Significance
Competition among cooccurring bacteria can change the structure and function of a microbial community. However, little is known about the molecular mechanisms that impact such interactions in vivo. We used the association between bioluminescent bacteria and their squid host to study how environmentally transmitted bacteria compete for a limited number of host colonization sites. Our work suggests that Vibrio fischeri use a type VI secretion system, acting as a contact-dependent interbacterial “weapon,” to eliminate competing strains from cooccupying sites in the host. This work illuminates a mechanism by which strain-specific differences drive closely related bacteria to engage in lethal battles as they establish a beneficial symbiosis, revealing how genetic variation among potential colonizers directly impacts the spatial structure of the host-associated population.
Keywords: type VI secretion, Aliivibrio fischeri, Euprymna scolopes, squid, symbiosis
Abstract
Intraspecific competition describes the negative interaction that occurs when different populations of the same species attempt to fill the same niche. Such competition is predicted to occur among host-associated bacteria but has been challenging to study in natural biological systems. Although many bioluminescent Vibrio fischeri strains exist in seawater, only a few strains are found in the light-organ crypts of an individual wild-caught Euprymna scolopes squid, suggesting a possible role for intraspecific competition during early colonization. Using a culture-based assay to investigate the interactions of different V. fischeri strains, we found “lethal” and “nonlethal” isolates that could kill or not kill the well-studied light-organ isolate ES114, respectively. The killing phenotype of these lethal strains required a type VI secretion system (T6SS) encoded in a 50-kb genomic island. Multiple lethal and nonlethal strains could be cultured from the light organs of individual wild-caught adult squid. Although lethal strains eliminate nonlethal strains in vitro, two lethal strains could coexist in interspersed microcolonies that formed in a T6SS-dependent manner. This coexistence was destabilized upon physical mixing, resulting in one lethal strain consistently eliminating the other. When juvenile squid were coinoculated with lethal and nonlethal strains, they occupied different crypts, yet they were observed to coexist within crypts when T6SS function was disrupted. These findings, using a combination of natural isolates and experimental approaches in vitro and in the animal host, reveal the importance of T6SS in spatially separating strains during the establishment of host colonization in a natural symbiosis.
Many eukaryotes establish important symbiotic associations with environmental bacteria (1). During the colonization process, these bacteria must transition from a free-living to a host-associated lifestyle. Although different natural environments can contain thousands or more bacterial taxa, host-associated bacterial communities that arise from the surrounding community are often far less diverse, sometimes composed of only a few species or strains in a given colonization site (2–5). For example, open wounds that are infected following exposure to seawater typically contain a lower diversity of bacteria relative to the diversity in seawater (6, 7). Human skin is also exposed to diverse environmental bacteria during the natural course of development, and recent work has revealed that hair follicle microbiomes are dominated by a single species of bacteria, Propionibacterium acnes (8), with individual humans being colonized by different strains of P. acnes that likely evolved from a common ancestor via clonal expansion (9). This dramatic “winnowing” of many bacterial genotypes to only a few successful colonizers could be explained in part by partner specificity through coevolution (1, 10, 11), resulting in select bacterial taxa capable of colonizing the host. These bacterial colonizers theoretically represent genotypes that could either stably coexist, or engage in competition to fill the same host niche. The genomes of many host-associated bacteria encode interbacterial killing mechanisms predicted to facilitate inter- and intraspecific competition (12–19), which may substantially alter the abundance and diversity of species or strains within the host, as well as their spatial distributions.
Mechanisms for interbacterial competition are diverse, and both diffusible and contact-dependent strategies have been described (20, 21). Diffusible mechanisms include bactericidal chemicals (22, 23), as well as secreted antimicrobial proteins (17). By contrast, contact-dependent mechanisms require direct cell–cell contact for transfer of a toxic protein from inhibitor to target cells (18, 24–28). The type VI secretion system (T6SS) is a broadly distributed contact-dependent killing mechanism that is commonly found among gammaproteobacteria (27, 28). T6SSs have been studied extensively in Vibrio cholerae (12, 29–33) and Pseudomonas aeruginosa (34, 35), as well as many other bacteria (13–16, 36–43), where they have been shown to act as molecular syringes that translocate effector molecules directly into target bacteria (27, 28, 44).
The T6SS uses a toxin/immunity mechanism to inhibit competitor cells (45). The inhibitor cell encodes both a toxic effector and its cognate immunity protein (46–49). The effector is translocated through the syringe-like T6SS apparatus and expression of the immunity protein, which remains within the inhibitor cell and is not predicted to be translocated, prevents self-intoxication and also protects isogenic cells from growth inhibition or cell lysis. However, if the toxic effector is translocated into a nonisogenic or competitor cell that does not encode the appropriate immunity gene, the competitor cell is inhibited or eliminated. Despite the broad distribution of interbacterial killing mechanisms, few studies have investigated their ecological roles (14, 17, 43, 50, 51). We hypothesize that in addition to encoding necessary host colonization factors, successful colonizers may also employ mechanisms to exclude closely related competitors and occupy a limited number of colonization sites.
To test this hypothesis, a tractable model system is required. The Vibrio–squid symbiosis is a simplified model for studying bacterial colonization in an animal host (52). Euprymna scolopes squid house multiple strains of bioluminescent Vibrio fischeri bacteria in a structure called the light organ (53). Juvenile squid hatch without their bacterial symbionts, which they must acquire from the surrounding seawater (54). Although studies addressing the strain-level diversity of V. fischeri in the seawater are limited (55), it is predicted to be much higher than the diversity found in a naturally colonized squid, which harbors only a few strains per light organ (53). Potential colonizers first aggregate on the surface of the light organ before entering pores that lead to six physically separated crypt spaces, where cells rapidly proliferate (56–58). Importantly, the passageway leading to the crypt entrance is physically constricted to the size of approximately one bacterial cell, restricting access to a single cell at a time (53). Because on average only a few cells enter each crypt space, the Vibrio–squid symbiosis offers a unique opportunity to answer questions about intraspecific competition that arises during the natural host colonization process.
Previous work suggests that V. fischeri strains employ competitive mechanisms as they colonize the host. For example, pairwise cocolonization experiments of juvenile squid using different combinations of light-organ isolates revealed that cocolonized animals often possess a light-organ population dominated by a single strain, based on counts of colony-forming units (CFUs) (59, 60). Recently, we showed competition that appears to occur within individual crypts of the light organ: when juveniles were exposed to an inoculum containing two different strains, the resulting crypts were colonized exclusively with one strain type, suggesting these strains are unable to coexist within the same crypt space (61). Thus, V. fischeri strains appear to have the ability to compete for light-organ dominance and segregate within individual crypts, although the mechanisms underlying these competitive interactions are not yet known. This work aims to identify the mechanism underlying intraspecific competition among naturally coisolated symbionts and determine how strain-specific genotypic differences among competitors drive these interactions to influence host colonization outcomes, including the spatial distribution of strains within the host.
Results
V. fischeri Isolates Are Capable of Strain-Specific Competitive Interactions.
To begin exploring potential competitive interactions among V. fischeri isolates, we first examined how strains coisolated from the same light organ interact with the commonly studied symbiotic isolate, ES114. V. fischeri ES114 was isolated in 1989 from an adult E. scolopes host collected in Kaneohe Bay, Hawaii (62). For more recently isolated strains, we selected six additional strains: three coisolated strains that each came from two different hosts. We chose to examine interactions using two groups of recently coisolated strains because these isolates may have physically encountered one another in aggregates on the light-organ surface or within crypts during the natural host colonization process in wild Euprymna juveniles. Moreover, some microorganisms lose competitive genes when kept in isolation (63); therefore, we reasoned that newly isolated strains may be less likely to have lost important genes that mediate interstrain interactions.
