Significance
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor(SNARE) proteins drive membrane fusion by zippering into coiled-coil bundles between membrane bilayers. In this work, we showed that certain layers in the SNARE bundle are dispensable for SNARE-mediated membrane fusion. However, these layers are required for fusion reactions activated by the cognate Sec1/Munc18 (SM) protein or a synthetic Vc peptide derived from the vesicular SNARE. Strikingly, we identified a conserved SNARE-like peptide (SLP) in SM proteins that structurally and functionally resembles Vc peptide. Like Vc peptide, SLP binds and activates target SNAREs, stimulating the fusion reaction. These data suggest that the SM protein uses its SNARE-like sequence to promote the SNARE zippering pathway, ensuring the efficiency of vesicle fusion.
Keywords: SNARE, membrane fusion, vesicle fusion, exocytosis, SM protein
Abstract
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) catalyze membrane fusion by forming coiled-coil bundles between membrane bilayers. The SNARE bundle zippers progressively toward the membranes, pulling the lipid bilayers into close proximity to fuse. In this work, we found that the +1 and +2 layers in the C-terminal domains (CTDs) of SNAREs are dispensable for reconstituted SNARE-mediated fusion reactions. By contrast, all CTD layers are required for fusion reactions activated by the cognate Sec1/Munc18 (SM) protein or a synthetic Vc peptide derived from the vesicular (v-) SNARE, correlating with strong acceleration of fusion kinetics. These results suggest a similar mechanism underlying the stimulatory functions of SM proteins and Vc peptide in SNARE-dependent membrane fusion. Unexpectedly, we identified a conserved SNARE-like peptide (SLP) in SM proteins that structurally and functionally resembles Vc peptide. Like Vc peptide, SLP binds and activates target (t-) SNAREs, accelerating the fusion reaction. Disruption of the t-SNARE–SLP interaction inhibits exocytosis in vivo. Our findings demonstrated that a t-SNARE–SLP intermediate must form before SNAREs can drive efficient vesicle fusion.
Intracellular vesicle fusion is mediated by cognate soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) that assemble into SNARE complexes between the vesicle and target membrane, releasing free energy to merge the membrane bilayers (1–4). Vesicle fusion is accompanied by the conversion of the SNARE complex from the trans (anchored to two apposed membranes) to the cis (on a single merged membrane) configuration (3, 5–7). The fully assembled cis-SNARE complex consists of a parallel four-helix coiled-coil bundle held together by 15 hydrophobic layers of interacting side chains (numbered −7 to −1 and +1 to +8), and a hydrophilic 0 layer, which consists of one arginine (R) and three glutamines (Q) (Fig. 1A) (8, 9). One helix of the bundle is contributed by the vesicle-anchored v-SNARE (R-SNARE) whereas three helices are from the target membrane-associated t-SNAREs (Q-SNAREs) (4, 8, 9).
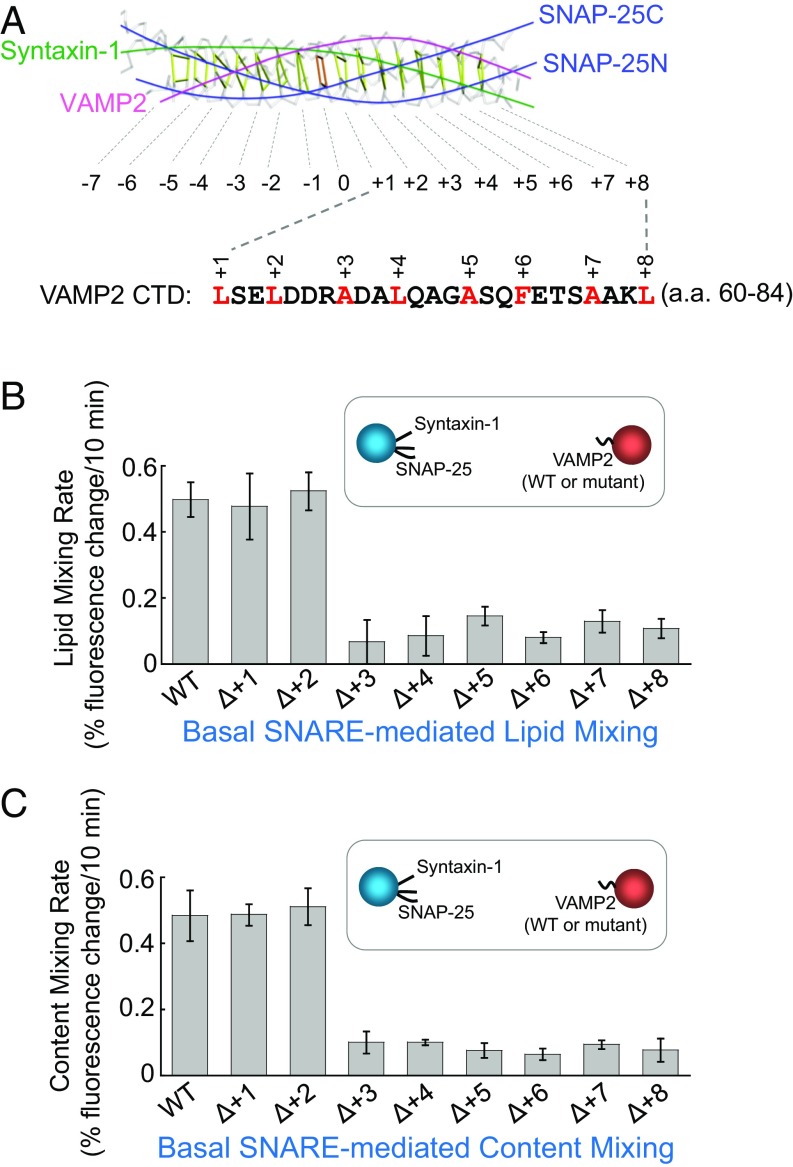
Fig. 1.
The +1 and +2 layers of the v-SNARE are dispensable for SNARE-mediated liposome fusion. (A, Top) Backbone view of the SNARE core bundle with individual layers indicated. (A, Bottom) The sequence of VAMP2 CTD (amino acids 60–84). Layer residues are numbered and highlighted. The model is based on the crystal structure of the synaptic SNARE complex (PDB ID code 1SFC) (8). (B) Initial lipid-mixing rates of the liposome fusion reactions reconstituted with WT synaptic exocytic t-SNAREs and WT or mutant VAMP2. In the layer deletion mutants, the layer residues were individually removed. Each fusion reaction contained 5 μM t-SNAREs, 1.5 μM v-SNARE, and 100 mg/mL of the macromolecular crowding agent Ficoll 70. Data are presented as the percentage of fluorescence change per 10 min. Error bars indicate SD. (C) Initial content-mixing rates of the above liposome fusion reactions.
While the postfusion cis-SNARE complex is well characterized, it remains poorly understood how the SNARE bundle zippers between membrane bilayers during the fusion reaction (8, 9). In this work, we demonstrated that the +1 and +2 layers in SNARE CTDs are dispensable for SNARE-mediated membrane fusion. However, all CTD layers are required for the fusion reactions activated by the cognate Sec1/Munc18 (SM) protein or a synthetic Vc peptide derived from the vesicular (v-) SNARE. These results suggest that the stimulatory functions of SM proteins and the Vc peptide likely involve a similar mechanism. Strikingly, we identified a conserved SNARE-like peptide (SLP) in SM proteins that structurally and functionally resembles Vc peptide. Like the Vc peptide, SLP binds and activates target (t-) SNAREs, strongly accelerating the fusion kinetics. Genetic analysis showed that inhibition of the t-SNARE–SLP interaction impairs synaptic exocytosis in cultured neurons. Together, these findings established that a t-SNARE–SLP intermediate must form before SNAREs can efficiently zipper to drive physiological levels of vesicle fusion.
