Significance
The Notch-signaling pathway is normally activated by receptor–ligand interactions. Extracellular domains (ECDs) of Notch receptors are heavily modified with O-linked glycans, such as O-glucose (O-Glc), O-fucose (O-Fuc), and O-GlcNAc. The significance of multiple types of O-glycans on Notch is not understood. NOTCH1 ECD interacts with ligands at multiple points, including an O-Glc monosaccharide on the 11th Epidermal Growth Factor (EGF) repeat (EGF11). Here, we identify two novel protein O-glucosyltransferases that modify NOTCH1 EGF11 with O-Glc. Combined deletion of the O-Glc site on EGF11 with O-Fuc modification sites on EGF8 or EGF12 markedly reduced NOTCH1 cell-surface expression or activation of NOTCH1 by Delta-like ligand 1, respectively. This study identifies a cooperative mechanism for fine-tuning the Notch-signaling pathway by different types of O-glycans.
Keywords: glycosyltransferases, Notch, signal transduction, development, O-glucose
Abstract
The Notch-signaling pathway is normally activated by Notch–ligand interactions. A recent structural analysis suggested that a novel O-linked hexose modification on serine 435 of the mammalian NOTCH1 core ligand-binding domain lies at the interface with its ligands. This serine occurs between conserved cysteines 3 and 4 of Epidermal Growth Factor-like (EGF) repeat 11 of NOTCH1, a site distinct from those modified by protein O-glucosyltransferase 1 (POGLUT1), suggesting that a different enzyme is responsible. Here, we identify two novel protein O-glucosyltransferases, POGLUT2 and POGLUT3 (formerly KDELC1 and KDELC2, respectively), which transfer O-glucose (O-Glc) from UDP-Glc to serine 435. Mass spectrometric analysis of NOTCH1 produced in HEK293T cells lacking POGLUT2, POGLUT3, or both genes showed that either POGLUT2 or POGLUT3 can add this novel O-Glc modification. EGF11 of NOTCH2 does not have a serine residue in the same location for this O-glucosylation, but EGF10 of NOTCH3 (homologous to EGF11 in NOTCH1 and -2) is also modified at the same position. Comparison of the sites suggests a consensus sequence for modification. In vitro assays with POGLUT2 and POGLUT3 showed that both enzymes modified only properly folded EGF repeats and displayed distinct acceptor specificities toward NOTCH1 EGF11 and NOTCH3 EGF10. Mutation of the O-Glc modification site on EGF11 (serine 435) in combination with sensitizing O-fucose mutations in EGF8 or EGF12 affected cell-surface presentation of NOTCH1 or reduced activation of NOTCH1 by Delta-like1, respectively. This study identifies a previously undescribed mechanism for fine-tuning the Notch-signaling pathway in mammals.
The Notch-signaling pathway regulates cell-fate decisions in a wide variety of cellular contexts in metazoans (1, 2). Aberrant activation or inactivation of this signaling pathway can lead to human diseases including developmental disorders, such as congenital heart defects and Alagille syndrome, and adult-onset diseases, including many different cancer types (3, 4). In mammals, there are four Notch receptors (NOTCH1–4) and four canonical ligands [DELTA-LIKE (DLL) 1 and 4; JAGGED (JAG) 1 and 2], all of which are type-I transmembrane proteins. Unlike other developmental signaling pathways, the Notch-signaling pathway is triggered by intercellular receptor–ligand interactions. The mammalian Notch receptors have 29–36 tandemly connected epidermal growth factor (EGF)-like repeats in their extracellular domain (ECD) that are involved in ligand binding. Ligand endocytosis exerts a pulling force on the Notch ECD, leading to unfolding of a negative regulatory domain and exposure of protease sites, resulting in release of the Notch intracellular domain from the membrane, which travels to the nucleus and induces transcription of downstream genes (1, 2).
The EGF repeats in the extracellular domain of Notch are posttranslationally modified with several types of O-linked glycans in the secretory pathway during Notch receptor biosynthesis (5–8). O-fucose (O-Fuc), O-glucose (O-Glc), and O-GlcNAc modifications can occur at specific positions within an EGF repeat if the appropriate O-glycosylation consensus sequence is present (Fig. 1A). Each EGF repeat has six conserved cysteine residues that form three disulfide bonds in a distinct pattern that stabilize the 3D structure of EGF repeats. O-Glc is added to serine residues within the consensus sequence C1-X-S-X-(P/A)-C2 (where the modified amino acid is underlined, X represents any amino acid, and C1 and C2 are the first and second cysteine residues of the EGF repeat) by protein O-glucosyltransferase 1 (POGLUT1, Rumi in flies) (9, 10). O-Fuc is added to serine or threonine residues within the consensus sequence C2-X-X-X-X-(S/T)-C3 by protein O-fucosyltransferase 1 (POFUT1) (11, 12), and O-GlcNAc is added to a serine or threonine residue within the putative consensus sequence C5-X-X-G-X-(T/S)-G-X-X-C6 by the EGF-specific O-GlcNAc transferase (EOGT) (13, 14). Interestingly, all three of these enzymes are soluble proteins localized in the endoplasmic reticulum (ER) that modify only properly folded EGF repeats containing the appropriate consensus sequence (9, 11–15).
Fig. 1.
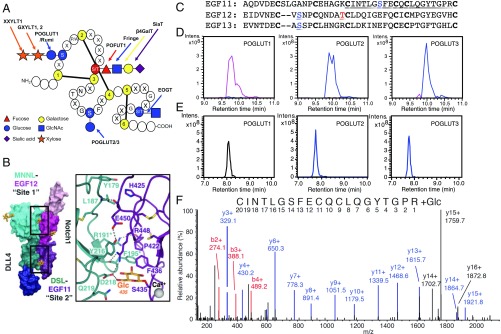
KDELC1 and KDELC2 are protein O-glucosyltransferases (POGLUTs). (A) Novel hexose exists at a distinct site from previously described O-Glc, O-Fuc, or O-GlcNAc modifications. Adapted from ref. 39. Copyright (2011), with permission from Elsevier. (B) Recent cocrystal structure of NOTCH1-DLL4 reveals a hexose modification on S435 of NOTCH1 EGF11 in contact with DLL4. From ref. 22. Reprinted with permission from AAAS. (C) Amino acid sequence of human NOTCH1 EGF11–13 with each EGF aligned based on conserved cysteines (boldface type). Known O-Glc and O-Fuc sites in EGF12 and -13 are indicated in blue and red, respectively. The O-Glc site in EGF11 (S435) is in blue, and the tryptic peptide containing this site described in E and F is underlined. (D) EICs of the reaction products of POGLUT1, POGLUT2, or POGLUT3 after overnight incubation with bacterially expressed human NOTCH1 EGF11–13 and UDP-Glc. Black line, unmodified EGF11–13 (+12, 1,236.5 m/z); blue line, EGF11–13 + 1 hexose (+12, 1,250.0 m/z); pink line, EGF11–13 + 2 hexoses (+12, 1,263.5 m/z). (E) The products from D were reduced/alkylated, digested with trypsin, and analyzed by LC-MS/MS. EICs were generated for different glycoforms of the triply charged peptide from EGF11 underlined in C. Black line, unmodified peptide (788.2 m/z). Blue line, peptide + 1 hexose (842.2 m/z). (F) HCD-MS/MS spectra of the O-glucosylated peptide from EGF11 confirms that S435 (equivalent to y-14 ion) is modified with O-Glc. Numerous fragment b-ions (red) and y-ions (blue) were detected. Note that y-ions 14 and 15 correspond to the peptide retaining a hexose (blue) although y-ions 14, 15, and 16 corresponding to the peptide ions that lost the hexose during fragmentation were also detected (black).
