Significance
Adult mammalian hearts have limited self-renewal capacity. Although mounting evidence indicates that new cardiomyocytes are derived from dedifferentiation and proliferation of existing cardiomyocytes, the contribution of adult cardiac progenitors to cardiomyocyte renewal during homeostasis and upon injury remains under debate. The basic helix–loop–helix transcription factor Twist2 is expressed in interstitial cells in the adult myocardium. Using genetic lineage tracing, we identified a Twist2-expressing cell population that gives rise to a small number of adult cardiomyocytes in vivo. These Twist2-expressing cells can differentiate into cardiomyocytes, endothelial cells, and fibroblasts in culture and contribute to cardiac renewal through cell fusion and de novo differentiation. Our findings add Twist2-expressing cells to the cellular constituents involved in adult cardiac maintenance and remodeling.
Keywords: Twist2, cardiac progenitors, cardiomyocytes, differentiation, cell fusion
Abstract
Twist transcription factors function as ancestral regulators of mesodermal cell fates in organisms ranging from Drosophila to mammals. Through lineage tracing of Twist2 (Tw2)-expressing cells with tamoxifen-inducible Tw2-CreERT2 and tdTomato (tdTO) reporter mice, we discovered a unique cell population that progressively contributes to cardiomyocytes (CMs), endothelial cells, and fibroblasts in the adult heart. Clonal analysis confirmed the ability of Tw2-derived tdTO+ (Tw2-tdTO+) cells to form CMs in vitro. Within the adult heart, Tw2-tdTO+ CMs accounted for ∼13% of total CMs, the majority of which resulted from fusion of Tw2-tdTO+ cells with existing CMs. Tw2-tdTO+ cells also contribute to cardiac remodeling after injury. We conclude that Tw2-tdTO+ cells participate in lifelong maintenance of cardiac function, at least in part through de novo formation of CMs and fusion with preexisting CMs, as well as in the genesis of other cellular components of the adult heart.
Adult mammalian hearts have limited capacity for self-renewal. In the adult mouse, new cardiomyocytes (CMs) are produced at a rate of 1.3–4% per year (1). In humans, only ∼1% of CMs renew each year before age 20 y, declining later in life to 0.4%/y (2). Upon myocardial injury, such as myocardial infarction (MI), the rate of CM turnover increases but is insufficient to offset CM loss, resulting in contractile demise and eventual heart failure (3–6).
Studies combining genetic lineage tracing and radioactive isotope labeling revealed that the few myocytes that are generated after birth arise largely from the proliferation of existing CMs (2, 5, 7, 8), whereas resident c-kit+ cardiac progenitor cells (CPCs) were originally reported to contribute significantly to CM renewal (9). Recent genetic lineage-tracing studies question these conclusions by showing only a minimal contribution of c-kit+ CPCs to CM renewal in the adult heart both during homeostasis and after injury (10–12). Although the contribution of c-kit+ cells to adult CMs appears minimal, a low level of renewal activity from CPCs is detectable, especially during cardiac remodeling after injury (3). In this regard, other types of CPCs have been identified in mice and humans based on the expression of specific cell-surface markers or cellular phenotypes; these include resident Sca1+ CPCs, cardiac side population (SP) cells, WT1+ epicardial-derived cells, Islet1 (Isl1)+ CPCs, endothelial-derived CPCs, and W8B2+ CPCs (9, 13–23). Although most of these CPCs have been reported to contribute to CM self-renewal to various extents, there is no consensus as to the set of markers that specifically identify CPCs, nor is there an understanding of the potential lineage relationships among the CPC populations.
Members of the Twist family of basic helix–loop–helix transcription factors function as ancestral regulators of mesodermal cell fates in organisms ranging from Drosophila to mammals (24–27). In adult Drosophila, Twist expression is restricted to muscle precursors, which are similar to muscle stem cells in vertebrates (28) and are required for the formation of the adult musculature (29). There are two Twist genes in vertebrates, Twist1 (Tw1) and Twist2 (Tw2), which are highly homologous and evolutionarily conserved (30). It has been shown that Twist overexpression can revert the terminal differentiation status of skeletal myotubes to undifferentiated myoblasts (31). Tw2 global-knockout mice failed to thrive and died by postnatal day (P) 15. Before death, homozygous mutant mice were underweight and frail and showed signs of impaired movement and wasting. The mutant mice also showed notable skin abnormalities and severe fat deficiency. A cardiac phenotype was not observed in Tw2 global-knockout mice by P15, possibly due to redundancy with its close family member Tw1 (32). Recently, we discovered an interstitial myogenic progenitor, marked by the expression of Tw2, which gives rise to type IIb/x skeletal muscle fibers (33). Tw1 has also been shown to promote epithelial–mesenchymal transition (EMT), metastasis, and tumor stemness in many cancer models (34–36). Collectively, these studies support the premise that Twist expression influences the stem cell state as well as cell-fate determination.
Within the developing heart, Tw1 controls proliferation, migration, and differentiation of the cardiac cushions (37, 38), but the potential involvement of Twist genes in the adult mouse heart has not been explored. Here, by lineage tracing using inducible Tw2-CreERT2 and tdTomato (tdTO) reporter mice, we discovered a Tw2-tdTO+ cell population that contributes to a subset of CMs as well as endothelial cells (ECs) and fibroblasts. Intriguingly, this cell population fuses with preexisting CMs and, to a lesser extent, gives rise to new CMs de novo. Our findings reveal a unique population of interstitial heart cells that contributes to cardiac homeostasis and regeneration.
Results
Tw2-tdTO+ Cells Contribute to CMs in the Adult Heart.
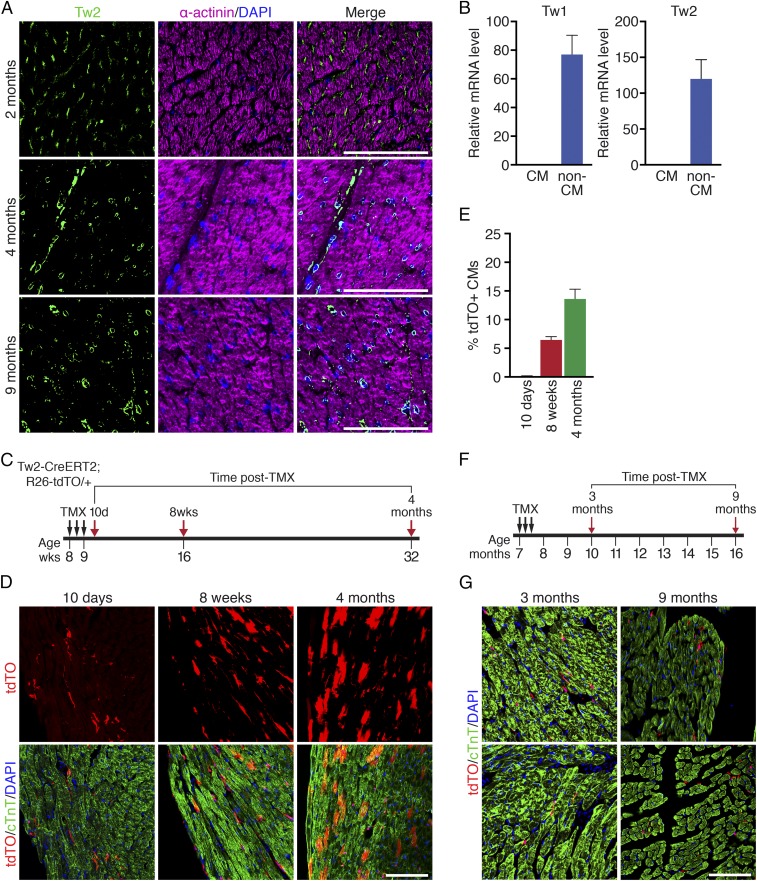
To explore the role of Tw2 in the adult heart, we first examined its expression in the adult mouse myocardium by immunostaining. Tw2 protein was detected in interstitial cells throughout the adult heart but not in CMs at 2–17 mo of age (Fig. 1A and SI Appendix, Fig. S1A). Consistent with these findings, Tw1 and Tw2 mRNA transcripts were readily detected in non-CM populations but not in CM populations separated by Langendorff perfusion of the adult ventricles (Fig. 1B).
Fig. 1.
Progressive contribution of Tw2-tdTO+ cells to adult CMs. (A) Immunostaining of Tw2 (green) and α-actinin (violet) proteins on heart sections of 2-, 4-, and 9-mo-old wild-type mice. (Scale bars: 100 μm.) (B) Expression of Tw1 and Tw2 transcripts in CM-enriched and non–CM-enriched populations in adult mouse heart as detected by real-time RT-PCR (n = 3). (C) Schematic of TMX treatment. Tw2-CreERT2; R26-tdTO mice were injected with 1 mg of TMX on three alternative days at 8 wk of age. Mice were analyzed at 10 d, 8 wk, and 4 mo following the first TMX injection. (D) Progressive tdTO labeling of CMs at the indicated time points following TMX treatment. Sections were costained with cTnT (green) to detect CMs. (Scale bar: 100 μm.) (E) The percentage of Tw2-tdTO+ CMs in ventricles was quantified and averaged at the indicated time points. n = 3 mice for each time point. (F) Schematic of TMX treatment of aged Tw2-CreERT2; R26-tdTO mice. (G) Hearts from aged Tw2-CreERT2; R26-tdTO mice at the indicated times post-TMX treatment were costained with cTnT (green). (Scale bar: 100 μm.) Data in B and E are expressed as mean ± SEM.