Results
The +1 and +2 Layers of the v-SNARE Are Dispensable for SNARE-Mediated Membrane Fusion.
To dissect the SNARE zippering pathway, we took advantage of in vitro membrane fusion assays reconstituted using defined components. SNAREs mediating synaptic exocytosis (neurotransmitter release)—syntaxin-1, SNAP-25, and VAMP2/synaptobrevin—were incorporated into proteoliposomes at physiologically relevant densities (10, 11). The SNAREs drove a basal level of liposome fusion as measured by lipid-mixing and content-mixing assays (Fig. 1 B and C). Next, the layer residues of the v-SNARE VAMP2 were individually deleted to disrupt SNARE zippering at the respective stages. Here, we focused on layers in the C-terminal domain (CTD, +1 to +8 layers) because these layers directly output the energy required for membrane merging, whereas the N-terminal domain (NTD) mediates upstream vesicle docking (12–14). We observed that most of the CTD layer deletions (+3 to +8) abrogated SNARE-mediated liposome fusion (Fig. 1 B and C), consistent with the central role of SNARE CTDs in driving membrane fusion. However, deletion of the +1 (L60) or +2 (L63) layer of VAMP2 had no effect on the fusion kinetics (Fig. 1 B and C), suggesting that these two CTD layers are dispensable for SNARE-mediated membrane fusion. In a trans-SNARE assembly assay, deletion of the +1 or +2 layer of VAMP2 did not alter the assembly of trans-SNARE complexes between liposomes (SI Appendix, Fig. S1), in agreement with the liposome fusion data.
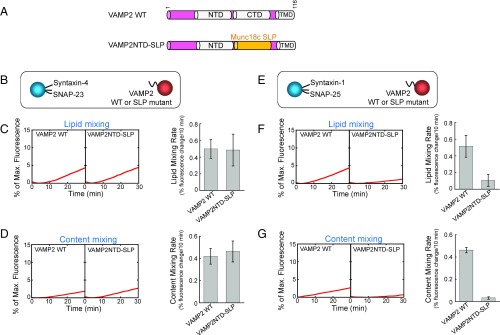
We next determined the effects of v-SNARE layer deletions on an unrelated vesicle fusion pathway. SNAREs involved in insulin-stimulated GLUT4 exocytosis—syntaxin-4, SNAP-23, and VAMP2—were reconstituted into proteoliposomes (SI Appendix, Fig. S2A). We observed that deletion of the +1 or +2 layer of VAMP2 resulted in normal fusion kinetics whereas other CTD layer deletions abrogated SNARE-mediated liposome fusion (SI Appendix, Fig. S2 B and C), similar to the findings of synaptic exocytic SNAREs. Thus, the +1 and +2 layers are also dispensable for the fusion reaction driven by GLUT4 exocytic SNAREs.
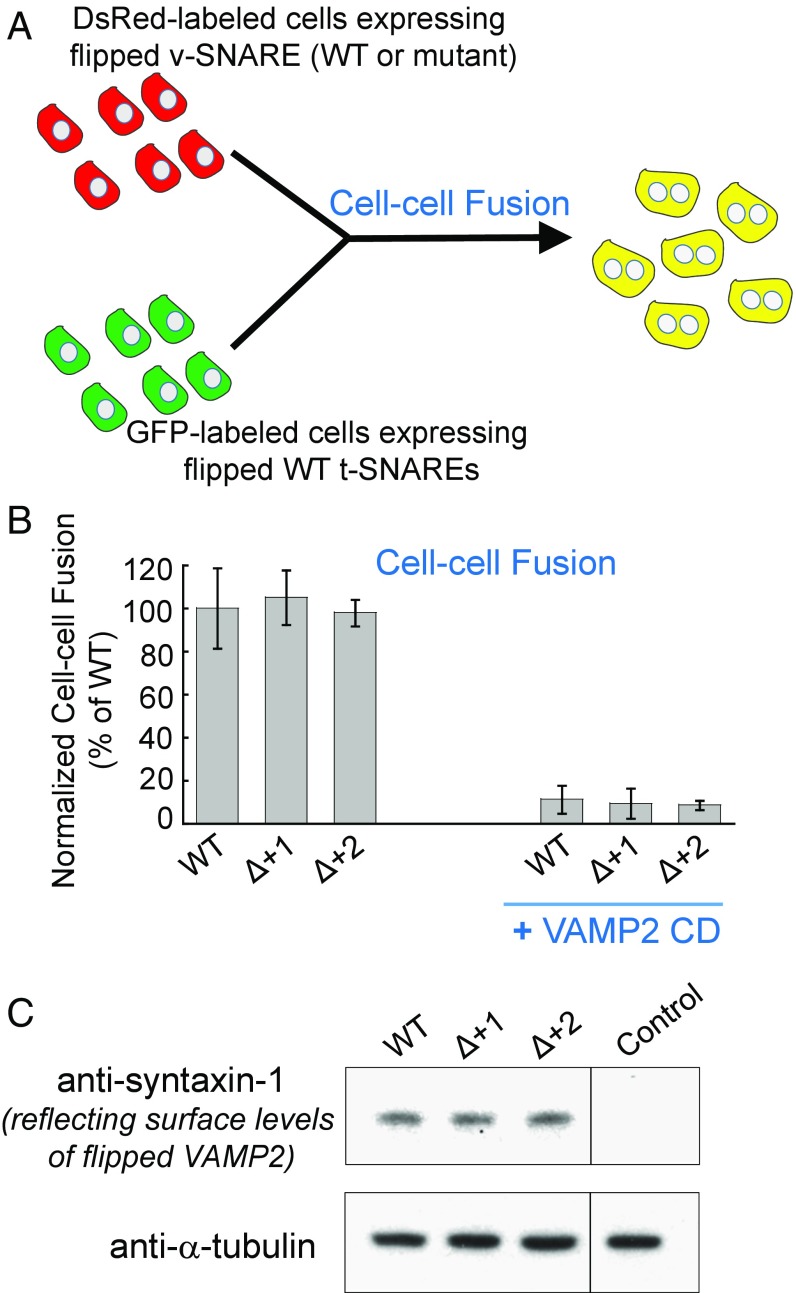
We then examined the functional roles of the +1 and +2 layers of VAMP2 in a “flipped” SNARE-mediated fusion assay (Fig. 2A) (15). When ectopically expressed on cell surfaces, flipped synaptic exocytic SNAREs drove the fusion of fluorescently labeled COS-7 cells as measured by flow cytometry (Fig. 2B). We observed that the flipped SNARE-mediated cell-cell fusion remained intact when the +1 or +2 layer of VAMP2 was deleted (Fig. 2B), confirming the liposome fusion data (Fig. 1 B and C). Flipped SNAREs were synthesized in the endoplasmic reticulum, which possesses a stringent quality control mechanism preventing the exit of unfolded or misfolded proteins (16). The flipped VAMP2 mutants were transported to the cell surface as efficiently as flipped wild-type (WT) VAMP2 (Fig. 2C), suggesting that the layer deletions did not compromise the overall architecture of the VAMP2 protein. Together, these liposome-liposome and cell-cell fusion data demonstrated that the +1 and +2 layers are dispensable for SNARE-mediated membrane fusion reactions.
Fig. 2.
