Abstract
Purpose
Zinc finger SWIM-type containing 5 (ZSWIM5) is a newly discovered protein, which contains a novel zinc-chelating domain SWIM (CxCxnCxH), and is predicted to interact with DNA or proteins. Currently, the knowledge of functions of ZSWIM5 remains limited. In this study, we aimed to elucidate the biological functions of ZSWIM5 and their mechanisms.
Patients and methods
We detected the expression of ZSWIM5 in samples from 139 cases of non-small-cell lung cancer (NSCLC) patients and six cell lines using immunohistochemistry and Western blot. Moreover, we explored the biological functions of ZSWIM5 in lung cancer cells by siRNA interference and cDNA transfection of ZSWIM5.
Results
The results showed that compared with adjacent non-tumor lung tissues, ZSWIM5 expression was significantly decreased in NSCLC tissues (P=0.0199) and that the ZSWIM5-positive rate in non-tumor tissues (76.67%) was notably higher than that in NSCLC tissues (40.29%). ZSWIM5 expression in human normal bronchial epithelial cells was also much higher than that in lung cancer lines (P<0.001). ZSWIM5-negative expression was significantly related to TNM stage (P<0.001), lymph node metastasis (P=0.002), and poor prognosis (P<0.001) of NSCLC patients. MTT and colony formation assays showed that ZSWIM5 could inhibit the proliferation and colony formation abilities of lung cancer cells. Meanwhile, the results of transwell and wound healing assays showed that ZSWIM5 could suppress the invasion and migration of lung cancer cells. Further investigation revealed that ZSWIM5 could downregulate cyclin D1, cyclin E, cyclin A2, MMP2, and MMP9 expression, which affected the proliferation, invasion, and migration abilities of lung cancer cells.
Conclusion
ZSWIM5 could inhibit the malignant progression of NSCLC by affecting the expression of cyclins and MMPs.
Keywords: NSCLC, zinc finger, proliferation, invasion, migration
Introduction
Zinc finger has a stable finger-like spatial conformation formed by a short protein structural motif coordinating with one or more zinc ions (Zn2+).1 Proteins containing the zinc finger (known as zinc finger proteins) were first identified in a study conducted on the African clawed frog and have now been proved to be ubiquitous in the biosphere.2,3 The vast majority of zinc finger proteins could bind to specific DNA, RNA, and proteins through their zinc fingers and play vital roles in a variety of biological processes.4–10
Currently, zinc finger proteins are classified into several classes based on the overall shape of the protein backbone in the folded domain,11 and SWIM is a novel Zn-finger-like domain with a predicted Zn-chelating CxCxnCxH motif.12 Zinc finger SWIM-type containing 5 (ZSWIM5) is a recently discovered 1188-amino acid-long protein with the SWIM domain. There is a 96% homology between mouse and human ZSWIM5 (1,138 of 1,188 amino acids similar). ZSWIM5 was predicted to localize to the nucleus and interact with DNA or other proteins in different contexts.13–15 However, we have limited information regarding the expression and functions of ZSWIM5, especially in malignant tumors.
In this study, we investigated ZSWIM5 expression in non-small-cell lung cancer (NSCLC) tissues and other cell lines and preformed a statistical analysis of the correlation between ZSWIM5 expression and clinicopathological characteristics of NSCLC patients. We also explored the effects of ZSWIM5 on the biological behavior of NSCLC cells and discussed its possible mechanism of action. Our results revealed the tumor suppressor function of ZSWIM5 for the first time and enriched our understanding of SWIM domain containing zinc finger protein. These findings also suggest ZSWIM5 as a novel potential molecular target for the diagnosis and therapy of NSCLC.
Patients and methods
Human specimens
The surgeries of 139 NSCLC patients (82 males and 57 females) were performed in the Cancer Hospital of China Medical University and Liaoning Cancer Hospital and Institute between 2009 and 2011. No neoadjuvant radiotherapy or chemotherapy was done before the surgery, and all the patients received standard chemotherapy after surgery. Histological diagnosis and grading were evaluated according to the 2015 World Health Organization (WHO) classification of tumors of lung. Only lung squamous cell carcinoma and lung adenocarcinoma specimens were selected. Tumor staging was performed according to the seventh edition of the International Union against Cancer TNM Staging System for Lung Cancer. In addition, 20 fresh specimens, including both tumor and paired adjacent non-tumor tissues, were stored at −80°C immediately after resection for protein extraction. Primary tumor specimens were obtained as approved by the local institutional review board at the Cancer Hospital of China Medical University and Liaoning Cancer Hospital and Institute. The patients whose tissue samples were used in this research had provided written informed consent.
Cell culture
Human normal bronchial epithelial (HBE) cell line and lung cancer cell lines H460, LK2, A549, H1299, and SPC-A1 were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and were cultured in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; [Thermo Fisher Scientific]). All cell lines were cultured in a humidified incubator containing 5% CO2 at 37°C.