Genetic studies indicate that POFUT1 and POGLUT1 are essential for Notch receptor activation (9, 16–19). Biochemical studies have suggested that O-Fuc glycans regulate Notch-ligand interactions while O-Glc glycans added by POGLUT1 do not (9, 19–21). Recent structural analysis of a NOTCH1–DLL4 complex indicated that the O-Fuc on threonine 466 within the O-Fuc consensus site of NOTCH1 EGF12 interacts directly with amino acids on the DLL4 ligand, providing structural support for the importance of the O-Fuc at this site for ligand binding (Fig. 1B) (22). Surprisingly, the same structure also showed a hexose modification on serine 435 of NOTCH1 in contact with DLL4, suggesting that it could also affect Notch–ligand interactions (Fig. 1B). Serine 435 occurs between cysteines 3 and 4 of EGF11, a location distinct from that modified by POGLUT1 (Fig. 1A). A hexose modification at this site on EGF11 had been previously identified using mass spectrometry by another group (23).
Our prior work demonstrated that POGLUT1 is highly specific for modification of a serine residue within the consensus sequence between cysteines 1 and 2 of an EGF repeat (Fig. 1A) (15, 24). The finding of an O-linked hexose at a distinct position within an EGF repeat suggested that a previously unidentified enzyme must be responsible for its addition. KDELC1 and KDELC2 had been identified as mammalian POGLUT1 homologs with unknown function (10). Both contain a signal peptide, a CAP10 domain (a fungal glycosyltransferase domain), and a C-terminal KDEL-like ER retention signal, but neither can modify EGF repeats at classical POGLUT1 consensus sequences or rescue rumi mutations in flies (10). KDELC1 has recently been suggested as playing a role in hepatic dysfunction (25), but the function of these proteins remains poorly understood.
Here, we confirm that the O-linked hexose on serine 435 of NOTCH1 is an O-linked Glc. Additionally, we identify an O-Glc modification at a similar site between cysteines 3 and 4 of EGF10 (equivalent to EGF11 from NOTCH1) of NOTCH3. We demonstrate that KDELC1 and KDELC2 are protein O-glucosyltransferases that transfer O-Glc to these sites in vitro and in cells, which we are renaming POGLUT2 and POGLUT3, respectively. Finally, we used site-directed mutagenesis of NOTCH1 to demonstrate that this novel O-Glc modification affects NOTCH1 localization and activity in combination with sensitizing mutations in O-Fuc sites, suggesting that POGLUT2 and POGLUT3 are previously unknown regulators of Notch activity.
Results
KDELC1 and KDELC2 Are POGLUTs That Add O-Glc to NOTCH1 EGF11.
Recent structural and mass spectral studies showed that S435 of NOTCH1 EGF11 is modified with an O-hexose monosaccharide that interacts with Notch ligands (Fig. 1B) (22, 23, 26). The identity of the hexose could not be clearly determined from the electron density (22, 26) or mass spectral data (23). The fact that S435 is located between the third and the fourth cysteine residues of the EGF repeat, distinct from the classical O-Glc modification site (Fig. 1A), strongly suggested that POGLUT1 does not modify this site. These findings prompted us to look for other POGLUT(s) that might be responsible for the O-glucosylation at NOTCH1 EGF11. We previously identified two genes that are homologous to POGLUT1 in mammals, KDELC1 and KDELC2, neither of which can add O-Glc to classical O-Glc sites on EGF repeats (10). To examine whether POGLUT1, KDELC1, or KDELC2 can modify NOTCH1 EGF11, we incubated bacterially expressed and in vitro refolded NOTCH1 EGF11–13 (Fig. 1C) with purified POGLUT1, KDELC1, or KDELC2 (SI Appendix, Fig. S1A) in the presence of UDP-Glc (Fig. 1D) or UDP-galactose (SI Appendix, Fig. S1B). EGF11 contains the novel O-glucosylation site (S435) while EGF12 and EGF13 contain classical O-glucosylation sites (Fig. 1 A and C). As previously reported (21), POGLUT1 transferred two Glc residues from UDP-Glc to serine residues S458 and S496 in EGF12 and EGF13, respectively (Fig. 1D). In contrast, both KDELC1 and KDELC2 transferred a single Glc from UDP-Glc (Fig. 1D) to NOTCH1 EGF11–13. None of the enzymes transferred galactose from UDP-galactose to NOTCH1 EGF11–13 (SI Appendix, Fig. S1 B and C). To determine which amino acid(s) is modified with O-Glc by KDELC1 or KDELC2, we digested the products with trypsin and analyzed the resulting tryptic digests by mass spectrometry. We found that the tryptic peptide 429-CINTLGSFECQCLQGYTGPR-448 from EGF11 was efficiently modified with a single O-Glc by KDELC1 or KDELC2, whereas the same peptide was not modified by POGLUT1 (Fig. 1E). These data strongly suggest that the only target site modified by KDELC1 or KDELC2 within NOTCH1 EGF11–13 is contained within this peptide (429-CINTLGSFECQCLQGYTGPR-448) and that KDELC1 and KDELC2 show distinct acceptor specificity from POGLUT1. Further mass spectral analysis revealed that S435 in the peptide was modified by KDELC1 and KDELC2 (Fig. 1F). These results indicate that KDELC1 and KDELC2 are POGLUTs. We designate them as POGLUT2 and POGLUT3, respectively.
Novel O-Glc Monosaccharide Modifications Were Found in the Ligand-Binding Domain of NOTCH1 and NOTCH3.
