have demonstrated the dynamic quality of histone lysine methylation during various processes in normal development and disease, suggesting critical roles for the corresponding methyltransferases and demethylases during these processes.4, 5
Specifically, embryonic development is a highly dynamic process, coupled with an orchestrated re-organization of the epigenome that shapes the chromatin environment to prepare for the subsequent development stage. In this review, we will first focus on the regulatory actions of histone demethylases on specific chromatin elements, including promoters, enhancers and repetitive elements, and then discuss their involvement in the regulation of mammalian embryonic development, including embryonic stem cell (ESC) pluripotency, primordial germ cell (PGC) formation and maternal-to-zygotic transition.
Regulation of histone demethylation at regulatory regions
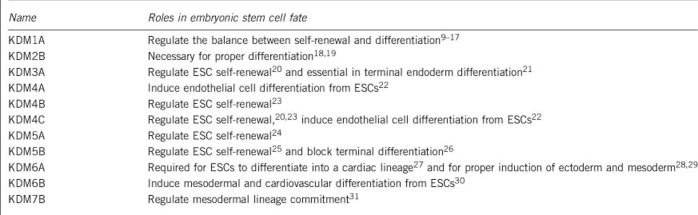
Recent epigenomic studies of human and mouse embryonic stem cells (hESCs and mESCs) have revealed certain characteristic features that are in contrast to differentiated cells, especially at regulatory regions, such as bivalent promoters,6 poised enhancers,7 and super enhancers8 as well as at some repetitive regions. Various demethylases have been reported to be involved in regulating these chromatin elements (Box 2).
Box 2 Histone demethylases and embryonic stem cell fate.
Regulation of bivalent promoters
Promoters are regulatory DNA elements that initiate gene transcription. Two of the most studied lysine methylations at promoters are histone H3 lysine 4 tri-methylation (H3K4me3) and H3 lysine 27 tri-methylation (H3K27me3), which generally represent active and repressed transcriptional states, respectively. In 2006, the Beinstein group identified thousands of promoters in mESCs that contain both H3K4me3 and H3K27me3 marks and called them ‘bivalent promoters’.6 Though bivalent promoters have recently been identified in differentiated cells, ESCs have a much larger number of bivalent promoters,32 suggesting the unique role of these promoters in regulating ESC function. Consistently, many bivalent promoters were found to be associated with developmental genes in ESCs and are believed to keep these genes in a poised state, which is ready for gene activation or repression upon differentiation by the removal of either H3K4me3 or H3K27me3, respectively.33, 34 The resolution of the bivalent state is believed to involve active demethylation processes.
Of the H3K4me3 demethylases, KDM5B is the most studied due to its regulation of bivalent promoters. Loss of KDM5B causes a number of bivalent genes, including Hox genes, to show elevated H3K4me3 levels during differentiation, resulting in enhanced self-renewal and compromised differentiation.35, 36, 37 On the other hand, during lineage specification, H3K27me3 demethylases are recruited to resolve H3K27me3-mediated repression and activate lineage-specific genes. For example, upon Nodal signaling, KDM6B is recruited to the bivalent promoter of Smad2, resulting in demethylation of H3K27me3 and gene activation.38 Another demethylase, KDM6A, has also been shown to be required for the activation of bivalent genes, such as Hox genes, during mESC differentiation.39
Interestingly, mammalian sperm also contain thousands of bivalent promoters.40 Despite the occurrence of global H3K4me3 and H3K27me3 demethylation at the two-cell stage after fertilization (also discussed in the section ‘Histone demethylation in fertilized oocytes’), after re-establishment, half of the sperm bivalent promoters regain the bivalent state in mESCs, but show paternal and maternal asymmetry.40, 41, 42 Further investigations are required to understand how bivalent promoters are regulated in gametic and zygotic cells and whether the bivalent promoters in the gametic genome have a preset unique chromatin environment for later stages after fertilization.
Regulation of enhancers
Enhancers are distinct genomic regions required for proper activation of their target gene(s), especially lineage-specific genes. Therefore, their activities need to be differentially regulated in each cell type. In fact, recent studies have revealed that enhancer activities are very dynamic, even in a given cell type.43, 44 Enhancers are generally considered to have silent, primed, poised and activated states, which are marked by different kinds of histone modifications. For instance, primed enhancers are marked by H3K4me1, active enhancers are marked by both H3K4me1 and H3K27Ac, and poised enhancers are marked by H3K4me1 and H3K27me3.45 Interestingly, in ESCs, many highly expressed pluripotency factors are controlled by a stretch of active enhancers in close proximity, which are also called super enhancers.8 During differentiation, enhancers of pluripotency genes need to be silenced, and in contrast, enhancers of differentiation genes need to be activated. In these processes, demethylases of H3K4me1, H3K4me3 and H3K27me3 play critical roles in regulating enhancer activities.
