Abstract
Esophageal squamous cell carcinoma (ESCC) is a deadly malignancy with regard to mortality and prognosis, and the 5-year survival rate for all patients diagnosed with ESCC remains poor. A better understanding of the biological mechanisms of ESCC tumorigenesis and progression is of great importance to improve treatment of this disease. In this study, we demonstrated that the glutathione metabolism pathway is highly enriched in ESCC cells compared with normal esophageal epithelial cells in an in vivo mouse model. In addition, treatment with L-buthionine-sulfoximine (BSO) to deplete glutathione decreased the ESCC tumor burden in mice, thus demonstrating the critical role of glutathione metabolism in ESCC progression. BSO treatment also led to decreased cell proliferation and activation of cell apoptosis in ESCC. Finally, BSO treatment blocked NF-kB pathway activation in ESCC. Our study reveals a new pathway that regulates ESCC progression and suggests that inhibition of glutathione metabolism may be a potential strategy for ESCC treatment.
Introduction
Esophageal squamous cell carcinoma (ESCC) is one of the most prevalent cancers in China, with an incidence of 43 cases per 100 000 persons annually.1 Current standard treatments for ESCC include surgery, chemotherapy and radiotherapy. However, the 5-year survival rate of ESCC patients remains relatively low at ~10%.2 Hence, identification of effective therapeutic targets is essential for ESCC treatment.
Glutathione, the most prevalent cellular antioxidant in mammals, participates in two metabolic processes, including reducing glutathione to its oxidized product and a conjugation reaction mediated by glutathione S-transferase (GST).3 The liver is typically the site with the highest levels of GSTs. However, certain isozymes of GSTs are also present in high amounts in the esophagus.4 Proteins associated with glutathione metabolism have long been associated with ESCC. For example, GSTO1, GSTP1 and GST-pi have been recognized as tumor-associated antigens and act as indicators of prognosis in human ESCC.5, 6, 7 Moreover, previous findings have demonstrated that a GSTM1 deletion polymorphism is associated with an increased ESCC risk.8 In contrast, another study has demonstrated that the GSTM1 non-null genotypes are highly represented in ESCC patients compared with control patients in China.9 However, the functional role of glutathione metabolism in ESCC remains largely uncharacterized. In skin SCC, the glutathione metabolism pathway is specifically enriched in the TGF-β-activating cancer stem cell population and is responsible for the chemoresistant property of these cells,10 thus suggesting that glutathione metabolism might be critical for ESCC tumor progression.
NF-κB has a critical role in a various stages of cancer initiation and progression. A previous study has demonstrated that the levels of NF-kB p65 in ESCC samples correlate with clinical staging, lymph node metastasis and tumor differentiation.11 Furthermore, survivin and NF-kB p65 have important roles in ESCC tumor progression.12 Importantly, depletion of glutathione with diethyl maleate leads to inhibition of NF-kB target gene expression induced by TNFα,13 thus indicating that glutathione metabolism block ESCC tumor progression by functioning through the NF-kB signaling pathway.
In this study, we used an ESCC mouse model to examine the unique gene profiles in ESCC. We found that glutathione metabolism was among the top enriched pathways in ESCC. To characterize the role of glutathione metabolism in ESCC progression, we treated mice with L-buthionine-sulfoximine (BSO), which depleted the glutathione levels in ESCC. BSO treatment led to a decreased tumor burden, inhibition of cell proliferation and activation of cell apoptosis. Finally, we demonstrated that depletion of glutathione caused inhibition of the NF-kB pathway. Our in vivo findings provide evidence that the glutathione metabolism pathway contributes to ESCC progression via inhibition of the NF-kB signaling pathway and provides a groundwork for further development of a glutathione metabolism inhibitor to treat ESCC.