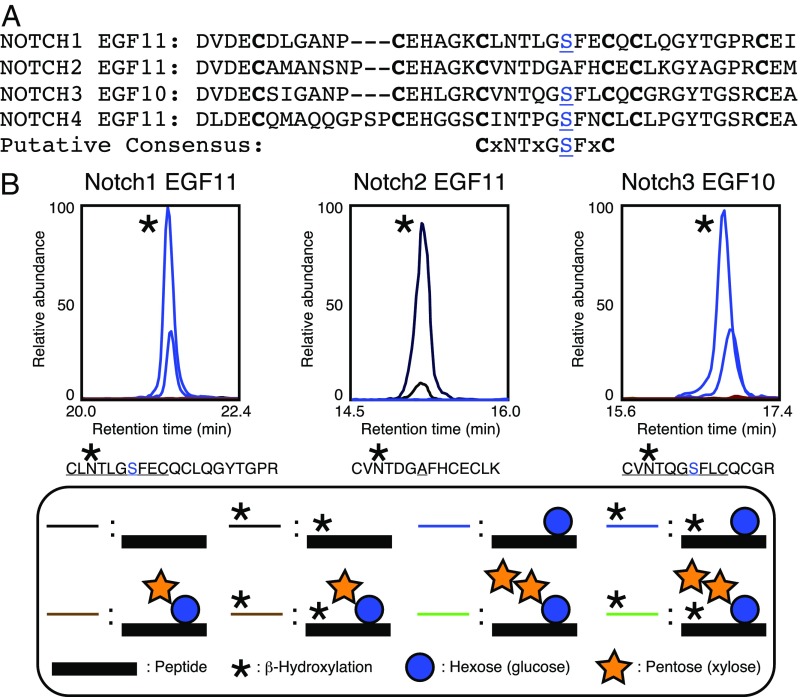
Sequence alignment of four mammalian Notch receptors suggested that the O-Glc site found within EGF11 of NOTCH1 is conserved on the corresponding EGF repeat of NOTCH3 and NOTCH4, but not of NOTCH2 (Fig. 2A) (27). To support this notion, we used a mass spectral approach to analyze the O-glucosylation status of these sites on NOTCH1, NOTCH2, and NOTCH3. Tagged, soluble NOTCH1, -2, and -3 ECD constructs were expressed in HEK293T cells, purified from media, digested with proteases, and analyzed by nano LC-MS/MS. Both the peptide from EGF11 of NOTCH1 and the peptide from EGF10 of NOTCH3 were found to be modified with O-Glc at high stoichiometry, whereas the peptide encompassing this region between cysteines 3 and 4 of NOTCH2 EGF11 was not modified (Fig. 2B and SI Appendix, Fig. S2), consistent with the lack of a serine or threonine at the same location as in NOTCH1 and -3 (Fig. 2A). Fragmentation of the peptides from NOTCH1 and NOTCH3 confirm the presence of the O-Glc on the indicated site (SI Appendix, Fig. S2). The similarities between the modification sites on NOTCH1 and -3 suggest a putative consensus sequence: C3-X-N-T-X-G-S-F-X-C4 (Fig. 2A). Interestingly, the major form of these peptides includes the β-hydroxylation of an asparagine residue (indicated by a single asterisk in Fig. 2) that is catalyzed by aspartyl/asparaginyl β-hydroxylase at D/N residues in EGF repeats within the overlapping consensus sequence C3-X-D/N-X-X-X-X-F/Y-X-C4 (28, 29). Lack of O-glucosylation of several other NOTCH1 EGF repeats with a serine or threonine residue at or near the same position between cysteines 3 and 4 [C3-X-X-X-X-X-(S/T)-X-X-C4, SI Appendix, Fig. S3] suggests that not just position, but also sequence context is important for modification. Each of the unmodified sites is missing one or more residues in the putative consensus sequence (SI Appendix, Table S1). Searches for Xyl-elongated forms of the novel O-Glc modifications, as classically found on O-Glc added to EGF repeats by POGLUT1 (Fig. 1A), revealed no elongation (Fig. 2B), suggesting that the major form added in HEK293T cells is the monosaccharide. A database search for proteins containing this putative consensus sequence suggests that as many as 86 distinct sites on 34 different proteins may be modified by POGLUT2 and POGLUT3 (SI Appendix, Table S2).
Fig. 2.
Ligand-binding domain EGF repeats from NOTCH1 and -3 are modified with novel O-Glc. (A) Amino acid sequence alignment of EGF11 from mouse NOTCH1, -2, and -4 and of EGF10 from mouse NOTCH3. EGF repeats aligned based on conserved cysteines (boldface type). Modified serine is in blue. Putative consensus sequence is at the bottom. (B) EICs of glycoforms of the peptides from EGF11 of mouse NOTCH1 EGF1-36-Myc-His6, EGF11 of mouse NOTCH2 EGF1-36-Myc-His6, and EGF10 of mouse NOTCH3 EGF1-34-Myc-His6. Each was expressed in HEK293T cells, purified from the medium, reduced, alkylated, digested with trypsin, and analyzed by nano LC-MS/MS. Black line, unmodified peptide; blue line, peptide + O-Glc. A single asterisk indicates β-hydroxylation of N in peptide.
Both POGLUT2 and POGLUT3 Are Capable of Adding O-Glc to NOTCH1 EGF11.
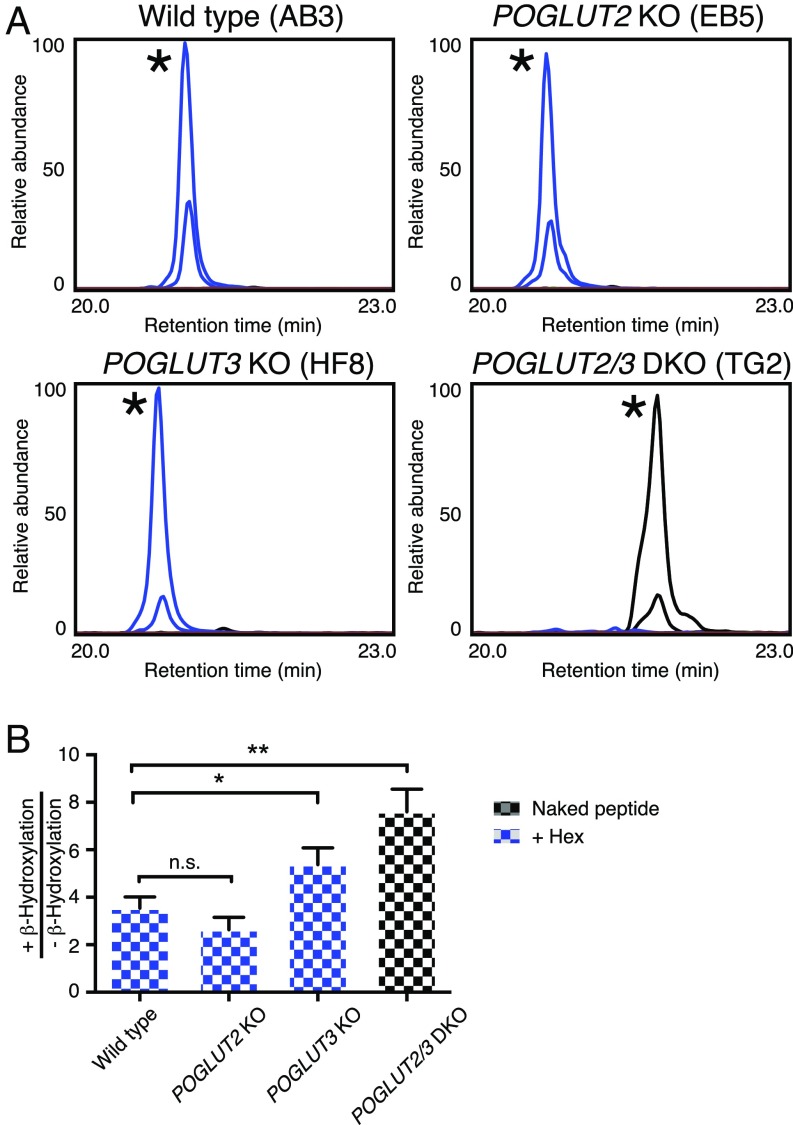
Our in vitro data suggested that both POGLUT2 and POGLUT3 can add O-Glc to NOTCH1 EGF11 (Fig. 1). Quantitative RT-PCR analysis shows that both POGLUT2 and POGLUT3 are expressed at comparable levels in HEK293T and NIH 3T3 cells (SI Appendix, Fig. S4 A and B). To determine whether both participate in addition of the novel O-Glc in HEK293T cells, we generated deletions of POGLUT2, POGLUT3, or both genes using CRISPR/Cas9 technology. Successful biallelic gene knockouts were confirmed by genomic DNA sequencing of HEK293T single KOs and POGLUT2 and POGLUT3 double KO cells (SI Appendix, Fig. S4C). NOTCH1 EGF1–36 was expressed in these cells, purified from the medium, and processed for mass spectral analysis to evaluate the glycosylation status of EGF11. O-glucosylation of NOTCH1 EGF11 was still observed in either POGLUT2 or POGLUT3 single-KO cells. However, O-glucosylation at EGF11 was completely eliminated in double KO cells (Fig. 3A). There is a slight but statistically significant enhancement in β-hydroxylation of N431 observed in the POGLUT3 single and POGLUT2/POGLUT3 double-KO cells (Fig. 3B). In addition, classical O-glucosylation at EGF10 was unaffected in wild-type (WT) and double-KO cells (SI Appendix, Fig. S5). Expression of either POGLUT2 or POGLUT3 partially rescued O-glucosylation at NOTCH1 EGF11 in double KO cells (SI Appendix, Fig. S6). These results indicate that both POGLUT2 and POGLUT3 are responsible for site-specific O-glucosylation at EGF11 of NOTCH1, that both enzymes are independently capable of adding this modification, and that the addition of this novel modification does not affect O-Glc addition or extension at classical O-Glc sites.