Immunohistochemistry
The tumor tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 4-µm thickness. The slides were then incubated with primary rabbit polyclonal antibody against ZSWIM5 (dilution, 1:400; Thermo Fisher Scientific), followed by incubation with biotin-labeled secondary antibodies (MaiXin, Fuzhou, China). Finally, the staining procedures were performed using diaminobenzidine reagent (MaiXin) and counterstaining was done using hematoxylin. The staining intensity and percentage of stained cells were evaluated by two investigators. The staining intensity was scored as 0 (no signal), 1 (weak), 2 (moderate), or 3 (high). The percentage of cells stained was scored as 1 (1%–25%), 2 (26%–50%), 3 (51%–75%), or 4 (76%–100%). The intensity scores multiplied by the area scores represent the final scores. A score of 4 was chosen as the cutoff value of ZSWIM5-positive expression, that is, the scores of ≥4 implied positive expression of ZSWIM5, and scores of 0–3 implied negative expression of ZSWIM5.
RNA interference and transfection of plasmids
The siRNAs (siNC, sc-37007, mixed siRNAs targeting ZSWIM5, sc-78822; Santa Cruz Biotechnology Inc., Dallas, TX, USA) were used for siRNA experiments. The plasmid vector containing ZSWIM5 (RC221559) and the empty vector were manufactured by OriGene Technologies, Inc. (Rockville, MD, USA). The cells were treated with either siRNAs or plasmids using Lipofectamine 3,000 (Thermo Fisher Scientific) for 48 hours.
Western blotting
The whole cell protein extracts were homogenized using a lysis buffer followed by centrifugation at 14,000 × g for 15 minutes. Bradford’s method was used to quantify the protein concentrations. After immunoblotting, the nitrocellulose membranes were incubated with specific antibodies. Rabbit polyclonal antibody against ZSWIM5 (dilution, 1:400) was purchased from Thermo Fisher Scientific; Myc-tag, cyclin D1, cyclin E, cyclin A2, cyclin B1, MMP2, and MMP9 (dilution 1:500) were purchased from Cell Signaling Technology (Danvers, MA, USA). e-cadherin (dilution 1:500) was obtained from BD Biosciences (San Jose, CA, USA); ZO1 (dilution 1:500) was bought from Proteintech (Rosemont, IL, USA), and GAPDH (dilution 1:5,000) was purchased from Santa Cruz Biotechnology Inc. The nitrocellulose membranes were then incubated with peroxidase-conjugated secondary antibodies at room temperature for 2 hours, and blots were detected by a bio-imaging scanner system (DNR Bio-Imaging Systems, Israel).
MTT assay
Cells were plated in 96-well plates in a medium containing 10% FBS at a density of ~3,000 cells per well. For evaluating cell viability, cultures were subjected to MTT assay 5 days post-transfection. Briefly, 20 µL of 5 mg/mL MTT (thiazolyl blue) solution was added to each well and incubated for 4 hours at 37°C. Then, the medium was aspirated from each well, and the resultant MTT formazan was solubilized in 150 µL of dimethyl sulfoxide. The results were obtained spectrophotometrically at a wavelength of 570 nm.
Colony formation assay
The A549 and H1299 cells were transfected with either siRNAs or plasmid of ZSWIM5 for 24 hours. Cells were seeded into three 6 cm cell culture dishes (1,000 per dish) and incubated at 37°C. After 12 days, the plates were washed with PBS and stained with Giemsa staining solution. The number of colonies with more than 50 cells was counted.
Transwell assay
For the invasion assay, 5×104 cells were plated in 24-well transwell plates containing inserts (Costar, Cambridge, MA, USA), which were coated with 20 µL Matrigel (dilution 1:3; BD Bioscience, San Jose, CA, USA). The 100-µL medium in inserts was FBS-free. The 600-µL medium poured in 24 wells was supplemented with 10% normal FBS. After 18 hours, the cells of inserts were fixed and stained. Representative fields were photographed. The number of invading cells in every field was counted.
Wound healing assay
Equal numbers of cells were seeded in six-well plates. When cell density was ~90%, the cell monolayers were wounded with a 1,000 µL pipette tip to draw a gap on the plates. The cells that migrated into the cleared section were observed using a microscope at 24 hours. The distances of the optical wound were measured by using the Image J software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Statistical analysis was performed with the SPSS software (version 23.0 for Mac; IBM Corporation, Armonk, NY, USA). The Pearson’s chi-squared test was performed to analyze the relationship between ZSWIM5 expression and clinicopathological factors. Values of P<0.05 were considered statistically significant.