Materials and methods
Mice and chemical administration
Six- to eight-week-old Balb/c mice were purchased from Guangdong Laboratory Animal Center, Guangzhou, China. All animal procedures were performed by following the Institutional Animal Care and Use Committee (IACUC) approved protocols. For ESCC induction, 4-nitroquinoline 1-oxide (Sigma, Shanghai, China) was dissolved in propylene glycol (Sigma) and diluted in the drinking water at a final concentration of 100 μg ml−1. For glutathione depletion in mice, BSO (Sigma) was dissolved in drinking water at a concentration of 20 mM. Control and BSO-treated esophagi were dissected from mice, flash frozen in liquid nitrogen and homogenized. Approximately 50 mg of tissue was collected for determination of GSH content, as previously described.14 GST and GPX activity were measured using a GST Activity Assay Kit (Abcam, Shanghai, China) and Glutathione Peroxidase Cellular Activity Assay Kit (Sigma), respectively.
Immunohistochemistry
Paraffin sections of ESCC tissue samples from mice were antigen retrieved, blocked and processed as previously described.15, 16 Primary antibodies to mouse K14 (ab7800, 1:500), p63 (ab53039, 1:100), PCNA (ab29, 1:200) and cleaved Caspase3 (ab13847, 1:100) were purchased from Abcam.
RNA isolation and quantitative RT-PCR
ESCC samples were flash frozen in liquid nitrogen and homogenized with a mortar and pestle, as previously described.17, 18, 19 Total RNA was extracted using TRIzol (Invitrogen, China).20 Quantitative RT-PCR was performed as previously described.19 The primers used for RT-PCR is listed below:
GAPDH F: 5′-TGGCCTTCCGTGTTCCTAC-3′
GAPDH R: 5′-GAGTTGCTGTTGAAGTCGCA-3′
IL1A F: 5′-CGAAGACTACAGTTCTGCCATT-3′
IL1A R: 5′-GACGTTTCAGAGGTTCTCAGAG-3′
IL1B F: 5′-GCAACTGTTCCTGAACTCAACT-3′
IL1B R: 5′-ATCTTTTGGGGTCCGTCAACT-3′
IL6 F: 5′-TAGTCCTTCCTACCCCAATTTCC-3′
IL6 R: 5′-TTGGTCCTTAGCCACTCCTTC-3′
MMP3 F: 5′-ACATGGAGACTTTGTCCCTTTTG-3′
MMP3 R: 5′-TTGGCTGAGTGGTAGAGTCCC-3′
MMP9 F: 5′-CTGGACAGCCAGACACTAAAG-3′
MMP9 R: 5′-CTCGCGGCAAGTCTTCAGAG-3′
CCL2 F: 5′-TTAAAAACCTGGATCGGAACCAA-3′
CCL2 R: 5′-GCATTAGCTTCAGATTTACGGGT-3′
CCL4 F: 5′-TTCCTGCTGTTTCTCTTACACCT-3′
CCL4 R: 5′-CTGTCTGCCTCTTTTGGTCAG-3′
BCL2 F: 5′-GTCGCTACCGTCGTGACTTC-3′
BCL2 R: 5′-CAGACATGCACCTACCCAGC-3′
BCL2A1A F: 5′-GGCTGAGCACTACCTTCAGTA-3′
BCL2A1A R: 5′-TGGCGGTATCTATGGATTCCAC-3′
CCND2 F: 5′-GAGTGGGAACTGGTAGTGTTG-3′
CCND2 R: 5′-CGCACAGAGCGATGAAGGT-3′
RNA-Sequencing (RNA-Seq) library preparation and data analysis
For RNA-Seq library preparation, total RNA was first extracted using Trizol, as previously described,21, 22 from control and ESCC tissues (n=3 each). mRNA was then isolated, fragmented and reverse-transcribed into the cDNA libraries with an mRNA-Seq preparation kit (Illumina, San Diego, CA, USA). Library quality was assessed on an Agilent Bioanalyzer 2100 (Agilent, Shanghai, China). The Illumina Hiseq2000 platform was used for single-end sequencing of cDNA libraries. Alignment of reads was performed using Tophat2 with the mm10 build of the mouse genome. Transcript assembly and differential expression genes were identified via Cufflinks software (Seattle, WA, USA).23 RNA-Seq data were analyzed using the CummeRbund package in R.24 Functional enrichment analysis of differential expression genes was performed using the DAVID Functional Annotation Clustering Tool. A heat map of differentially expressed genes involved in glutathione metabolism was generated using Heatmap Builder.25
Western blot
Western blot analysis was performed as previously described.26, 27 Primary antibodies included anti-phospho-p65 antibody (Cell signaling #3033, 1:1000), anti-p65 antibody (Cell Signaling #6956, 1:1000), anti-IκBα antibody (Cell Signaling #3033, 1:1000), anti-IκBα antibody (Cell Signaling #9242, 1:1000), anti-phospho- IκBα antibody (Cell Signaling #6956, 1:2859) and anti-α-tubulin (Sigma-Aldrich #T-9026, 1:5000).