Fig. 3.
POGLUT2 and POGLUT3 are responsible for O-glucosylation of mouse NOTCH1 EGF11. (A) EICs of glycoforms of the EGF11 peptide from NOTCH1 EGF1-36-Myc-His6 produced in WT HEK293T cells (clone AB3), POGLUT2 KO cells (clone EB5), POGLUT3 KO cells (clone HF8), or POGLUT2 and POGLUT3 double-KO cells (clone TG2). Black line, unglucosylated peptide; blue line, O-glucosylated peptide. A single asterisk indicates β-hydroxylation of N in peptide. (B) Ratios of peak areas of β-hydroxylated versus non–β-hydroxylated peptides were quantified by determining the total peak area from each EIC. The data were from three independent transfection experiments. Error bars show SEM. *P < 0.05; **P < 0.01. n.s., not significant.
POGLUT2 and POGLUT3 Modify Only Folded EGF Repeats, Show Distinct Preferences for Different EGF Repeats, and Can Transfer Xyl as Well as Glc to EGF Repeats.
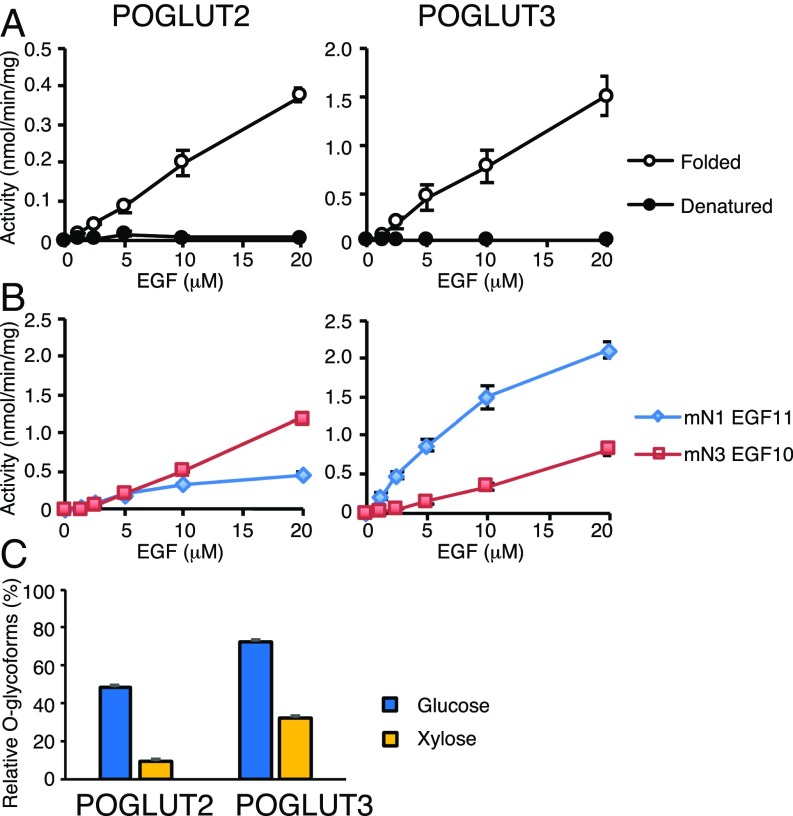
All other EGF-modifying glycosyltransferases (POFUT1, POGLUT1, EOGT) recognize folded EGF repeats and do not modify linear peptides (11, 14, 15). To test whether POGLUT2 and -3 share this requirement, we generated properly folded NOTCH1 EGF11 in bacteria and developed an in vitro assay for both enzymes (Fig. 4A). Neither enzyme required divalent cations for activity (SI Appendix, Fig. S7A), similar to POGLUT1 (24, 30). Both enzymes showed enhanced activity below pH 6 (SI Appendix, Fig. S7B), suggesting that protonation of a residue in the enzyme or EGF repeat enhances activity. Nonetheless, we performed all assays under more physiological conditions at pH 6.8. Denaturing EGF11 (by reducing and alkylating the disulfide bonds; SI Appendix, Fig. S7C) completely eliminated the ability of EGF11 to serve as an acceptor, showing that POGLUT2 and -3 recognize only folded EGF repeats (Fig. 4A). Identification of two enzymes that catalyze the same reaction raised the question of whether they have distinct specificities, similar to what we have seen with the three Fringe enzymes (31). To test for differences in EGF specificity, we generated a second folded acceptor substrate in bacteria, NOTCH3 EGF10. Comparison of these two EGF repeats revealed that POGLUT2 shows a slight preference for NOTCH3 EGF10 and that POGLUT3 shows a marked preference for NOTCH1 EGF11 (Fig. 4B). These results suggest that POGLUT2 and POGLUT3 differentially contribute to the site-specific O-glucosylation on NOTCH1 and NOTCH3. Finally, both POGLUT2 and -3 can transfer Xyl from UDP-Xyl to NOTCH1 EGF11, although less efficiently than Glc (Fig. 4C), suggesting that they also have protein O-xylosyltransferase activity such as that of POGLUT1 (10). Searches of the mass spectral data did not reveal any O-Xyl modification on NOTCH1 EGF11 from protein produced in HEK293T cells, suggesting that there may be specificity for which EGF repeats are modified by O-Xyl or that there is insufficient UDP-Xyl in HEK293T cells to result in significant levels of xylosylation.
Fig. 4.
POGLUT2 and POGLUT3 require folded EGF repeats, show distinct acceptor specificities, and can also transfer Xyl. (A) POGLUT enzymatic assays with folded and denatured mouse NOTCH1 EGF11. (B) Kinetic studies with mouse NOTCH1 EGF11 and mouse NOTCH3 EGF10. The data are from three independent assays. The values show the average and error bars represent SEM. (C) NOTCH1 EGF11 was incubated with POGLUT2 or -3 in the presence of either UDP-Glc or UDP-Xyl for 1 h, and the products were analyzed by mass spectrometry.
O-Glc on S435 Modulates Cell-Surface Presentation and DLL1 Activation of NOTCH1.