The +1 and +2 layers of the v-SNARE are not required in a flipped SNARE-mediated cell-cell fusion. (A) Diagram illustrating the cell-cell fusion assay. (B) COS-7 cells expressing flipped t-SNAREs were labeled with GFP while cells expressing flipped v-SNAREs were labeled with DsRed. After incubation, the fused cells (displaying both GFP and DsRed) were measured by flow cytometry. In the negative control, 20 μM VAMP2 cytosolic domain (CD, amino acids 1–95) was added at the beginning of the fusion reactions. Data are presented as normalized percentage of cell-cell fusion driven by WT-flipped SNAREs. Error bars indicate SD. (C) Immunoblots showing the surface expression levels of WT or mutant-flipped VAMP2. Since VAMP2 proteins could not be efficiently biotinylated (15), surface levels of VAMP2 were measured indirectly through its binding to t-SNAREs. COS-7 cells expressing flipped VAMP2 were incubated with 5 μM recombinant t-SNARE CD (syntaxin-1 CD and SNAP-25). After washing, surface-bound syntaxin-1 CD (amino acids 1–265) was measured by immunoblotting to reflect the surface expression levels of flipped VAMP2. Untransfected cells were used as the control.
The +1 and +2 Layers of the v-SNARE Are Essential to Fusion Reactions Containing the Cognate SM Protein or the Vc Peptide.
While dispensable for reconstituted SNARE-mediated membrane fusion, the +1 and +2 layers of VAMP2 are critical to exocytosis in vivo (11). We reasoned that the discrepancy could be due to the presence of SNARE-binding regulatory factors in the cell that alter the SNARE zippering pathway. SM proteins are prominent candidates in this regard because they are essential to every SNARE-dependent vesicle fusion pathway (2, 3). Soluble factors of 60–70 kDa, SM proteins bind to their cognate SNAREs and promote vesicle fusion through an incompletely understood mechanism (10, 17–20). The synaptic SM protein Munc18-1 (also known as nSec1 or STXBP1) strongly accelerated membrane fusion kinetics when added to the liposome fusion reaction driven by synaptic exocytic SNAREs (10, 21, 22). We observed that the Munc18-1–stimulated fusion reaction was highly sensitive to v-SNARE layer deletions (Fig. 3A). The fusion was reduced to the basal level when any of the VAMP2 CTD layers—including the +1 and +2 layers—were deleted (Fig. 3A), in stark contrast to the basal fusion reaction mediated by SNAREs alone (Fig. 1 B and C). Thus, while dispensable for the basal SNARE-mediated fusion, the +1 and +2 layers are essential for the SM protein-stimulated fusion reaction. Since intracellular vesicle fusion is driven by the concerted action of SNAREs and SM proteins rather than by SNAREs alone (3), our findings provide a molecular explanation for the crucial roles of the +1 and +2 layers of the v-SNARE in vivo (11).
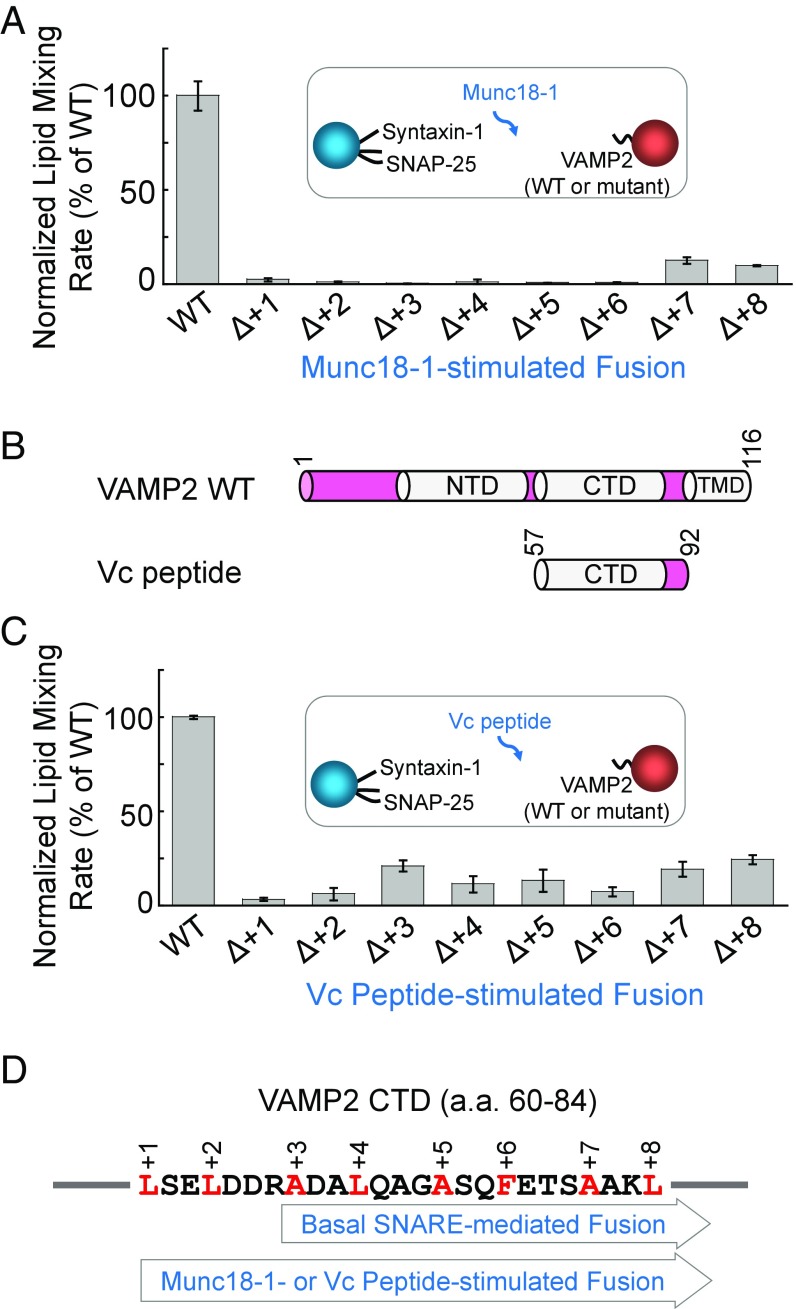
Fig. 3.
The +1 and +2 layers of the v-SNARE are essential to fusion reactions containing the cognate SM protein or the Vc peptide. (A) Normalized initial lipid-mixing rates of reconstituted fusion reactions containing 5 µM WT Munc18-1, 5 µM WT synaptic exocytic t-SNAREs, and 1.5 µM WT or mutant VAMP2. Data are presented as the percentage of initial lipid-mixing rate of WT reactions. The reconstituted fusion reactions contained 100 mg/mL of the macromolecular crowding agent Ficoll 70. In the presence of macromolecular crowding agents, the SNARE-SM–mediated fusion reaction was faithfully recapitulated without requiring a low-temperature preincubation step (11). Error bars indicate SD. (B) Diagrams of WT VAMP2 and VAMP2-derived Vc peptide. (C) Normalized initial lipid-mixing rates of reconstituted fusion reactions containing 5 µM Vc peptide, 5 µM WT synaptic exocytic t-SNAREs, 1.5 µM WT or mutant VAMP2, and 100 mg/mL Ficoll 70. Data are presented as the percentage of initial lipid-mixing rate of WT reactions. Error bars indicate SD. (D) The basal SNARE-mediated fusion reaction involves the +3 to +8 layers in CTDs, whereas all of the CTD layers (+1 to +8) are required for the Munc18-1– or Vc peptide-stimulated fusion reaction.
The CTDs of t-SNAREs are partially unstructured (23, 24), which may prevent proper SNARE zippering such that the +1 and +2 layers are dispensable. This hypothesis predicts that restructuring t-SNARE CTDs would achieve the same effect on SNARE zippering as the SM protein. To test this possibility, we examined the regulatory activity of an engineered peptide corresponding to the CTD of the v-SNARE (Vc peptide) (Fig. 3B), which binds t-SNARE CTDs and converts the latter into a helical configuration (23, 25, 26). We observed that Vc peptide markedly accelerated SNARE-mediated liposome fusion, and the Vc peptide-stimulated fusion reaction was highly sensitive to deletions in all CTD layers including the +1 and +2 layers (Fig. 3C). These data suggest that the stimulatory functions of Munc18-1 and Vc peptide involve a similar mechanism (Fig. 3D).
