Significance
Quantifying the preference for particular foods is difficult in Drosophila because of the animal’s small meal size. It is common for fly feeding assays to induce or require starvation conditions. We have developed BARCODE, a method of tracking consumption preference by tagging foods with different oligonucleotides and later performing qPCR from the fly body to determine its feeding history. Because a fraction of the ingested oligomer persists for about a week, this assay provides a long-term record of the feeding pattern. We use this assay to study sexually dimorphic differences in ethanol consumption preference in Drosophila. BARCODE is also suitable for multiplex feeding studies in which the preference between more than two foods is simultaneously compared.
Keywords: alcohol, behavior, Drosophila, preference, feeding
Abstract
Drosophila melanogaster is a powerful model organism for dissecting the neurogenetic basis of appetitive and aversive behaviors. However, some methods used to assay food preference require or cause starvation. This can be problematic for fly ethanol research because it can be difficult to dissociate caloric preference for ethanol from pharmacological preference for the drug. We designed BARCODE, a starvation-independent assay that uses trace levels of oligonucleotide tags to differentially mark food types. In BARCODE, flies feed ad libitum, and relative food preference is monitored by qPCR of the oligonucleotides. Persistence of the ingested oligomers within the fly records the feeding history of the fly over several days. Using BARCODE, we identified a sexually dimorphic preference for ethanol. Females are attracted to ethanol-laden foods, whereas males avoid consuming it. Furthermore, genetically feminizing male mushroom body lobes induces preference for ethanol. In addition, we demonstrate that BARCODE can be used for multiplex diet measurements when animals are presented with more than two food choices.
An invaluable model organism for the study of alcohol-use disorder (AUD) is Drosophila melanogaster. Drosophila shares substantial genetic homology and conserved ethanol responses with mammals, meaning that the genetics of ethanol responses identified in Drosophila are likely to be conserved in mammals. Like humans, Drosophila have a natural relationship with ethanol (1, 2) and show adaptations when exposed to ethanol. In humans, these adaptations are associated with AUD and include adaptations such as functional ethanol tolerance and ethanol-withdrawal hyperexcitability/reduced seizure threshold (3, 4). Consumptive ethanol preference—another behavior associated with AUD—has been difficult to quantify in flies due to their small meal size. The most commonly used method to measure ethanol drinking preference in flies has been the flexible and powerful capillary feeding (CAFE) assay (5, 6) in which flies select between ethanol-containing or ethanol-free liquid food delivered in 5-μL capillary pipettes (7). However, there is evidence that CAFE feeding negatively affects nutrient uptake and longevity (8–10) and that the caloric value of ethanol contributes to preference for ethanol food in this assay (11, 12).
The BARCODE assay does not cause or require calorie restriction. BARCODE monitors ethanol preference in a population of animals that are feeding ad libitum from a plentiful and easy-to-consume supply of solid fly food. In the behavioral chamber, two independent measures of feeding preference can be simultaneously applied—a computer-mediated observational method that monitors fly location on the food patches throughout the assay and the BARCODE qPCR-consumption method that measures the relative consumption preference of the fly for particular foods. In the qPCR-consumption method, two or more DNA oligomers with sequences not found in the fly or human genomes are added to ethanol and nonethanol fly-food patches at trace levels—a concentration that the flies cannot taste but that can still be used to determine their recent drinking history by qPCR of the oligomer. A representative fraction of the oligomer persists in the fly, making oligomer level a good biomarker for monitoring food choice. Using BARCODE, we document a sexually dimorphic difference in ethanol preference.
Results
We conceived of the BARCODE qPCR-consumption method as an assay in which flies have free access to large quantities of food and in which the ethanol-containing food is labeled with one DNA oligomer and the nonethanol food is labeled with another DNA oligomer. We anticipated that ethanol preference could then be measured by quantifying the relative concentration of oligomer in the fly body (Fig. 1A). We designed DNA oligomers suitable for quantification by qPCR and used them as a food additive. These ∼100-nt oligomers were designed to be recognized by qPCR primers that do not amplify sequence from any relevant genome (Drosophila, human, or common skin bacteria genomes).
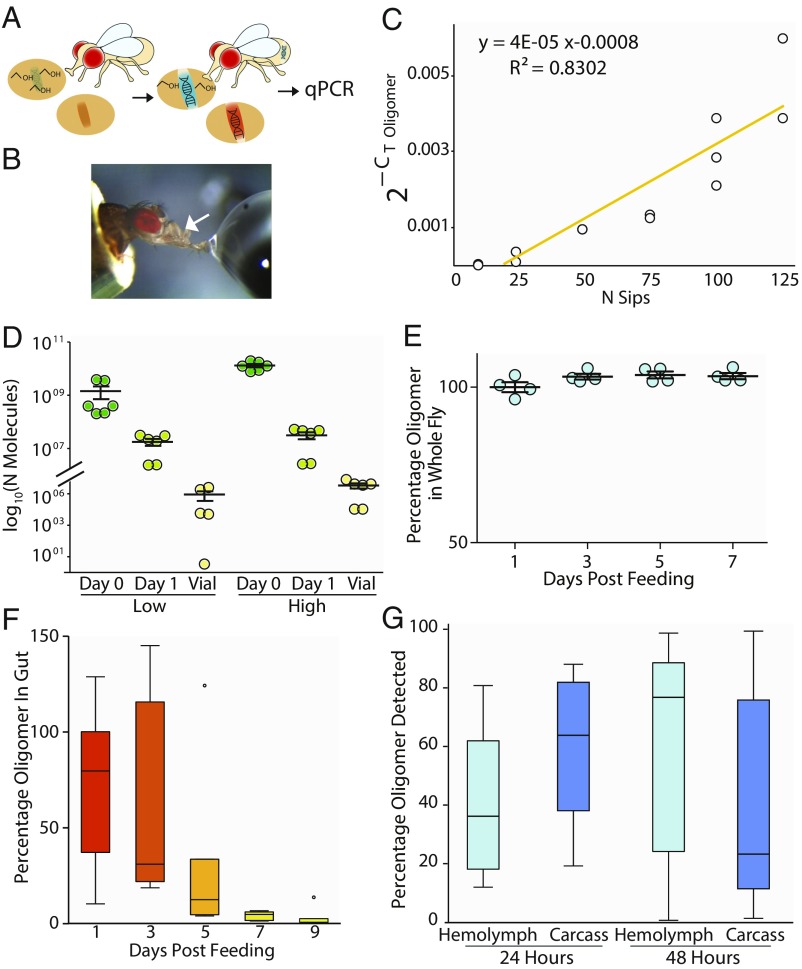
Fig. 1.