Results
ZSWIM5 exhibits decreased expression in NSCLC tissues and other cell lines
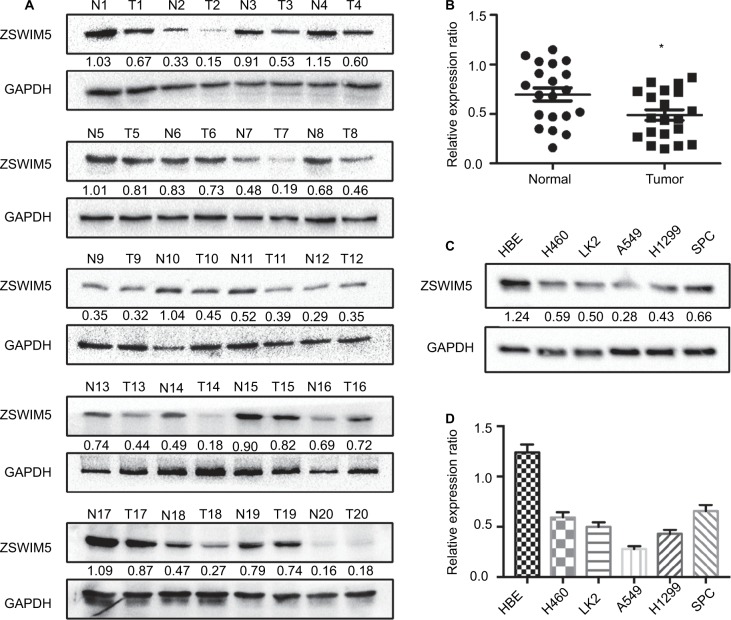
In order to explore the function of ZSWIM5 in NSCLC, we first detected its expression in 20 pairs of NSCLC tissues and adjacent non-tumor lung tissues by Western blot (Figure 1). Our results showed that in 17 pairs of fresh tissues (17/20, 85%), ZSWIM5 expression in adjacent non-tumor lung tissues was higher than that in NSCLC tissues (Figure 1A). The relative expression rate of ZSWIM5 in normal lung tissues was also significantly higher than that in NSCLC tissues (P<0.02, Figure 1B). We further evaluated ZSWIM5 expression in HBE cell line and five other types of lung cancer cell lines. Western blot results showed that ZSWIM5 expression in HBE was notably higher than that in the five lung cancer cell lines (P<0.001, Figure 1C and D).
Figure 1.
The ZSWIM5 expression is decreased in NSCLC tissues and cell lines.
Notes: (A) Western blot detected 20 pairs of NSCLC tissues and adjacent non-tumor lung tissues. ZSWIM5 expression in 17 cases of adjacent non-tumor lung tissues was higher than that in the corresponding NSCLC tissues (17/20, 85%). (B) The relative expression rate of ZSWIM5 in normal lung tissues was significantly higher than that in the NSCLC tissues (P<0.02). (C, D) ZSWIM5 expression in HBE was notably higher than that in five lung cancer cell lines (all P<0.001). *P<0.05.
Abbreviations: ZSWIM5, zinc finger SWIM-type containing 5; NSCLC, non-small-cell lung cancer; HBE, human normal bronchial epithelial.
ZSWIM5 expression is closely related to the malignant progression and poor prognosis of NSCLC patients
We used immunohistochemical staining to evaluate ZSWIM5 expression in 139 cases of NSCLC specimens and 30 cases of adjacent non-tumor lung tissue specimens. In adjacent non-tumor lung tissues, ZSWIM5 exhibited moderate to strong staining and was mainly located in the cytoplasm and nucleus (Figure 2A and B). ZSWIM5 staining was considerably weaker in adenocarcinoma tissues (Figure 2C and D) and squamous cell carcinoma tissues (Figure 2E and F) and was mainly located in the cytoplasm of the cancer cells. Especially in the same field of view, ZSWIM5 staining in lung cancer cells was notably weaker than that in the adjacent non-tumor lung tissue and bronchial epithelial cells (Figure 2G–I). The positive rate in adjacent non-tumor lung tissues (76.67%) was markedly higher than that in NSCLC tissues (40.29%, P<0.001, Table 1). Statistical analysis showed that the decreased expression of ZSWIM5 was significantly correlated with TNM stage (P<0.001) and lymph node metastasis (P=0.002), but had no significant association with age (P>0.1), gender (P>0.1), histology type (P>0.6), and differentiation stages (P>0.3, Table 2) of NSCLC patients. Kaplan–Meier survival analysis showed that decreased expression of ZSWIM5 was significantly correlated with poor prognosis of NSCLC patients (P<0.001, Figure 2J). We also used the online database KM Plotter Online Tool (http://www.kmplot.com) to analyze the relationship between ZSWIM5 gene expression and survival prognosis of lung cancer patients. ZSWIM5 gene expression showed a significant correlation with the overall survival of lung cancer patients (overall survival [OS], n=1145, P=3e−09, Figure S1), which is consistent with our Kaplan–Meier survival analysis results based on immunohistochemical staining data.
Figure 2.