SM Proteins Possess a SNARE-Like Peptide That Structurally and Functionally Resembles Vc Peptide.
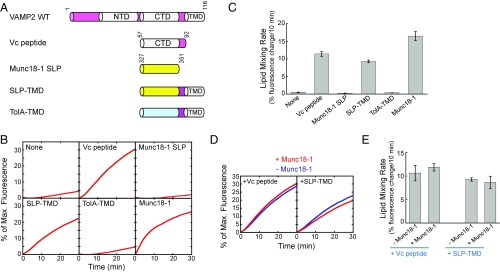
How do these seemingly unrelated molecules—SM proteins and Vc peptide—achieve a similar effect on SNARE zippering? In an unbiased global alignment of protein sequences, we unexpectedly identified a sequence in SM proteins that strikingly resembles Vc peptide (VAMP2 CTD). Located within domain 3a of SM proteins, this SNARE-like peptide (SLP) contains heptad repeats of hydrophobic residues that align with the CTD layers of VAMP2 (Fig. 4A and SI Appendix, Fig. S3). Found in all SM proteins we analyzed (SI Appendix, Fig. S3), SLP adopts a helical configuration upon SNARE binding (27), a characteristic feature of SNARE core domains (3). The stimulatory function of Munc18-1 in the fusion reaction was abolished by SLP deletion or point mutations in the layer-like residues of SLP (SI Appendix, Figs. S4 and S5). Likewise, the stimulatory activity of Munc18c, the cognate SM protein in GLUT4 exocytosis (21), was abrogated when its SLP was removed (SI Appendix, Fig. S6). The circular dichroism (CD) spectra of the SLP deletion mutants were similar to those of WT proteins (SI Appendix, Fig. S7), suggesting that the overall folding of SM proteins is not compromised by SLP deletion. Together, these results confirmed the importance of SLP to SM protein function.
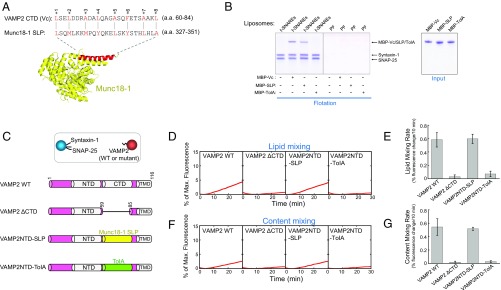
Fig. 4.
SM proteins possess a SNARE-like peptide that structurally and functionally resembles Vc peptide. (A, Top) Alignment of VAMP2 CTD (Vc peptide) and the SNARE-like peptide (SLP) from Munc18-1 (identity: 20.0%; similarity: 56.0%). The alignment was performed using the LALIGN program (https://embnet.vital-it.ch/software/LALIGN_form.html). Alignment method, global; scoring matrix, PAM250; other parameters, default. Layer residues of Vc peptide are numbered and highlighted. (A, Bottom) Crystal structure of Munc18-1 with SLP shown in red (PDB ID code 3PUJ) (27). (B) Coomassie blue-stained SDS/PAGE gels showing the binding of Vc peptide, Munc18-1–derived SLP, and the bacterial TolA helix to the indicated liposomes. The t-SNARE complex was composed of syntaxin-1 and SNAP-25. PF, protein free. Each binding reaction contained 5 μM SNAREs and 5 μM MBP-tagged peptides. (C, Top) Illustration of the liposome fusion pairs. (C, Bottom) Diagrams of WT and mutant VAMP2 proteins used in the liposome fusion assays. The sequences of the chimeras are included in Materials and Methods. (D) Lipid mixing of the reconstituted liposome fusion reactions containing WT synaptic exocytic t-SNAREs and WT or mutant VAMP2. Each fusion reaction contained 5 μM t-SNAREs, 1.5 μM v-SNARE, and 100 mg/mL Ficoll 70. (E) Initial rates of the lipid mixing reactions shown in D. (F) Content mixing of the liposome fusion reactions described in D. (G) Initial rates of the lipid mixing reactions shown in F. Data in E and G are presented as percentage of fluorescence change per 10 min. Error bars indicate SD.
In a liposome coflotation assay, recombinant SLP derived from Munc18-1 bound to t-SNARE liposomes but not to protein-free liposomes, similar to the binding property of Vc peptide (Fig. 4B). We next examined the ability of SLP to pair with the t-SNAREs to drive membrane fusion (in the absence of full-length SM proteins). To this end, we created a VAMP2-SLP chimera in which the CTD of VAMP2 was replaced with the SLP from Munc18-1 (Fig. 4C). Strikingly, we observed that the v-SNARE-SLP chimera paired with synaptic exocytic t-SNAREs and drove a basal liposome fusion reaction at a level comparable to that mediated by WT SNAREs (Fig. 4 D–G). By contrast, the bacterial TolA helix failed to drive liposome fusion when introduced into VAMP2 (Fig. 4 D–G). Similarly, a v-SNARE–SLP chimera containing the Munc18c-derived SLP paired with the cognate t-SNAREs of GLUT4 exocytosis to mediate a WT level of liposome fusion (Fig. 5 A–D).
Fig. 5.
The SLP of Munc18c is capable of driving membrane fusion when introduced into VAMP2. (A) Diagrams of WT VAMP2 and VAMP2-SLP chimera. SLP corresponds to amino acids 327–351 of Munc18c. The sequence of the chimera is included in Materials and Methods. (B) Illustration of the liposome fusion pairs. (C, Left) Lipid mixing of the reconstituted fusion reactions described in B. (C, Right) Initial lipid-mixing rates of the reconstituted fusion reactions. Data are presented as the percentage of fluorescence change per 10 min. Error bars indicate SD. (D, Left) Content mixing of the reconstituted fusion reactions described in B. (D, Right) Initial content-mixing rates of the reconstituted fusion reactions. Data are presented as the percentage of fluorescence change per 10 min. Error bars indicate SD. (E) Illustration of the liposome fusion pairs. (F, Left) Lipid mixing of the reconstituted fusion reactions described in E. (F, Right) Initial lipid-mixing rates of the reconstituted fusion reactions. Data are presented as the percentage of fluorescence change per 10 min. Error bars indicate SD. (G, Left) Content mixing of the reconstituted fusion reactions described in E. (G, Right) Initial content-mixing rates of the reconstituted fusion reactions. Data are presented as the percentage of fluorescence change per 10 min. Error bars indicate SD.
Interestingly, Munc18c-derived SLP failed to drive liposome fusion when paired with the noncognate t-SNAREs involved in synaptic exocytosis (Fig. 5 E–G). Likewise, the speed of the fusion reaction driven by Munc18-1–derived SLP was markedly reduced when the cognate t-SNARE complex was replaced with the noncognate GLUT4 exocytic t-SNAREs (SI Appendix, Fig. S8). These results suggest that SLP retains the compartmental specificity of the corresponding SM protein. Next, we created a v-SNARE chimera in which the NTD of VAMP2 was replaced with the SLP of Munc18-1. This chimera, however, failed to drive liposome fusion when paired with synaptic t-SNAREs (SI Appendix, Fig. S9), demonstrating that SLP is unable to functionally replace v-SNARE NTD. Together, these data strongly suggest that SLP-mediated fusion results from the formation of specific, energy-generating t-SNARE–SLP complexes, rather than nonselective helical interactions, supporting the notion that t-SNARE CTDs are a biologically relevant target of SLP in the SNARE-SM–mediated fusion reaction. Thus, despite its origin in SM proteins, SLP is capable of pairing with the t-SNAREs to drive membrane fusion, establishing that SLP structurally and functionally resembles Vc peptide.