Consumed DNA oligomers provide a long-lasting quantitative measure of the amount of food eaten. (A) Schematic of the BARCODE assay. (B) Flies are hand fed an oligomer containing solution while oscillations (sips) of the cibarial pump (arrow) are counted (Movie S1). (C) Sips of oligomer solution (35 ng/µL) and oligomer in the fly correlate linearly (qPCR normalized to the genomic Cyp1 gene; n = 4, three flies each; experiment has been repeated three times with equivalent outcome). In D–G flies are hand fed 100 sips of oligomer solution. Oligomer is quantified by qPCR. (D) Number of oligomers in flies immediately after (day 0), 24 h after (day 1), and in feces deposited over 24 h after consumption (vial). Low = 3.5 ng/µL; high = 35 ng/µL, center line is mean, error bars are SEM, n = 6, four flies each. (E) Oligomer persists within the fly for days. Internal oligomer concentration was monitored for 7 d after feeding by qPCR; n = 4. Related control is found in SI Appendix, Fig. S1. (F) Oligomer persists within gut for many days. Dissected guts assayed over 7 d after feeding; n = 4, three flies each (experiment was repeated twice with equivalent outcome). (G) Oligomer partitions into the hemolymph. For F and G, boxes are Q1–Q3, center line is mean, whiskers are 95% CIs; n = 4, six flies each.
DNA Oligomers in Food Can Be Used to Quantitatively Measure Consumption.
To evaluate the utility of using DNA oligomers to monitor food preference in flies, we hand fed immobilized flies the DNA oligomer in a sucrose solution. The flies were allowed to take a predetermined number of sips of the oligomer solution (Fig. 1B and Movie S1), and then the relative abundance of the oligomer within the fly was measured by qPCR. We observed a linear relationship between sips of oligomer-laced food and the amount of oligomer assayed in a postmortem qPCR assay (Fig. 1C). During the first 24 h after oligomer consumption, 99% of the oligomer is lost from the fly, 1% of the oligomer remains in the fly, and 0.1% of the oligomer is passed through to the food (Fig. 1D and SI Appendix, Fig. S10). Most of the oligomer is destroyed by digestion within the first 60 min following feeding. However, the remaining 1% in the fly is surprisingly stable, persisting for about 7 d after consumption (Fig. 1E and SI Appendix, Fig. S1). The oligomer that persists in the fly was found in the gut (Fig. 1F) and hemolymph of the fly (Fig. 1G). Overall, these data show that ∼1% of the consumed oligomer remained within the fly and could be used as a proxy measure for food consumption. Finally, a pulse chase experiment (SI Appendix, Fig. S12) demonstrates that the consumed oligomer is stable over multiple days and that recently consumed oligomer does not displace previously consumed oligomer.
Removal of DNA Oligomer from Fly Exterior.
In a large behavioral chamber, flies have unrestricted access to the oligomer-laced food, and therefore it seemed likely that oligomer might contaminate the exterior of the fly body. To evaluate the extent of this problem and identify a way to remove external oligomer, we coated dead flies with oligomer by rolling them for 2 h on fly food containing 3.5 ng/μL oligomer, and then we evaluated methods for removing the oligomer. We observed that a substantial amount of oligomer accumulated on the fly body (Fig. 2).
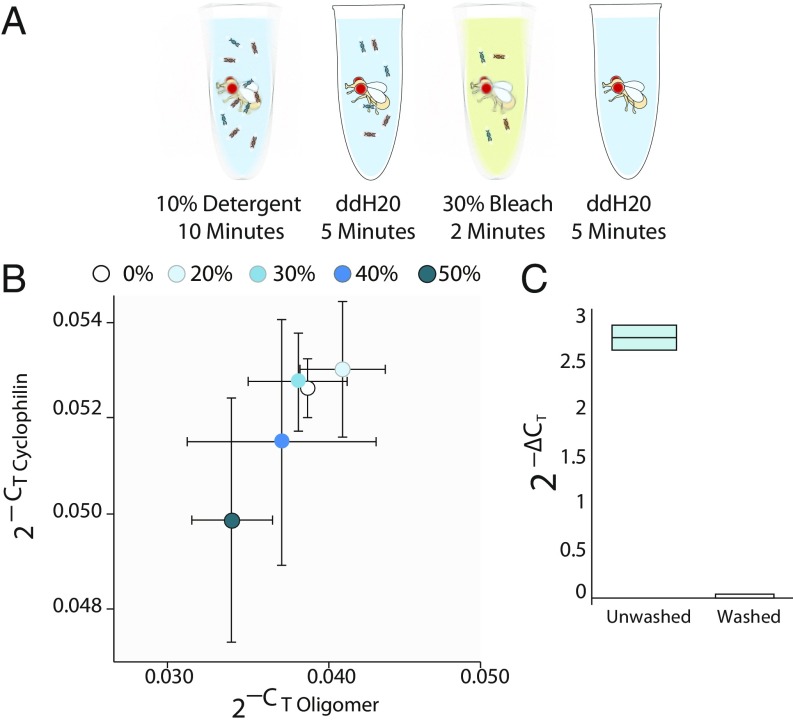
Fig. 2.
