Significance
Regulatory Factor X 3 (RFX3) is one of the key transcription factors involved in cilia formation and functions. Our study reveals that auto-fatty acylation is a critical regulatory mechanism for RFX3 transcriptional activities. Fatty acylation of RFX3 is required for its dimerization, leading to transcription of cilia-associated genes. More importantly, fatty acylation of RFX3 regulates ciliogenesis and Hedgehog signaling pathways, which are associated with developmental and degenerative disorders known as ciliopathies. Our results indicate a major role of auto-fatty acylation in the regulation of RFX3 function and ciliogenesis, providing a potential link between deregulation of fatty acid metabolism to ciliopathies and diabetes.
Keywords: RFX3, fatty acylation, ciliogenesis, autoacylation, palmitoylation
Abstract
Defects in cilia have been associated with an expanding human disease spectrum known as ciliopathies. Regulatory Factor X 3 (RFX3) is one of the major transcription factors required for ciliogenesis and cilia functions. In addition, RFX3 regulates pancreatic islet cell differentiation and mature β-cell functions. However, how RFX3 protein is regulated at the posttranslational level remains poorly understood. Using chemical reporters of protein fatty acylation and mass spectrometry analysis, here we show that RFX3 transcriptional activity is regulated by S-fatty acylation at a highly conserved cysteine residue in the dimerization domain. Surprisingly, RFX3 undergoes enzyme-independent, “self-catalyzed” auto-fatty acylation and displays preferences for 18-carbon stearic acid and oleic acid. The fatty acylation-deficient mutant of RFX3 shows decreased homodimerization; fails to promote ciliary gene expression, ciliogenesis, and elongation; and impairs Hedgehog signaling. Our findings reveal a regulation of RFX3 transcription factor and link fatty acid metabolism and protein lipidation to the regulation of ciliogenesis.
Cilia are conserved organelles present on the surface of most vertebrate cells, sensing diverse extracellular signals crucial for cell proliferation, differentiation, polarity, and tissue homeostasis (1–4). For example, cilia are critical for Hedgehog (Hh) pathway, and many pathway components are required to localize to cilia for signal transduction. Therefore, loss of cilia leads to impairment of Hh signaling activities (2). An increasing number of human developmental and degenerative disorders have been attributed to defects in cilia formation and functions, which are collectively termed ciliopathies (5–8). Many ciliopathies are caused by disruption of ciliogenesis that is transcriptionally regulated by Regulatory Factor X (RFX) transcription factors (8–11). RFX was identified initially as a regulatory protein that binds to the X-box of a major histocompatibility complex (MHC) class II promoter (12). RFX family transcription factors are conserved across a wide range of species, including Saccharomyces cerevisiae, Schizosaccharomyces prombe, Caenorhabditis elegans, Drosophila melanogaster, and vertebrates (13). Eight members of human RFX transcription factors (RFX1–8) have been identified (14). RFX factors have conserved and critical roles in controlling ciliary gene expression and cilia formation, including RFX1–4 in vertebrates, DAF19 in C. elegans, and RFX in D. melanogaster (9, 10, 15–21). RFX1, RFX2, and RFX3 are the closest homologs of DAF19 in C. elegans and RFX in D. melanogaster (13). In particular, RFX3 plays a critical role in ciliogenesis. It has been shown that RFX3 is broadly expressed in ciliated cells and is essential for growth and function of cilia in ciliated cells, including embryonic node cells, brain ependymal cells, pancreatic islet cells, neuronal cells, and basal cells (10, 11, 17, 18, 22–24). RFX3-deficient mice exhibit defective cilia in the brain and pancreas, with hallmarks of ciliopathies including left–right asymmetry defects, hydrocephalus, corpus callosum agenesis, and abnormal thalamocortical tract formation (17, 22, 23, 25, 26).