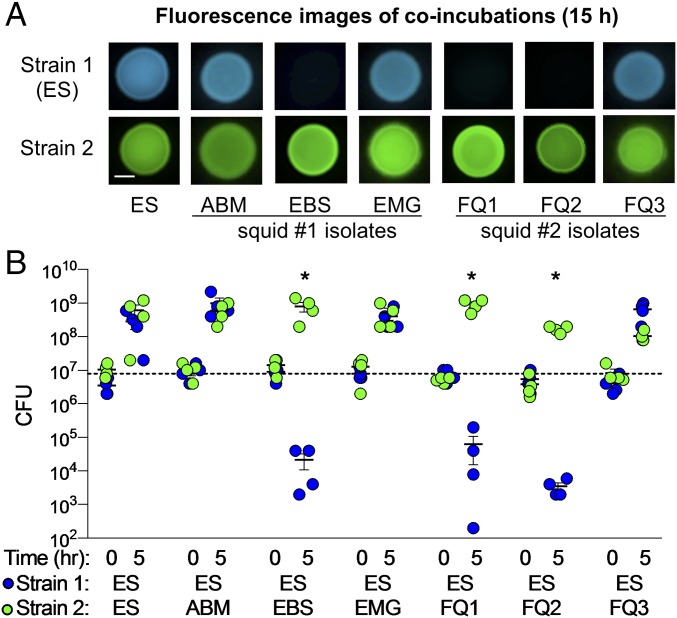
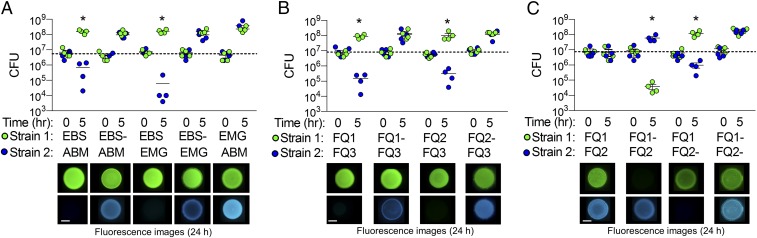
To identify possible competitive behaviors among different symbiotic strains, we used a culture-based coincubation assay to reflect the close interactions experienced by symbiotic cells in the host. These assays were optimized for V. fischeri and are based on similar coincubation assays described previously (13, 16). Briefly, strains were transformed with stable plasmids expressing different fluorescent proteins and antibiotic resistance genes. These differentially labeled coincubated strains could be spatially distinguished within a mixed colony and directly quantified by plating onto selective medium. Based on CFU measurements, each strain was present at equal abundance (1:1 ratio) at the start of the coincubation. When strain ES114 was coincubated 1:1 with each of our six V. fischeri isolates, we found there was a strain-specific difference in the ability of ES114 to grow in the presence of another strain (Fig. 1A). ES114 was able to grow in the presence of strains ABM004, EMG003, and FQ-A003: both strains were visibly present in the mixed colony after 15 h. In contrast, ES114 was not visibly detectable after 15 h when coincubated with strains EBS004, FQ-A001, and FQ-A002, suggesting that the growth of ES114 was inhibited. These findings suggest that certain light-organ isolates are capable of strain-specific competitive interactions, and that the interactions can be captured using an in vitro assay.
Fig. 1.
Impact of coincubation on growth of V. fischeri ES114. (A) Fluorescence microscopy images and (B) CFU counts for each coincubation spot for coincubations of ES114 (ES) with itself, or squid 1 and 2 coisolates. Squid 1 coisolates: ABM004 (ABM), EBS004 (EBS), EMG003 (EMG). Squid 2 coisolates: FQ-A001 (FQ1), FQ-A002 (FQ2), FQ-A003 (FQ3). Fluorescence images show ES114 (blue) and coincubated strain (green) after 15 h. (Scale bar, 2 mm.) CFU counts are reported for 0 and 5 h coincubation of individual biological reps (n = 4). Asterisks indicate P < 0.03 using a Student’s t test comparing CFUs of ES114 at 5 h between the control (strain 2 = ES114) and treatment coincubations. One of three representative experiments is shown.
ES114 Is Killed in a Contact-Dependent Manner.
We first hypothesized that these strain-specific competitive outcomes could be explained by differences in growth rates among our isolates. For example, if EBS004, FQ-A001, and FQ-A002 grow significantly faster than ES114, then these three strains could competitively exclude ES114 after only 15 h. To test this hypothesis, we quantified the growth rates of ES114 and the competitive strains (EBS004, FQ-A001, and FQ-A002) by measuring the CFUs of each strain when grown under the same conditions used in the coincubation assays (SI Appendix, Fig. S1A). The doubling time of ES114 (39 ± 5.8 min per generation) was not statistically significantly different (Student’s t test, P > 0.05) from those of the three competitive strains: EBS004 (36 ± 4.7 min per generation), FQ-A001 (43 ± 7.6 min per generation ), and FQ-A002 (43 ± 10 min per generation ), suggesting their ability to outcompete ES114 in our coincubation assay is not due to differences in growth rate.
To determine whether the competitive strains outcompete ES114 through growth inhibition or physical elimination of ES114 cells, we measured the change in ES114 abundance in coincubations with our V. fischeri isolates using two methods: measuring the total CFUs of ES114 remaining after 5 h and direct cell counts by flow cytometry. When ES114 was coincubated with ABM004, EMG003, or FQ-A003, the CFUs of all strains increased after 5 h (Fig. 1B) and were uniformly mixed in colonies after 15 h (Fig. 1A), suggesting these strains were able to coexist. However, when ES114 was coincubated with the three competitive strains, CFU counts for all three competitor strains increased after 5 h, but ES114 CFU counts decreased by ∼3 logs (Fig. 1B). The decrease in ES114 CFUs was confirmed by direct cell counts of GFP-tagged ES114 using a flow cytometer, which showed that >90% of ES114 cells are eliminated after a 5-h coincubation with competitive strains (SI Appendix, Fig. S1B).
To determine whether the ability of competitive strains to eliminate ES114 is dependent on direct cell–cell contact, or is mediated by a diffusible antimicrobial compound, we performed coincubations in which a 0.22-µm filter was placed between ES114 and the coincubated strain. This filter allows for diffusion of antimicrobial molecules, but prevents direct cell–cell contact between strains. With the filter, ES114 was able to grow and comprised at least 50% of the cells after 5 h (SI Appendix, Fig. S1C), suggesting that the filter protected ES114 cells from being killed by the competitive strains. As a positive control for a diffusible antimicrobial, an antibiotic was used in a parallel experiment. In this case, ES114 was killed (SI Appendix, Fig. S1D), showing that the filter did not protect ES114 from killing by a diffusible compound. Taken together, these findings indicate that some V. fischeri strains are capable of contact-dependent killing of ES114. We term isolates with the ability to kill ES114 as “lethal” strains and isolates that did not inhibit the growth of ES114 as “nonlethal.”
The Ability to Kill Is Independent of Strain Phylogeny.
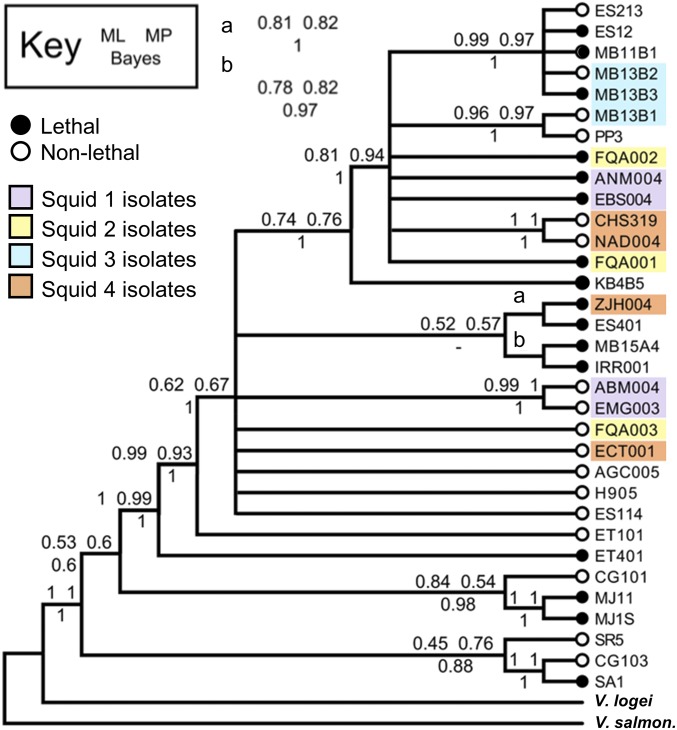
Given that our initial assays included only six light-organ isolates, we wondered how prevalent lethal and nonlethal strains are among V. fischeri isolates and if there was any correlation between phylogeny and killing ability. To determine the relative abundance of lethal and nonlethal strains, we repeated the coincubation assays with ES114 and 32 other differentially tagged V. fischeri strains, isolated primarily from light-organ symbioses, and tested whether they could prevent growth of ES114 in a manner similar to what was observed for lethal strains in Fig. 1A. Because some of these strains have slower growth rates than ES114, we modified the assay so that the ES114 target was outnumbered 5:1 by the coincubating strain. Mixed colonies were imaged for fluorescence after ∼24 h to score for the presence or absence of ES114 (SI Appendix, Fig. S2), which would indicate whether the coincubated strain is nonlethal or lethal, respectively, as defined above. The presence of the coincubated strain was also confirmed by fluorescence (SI Appendix, Fig. S2). This lethal or nonlethal behavior was then mapped to a consensus phylogenetic tree built using four concatenated housekeeping genes (60). Of the 32 V. fischeri strains tested, 16 isolates are lethal (little to no ES114 observed) and 16 isolates are nonlethal (ES114 observed) (SI Appendix, Fig. S2). Moreover, the ability to kill was not correlated with phylogeny: lethal strains (black circles in Fig. 2) are found throughout the tree and do not comprise a monophyletic group, suggesting the killing mechanism is not a trait shared only among closely related strains. Finally, when we considered strains coisolated from four different E. scolopes light organs (purple, yellow, blue, and orange in Fig. 2), each set of coisolates consisted of both lethal and nonlethal strains. Taken together, these findings yielded three important observations: (i) both lethal and nonlethal strains are prevalent, (ii) killing does not correlate with phylogeny, and (iii) strains coisolated from the same light organ are a mix of lethal and nonlethal strains.