A protocol to remove oligomer from the exterior of the fly body. (A) Protocol schematic for removing oligomer from the fly exterior. (B) Flies were subjected to the washing protocol varying the bleach concentration from 0 to 50%. The abscissa is relative oligomer determined by qPCR and the ordinate is the relative concentration of the genomic Cyp 1 gene (abundance = 2−CT). Bleach washes of 0%, 20%, and 30% do not damage consumed oligomer or genomic DNA. Error bars are SD. Horizontal error bar for zero is eclipsed by the data point; n = 5. (C) Dead flies were rolled on 3.5 ng/µL oligomer food. qPCR shows that the 10% detergent/30% bleach washing protocol removes oligomer from the fly exterior {relative concentration is oligomer normalized to the genomic Cyp 1 control [2−ΔCT]}. Box is Q1–Q3s and center line is mean; n = 5. The experiment was repeated three times with equivalent results.
This oligomer was tightly bound to the chitinous exoskeleton as indicated by the fact that treatments that removed external oligomer without damaging internal DNA were difficult to find. Neither DNase incubation, UV irradiation, or washing with acid, detergent, or bleach was individually effective. We eventually found that a four-step protocol consisting of (i) a detergent wash, (ii) a water rinse, (iii) a dilute bleach wash, and (iv) a second water rinse could effectively remove the oligomer from the fly’s exterior.
To find the highest concentration of bleach that would remove oligomer from the exterior of the fly but not damage consumed oligomer or genomic DNA, we hand fed flies 100 sips of a 3.5-ng/μL oligomer solution and then processed the flies using the four-step protocol with varying concentrations of bleach (0–50%; Fig. 2A). We then determined the relative abundance of the oligomer (Fig. 2B). We observed that 40% and 50% bleach damaged both the internalized oligomer and the Cyclophilin 1 genomic DNA marker, while external oligomer was removed by a 20% or higher bleach wash. We chose to standardize on 30% bleach because it was the strongest concentration tested that did not affect internal oligomer abundance or genomic DNA abundance (Fig. 2B) and because it was more than sufficient to remove external oligomer (Fig. 2C).
DNA Oligomer Is Not Transferred Between Flies.
In the restricted environment of a fly vial or assay chamber, flies may be coprophagic. This behavior could facilitate oligomer transfer between flies, which would confound the use of oligomers for monitoring individual feeding patterns in a population of flies. To determine whether cohabitation enables transfer of the oligomer between flies, we hand fed flies and then housed each of them with a fly that had not been fed oligomer. After 2 d, the flies were separately assayed for the oligomer to determine the amount of transfer between animals. We observed no meaningful transfer between flies during this period (Fig. 3A).
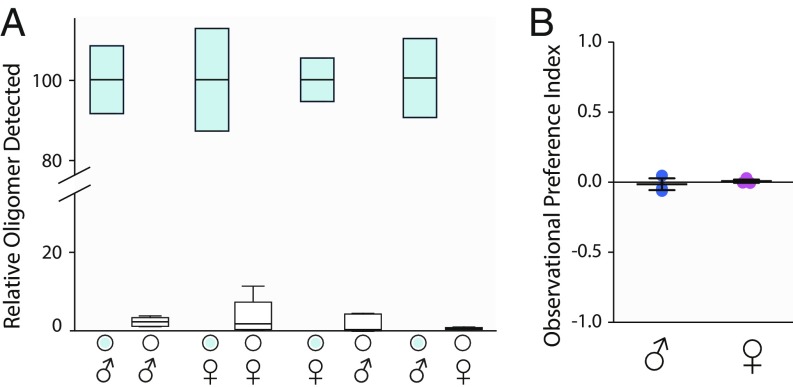
Fig. 3.
DNA oligomers do not transfer well between flies and DNA oligomer does not influence food preference. (A) One fly was hand fed 100 sips of oligomer solution (3.5 ng/µL; blue filled) and housed for 2 d in a food vial with a second nonfed fly. Values have been normalized to the number of oligomer molecules in the fed fly (determined by qPCR to be ∼20,000 molecules). Boxes are Q1–Q3; center line is mean, whiskers are 95% CIs; n = 6. There is no significant transfer of oligomer between most animals (one-sampled t test µo = 0; n = 6; µ = 2.3%, 3.4%, and 1.6%, respectively. P = 0.19, 0.22, and 0.32, respectively), except for slight transfer from males to females (n = 6; µ = 0.9%, P = 0.00418). (B) Oligomer added to standard cornmeal/molasses/agar media does not alter preference. Preference was measured using an observation assay (computer photography). Positive preference indexes indicate attraction, whereas negative values indicate aversion for the oligomer-laden food. Data are from three experiments in which 50 flies were allowed to feed for 2 d. Each data point is one experiment. Related controls are in SI Appendix, Figs. S2 and S3.
BARCODE Behavioral Chamber.