In addition to regulating ciliogenesis, RFX3 plays an important role in regulating pancreatic endocrine cell differentiation, mature β-cell function, and glucokinase gene expression (22, 27). Interestingly, RFX3- and RFX6-knockout mice share many common defects in islet cell differentiation and mature β-cell function, suggesting that RFX3 and RFX6 may coordinately regulate a set of genes associated with pancreas development and β-cell function (22, 28). A recent study observed a remarkably strong correlation between RFX transcription factors and type 2 diabetes (T2D) risk, suggesting that RFX factors may play a significant role in the genetic component of T2D predisposition (29). Furthermore, a recent genome-wide association study (GWAS) revealed that RFX3 is highly associated with gout arthritis, which is one of the most common types of inflammatory arthritis, suggesting that RFX3 might also play a role in the autoimmune regulation (30). RFX1/3 double-knockout mice developed rapidly progressive hearing loss, indicating that RFX3 and RFX1 may coordinately regulate genes involved in mice hearing (31). This is supported by the ChIP-sequencing (ChIP-seq) analysis, which showed significant overlap of target genes between RFX1 and RFX3 (27, 31).
RFX family proteins have a conserved winged-helix DNA-binding domain in the N terminus, and most members contain a conserved dimerization domain in the C terminus (10, 13, 32, 33). Although RFX family proteins can bind to DNA as a monomer in vitro, numerous studies have suggested that RFX dimerization or interactions with partner proteins is critical for its transcriptional activity (10, 33–42). Various lines of evidence have shown that RFX1–4 and RFX6 can form homodimers and diverse heterodimers (14, 37, 40, 43), supporting the notion that dimerization is essential for RFX transcription factors to cooperatively regulate target genes. However, how different RFX factors cooperate with each other in regulating target gene expression remains elusive. In addition, how RFX factors are regulated by posttranslational modifications is largely unknown.
Protein fatty acylation is an important cotranslational or posttranslational modification of proteins, in which various fatty acids are covalently attached to proteins. Fatty acylation plays important roles in protein trafficking and localization, stability, and binding to cofactors (44). Bioorthogonal chemical reporters have been widely used to detect potential fatty acylated proteins in previous studies (45, 46). Here we show that RFX1 and RFX3 are S-fatty acylated proteins using chemical reporter and mass spectrometry (MS) analysis. RFX3 is fatty acylated through an enzyme-independent and self-catalyzed autoacylation process and displays preferences toward 18-carbon fatty acids, such as stearic acid and oleic acid. Furthermore, we identified that RFX3 is fatty acylated at an evolutionarily conserved cysteine residue in the dimerization domain. Auto-fatty acylation of RFX3 is required for its dimerization and transcriptional activity, thereby regulating target genes involved in ciliogenesis. Our studies thus revealed RFX3 as an example of autoacylated transcription factors and provided mechanistic insight into how intracellular fatty acid pool and dynamic fatty acylation of transcription factors regulate ciliogenesis.
Results
RFX3 Is Fatty Acylated at a Conserved Cysteine Residue in the Dimerization Domain.
To identify potential fatty acylated proteins, we performed large-scale proteomic profiling of fatty acylated proteins in HEK293A cells and pancreatic β-cell line MIN6 using saturated fatty acid (15-hexadecynoic acid, Alk-C16) or unsaturated fatty acid chemical reporter (cis-9-hexadecen-15-ynoic acid, Alk-C16:1) (SI Appendix, Fig. S1), followed by copper-mediated 1,3-dipolar cycloaddition (Click reaction) with biotin-azide, enriched by streptavidin beads, and subjected for proteomic analysis by liquid chromatography–MS/MS (LC-MS/MS) (47–49). The MS data revealed both probes could label endogenous RFX1 and RFX3 (SI Appendix, Fig. S1). These results are consistent with previous proteomic studies using chemical reporters of fatty acylation (17-ODCA), where RFX1 was detected in T cells and neuronal cells and RFX3 was detected in neuronal cells (50–53). Taken together, the proteomic studies suggested that RFX1 and RFX3 could be fatty acylated, but detailed biochemical follow-up studies are needed to validate these findings.