To further assess the potential contribution of Tw2-expressing cells to adult tissues, we generated tamoxifen (TMX)-inducible Tw2-CreERT2; R26-tdTO reporter mice (33) in which Tw2-expressing cells and their derivatives were terminally labeled with tdTO. Following treatment of 8-wk-old Tw2-CreERT2; R26-tdTO mice with TMX on three alternative days, we monitored tdTO labeling as a marker for Tw2-expressing cells in the heart at various time points (Fig. 1C). At 10 d post-TMX treatment we observed tdTO labeling of interstitial cells but rarely of CMs in the adult heart ventricles (Fig. 1D). Colocalization of tdTO labeling and Tw2 protein was also observed in interstitial cells at 10 d post-TMX treatment (SI Appendix, Fig. S1B). In addition, tdTO-labeled CMs were significantly more abundant in atria than in ventricles at 10 d post-TMX treatment (SI Appendix, Fig. S1 C–E).
By 8 wk post-TMX treatment strong tdTO labeling was observed in the CM population in both ventricles and atria as well as in the interventricular septum (Fig. 1D and SI Appendix, Fig. S1E). The number of tdTO+ CMs continued to increase for up to 4 mo (Fig. 1D), when ∼13% of CMs in the ventricles were labeled by tdTO (Fig. 1E). No tdTO+ labeling was detected in the hearts of Tw2-CreERT2; R26-tdTO mice without TMX treatment even at 9 mo of age, indicating that there was no leakiness of the reporter (SI Appendix, Fig. S1F).
To determine whether the contribution of Tw2-tdTO+ cells to CMs is age dependent, we performed TMX treatment on 7-mo-old Tw2-CreERT2; R26-tdTO mice (Fig. 1F). At 3 and 9 mo post-TMX treatment, only non-CMs were labeled by Tw2-tdTO in these mice (Fig. 1G). This is in contrast to the observation that in young adult (8-wk-old) mice both CMs and non-CMs were labeled by Tw2-tdTO (Fig. 1D). These findings suggest that Tw2-tdTO+ cells contribute to ventricular CMs in young adult mice at 2 mo of age but lose this ability by 7 mo of age. Furthermore, in comparison with the Tw2-tdTO labeling in CMs of young adult mice (Fig. 1 C and D), our observations in older mice (Fig. 1 F and G) indicate that tdTO labeling in CMs at 4 mo post-TMX treatment is not due to Tw2 expression in adult CMs.
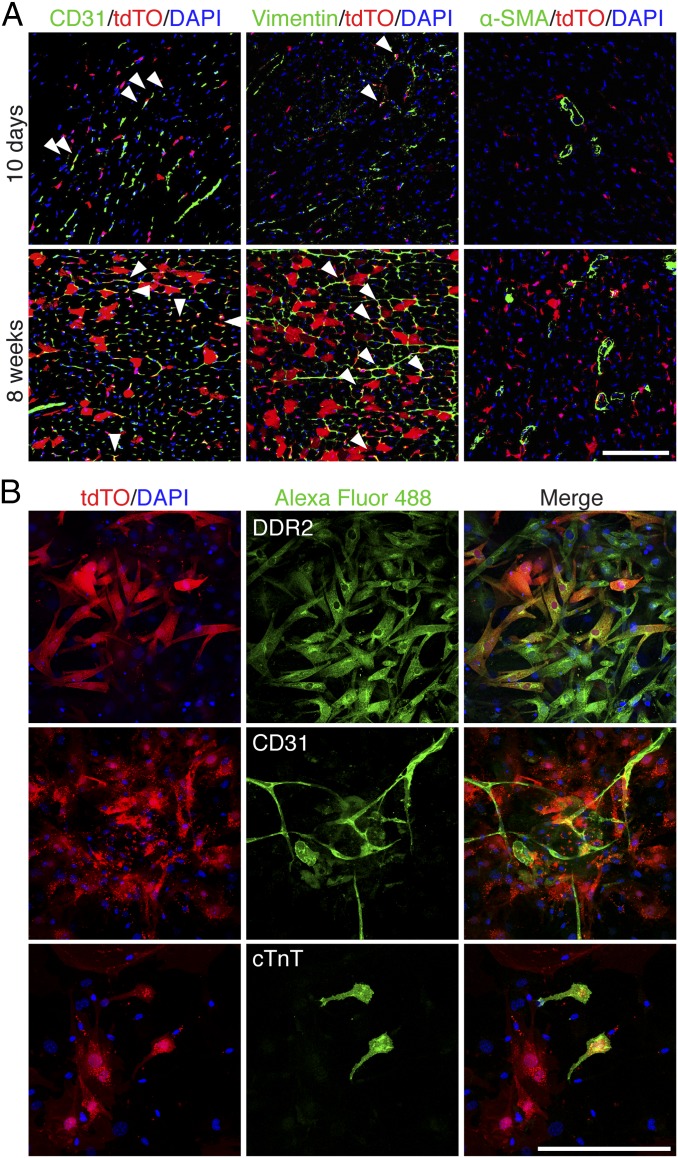
In addition to labeling CMs, ∼7.5% of non-CMs were Tw2-tdTO+ in adult ventricles at 10 d post-TMX treatment and by 4 mo post-TMX treatment ∼20.3% of total non-CMs were Tw2-tdTO+ (SI Appendix, Fig. S1G), indicating a substantial contribution of Tw2-tdTO+ cells to lineages other than CMs. Specifically, Tw2-tdTO–labeled ECs and fibroblasts but not smooth muscle cells, as evidenced by costaining with cell-specific markers at 10 d post-TMX treatment (Fig. 2A). At 8 wk post-TMX treatment, tdTO labeling continued to be observed in ECs and fibroblasts, suggesting that Tw2 tdTO+ cells gradually contribute to these cell lineages.
Fig. 2.
Tw2-tdTO+ cells express markers of fibroblasts and ECs but not smooth muscle cells. (A) Ventricles from 10 d (Upper) and 8 wk (Lower) post-TMX treatment were immunostained with antibodies against CD31, vimentin, or smooth muscle α-actin (α-SMA). White arrowheads indicate colocalization with tdTO signals. (Scale bar: 100 μm.) (B) Ventricles from 10 d post-TMX treatment were dissociated, and the non–CM-enriched population was cultured for a week and immunostained with antibodies against DDR2 (Upper), CD31 (Middle), or cTnT (Lower). (Scale bar: 100 μm.)
Differentiation Potential of Tw2-tdTO+ Cells in Vitro.
To study their differentiation potential in vitro, Tw2-tdTO+ cells were isolated together with other non-CMs from ventricles of adult Tw2-CreERT2; R26-tdTO/+ mice at 10 d post-TMX treatment. Tw2-tdTO+ cells in regular culture medium spontaneously differentiated into fibroblasts, as indicated by the expression of DDR2, a fibroblast marker (Fig. 2B). Under culture conditions that favor EC formation, Tw2-tdTO+ cells differentiated into ECs and expressed the EC marker CD31 (Fig. 2B). Tw2-tdTO+ cells differentiated into CMs when maintained in CM induction medium, as identified by the expression of the CM marker cardiac troponin T (cTnT) (Fig. 2B). Some of the differentiated CMs began beating after 2 wk of culture (SI Appendix, Fig. S2 and Movie S1). These results are consistent with the cardiogenic potential of Tw2-tdTO+ cells in vivo.
Given our prior findings that Tw2-tdTO+ cells within the interstitial region of skeletal muscle can form skeletal muscle and bone cells under appropriate conditions (32), we also exposed Tw2-tdTO+ cells isolated from the heart to culture conditions that favor the formation of myotubes and osteoblasts. However, we observed no myotube or osteoblast formation, suggesting that the potential fates of Tw2-tdTO+ cells differ depending on the tissue niche and/or lineage of origin.
Tw2-tdTO+ Cells Contribute to CMs by both Fusion and de Novo Differentiation.