Fig. 2.
Prevalence and phylogeny of lethal vs. nonlethal strains. Consensus phylogenic tree constructed using four concatenated housekeeping genes (recA, mdh, katA, pyrC) of 35 Vibrio isolates; open circles indicate nonlethal strains and black circles indicate lethal strains; colored boxes indicate strains isolated from the same light organ. Node values were calculated by maximum likelihood (ML) and maximum parsimony (MP) bootstrap values or Bayesian (Bayes) posterior probability.
Whole-Genome Comparison Reveals a Correlation Between Killing and T6SS2.
To identify the genetic determinants of interstrain killing, we performed comparative genomics using the complete genomes of two isolates: a nonlethal strain (ES114) and a lethal strain (MJ11) (64–66). We searched the MJ11 genome for genes encoding putative contact-dependent killing mechanisms that are not present in the ES114 genome and found strain-specific differences in T6SS genes.
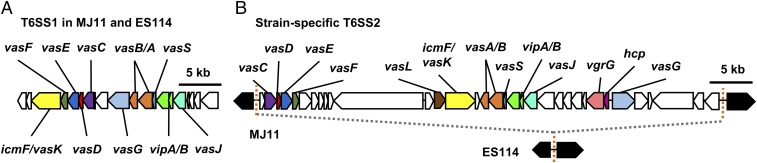
Like most vibrios, V. fischeri has two chromosomes, and although both of the genomes examined encode a predicted T6SS on chromosome I (T6SS1), strain MJ11 encodes a second predicted T6SS on chromosome II (T6SS2) that is absent in ES114 (Fig. 3 and SI Appendix, Fig. S3). T6SS1 and T6SS2 gene clusters show very different genetic organization and essential T6SS structural proteins share low levels of homology: IcmF_1 and IcmF_2 are 24% identical, and VasA_1 and VasA_2 are 34% identical (Fig. 3 and SI Appendix, Table S1). Moreover, when the presence of IcmF_1 and IcmF_2 homologs was mapped onto a phylogenetic tree consisting of multiple Vibrio species, IcmF_1 homologs were restricted to a monophyletic group consisting of V. fischeri isolates and closely related species, while IcmF_2 homologs were found throughout the tree (SI Appendix, Fig. S4). Similar to other organisms encoding multiple T6SSs (67–69), these findings suggest the two T6S systems in V. fischeri are likely not the result of a recent duplication event.
Fig. 3.
T6SS2 is located on a strain-specific genomic island. T6SSs located on (A) chromosome I and (B) chromosome II of V. fischeri strains: MJ11 (lethal), ES114 (nonlethal). Both MJ11 and ES114 encode putative T6SS genes on chromosome I (T6SS1). MJ11 carries a 50-kb gene cluster encoding predicted T6SS genes on chromosome II (T6SS2) that is lacking in ES114 (B). Conserved T6SS genes are the same color in T6SS1 and T6SS2 and genes of unknown function are indicated in white. Orange dashed lines indicate a 16-bp sequence (GTTTAAAAAAGCAG/CAA) flanking the MJ11 genomic island and corresponding sequence location in ES114. Black arrows indicate conserved genes flanking the MJ11 genomic island.
The MJ11 T6SS2 encodes all necessary components for a functional secretion apparatus, as well as three auxiliary gene clusters encoding putative effector/immunity genes (Fig. 3B and SI Appendix, Tables S1 and S3). Further examination of the T6SS2-encoding gene cluster in the MJ11 genome revealed two hallmarks of genomic islands: the 50-kb region is located next to a tRNA gene (SI Appendix, Table S1) and is flanked by 16-bp direct repeat sequences (Fig. 3B) (70). ES114 contains a single copy of the 16-bp repeat sequence between the conserved flanking genes (Fig. 3B), and also encodes two of the three auxiliary gene clusters found in MJ11 (SI Appendix, Table S3). Moreover, the T6SS2 gene cluster appears to be broadly conserved among other Vibrio species that have been isolated from marine and human hosts (SI Appendix, Fig. S4 and Table S2). Taken together, these findings suggest strain MJ11 contains genomic features that may facilitate horizontal transfer and/or loss of the genomic island. Therefore, we were interested in assaying additional strains for presence or absence of the island.
To determine whether other V. fischeri strains harbor this genomic island, we performed PCR using primers specific to two essential T6SS2 genes, icmF_2 and vasA_2 (27), as well as the left junction of this genomic island. We obtained products for icmF_2, vasA_2, and the left junction for all three lethal strains (EBS004, FQ-A001, and FQ-A002), as well as MJ11, which served as a positive control (SI Appendix, Fig. S5A). In contrast, products from the three nonlethal strains (ABM004, EMG003, and FQ-A003), as well as ES114, were only observed when using primers for the recA control (SI Appendix, Fig. S5A), suggesting the T6SS2-encoding genomic island is not present in these strains. When we screened the remaining isolates from Fig. 2 using the left junction primers, we did not observe PCR products for most of the nonlethal strains (Fig. 2, white circles, and SI Appendix, Fig. S5B); however, we did observe left junction PCR products for a few nonlethal strains (ES213, MB13B2, ANM004, H905), suggesting some strains may encode the genomic island but their T6SS2 is not functional. We observed PCR products for the left junction for all lethal strains (SI Appendix, Fig. S5B), suggesting the presence of the genomic island in these strains. Moreover, we observed products using primers specific to recA and vasA_1 for all strains (SI Appendix, Fig. S5). Taken together, these results indicate that although all strains appear to encode T6SS1, the ability to kill is correlated with the presence of genes associated with the T6SS2-encoding genomic island.
Killing Is Dependent on T6SS2 Function.
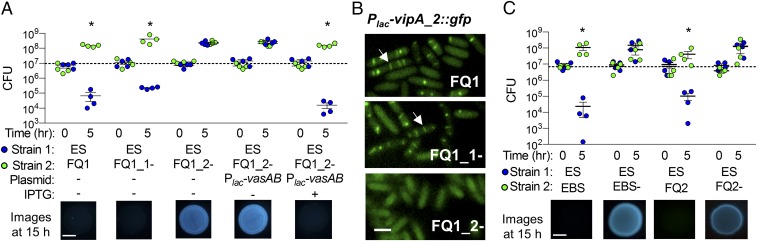
To directly test if either T6SS is required for interstrain killing, we introduced a disruption mutation into the vasA_1 or vasA_2 gene in the lethal strain, FQ-A001. The vasA gene is predicted to encode a homolog of TssF, which is an essential inner membrane protein of the structural machinery of T6SSs that is required for sheath assembly in V. cholerae and Serratia marcescens; disruption of this gene has previously been shown to eliminate T6SS function in these bacteria (71, 72). We next conducted coincubation assays with ES114 and the wild-type, vasA_1, or vasA_2 mutant strains of FQ-A001. When incubated with either wild-type FQ-A001 or the vasA_1 mutant, ES114 was killed by 5 h and not visibly detected at 15 h (Fig. 4A). In contrast, when incubated with the vasA_2 mutant, ES114 was able to grow after 5 h and was visibly detectable in the mixed colony after 15 h (Fig. 4A). When vasA_2 was complemented in trans using an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression vector, killing was restored in an IPTG-dependent manner (Fig. 4A). Together, these results suggest that FQ-A001 uses T6SS2 to kill ES114.
Fig. 4.