To test whether the O-Glc on S435 affects NOTCH1 activity, we mutated the serine residue to alanine and tested the mutant in cell-based NOTCH1 activation assays using DLL1 as the activating ligand (Fig. 5A). The S435A mutation alone caused no significant reduction in NOTCH1 activity compared with WT NOTCH1 activated by DLL1-expressing cells. Since the O-Fuc residues on T311 in EGF8 and T466 in EGF12 are known to be important for NOTCH1 activation by DLL1 (31), we generated two double mutants to test whether the S435A mutant exacerbated the inhibitory effect of the O-Fuc site mutants. Both double mutants caused a significant reduction in DLL1-mediated NOTCH1 activation relative to the O-Fuc mutants alone. In contrast to the EGF12/S435A mutant, the EGF8/S435A double mutant caused a significant reduction in cell-surface expression of NOTCH1 (Fig. 5 B and C), offering an explanation for the reduced levels of DLL1-mediated NOTCH1 activation with this double mutant. A similar reduction in DLL1 binding to the EGF8/S435A double-mutant NOTCH1 was observed in a cell-based binding assay (Fig. 5D). In contrast to the effect seen in the DLL1-NOTCH1 activation assay (Fig. 5A), the EGF12/S435A double mutant did not reduce DLL1-NOTCH1 binding significantly relative to the EGF12 mutant alone (Fig. 5D). Taken together, these results suggest that the O-Glc on S435A affects cell-surface presentation of NOTCH1 in combination with an EGF8 O-Fuc mutation and, in combination with an EGF12 O-Fuc mutation, modulates the ability of DLL1 to activate NOTCH1, apparently through a mechanism other than reducing binding.
Fig. 5.
O-Glc on S435 of NOTCH1 EGF11 affects cell-surface presentation and DLL1 activation of NOTCH1. (A) Cell-based NOTCH1 (WT and mutant) activity assays with DLL1 as activating ligand. (B and C) Cell-surface expression levels of NOTCH1 based on cell-surface antibody staining analyzed by (B) flow cytometry and (C) Western blot analysis showing levels of the furin-cleaved form of NOTCH1 (TM, for transmembrane), a measure of NOTCH1 that has passed through the Golgi to the cell surface (31). (D) Cell-based NOTCH1 (WT and mutant) ligand-binding assays with soluble DLL1-Fc as analyzed by flow cytometry. One-way ANOVAs with Tukey’s post hoc analysis were used to assess statistical significance; *P < 0.05, ***P < 0.001 for all experiments. ns, not significant.
Discussion
O-glycosylation of EGF repeats plays an extremely important role in the regulation of Notch function. The data shown here confirm the presence of a novel O-Glc on EGF repeats in the ligand-binding domains of NOTCH1 and -3. The O-Glc occurs as a monosaccharide modification at high stoichiometries on serines within the putative consensus C3-X-N-T-X-G-S-F-X-C4, a novel site for O-Glc addition (Fig. 1A). Furthermore, we show that POGLUT2 and POGLUT3 (formerly KDELC1 and KDELC2, respectively) are the enzymes responsible for these modifications. Both POGLUT2 and -3 require folded EGF repeats for recognition, although they differ in their preference for EGF repeat acceptors, and they can transfer Xyl as well as Glc to EGF repeats in vitro. Finally, our data indicate that these novel O-Glc modifications play a role in modulating Notch signaling.
POGLUT2 and POGLUT3 are the fourth and fifth enzymes demonstrated to modify properly folded EGF repeats at a consensus sequence (Fig. 1A). Recent cocrystal structures of two of these enzymes [POGLUT1 (24, 30) and POFUT1 (32)], each in complex with an EGF repeat, highlight the reason why the EGF repeats need to be folded and have an appropriate consensus sequence to be modified. The combination of the consensus sequence and the 3D fold of the EGF repeat, bound in a particular orientation within a large pocket on the surface of these enzymes, places the hydroxyl group to be modified precisely in the proper place to serve as a nucleophile in the transfer reaction. Movement of the position of the modified hydroxy amino acid in the sequence, or unfolding of the EGF repeat, disrupts recognition and modification by these enzymes. Our data also show that the O-Glc on EGF11 is not elongated by Xyl, suggesting that neither GXYLT1 nor GXYLT2, the enzymes that elongate classical O-Glc modifications added by POGLUT1, recognize an O-Glc in this position of an EGF repeat, consistent with our data that those enzymes also modify properly folded EGF repeats with a classical O-Glc modification (15).
The existence of consensus sequences has been quite useful in determining the biological function of the O-Fuc and the classical O-Glc modifications on EGF repeats. For example, early studies showed that the Notch family of receptors had more consensus sequences for these modifications than any other proteins in the databases (19, 33, 34). Searches of the human proteome with the C-X-N-T-X-G-S-F-X-C sequence identify over 30 potentially modified proteins involved in a wide variety of biological events, including signaling, extracellular matrix formation, complement activation, and homeostasis (SI Appendix, Table S2). Future studies are needed to refine the consensus sequence, determine whether these proteins are modified, and evaluate how the modification affects their function.
Both POGLUT2 and POGLUT3 contain C-terminal KDEL-like ER retention signals, suggesting that they localize to the ER, and prior work has demonstrated that POGLUT2 (KDELC1) is localized to the ER (25, 35). However, further work will be needed to confirm the subcellular localization of POGLUT3. The fact that these enzymes likely reside in the ER and recognize a folded structure suggests that they could play a role in quality control of EGF repeat folding. Our recent demonstration that POFUT1 and POGLUT1 participate in folding of NOTCH1 EGF repeats supports this idea (36). Although the S435A mutation in EGF11 alone did not reduce cell-surface expression of NOTCH1, the EGF8 O-Fuc/S435A double mutation showed a significant loss in cell-surface expression, consistent with a role of this O-Glc in quality control. This result suggests that this region of NOTCH1 (EGF8-11) is particularly sensitive to folding defects and may need O-Fuc and O-Glc modifications on EGF8 and -11 to assist in proper folding. The recently described flexibility of this region, especially between EGF9 and -10 (37), may make folding more difficult. In biological contexts where folding of Notch is compromised, the S435 O-Glc modification may play a critical role in regulating cell-surface expression of Notch. Additionally, proteins containing multiple EGF repeats modified by POGLUT2 and -3 would likely have a larger dependence on these enzymes for proper folding and secretion. For example, assuming that Y can substitute for F in the putative consensus sequence, Fibrillin 1 could have almost half of its 47 EGF repeats modified by POGLUT2 and -3.
Finally, we demonstrate that the O-Glc modification on EGF11 plays an important role in regulating NOTCH1 activity. While elimination of the O-Glc on EGF11 alone had little effect on NOTCH1 activation from DLL1, loss of this modification significantly exacerbated the reduction in DLL1-NOTCH1 activation caused by mutation in the O-Fuc site of EGF12. Both the O-Fuc on EGF12 and the O-Glc on EGF11 of NOTCH1 are located at the interface between Delta-like ligands and the NOTCH1 receptor (22), and our data suggest that they both contribute to enhancing NOTCH1 activation by DLL1. Surprisingly, the reduced DLL1-NOTCH1 activation observed in the EGF12/S435A double mutant does not appear to be mediated by a significant decrease in DLL1-NOTCH1 binding. Since Fringe modifications inhibit JAG1-NOTCH1 signaling without reducing JAG1-NOTCH1 binding (31), we have proposed that Fringe affects an event downstream of initial JAG1-NOTCH1 binding such as the catch bond recently shown to be formed by the tension generated upon ligand endocytosis (26). Perhaps the O-Glc on EGF11 of NOTCH1 has a similar effect on formation of a catch bond between DLL1 and NOTCH1.