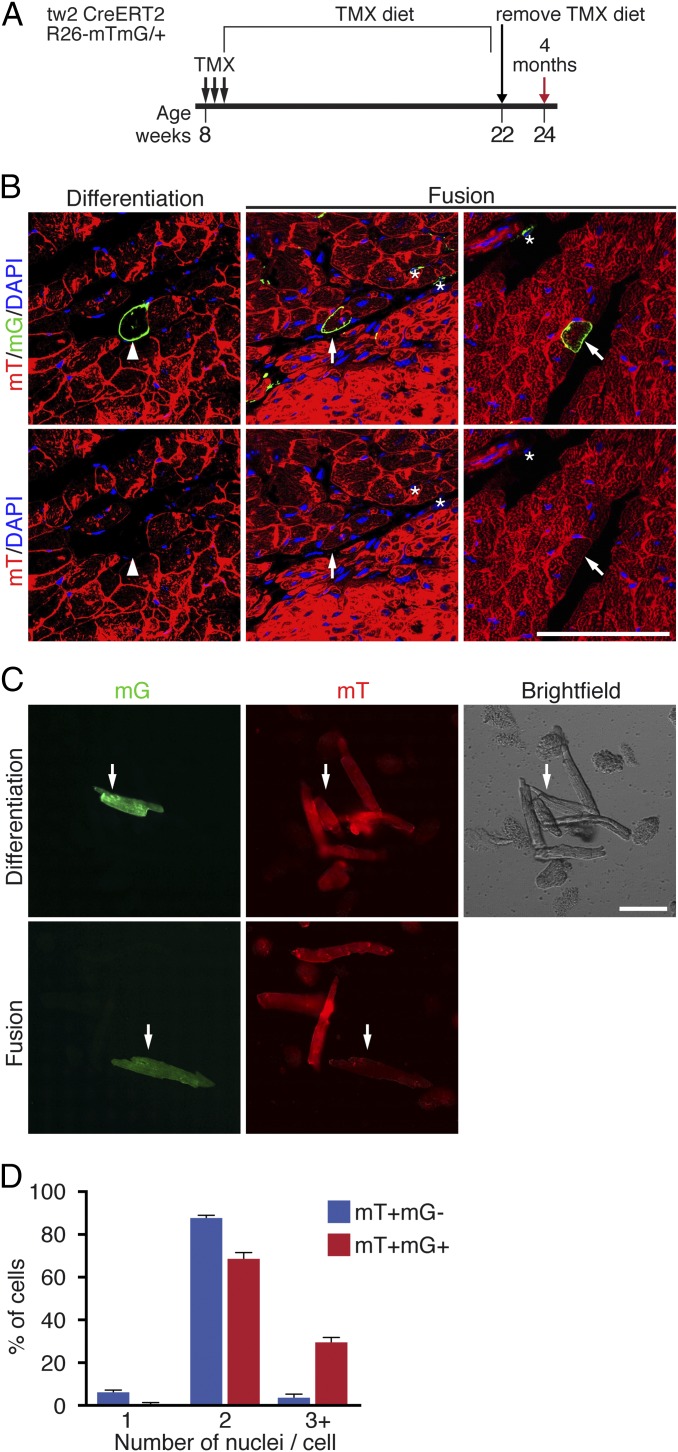
The progressive labeling of CMs in the young adult heart (Fig. 1D) could result from either de novo generation of CMs from Tw2-tdTO+ cells or fusion of Tw2-tdTO+ cells with existing CMs. To distinguish between these possibilities, we generated Tw2-CreERT2; ROSA26-mT/mG/+ reporter mice by breeding the Tw2-CreERT2 mice with Rosa26-mT/mG mice. The R26-mT/mG mice harbor a membrane Tomato (mT) cassette and a polyA signal flanked by two loxP sites at the Rosa26 locus, followed by a membrane eGFP (mG) cassette (39). In the absence of Cre activity, every cell in the Tw2-CreERT2; R26-mT/mG reporter mice expresses mT but not mG. Upon Cre activation by TMX, the mT cassette is excised, resulting in mG expression and loss of mT expression. Therefore, if a Tw2-expressing interstitial cell fuses with an existing CM, both the mT and mG signal will be present (SI Appendix, Fig. S3). In contrast, if a Tw2-expressing cell differentiates de novo into a CM, that CM would be mG+ and mT− (SI Appendix, Fig. S3).
To activate Cre expression, Tw2-CreERT2; R26-mTmG mice were treated with three doses of TMX at 8 wk of age and were kept on a TMX-containing diet for 14 wk. Two weeks before the hearts were harvested, the TMX diet was removed (Fig. 3A). As shown in Fig. 3B, the majority of mG+ CMs retained mT expression, indicating fusion of Tw2-mG+ interstitial cells with existing mT+ CMs. To a lesser extent, we observed mG+ CMs without mT expression, indicating de novo formation of CMs from a Tw2-mG+ interstitial cell (Fig. 3B). All mG+ non-CMs were mT−, validating the fidelity of the labeling strategy (Fig. 3B).
Fig. 3.
Tw2-tdTO+ cells fuse with CMs in adult hearts. (A) Schematic of TMX treatment of Tw2-CreERT2; R26-mT/mG mice. (B) Representative images of differentiation and fusion events detected in the hearts of Tw2-CreERT2; R26-mT/mG mice. (Left) The mG+ CMs no longer express the mT signal, indicating de novo CM formation (white arrowhead). (Center) The Tw2-mG+ CMs are mT− (white asterisks). (Right) The Tw2-mG+ CMs retain mT expression, indicating that Tw2-mG+ non-CMs fused with existing CMs (white arrow). Nuclei were stained with DAPI (blue). (Scale bar: 100 μm.) (C) CMs from ventricles of the Tw2-CreERT2; R26-mT/mG mice were dissociated and plated in culture. (Upper) A de novo CM that is mG+ but mT−. (Lower) Evidence of a fusion event in which the CM is mG+ and mT+ double positive. (Scale bar: 100 μm.) (D) Percent of cells that contain 1, 2, or >3 nuclei among all mT+ and mG− CMs or mT+ and mG+ CMs. n = 3 mice. Data in D are expressed as mean ± SEM.
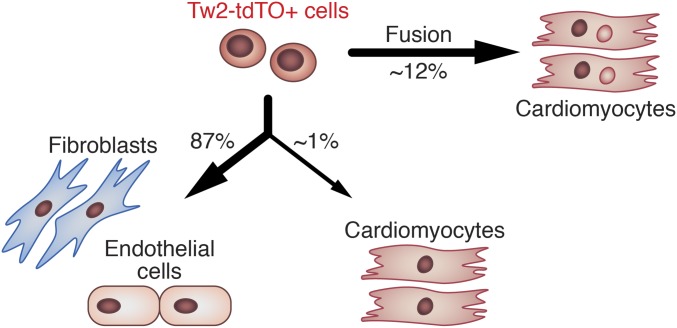
To quantify the frequency of differentiation versus fusion events, hearts from Tw2-CreERT2; R26-mT/mG mice were dissociated by Langendorff perfusion, and CMs of ventricles were plated in culture. mG+/mT− and mG+/mT+ CMs were imaged and quantified (Fig. 3C). Among the mG+ CMs, 11% had lost mT expression, indicating that they were derived from de novo differentiation, whereas 89% of mG+ CMs were derived from fusion, as they retained mT expression. We further quantified the number of nuclei in the fusion-derived mG+/mT+ CMs and found that 99.3% of mT and mG double-positive cells had more than one nucleus, with 69.2% having two nuclei and 30.1% having three or more nuclei per cell (Fig. 3D). The percentage of cells with three or more nuclei was 6.9-fold higher in mG+/mT+ CMs than in mG−/mT+ cells. These results suggest that the majority of Tw2-mG+ cells fuse with CMs in young adult mice and that a small subset of these Tw2-mG+ can differentiate into CMs.
Cardiogenic Potential of Single Tw2-tdTO+ Cells.
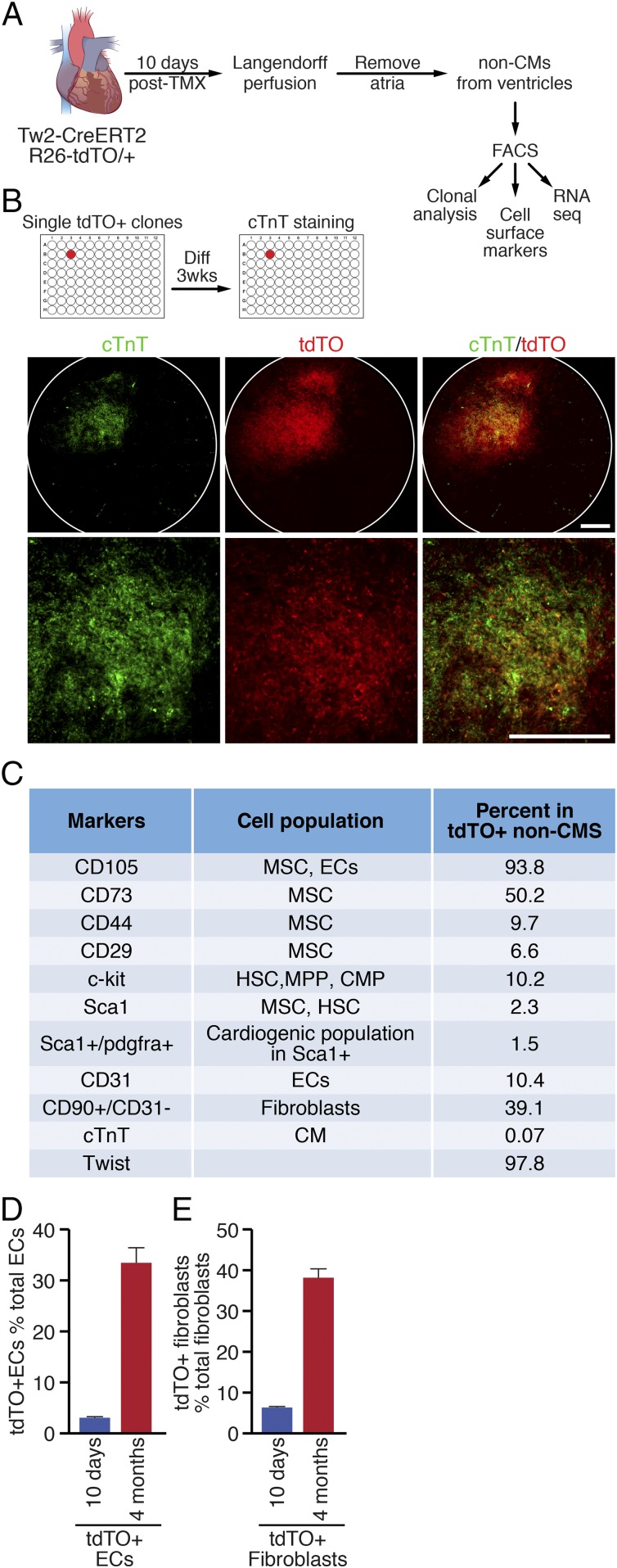
The finding that a small but significant number of CMs were derived de novo from Tw2-tdTO+ cells suggests that Tw2-tdTO+ cells in the adult heart include a potential cardiogenic population. To determine whether single Tw2-tdTO+ cells can differentiate into CMs, we performed clonal analysis of Tw2-tdTO+ cells. Briefly, Tw2-CreERT2; R26-tdTO/+ mice were treated with TMX at 8 wk of age as described in Fig. 1C. Ten days after the first dose of TMX, cells from the heart ventricle were dissociated, and Tw2-tdTO+ interstitial cells were isolated by FACS (Fig. 4A). To avoid contamination of CMs in the isolation, we stained cells with the mitochondrial dye Mitoview 633, which preferentially labels CMs because of their high mitochondria content. Single clones of Tw2-tdTO+ interstitial cells, identified as tdTO+ and Mitoview 633low, were sorted into individual wells on 96-well plates and were cocultured with murine embryonic fibroblasts (MEFs) (Fig. 4 A and B and SI Appendix, Fig. S4A). Tw2-tdTO+ clones were then cultured for 3 wk in growth medium before being switched to differentiation medium to induce cardiogenesis (Fig. 4B). CMs derived from individual Tw2-tdTO+ clones were identified by immunostaining for cTnT and were captured by high-throughput confocal microscopy. Of 4,512 Tw2-tdTO+ clones, 43 were positive for cTnT staining (Fig. 4B), while other clones were negative for cTnT (SI Appendix, Fig. S4B), demonstrating that 0.95% of the Tw2-tdTO+ single clones differentiate into CMs in vitro. Taken together with the in vivo observations, these findings suggest that a subset of Tw2-tdTO+ cells has the potential to differentiate into CMs.