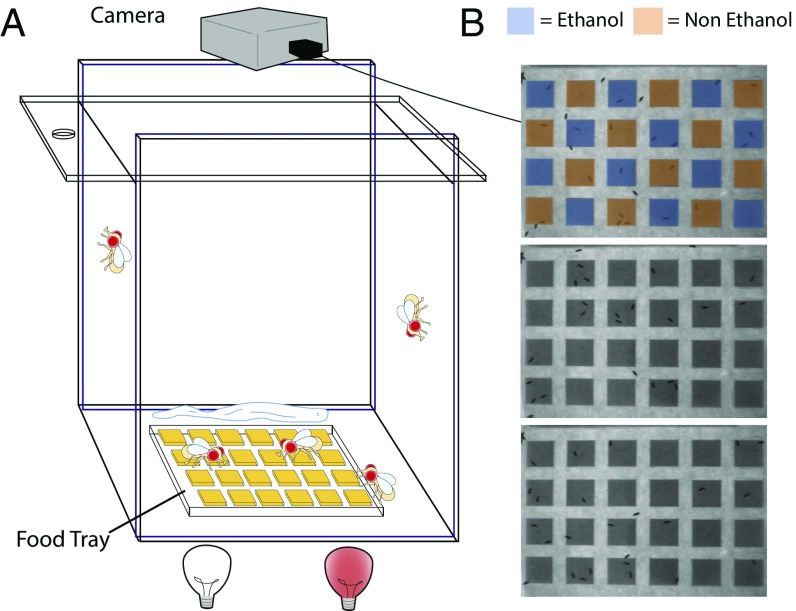
For our BARCODE ethanol-preference experiments, we use a 14 × 14.5 × 22.5 cm clear chamber that contains a food tray (Fig. 4). The chamber and tray were constructed with Plexiglas using a laser cutter (templates available in SI Appendix, Supplemental Files 1–4). Food trays contain 24 wells, and each well is 1.5 × 1.5 × 0.5 cm. In an ethanol-preference assay, flies feed from trays whose wells are loaded in an interdigitated pattern with either ethanol food or nonethanol food. Each food type contains a DNA oligomer specific for that food type at a concentration of 3.5 ng/μL (Fig. 4B). Fifty flies are tested at a time in the chamber.
Fig. 4.
Barcode behavioral assay chamber. (A) A 14 × 14.5 × 22.5 cm acrylic chamber with a sliding top. Camera on top of the chamber is used to report the occupancy of flies on particular wells in the 24-well food plate at the bottom. The top of the chamber can be slid to engage the fly transfer hole. Flies are added, and samples of flies are taken, through this hole at the end of day 1 and day 2 using a flypette. Illumination is provided by white light during the day and red light at night. (B) Representative photos of plates show the arrangement of ethanol-containing and nonethanol-containing food (and the associated DNA oligomers). Flies are easily seen in these pictures.
We use two distinct methods to measure ethanol preference in this assay chamber (Fig. 4A). One is the qPCR-based method, that we call BARCODE, in which the relative abundance of the consumed oligomer 1 and oligomer 2 within the flies is measured (five groups of three to five flies are assayed by qPCR, and the data are normalized to the relative abundance of the genomic Cyp1 gene). The second method is an observational method in which snapshots of the food grid are taken every 5 min and the number of flies over each well of food is recorded (Fig. 4B). The second method is used to help validate the BARCODE method. Preference indices are calculated from both of these methods.
DNA Oligomers Do Not Influence Preference.
This was demonstrated in three ways—a proboscis extension response assay showed that an oligomer added to the food does not alter sip probability except at concentrations ∼450× that used to measure preference (SI Appendix, Fig. S2); an observational preference assay (computer photographically recorded food preference) that showed that flies do not distinguish between oligomer-containing food patches and nonoligomer-containing food patches (Fig. 3B). We have observed that the BARCODE qPCR-consumption assay and the observational assay show strong agreement for 5% alcohol food preference (e.g., Figs. 5 A and B and 6 and SI Appendix, Fig. S8 A and C). The concordance in these independent measures helped validate the BARCODE assay.
Fig. 5.
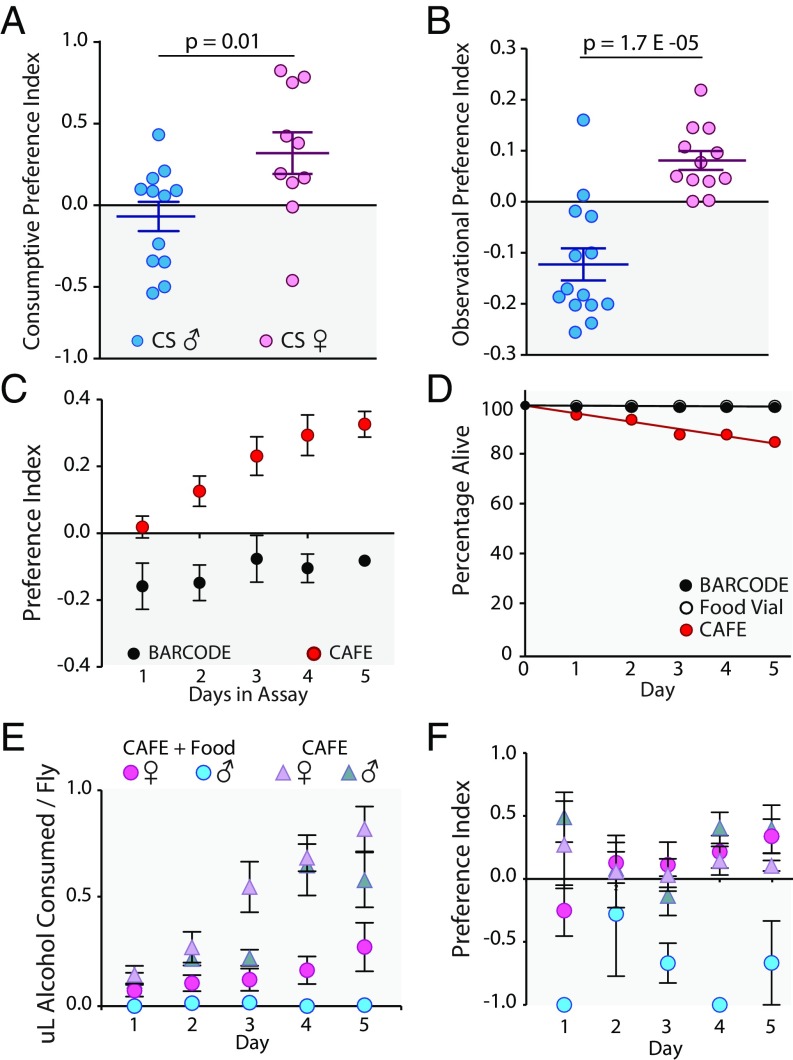
(A) BARCODE qPCR-consumption assay shows sexually dimorphic ethanol preference for 5% ethanol food (five flies per data point, n = 12 for males, n = 10 for females, two-tailed t test). (B) Sexually dimorphic ethanol preference is validated in the observation assay (each data point is one 2-d test); n = 14 for males; n = 10 for females. (C) In CAFE, males show increasing preference for 15% ethanol food (n = 16), whereas in BARCODE, males show no attraction for 5% ethanol food (n = 3). (D) CAFE shows higher mortality than BARCODE whose data points overlap with those for vial; n = 8 CAFE, n = 4 BARCODE, n = 1 fly vial. (E) When food is added to the bottom of CAFE vials, males do not consume any ethanol (blue circles), whereas females do (pink circles); absence of solid food causes males to readily consume the ethanol liquid (blue triangle). (F) CAFE + food eliminates male preference for alcohol (blue circles), whereas females stay attracted to alcohol regardless of the presence of solid food (pink circles and pink triangles). Error bars are SD of the mean and the data point or the center line is positioned at the mean. In box and whisker plot, box is Q1–Q3, and whiskers are 95% CIs. Additional related controls are found in SI Appendix, Figs. S7 and S8.