The SLP of SM Proteins and Vc Peptide Use a Similar Mechanism to Activate SNARE-Dependent Membrane Fusion.
Identification of the Vc peptide-like SLP in SM proteins raises the intriguing possibility that SM proteins and Vc peptide may use a similar mechanism to activate SNARE-dependent membrane fusion. To test this possibility, recombinant SLP derived from Munc18-1 was introduced into the liposome fusion reaction mediated by WT synaptic exocytic SNAREs (Fig. 6A). Soluble SLP, however, had little effect on the kinetics of the SNARE-mediated liposome fusion (Fig. 6 B and C). Since SM proteins possess other SNARE-binding motifs (2), we reasoned that SLP may require recruitment to the vicinity of the t-SNAREs by non-SLP interactions before it can activate membrane fusion. Indeed, when tethered to t-SNARE liposomes through an engineered membrane anchor, SLP markedly accelerated SNARE-dependent liposome fusion, comparable to the stimulatory activity of Vc peptide (Fig. 6 B and C). By contrast, the liposome-anchored TolA helix was unable to accelerate fusion kinetics (Fig. 6 B and C). Thus, the SM protein-derived SLP is capable of stimulating SNARE-dependent membrane fusion, further demonstrating its functional similarity to Vc peptide. Our findings also suggest that previous reconstituted fusion reactions containing the synthetic Vc peptide recapitulated the SM protein-stimulated fusion reaction, rather than the basal SNARE-mediated fusion (14, 24, 26).
Fig. 6.
SLP and Vc peptide activate SNARE-mediated membrane fusion through a similar mechanism. (A) Diagrams of the molecules added to the reconstituted fusion reaction: Vc peptide, Munc18-1–derived SLP, Munc18-1–derived SLP with an engineered transmembrane domain (TMD, from VAMP2), and the TolA helix with a VAMP2-derived TMD. The sequences of the chimeras are included in Materials and Methods. (B) Reconstituted SNARE-dependent fusion reactions containing 5 µM WT synaptic exocytic t-SNAREs, 1.5 µM WT VAMP2, 100 mg/mL Ficoll 70, and 5 μM of the indicated molecules. (C) Initial lipid-mixing rates of the reconstituted fusion reactions shown in B. (D) Reconstituted liposome fusion reactions were performed as described in B in the absence or presence of 5 μM Munc18-1. (E) Initial lipid-mixing rates of the reconstituted fusion reactions shown in D. Data in C and E are presented as the percentage of fluorescence change per 10 min. Error bars indicate SD.
Next, we sought to further delineate the molecular mechanism of SLP. Using a trans-SNARE assembly assay, we observed that membrane-anchored SLP strongly enhanced the assembly of trans-SNARE complexes between liposome membranes (SI Appendix, Fig. S10A), consistent with its stimulatory effect on fusion kinetics. By contrast, SLP did not significantly increase the docking of SNARE liposomes (SI Appendix, Fig. S10 B and C), indicating that it is not involved in the initial pairing of SNAREs. These results suggest that SLP regulates a relatively late stage of SNARE assembly.
While the stimulatory activities of Vc peptide and SLP in the fusion reactions are robust, they are still less potent than that of full-length Munc18-1 (Fig. 6 B and C), in agreement with the roles of non-SLP SM protein domains and the additional regulatory activities of SM proteins in SNARE-mediated fusion (17, 28). Notably, fusion reactions containing Vc peptide or SLP were not further stimulated by Munc18-1 (Fig. 6 D and E). Thus, instead of acting synergistically with Munc18-1, Vc peptide and SLP are competitive inhibitors of Munc18-1, confirming that the stimulatory function of SM proteins requires the binding of their SLPs to t-SNARE CTDs, the same region recognized by Vc peptide. These results demonstrated that SM proteins use their SLPs to activate fusion in a similar way as Vc peptide, and established t-SNARE–SLP interaction as an essential step in the fusion reaction.
Inhibition of the t-SNARE–SLP Interaction Impairs Synaptic Exocytosis in Cultured Neurons.
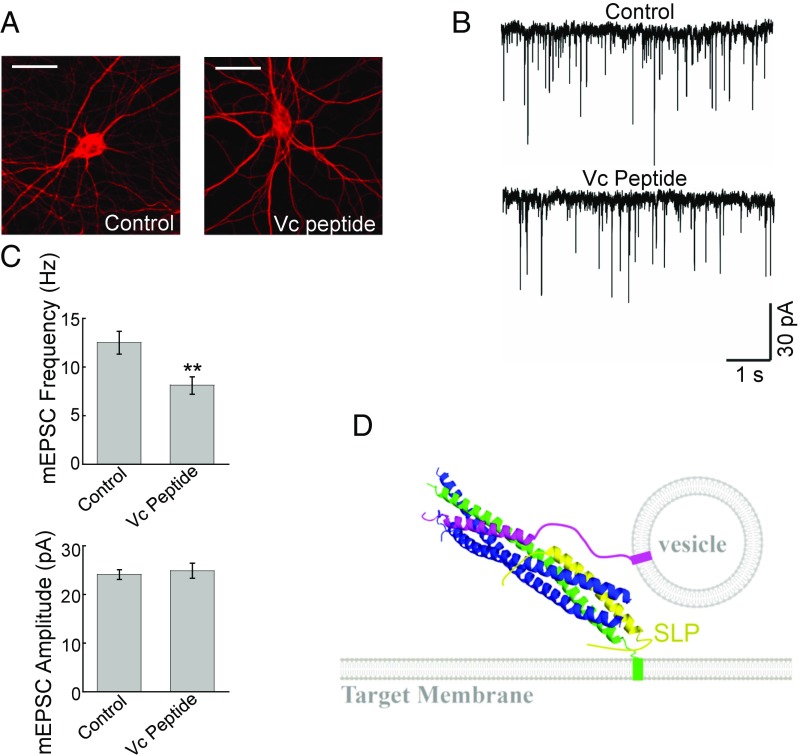
The outcomes of our reconstitution experiments correlate well with previous genetic observations. For example, mutations of residues within SLP abrogated SM protein functions in vesicle fusion (29–32). To further explore the physiological relevance of our findings, we examined how synaptic exocytosis is influenced by the expression of Vc peptide. Our reconstitution studies showed that the Vc peptide interferes with Munc18-1 function by inhibiting the t-SNARE-SLP–binding mode in the fusion reaction (Fig. 6 D and E). Thus, expression of the Vc peptide is expected to decrease synaptic exocytosis rather than enhance it as previous models suggested. To test this prediction, Vc peptide was expressed in primary cortical neurons isolated from newborn mice. Vc peptide expression did not alter neuronal morphology or levels of vesicle fusion proteins (Fig. 7A and SI Appendix, Fig. S11A). Electrophysiological recordings showed that expression of the Vc peptide significantly lowered the frequency of miniature excitatory postsynaptic currents (mEPSCs) (Fig. 7 B and C), reflecting reduced spontaneous vesicle fusion events. The amplitude of mEPSCs remained unaffected (Fig. 7 B and C), suggesting that the biogenesis of synaptic vesicles was intact. Stimulus-evoked EPSCs were also inhibited in Vc peptide-expressing neurons, albeit to a lesser degree (SI Appendix, Fig. S11 B and C). The reduced synaptic exocytosis observed in Vc peptide-expressing neurons is in agreement with the ability of Vc peptide to impair Munc18-1 function in reconstituted fusion reactions, further supporting the physiological relevance of our results.
Fig. 7.