Fig. 4.
Clonal and marker analysis of Tw2-tdTO+ cells. (A) Schematic of the analysis of Tw2-tdTO+ cells in vitro. Following Langendorff perfusion and FACS isolation, Tw2-tdTO+ cells from ventricles were subjected to cell-surface marker analysis and RNA-seq. In addition, single Tw2-tdTO+ clones were plated on 96-well plates for clonal analysis. (B, Upper) Representative images of a well containing cTnT+ cells from clonal analysis. The images were acquired using an IN Cell high-throughput confocal microscope under a 4× lens. (Lower) Magnified images. (Scale bars: 100 μm.) (C) Summary of cell marker expression of Tw2-tdTO+ cells by FACS analysis. Hematopoietic stem cell (HSC), multipotent progenitor (MPP), and common myeloid progenitor (CMP). (D) Quantification of Tw2-tdTO+ ECs as a percentage of total cardiac ECs at 10 d and 4 mo post-TMX treatment as determined by FACS analysis. n = 3 mice for each time point. (E) Quantification of Tw2-tdTO+ fibroblasts as a percentage of total cardiac fibroblasts at 10 d and 4 mo post-TMX treatment as determined by FACS analysis. n = 3 mice for each time point. Data in D and E are expressed as mean ± SEM.
Molecular Signature of Tw2-tdTO+ Cells.
To further define the molecular identity of Tw2-tdTO+ cells, we examined by FACS analysis the expression of cell-surface markers on Tw2-tdTO+ cells freshly isolated from 8-wk-old Tw2-CreERT2; R26-tdTO/+ mice at 10 d post-TMX treatment (Fig. 4A). Tw2-tdTO+ cells were identified as tdTO+ and Mitoview 633low cells (SI Appendix, Fig. S4A). FACS analysis revealed that the CPC marker c-kit was expressed in 10.2% of Tw2-tdTO+ cells, while Sca1 was expressed in 2.3% of the cells. In addition, consistent with the in vivo findings, 10.4% of the Tw2-tdTO+ cells expressed the EC marker CD31, and 39.1% of them were identified as fibroblasts by CD90+ and CD31− surface maker staining (Fig. 4C).
To confirm the fidelity of the FACS analysis, we immunostained Tw2-tdTO+ cells for cTnT and Twist at 10 d post-TMX treatment and found that 97.8% of the Tw2-tdTO+ cells were Twist+ and 99.93% were cTnT− (SI Appendix, Fig. S4C). Furthermore, 93.8% of the Tw2-tdTO+ cells expressed the mesenchymal stem cell (MSC) marker CD105, while other MSC markers, such as CD73, CD44, and CD29, were expressed at varying levels in Tw2-tdTO+ cells (Fig. 4C and SI Appendix, Fig. S4D). Multicolor FACS analysis using an MSC antibody panel revealed that 1.2% of the Tw2-tdTO+ cells expressed the consensus MSC markers CD105, CD29, and Sca-1 but not CD45 (SI Appendix, Fig. S4E). MSCs have the potential to differentiate into CMs both in vitro and in vivo (40, 41). Therefore, it is possible that the cardiogenic population of Tw2-tdTO+ cells represents a subset of resident MSCs.
We also quantified the contribution of Tw2-tdTO+ cells to ECs and fibroblasts over time by FACS analysis. At 10 d post-TMX treatment, 3.1% of the Tw2-tdTO+ cells were identified as ECs, and the number increased to 33.4% at 4 mo post-TMX treatment (Fig. 4D). In addition, Tw2-tdTO+ cells accounted for 6.4% of all cardiac fibroblasts at 10 d, and the population increased to 38.1% at 4 mo post-TMX treatment (Fig. 4E). We conclude that Tw2-tdTO+ cells contribute to a substantial proportion of ECs and fibroblasts in the heart. However, since 3.1% of ECs and 6.4% of cardiac fibroblasts are labeled by tdTO at 10 d post-TMX treatment, we cannot exclude the possibility that Tw2 is expressed in a small population of ECs and fibroblasts which give rise to more ECs and fibroblasts over time.
Single-Cell RNA Sequencing of Tw2+ Cells.
To determine whether Tw2-tdTO+ cells are a homogenous or heterogeneous cell population, we performed single-cell RNA-sequencing (scRNA-seq) on the Tw2-tdTO+ cells. Tw2-tdTO+ cells were isolated by FACS based on the expression of tdTO from ventricles of Tw2-CreERT2; R26-tdTO/+ mice at 10 d post-TMX treatment. Graphic clustering based on transcriptional similarities revealed four main clusters of 232 Tw2-tdTO+ cells, indicating that they are a heterogeneous population (SI Appendix, Fig. S5A). Each cluster exhibited distinct gene-expression patterns (SI Appendix, Fig. S5B), which could be categorized as ECs, fibroblasts, immune cells, and smooth muscle cells based on the top 10 marker genes. Further clustering using MSC cell markers revealed that a small population of Tw2-tdTO+ cells expresses MSC marker genes, correlating with the results of FACS analysis (Fig. 4C and SI Appendix, Figs. S4 D and E and S5C).