Among the H3K4 demethylases, KDM1A, a demethylase of H3K4me1 and H3K4me2, has been extensively studied. It is found to occupy ESC-specific enhancers and catalyze H3K4me1 demethylation at these enhancers upon differentiation. This process is required for the decommissioning of pluripotency-specific enhancers to facilitate differentiation,9, 10, 11, 12, 13 while other groups have also reported that KDM1A is required for the maintenance of the ESC self-renewal ability.14, 15, 16, 17
Moreover, recent studies have found that active enhancers are also regulated by the H3K4me3 demethylase KDM5C.46, 47, 48 In Shen’s study, active enhancers were demonstrated to be marked by H3K4me3 and H3K27Ac, and interestingly, these enhancers show higher activity than enhancers marked by H3K4me1 and H3K27Ac. They have termed such enhancers ‘overly activated enhancers’. Importantly, a chromatin complex containing RACK7/ZMYND8 and KDM5C was identified to control the balance of H3K4me3 and H3K4me1 at these ‘overly activated enhancers’ to avoid inappropriate activation of downstream genes. This finding not only indicated that a subset of the H3K4me1 marked enhancers could be the demethylation products but also revealed a previously unknown enhancer state. Although the study was primarily done in a breast cancer cell line, RACK7 was also shown to bind thousands of active enhancers in mESCs in the same study; whether it and the KDM5C complex play a role in regulating ESC pluripotency is an interesting question for further study. Notably, KDM5B also binds and regulates H3K4 methylation at active enhancers in mESCs, and KDM5B loss leads to the spread of H3K4 methylation to enhancer shores,36 which appears to indicate a different mechanism compared to KDM5C-mediated regulation.
Regulation of repetitive elements
Retrotransposons, which encompass more than half of the human genome, are usually epigenetically silent and packed into heterochromatin, to avoid insertional mutagenesis or transcriptional perturbation. However, it has recently been reported that they could also be transcriptionally active in ESCs, and this mechanism may create genetic diversity at genic and regulatory regions. Conceivably, this mechanism needs to be tightly regulated during embryonic development to avoid the deleterious effects of potential retrotransposition.
Consistent with the fact that they are controlled at a low transcriptional state, the constitutive heterochromatin marks H3K9me3 and H4K20me3 are enriched at telomeric, satellite and long terminal repeats in mESCs,32 with similar patterns. In addition, H3K9me3 has been reported to specifically repress intact LINE elements in mESCs.49 The demethylation of H3K9me3 by the JmjC family of histone demethylases such as KDM4A and KDM4B is required to control heterochromatic organization.50, 51, 52
Moreover, H3K27me3, a facultative heterochromatin mark, has also been shown to be regulated at certain retrotransposons in mESCs. When cultured in serum and LIF conditions, mESCs display a great degree of intercellular heterogeneity, consisting of a ‘ground’ state with robust self-renewal ability and a more committed ‘primed’ state. Interestingly, repetitive elements are differentially regulated in these two states. Compared to the primed state, a lower level of H3K27me3 at retrotransposons, such as HERV-H, was observed in the ground state,53 suggesting that H3K27 demethylases are potentially involved in regulating these repetitive elements in the transition between the ground and primed states.
As the long terminal repeat elements were originally retroviral promoters, it is conceivable that they may escape the repression mechanisms discussed above and regain transcriptional activities. Supporting this notion, the active histone mark H3K4 methylation has been found at MERV-L elements and correlates with their transcriptional activities. Importantly, KDM1A plays a role in safeguarding de-repressed MERV-L. This mechanism, when compromised, leads to development arrest at gastrulation.54
Regulation of histone demethylation in embryonic development
In this section, we will discuss the involvement of histone lysine demethylases in various embryonic development processes. We have primarily summarized the processes using ESCs and KO mice as models. In addition, due to the uniqueness of PGC formation and maternal-to-zygotic transition, we have discussed them separately at the end of this section.