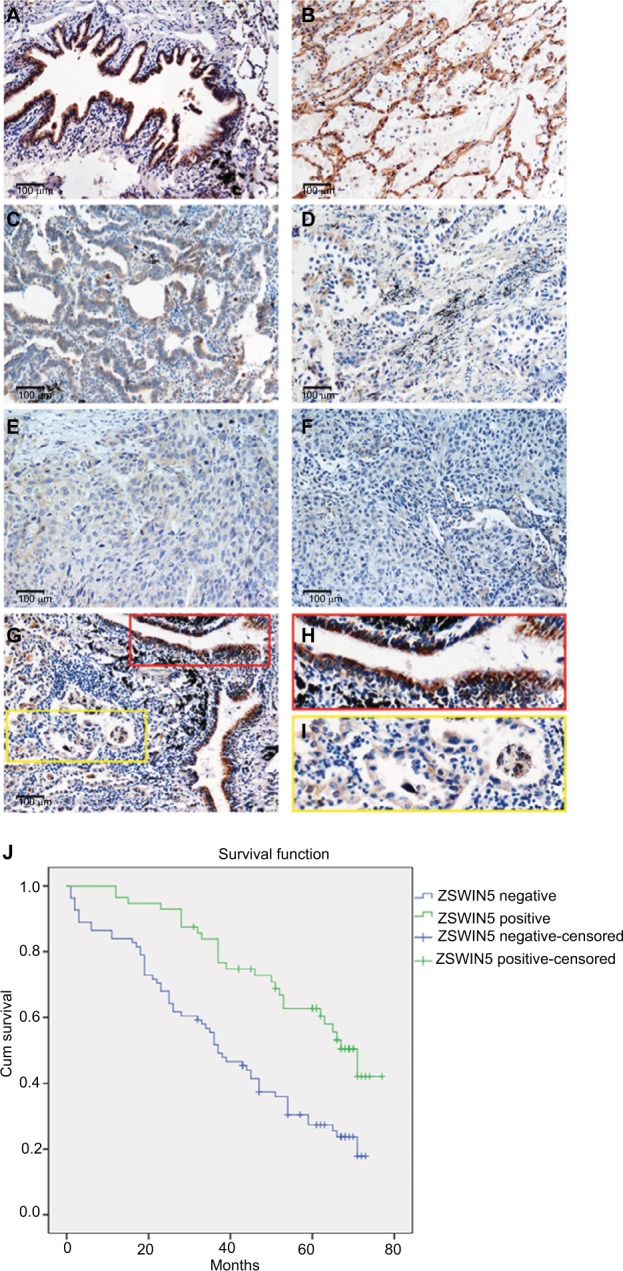
ZSWIM5 expression is decreased in NSCLC tissues and closely related to the poor prognosis of NSCLC patients.
Notes: (A, B) ZSWIM5 exhibited moderate to strong staining in adjacent non-tumor lung tissues and was located in the cytoplasm and nucleus (200× magnification). (C, D) ZSWIM5 showed weak staining in adenocarcinoma tissues and was mainly located in the cytoplasm of cancer cells (200× magnification). (E, F) ZSWIM5 also showed weak staining in squamous cell carcinoma tissues and was located in the cytoplasm of cancer cells (200× magnification). (G–I) ZSWIM5 staining in lung cancer cells was considerably weaker than that in adjacent non-tumor lung tissue bronchial epithelial cells in the same field of view (G, 200× magnification). (J) Kaplan–Meier survival analysis shows that the decreased expression of ZSWIM5 was significantly correlated with poor prognosis of NSCLC patients (P<0.001).
Abbreviations: ZSWIM5, zinc finger SWIM-type containing 5; NSCLC, non-small-cell lung cancer.
Table 1.
ZSWIM5 expression in adjacent non-tumor tissues and lung cancer
| Negative | Positive | |
|---|---|---|
| Adjacent non-tumor tissues | 7 (23.33%) | 23 (76.67%) |
| Lung cancer | 83 (59.12%) | 56 (40.29%)a |
Notes:
P<0.001, indicates statistical significance.
Abbreviation: ZSWIM5, zinc finger SWIM-type containing 5.
Table 2.
Summary of correlation of ZSWIM5 expression with clinicopathological characteristics of NSCLC patients
| Clinicopathological feature | N | ZSWIM5
|
χ2 | P-value | |
|---|---|---|---|---|---|
| Negative | Positive | ||||
| All cases | 139 | 83 | 56 | ||
| Age (years) | |||||
| ≤60 | 73 | 39 | 34 | 2.527 | 0.112 |
| >60 | 66 | 44 | 22 | ||
| Gender | |||||
| Male | 82 | 53 | 29 | 2.014 | 0.156 |
| Female | 57 | 30 | 27 | ||
| Histology | |||||
| Squamous cell carcinoma | 55 | 34 | 21 | 0.168 | 0.682 |
| Adenocarcinoma | 84 | 49 | 35 | ||
| TNM stage | |||||
| I+II | 74 | 30 | 44 | 24.178 | 0.000 |
| III | 65 | 53 | 12 | ||
| Differentiation | |||||
| Well | 33 | 17 | 16 | 1.209 | 0.312 |
| Moderate or poor | 106 | 66 | 40 | ||
| Lymph node metastasis | |||||
| Negative | 85 | 42 | 43 | 9.650 | 0.002 |
| Positive | 54 | 41 | 13 | ||
Abbreviation: ZSWIM5, zinc finger SWIM-type containing 5.