Statistics
Data are presented as the means±s.d. or s.e. The two-tailed Student’s t-test and one-way analysis of variance were used to calculate statistical significance. A P-value of <0.05 was considered significant.
Results
Mice developed ESCC after carcinogen treatment
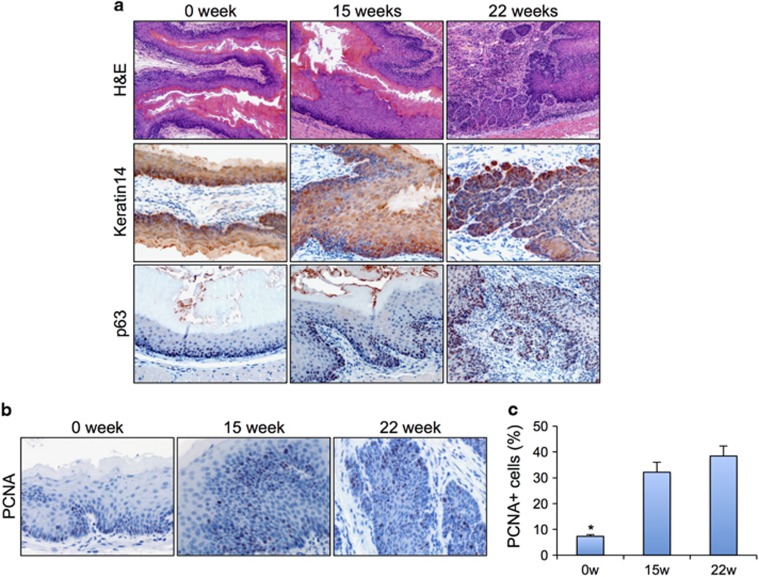
To study ESCC in vivo, we modified a previously described carcinogen-induced mouse model.28 Mice were administered drinking water containing 100 μg ml−1 carcinogen 4-nitroquinoline 1-oxide for 15 weeks and then were switched back to dH2O. We analyzed the changes in the histopathology of the esophageal epithelia 15 weeks and 22 weeks after the initial carcinogen treatment. H&E staining results showed the esophageal epithelia from carcinogen-induced mice were noticeably thicker than those in the controls (Figure 1a). Immunohistochemical results revealed that the expression pattern of Keratin14 (K14) was expanded at both dysplasia and ESCC compared with that in normal controls (Figure 1a). K14 is typically specifically expressed in the proliferative basal layer. However, we observed expression of K14 in the suprabasal layer of dysplasia and SCC. Furthermore, we examined the expression pattern of p63, which is elevated in human ESCCs.29 In the control esophageal epithelia, we detected weak p63 protein expression levels in the basal cell layer. However, we observed upregulated p63 expression in both basal and suprabasal layers of dysplasia and ESCC samples (Figure 1a). Next, we quantified the proliferation index of the epithelial cells in the esophagus on the basis of the results from immunohistochemical, by using an antibody against PCNA (Figure 1b). We detected significant increases in cellular proliferation rates in both dysplasia (32.1±3.9%, P<0.05) and SCC (38.4±3.8%, P<0.05) compared with the normal control (7.3±0.7% Figure 1c).
Figure 1.
Mice treated with carcinogen developed dysplasia and ESCC. (a) H&E staining and K14 and p63 IHC staining of normal esophageal epithelium, dysplasia and SCC. (b) PCNA IHC for cell proliferation in normal esophageal epithelium, dysplasia and SCC. (c) Quantification of cell proliferation in normal esophageal epithelium, dysplasia and SCC. H&E, hematoxylin and eosin. IHC, immunohistochemical.