Demethylases regulating ESC pluripotency and embryonic development
H3K4 demethylases
Of the four H3K4me3 demethylases (Box 1), KDM5B is the most studied in ESCs. As mentioned above, KDM5B acts as an important regulator at promoters and enhancers in mESCs and is required for proper differentiation. KDM5B expression has been reported in various tissues during mouse embryogenesis, whereas its expression becomes restricted to testis in adults, indicating a role in testis-related functions.55, 56 As mentioned above, KDM5B is essential for ESC pluripotency and is required for the resolution of the bivalent state of developmental genes during differentiation,25, 26 and its loss leads to early embryonic lethality between E4.5 and E7.5.57 However, another group reported that the loss of KDM5B only resulted in post-natal lethality of the majority of pups.58 This discrepancy could be due to the differences in genetic backgrounds and experimental approaches. In contrast, its closely related paralog, KDM5A, is not an essential gene in mice,57 although KDM5A loss in mESCs leads to downregulation of stem cell markers and activation of differentiation genes.24, 59, 60 A KDM5C gene trap mouse model was reported to exhibit abnormal social behavior, consistent with the observation that it is a frequently mutated gene in patients with mental retardation and autism.61 The H3K4me1 and H3K4me2 demethylase KDM1A is important for enhancer and ERV silencing in ESCs and essential for mouse embryonic development beyond E6.5 and extra-embryonic tissue development.54
H3K27 demethylases
On the H3K27me3 side, both KDM6A and KDM6B have been found to be required for proper lineage specification. Loss of KDM6A was reported to compromise mESC differentiation but not the self-renewal ability.27, 28, 62, 63 Although still capable of retinoic acid-induced differentiation,63 KDM6A KO mESCs displayed impaired cardiac lineage differentiation,27 and improper ectoderm and mesoderm formation.28, 29 Interestingly, while KDM6A-deficient females develop complete embryonic lethality, males only display partial embryonic lethality.27, 29, 63, 64, 65
Regarding KDM6B, there are discrepancies among different studies. In some studies, KDM6B was required for blastocyst development66 and essential for embryo development.30 However, in other studies, KDM6B KO fetuses could survive until birth but with several defects in lung development,67, 68, 69 possibly due to compromised regulation of the WNT pathway.70
H3K9 demethylases
H3K9 demethylases have also been shown to regulate ESC function. For instance, KDM3A was reported to demethylate H3K9me2 at the promoter regions of several pluripotency-associated genes and positively regulate their expression.20 It also acts as a key modulator of cell fate decisions in terminal endoderm differentiation.21 KDM3A directly regulates the promoter H3K9 demethylation and the expression of Sry, causing XY sex reversal.71 In another study, the Zhang group showed that KDM3A knockout mice exhibit spermatogenesis defects and develop obesity in adulthood.72 In addition, it has been reported that the H3K9me3 demethylases KDM4A and KDM4C are both required for mESC differentiation into endothelial cells.22 KDM4A initiates differentiation by targeting the Flk1 promoter, whereas KDM4C promotes endothelial cell fate by targeting the VE-cadherin promoter.22 In addition, KDM4B and KDM4C are both required for ESC self-renewal. Mechanically, KDM4C has been shown to regulate Nanog expression,20 while KDM4B regulates a different set of target genes,23 indicating non-overlapping functions between the two paralogs. Moreover, another H3K9 demethylase, KDM7B, has also been reported to regulate mesodermal lineage commitment and cardiomyocyte differentiation of mESCs.31
H3K36 demethylases
As an accessory factor of PRC1 (Polycomb repressive complex 1), KDM2B was reported to be enriched at PRC1-repressed genes related to embryo development, morphogenesis and cellular differentiation in ESCs. In addition, depletion of KDM2B induced expression of a subset of these genes and, similar to PRC1 loss, caused a defect in embryoid body (EB) formation.18, 19 Interestingly, the function of KDM2B in ESCs is dependent on the non-methylated CpG island binding ability through its CXXC domain, but not the H3K36me2 demethylase activity.18, 19 Consistently, KDM2B prevents DNA hypermethylation of target CpG islands, and its loss leads to increased embryonic lethality, especially in females.73, 74 This may be because KDM2B is required for the coordinated expression of Xist, Tsix and Xist RNA-associated factors.74 On the other hand, KDM2A, a closely related paralog but without a CXXC domain, is required for proper regulation of cell-cycle genes, and its loss leads to embryonic lethality at E10.5–E12.5.75
Many demethylases have been shown to be required for ESC pluripotency and embryonic development, suggesting that active demethylation plays critical roles in these processes. Interestingly, despite a high level of sequence similarities, subfamily members usually possess non-redundant roles, and their KO mice develop different phenotypes (Boxes 2 and 3).