ZSWIM5 inhibits the proliferation, colony formulation, invasion, and migration abilities of lung cancer cells
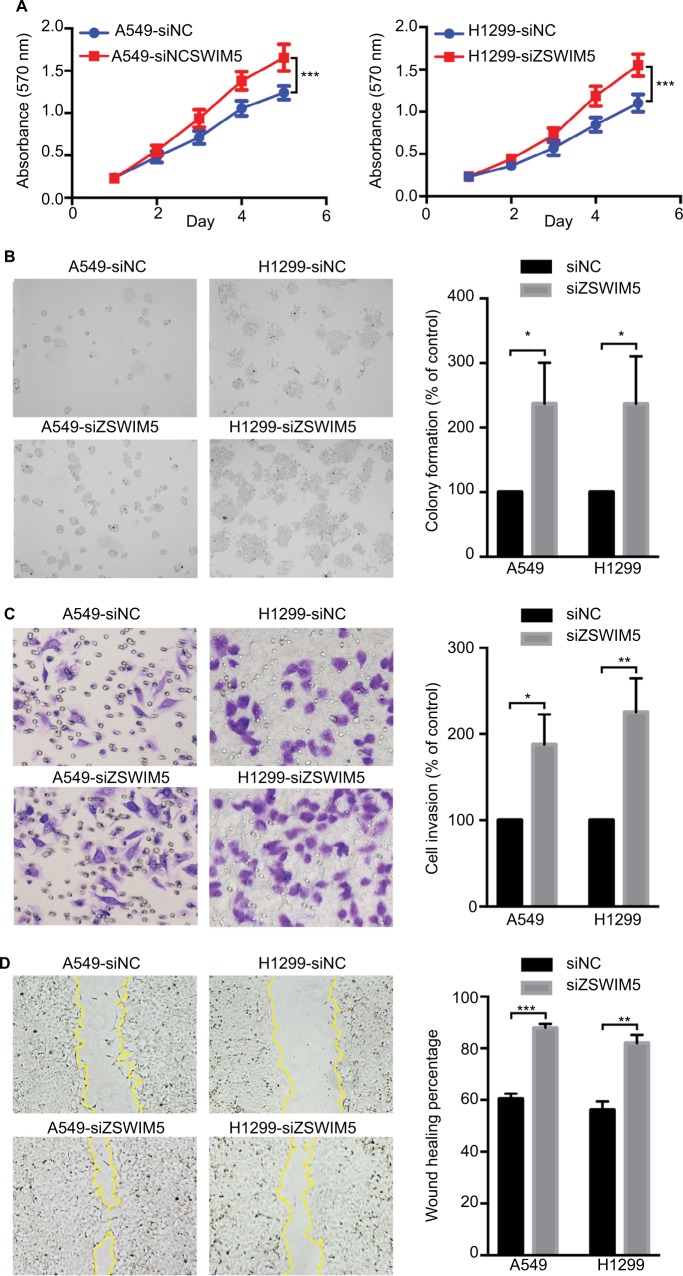
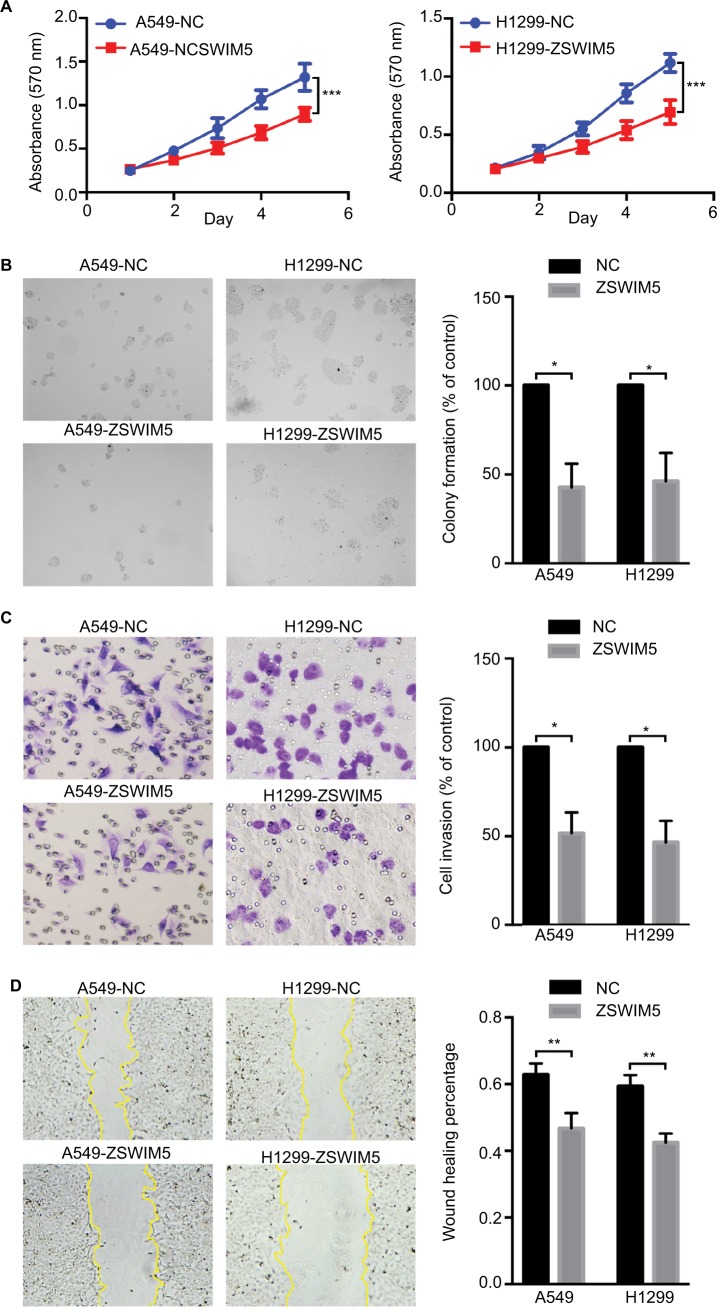
Next, to investigate the biological functions of ZSWIM5 in NSCLC, we examined the effects of siRNA interference and cDNA transfection of ZSWIM5 on the biological behaviors of the two kinds of lung adenocarcinoma cells A549 and H1299. MTT and clone formation assays showed that ZSWIM5 interference significantly promoted the proliferation (the fifth day, both P<0.001) and clone formation abilities (both P<0.01) of A549 and H1299 cells (Figure 3A and B). In contrast, the transfection of ZSWIM5 in A549 and H1299 cells inhibited cell proliferation (the fifth day, both P<0.001) and clone formation abilities (both P<0.01, Figure 4A and B). Transwell and wound healing assays showed that after ZSWIM5 siRNA treatments, the invasion (both P<0.05) and migration (both P<0.01) abilities of A549 and H1299 cells were stronger (Figure 3C and D). Mean while, the A549 and H1299 cells transfected with ZSWIM5 showed considerably weaker invasion (both P<0.05) and migration (both P<0.05) abilities compared to the control cells (Figure 4C and D).