Components of the glutathione metabolism pathway were enriched in ESCC
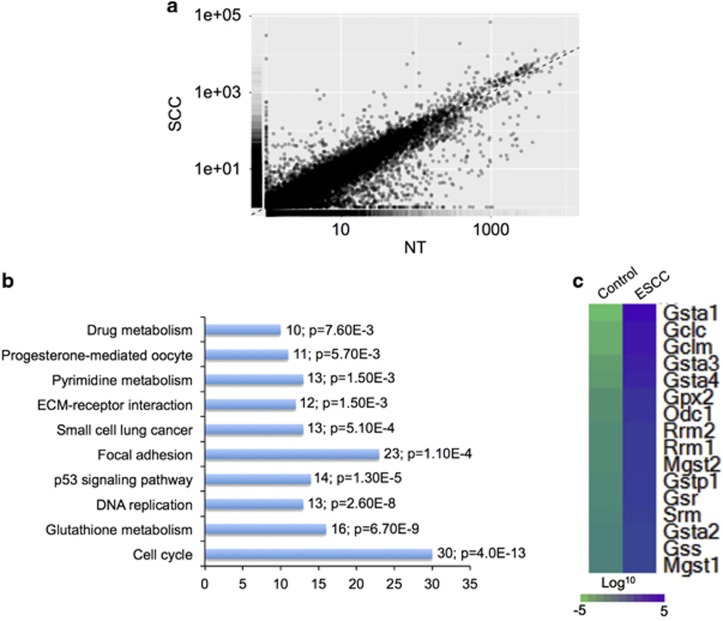
To probe the molecular events responsible for ESCC development, we extracted RNA from normal control and ESCC samples (n=3 each) and performed RNA-Seq. To determine the differences between control and ESCC, we generated density plots of transcriptome-wide differential expression in both conditions (Figure 2a). Our data indicated that 797 transcripts were upregulated, and 1383 transcripts were downregulated in ESCC compared with the control (Figure 2a and Supplementary Table 1). We performed KEGG pathway analysis to obtain a more comprehensive understanding of the transcripts that were enriched in the ESCC sample. Our results revealed that the specific upregulated pathways in ESCC included cell cycle, glutathione metabolism, DNA replication, p53 signaling pathway, focal adhesion, small cell lung cancer, ECM-receptor interaction, pyrimidine metabolism, progesterone-mediated oocyte and drug metabolism (Figure 2b). We generated a heat map of 16 differentially expressed transcripts involved in glutathione metabolism, including Gsta1, Gclc, Gclm, Gsta3, Gsta4, Gpx2, Odc1, Rrm2, Rrm1, Mgst2, Gstp1, Gsr, Srm, Gsta2, Gss and Mgst1 (Figure 2c).
Figure 2.
RNA-Seq revealed the upregulation of the glutathione metabolism pathway in ESCC. (a) Scatter plot generated from RNA-Seq data demonstrates that a large number of transcripts were altered in ESCC compared with control. (b) Selected enriched KEGG pathways in ESCC, ordered from top to bottom from highest to lowest P-value. Numbers of transcripts and P-value in each pathway are indicated in the text next to each pathway. (c) Heat map of 16 differentially expressed genes involved in glutathione metabolism.