T6SS2 is necessary for killing. (A) CFU counts for each coincubation spot for coincubations of ES114 with FQ1 wild-type, vasA_1 mutant, vasA_2 mutant, vasA_2 mutant with IPTG-inducible vasAB_2 complement plasmid, and (C) ES114 with FQ2 and EBS wild-type and vasA_2 mutant strains and corresponding fluorescence microscopy images of ES114 at 15 h. (Scale bars, 2 mm.) All experiments were performed at least three times and a representative experiment (n = 4) is shown. Asterisks indicate P < 0.05 (Student’s t test) indicating a statistically significant decrease in ES114 CFUs at 5 h compared with 0 h for each coincubation. (B) Fluorescence microscopy images of FQ-A001 strains (wild-type, vasA_1 mutant, and vasA_2 mutant) with IPTG-inducible vipA_2::gfp fusion plasmid were taken after 2 h on LBS agar pads supplemented with 0.5 mM IPTG. White arrows indicate T6SS2 sheaths. (Scale bar, 1 µm.)
To determine whether the T6SS2 genes encode the proteins required for constructing a T6SS sheath, we next constructed a VipA_2-GFP expression vector to visualize possible T6SS sheaths in wild-type and mutant FQ-A001 strains. Previous work has shown that tagging the VipA subunit of a T6SS sheath allows for direct visualization of sheath assembly in V. cholerae (73, 74). We moved the VipA_2-GFP expression vector into wild-type, vasA_1, and vasA_2 mutants of FQ-A001 and used single-cell fluorescence microscopy to visualize sheath assembly. We observed GFP-tagged sheaths for the VipA_2 fusion in the wild-type and vasA_1 mutant, but not in the vasA_2 mutant (Fig. 4B), suggesting that T6SS2 encodes the necessary components to assemble a sheath, which is dependent on the baseplate component VasA_2.
To determine whether T6SS2 is required for killing in the other coisolated lethal strains, we made vasA_2 mutations in strains FQ-A002 and EBS004. Disruption of vasA_2 also abrogated killing of ES114 in these strains (Fig. 4C). Moreover, the vasA_2 mutant strains were able to coexist with their wild-type parent, suggesting the vasA_2 mutant is immune to the parental strain’s T6SS2-delivered effectors (SI Appendix, Fig. S1E). These findings suggest that interstrain killing among light-organ isolates is mediated by the T6SS on chromosome II.
Coisolated Strains Are Incompatible.
The findings from our coincubation assays with ES114 indicate lethal strains can eliminate a nonlethal strain; however, both lethal and nonlethal strains are commonly isolated from the same light organ (Fig. 2). Given this finding, we wondered how coisolated strains might be able to coinhabit the same host. Because light-organ colonization can only occur within a window of time after the animal hatches (75), we considered two alternative hypotheses: (i) the natural host colonization process selects for compatible strains that are immune to each other’s T6SS2 effectors or (ii) coisolated strains are not compatible and their ability to coinhabit the same light organ is dependent on physical separation within the host.
To test whether naturally coisolated strains are compatible, we performed pairwise coincubation assays using both sets of squid isolates. For squid 1 isolates, we found the lethal strain EBS004 kills both nonlethal coisolates (ABM004 and EMG003) in a T6SS2-dependent manner after 5 h, resulting in no visibly detectable nonlethal strain after 24 h (Fig. 5A). When the two nonlethal isolates were coincubated, each strain was able to grow after 5 h, resulting in a well-mixed colony after 24 h (Fig. 5A). Similar results were obtained for squid 2 isolates: both lethal strains (FQ-A001 and FQ-A002) eliminate the nonlethal strain (FQ-A003) in a T6SS2-dependent manner (Fig. 5B). These results suggest that nonlethal strains lack the appropriate immunity genes required to coexist with their coisolated lethal strains.
Fig. 5.
Coisolated strains are not compatible. CFU counts for each coincubation spot for pairwise coincubations with squid 1 (A) and squid 2 (B and C) isolates. Asterisks indicate P < 0.02 (Student’s t test) indicating a statistically significant decrease in a strain’s CFUs at 5 h compared with 0 h for each coincubation. A dash (“—”) indicates vasA_2 mutants. Fluorescence microscopy images were taken after 24 h. (Scale bars, 2 mm.) All experiments were performed at least three times and a representative experiment is shown (n = 4).
When the two lethal strains from squid 2 were coincubated, they maintained a 1:1 ratio and did not increase in CFUs after 5 h, and they were both observed in the mixed colony after 24 h (Fig. 5C). When the FQ-A001 vasA_2 mutant was coincubated with wild-type FQ-A002, or if wild-type FQ-A001 was coincubated with the FQ-A002 vasA_2 mutant, the wild-type strain always eliminated the vasA_2 mutant (Fig. 5C). However, when the two vasA_2 mutants were coincubated, these strains maintained a 1:1 ratio and increased in CFUs after 5 h and were both visibly detectable after 24 h (Fig. 5C), consistent with what is observed when coincubating nonlethal strains (Fig. 5A). These results suggest that FQ-A001 and FQ-A002 encode unique T6SS2-exported effectors that allow for T6SS2-mediated elimination of the other strain when it lacks a functioning T6SS2 for defense. Moreover, these data suggest that when coincubating strains are incompatible lethal strains, they actively kill one another, which can initially restrict their growth (Fig. 5C).
Strain Compatibility Is only Partly Predicted by T6SS Toxin Genotype.
Next, we expanded our search for compatible strains by examining the predicted toxin/immunity pairs encoded in our strain collection for isolates with available draft genomes. Because MJ11 is the only lethal V. fischeri strain with a completely sequenced genome, we searched draft genomes of strains ES213, MB11B1, MB13B2, MB13B3, MB13B1, KB4B5, MB15A4, ES114, and SR5 for homologs of the putative T6SS toxins found in the MJ11 genome (SI Appendix, Fig. S6). Previous work in V. cholerae indicates that strain compatibility can be inferred based on predicted T6SS toxin/immunity genes: strains that share the same toxin/immunity genes are compatible and those that have different toxin repertoires are not compatible (47). Moreover, recent work has shown that the C-terminal toxin/immunity genes can diversify via allelic exchange, resulting in varying alleles of toxins among closely related strains (76–78).
The MJ11 genome encodes putative T6SS toxins in the primary T6SS2 gene cluster and has one predicted toxin in each of three auxiliary gene clusters (Fig. 3B and SI Appendix, Table S3). We compared the sequences for these toxins across the available 10 draft genomes and found two alleles for auxiliary toxin 1 (A1), two alleles for auxiliary toxin 2 (A2), three alleles for auxiliary toxin 3 (A3), and one allele for the predicted toxin associated with the primary T6SS2 gene cluster (P) (SI Appendix, Fig. S6A). In addition to diverse toxin alleles, some strains were missing certain gene clusters altogether. Based on the toxin alleles we were able to divide these 10 strains into 6 predicted compatibility groups (SI Appendix, Fig. S6B). Only two groups contained more than one strain: group 1 (ES213, MB11B1, MB13B2, and MB13B3) and group 3 (KB4B5 and MB15A4).
We next performed pairwise coincubation assays to test whether any strains within these groups were in fact compatible. For group 1, we found that only two sets of strains were able to coexist in our assays: MB13B2 and ES213 (both nonlethal strains) and MB11B1 and MB13B3 (both lethal strains) (SI Appendix, Fig. S6C). Although strains MB11B1 and MB13B3 can kill other group 1 nonlethal strains ES213 and MB13B2, they were able to coexist with one another even if the other strain was lacking a functional T6SS2, suggesting both MB11B1 and MB13B3 contain the same toxin/immunity genes and are indeed compatible (SI Appendix, Fig. S6C). This result is perhaps not surprising given that these two strains are closely related, according to our phylogenetic tree (Fig. 2). For group 3 strains, both MB15A4 and KB4B5 are lethal; however, when the two are coincubated, MB15A4 eliminates KB4B5, indicating these strains are not compatible. Taken together, these results suggest that predicted T6SS toxin genotypes can only partly predict strain compatibility among V. fischeri strains, and strains appear to diversify their toxins rapidly and are largely incompatible, except for nonlethal strains.
Lethal Strains Spatially Separate in a T6SS2-Dependent Manner.