Figure 3.
siRNA interference of ZSWIM5 promotes lung cancer cell proliferation, colony formation, invasion, and migration.
Notes: (A) MTT assays showed that ZSWIM5 interference significantly promoted the proliferation abilities (the fifth day, both P<0.001) of A549 and H1299 cells. (B) Clone formation assays showed that the clone formation abilities of A549 and H1299 cells after siRNA interference were stronger (both P<0.01). (C) Transwell assays showed that siRNA treatment of ZSWIM5 promoted the invasion abilities of A549 and H1299 cells (both P<0.05). (D) Wound healing assays showed that the migration abilities of A549 and H1299 cells with knocked-down ZSWIM5 were stronger (both P<0.01). *P<0.05; **P<0.01; ***P<0.001.
Abbreviation: ZSWIM5, zinc finger SWIM-type containing 5.
Figure 4.
Transfection with ZSWIM5 inhibits lung cancer cell proliferation, colony formation, invasion, and migration.
Notes: (A, B) A549 and H1299 cells transfected with ZSWIM5 showed decreased proliferation (the fifth day, both P<0.001) and clone formation abilities (both P<0.01). (C, D) Transfection with ZSWIM5 also inhibited invasion (both P<0.05) and migration (both P<0.05) of A549 and H1299 cells. *P<0.05; **P<0.01; ***P<0.001.
Abbreviation: ZSWIM5, zinc finger SWIM-type containing 5.
ZSWIM5 downregulates the expression of cyclins and MMPs
In order to explore the mechanism of action of ZSWIM5, we identified many important factors related to the proliferation, invasion, and migration of cancer cells. We found that siRNA treatment of ZSWIM5 in A549 and H1299 cells upregulated the expressions of cyclin D1, cyclin E, cyclin A2, MMP2, and MMP9 (Figure 5). In contrast, transfection of ZSWIM5 in A549 and H1299 cells inhibited the expressions of cyclin D1, cyclin E, cyclin A2, MMP2, and MMP9 (Figure 5). However, ZSWIM5 did not affect the expressions of cyclin B1, e-cadherin, and ZO-1 in A549 or H1299 cells (Figure 5).
Figure 5.
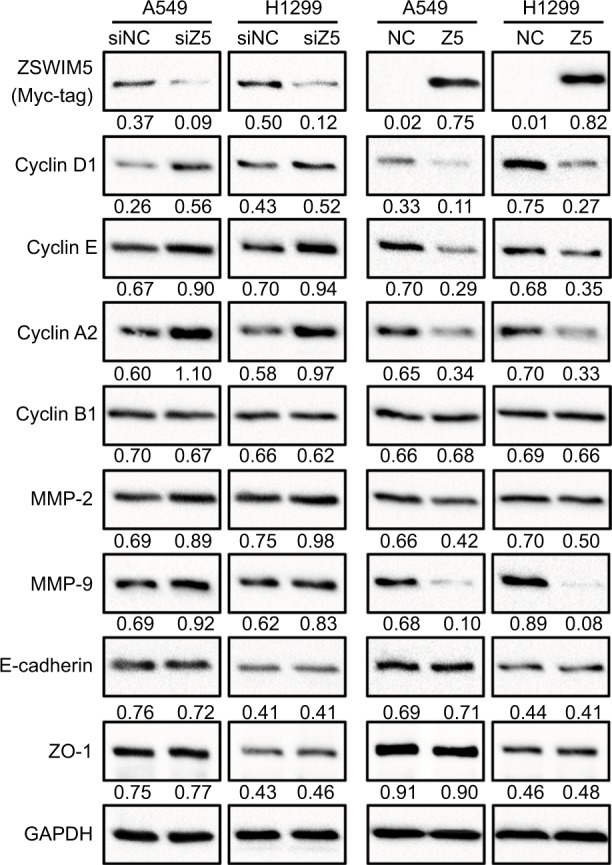
ZSWIM5 downregulates the expression of cyclins and MMPs.