Box 3 The reported phenotypes of KDM knockout animals.

Demethylases regulating PGC development
Reprogramming of histone methylation is a critical process during mammalian germline development. It has been shown that the epigenome of early gonadal human PGCs is characterized by low H3K9me2 and high H3K27me3 levels.77 Accordingly, various histone demethylases have been reported to be involved in this process.
KDM1B, an H3K4me2 demethylase, is required to establish maternal genomic imprinting.78 In the same study, conditional deletion of KDM1A in growing oocytes resulted in precocious resumption of meiosis and spindle, and chromosomal abnormalities.78 The reprogramming of H3K27me3 in E10.5–E11 primordial germ cells is safeguarded by KDM6A.62 This study also reported that KDM6A-deficient PGCs showed aberrant epigenetic reprogramming in vivo, which led to germline transmission failure in mouse chimaeras generated from KDM6A KO mESCs.62 Moreover, it has also been reported that the H3K9me2 demethylase KDM3A is critical for Tnp1 and Prm1 transcription, and mouse spermatogenesis.79, 80 Last but not the least, KDM2B has been reported to regulate the proliferation of spermatogonia and to ensure long-term sustainable spermatogenesis in mice.81
Histone demethylation in oocytes
Embryogenesis starts with a fertilized oocyte, which undergoes multiple mitotic divisions and cellular differentiation without growing in size, leading to the development of a multicellular embryo. Due to the limitation in obtaining sufficient material for epigenomic analyses, the dynamic regulation of histone lysine methylation in this process had remained unclear until recent progress in small-cell-number ChIP technology.82, 83, 84 Importantly, these studies uncovered a highly dynamic nature of histone methylation from the fertilized oocyte to ICM.
In differentiated cells, trimethylation of histone H3 at lysine 4 (H3K4me3) primarily forms sharp peaks (~1000 bp) at transcriptional start sites (TSSs);85 however, two different kinds of ‘broad’ H3K4me3 domains have recently been discovered, and the involvement of KDM5A and KDM5B in regulating these broad H3K4me3 domains has also been illustrated. Specifically, one kind of broad H3K4me3 domain (hereafter type A) mainly occurs at TSSs and is positively correlated with transcription level in two-cell embryos.82, 83 The other kind (hereafter type B), however, is not restricted to promoters and exists at a large number of intergenic loci84 and coincides with gene silencing in metaphase II (MII) oocytes. Type A broad H3K4me3 domains have been shown to be inversely correlated with DNA methylation but positively correlated with transcriptional activity.82 In contrast, type B broad H3K4me3 domains overlap almost exclusively with partially methylated DNA domains,84 and their removal leads to improper activation of the target genes. Despite the different features of these two kinds of broad H3K4me3 domains, active removal of H3K4me3 by the corresponding demethylases, KDM5A and KDM5B, has been shown to be required for the integrity of both kinds of broad H3K4me3 domains and normal early embryo development.82, 83, 84
In addition to H3K4me3, global demethylation of H3K27me3 was also found at the two-cell stage right after fertilization and was then re-established asymmetrically at the maternal and paternal bivalent genes at the ICM stage.84 The underlying mechanisms and the purpose of such global erasure and re-establishment are interesting subjects for future investigations.
Concluding remarks
In this review, we first reviewed the dynamic regulation by histone demethylases at characteristic chromatin regions and then the contributions of these demethylases to mammalian embryonic development processes, including ESC pluripotency, PGC formation and maternal-to-zygotic transition. Insights into these processes have accumulated rapidly in recent years. Along with technological advances, we can expect that more epigenomic features and the involvement of demethylation process, such as broad H3K4 domains in the oocyte and two-cell stage after fertilization and the global H3K4me3 and H3K27me3 demethylation after fertilization, will be revealed.
Future investigations of histone demethylases are needed to advance our understanding of their chromatin recruitment mechanism, as well as the crosstalk with cellular metabolic states and their roles in regulating cell fate decisions. In addition, demethylases targeting H3K79, as well as histone arginine methylation, remain to be discovered.