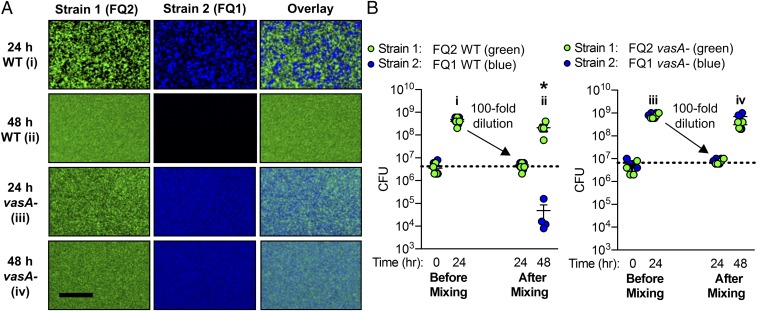
Our initial characterization of strain compatibility indicates that although nonlethal strains can coexist, competitive outcomes of two lethal strains include: (i) one strain killing the other (MB15A4 vs. KB4B5), (ii) stable coexistence of compatible killers (MB11B1 and MB13B3), or (iii) coexistence by mutual killing (FQ-A001 vs. FQ-A002). Because our initial coincubations with FQ-A001 and FQ-A002 resulted in coexistence that was T6SS2-dependent, we more closely examined the mixed colonies after 24 h. Interestingly, we found these lethal strains coexisted as spatially separated microcolonies that were T6SS2-dependent (Fig. 6 A, i and iii). Although this mechanism of strain separation has been observed by others (31, 79, 80), it is not yet known if this spatial segregation is stable or if it can be destabilized by ecologically relevant conditions.
Fig. 6.
Spatial separation of lethal strains is not stable in the presence of physical mixing. (A) Fluorescence microscopy images for pairwise coincubations with FQ1 and FQ2 wild-type (WT) and vasA_2 mutant (vasA-) strains taken at 24 and 48 h (A); an overlay of images from columns 1 and 2 is shown in column 3; image will be light blue when strain 1 (green) and strain 2 (blue) cells are present in the same pixel. (Scale bar, 25 µm.) (B) Corresponding CFU counts for each coincubation spot. Coincubations were set up in the standard method (“before mixing”) and at 24 h were resuspended, mixed, diluted 100-fold, and spotted onto fresh LBS plates for the remainder of the assay (“after mixing”). Asterisk indicates P < 0.01 (Student’s t test) indicating a statistically significant decrease in a strain’s CFUs at 48 h compared with 24 h for each coincubation. All experiments were performed at least three times and a representative experiment is shown (n = 4).
We next considered whether physical disruption impacts the spatial separation of incompatible lethal strains observed in vitro. Such physical disruption is expected to occur within the light organ, which experiences daily venting of the crypt spaces: a light cue at dawn stimulates the light organ to contract and forcefully vent ∼90–95% of the crypt contents through the pores (81, 82). In addition to diluting the bacterial cell numbers, contraction of the light organ to force out its contents could also act to destabilize the spatial structure of symbiotic cells. We wondered whether incompatible lethal strains could continue to coexist as spatially separated microcolonies in our in vitro assay if physical disruption was introduced. We predicted two possible outcomes: (i) both lethal strains would be present after disruption and would again be spatially separated in a T6SS2-dependent manner, or (ii) disruption of the initial spatial separation of strains would allow one lethal strain to dominate and eliminate the other.
To test these predictions, we coincubated the two lethal strains FQ-A001 and FQ-A002 and physically disrupted the cells after the initial coincubation. After the initial 24-h coincubation, both strains grew equally and remained at a 1:1 ratio (Fig. 6B) and spatially separated in a T6SS2-dependent manner (Fig. 6A). We then resuspended these coincubation spots in 1 mL broth, physically mixed the cells by pipetting up and down, and spotted 10 µL of this cell suspension onto agar plates. When cells were spotted after this mixing step, both strains were present in a 1:1 ratio, yet after an additional 24-h incubation, FQ-A002 was consistently able to eliminate FQ-A001 while the two T6SS2 mutant strains grew equally well and were spatially well-mixed (Fig. 6). Taken together, these findings suggest that lethal coisolated strains do not intermix under dynamic conditions: although T6SS2-mediated killing can initially spatially separate competing strains, this coexistence is not stable in the presence of physical disruption.
T6SS2 Is Necessary for Strain Incompatibility Within the Host.
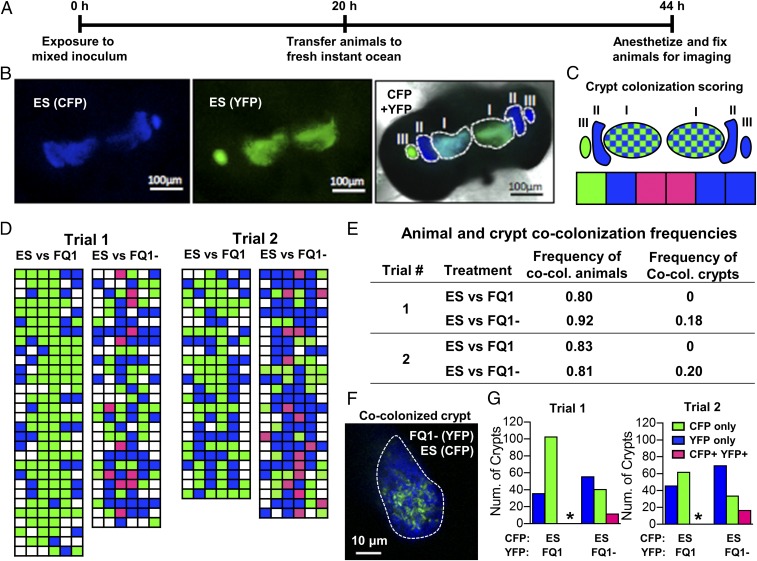
Our in vitro assay indicates that T6SS2 mediates strain incompatibility among light-organ isolates. Specifically, lethal strains (FQ-A001) are able to kill nonlethal strains (ES114) in a T6SS2-dependent manner. Moreover, previous work has shown that squid exposed to an inoculum of FQ-A001 and ES114 do not exhibit crypt spaces cocolonized with both strain types (61). To investigate the impact of T6SS2 on the intercellular interactions that V. fischeri cells experience in vivo, we turned to the colonization model involving the light organ of E. scolopes. The nascent light organ features six crypt spaces that are each independently colonized by V. fischeri cells in direct contact with each other (58). Each crypt space has a physical bottleneck that permits entry of only one to two cells at a time, which proliferate into the resulting symbiotic population (53). By labeling two strains with different fluorescent proteins, it is possible to use confocal fluorescence microscopy to identify the strain types of each colonized crypt (Fig. 7 A–C) (61, 83). For example, in the light organ shown in Fig. 7B, which was exposed to an inoculum mixed evenly with ES114 cells labeled with either YFP or CFP, there are three crypts colonized with only CFP (CFP+ YFP−), one crypt with only YFP (CFP− YFP+), and two crypts cocolonized with both strain types (CFP+ YFP+) (Fig. 7C). Thus, imaging light organs in this way provides insight into the strain types that initially colonized the crypt spaces.
Fig. 7.
Impact of T6SS2 on crypt cocolonization. (A) Timeline for cocolonization experiment. Animals were exposed to one of two different inocula that compete the CFP-tagged ES114 strain against the YFP-tagged FQ-A001 wild-type (FQ1) or vasA_2 mutant (FQ1−). Animals were exposed to the inoculum for ∼22 h, and animals were fixed for imaging ∼44 h after the initial inoculum exposure. (B) Image of the light organ of an animal exposed to ES114 differentially expressing CFP or YFP: (Left) CFP+ YFP− crypts; (Center) CFP−YFP+ crypts; overlay of CFP, YFP, and bright-field images of light organ. (C) Map of crypt colonization counts and how images are scored. Crypt colonizations are labeled according to strain type: singly colonized, either YFP (green) or CFP (blue) or cocolonized, YFP+ CFP+ (checkered/pink). (D) Individual animal crypt colonization scores for indicated competitions between CFP-tagged ES114 (ES) and YFP-tagged FQ-A001 wild-type (FQ1) or vasA_2 mutant (FQ1−); each row represents an animal and each box represents a crypt. Blue boxes represent crypts that were scored as CFP+ YFP−. Green boxes represent crypts that were scored as CFP− YFP+. Pink boxes represent crypts that were scored as CFP+ YFP+. (E) Table summarizing animal and crypt cocolonization frequencies for each trial of each treatment. (F) High-magnification, confocal image of an individual crypt cocolonized with ES114 and FQ1−. (G) Number of crypts that were scored as CFP+ YFP−, CFP− YFP+, and CFP+ YFP+ for competitions between ES114 and FQ1 or between ES114 and the FQ1 vasA_2 mutant. Data from two different trials are shown for each competition. The proportion of CFP+ YFP+ crypts between different types of competitions were compared using a two-proportion z-test and an asterisk indicates P < 0.001. The inoculum levels and YFP/CFP ratios for trial 1 were 7,760 CFU/mL and 1.15, respectively, for ES vs. FQ1, and 16,160 CFU/mL and 1.04, respectively, for ES vs. FQ1−. The inoculum levels and YFP/CFP ratios for trial 2 were 6,020 CFU/mL and 0.87, respectively, for ES vs. FQ1, and 8,400 CFU/mL and 1.14 for ES vs. FQ1−.