Fig. 6.
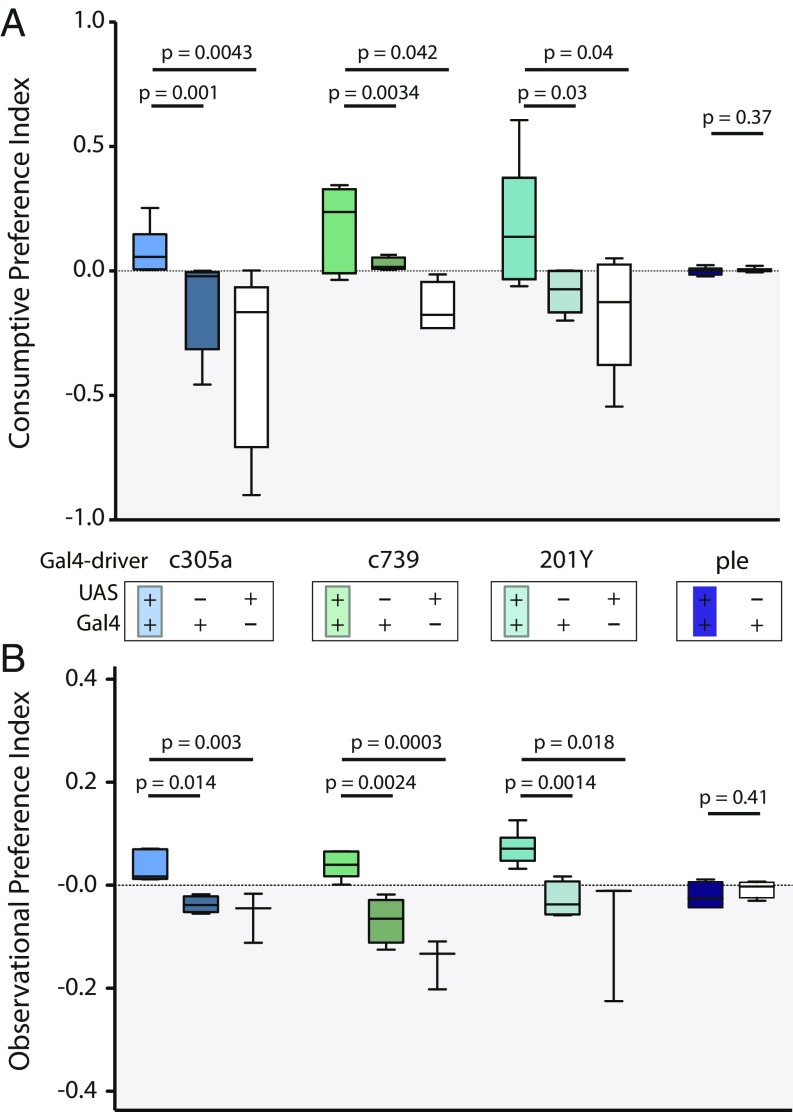
Feminization of male mushroom body lobes alters male ethanol preference. (A) Behavior in BARCODE was assayed using the qPCR-consumption assay. γ-Lobe feminization (201Y, n = 18), ɑβ lobe feminization (c739, n = 12), and ɑ′β′ feminization (c305a, n = 16). Feminization of MB lobes increases preference for 5% ethanol food compared with controls. Feminization of TH-expressing neurons does not promote ethanol preference in males; n = 6. (B) Behavior in the chamber was also assayed using the observation assay. MB driver c305a (n = 6; 50 flies used per number), c739 (n = 3; 50 flies used per number) and, 201Y (n = 4; 50 flies used per number) all showed significant increases in observational preference compared with controls. Feminization of TH-expressing neurons does not promote ethanol preference in males; n = 3. For both A and B, statistical significance was determined using a two-tailed t test. Note that the Gal4 drivers used are more broadly expressed. Refer to ref. 40 for expression patterns.
Flies Are Sensitive to Quinine in a Dose-Dependent Manner in BARCODE.
To determine the sensitivity of the BARCODE qPCR assay we performed a quinine dose–response assay. Quinine is a naturally bitter tasting compound that has been previously shown to be aversive to flies (13). Flies have been shown to be able to distinguish concentrations of quinine up to 1 mM in CAFE (14). We used concentrations of 0.1 mM, 1 mM, and 10 mM and observed dose-dependent aversion down to 0.1 mM in both the consumptive BARCODE qPCR assay and the observational preference assay (SI Appendix, Fig. S8 A and C).
BARCODE Measurement of Ethanol Preference Is Not Sensitive to Ethanol Calories.