Expression of Vc peptide impairs the synaptic exocytosis in cultured neurons. (A) Representative immunofluorescence images showing cultured mouse cortical neurons expressing GFP (control) or GFP and Vc peptide. The cells were permeabilized and stained with anti-MAP2 antibodies. (Scale bars: 20 µm.) (B) Representative traces of miniature excitatory postsynaptic currents (mEPSCs) recorded in the control or Vc peptide-expressing cortical neurons. (C, Top) Summary graph of mEPSC frequency. (C, Bottom) Summary graph of mEPSC amplitudes. Numbers of neurons and independent cultures are listed in SI Appendix, Table S1. **P < 0.01. (D) Model illustrating the functional role of SLP in vesicle fusion reaction. SLP recognizes and restructures t-SNARE CTDs in the context of the half-zippered trans-SNARE complex. Green, Qa SNARE; blue, Qb and Qc SNAREs; magenta, v-SNARE (R-SNARE); yellow, SLP of the SM protein. For clarity, the rest of the SM protein is not shown. The model is based on the crystal structure of the synaptic exocytic SNARE complex (PDB ID code 1SFC).
Discussion
Based on our in vitro and in vivo observations, we propose that the current model of SNARE zippering needs to be revised to include the t-SNARE–SLP complex as a crucial intermediate. Mimicking the four-helix SNARE bundle (Fig. 7D), the SNARE–SLP complex restructures t-SNARE CTDs into a helical conformation competent for optimal v-SNARE zippering. SLP may also prevent the dead-end 2:1 configuration of the t-SNARE complex as previously demonstrated for the Vc peptide (14, 33). When the SLP is subsequently displaced by the vesicle-anchored v-SNARE, SNARE CTDs can fully and optimally zipper to drive membrane fusion, requiring all CTD layers. Without the assistance of SLP, however, SNARE CTDs adopt a partial zippering route, skipping the +1 and +2 layers. Partial SNARE zippering reduces free energy change and disrupts zippering cooperativity, demanding greater numbers of SNARE complexes to accumulate at the fusion site to overcome the energy barrier of membrane merging. As a result, the probability of membrane fusion events is reduced to a basal level that cannot meet physiological demands. We note that, while this hypothetical model is supported by our biochemical and genetic data, additional studies (e.g., optical tweezer measurements) will be needed to further test this model by directly examining SNARE zippering.
SLP and its adjacent sequences in domain 3a are ideally suited to interact with SNARE helices in a regulatory manner. Besides t-SNARE CTDs, this region also recognizes v-SNARE, syntaxin (Qa-SNARE), and cis-SNARE complexes (17, 20, 27, 34, 35). In particular, the association of SLP and adjacent regions with v-SNARE and syntaxin guides the initial pairing of SNARE NTDs to form a partially zippered trans-SNARE complex (17). Subsequently, with the SM protein remaining associated with the trans-SNARE complex, SLP shifts its binding partner to t-SNARE CTDs, enabling SNARE CTDs to fully zipper to drive efficient membrane fusion. Thus, the t-SNARE–regulating function of SLP lies downstream of initial vesicle docking but upstream of vesicle fusion execution. Besides these direct SNARE-regulating functions, SM proteins also indirectly promote SNARE assembly by shielding SNAREs from the negative activities of NSF and αSNAP (19). It would be interesting to determine whether this shielding function also requires SLP. Together, these findings provide molecular explanations for the observations that SM proteins control both the early and late steps of the SNARE-zippering pathway (10, 17, 20, 21, 28), correlating well with the roles of SM proteins at both the docking and postdocking stages of intracellular vesicle fusion (36–38). The binding of SLP to multiple SNARE targets requires SLP to shift binding partners as SNARE zippering progresses. We suggest that sequential binding of SLP and adjacent sequences to multiple SNARE targets provides the driving force for SM proteins to guide the entire SNARE-zippering pathway. In particular, free energy derived from SLP binding to t-SNARE CTDs facilitates the release of SLP and adjacent sequences from SNARE NTDs, a binding mode that initiates SNARE assembly but must be released before SNARE zippering can proceed further.
In constitutive vesicle fusion pathways, SNARE CTDs directly proceed to drive membrane fusion following SLP displacement. In a regulated vesicle fusion pathway, however, the conserved SNARE-SM machinery is superimposed by specialized regulatory factors to achieve an integrated response. For example, in Ca2+-triggered synaptic exocytosis, synaptotagmin-1, and complexin bind and clamp the half-zippered trans-SNARE complex, allowing SNARE CTD zippering to be coupled to Ca2+ stimulation (39). It is possible that synaptotagmin-1, complexin, and the SLP of Munc18-1 bind simultaneously to the half-zippered trans-SNARE complex to form a prefusion supracomplex. Alternatively, SLP may restructure t-SNAREs and then dissociate before synaptotagmin-1 and complexin can recognize and clamp the half-zippered trans-SNARE complex. Interestingly, the exocytic regulator Munc13 also modulates the conformational states of SNARE subunits (40, 41), likely at a different stage as SM proteins, to further accelerate the fusion speed as demanded by synaptic transmission. A key future research direction is to investigate how these specialized factors cooperate with the conserved SNARE-SM machinery to control SNARE zippering in a stimulus-dependent manner.
Materials and Methods
Recombinant Protein Expression and Purification.
Recombinant v- and t-SNAREs were expressed in Escherichia coli and purified by nickel affinity chromatography. The synaptic exocytic t-SNARE complex was composed of untagged rat syntaxin-1 and mouse SNAP-25 with an N-terminal His6 tag. The GLUT4 exocytic t-SNARE complex was composed of untagged rat syntaxin-4 and mouse His6-tagged SNAP-23. Recombinant v-SNARE proteins had no extra residues left after the tags were removed (42). Recombinant untagged Munc18-1 and Munc18c proteins were produced in E. coli and Sf9 insect cells, respectively, using procedures we established (10, 11, 21, 43). MBP (maltose-binding protein)-tagged Vc peptide (amino acids 57–92 of VAMP2), SLP (amino acids 327–351 of Munc18-1), and TolA were expressed and purified in the same way as Munc18-1. SNARE and Munc18-1 mutants were generated by site-directed mutagenesis (Stratagene/Agilent Technologies) and purified in similar ways as the corresponding WT proteins.