To determine the frequency in which a light organ becomes colonized by otherwise isogenic but differentially tagged strains, we conducted a series of colonization assays using inocula mixed evenly with either YFP- and CFP-labeled ES114, or with YFP- and CFP-labeled FQ-A001, with the total inoculum sizes varied among trials. For total inoculum sizes ranging from 3,560–32,560 CFU/mL, the majority of animals (>50%) for both strain types were cocolonized (SI Appendix, Fig. S7A), suggesting that differentially tagged strains can access one or more crypts of a given light organ. When the frequency of cocolonized crypts was calculated for these animals, we found that 3–70% of crypts were cocolonized for any given trial (SI Appendix, Fig. S7B). Importantly, the frequency of observed cocolonized animals or cocolonized crypts did not appear to change as a function of total inoculum size (SI Appendix, Fig. S7). Finally, using a two-proportion z-test, we were able to show that statistically significant differences in cocolonization frequencies can be detected between treatments using this assay (SI Appendix, Fig. S7C). Taken together, these results suggest that the colonization of crypt spaces by more than one cell occurs at a frequency that is detectable during squid colonization assays involving differentially tagged ES114 or FQ-A001 inoculums, and differences in crypt cocolonization frequencies between treatments can be statistically validated.
Previously, we reported that squid exposed to an inoculum mixed with ES114 and FQ-A001 do not exhibit crypt spaces cocolonized with both strains (61). Our results from the culture-based coincubation assay described above suggest that T6SS2 mediates the killing of ES114 by FQ-A001 in vitro. To test the hypothesis that T6SS2 prevents the establishment of cocolonized crypts mixed with FQ-A001 and ES114 in the light organ, we conducted squid colonization assays with CFP-labeled ES114 and either YFP-labeled wild-type FQ-A001 or the vasA_2 mutant, and scored each animal for light organ and crypt colonization frequencies of each strain after 44-h postinoculation (Fig. 7D). The frequency of animals that were cocolonized by ES114 and wild-type FQ-A001 ranged between 80% and 83%, which was comparable to that observed for animal groups exposed to ES114 and the FQ-A001 vasA_2 mutant (81% and 92% in each trial, two-proportion z-test, α = 0.05) (Fig. 7E and SI Appendix, Table S4). These results suggest that a functional T6SS2 in FQ-A001 does not impact the frequency of an animal to become colonized with incompatible strains or access host colonization sites. However, when the frequency of cocolonization of individual crypts was scored, 18–20% of the colonized crypts in animals exposed to the inoculum mixed with FQ-A001 vasA_2- and ES114 were cocolonized (CFP+ YFP+) (Fig. 7E). In these cocolonized crypts, the cells of each strain type were mixed throughout the crypt space (Fig. 7F), showing that ES114 and the FQ-A001–derived cells are in direct contact within cocolonized crypts. In contrast, none of the colonized crypts resulting from animals exposed to wild-type FQ-A001 and ES114 were CFP+ YFP+ (Fig. 7 E and G), which was consistent with our previous report (61). This effect of T6SS2 on crypt cocolonization frequencies was statistically significant according to a two-proportion z-test (SI Appendix, Table S5). To ensure that our approach was sensitive enough to detect significant differences in crypt cocolonization frequencies, we performed a power analysis that estimated the effect size to be greater than 0.37 (two-proportion z-test, α = 0.05, power = 0.80) (SI Appendix, Table S5), suggesting our crypt colonization assay sampled a sufficient number of colonization sites (n > 107 per treatment, per trial) to statistically support the conclusion that VasA_2, and thus T6SS2, prevents the establishment of crypts cocolonized with FQ-A001 and ES114. Taken together, these data suggest that T6SS2 promotes the separation of incompatible strains within the natural host.
Discussion
This work demonstrates that the Vibrio–squid symbiosis is a valuable system for investigating intraspecific bacterial interactions and dynamics as well as their impact on host colonization. Our evidence suggests these interactions are relevant in a natural host, where they are capable of shaping the spatial distributions of host-associated microbial populations. This study shows that the T6SS prevents coexistence of strains in the context of a natural, beneficial bacteria–host interaction. Moreover, these interactions can be recapitulated using culture-based assays, which provide a powerful system to probe molecular mechanisms of interbacterial competition and their ecological and evolutionary roles.
Based on our results, we propose a model for the role of T6SS2 during host colonization. Juvenile squid hatch without their bacterial symbionts, and their crypts initially are colonized by one to two cells of different genotypes. During their growth, the initial populations within cocolonized crypt spaces come into contact with each other. In this context, our data suggest that T6SS2+ strains begin to kill competitor cells in a contact-dependent manner. By 44-h postinoculation, T6SS2-deficient cells within these initially cocolonized crypts have been eliminated, resulting in crypts colonized exclusively by the T6SS2+ genotype.
Further investigation should examine the details of when and where competitors use T6SS2 during the first 44 h of host colonization, as well as the role of the host in influencing these interactions. Although this work did not directly test the impact of venting on T6SS2-mediated competition within crypts, our in vitro data suggest that physical mixing and dilution of competing populations promotes exclusion of a less-competitive lethal strain (Fig. 6), and therefore warrants further study.
Because this work focused on T6SS2, the functional role of T6SS1 remains unclear. Although we did not observe vasA_1-dependent killing of ES114 (Fig. 4A), T6SS1 may be active under different conditions compared with T6SS2. Indeed, other Vibrio species show T6SS activity that is conditionally regulated (13, 33, 84), and future work will need to explore how free-living versus host-like conditions modulate V. fischeri T6SS gene expression and function. Alternatively, T6SS1 may be used to interact with eukaryotic cells (15, 28, 72). In a previous transposon sequencing study during squid colonization, the genes in the predicted T6SS1 locus contained abundant transposon insertions (i.e., are not essential); however, a role for T6SS1 in host colonization was not apparent from that work (85).
Our observations also raise many questions from an evolutionary perspective. The distribution of killing activity and the T6SS2-encoding genomic island among extant V. fischeri does not follow the pattern of a shared, derived trait in this clade (Fig. 2). Future work may determine how and why such a powerful trait as killing exhibits a random distribution among sampled isolates’ phylogeny. For example, further research may demonstrate whether there are trade-offs associated with carrying the T6SS2-encoding genomic island, and if its presence influences survival in both the host and greater marine environments.
One model for the evolutionary dynamics of the T6SS2-containing island is that it is frequently transferred among strains by horizontal gene transfer, and the noted hallmarks of flanking repeats and tRNA locus support such a model. Given the presence of the V. fischeri T6SS2 homologs in closely related Vibrio spp. (SI Appendix, Fig. S4), an alternate model may be that the locus was present in a common ancestor of V. fischeri and closely related Vibrios, and has since been lost among V. fischeri strains in environments where interbacterial killing is no longer advantageous. Additional work is required to evaluate these options, and pursuing these ideas may provide evidence as to how selective pressure on the T6SS influences V. fischeri evolution.
If V. fischeri strains use T6SS2 to eliminate competitor populations when the symbiosis is first established, it is possible that once a crypt becomes clonally colonized, the T6SS2 no longer provides the population with an advantage. Moreover, maintaining a 50-kb genomic island could be a fitness cost to the cell, in addition to the energetic costs associated with synthesizing and using the T6SS apparatus (86). Given the presence of direct repeat sequences flanking the genomic island, it is possible that the island could be lost by excision through homologous recombination, leaving behind a single copy of this sequence as observed in strain ES114 (Fig. 3B). Because a functional T6SS is not needed for immunity to T6SS-exported effectors of isogenic strains (SI Appendix, Fig. S1E), which are encoded in auxiliary gene clusters elsewhere on the chromosome (SI Appendix, Table S3), loss of T6SS2 would not be detrimental to the cell’s survival. Such a scenario could explain the consistent mix of lethal and nonlethal strains isolated among naturally cooccurring light-organ symbionts, as well as the presence of auxiliary gene clusters in nonlethal strains lacking the T6SS2-encoding genomic island (ES114, MB13B1, SR5) (SI Appendix, Fig. S6). Further research may reveal more details underlying the mechanism and selective pressures that drive the strain-specific nature of T6SSs among closely related isolates.