Materials and Methods
Cells Culture.
HEK293T cells and NIH 3T3 cells were purchased from ATCC and cultured in DMEM-high glucose supplemented with 10% bovine calf serum and 1% penicillin/streptomycin.
Preparation of Recombinant Protein O-Glucosyltransferases and EGF Repeats.
Cloning of the full-length ORFs of human POGLUT1, POGLUT2, or POGLUT3 into mammalian the expression vector pcDNA4/TO/myc-6xHis was previously described (10). Recombinant human POGLUT1, POGLUT2, and POGLUT3 were expressed and purified as previously described (10). Briefly, the expression plasmids were transfected in HEK293T cells, and the proteins secreted to the culture media were purified by Ni-NTA affinity chromatography. The proteins were eluted in Tris-buffered saline (TBS) containing 250 mM imidazole and dialyzed against TBS containing 20% glycerol. The samples were stored at −80 °C until use. The concentrations of purified proteins were determined by 10% SDS/PAGE followed by Coomassie stain with BSA standards.
Expression of EGF11–13 from human NOTCH1 in Escherichia coli and purification were performed as previously described (21). For expression and purification of single EGF repeats in E. coli, the cDNA encoding EGF11 from mouse NOTCH1 was amplified with Herculase polymerase, the primers 5′-ATATATGAATTCGGACGTGGATGAGTGTGATCTG-3′ and 5′-ATCTAG GCGGCCGCCTCATTAACATCAATCTCACAGCC-3′, and pSecTag2c encoding mouse NOTCH1 EGF1–36 as a template. The cDNA encoding EGF10 from mouse NOTCH3 was amplified with Herculase polymerase, the primers 5′-ATATATGAATTCGGATGTGGATGAGTGCTCGATT-3′ and 5′-ATCTAGGCGGCCGCCTCATTGACATCAGTCTCACAGC-3′, and pSecTag2c encoding mouse NOTCH3 EGF1–34 as a template. The amplified cDNAs were digested with EcoRI and NotI and cloned into the pET20b(+) bacterial expression vector. Successful cloning was confirmed by direct DNA sequencing. Expression and purification of single EGF repeats were performed as previously described (15). Briefly, BL21(DE3) was transformed with the expression plasmids, and protein was induced with isopropyl β-d-1-thiogalactopyranoside at 20 °C overnight. After the cultured E. coli was suspended in 50 mM Tris⋅HCl with 1 mM PMSF and sonicated, the expressed proteins were purified from the soluble fractions using Ni-NTA affinity chromatography. The EGF proteins were eluted in TBS containing 250 mM imidazole and further purified by reverse-phase HPLC. Properly folded isomers of the EGF repeats were identified by their ability to be modified by POGLUT2 or POGLUT3 in the presence of UDP-Glc.
Denaturing of EGF proteins was performed as previously described (36) with slight modification. Briefly, ∼100 μg of EGF11 from mouse NOTCH1 was dissolved in 250 μL of 0.4 M ammonium bicarbonate buffer containing urea with or without 20 mM DTT and then incubated at 50 °C for 15 min. Fifty μl of 200 mM iodoacetamide or water was added to the DTT-treated or control sample, respectively, and incubated at room temperature for 15 min in the dark. The samples were purified by reverse-phase HPLC.
Enzymatic Assays for Protein O-Glucosyltransferase Activity.
Radioactive protein O-glucosyltransferase assays were performed as previously described (15). Briefly, recombinant POGLUT2 and POGLUT3 proteins (prepared as described above) were incubated at 37 °C for 20 min in 10-μL standard reaction mixtures containing 50 mM Hepes (pH 6.8), 0.16 μM UDP-[6-3H]Glc (60 Ci/mmol; American Radiolabeled Chemicals), 10 μM UDP-Glc, 10 μM EGF repeats, and 0.5% Nonidet P-40. Reactions were stopped by adding 900 μL of 100 mM EDTA (pH 8.0). The sample was loaded onto a C18 cartridge (100 mg; Agilent Technologies). After the cartridge was washed with 5 mL of H2O, the EGF repeat was eluted with 1 mL of 80% methanol. Incorporation of [6-3H]Glc into the EGF repeats was determined by scintillation counting of the eluates. Reactions without substrates were used as a background control. Data are from three independent assays. Values indicate mean ± SEM. For mass spectral protein O-glucosyltransferase assays, the reactions were performed in a 30-μL reaction volume (50 mM Hepes, pH 6.8) with 500 ng POGLUT2 or -3, 10 μM EGF repeat, and 200 μM UDP-Glc, UDP-Gal, or UDP-Xyl for the indicated times. Reactions were stopped by addition of 0.1% formic acid, and samples were analyzed by nano LC-MS using a Q-Exactive (Thermo Fisher Scientific) to determine relative amounts of unmodified and glycosylated EGF repeat. Determining the pH dependence was performed using the mass spectral assays in 50 mM MES buffer at the indicated pH value.
Mass Spectral Analysis of O-Glucosylation on Notch.
Recombinant mouse NOTCH1 EGF1-36-Myc-6xHis, mouse NOTCH2 EGF1-36-Myc-6xHis, and mouse NOTCH3 EGF1-34-Myc-6xHis were expressed in HEK293T cells and purified as previously described (10). Briefly, the expression plasmids were transfected in HEK293T cells, and the proteins secreted to the culture media were purified by Ni-NTA affinity chromatography. The proteins were eluted in TBS containing 250 mM imidazole. For in-gel digestion, the purified samples (∼1 μg) were acetone-precipitated and reduced and alkylated using 10 mM tris(2-carboxyethyl)phosphine (Thermo Fisher Scientific) and iodoacetamide. The samples were run on 10% SDS/PAGE and stained with Gel Code Blue (Thermo Fisher Scientific). The stained bands containing Notch proteins were excised, sliced into ∼1-mm3 cubes, and washed thoroughly with 20 mM ammonium phosphate with 50% methanol at room temperature overnight. After the gel pieces were dehydrated with 100% acetonitrile, protease solutions were added to the gel pieces. Protease digestion was performed at 37 °C for 4 h. The resulting digests were applied to Zip-Tip and eluted with 50% methanol and 0.1% formic acid. Five μl of ∼2-ng/μL digests diluted in 0.1% formic acid were injected into a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with an Easy nano-LC HPLC system with reverse-phase column (Thermo Fisher Scientific). Separation of (glyco)peptides was performed with a binary gradient solvent system that consists of solvent A (0.1% formic acid in water) and solvent B (90% acetonitrile and 0.1% formic acid in water) with a constant flow rate of 300 nL/min. Spectra were recorded with a resolution of 35,000 in the positive polarity mode over the range of m/z 350–2,000 and an automatic gain control target value was 1 × 106. The 10 most prominent precursor ions in each full scan were isolated for higher energy collisional dissociation-tandem mass spectrometry (HCD-MS/MS) fragmentation with normalized collision energy of 35%, an automatic gain control target of 2 × 105, an isolation window of m/z 3.0, dynamic exclusion enabled, and fragment resolution of 17,500. Raw data files were analyzed using Proteome Discoverer v2.1 (Thermo Fisher Scientific) with Byonic v (Protein Metrics) as a module for automated identification of (glyco)peptides. Extracted ion chromatograms (EICs) of all identified (glyco)peptides were generated using Xcalibur v (Thermo Fisher Scientific).
qRT-PCR Assays for Fibroblast Extracts.