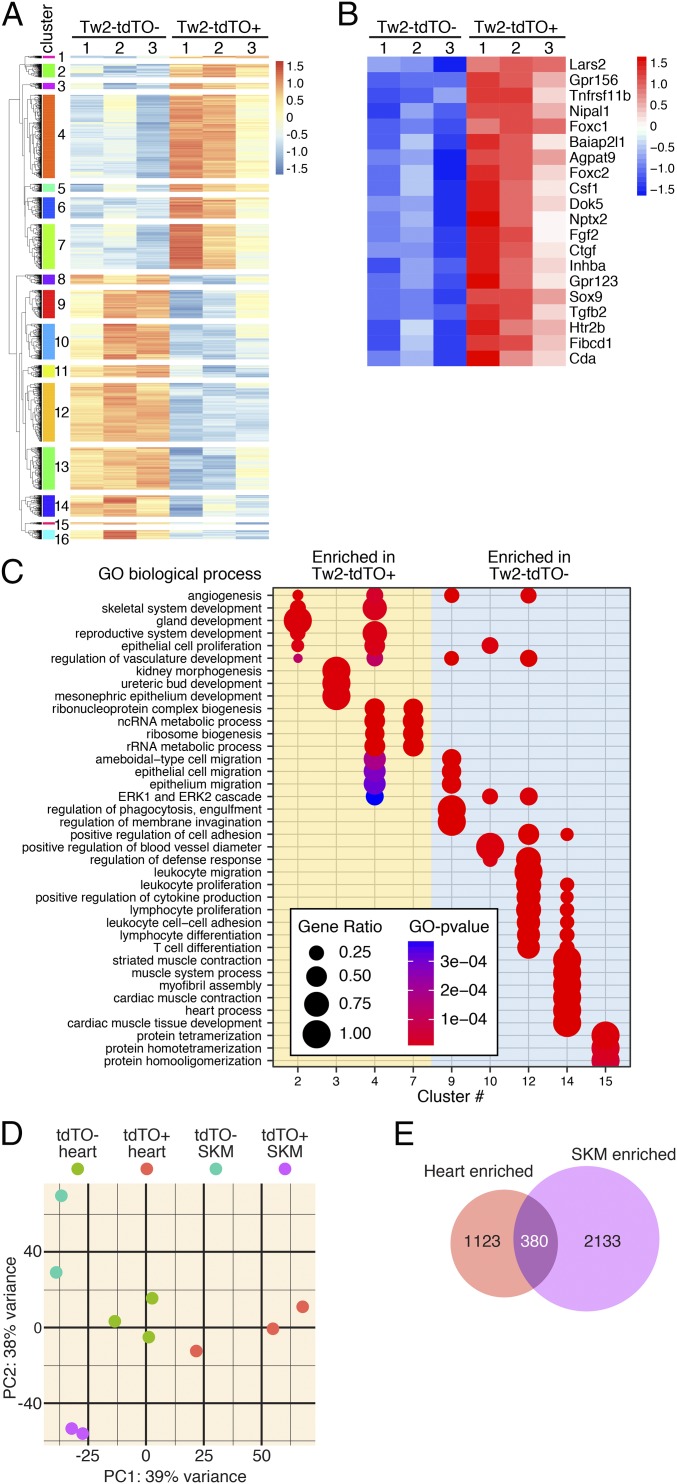
To determine the molecular signature of Tw2-tdTO+ cells, we performed unbiased transcriptome profile analysis by RNA-seq. Tw2-tdTO+ and Tw2-tdTO− cells were isolated by FACS based on the expression of tdTO from ventricles of Tw2-CreERT2; R26-tdTO/+ mice at 10 d post-TMX treatment (SI Appendix, Fig. S6A). Transcriptome analysis revealed that Tw2-tdTO+ cells and Tw2-tdTO− cells displayed highly distinct gene-expression patterns (Fig. 5A). Bioinformatics analysis identified 3,365 genes that were differentially expressed between the two populations, with 1,502 genes being enriched in Tw2-tdTO+ cells and 1,863 genes being down-regulated (Fig. 5A). A list of the top 20 enriched genes in Tw2-tdTO+ cells is shown in Fig. 5B, including the known Tw2 target genes Ctgf, Htr2b, and Sox9, the EMT regulators Foxc1, Foxc2, and Tgfb2, and Fgf2, an upstream regulator of Tw2 (Fig. 5B). Sixteen gene-expression clusters were assigned according to the hierarchy correlations of the differentially expressed genes (Fig. 5A). These clusters were functionally annotated using Gene Ontology (GO) analysis and compared using clusterProfiler (Fig. 5C). Clusters 2–7 represent the enriched genes in the Tw2-tdTO+ cells, and clusters 9–15 represent the enriched genes in the Tw2-tdTO− cells. Clusters for enriched genes in Tw2-tdTO+ cells were grouped into several GO biological functions (Fig. 5C). Angiogenesis-, mesenchymal tissue development-, and epithelial cell proliferation-related genes were enriched in clusters 2 and 4. In addition, cluster 4 includes genes involved in epithelial cell migration. These results are consistent with the known functions of Tw2 in these pathways. Cluster 3 represents kidney morphogenesis- and renal developmental-related genes, which is consistent with reports showing that Tw1 activation is a common mechanism in the pathophysiology of a wide range of chronic renal diseases (42, 43). Ribosomal RNA processing- and ribosome biogenesis-related genes were represented in clusters 4 and 7. Enriched genes in the Tw2-tdTO− cells correlated with immune cell activation, migration, and proliferation pathways, as depicted in clusters 9, 10, 12, and 14 (Fig. 5C).
Fig. 5.
Transcriptome analysis of Tw2-tdTO+ cells in adult heart. (A) Heat map of 3,365 genes in freshly sorted Tw2-tdTO+ and Tw2-tdTO− cells in the heart as identified by RNA-seq. Cells were isolated by FACS from ventricles of Tw2-CreERT2; R26-tdTO mice at 10 d post-TMX treatment. n = 3 for each sample. Sixteen clusters of genes were identified and are indicated on the left. (B) The top 20 most enriched genes in Tw2-tdTO+ cells, as identified by RNA-seq. (C) The GO enrichment by differentially expressed genes in Tw2-tdTO+ and Tw2-tdTO− cells analyzed using the clusterProfiler R package. The size of the circle corresponds to the number of genes enriched for a particular GO term (gene ratio). The enrichment term is represented by colored dots (red indicates high enrichment, and blue indicates low enrichment). (D) PCA of Tw2-tdTO+ and Tw2-tdTO− cells in skeletal muscle (SKM) and hearts from Tw2-CreERT2; R26-tdTO mice identified by RNA-seq. (E) The Venn diagram indicates overlapping genes and genes that are differentially in Tw2-tdTO+ cells in heart and skeletal muscle.
Ingenuity Pathway Analysis (IPA) also revealed that genes with enriched expression in Tw2-tdTO+ cells were involved in EIF2 signaling, mTOR signaling, embryonic stem cell pluripotency, EMT, and cancer metastasis, consistent with the known functions of Tw2 in these pathways (SI Appendix, Fig. S6A). GO analysis revealed similar pathways for genes with enriched expression in Tw2-tdTO+ cells (SI Appendix, Fig. S6B).
Comparison of Cardiac and Skeletal Muscle Tw2+ Cells.
Within adult skeletal muscle, Tw2 marked an interstitial myogenic progenitor population that contributes to type IIb/x myofibers (33). To determine the similarity of Tw2-tdTO+ cells in heart and skeletal muscle, we compared the transcriptome of these two cell populations and found that they were significantly different by principal component analysis (PCA) (Fig. 5D). Whereas 380 genes were enriched in Tw2-tdTO+ cells from both heart and skeletal muscle, 1,123 genes were enriched only in cardiac Tw2-tdTO+ cells, and 2,133 genes were expressed only in Tw2-tdTO+ cells from skeletal muscle. These results are consistent with our in vitro differentiation assays and indicate that Tw2-tdTO+ cells from the heart and skeletal muscle are largely distinct populations derived from different origins. Interestingly, IPA analysis of the 380 genes that were coexpressed in the two Tw2-tdTO+ populations revealed that they are involved in pathways associated with stemness, such as cancer metastasis, embryonic stem cell pluripotency, and cardiogenesis (SI Appendix, Fig. S6C). Since Tw2-tdTO+ cells have fusogenic potential, we looked for known muscle cell fusion-related genes, such as myomaker and myomixer in the Tw2-tdTO− and Tw2-tdTO+ populations. No significant differences in expression of myomaker and myomixer were detected between the two populations (SI Appendix, Fig. S6D).
Tw2-tdTO+ Cells Contribute to Cardiac Remodeling After Myocardial Infarction.
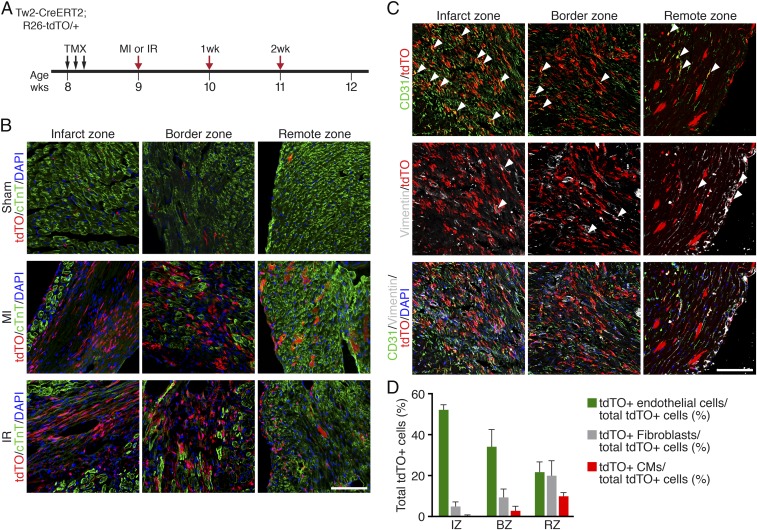
Our results showed that Tw2-tdTO+ cells give rise to CMs in the young adult heart. To determine whether Tw2-tdTO+ cells contribute to cardiac remodeling after injury, we performed permanent ligation of the left anterior descending (LAD) artery to induce MI in 9-wk-old Tw2-CreERT2;R26-tdTO mice that had been treated with TMX 1 wk before surgery, as schematized in Fig. 6A. One week post-MI, Tw2-tdTO+ cells were strongly activated at the infarct and border zones of infarcted hearts (Fig. 6B), and there were more Tw2-tdTO+ CMs in the remote zone following MI than in mice treated with sham surgery (Fig. 6B and SI Appendix, Fig. S7A). We also performed transient LAD artery ligation followed by release of the ligation to induce ischemia–reperfusion (I/R) injury and observed similar accumulation of Tw2-tdTO+ interstitial cells and CMs at 1 wk postinjury (Fig. 6B). Injury-induced Tw2-tdTO+ interstitial cells expressed markers of ECs (CD31) or of fibroblasts (Vimentin) in the infarct zone, border zone, and remote zone of MI or I/R hearts (Fig. 6C and SI Appendix, Fig. S7B). More Tw2-tdTO+ cells differentiated into ECs than into fibroblasts after injury (Fig. 6 C and D). Tw2-tdTO+ ECs were more abundant in the infarct and border zones than in the remote zone (Fig. 6 C and D). Tw2-tdTO+ cells continued to accumulate in the infarct zone of MI or I/R hearts at 2 wk postinjury (SI Appendix, Fig. S7C). Together, these results indicate that Tw2-tdTO+ cells participate in cardiac remodeling during injury.