In Pohl et al. (11), it was shown that in the CAFE assay, flies had strong preference for ethanol and that this preference was completely eliminated once the caloric content of ethanol- and nonethanol-containing foods was balanced. However, in BARCODE, balancing the caloric content of the ethanol food and nonethanol food does not eliminate preference for ethanol (SI Appendix, Fig. S7), indicating that ethanol preference measured here is not dependent on the caloric value of ethanol. Consistent with the idea that CAFE is a calorie-restrictive assay, we observed 16% death in the CAFE assay over a 5-d period. In the BARCODE assay, we observed just 0.25% death (averaged over four assays) and none in food-vial-housed flies over a 5-d period (Fig. 5D). Furthermore, CAFE-housed males showed a 40% reduction in mean body weight (SI Appendix, Fig. S4A) compared with vial-housed flies. Finally, adding solid fly food to the bottom of the CAFE vials to reduce starvation eliminated male preference for alcohol but did not change female preference for alcohol (Fig. 5 E and F). These results with food-supplemented CAFE more closely emulate our BARCODE alcohol-preference results.
Genetic Feminization Modulates Ethanol Behaviors.
In BARCODE, we observed that wild-type male Canton S (CS) flies are not attracted to 5% ethanol by volume (ABV), whereas female CS flies prefer ethanol-laden foods up to 10% ABV (Fig. 5 A and B and SI Appendix, Fig. S5B). We hypothesized that genetic sex differences contribute to sexually dimorphic ethanol preference. To test this, we genetically feminized male neuropil by inhibiting FruM production using an inducible UAS-Transformer (TraF) transgene. Expression of this splicing factor induces female-specific cellular identity and when ectopically expressed in males, feminizes the cells by suppressing production of the male-specific FruM splice variant (15). Fru is endogenously expressed in the mushroom bodies (MBs) (16). Using three Gal4 lines, we individually drove UAS-TraF in the MBs. The resultant feminization of male flies made males relatively more attracted to 5% ethanol food (Fig. 6). Interestingly, when we genetically feminized tyrosine hydroxylase (TH)-producing neurons (ple-Gal4) we saw no such increase in preference for ethanol. In addition, male MB feminization also reduced the male’s capacity to acquire 24-h functional alcohol tolerance (alcohol-induced alcohol resistance) but had no effect on tolerance on females (SI Appendix, Fig. S9).
Discussion
The BARCODE assay was developed as a way to measure preference for ethanol in D. melanogaster. However, because it uses DNA oligomers of unique sequence to label foods, the BARCODE assay is not limited to binary food choices and can also be used for measuring feeding patterns when more than two food choices are simultaneously presented. In the context of ethanol research, this might prove useful for monitoring the progression from low- to high-ethanol-containing foods.
In parallel with the BARCODE qPCR-based assay, we also collected photographic data of the feeding preference of flies in the same chamber. We observed that flies tend to occupy food patches that match their food of choice as measured by the BARCODE qPCR assay even when they are not feeding—this is likely because flies have taste receptors on their feet.
CAFE for alcohol preference is typically performed in the absence of extra food; this reduces nutrient uptake (8) as evidenced here by reduced body weight and longevity. Typically, rodent alcohol-preference paradigms include readily abundant solid food as well as ethanol and nonethanol liquid choices. Abundant food serves to keep the animals healthy and reduces the possibility that animals consume alcohol for its caloric value so that the experimenter can more accurately measure pharmacological preference for alcohol. To emulate this paradigm, we added food to the bottom of vials in the CAFE assay. We found that male flies no longer preferred ethanol food, replicating the results obtained in BARCODE.
A surprising observation was the perdurance of the consumed BARCODE oligomer in the fly. We believe that a fraction of the consumed oligomer is in some manner protected from digestion in the intestine. This might arise from transport of some of the oligomer from the lumen of the intestine into the hemolymph. Alternatively, a fraction of the oligomer may bind to chitin within the digestive tract, making that oligomer fraction unavailable for digestion. Impermeable cuticle lines the foregut and hindgut, while the midgut is lined by a type II peritrophic membrane that is made of chitin and glycoproteins (17, 18). The nonspecific binding of DNA probes to cuticle has been previously noted during the course of Drosophila in situ hybridization experiments (19–22). While some insects lack a peritrophic membrane, they still retain other cuticular structures within the digestive tract. Thus, the BARCODE oligomer assay may generalize to other insects.
The ethanol-preference behavior of flies in BARCODE is substantially different from that observed in the CAFE assay. In CAFE, we have observed that both sexes show the same bias for consumption of ethanol-containing food [SI Appendix, Fig. S5A and also figure 7 in Pohl et al. (11)], while in BARCODE we observed that males and females have the opposite response to ethanol in their food, with males showing avoidance of ethanol in their food (even at 5%) and females showing preference for ethanol in their food (up to 10%). There is also a difference in that CAFE-housed flies typically show a day-by-day increase in ethanol preference whereas BARCODE-housed flies show the same ethanol preference on each day of the assay (Fig. 5C and SI Appendix, Fig. S5B).
We genetically manipulated the sex of the MB lobes to determine if the sexually dimorphic ethanol preference is genetically encoded. The MBs receive numerous inputs from various areas of the brain. They receive sensory inputs as well as inputs that modulate valence of a stimulus such as those from dopaminergic neuropil (23). The MBs have also been shown to be necessary for expression of appetitive and aversive behaviors (24–26). Others have also shown that the mushroom body is important in modulating ethanol preference (27) and a subset of the MB output neurons have been demonstrated to be necessary for expressing olfactory ethanol attraction (28). Induction of TraF expression was used to feminize the MB lobes of males. TraF is a splicing factor that causes female-specific splicing of both fruitless (fru) and doublesex (dsx) transcripts. In males, TraF is not active, resulting in expression of the “default” male-specific form of both fru (fruM) and dsx (dsxM) (29, 30). The encoded FruM and DsxM proteins are transcription factors. FruM is neural specific, whereas DsxM is expressed in male neuronal and nonneuronal tissues (15, 31). FruM has been shown to be necessary for sex-specific dendritic arborization patterns (32, 33) and has also been associated with male-specific behavior such as courtship and aggression (34). Previously, the fru gene was implicated in ethanol-induced disinhibition of courtship (35) as well as in modulating the sensitivity to sedation with ethanol vapor (36). Feminization of any one of the three male MB lobes causes males to increase preference for ethanol in the BARCODE assay chamber. We propose that FruM expression in the MB modulates male ethanol preference. We do not think that DsxM plays this role because DsxM has primarily been associated with male physical attributes (37). The effect of TraF on ethanol tolerance only in males may arise from a change in neural circuitry during development. Alternatively, the FruM or DsxM transcription factors themselves may modulate ethanol-induced genes that are required for functional ethanol tolerance. The absence of an effect in females suggests that the mechanism generating ethanol tolerance has a sexually dimorphic component.