RNA was isolated from HEK293T and NIH 3T3 cells using TRIzol (Thermo Fisher Scientific) according to the manufacturer’s protocol. RNA was quantified following purification, and SuperScript II Reverse Transcriptase (Thermo Fisher Scientific) was used to generate cDNA according to the manufacturer’s protocol. cDNA was quantified and used for RT-qPCR analysis. The SYBR Green Real-Time PCR kit was used with 1 μg cDNA and 1 μg primers (see below for sequences). For human POGLUT2 (KDELC1), the forward primer was 5′-ATTCAGGCAGTGGATACATCAG-3′ and the reverse primer was 5′-TGAAGGACCCATCTTTTCGG-3′. For human POGLUT3 (KDELC2), the forward primer was 5′-CCCGGAGGTGCTGGTCA-3′ and the reverse primer was 5′-GACCGCCTGCAGGTAGAAAT-3′. For human GAPDH, the forward primer was 5′-ACATCGCTCAGACACCATG-3′ and the reverse primer was 5′-ATGACAAGCTTCCCGTTCTC-3′. For mouse POGLUT2 (KDELC1), the forward primer was 5′- CGTTGGCTTTAGGATTTTCATGG-3′ and the reverse primer was 5′-AGATCGGCTGAATGTTGGAG-3′. For mouse POGLUT3 (KDELC2), the forward primer was 5′-TGAAATTTTGCTGTCACTGGC-3′ and the reverse primer was 5′-AGCCACACCAGGAAATGATAG-3′. For mouse GAPDH, the forward primer was 5′-CCAATGTGTCCGTCGTGGATCT-3′ and the reverse primer was 5′-GTTGAAGTCGCAGGAGACAACC-3′. Mixtures were added to wells of a 384-well plate and run on the Light Cycler 480 (Roche) for 40 cycles. The 2−ΔΔCT method was used for analysis of results, and samples were normalized to GAPDH. Three independent experiments were performed for each sample (n = 9).
CRISPR/Cas9-Mediated Targeting of POGLUT2 and POGLUT3 in HEK293T Cells.
Gene-specific guide RNAs (gRNAs) were inserted into the CAG-Cas9-2A-GFP vector (38). The gRNA sequence for targeting POGLUT2 exon 1 was 5′-CCAGCACTCGCCGAGACCGG-3′. The gRNA sequence for targeting POGLUT3 exon 1 was 5′-CTGCAGGCGGTCAACTCGGA-3′. The vector (2 μg) was transfected in HEK293T cells grown in 10-cm dishes using polyethylenimine (PEI). Single-cell sorting was performed in 96-well plates using the MoFlo XDP Cell Sorter (Beckman Coulter) at the Center for Tropical and Emerging Global Diseases (CTEGD) Cytometry Resource Laboratory at the University of Georgia. Successful gene editing was confirmed by genomic PCR with Q5 polymerase (New England Biolabs) and DNA sequencing. For establishing double knockout cells, POGLUT2 KO cells (clone EB5) were transfected with CAG-Cas9-2A-GFP vector encoding the gRNA targeting POGLUT3 exon 1. After a 24-h transfection, GFP-positive cells were sorted into 96-well plates. Successful gene editing was confirmed by genomic PCR with Q5 polymerase and DNA sequencing.
Site-Directed Mutagenesis of Mouse NOTCH1 Expression Vector.
The pcDNA-I expression plasmids encoding C-terminally Myc-tagged WT and two O-fucosylation site mutants (T311V in EGF8 and T466V in EGF12) of mouse NOTCH1 were described previously (31). The S435A mutation in mouse NOTCH1 was introduced by standard PCR-based site-mutagenesis with this plasmid and two primers. The sequences of the primers were 5′-CACTGGGTGCTTTTGAGTGCCAGTGTCTAC-3′ and 5′- CTCAAAAGCACCCAGTGTGTTGAGGCATTTGC-3′. Successful incorporation of the mutation was confirmed by direct DNA sequencing.
Notch Ligand-Binding Assays with Flow Cytometry.
Cell-based Notch ligand-binding assays were performed as previously described (31). Briefly, HEK293T cells were cotransfected with 1.5 μg of pcDNAI-mouse NOTCH1-Myc WT or mutants along with 0.4 μg of pEGFPN1 to a 3.5-cm plate using PEI. At 28–30 h posttransfection, the cells were dissociated with cold PBS containing 1% BSA and resuspended in a binding buffer (1 mM CaCl2, 1% BSA, and 0.05% NaN3 in Hanks’ balanced salt solution, pH 7.4; GIBCO). Cells (1 × 106) were incubated with soluble Notch ligand DLL1-Fc (R&D Systems) for 1 h at 4 °C. After washing with binding buffer twice, the cells were incubated with R-phycoerythrin (PE)-conjugated goat-anti human IgG (1:100) (Jackson ImmunoResearch) for 30 min at 4 °C. For clustered ligand-binding assays, DLL1-Fc was incubated with secondary PE-conjugated antibody for 30 min at room temperature, and the clustered ligands were added to the cells for 1 h at 4 °C. After binding, cells were washed two times with binding buffer and analyzed FACSCalibur (BD Bioscience). Gates were set to collect a GFP-positive population of 20,000 events for each sample and analyzed using FlowJo Data Analysis Software v10 (FlowJo).
Flow Cytometry Based NOTCH1 Cell-Surface Expression Assays.
HEK293T cells were cotransfected 1.5 μg of EV, pcDNA-N1-MycHis, or pcDNA-N1-MycHis with the indicated site-directed mutagenesis, and 0.4 μg of GFP (pEGFP-N1) in a 3.5-cm plate using PEI transfection reagent. After 4 h in culture with transfection reagents, media were changed to fresh DMEM with 10% BCS. At 28–30 h posttransfection, the cells were dissociated with cold PBS (pH 7.4) containing 1% BSA and resuspended in binding buffer [1 mM CaCl2, 1% BSA, and 0.05% NaN3 in Hanks’ balanced salt solution, pH 7.4 (Gibco)]. Cells were incubated with 100 μL of anti-mN1 (ECD) antibody (R&D Systems, AF5267) at 10 μg/mL for 1 h at 4 °C. Cells were washed with binding buffer and then incubated with PE–anti-sheep IgG (1:100; Santa Cruz, SC-3757) for 30 min at 4 °C. After two washes with binding buffer, the cells then were analyzed with a FACSCalibur (BD, Bioscience) flow cytometer. The gate was set to collect the GFP-positive population of 30,000 events for each sample and analyzed using FlowJo software (version 9.4.10). Three independent experiments were performed (n = 3).
Western-Blot–Based NOTCH1 Cell-Surface Expression Assay.
HEK293T cells were cotransfected 1.5 μg of EV, pcDNA-N1-MycHis, or pcDNA-N1-MycHis with the indicated site-directed mutagenesis, and 0.4 μg of GFP (pEGFP-N1) in a 3.5-cm plate using PEI transfection reagent. After 4 h in culture with transfection reagents, media were changed to fresh DMEM with 10% BCS. At 28–30 h posttransfection cells were lysed using 1% Nonidet P-40 in 20 mM Tris (pH 7.4), 0.15 M NaCl, and protease inhibitor (Complete mini, EDTA free; Roche). Western blot was performed as previously described (31). β-Actin was used as a loading control for all samples.
Cell-Based Notch Signaling Luciferase Reporter Assays.