Fig. 6.
Tw2-tdTO+ cells contribute to cardiac remodeling after MI. (A) Schematic of TMX treatment and LAD artery ligation to induce MI or I/R in Tw2-CreERT2; R26-tdTO mice. (B) Different areas of the heart after MI and I/R were immunostained with cTnT (green). Shown are the infarct zone (Left), border zone (Center), and remote zone (Right) 1 wk after MI. (Scale bar: 100 μm.) (C) One week after MI different areas of the heart were costained with vimentin (gray) and CD31 (green). White arrowheads indicate colocalization with tdTO signals. Shown are the infarct zone (Left), border zone (Center), and remote zone (Right) after 1 wk of MI. (Scale bar: 100 μm.) (D) Quantification of tdTO+ ECs, fibroblasts, and CMs as a percentage of total of total tdTO+ cells in the infarct zone (IZ), boarder zone (BZ), and remote zone (RZ) of the heart 1 wk after MI. n = 3. Data are expressed as mean ± SEM. (D) Quantification of Tw2-tdTO+ ECs, fibroblasts, and CMs as a percentage of total tdTO+ cells 1 wk after MI in Tw2-CreERT2; R26-tdTO mice. n = 3. Data are expressed as mean ± SEM.
Discussion
Using genetic lineage tracing, we identified a previously unrecognized population of cardiogenic cells in the adult heart marked by the expression of Tw2. Tw2-tdTO+ cells can contribute to CMs, ECs, and fibroblasts in vivo and in vitro (Fig. 7). We did not exclude the possibility that the population of Tw2-tdTO+ cells contains a subset of ECs and fibroblasts before 10 d post-TMX treatment, thus continuously giving rise to subsequent EC and fibroblast populations. Although the majority of Tw2-tdTO+ cells contribute to CMs via fusion, a small subset of Tw2-tdTO+ cells can differentiate into CMs in vivo and in vitro, confirming their cardiogenic potential. Tw2-tdTO+ cells are activated in response to MI and I/R injury. Tw2-tdTO+ cells appear to represent a subset of MSCs and are distinct from other CPCs that have been reported to give rise to CMs, including Sca1+, c-kit+, SPs, and endothelial-derived CPCs.
Fig. 7.
Model for the role of Tw2-tdTO+ cells in heart maintenance. In the adult heart, Tw2-tdTO+ cells can give rise to CMs and maintain a group of fibroblasts and ECs. Tw2-tdTO+ cells contribute to CMs mainly by fusion.
Comparison with Other CPCs.
There is intense debate in the field as to whether adult CPCs exist and make functional contributions to the adult heart (10, 11, 44). Sca1-derived cells have been reported to make a substantial and lasting contribution to CMs during normal aging (17). These cells are a source for CM renewal rather than fusion. Sca1+ cells were further subdivided based on marker gene expression, among which the PDGFRα+/Sca1+ subgroup displayed both cardiogenic potential and clonogenicity (18). In addition, ECs retain cardiogenic potential in specific areas of the adult heart (23). Endothelial-derived CPCs express GATA4 and Sca1, and the majority of Sca1+ CPCs are derived from cells with endothelial characteristics, indicating substantial overlap between these CPC populations (23). Tw2-tdTO+ cells from the adult heart share similarities with these cells in terms of cardiogenic potential and clonogenicity. However, based on FACS analysis, less than 3% of Tw2-tdTO+ cells express Sca1 and Sca1/PDGFRα, and only 10% are CD31+. In addition, our study shows that Tw2-tdTO+ cells contribute to 13% of CMs after 4 mo of lineage tracing, which is a higher level than reported by lineage tracing with other CPC markers (10, 17). These results indicate that Tw2-tdTO+ cells are distinct from Sca1+, c-kit+, and EC-derived CPCs. Our study also showed that Tw2-tdTO+ cells are a heterogeneous population, as assessed by FACS analysis and scRNA-seq. These results indicated that there is a potential subpopulation within the Tw2-tdTO+ population that contributes to specific cell types. MSCs have been shown to differentiate into CMs both in vitro (40) and in vivo (41). In humans, a novel population of MSCs marked by expression of W8B2 has been found in adult atrial appendages (22). These W8B2+ CPCs are clonogenic, are capable of self-renewal in culture, and can give rise to several cell lineages within the heart, including CMs, ECs, and smooth muscle cells (22). It will be interesting to examine whether Tw2-tdTO+ cells also exist in humans and whether there is overlap between Tw2-tdTO+ cells and W8B2+ CPCs.
Fusion and Ploidy.
While adult CMs are generally multinucleated, the potential contributions of fusion versus nuclear division without cytokinesis to this phenomenon are not fully understood. A recent study reported transient membrane fusion events in the zebrafish heart (45). Fusion between CMs and resident cells or CMs within the adult heart has not been characterized in mice, possibly due to the lack of knowledge of the cell types and/or markers for this event. Our findings show that resident Tw2-tdTO+ cells can fuse with existing CMs during normal aging in mice. Interestingly, fusion of Tw2-tdTO+ cells with CMs occurs only in young adult but not in aged mice. Cell fusion can provide specific proteins or change gene expression of CMs during aging. It would be interesting to understand the downstream gene-expression differences between the Tw2-tdTO+ cells in 2-mo-old and 7-mo-old mouse hearts, which may provide insights into the potential mechanism for CM maintenance under homeostasis in young and old adult hearts. Cell fusion can offer substantial beneficial effects in the setting of heart disease. For example, transplanted bone marrow cells have been shown to fuse with CMs to rescue them from apoptosis and restore cardiac function following ischemic injury (46–48). Thus, enhancing cell fusion may provide a means of preserving CMs following MI. Since Tw2 tdTO+ cells are activated following MI, they might play a role in maintaining CM survival and/or function after injury.
Twisting Cell Fate.
Twist proteins can influence cell-fate decisions by inhibiting lineage-specific transcription factors, such as MyoD in the muscle lineage (49). Twist proteins can also reverse the terminal differentiation of myotubes and promote their proliferation to become undifferentiated myoblasts (33, 50). RNA-seq analysis confirmed that Tw2 overexpression inhibits myogenesis and activates genes involved in cell-cycle regulation, cancer metastasis, many signaling pathways, and EMT (33). Generating heart-specific Tw2 and/or Tw1 conditional-knockout mice would reveal the cardiac lineage contribution of Twist proteins. In addition, Twist has been reported to inhibit chondrogenesis (51) and osteogenesis (52) but to promote adipogenesis (53). Similar results were observed when Twist proteins were overexpressed in human bone marrow MSCs. MSCs overexpressing Tw1 and Tw2 showed a higher proliferation rate and extended life span and maintained an immature mesenchymal precursor phenotype (53). In this study, Tw2 was used as a marker for lineage tracing in the heart. However, given the involvement of Twist proteins in differentiation of several MSC lineages, it is conceivable that Tw2 plays a role in regulating cardiac lineage and CM homeostasis in adult mice.
Materials and Methods
Mouse Lines.
Tw2-CreERT2 mice were previously described (33). R26-tdTO [(ROSA)26Sortm14(CAG-tdTomato), stock no. 007914], and ROSA26-mT/mG [(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J, stock no. 007576)] mice were obtained from the Jackson Laboratory. Both male and female adult mice were used in the studies. Mice were maintained on a mixed genetic background. Mice were maintained in 12-h light:dark cycles (light: 6:00 AM to 6:00 PM) at 22 °C. Animal work described in this paper was approved by and conducted under the oversight of the University of Texas Southwestern Institutional Animal Care and Use Committee.
Primary Cultures.
Tw2-CreERT2;R26-tdTO mice were treated with TMX at 8 wk of age. At 10 d after the first dose of TMX, a pool of cardiac cells from ventricles (the atria were removed) was isolated using Langendorff perfusion. Cells were then further separated by gravity and a BSA gradient into CM-enriched and non–CM-enriched populations. Staining with Mitoview 633 (Biotium) was performed on the non-CM population, which was then sorted on a BD Aria cell sorter based on tdTO and Mitoview633 signals. Tw2-TdTO+ cells and Tw2-TdTO− cells were harvested for subsequent culturing, staining, clonal analysis, or RNA-seq analysis.
Tw2-tdTO+ cells were isolated by FACS sorting as the tdTO+ and Mitoview 633low population. Cells were maintained in cardiac stem cell maintenance medium (catalog no. SCM101; Millipore). For CM differentiation, cells were switched to cardiomyocyte differentiation medium (catalog no. SCM102; Millipore). For EC differentiation, cells were cultured in EGM-2 BulletKit EC growth medium (catalog no. CC-3162; Lonza). For culturing fibroblasts, cell were maintained in DMEM and 10% FBS.
TMX Treatment.