Inhibition of glutathione metabolism by BSO decreased the ESCC tumor burden
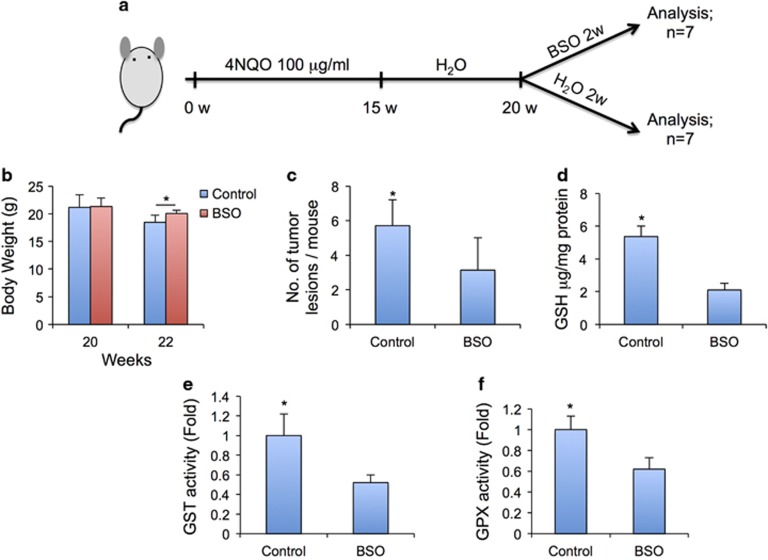
To investigate whether glutathione metabolism contributed to the progression of ESCC development, we treated mice bearing ESCC 20 weeks after initial carcinogen treatment with buthionine sulfoximine (BSO), a commonly used chemical inhibitor of γ -glutamylcysteine synthetase (γ-GCS) that decreases tissue glutathione concentrations (Figure 3a). Two weeks after BSO treatment, mice were killed for analysis. Mice treated with BSO, compared with control mice, exhibited a significant recovery of body weight (Figure 3b). In addition, the tumor lesion numbers per mouse were significantly decreased in mice treated with BSO (Figure 3c). Finally, we validated that glutathione concentrations, GSH transferase and GSH peroxidase activity were inhibited in the esophageal tissues treated with BSO (Figures 3d–f).
Figure 3.
Depletion of glutathione by BSO reduced the ESCC tumor burden. (a) Experimental schematic representation. (b) Body weights of control and BSO-treated mice before and 2 weeks after BSO treatment. (c) Incidence of esophageal tumors after BSO treatment in ESCC-bearing mice. (d) Glutathione concentration in BSO-treated and control esophagi. (e) BSO treatment decreases cellular GSH activity in ESCC cells. (f) BSO treatment decreases cellular GPX activity in ESCC cells.
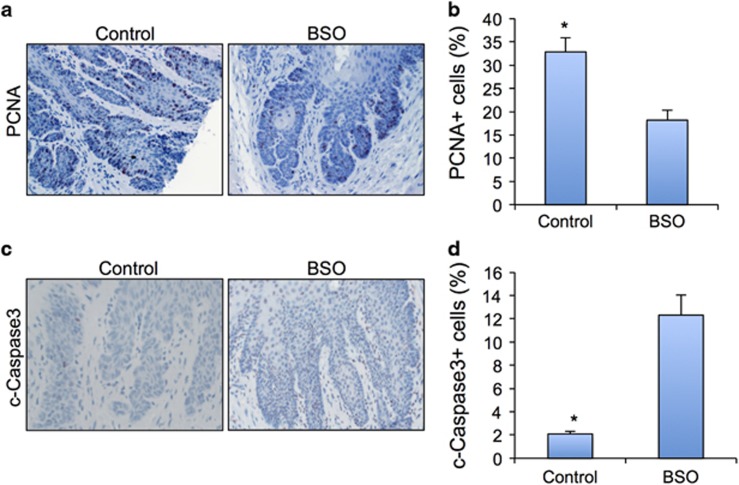
Next, we examined the cell proliferation rates of ESCC cells after BSO treatment by examining PCNA expression. Our immunohistochemical and quantification results demonstrated a clear inhibition of cell proliferation in ESCC treated with BSO (Figures 4a and b). Furthermore, the apoptotic cells in ESCC treated with BSO, compared with the control samples, exhibited a dramatic increase (Figures 4c and d).
Figure 4.
BSO decreased cell proliferation and induced cell apoptosis in ESCC. (a) PCNA IHC for cell proliferation in control and BSO-treated ESCC. (b) Quantification of cell proliferation in control and BSO-treated ESCC. (c) Cleaved caspase3 indicating cell apoptosis in control and BSO-treated ESCC. (d) Quantification of cell apoptosis in control and BSO-treated ESCC. IHC, immunohistochemical.