Interestingly, a transition from T6SS+ strains to T6SS− strains among beneficial symbionts within a host is not without precedent. Recent work exploring the occurrence of T6SSs among Bacteroides fragilis strains in the guts of human infants and adults suggests one particular T6SS may provide B. fragilis with a competitive advantage during initial colonization but is less prominent in adult microbiomes, once colonization is established (87). Although the mammalian gut and squid light organ represent distinct host habitats, these organs share commonalities in how they become colonized by their bacterial symbionts, which face similar physical conditions, including epithelium-lined crypts, as well as physical disruption due to hosts’ daily rhythms. Moreover, Vibrios are uniquely adapted to both colonizing marine hosts and causing disease in humans by way of the gut (88). Given that homologs of the V. fischeri T6SS2 are broadly distributed among other Vibrio species (SI Appendix, Fig. S4), we believe the Vibrio–squid model system will provide broad, comparative knowledge about how competitors of a host habitat use the T6SS to influence colonization outcomes and the spatial structure of host-associated communities.
Materials and Methods
The laboratory practices were carried out under the general principles described in the Guide for the Care and Use of Laboratory Animals (89) in communication with the Institutional Animal Care and Use Committee Office at Pennsylvania State University. See SI Appendix for additional experimental details, including media and growth conditions, isolation of symbiotic V. fischeri, strain and plasmid construction, contact-dependence assays, fluorescence microscopy, and phylogenetic analysis. Bacterial strains, plasmids, and oligonucleotides used in this study are in SI Appendix, Tables S6 and S7.
Coincubation Assays.
V. fischeri strains were grown overnight on LBS agar plates supplemented with the appropriate antibiotic at 24 °C. Cells were scraped from agar surfaces, resuspended in LBS medium, and diluted to an OD600 of 1.0. For each coincubation, strains were mixed in a 1:1 ratio and 10 µL of the mixture was spotted on LBS agar plates and incubated at 24 °C. After 5–24 h, each coincubation was resuspended in 1 mL LBS medium. Strains were quantified by plating serial dilutions onto LBS plates supplemented with antibiotics selective for each strain.
Fluorescence Microscopy.
For coincubation assays, fluorescence microscopy images were taken with a stereo microscope equipped with a Nightsea fluorescence adapter kit for green and red fluorescence detection. Images were taken using an OMAX 14MP camera with ToupView software and color changes were made by adjusting the HLS color module. No brightness or contrast adjustments were made. For high-magnification images of coincubations (Fig. 6A), spots were imaged using using an Olympus BX61 microscope outfitted with a Hammatsu ORCA RC camera and either a 4×/0.13 UPlanFLN or 10×/0.3 UPlanFLN objective lens. Images were captured using Improvision’s Velocity software. To visualize GFP-tagged T6SS2 sheath formation, strains carrying the IPTG-inducible vipA_2-gfp fusion expression vector were spotted onto a thin pad of 2% LBS agar with 0.5 mM IPTG and imaged using an Olympus BX51 microscope outfitted with a Hammatsu C8484-03G01 camera and a 100×/1.30 Oil Ph3 objective lens. Images were captured using MetaMorph software. Contrast on images was adjusted uniformly across images by subtracting background using ImageJ software.
Phylogenetic Analysis.
A multilocus phylogenetic analysis was performed using partial sequences of four loci: recA, mdh, katA, and pyrC. Published sequence data and newly amplified sequences of 35 total Vibrio isolates were collected, combined into a single concatenated sequence (ordered recA mdh katA pyrC – ∼2,880 nucleotides), and aligned with ClustalX 2.1 (90). The concatenated sequence alignment was analyzed by jModelTest 2.1 v20160303 (53) via three information criteria methods (Akaike, Bayesian, and Decision Theory). Construction of the majority-rule consensus tree and statistical analysis of clade membership/presence was assessed by sampling an “appropriately stationary” posterior probability distribution (91). Sequences associated with this analysis were submitted to the GenBank database and their accession numbers are listed in SI Appendix, Table S6.
Squid Colonization Assays.
For each treatment, 24–30 freshly hatched juvenile squid were exposed to the inoculum containing an even mix of YFP- and CFP-labeled strains at a final concentration ranging from 1,200–45,760 CFU/mL. Squid were exposed to this mixed inoculum for 20 h and then washed in fresh filter-sterilized seawater. After 44 h, animals were fixed, washed, and prepared for fluorescence microscopy by dissecting the ventral side of the mantle to reveal the light organ. YFP and CFP images were taken using a Zeiss 780 confocal microscope equipped with a 10× or 40× water lens. Each crypt space was scored separately for CFP and YFP fluorescence.
Supplementary Material
Acknowledgments
We thank Peggy Cotter, Anne Dunn, and Barbara MacGregor for helpful discussions; Ned Ruby and Eric Stabb for supplying previously isolated Vibrio fischeri strains and plasmids; and Scott Gifford and Andreas Teske for technical assistance. Emily Grandinette, Zack Houston, Eli LaSota, Aleia Mouchref, Andrew Murtha, Nadia Ortega, Imperio Real Ramirez, Emma Schwendeman, Caroline Steingard, and Elli Tatsumi isolated V. fischeri strains reported in this study as part of undergraduate research projects in the laboratory of T.M. A.N.S. was supported by the Gordon and Betty Moore Foundation through Grant GBMF 2550.03 to the Life Sciences Research Foundation. T.M. was supported by National Institutes of Health Grant R00GM097032. M.J.M. was supported by the National Institutes of Health Grants R35GM119627 and R21AI117262 and National Science Foundation Grant IOS-1757297.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. MF076793–MF076804, MF076806–MF076817, MF076819–MF076830, and MF076832–MF076843).
See Commentary on page 8855.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808302115/-/DCSupplemental.