Mice were randomly divided into treatment groups while satisfying the criterion that the average body weight in each group would be about the same. TMX (Sigma-Aldrich) was dissolved at 10 mg/mL in a mixture of sesame oil and ethanol (9:1) and was administered by i.p. injection as schematized in the figures. Tw2-CreERT2; R26-mTmG mice were placed on a TMX-containing diet (250 mg/kg) (Harlan Laboratories) immediately following TMX injection.
Histology and Immunohistochemistry.
For immunohistochemistry, hearts were harvested and fixed in 4% paraformaldehyde at 4 °C for 1 h. Tissues were then switched to 10% sucrose/PBS overnight followed by 18% sucrose/PBS at 4 °C overnight before they were frozen, embedded, and sectioned. For immunostaining, slides were rinsed three times in PBS and blocked in 10% goat serum for 20 min followed by three rinses in PBS. This was followed by overnight incubation with primary antibodies (see SI Appendix, Table S1 for antibody information). The following day, slides were washed three times in PBS and incubated with anti-mouse and anti-rabbit secondary antibodies conjugated to Alexa Fluor 488 or 555 (1:400 dilution; Invitrogen) for 1 h at room temperature. Slides were washed three times in PBS and mounted in VECTASHIELD containing DAPI (Vector Laboratories). For a list of primary antibodies, please refer to SI Appendix, Table S1.
FACS.
Following isolation, mononuclear cells were resuspended in PBS/2% BSA at 1 × 106 cells/50 µL, aliquoted into 100 µL per tube, and incubated on ice for 1 h with Fc blocking agent and one of the following fluorophore-conjugated antibodies: CD73-Alexa Fluor 488 rat IgG2A (1:50), CD44-APC rat IgG2B (1:50), CD117-APC rat IgG2B (1:50), CD31-APC rat IgG2A (1:50), CD90.2-APC rat IgG2A (1:50), Ly-6A/E-APC-Cy7 rat IgG2A (1:20) (all from BD Biosciences-Pharmingen), CD105-APC rat IgG2A (1:50; BioLegend), pdgfra-Alexa Fluor 488 goat IgG (1:50; R&D Systems), CD45-PerCP rat IgG2B (1:20; R&D Systems), CD29-APC hamster IgG (1:20; eBioscience), or CD105/Endoglin-CFS rat IgG2A (1:20; R&D Systems). Isotype-specific controls were also performed on these cells. Unstained cells isolated from Cre+ and Cre− mice were used as unstained controls. Cells were analyzed on a FACSCalibur (BD) flow cytometer, and FACS data were analyzed using FlowJo Software (Tree Star). For a list of primary antibodies, please refer to SI Appendix, Table S1.
Immunostaining of Cultured Cells.
Cells grown on plates or chamber slides were fixed with 4% paraformaldehyde at room temperature for 15 min and subsequently permeabilized with 0.3% Triton X-100/PBS. Cells were blocked in 10% goat serum diluted in 0.1% Triton X-100/PBS at room temperature for 1 h. Primary antibodies were diluted in 1% goat serum/0.1% TritonX-100 and were incubated at room temperature for 2 h (see SI Appendix, Table S1 for antibody information). Cells were washed, and secondary antibodies were diluted in 1% goat serum/0.1%Triton X-100 and were incubated at room temperature for 1 h. After washing with PBS, coverslips were mounted on glass slides with VectoLab Mounting Medium (with DAPI) (Vector Laboratories).
Clonal Analysis.
MEFs were used as feeder cells for clonal analysis. One day before FACS sorting, confluent MEFs were treated with 8 µg/mL of mitomycin C for 2.5 h at 37 °C to inhibit cell proliferation. Cells were then washed twice with PBS followed by trypsinization. MEFs were then seeded into 96-well plates at a density of 1.5 × 106 cells per plate in DMEM/10% FBS/penicillin and streptomycin and were allowed to attach overnight at 37 °C. Two hours before sorting, the medium was replaced with cardiac stem cell maintenance medium (catalog no. SCM101; Millipore). Single clones of Tw2-tdTO+ cells were sorted by FACS onto 96-well plates coated with MEFs, were cocultured for 3 wk in cardiac stem cell maintenance medium, and then were switched to CM differentiation medium (catalog no. SCM102; Millipore) to promote CM differentiation. Cells were then stained with cTnT and Hoechst. High-throughput imaging was performed using an IN Cell Analyzer 600 Cell Imaging System (GE Healthcare Life Sciences).
Adult MI and I/R.
Mice (8–12 wk old) were anesthetized with 2.4% isoflurane and placed in a supine position on a heating pad (37 °C). Animals were intubated with a 19-gauge stump needle and were ventilated with room air using a MiniVent mouse ventilator (Hugo Sachs Elektronik; stroke volume, 250 μL; respiratory rate, 210 breaths/min). Via left thoracotomy between the fourth and fifth ribs, the LAD artery was visualized under a microscope and ligated by using a 6–0 PROLENE suture. Regional ischemia was confirmed by visual inspection under a dissecting microscope (Leica) by discoloration of the occluded distal myocardium. For I/R, the ligation was released after 45 min of ischemia, and tissue was allowed to reperfuse as confirmed by visual inspection. Sham-operated animals underwent the same procedure without occlusion of the LAD artery.
Single-Cell RNA-Seq and Analysis.
Tw2-tdTO+ and Tw2-tdTO− cells were isolated from 8-wk-old mouse hearts 10 d after TMX treatment using the protocol described in Primary Cultures section above. FACS-sorted cells were washed in PBS with 0.04% BSA and resuspended at a concentration of ∼1,000 cells/µL. Library generation for 10× Genomics v2 chemistry was performed following the Chromium Single Cell 3ʹ Reagents Kit v2 User Guide: CG00052 Rev B. Sequencing was performed on an Illumina NextSeq 500 sequencing platform using the 75-cycle kit. On average more than 500,000 reads per cell were sequenced.
Fastq files were generated using cellranger mkfastq and were mapped to the mm10 genome using cellranger count with standard parameters. For cluster identification, R package Seurat (https://www.nature.com/articles/nbt.3192) was used. Briefly, the data matrices were imported into R and were processed with the Seurat R package version 1.2.1. A minimum cutoff of 200 genes per cell and a maximum cutoff of 5,000 genes per cell were used for this dataset. In addition, cells in which >25% of total reads aligned to the mitochondrial genome were removed. After data filtering, PCA was performed using highly variable genes. The first eight principal components were used to find clusters with the resolution parameter set to 1.0. Identified clusters were visualized with t-distributed statistical neighbor embedding (tSNE) plots. Marker genes for each cluster were identified using the FindMarkers function in Seurat. The scRNA-seq data have been deposited in the Gene Expression Omnibus database under the ID code GSE118411 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118411).
RNA-Seq and Real-Time RT-PCR Analysis.
Cells were resuspended in 1 mL of TRIzol and homogenized using 20-gauge needles. Following chloroform extraction, the supernatant was mixed with an equal volume of 75% ethanol and loaded on RNeasy minicolumns (Qiagen). Total RNA was isolated according to the manufacturer’s instructions (Qiagen). RNA quality was verified by the Agilent 2100 Bioanalyzer, and RNA-seq was performed using an Illumina HiSeq 2500 by the University of Texas Southwestern Genomics and Microarray Core Facility.
For real-time RT-PCR analysis, total RNA was extracted from sorted cells with TRIzol (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen), and the expression of selected genes was analyzed by real-time RT-PCR using Taqman probes.
Analysis of RNA-Seq Data.
Quality assessment of the RNA-seq data was performed using the NGS-QC-Toolkit (v2.3.3) (journals.plos.org/plosone/article?id=10.1371/journal.pone.0030619). Reads with more than 30% nucleotides with phred quality scores less than 20 were removed from further analysis. Quality-filtered reads were then aligned to the mouse reference genome GRCm38 (mm10) using the Tophat2 (v 2.0.0) aligner using default settings except for –library-type = fr-firststrand. Aligned reads were counted using featurecount (v1.4.6) (https://academic.oup.com/bioinformatics/article/30/7/923/232889) per gene ID. Differential gene-expression analysis was done using the R package DEseq2 (v 1.6.3). Cutoff values of absolute fold change >1.5 and a false-discovery rate ≤0.05 were then used to select for differentially expressed genes between sample group comparisons. Normalized gene-count values were averaged within groups for heat map generation and were clustered using the R package clusterProfiler (v3.6.0).
GO Analysis.
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (v6.8) gene functional annotation and classification tool [PubMed ID (PMID):19131956, PMID:19033363] was used to annotate the list of differentially expressed genes with respective GO terms and to perform GO enrichment analysis for biological function category. The enrichGO function in clusterProfiler (v3.6.0) was used to perform GO enrichment analysis for each cluster. GO groups were selected for significance by using a P value cutoff of 1%.
Pathway Analysis.
Significant pathway enrichment analysis was performed using IPA (Ingenuity Systems). Statistically significant biological pathways were then identified by using a P value cutoff of 0.05.
Statistical Analysis.
All statistical analyses were performed using GraphPad Prism 7 (GraphPad Software). Data are presented as mean ± SEM. Differences between groups were tested for statistical significance by using the unpaired two-tailed Student’s t test. P < 0.05 was considered significant. For analysis of multiple groups, we utilized the Holm–Sidak correction for multiple comparisons with a false-discovery rate of 0.05. The number of biological (nontechnical) replicates for each experiment is indicated.