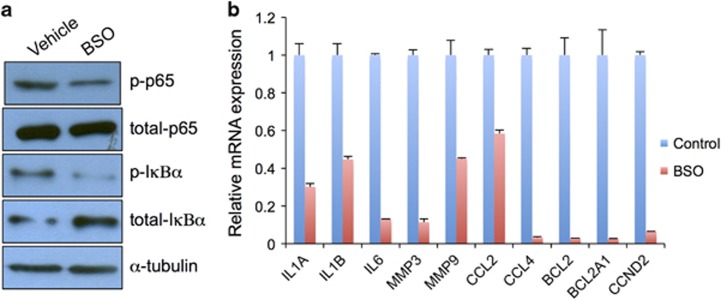
Treatment of BSO inhibited the NF-κB signaling pathway in ESCC
A previous report has demonstrated that the NF-kB signaling pathway is constitutively activated in ESCC cell lines and is a potential target for ESCC treatment.30 Moreover, depletion of glutathione leads to decreased expression of NF-kB genes in hepatocytes. However, whether BSO treatment inhibits NF-kB signaling transduction in ESCC in vivo remains unknown. To test this possibility, we examined the expression of total p65 and phosphorylated-p65 (p-p65). As expected, we detected decreased levels of p-p65 in ESCC samples treated with BSO (Figure 5a). Furthermore, p-IκBα was significantly repressed by BSO (Figure 5a), thus suggesting that BSO blocks NF-kB signaling transduction. Moreover, mRNA expression levels of NF-kB downstream targets, including Il1a, Il1b, Il6, Mmp3, Mmp9, Ccl2, Ccl4, Bcl2, Bcl2a1a and Ccnd2, were downregulated in ESCC by BSO (Figure 5b).
Figure 5.
BSO downregulated the NF-κB signaling pathway. (a) Western blot analysis of p-p65, total p65, p-IκBα and total IκBα proteins. Equal protein loading was controlled for with an anti-α-tubulin antibody. (b) mRNA expression levels of target genes of the NF-κB signaling pathway.
Discussion
ESCC is often refractory to current therapeutic treatment and has poor outcomes. Therefore, effective approaches for treating esophageal cancer are urgently needed. To explore the cellular and molecular events that regulate ESCC development, we utilized a previously established mouse model combined with a high-throughput sequencing assay. Glutathione metabolism was among the top pathways enriched in ESCC. Our study provides the first report that glutathione metabolism is required for ESCC progression in an animal model, thus suggesting that this model is an excellent tool for testing glutathione metabolism inhibitors in the prevention and treatment of ESCC.
Numerous malignancies contain higher glutathione levels than those found in normal tissues. Glutathione content is involved in mutagenic mechanisms, DNA synthesis and drug resistance.10 Although it has long been proposed that application of selective GSH-depleting approaches will be beneficial in cancer therapy, the strategy remains elusive.31 In our study, we demonstrated inhibition of the glutathione metabolism pathway by BSO, which decreased the ESCC tumor burden in mice. However, the full toxicity of this treatment must be more rigorously tested in the future to prevent any major side effects.
The activation of key NF-kB signaling pathway components was inhibited by BSO treatment, thus suggesting that NF-kB might have an important role in ESCC progression. Consistently with these results, a recent report has established a positive correlation among the expression of NF-kB p65 and clinical staging, lymph node metastasis and tumor differentiation of ESCC.11 In addition, the NF-κB pathway was constitutively activated in the ESCC cell lines. Silencing of p65 exhibits anti-proliferative effects and confers the chemo-drug sensitivity of the ESCC cells.30 NF-κB is a positive regulator of GSTP1-1, thus suggesting the importance of NF-κB in the GSH pathway.32 However, the detailed mechanism regarding how glutathione depletion leads to inhibition of the NF-kB signaling pathway remains largely uncharacterized. A previous study using hepatocytes has shown that glutathione depletion represses TNFα-triggered NF-kB activity through both IκB kinase-dependent and -independent routes.33 In addition, endothelial cells treated with GSH exhibit blockade of NF-κB activity decreased expression of NF-κB downstream target gene inter-cellular adhesion molecule-1 (ICAM-1).34 Our data showed that BSO treatment indeed caused inhibition of the phosphorylation status of IkBα in ESCC, suggesting that inhibition of the NF-kB signaling pathway in ESCC by BSO depends on IkBα. However, we could not exclude the possibility that other alternative pathways might be involved in this inhibition.