Another interesting sexually dimorphic behavior we observed was fly clustering. The photographic observational method that we used records the number of flies per well over each 5-min time increment throughout the duration of the assay. We noticed that males clustered less frequently than females as evidenced by a lower ClusterF index (SI Appendix, Fig. S6). This could reflect an important social dynamic and may influence which food patches the females occupy and feed on. This behavior cannot be observed in CAFE because a micropipette does not provide the physical space for clustering. Previous studies on social clustering did not show sexual dimorphism. However, this may be because the studies were performed in a small chamber in the absence of food (38).
The BARCODE qPCR assay is a food consumption assay that is well suited for determining the relative food preference of flies. For our purposes, we use the ∆∆CT qPCR method to compare the preference of flies for ethanol food versus nonethanol food (39). Determination of the absolute volume of food consumed is complicated by the fact that only a representative amount of oligomer survives digestion. This limitation could be resolved by using nonhydrolyzable DNA oligomers to prevent oligo digestion and allow the experimenter to determine the absolute volume of food consumed. Furthermore, nonhydrolyzable oligomers might also increase the sensitivity of the assay—we have not yet tested these ideas.
The BARCODE assay is very flexible and can be used to conduct experiments that were previously not possible. The availability of a huge number of distinctly assayable DNA oligomers should enable one to monitor the selection between many different food choices. In SI Appendix, Fig. S11 we demonstrate the feasibility of using BARCODE to simultaneously measure the consumption of three different food sources. In addition, when we sequentially fed two oligomers to flies, we observed that the second oligomer did not displace the first oligomer (SI Appendix, Fig. S12). Thus, if one switched feeding plates each day, and each plate contained a unique oligomer set, one should be able to use qPCR to determine the relative consumption of each food during each day of a given period. Moreover, because the BARCODE assay quantifies DNA oligomers in a small number of flies (we used three to five flies per PCR for genetic feminization experiments and single flies for the quinine aversion experiment) (SI Appendix, Fig. S8B), BARCODE could lend itself to monitoring mixed populations of flies to determine how different types (genotypes, sexes, species) of flies influence each other. Finally, it should not be overlooked that males and females have neurally based differences in ethanol preference. In Drosophila because the gender of individual neurons can be manipulated, this should prove useful as a tool for mapping circuits and identifying genes that modulate ethanol preference.
Materials and Methods
Oligomers were designed using an Excel spreadsheet (Microsoft) using randomly generated numerical sequences with corresponding nucleotides: 1 = A, 2 = T, 3 = G, 4 = C. Primers were designed using IDT PrimerQuest Tool (idtdna.com) and screened using BLAST (National Center for Botechnology Information) to identify those that did not match Drosophila, human, or skin flora bacteria (SI Appendix, Supplemental Materials and Methods). Preference was determined two ways: (i) consumptive preference using the qPCR method and (ii) observational preference using photographic data. For BARCODE qPCR-preference assays flies were anesthetized with CO2 and homogenized as described (SI Appendix, Supplemental Materials and Methods). Homogenate was amplified using SYBR Green PCR Master Mix with a Tm = 60 °C and 40 cycles per run. Observational preference index was determined using an image processing script that counts flies on each food well (41). See SI Appendix, Supplemental Materials and Methods for more details. For consumptive qPCR preference, animals were harvested at the end of the second day except for SI Appendix, Fig. S8, which were sampled at the end of day 1 and day 2. For observational preference, the occupancy of each food substrate was recorded every 5 min and plotted (SI Appendix, Fig. S8C), or binned into groups of 50 min (SI Appendix, Figs. S3B, S5C, and S7B), or averaged into a single value for a 2-d period (Figs. 3B, 5B, and 6B and SI Appendix, Fig. S7A), or averaged per day (SI Appendix, Figs. S3A and S5B).
Supplementary Material
Acknowledgments
We thank Christopher Stojanik for assistance with primer design. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This work was supported by NIH Grants 2R01AA018037-06A1 (to N.S.A.) and NIH T32AA07471 (to A.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716880115/-/DCSupplemental.