Notes: siRNA treatment of ZSWIM5 in A549 and H1299 cells upregulated the expressions of cyclin D1, cyclin E, cyclin A2, MMP2, and MMP9. In contrast, transfection A549 and H1299 cells with ZSWIM5 inhibited the expressions of cyclin D1, cyclin E, cyclin A2, MMP2, and MMP9. There was no obvious change in the expressions of cyclin B1, e-cadherin, and ZO-1 in A549 or H1299 cells.
Abbreviation: ZSWIM5, zinc finger SWIM-type containing 5.
Discussion
NSCLC is a common malignancy with high global incidence and mortality. Discovery of novel tumor-related factors and revelation of their roles and mechanisms in NSCLC are urgently needed for the identification and development of new molecular targets for NSCLC diagnosis and therapy. Many previous studies have demonstrated significant roles of a variety of zinc finger proteins in the development of various malignancies.5–8,10 ZSWIM5 is a novel SWIM domain containing zinc finger protein, which is predicted to interact with DNA and other proteins.12 Current data show that ZSWIM5 expression is high in normal brain and malignant gliomas.13–15 Besides this, we have acquired limited knowledge on the function of ZSWIM5.
In this study, we first used Western blotting to evaluate the expression levels of ZSWIM5 and proved that ZSWIM5 expression was decreased in NSCLC fresh tissues and lung cancer cells. This was further confirmed by the immunohistochemical staining of 139 NSCLC specimens. ZSWIM5 exhibited intense staining in adjacent non-tumor lung tissues and was located in both the nucleus and the cytoplasm. In contrast, in NSCLC tissues, ZSWIM5 was mainly located in the cytoplasm of cancer cells and showed weak staining. Based on the above results, we inferred that the expression of ZSWIM5 is decreased in NSCLC.
Further statistical analysis showed that low expression of ZSWIM5 was significantly correlated with the TNM stage, lymph node metastasis, and poor prognosis of NSCLC patients, which is consistent with the gene expression result of KM Plotter database (http://www.kmplot.com). In case of siRNA interference and cDNA transfection of ZSWIM5 in A549 and H1299 lung cancer cell lines, we found that ZSWIM5 could significantly inhibit the proliferation, colony formation, invasion, and migration abilities of the lung cancer cells. These results indicated that ZSWIM5 exhibited a tumor suppressor function and inhibited the malignant progression of NSCLC.
Next, we tried to further explore the relevant mechanisms where ZSWIM5 could play the above biological functions. We found that ZSWIM5 downregulated many cyclins, including cyclin D1, cyclin E, and cyclin A2, which play a key role in cell cycle progression.16–19 Downregulation of these cyclins could significantly inhibit cellular proliferation and colony formation abilities. Meanwhile, we found that although the expression of adhesion molecules e-cadherin and ZO-1 remained unaltered, the expression of MMP2 and MMP9 was significantly downregulated by ZSWIM5. MMP2 and MMP9 are involved in the degradation of extracellular matrix and promote cancer cell invasion and migration.20,21 However, the exact mechanism of action of ZSWIM5 on these cyclins and MMPs is yet unknown.
The data from the Biological General Repository for Interaction Datasets (BioGRID), which is a biological database of protein–protein interactions, showed that ZSWIM5 may interact with CDC42.22 CDC42 is a small GTPase of the Rho family, which regulates diverse cellular functions including cell morphology, polarity, invasion, migration, cell cycle, proliferation, differentiation, and endocytosis.23–25 Therefore, ZSWIM5 is likely to regulate the cyclins and MMPs by interacting with CDC42. Meanwhile, this also implies that besides the regulation of cyclins and MMPs, ZSWIM5 may use other mechanisms to regulate the biological behavior of tumor cells. It is likely to be able to regulate the cytoskeleton and cell polarity, which could also greatly affect the biological behavior of cancer cells. We hope to further clarify the specific mechanisms of action of ZSWIM5 in our future studies.
Conclusion
ZSWIM5 expression was decreased in NSCLC, and its low expression was significantly correlated with TNM stage, lymph node metastasis, and poor prognosis of NSCLC patients. ZSWIM5 exhibited a tumor suppressor function and inhibited the malignant progression of NSCLC.
Supplementary material
ZSWIM5 gene expression is significantly correlated with the overall survival of lung cancer patients.
Notes: Data from KM Plotter Online Tool (http://www.kmplot.com) showed that ZSWIM5 gene expression was significantly correlated with the overall survival of lung cancer patients (OS, n=1,145, P=3e–09), which is consistent with our Kaplan–Meier survival analysis results based on immunohistochemical staining data.
Abbreviation: ZSWIM5, zinc finger SWIM-type containing 5.