NIH 3T3 cells were seeded in 24-well plates (5 × 104 cells/well) and transfected with plasmids expressing WT mouse NOTCH1, the mutant Notch receptors, or empty vector (0.1 μg/well). Cells were cotransfected with 0.12 μg of TP-1 luciferase Notch-signaling reporter construct and 0.06 μg of gWIZ β-galactosidase construct for transfection efficiency normalization using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. After 4 h, media were removed and L-cells stably expressing DLL1 (1.5 × 105 cells/well in 1 mL media) were overlaid on the NIH 3T3 transfectants. After an additional 24 h, cells were lysed and luciferase (Luciferase Assay System; Promega) and β-galactosidase assays were performed, as previously described (31). At least nine biological replicates and three independent experiments were performed for each sample in all signaling assays.
Supplementary Material
Acknowledgments
We thank Drs. Steve Dalton and Amar M. Singh (University of Georgia) for technical advice; Julie Nelson (CTEGD Cytometry Shared Resource Laboratory, University of Georgia) for technical assistance; and members of the R.S.H. laboratory for technical advice, discussion, and critical comments on this manuscript. This work was supported by NIH Grant GM061126 (to R.S.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804005115/-/DCSupplemental.
References
- 1.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17:722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 3.Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin Cell Dev Biol. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aster JC, Pear WS, Blacklow SC. The varied roles of Notch in cancer. Annu Rev Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 2010;92:131–164. doi: 10.1016/S0070-2153(10)92004-8. [DOI] [PubMed] [Google Scholar]
- 6.Jafar-Nejad H, Leonardi J, Fernandez-Valdivia R. Role of glycans and glycosyltransferases in the regulation of Notch signaling. Glycobiology. 2010;20:931–949. doi: 10.1093/glycob/cwq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi H, Haltiwanger RS. Significance of glycosylation in Notch signaling. Biochem Biophys Res Commun. 2014;453:235–242. doi: 10.1016/j.bbrc.2014.05.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider M, Al-Shareffi E, Haltiwanger RS. Biological functions of fucose in mammals. Glycobiology. 2017;27:601–618. doi: 10.1093/glycob/cwx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Acar M, et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132:247–258. doi: 10.1016/j.cell.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi H, et al. Rumi functions as both a protein O-glucosyltransferase and a protein O-xylosyltransferase. Proc Natl Acad Sci USA. 2011;108:16600–16605. doi: 10.1073/pnas.1109696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 12.Luo Y, Haltiwanger RS. O-fucosylation of notch occurs in the endoplasmic reticulum. J Biol Chem. 2005;280:11289–11294. doi: 10.1074/jbc.M414574200. [DOI] [PubMed] [Google Scholar]
- 13.Sakaidani Y, et al. O-linked-N-acetylglucosamine on extracellular protein domains mediates epithelial cell-matrix interactions. Nat Commun. 2011;2:583. doi: 10.1038/ncomms1591. [DOI] [PubMed] [Google Scholar]
- 14.Sakaidani Y, et al. O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochem Biophys Res Commun. 2012;419:14–19. doi: 10.1016/j.bbrc.2012.01.098. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi H, Kantharia J, Sethi MK, Bakker H, Haltiwanger RS. Site-specific O-glucosylation of the epidermal growth factor-like (EGF) repeats of notch: Efficiency of glycosylation is affected by proper folding and amino acid sequence of individual EGF repeats. J Biol Chem. 2012;287:33934–33944. doi: 10.1074/jbc.M112.401315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okajima T, Irvine KD. Regulation of notch signaling by O-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 17.Shi S, Stanley P. Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways. Proc Natl Acad Sci USA. 2003;100:5234–5239. doi: 10.1073/pnas.0831126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasamura T, et al. Neurotic, a novel maternal neurogenic gene, encodes an O-fucosyltransferase that is essential for Notch-Delta interactions. Development. 2003;130:4785–4795. doi: 10.1242/dev.00679. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Valdivia R, et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138:1925–1934. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu A, et al. In vitro reconstitution of the modulation of Drosophila Notch-ligand binding by Fringe. J Biol Chem. 2007;282:35153–35162. doi: 10.1074/jbc.M707040200. [DOI] [PubMed] [Google Scholar]
- 21.Taylor P, et al. Fringe-mediated extension of O-linked fucose in the ligand-binding region of Notch1 increases binding to mammalian Notch ligands. Proc Natl Acad Sci USA. 2014;111:7290–7295. doi: 10.1073/pnas.1319683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luca VC, et al. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347:847–853. doi: 10.1126/science.1261093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrawes MB, et al. Intrinsic selectivity of Notch 1 for Delta-like 4 over Delta-like 1. J Biol Chem. 2013;288:25477–25489. doi: 10.1074/jbc.M113.454850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu H, et al. Structural analysis of Notch-regulating Rumi reveals basis for pathogenic mutations. Nat Chem Biol. 2016;12:735–740. doi: 10.1038/nchembio.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. KDELC1, a novel endoplasmic reticulum resident glycoprotein in hepatic dysfunction. Int J Clin Exp Med. 2016;9:10529–10536. [Google Scholar]
- 26.Luca VC, et al. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science. 2017;355:1320–1324. doi: 10.1126/science.aaf9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi H, Haltiwanger RS. Role of glycosylation of Notch in development. Semin Cell Dev Biol. 2010;21:638–645. doi: 10.1016/j.semcdb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris RJ, Spellman MW. O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology. 1993;3:219–224. doi: 10.1093/glycob/3.3.219. [DOI] [PubMed] [Google Scholar]
- 29.Dinchuk JE, et al. Absence of post-translational aspartyl beta-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. J Biol Chem. 2002;277:12970–12977. doi: 10.1074/jbc.M110389200. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, et al. Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1) Nat Commun. 2017;8:185. doi: 10.1038/s41467-017-00255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kakuda S, Haltiwanger RS. Deciphering the Fringe-mediated Notch code: Identification of activating and inhibiting sites allowing discrimination between ligands. Dev Cell. 2017;40:193–201. doi: 10.1016/j.devcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, et al. Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1. Nat Chem Biol. 2017;13:757–763. doi: 10.1038/nchembio.2381. [DOI] [PubMed] [Google Scholar]
- 33.Rampal R, Luther KB, Haltiwanger RS. Notch signaling in normal and disease states: Possible therapies related to glycosylation. Curr Mol Med. 2007;7:427–445. doi: 10.2174/156652407780831593. [DOI] [PubMed] [Google Scholar]
- 34.Rana NA, et al. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem. 2011;286:31623–31637. doi: 10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimata Y, et al. Identification of a novel mammalian endoplasmic reticulum-resident KDEL protein using an EST database motif search. Gene. 2000;261:321–327. doi: 10.1016/s0378-1119(00)00499-6. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi H, et al. O-glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking. J Biol Chem. 2017;292:15964–15973. doi: 10.1074/jbc.M117.800102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisshuhn PC, et al. Non-linear and flexible regions of the human Notch1 extracellular domain revealed by high-resolution structural studies. Structure. 2016;24:555–566. doi: 10.1016/j.str.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger RP, et al. ST8SIA4-dependent polysialylation is part of a developmental program required for germ layer formation from human pluripotent stem cells. Stem Cells. 2016;34:1742–1752. doi: 10.1002/stem.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rana N, Haltiwanger RS. Fringe benefits: Functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr Opin Struc Biol. 2011;21:583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.