Supplementary Material
Acknowledgments
We thank Angela Mobley of the University of Texas Southwestern (UTSW) Flow Cytometry Core Facility, Hanspeter Niederstrasser of the UTSW High Throughput Screening Core, Beibei Chen of the Department of Clinical Sciences and the Bioinformatics Core Facility, (UTSW), John M. Shelton of the UTSW Histopathology Core, and the UTSW Genomics and Microarray Core Facility for technical help and service; Cheryl Nolen for technical assistance; Wei Tan for assistance with rodent surgery; and Drs. Hesham Sadek and Nikhil Munshi for constructive suggestions and advice. This work was supported by NIH Grants AR-067294, HL-130253, HL-138426, and AR-071980; Fondation Leducq Networks of Excellence; and Robert A. Welch Foundation Grant 1-0025 (to E.N.O.). N.L. was supported by Beginning-Grant-In-Aid 13BGIA17150004 from the American Heart Association. Y.-L.M. was supported by a Hamon Center for Regenerative Science and Medicine Trainee Fellowship. V.S.M. was supported by Cancer Prevention and Research Institute of Texas Grant RP150596.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNA-sequencing data have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE118411).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800526115/-/DCSupplemental.
References
- 1.Malliaras K, et al. Cardiomyocyte proliferation and progenitor cell recruitment underlie therapeutic regeneration after myocardial infarction in the adult mouse heart. EMBO Mol Med. 2013;5:191–209. doi: 10.1002/emmm.201201737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh PC, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton MGSJ, Sharpe N. Left ventricular remodeling after myocardial infarction: Pathophysiology and therapy. Circulation. 2000;101:2981–2988. doi: 10.1161/01.cir.101.25.2981. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann O, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 7.Mollova M, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci USA. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura W, et al. Hypoxia fate mapping identifies cycling cardiomyocytes in the adult heart. Nature. 2015;523:226–230. doi: 10.1038/nature14582. [DOI] [PubMed] [Google Scholar]
- 9.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 10.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Berlo JH, Molkentin JD. An emerging consensus on cardiac regeneration. Nat Med. 2014;20:1386–1393. doi: 10.1038/nm.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sultana N, et al. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat Commun. 2015;6:8701. doi: 10.1038/ncomms9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anversa P, Kajstura J, Rota M, Leri A. Regenerating new heart with stem cells. J Clin Invest. 2013;123:62–70. doi: 10.1172/JCI63068. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Bearzi C, et al. Human cardiac stem cells. Proc Natl Acad Sci USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison GM, et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–842. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 16.Oh H, et al. Cardiac progenitor cells from adult myocardium: Homing, differentiation, and fusion after infarction. Proc Natl Acad Sci USA. 2003;100:12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchida S, et al. Sca1-derived cells are a source of myocardial renewal in the murine adult heart. Stem Cell Reports. 2013;1:397–410. doi: 10.1016/j.stemcr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noseda M, et al. PDGFRα demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat Commun. 2015;6:6930. doi: 10.1038/ncomms7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfister O, et al. CD31- but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circ Res. 2005;97:52–61. doi: 10.1161/01.RES.0000173297.53793.fa. [DOI] [PubMed] [Google Scholar]
- 20.Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR. Islet1 cardiovascular progenitors: A single source for heart lineages? Development. 2008;135:193–205. doi: 10.1242/dev.001883. [DOI] [PubMed] [Google Scholar]
- 21.Smart N, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Cardiac repair with a novel population of mesenchymal stem cells resident in the human heart. Stem Cells. 2015;33:3100–3113. doi: 10.1002/stem.2101. [DOI] [PubMed] [Google Scholar]
- 23.Fioret BA, Heimfeld JD, Paik DT, Hatzopoulos AK. Endothelial cells contribute to generation of adult ventricular myocytes during cardiac homeostasis. Cell Rep. 2014;8:229–241. doi: 10.1016/j.celrep.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Currie DA, Bate M. The development of adult abdominal muscles in Drosophila: Myoblasts express twist and are associated with nerves. Development. 1991;113:91–102. doi: 10.1242/dev.113.1.91. [DOI] [PubMed] [Google Scholar]
- 25.Borkowski OM, Brown NH, Bate M. Anterior-posterior subdivision and the diversification of the mesoderm in Drosophila. Development. 1995;121:4183–4193. doi: 10.1242/dev.121.12.4183. [DOI] [PubMed] [Google Scholar]
- 26.Baylies MK, Bate M. Twist: A myogenic switch in Drosophila. Science. 1996;272:1481–1484. doi: 10.1126/science.272.5267.1481. [DOI] [PubMed] [Google Scholar]
- 27.Cripps RM, et al. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figeac N, Daczewska M, Marcelle C, Jagla K. Muscle stem cells and model systems for their investigation. Dev Dyn. 2007;236:3332–3342. doi: 10.1002/dvdy.21345. [DOI] [PubMed] [Google Scholar]
- 29.Cripps RM, Olson EN. Twist is required for muscle template splitting during adult Drosophila myogenesis. Dev Biol. 1998;203:106–115. doi: 10.1006/dbio.1998.9040. [DOI] [PubMed] [Google Scholar]
- 30.Castanon I, Baylies MK. A twist in fate: Evolutionary comparison of twist structure and function. Gene. 2002;287:11–22. doi: 10.1016/s0378-1119(01)00893-9. [DOI] [PubMed] [Google Scholar]
- 31.Mastroyiannopoulos NP, Antoniou AA, Koutsoulidou A, Uney JB, Phylactou LA. Twist reverses muscle cell differentiation through transcriptional down-regulation of myogenin. Biosci Rep. 2013;33:e00083. doi: 10.1042/BSR20130068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Šošić D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 33.Liu N, et al. A Twist2-dependent progenitor cell contributes to adult skeletal muscle. Nat Cell Biol. 2017;19:202–213. doi: 10.1038/ncb3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beck B, et al. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16:67–79. doi: 10.1016/j.stem.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt JM, et al. Stem-cell-like properties and epithelial plasticity arise as stable traits after transient Twist1 activation. Cell Rep. 2015;10:131–139. doi: 10.1016/j.celrep.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Shelton EL, Yutzey KE. Twist1 function in endocardial cushion cell proliferation, migration, and differentiation during heart valve development. Dev Biol. 2008;317:282–295. doi: 10.1016/j.ydbio.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.VanDusen NJ, Firulli AB. Twist factor regulation of non-cardiomyocyte cell lineages in the developing heart. Differentiation. 2012;84:79–88. doi: 10.1016/j.diff.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 40.Makino S, et al. Cardiomyocytes can be generated from marrow stromal cells in vitro. J Clin Invest. 1999;103:697–705. doi: 10.1172/JCI5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toma C, Pittenger MF, Cahill KS, Byrne BJ, Kessler PD. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93–98. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- 42.Grande MT, et al. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med. 2015;21:989–997. doi: 10.1038/nm.3901. [DOI] [PubMed] [Google Scholar]
- 43.Lovisa S, et al. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med. 2015;21:998–1009. doi: 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leri A, Rota M, Pasqualini FS, Goichberg P, Anversa P. Origin of cardiomyocytes in the adult heart. Circ Res. 2015;116:150–166. doi: 10.1161/CIRCRESAHA.116.303595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawamiphak S, Kontarakis Z, Filosa A, Reischauer S, Stainier DYR. Transient cardiomyocyte fusion regulates cardiac development in zebrafish. Nat Commun. 2017;8:1525. doi: 10.1038/s41467-017-01555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alvarez-Dolado M, et al. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature. 2003;425:968–973. doi: 10.1038/nature02069. [DOI] [PubMed] [Google Scholar]
- 47.Nygren JM, et al. Bone marrow-derived hematopoietic cells generate cardiomyocytes at a low frequency through cell fusion, but not transdifferentiation. Nat Med. 2004;10:494–501. doi: 10.1038/nm1040. [DOI] [PubMed] [Google Scholar]
- 48.Yang WJ, Li SH, Weisel RD, Liu SM, Li RK. Cell fusion contributes to the rescue of apoptotic cardiomyocytes by bone marrow cells. J Cell Mol Med. 2012;16:3085–3095. doi: 10.1111/j.1582-4934.2012.01600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong XQ, Li L. Dermo-1, a multifunctional basic helix-loop-helix protein, represses MyoD transactivation via the HLH domain, MEF2 interaction, and chromatin deacetylation. J Biol Chem. 2002;277:12310–12317. doi: 10.1074/jbc.M110228200. [DOI] [PubMed] [Google Scholar]
- 50.Hjiantoniou E, et al. Twist induces reversal of myotube formation. Differentiation. 2008;76:182–192. doi: 10.1111/j.1432-0436.2007.00195.x. [DOI] [PubMed] [Google Scholar]
- 51.Reinhold MI, Kapadia RM, Liao Z, Naski MC. The Wnt-inducible transcription factor Twist1 inhibits chondrogenesis. J Biol Chem. 2006;281:1381–1388. doi: 10.1074/jbc.M504875200. [DOI] [PubMed] [Google Scholar]
- 52.Bialek P, et al. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 53.Isenmann S, et al. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.