Acknowledgments
The authors thank the Department of Pathology of Cancer Hospital of China Medical University and Liaoning Cancer Hospital and Institute for providing tissue samples and technical support. We would like to thank Editage (www.editage.cn) for English language editing.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Klug A, Rhodes D. Zinc fingers: a novel protein fold for nucleic acid recognition. Cold Spring Harb Symp Quant Biol. 1987;52:473–482. doi: 10.1101/sqb.1987.052.01.054. [DOI] [PubMed] [Google Scholar]
- 2.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 3.Hanas JS, Hazuda DJ, Bogenhagen DF, Wu FY, Wu CW. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J Biol Chem. 1983;258(23):14120–14125. [PubMed] [Google Scholar]
- 4.Arenzana TL, Schjerven H, Smale ST. Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev. 2015;29(17):1801–1816. doi: 10.1101/gad.266999.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L, Zhang W, Li Y, et al. SHP-2-upregulated ZEB1 is important for PDGFRα-driven glioma epithelial-mesenchymal transition and invasion in mice and humans. Oncogene. 2016;35(43):5641–5652. doi: 10.1038/onc.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L, Wang H, Kornblau SM, et al. Evidence of a role for the novel zinc-finger transcription factor ZKSCAN3 in modulating Cyclin D2 expression in multiple myeloma. Oncogene. 2011;30(11):1329–1340. doi: 10.1038/onc.2010.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai KP, Chen J, He M, et al. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014;135(8):1790–1799. doi: 10.1002/ijc.28819. [DOI] [PubMed] [Google Scholar]
- 8.Ma X, Huang M, Wang Z, Liu B, Zhu Z, Li C. ZHX1 inhibits gastric cancer cell growth through inducing cell-cycle arrest and apoptosis. J Cancer. 2016;7(1):60–68. doi: 10.7150/jca.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JG, Armstrong RC, V Agoston D, et al. Myelin transcription factor 1 (Myt1) of the oligodendrocyte lineage, along with a closely related CCHC zinc finger, is expressed in developing neurons in the mammalian central nervous system. J Neurosci Res. 1997;50(2):272–290. doi: 10.1002/(SICI)1097-4547(19971015)50:2<272::AID-JNR16>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 10.Jandrig B, Seitz S, Hinzmann B, et al. ST18 is a breast cancer tumor suppressor gene at human chromosome 8q11.2. Oncogene. 2004;23(57):9295–9302. doi: 10.1038/sj.onc.1208131. [DOI] [PubMed] [Google Scholar]
- 11.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31(2):532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarova KS, Aravind L, Koonin EV. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem Sci. 2002;27(8):384–386. doi: 10.1016/s0968-0004(02)02140-0. [DOI] [PubMed] [Google Scholar]
- 13.Tucker ES, Segall S, Gopalakrishna D, et al. Molecular specification and patterning of progenitor cells in the lateral and medial ganglionic eminences. J Neurosci. 2008;28(38):9504–9518. doi: 10.1523/JNEUROSCI.2341-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer MA. Highly expressed genes in human high grade gliomas: immunohistochemical analysis of data from the human protein atlas. Neurol Int. 2014;6(2):5348. doi: 10.4081/ni.2014.5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong TC, Rebbert M, Wang C, et al. Genes regulated by potassium channel tetramerization domain containing 15 (Kctd15) in the developing neural crest. Int J Dev Biol. 2016;60(4–6):159–166. doi: 10.1387/ijdb.160058id. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7(5):812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 17.Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70(6):993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 18.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11(3):961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung TK, Ma HT, Poon RY. Specialized roles of the two mitotic cyclins in somatic cells: cyclin A as an activator of M phase-promoting factor. Mol Biol Cell. 2007;18(5):1861–1873. doi: 10.1091/mbc.E06-12-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob A, Prekeris R. The regulation of MMP targeting to invadopodia during cancer metastasis. Front Cell Dev Biol. 2015;3:4. doi: 10.3389/fcell.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merchant N, Nagaraju GP, Rajitha B, et al. Matrix metalloproteinases: their functional role in lung cancer. Carcinogenesis. 2017;38(8):766–780. doi: 10.1093/carcin/bgx063. [DOI] [PubMed] [Google Scholar]
- 22.Hein MY, Hubner NC, Poser I, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163(3):712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 23.Qadir MI, Parveen A, Ali M. Cdc42: role in cancer management. Chem Biol Drug Des. 2015;86(4):432–439. doi: 10.1111/cbdd.12556. [DOI] [PubMed] [Google Scholar]
- 24.du DS, Yang XZ, Wang Q, et al. Effects of CDC42 on the proliferation and invasion of gastric cancer cells. Mol Med Rep. 2016;13(1):550–554. doi: 10.3892/mmr.2015.4523. [DOI] [PubMed] [Google Scholar]
- 25.Melendez J, Grogg M, Zheng Y. Signaling role of Cdc42 in regulating mammalian physiology. J Biol Chem. 2011;286(4):2375–2381. doi: 10.1074/jbc.R110.200329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ZSWIM5 gene expression is significantly correlated with the overall survival of lung cancer patients.
Notes: Data from KM Plotter Online Tool (http://www.kmplot.com) showed that ZSWIM5 gene expression was significantly correlated with the overall survival of lung cancer patients (OS, n=1,145, P=3e–09), which is consistent with our Kaplan–Meier survival analysis results based on immunohistochemical staining data.
Abbreviation: ZSWIM5, zinc finger SWIM-type containing 5.