In summary, we combined a carcinogen-induced ESCC mouse model with RNA-Seq to investigate the molecular changes associated with the progression of ESCC. We demonstrated that BSO treatment significantly decreased the ESCC tumor burden in mice. Additionally, BSO treatment attenuated NF-kB signaling transduction and repressed NF-kB target gene expression. Our model provides an excellent tool to determine the prognostic markers for ESCC and provides support for the development of potential drugs for prevention or treatment of ESCC.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no 81500354), Beijing Municipal Natural Science Foundation (grant no Z13110200400000) and Wu Jieping Medical Foundation (grant no 320.6750.14331).
Author contributions
LP and RL conceived the ideas, designed the experiments, collected the data and wrote the manuscript. DC, JY, XK, XW and YH participated in data analysis and manuscript editing. YH, YZJ and JY supervised the project.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Chen W, Zheng R, Zeng H, Zhang S. The updated incidences and mortalities of major cancers in China, 2011. Chin J Cancer 2015; 34: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. 2016CA Cancer J Clin 2016; 66: 7–30. [DOI] [PubMed] [Google Scholar]
- Lushchak VI. Glutathione homeostasis and functions: potential targets for medical interventions. J Amino Acids 2012; 2012: 736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JD, Nieves E, Listowsky I. Subunit diversity and tissue distribution of human glutathione S-transferases: interpretations based on electrospray ionization-MS and peptide sequence-specific antisera. Biochem J 1997; 325: 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Peng B, Shao Q, Qian W, Zhang JY. Identification of glutathione S-transferase omega 1 (GSTO1) protein as a novel tumor-associated antigen and its autoantibody in human esophageal squamous cell carcinoma. Tumour Biol 2014; 35: 10871–10877. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Konishi H, Ichikawa D, Arita T, Shoda K, Komatsu S et al. Significance of GSTP1 for predicting the prognosis and chemotherapeutic efficacy in esophageal squamous cell carcinoma. Oncol Rep 2013; 30: 1687–1694. [DOI] [PubMed] [Google Scholar]
- Huang JX, Li FY, Xiao W, Song ZX, Qian RY, Chen P et al. Expression of thymidylate synthase and glutathione-s-transferase pi in patients with esophageal squamous cell carcinoma. World J Gastroenterol 2009; 15: 4316–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao CM, Takezaki T, Wu JZ, Li ZY, Liu YT, Li SP et al. Glutathione-S-transferases M1 (GSTM1) and GSTT1 genotype, smoking, consumption of alcohol and tea and risk of esophageal and stomach cancers: a case-control study of a high-incidence area in Jiangsu Province, China. Cancer Lett 2002; 188: 95–102. [DOI] [PubMed] [Google Scholar]
- Tan W, Song N, Wang GQ, Liu Q, Tang HJ, Kadlubar FF et al. Impact of genetic polymorphisms in cytochrome P450 2E1 and glutathione S-transferases M1, T1, and P1 on susceptibility to esophageal cancer among high-risk individuals in China. Cancer Epidemiol Biomarkers Prev 2000; 9: 551–556. [PubMed] [Google Scholar]
- Oshimori N, Oristian D, Fuchs E. TGF-beta promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 2015; 160: 963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping FM, Liu GJ, Liu ZJ, Li HB, Zhai JW, Li SX et al. Expression of RKIP, E-cadherin and NF-kB p65 in esophageal squamous cell carcinoma and their correlations. Int J Clin Exp Pathol 2015; 8: 10164–10170. [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Li H, Chen Y, Lv H, Liu L, Ran J et al. Survivin activates NFkappaB p65 via the IKKbeta promoter in esophageal squamous cell carcinoma. Mol Med Rep 2016; 13: 1869–1880. [DOI] [PubMed] [Google Scholar]
- Nagai H, Matsumaru K, Feng G, Kaplowitz N. Reduced glutathione depletion causes necrosis and sensitization to tumor necrosis factor-alpha-induced apoptosis in cultured mouse hepatocytes. Hepatology 2002; 36: 55–64. [DOI] [PubMed] [Google Scholar]