Acknowledgments
FL was supported by China ‘Thousand Youth Talents’ (KHH1340001), NSFC (91419306, 31371303), Shanghai Ministry of Science and Technology (16XD1400500), ‘973’ State Key Development Program (2014CB943103) and ISTC (2014DFB30020). HS was supported by NSFC (31601060) and Project funded by China Postdoctoral Science Foundation (2016M601498).
Footnotes
The authors declare no conflict of interest.
References
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys 1972; 148: 558–567. [DOI] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambacher S, Hahn M, Schotta G. Epigenetic regulation of development by histone lysine methylation. Heredity 2010; 105: 24–37. [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006; 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011; 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jang H, Kim H, Kim ST, Cho EJ, Youn HD. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem Biophys Res Commun 2010; 401: 327–332. [DOI] [PubMed] [Google Scholar]
- Musri MM, Carmona MC, Hanzu FA, Kaliman P, Gomis R, Parrizas M. Histone demethylase LSD1 regulates adipogenesis. J Biol Chem 2010; 285: 30034–30041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 2009; 41: 125–129. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007; 446: 882–887. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 2012; 482: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ et al. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol 2011; 13: 652–659. [DOI] [PubMed] [Google Scholar]
- Foster CT, Dovey OM, Lezina L, Luo JL, Gant TW, Barlev N et al. Lysine-specific demethylase 1 regulates the embryonic transcriptome and CoREST stability. Mol Cell Biol 2010; 30: 4851–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Wang E, Huang Y, Guo X, Yu Y, Du Q et al. Inhibition of lysine-specific demethylase-1 (LSD1/KDM1A) promotes the adipogenic differentiation of hESCs through H3K4 methylation. Stem Cell Rev 2016; 12: 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Lan R, Zhang X, Zhu L, Chen F, Xu Z et al. LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol Cell Biol 2014; 34: 158–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 2013; 15: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 2013; 49: 1134–1146. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev 2007; 21: 2545–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog M, Josseaux E, Dedeurwaerder S, Calonne E, Volkmar M, Fuks F. The histone demethylase Kdm3a is essential to progression through differentiation. Nucleic Acids Res 2012; 40: 7219–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wary KK, Revskoy S, Gao X, Tsang K, Komarova YA et al. Histone demethylases KDM4A and KDM4C regulate differentiation of embryonic stem cells to endothelial cells. Stem Cell Rep. 2015; 5: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Shao Z, Beyaz S, Apostolou E, Pinello L, De Los Angeles A et al. Distinct and combinatorial functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in mouse embryonic stem cell identity. Mol Cell 2014; 53: 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Cao J, Liu J, Beshiri ML, Fujiwara Y, Francis J et al. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci USA 2011; 108: 13379–13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S et al. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 2011; 30: 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol 2008; 28: 5312–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell 2012; 22: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales Torres C, Laugesen A, Helin K. Utx is required for proper induction of ectoderm and mesoderm during differentiation of embryonic stem cells. PLoS ONE 2013; 8: e60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA 2012; 109: 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T et al. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res 2013; 113: 856–862. [DOI] [PubMed] [Google Scholar]
- Tang Y, Hong YZ, Bai HJ, Wu Q, Chen CD, Lang JY et al. Plant homeo domain finger protein 8 regulates mesodermal and cardiac differentiation of embryonic stem cells through mediating the histone demethylation of pmaip1. Stem Cells 2016; 34: 1527–1540. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007; 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A, Meshorer E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep 2015; 16: 1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev 2013; 27: 1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Hu G, Yu ZX, Liu C, Zhao K. Extended self-renewal and accelerated reprogramming in the absence of Kdm5b. Mol Cell Biol 2013; 33: 4793–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Hu G, Zhao K. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol 2014; 15: R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M et al. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J 2011; 30: 4586–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle O, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 2010; 3: ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Lee SH, Chen K, Zhu G, Oh W, Allton K et al. An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res 2016; 44: 3659–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009; 460: 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol 2012; 24: 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell 2016; 63: 1066–1079. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M et al. An atlas of active enhancers across human cell types and tissues. Nature 2014; 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi WF, Xing M, Xu C, Yao X, Ramlee MK, Lim MC et al. Epigenomic profiling of primary gastric adenocarcinoma reveals super-enhancer heterogeneity. Nat Commun 2016; 7: 12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 2013; 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outchkourov NS, Muino JM, Kaufmann K, van Ijcken WF, Groot Koerkamp MJ, van Leenen D et al. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep 2013; 3: 1071–1079. [DOI] [PubMed] [Google Scholar]
- Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 2011; 30: 4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Xu W, Guo R, Rong B, Gu L, Wang Z et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell 2016; 165: 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, De La Rosa-Velazquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell 2014; 55: 277–290. [DOI] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev 2006; 20: 1557–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 2006; 442: 312–316. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006; 125: 467–481. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009; 4: 487–492. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 2011; 25: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem 1999; 274: 15633–15645. [DOI] [PubMed] [Google Scholar]
- Madsen B, Tarsounas M, Burchell JM, Hall D, Poulsom R, Taylor-Papadimitriou J. PLU-1, a transcriptional repressor and putative testis-cancer antigen, has a specific expression and localisation pattern during meiosis. Chromosoma 2003; 112: 124–132. [DOI] [PubMed] [Google Scholar]
- Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M et al. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol 2011; 38: 1267–1277. [DOI] [PubMed] [Google Scholar]
- Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC et al. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet 2013; 9: e1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S et al. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell 2008; 31: 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-repressive complex 2. Genes Dev 2008; 22: 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Brookes E, Agarwal S, Badeaux AI, Ito H, Vallianatos CN et al. A mouse model of X-linked intellectual disability associated with impaired removal of histone methylation. Cell Rep 2016; 14: 1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 2012; 488: 409–413. [DOI] [PubMed] [Google Scholar]
- Welstead GG, Creyghton MP, Bilodeau S, Cheng AW, Markoulaki S, Young RA et al. X-linked H3K27me3 demethylase Utx is required for embryonic development in a sex-specific manner. Proc Natl Acad Sci USA 2012; 109: 13004–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 2012; 8: e1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieme S, Gyarfas T, Richter C, Ozhan G, Fu J, Alexopoulou D et al. The histone demethylase UTX regulates stem cell migration and hematopoiesis. Blood 2013; 121: 2462–2473. [DOI] [PubMed] [Google Scholar]
- Canovas S, Cibelli JB, Ross PJ. Jumonji domain-containing protein 3 regulates histone 3 lysine 27 methylation during bovine preimplantation development. Proc Natl Acad Sci USA 2012; 109: 2400–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK et al. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep 2012; 2: 1244–1258. [DOI] [PubMed] [Google Scholar]
- Li Q, Wang HY, Chepelev I, Zhu Q, Wei G, Zhao K et al. Stage-dependent and locus-specific role of histone demethylase Jumonji D3 (JMJD3) in the embryonic stages of lung development. PLoS Genet 2014; 10: e1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010; 11: 936–944. [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res 2013; 23: 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki S, Matoba S, Akiyoshi M, Matsumura Y, Miyachi H, Mise N et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science 2013; 341: 1106–1109. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 2009; 458: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulard M, Edwards JR, Bestor TH. FBXL10 protects polycomb-bound genes from hypermethylation. Nat Genet 2015; 47: 479–485. [DOI] [PubMed] [Google Scholar]
- Boulard M, Edwards JR, Bestor TH. Abnormal X chromosome inactivation and sex-specific gene dysregulation after ablation of FBXL10. Epigenetics Chromatin 2016; 9: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami E, Tokunaga A, Ozawa M, Sakamoto R, Yoshida N. The histone demethylase Fbxl11/Kdm2a plays an essential role in embryonic development by repressing cell-cycle regulators. Mech Dev 2015; 135: 31–42. [DOI] [PubMed] [Google Scholar]
- Cox BJ, Vollmer M, Tamplin O, Lu M, Biechele S, Gertsenstein M et al. Phenotypic annotation of the mouse X chromosome. Genome Res 2010; 20: 1154–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguizabal C, Herrera L, De Onate L, Montserrat N, Hajkova P, Izpisua Belmonte JC. Characterization of the epigenetic changes during human gonadal primordial germ cells reprogramming. Stem Cells 2016; 34: 2418–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009; 461: 415–418. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zhou S, Liao L, Chen X, Meistrich M, Xu J. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J Biol Chem 2010; 285: 2758–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007; 450: 119–123. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Fukuda T, Sakamoto R, Honda H, Yoshida N. The histone demethylase FBXL10 regulates the proliferation of spermatogonia and ensures long-term sustainable spermatogenesis in mice. Biol Reprod 2016; 94: 92. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016; 537: 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016; 537: 558–562. [DOI] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016; 537: 553–557. [DOI] [PubMed] [Google Scholar]
- Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet 2015; 47: 1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]