Sequences of chimeric proteins used in this work are listed below:
-
i)
VAMP2NTD-SLP (Munc18-1) (SLP is derived from amino acids 327–351 of rat Munc18-1, highlighted in bold):
MSATAATVPPAAPAGEGGPPAPPPNLTSNRRLQQTQAQVDEVVDIMRVNVDKVLERDQKLSQMLKKMPQYQKELSKYSTHLHLAKRKYWWKNLKMMIILGVICAIILIIIIVYFST
-
ii)
VAMP2NTD-SLP (Munc18c) (SLP is derived from amino acids 327–351 of mouse Munc18c, highlighted in bold):
MSATAATVPPAAPAGEGGPPAPPPNLTSNRRLQQTQAQVDEVVDIMRVNVDKVLERDQKLTQLMKKMPHFRKQISKQVVHLNLAKRKYWWKNLKMMIILGVICAIILIIIIVYFST
-
iii)
SLP-VAMP2CTD (SLP is derived from amino acids 327–351 of rat Munc18-1, highlighted in bold):
MSATAATVPPAAPAGEGGPPAPPPNLTSNRRLSQMLKKMPQYQKELSKYSTHLHLADQKLSELDDRADALQAGASQFETSAAKLKRKYWWKNLKMMIILGVICAIILIIIIVYFST
-
iv)
VAMP2NTD-TolA (the CTD of VAMP2 was replaced with a bacterial TolA sequence, which is shown in bold):
MSATAATVPPAAPAGEGGPPAPPPNLTSNRRLQQTQAQVDEVVDIMRVNVDKVLERDQKGGSSIDAVMVDSGAVVEQYKRMQSQKRKYWWKNLKMMIILGVICAIILIIIIVYFST
-
v)
Munc18-1-Vc (Vc is derived from amino acids 60–84 of mouse VAMP2, highlighted in bold):
MAPIGLKAVVGEKIMHDVIKKVKKKGEWKVLVVDQLSMRMLSSCCKMTDIMTEGITIVEDINKRREPLPSLEAVYLITPSEKSVHSLISDFKDPPTAKYRAAHVFFTDSCPDALFNELVKSRAAKVIKTLTEINIAFLPYESQVYSLDSADSFQSFYSPHKAQMKNPILERLAEQIATLCATLKEYPAVRYRGEYKDNALLAQLIQDKLDAYKADDPTMGEGPDKARSQLLILDRGFDPSSPVLHELTFQAMSYDLLPIENDVYKYETSGIGEARVKEVLLDEDDDLWIALRHKHIAEVSQEVTRSLKDFSSSKRMNTGEKTTMLQDQKLSELDDRADALQAGASQFETSAAKLGTCMKHYQGTVDKLCRVEQDLAMGTDAEGEKIKDPMRAIVPILLDANVSTYDKIRIILLYIFLKNGITEENLNKLIQHAQIPPEDSEIITNMAHLGVPIVTDSTLRRRSKPERKERISEQTYQLSRWTPIIKDIMEDTIEDKLDTKHYPYISTRSSASFSTTAVSARYGHWHKNKAPGEYRSGPRLIIFILGGVSLNEMRCAYEVTQANGKWEVLIGSTHILTPQKLLDTLKKLNKTDEEISS
-
vi)
SLP-TMD (the SLP sequence of rat Munc18-1 is shown in bold):
LSQMLKKMPQYQKELSKYSTHLHLAKRKYWWKNLKMMIILGVICAIILIIIIVYFST
-
vii)
TolA-TMD (the bacterial TolA sequence is shown in bold):
GGSSIDAVMVDSGAVVEQYKRMQSQKRKYWWKNLKMMIILGVICAIILIIIIVYFST
Proteoliposome Preparation.
Lipids used in this work were acquired from Avanti Polar Lipids. For t-SNARE reconstitution, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), and cholesterol were mixed in a molar ratio of 60:20:10:10. For v-SNARE reconstitution, POPC, POPE, POPS, cholesterol, N-(7-nitro-2,1,3-benzoxadiazol-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE), and N-(Lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (rhodamine-DPPE) were mixed at a molar ratio of 60:17:10:10:1.5:1.5. SNARE proteoliposomes were prepared by detergent dilution and isolated on a Nycodenz density gradient flotation (42). Complete detergent removal was achieved by overnight dialysis of the samples in Novagen dialysis tubes against the reconstitution buffer (25 mM Hepes [pH 7.4], 100 mM KCl, 10% glycerol, and 1 mM DTT). To prepare sulforhodamine-loaded liposomes, t-SNARE or v-SNARE liposomes were reconstituted in the presence of 50 mM sulforhodamine B (Sigma). Free sulforhodamine B was removed by overnight dialysis followed by liposome flotation on a Nycodenz gradient. The protein:lipid ratio was at 1:200 for v-SNAREs and 1:500 for t-SNARE liposomes.
Liposome Fusion Assay.
A standard liposome fusion reaction contained 5 μM t-SNAREs, 1.5 μM v-SNARE, and 100 mg/mL of the macromolecular crowding agent Ficoll 70. In lipid-mixing assays, v-SNARE liposomes were labeled with NBD and rhodamine and were directed to fuse with unlabeled t-SNARE liposomes. The fusion reactions were conducted in a 96-well microplate at 37 °C. The NBD fluorescence (excitation: 460 nm; emission: 538 nm) was measured every 2 min in a BioTek Synergy HT microplate reader. For content mixing assays, unlabeled t-SNARE liposomes were directed to fuse with sulforhodamine B-loaded v-SNARE liposomes. The sulforhodamine B fluorescence (excitation: 565; emission: 585 nm) was measured every 2 min. At the end of the reaction, 10 μL of 10% CHAPSO was added to each sample. The v-SNARE and t-SNARE liposomes were mixed with the regulators and immediately loaded into a preheated microplate (37 °C) to initiate fusion. The fusion reactions were performed as we described (11). Fusion data were presented as the percentage of maximum fluorescence change. The maximum fusion rate within the first 10 min of liposome fusion reaction was used to represent the initial rate of a fusion reaction. Full accounting of statistical significance was included for each figure based on at least three independent experiments.
Liposome Coflotation Assay.
MBP-tagged Vc peptide, SLP, or TolA peptide was individually incubated with protein-free (PF) liposomes or t-SNARE liposomes at 4 °C with gentle agitation. After 1 h, an equal volume of 80% Nycodenz (wt/vol) in the reconstitution buffer was added and transferred to 5 mm by 41 mm centrifuge tubes. The liposomes were overlaid with 200 μL each of 35% and 30% Nycodenz, and then with 20 μL of reconstitution buffer on the top. The gradients were centrifuged for 4 h at 52,000 rpm in a Beckman SW55 rotor. Samples were collected from the 0/30% Nycodenz interface (2 × 20 μL) and analyzed by SDS/PAGE.
Cell-Cell Fusion Assay.
Flipped SNARE-mediated fusion of COS-7 cells were carried out using a previously described procedure with modifications (15). COS-7 cells were transiently transfected with plasmids expressing flipped syntaxin-1, flipped SNAP-25, and sfGFP-C1 using FuGENE HD transfection reagents (Active Motif). Flipped VAMP2 (WT or mutants) and DsRed-NES were transiently expressed in a separate population of COS-7 cells. After 24 h of expression, the two populations of cells were detached by an EDTA-based dislodging solution and mixed at a 1:1 ratio. After incubated at 37 °C for 9 h, the cells were dislodged and the numbers of fused cells (expressing both GFP and DsRed) were measured by flow cytometry. The percentage of cell-cell fusion was calculated by the following formula: 100 × 2F/(V+T+2F). V represents the number of cells showing a DsRed signal, T represents the number of cells showing a GFP signal, and F represents the number of cells showing both GFP and DsRed signals. Data are presented as normalized percentage of cell-cell fusion mediated by WT-flipped SNAREs. To quantify surface levels of flipped VAMP2, cells expressing flipped VAMP2 were rapidly chilled on an ice bath and incubated with 5 μM soluble recombinant t-SNARE CD for 1 h. After washing with PBS, the cells were lysed in a SDS sample buffer and the proteins were resolved on SDS/PAGE. Syntaxin-1 CD was quantified by immunoblotting using monoclonal anti-syntaxin-1 antibodies (HPC-1; Synaptic Systems).
Primary Neuron Culture and Lentiviral Infection.
Cortical neurons were isolated from newborn C57BL/6 mice as described (11, 43). The cells were dissociated by papain (Worthington) digestion and plated on glass coverslips coated with Poly-d-lysine (Sigma). The neurons were maintained in the Neurobasal medium (Life Technologies) supplemented with B-27 (Life Technologies), glutaMAX (Life Technologies), and Ara-C (Sigma) according to Life Technologies’ instructions.