- Reliene R, Schiestl RH. Glutathione depletion by buthionine sulfoximine induces DNA deletions in mice. Carcinogenesis 2006; 27: 240–244. [DOI] [PubMed] [Google Scholar]
- Chen D, Jarrell A, Guo C, Lang R, Atit R. Dermal beta-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development 2012; 139: 1522–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnick I, Hamburg-Shields E, Chen D, Torre E, Jarrell A, Akhtar-Zaidi B et al. Defining the identity of mouse embryonic dermal fibroblasts. Genesis 2016; 54: 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Zhu F, Zhang H, Chen D, Zhang X, Gao Q et al. Conditional ablation of TGF-beta signaling inhibits tumor progression and invasion in an induced mouse bladder cancer model. Sci Rep 2016; 6: 29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Hu Y, Chen D, Linghu R, Wang Y, Kou X et al. Ubiquitin specific protease 21 upregulation in breast cancer promotes cell tumorigenic capability and is associated with the NOD-like receptor signaling pathway. Oncol Lett 2016; 12: 4531–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XX, Yan YW, Chen D, Ai CZ, Lu X, Xu SS et al. Long non-coding RNA HoxA-AS3 interacts with EZH2 to regulate lineage commitment of mesenchymal stem cells. Oncotarget 2016; 7: 63561–63570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei M, Chen D, Li J, Wei L. Histone deacetylase 4 promotes TGF-beta1-induced synovium-derived stem cell chondrogenesis but inhibits chondrogenically differentiated stem cell hypertrophy. Differentiation 2009; 78: 260–268. [DOI] [PubMed] [Google Scholar]
- Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016; 538: 477–482. [DOI] [PubMed] [Google Scholar]
- Dong Y, Liu W, Lei Y, Wu T, Zhang S, Guo Y et al. Effect of gelatin sponge with colloid silver on bone healing in infected cranial defects. Mater Sci Eng C Mater Biol Appl 2017; 70: 371–377. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics 2005; 23: 103–118. [DOI] [PubMed] [Google Scholar]
- Peng L, Hu Y, Chen D, Jiao S, Sun S. Ubiquitin specific peptidase 21 regulates interleukin-8 expression, stem-cell like property of human renal cell carcinoma. Oncotarget 2016; 7: 42007–42016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P, Song Z, Chen D, Linghu R, Wang Y, Zhang X et al. GINS2 regulates matrix metallopeptidase 9 expression and cancer stem cell property in human triple negative breast cancer. Biomed Pharmacother 2016; 84: 1568–1574. [DOI] [PubMed] [Google Scholar]
- Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res 2004; 10: 301–313. [DOI] [PubMed] [Google Scholar]
- Hu H, Xia SH, Li AD, Xu X, Cai Y, Han YL et al. Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int J Cancer 2002; 102: 580–583. [DOI] [PubMed] [Google Scholar]
- Tian F, Zang WD, Hou WH, Liu HT, Xue LX. Nuclear factor-kB signaling pathway constitutively activated in esophageal squamous cell carcinoma cell lines and inhibition of growth of cells by small interfering RNA. Acta Biochim Biophys Sin (Shanghai) 2006; 38: 318–326. [DOI] [PubMed] [Google Scholar]
- Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci 2006; 43: 143–181. [DOI] [PubMed] [Google Scholar]
- Xia C, Hu J, Ketterer B, Taylor JB. The organization of the human GSTP1-1 gene promoter and its response to retinoic acid and cellular redox status. Biochem J 1996; 313: 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Kaplowitz N. Glutathione depletion down-regulates tumor necrosis factor alpha-induced NF-kappaB activity via IkappaB kinase-dependent and -independent mechanisms. J Biol Chem 2007; 282: 29470–29481. [DOI] [PubMed] [Google Scholar]
- Liao BC, Hsieh CW, Lin YC, Wung BS. The glutaredoxin/glutathione system modulates NF-kappaB activity by glutathionylation of p65 in cinnamaldehyde-treated endothelial cells. Toxicol Sci 2010; 116: 151–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.