References
- 1.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson JK, Murphree RL, Tamplin ML. Evidence that mortality from Vibrio vulnificus infection results from single strains among heterogeneous populations in shellfish. J Clin Microbiol. 1997;35:2098–2101. doi: 10.1128/jcm.35.8.2098-2101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keymer DP, Miller MC, Schoolnik GK, Boehm AB. Genomic and phenotypic diversity of coastal Vibrio cholerae strains is linked to environmental factors. Appl Environ Microbiol. 2007;73:3705–3714. doi: 10.1128/AEM.02736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kueh CSW, Kutarski P, Brunton M. Contaminated marine wounds—The risk of acquiring acute bacterial infection from marine recreational beaches. J Appl Bacteriol. 1992;73:412–420. doi: 10.1111/j.1365-2672.1992.tb04997.x. [DOI] [PubMed] [Google Scholar]
- 5.Nyholm SV, McFall-Ngai MJ. The winnowing: Establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 6.Dechet AM, Yu PA, Koram N, Painter J. Nonfoodborne Vibrio infections: An important cause of morbidity and mortality in the United States, 1997-2006. Clin Infect Dis. 2008;46:970–976. doi: 10.1086/529148. [DOI] [PubMed] [Google Scholar]
- 7.Del Gigia-Aguirre L, Sánchez-Yebra-Romera W, García-Muñoz S, Rodríguez-Maresca M. First description of wound infection with Vibrio harveyi in Spain. New Microbes New Infect. 2017;19:15–16. doi: 10.1016/j.nmni.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitz-Gibbon S, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomida S, et al. Pan-genome and comparative genome analyses of Propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. MBio. 2013;4:e00003-13. doi: 10.1128/mBio.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraune S, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altura MA, Stabb E, Goldman W, Apicella M, McFall-Ngai MJ. Attenuation of host NO production by MAMPs potentiates development of the host in the squid-vibrio symbiosis. Cell Microbiol. 2011;13:527–537. doi: 10.1111/j.1462-5822.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One. 2013;8:e61086. doi: 10.1371/journal.pone.0061086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sana TG, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci USA. 2016;113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwarz S, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. MBio. 2013;4:e00374-13. doi: 10.1128/mBio.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. MBio. 2016;7:e01055-16. doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Bayona L, Guo MS, Laub MT. Contact-dependent killing by Caulobacter crescentus via cell surface-associated, glycine zipper proteins. eLife. 2017;6:e24869. doi: 10.7554/eLife.24869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefanic P, Kraigher B, Lyons NA, Kolter R, Mandic-Mulec I. Kin discrimination between sympatric Bacillus subtilis isolates. Proc Natl Acad Sci USA. 2015;112:14042–14047. doi: 10.1073/pnas.1512671112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stubbendieck RM, Straight PD. Multifaceted interfaces of bacterial competition. J Bacteriol. 2016;198:2145–2155. doi: 10.1128/JB.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cornforth DM, Foster KR. Antibiotics and the art of bacterial war. Proc Natl Acad Sci USA. 2015;112:10827–10828. doi: 10.1073/pnas.1513608112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Microbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dey A, et al. Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. J Bacteriol. 2016;198:994–1004. doi: 10.1128/JB.00964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danka ES, Garcia EC, Cotter PA. Are CDI systems multicolored, facultative, helping greenbeards? Trends Microbiol. 2017;25:391–401. doi: 10.1016/j.tim.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willett JLE, Ruhe ZC, Goulding CW, Low DA, Hayes CS. Contact-dependent growth inhibition (CDI) and CdiB/CdiA two-partner secretion proteins. J Mol Biol. 2015;427:3754–3765. doi: 10.1016/j.jmb.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016;24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Joshi A, et al. Rules of engagement: The type VI secretion system in Vibrio cholerae. Trends Microbiol. 2017;25:267–279. doi: 10.1016/j.tim.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altindis E, Dong T, Catalano C, Mekalanos J. Secretome analysis of Vibrio cholerae type VI secretion system reveals a new effector-immunity pair. MBio. 2015;6:e00075. doi: 10.1128/mBio.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernardy EE, Turnsek MA, Wilson SK, Tarr CL, Hammer BK. Diversity of clinical and environmental isolates of Vibrio cholerae in natural transformation and contact-dependent bacterial killing indicative of type VI secretion system activity. Appl Environ Microbiol. 2016;82:2833–2842. doi: 10.1128/AEM.00351-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong M, et al. Microbial herd protection mediated by antagonistic interaction in polymicrobial communities. Appl Environ Microbiol. 2016;82:6881–6888. doi: 10.1128/AEM.02210-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vettiger A, Basler M. Type VI secretion system substrates are transferred and reused among sister cells. Cell. 2016;167:99–110.e12. doi: 10.1016/j.cell.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 33.Metzger LC, et al. Independent regulation of type VI secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep. 2016;15:951–958. doi: 10.1016/j.celrep.2016.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeRoux M, et al. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci USA. 2012;109:19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hachani A, Allsopp LP, Oduko Y, Filloux A. The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J Biol Chem. 2014;289:17872–17884. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alcoforado Diniz J, Coulthurst SJ. Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol. 2015;197:2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alteri CJ, et al. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 2013;9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carruthers MD, Nicholson PA, Tracy EN, Munson RS., Jr Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One. 2013;8:e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wexler AG, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majerczyk C, Schneider E, Greenberg EP. Quorum sensing control of type VI secretion factors restricts the proliferation of quorum-sensing mutants. eLife. 2016;5:e14712. doi: 10.7554/eLife.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernal P, Allsopp LP, Filloux A, Llamas MA. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017;11:972–987. doi: 10.1038/ismej.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014;16:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: The bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: Poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ringel PD, Hu D, Basler M. The role of type VI secretion system effectors in target cell lysis and subsequent horizontal gene transfer. Cell Rep. 2017;21:3927–3940. doi: 10.1016/j.celrep.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 47.Unterweger D, et al. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alcoforado Diniz J, Liu YC, Coulthurst SJ. Molecular weaponry: Diverse effectors delivered by the type VI secretion system. Cell Microbiol. 2015;17:1742–1751. doi: 10.1111/cmi.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang JY, Bullen NP, Ahmad S, Whitney JC. Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J Biol Chem. 2018;293:1504–1514. doi: 10.1074/jbc.RA117.000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao W, Caro F, Robins W, Mekalanos JJ. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science. 2018;359:210–213. doi: 10.1126/science.aap8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Logan SL, et al. The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc Natl Acad Sci USA. 2018;115:E3779–E3787. doi: 10.1073/pnas.1720133115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mandel MJ, Dunn AK. Impact and influence of the natural Vibrio-squid symbiosis in understanding bacterial-animal interactions. Front Microbiol. 2016;7:1982. doi: 10.3389/fmicb.2016.01982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee KH, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–1571. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruby EG, Lee KH. The Vibrio fischeri-Euprymna scolopes light organ association: Current ecological paradigms. Appl Environ Microbiol. 1998;64:805–812. doi: 10.1128/aem.64.3.805-812.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nyholm SV, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: The first site of symbiont specificity. Appl Environ Microbiol. 2003;69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol. 2006;62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sycuro LK, Ruby EG, McFall-Ngai M. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol. 2006;267:555–568. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 59.Bongrand C, et al. A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J. 2016;10:2907–2917. doi: 10.1038/ismej.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wollenberg MS, Ruby EG. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J. 2012;6:352–362. doi: 10.1038/ismej.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y, et al. Intraspecific competition impacts Vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl Environ Microbiol. 2016;82:3082–3091. doi: 10.1128/AEM.04143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 64.Mandel MJ, Stabb EV, Ruby EG. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: Focus on point miscalls and artefactual expansions. BMC Genomics. 2008;9:138. doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ruby EG, et al. Complete genome sequence of Vibrio fischeri: A symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci USA. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernal P, Llamas MA, Filloux A. Type VI secretion systems in plant-associated bacteria. Environ Microbiol. 2018;20:1–15. doi: 10.1111/1462-2920.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics. 2016;17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darmon E, Leach DRF. Bacterial genome instability. Microbiol Mol Biol Rev. 2014;78:1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.English G, Byron O, Cianfanelli FR, Prescott AR, Coulthurst SJ. Biochemical analysis of TssK, a core component of the bacterial type VI secretion system, reveals distinct oligomeric states of TssK and identifies a TssK-TssFG subcomplex. Biochem J. 2014;461:291–304. doi: 10.1042/BJ20131426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sana TG, et al. Internalization of Pseudomonas aeruginosa strain PAO1 into epithelial cells is promoted by interaction of a T6SS effector with the microtubule network. MBio. 2015;6:e00712. doi: 10.1128/mBio.00712-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 75.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol. 2014;23:1624–1634. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salomon D, et al. Type VI secretion system toxins horizontally shared between marine bacteria. PLoS Pathog. 2015;11:e1005128. doi: 10.1371/journal.ppat.1005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Unterweger D, et al. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 2015;34:2198–2210. doi: 10.15252/embj.201591163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirchberger PC, Unterweger D, Provenzano D, Pukatzki S, Boucher Y. Sequential displacement of type VI secretion system effector genes leads to evolution of diverse immunity gene arrays in Vibrio cholerae. Sci Rep. 2017;7:45133. doi: 10.1038/srep45133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McNally L, et al. Killing by type VI secretion drives genetic phase separation and correlates with increased cooperation. Nat Commun. 2017;8:14371. doi: 10.1038/ncomms14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borenstein DB, Ringel P, Basler M, Wingreen NS. Established microbial colonies can survive type VI secretion assault. PLOS Comput Biol. 2015;11:e1004520. doi: 10.1371/journal.pcbi.1004520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boettcher KJ, Ruby EG, McFall-Ngai MJ. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J Comp Physiol A. 1996;179:65–73. [Google Scholar]
- 82.Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: Description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- 83.Verma SC, Miyashiro T. Niche-specific impact of a symbiotic function on the persistence of microbial symbionts within a natural host. Appl Environ Microbiol. 2016;82:5990–5996. doi: 10.1128/AEM.01770-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang Y, et al. Functional characterization and conditional regulation of the type VI secretion system in Vibrio fluvialis. Front Microbiol. 2017;8:528. doi: 10.3389/fmicb.2017.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brooks JF, 2nd, et al. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci USA. 2014;111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Basler M. Type VI secretion system: Secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci. 2015;370:20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Verster AJ, et al. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe. 2017;22:411–419.e4. doi: 10.1016/j.chom.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson JR, Marcelino LA, Polz MF. Diversity, sources, and detection of human bacterial pathogens in the marine environment. In: Belkin S, Colwell RR, editors. Oceans and Health: Pathogens in the Marine Environment. Springer; New York: 2005. pp. 29–68. [Google Scholar]
- 89.National Research Council 2011. Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC), 8th Ed.
- 90.Larkin MA, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 91.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.