The pLKO.1-based SHC001 plasmid was obtained from University of Colorado Functional Genomics Facility. The plasmid was digested with BamHI and KpnI to remove the puromycin resistance gene (puroR). A DNA fragment encoding GFP, P2A sequence, and the Vc peptide was subcloned into the BamHI and KpnI sites such that their expression was driven by the hPGK promoter. In a control construct, GFP and P2A were subcloned in the same way except that the Vc peptide sequence was omitted. To produce lentiviral particles, lentiviral expression plasmids were transfected into HEK 293T cells along with helper plasmids—pCMV-VSVG, psPAX2, and pAdVAntage. Forty-eight hours after transfection, cell culture supernatants containing lentiviral particles were collected and filtered through 0.45-µm syringe filters (Corning). The supernatants were concentrated by centrifugation at 25,000 rpm for 1 h in a Beckman SW28 rotor. Neurons were infected with the lentiviruses at days in vitro (DIV) 4 and analyzed by electrophysiological measurements at DIV 16–20. Syntaxin-1 expression was measured using the antibodies described above. VAMP2 and Munc18-1 expression levels were detected using anti-VAMP2 antibodies (clone Cl69.1; Synaptic Systems) and anti-Munc18-1 antibodies (clone 31/Munc-18; BD Biosciences), respectively. Anti-α-tubulin antibodies (clone TU-01) were acquired from BioLegend. Neuronal morphology was examined by immunostaining using anti-MAP2 antibodies (M9942; Sigma) as described (43). All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Colorado Boulder.
Electrophysiological Recordings.
The mEPSCs of the neurons were sampled at 10 kHz in the presence of 1 µM tetrodotoxin (TTX; Sigma). The resistance of pipettes was 3–5 megaohms. The series resistance was adjusted to 8–10 megaohms once the whole-cell configuration was established. About 70% of cultured neurons expressed GFP. Recordings were carried out on neurons expressing GFP and surrounded by GFP+ cells. Evoked synaptic transmission was triggered by 1-ms current injections using a concentric bipolar microelectrode (FHC; Model: CBAEC75) placed about 100–150 µm from the cell bodies of patched neurons. The extracellular stimuli were manipulated using an Isolated Pulse Stimulator (World Precision Instruments). The evoked responses were measured by whole-cell recordings using a Multiclamp 700B amplifier (Molecular Devices). The whole-cell pipette solution contained 135 mM CsCl, 10 mM Hepes-CsOH (pH 7.25), 0.5 mM EGTA, 2 mM MgCl2, 0.4 mM NaCl-GTP, and 4 mM NaCl-ATP. The bath solution contained 140 mM NaCl, 5 mM KCl, 2 mM CaCl2, 0.8 MgCl2, 10 mM Hepes-NaOH (pH 7.4), and 10 mM Glucose. EPSCs were distinguished by addition of 50 µM picrotoxin (all from Sigma) in the bath solution. The electrophysiological data were analyzed using the pClamp 10 software (Molecular Devices). For statistical calculations, all data are shown as means ± SEMs. The P values were calculated using Student’s t test.
Supplementary Material
Acknowledgments
We thank Drs. Jenny Martin, Frederick Hughson, Yongli Zhang, Josep Rizo, Bill Wickner, Gustav Lienhard, and Richard Baker for reagents, protocols, or suggestions. This work was supported by National Institutes of Health Grants DK095367 (to J.S.), GM126960 (to J.S.), and GM114496 (to M.H.B.S.), and a University of Colorado Seed Grant (to J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. Y.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802645115/-/DCSupplemental.
References
- 1.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Rizo J, Südhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices–Guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- 3.Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickner W. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 5.Tamm LK, Crane J, Kiessling V. Membrane fusion: A structural perspective on the interplay of lipids and proteins. Curr Opin Struct Biol. 2003;13:453–466. doi: 10.1016/s0959-440x(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 6.Reese C, Heise F, Mayer A. Trans-SNARE pairing can precede a hemifusion intermediate in intracellular membrane fusion. Nature. 2005;436:410–414. doi: 10.1038/nature03722. [DOI] [PubMed] [Google Scholar]
- 7.Lu X, Zhang Y, Shin YK. Supramolecular SNARE assembly precedes hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2008;15:700–706. doi: 10.1038/nsmb.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 9.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, et al. Reconstituting intracellular vesicle fusion reactions: The essential role of macromolecular crowding. J Am Chem Soc. 2015;137:12873–12883. doi: 10.1021/jacs.5b08306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, et al. A half-zippered SNARE complex represents a functional intermediate in membrane fusion. J Am Chem Soc. 2014;136:3456–3464. doi: 10.1021/ja410690m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 15.Hu C, et al. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- 16.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 17.Baker RW, et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114. doi: 10.1126/science.aac7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodkey TL, Liu S, Barry M, McNew JA. Munc18a scaffolds SNARE assembly to promote membrane fusion. Mol Biol Cell. 2008;19:5422–5434. doi: 10.1091/mbc.E08-05-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobingier BT, Nickerson DP, Lo SY, Merz AJ. SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18. eLife. 2014;3:e02272. doi: 10.7554/eLife.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, et al. Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLife. 2015;4:e09580. doi: 10.7554/eLife.09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, et al. Comparative studies of Munc18c and Munc18-1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 2013;110:E3271–E3280. doi: 10.1073/pnas.1311232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rathore SS, et al. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci USA. 2010;107:22399–22406. doi: 10.1073/pnas.1012997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, et al. Stability, folding dynamics, and long-range conformational transition of the synaptic t-SNARE complex. Proc Natl Acad Sci USA. 2016;113:E8031–E8040. doi: 10.1073/pnas.1605748113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paumet F, et al. A t-SNARE of the endocytic pathway must be activated for fusion. J Cell Biol. 2001;155:961–968. doi: 10.1083/jcb.200104092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F, Tiwari N, Rothman JE, Pincet F. Kinetic barriers to SNAREpin assembly in the regulation of membrane docking/priming and fusion. Proc Natl Acad Sci USA. 2016;113:10536–10541. doi: 10.1073/pnas.1604000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melia TJ, et al. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu SH, et al. Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation. Proc Natl Acad Sci USA. 2011;108:1040–1045. doi: 10.1073/pnas.0914906108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munch AS, et al. Extension of helix 12 in Munc18-1 induces vesicle priming. J Neurosci. 2016;36:6881–6891. doi: 10.1523/JNEUROSCI.0007-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyd A, et al. A random mutagenesis approach to isolate dominant-negative yeast sec1 mutants reveals a functional role for domain 3a in yeast and mammalian Sec1/Munc18 proteins. Genetics. 2008;180:165–178. doi: 10.1534/genetics.108.090423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin S, et al. The Munc18-1 domain 3a loop is essential for neuroexocytosis but not for syntaxin-1A transport to the plasma membrane. J Cell Sci. 2013;126:2353–2360. doi: 10.1242/jcs.126813. [DOI] [PubMed] [Google Scholar]
- 32.Han GA, Bin NR, Kang SY, Han L, Sugita S. Domain 3a of Munc18-1 plays a crucial role at the priming stage of exocytosis. J Cell Sci. 2013;126:2361–2371. doi: 10.1242/jcs.126862. [DOI] [PubMed] [Google Scholar]
- 33.Jakhanwal S, Lee CT, Urlaub H, Jahn R. An activated Q-SNARE/SM protein complex as a possible intermediate in SNARE assembly. EMBO J. 2017;36:1788–1802. doi: 10.15252/embj.201696270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisotto D, et al. An extended helical conformation in domain 3a of Munc18-1 provides a template for SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex assembly. J Biol Chem. 2014;289:9639–9650. doi: 10.1074/jbc.M113.514273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 36.Weimer RM, et al. Defects in synaptic vesicle docking in unc-18 mutants. Nat Neurosci. 2003;6:1023–1030. doi: 10.1038/nn1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verhage M, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 38.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, et al. The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis. Nature. 2017;548:420–425. doi: 10.1038/nature23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai Y, et al. Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron. 2017;95:591–607.e10. doi: 10.1016/j.neuron.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, et al. Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat Struct Mol Biol. 2015;22:547–554. doi: 10.1038/nsmb.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190:55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen C, et al. The trans-SNARE-regulating function of Munc18-1 is essential to synaptic exocytosis. Nat Commun. 2015;6:8852. doi: 10.1038/ncomms9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.