References
- 1.Dudley R. Ethanol, fruit ripening, and the historical origins of human alcoholism in primate frugivory. Integr Comp Biol. 2004;44:315–323. doi: 10.1093/icb/44.4.315. [DOI] [PubMed] [Google Scholar]
- 2.Nunney L. The colonization of oranges by the cosmopolitan Drosophila. Oecologia. 1996;108:552–561. doi: 10.1007/BF00333733. [DOI] [PubMed] [Google Scholar]
- 3.Ghezzi A, Krishnan HR, Atkinson NS. Susceptibility to ethanol withdrawal seizures is produced by BK channel gene expression. Addict Biol. 2014;19:332–337. doi: 10.1111/j.1369-1600.2012.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholz H, Ramond J, Singh CM, Heberlein U. Functional ethanol tolerance in Drosophila. Neuron. 2000;28:261–271. doi: 10.1016/s0896-6273(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 5.David JRR. Quantitative evaluation of liquid food intake by Drosophila. Drosophila Inf Serv. 1970;45:89–90. [Google Scholar]
- 6.Ja WW, et al. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci USA. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devineni AV, Heberlein U. Preferential ethanol consumption in Drosophila models features of addiction. Curr Biol. 2009;19:2126–2132. doi: 10.1016/j.cub.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande SA, et al. Quantifying Drosophila food intake: Comparative analysis of current methodology. Nat Methods. 2014;11:535–540. doi: 10.1038/nmeth.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KP, et al. Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc Natl Acad Sci USA. 2008;105:2498–2503. doi: 10.1073/pnas.0710787105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vigne P, Frelin C. Food presentation modifies longevity and the beneficial action of dietary restriction in Drosophila. Exp Gerontol. 2010;45:113–118. doi: 10.1016/j.exger.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 11.Pohl JB, et al. Ethanol preference in Drosophila melanogaster is driven by its caloric value. Alcohol Clin Exp Res. 2012;36:1903–1912. doi: 10.1111/j.1530-0277.2012.01817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekhon ML, Lamina O, Hogan KE, Kliethermes CL. Common genes regulate food and ethanol intake in Drosophila. Alcohol. 2016;53:27–34. doi: 10.1016/j.alcohol.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Tompkins L, Cardosa MJ, White FV, Sanders TG. Isolation and analysis of chemosensory behavior mutants in Drosophila melanogaster. Proc Natl Acad Sci USA. 1979;76:884–887. doi: 10.1073/pnas.76.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sellier MJ, Reeb P, Marion-Poll F. Consumption of bitter alkaloids in Drosophila melanogaster in multiple-choice test conditions. Chem Senses. 2011;36:323–334. doi: 10.1093/chemse/bjq133. [DOI] [PubMed] [Google Scholar]
- 15.Ferveur JF, Störtkuhl KF, Stocker RF, Greenspan RJ. Genetic feminization of brain structures and changed sexual orientation in male Drosophila. Science. 1995;267:902–905. doi: 10.1126/science.7846534. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto D, Koganezawa M. Genes and circuits of courtship behaviour in Drosophila males. Nat Rev Neurosci. 2013;14:681–692. doi: 10.1038/nrn3567. [DOI] [PubMed] [Google Scholar]
- 17.Lehane MJ. Peritrophic matrix structure and function. Annu Rev Entomol. 1997;42:525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- 18.Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster. Annu Rev Genet. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- 19.Becker MN, Brenner R, Atkinson NS. Tissue-specific expression of a Drosophila calcium-activated potassium channel. J Neurosci. 1995;15:6250–6259. doi: 10.1523/JNEUROSCI.15-09-06250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fechtel K, Fristrom DK, Fristrom JW. Prepupal differentiation in Drosophila: Distinct cell types elaborate a shared structure, the pupal cuticle, but accumulate transcripts in unique patterns. Development. 1989;106:649–656. doi: 10.1242/dev.106.4.649. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert LI. Insect Molecular Biology and Biochemistry. Elsevier/Academic; London: 2012. [Google Scholar]
- 22.Spletter ML, et al. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural Dev. 2007;2:14. doi: 10.1186/1749-8104-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keene AC, Waddell S. Drosophila olfactory memory: Single genes to complex neural circuits. Nat Rev Neurosci. 2007;8:341–354. doi: 10.1038/nrn2098. [DOI] [PubMed] [Google Scholar]
- 24.Heisenberg M, Borst A, Wagner S, Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J Neurogenet. 1985;2:1–30. doi: 10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- 25.Kim YC, Lee HG, Han KA. D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci. 2007;27:7640–7647. doi: 10.1523/JNEUROSCI.1167-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S, et al. The propensity for consuming ethanol in Drosophila requires rutabaga adenylyl cyclase expression within mushroom body neurons. Genes Brain Behav. 2012;11:727–739. doi: 10.1111/j.1601-183X.2012.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aso Y, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshijima K, Inoue K, Higuchi I, Sakamoto H, Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991;252:833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- 30.McKeown M, Belote JM, Boggs RT. Ectopic expression of the female transformer gene product leads to female differentiation of chromosomally male Drosophila. Cell. 1988;53:887–895. doi: 10.1016/s0092-8674(88)90369-8. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 2011;9:e1001131. doi: 10.1371/journal.pbio.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura K, Ote M, Tazawa T, Yamamoto D. Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature. 2005;438:229–233. doi: 10.1038/nature04229. [DOI] [PubMed] [Google Scholar]
- 33.Lee G, et al. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J Neurobiol. 2000;43:404–426. doi: 10.1002/1097-4695(20000615)43:4<404::aid-neu8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 34.Manoli DS, et al. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 35.Lee HG, Kim YC, Dunning JS, Han KA. Recurring ethanol exposure induces disinhibited courtship in Drosophila. PLoS One. 2008;3:e1391. doi: 10.1371/journal.pone.0001391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devineni AV, Heberlein U. Acute ethanol responses in Drosophila are sexually dimorphic. Proc Natl Acad Sci USA. 2012;109:21087–21092. doi: 10.1073/pnas.1218850110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shirangi TR, Taylor BJ, McKeown M. A double-switch system regulates male courtship behavior in male and female Drosophila melanogaster. Nat Genet. 2006;38:1435–1439. doi: 10.1038/ng1908. [DOI] [PubMed] [Google Scholar]
- 38.Simon AF, et al. A simple assay to study social behavior in Drosophila: Measurement of social space within a group. Genes Brain Behav. 2012;11:243–252. doi: 10.1111/j.1601-183X.2011.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Applied Biosystems Inc. 2001. User Bulletin #2 ABI PRISM 7700 Sequence Detection System (Applied Biosystems Inc., Foster City, CA), pp 1–36.
- 40.Aso Y, et al. The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet. 2009;23:156–172. doi: 10.1080/01677060802471718. [DOI] [PubMed] [Google Scholar]
- 41.Ramazani RB, Krishnan HR, Bergeson SE, Atkinson NS. Computer automated movement detection for the analysis of behavior. J Neurosci Methods. 2007;162:171–179. doi: 10.1016/j.jneumeth.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.