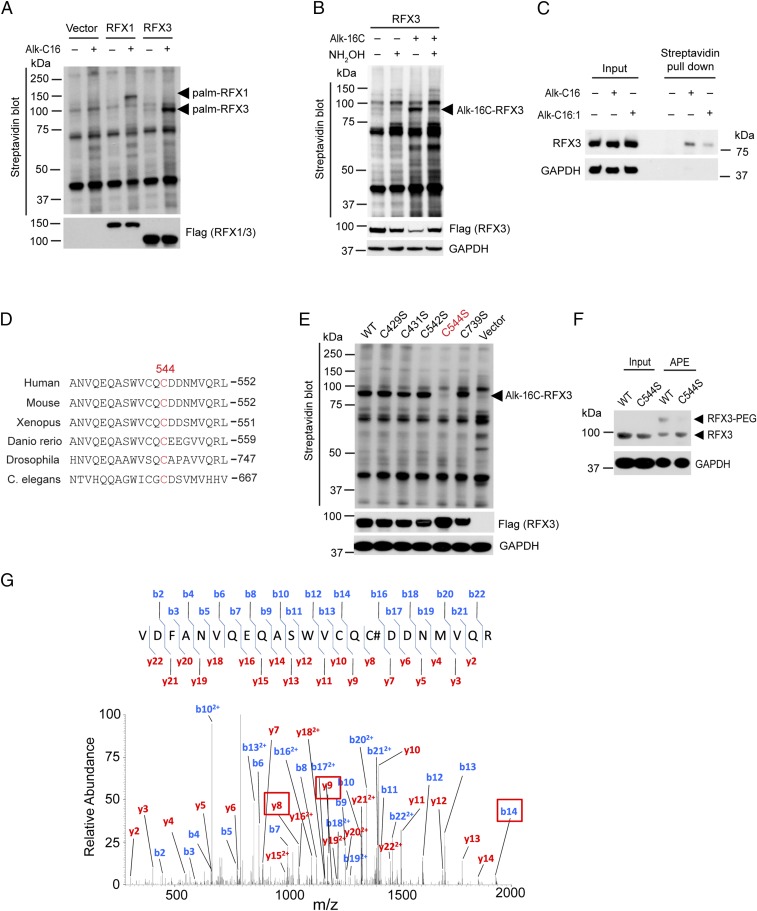
Therefore, we set out to confirm that RFX1 and RFX3 could indeed be fatty acylated. HEK293A cells were transfected with N-terminal Flag-tagged RFX1 and RFX3 and then labeled with 50 µM of Alk-C16, followed by Click reaction with biotin-azide. The streptavidin blot revealed that RFX1 and RFX3 could indeed be labeled by Alk-C16 (Fig. 1A). Treatment of Click reaction products with 2.5% (vol/vol) hydroxylamine (NH2OH) dramatically decreased fatty acylation levels of RFX3. Taken together, our results confirmed that RFX1 and RFX3 are S-fatty acylated through thioester bonds with cysteine residues (Fig. 1B and SI Appendix, Fig. S2). Previous studies suggested that RFX3 plays a significant role in ciliogenesis, pancreas development, and β-cell function (17, 18, 22, 24, 27), and we focused our studies on fatty acylation of RFX3. To test whether endogenous RFX3 protein can be fatty acylated, HEK293A cells were metabolically labeled with 100 µM Alk-C16, followed by Click reaction with biotin-azide. The fatty acylated proteome was then enriched by streptavidin beads pull-down. Western blotting for RFX3 in the pull-down samples indicated that endogenous RFX3 was indeed fatty acylated (Fig. 1C).
Fig. 1.
RFX3 is S-fatty acylated at an evolutionarily conserved cysteine residue. (A) Biochemical validation of RFX1 and RFX3 fatty acylation by metabolic labeling using chemical reporters of fatty acylation and streptavidin blot. (B) Treatment with hydroxylamine significantly decreased RFX3 fatty acylation. (C) Streptavidin pull-down and Western blotting confirmed that endogenous RFX3 is fatty acylated in human pancreatic β-cells. (D) Alignment of dimerization domain sequences of RFX3 family proteins by ClustalW, including human, mouse, Xenopus, Danio rerio, Drosophila, and C. elegans. (E) Mutation of cysteine 544 to serine completely abolished fatty acylation of RFX3. (F) APE assay confirmed S-fatty acylation of RFX3. (G) MS/MS spectral of RFX3 modified peptide showing S-fatty acylation on Cys544. #, IAA modification.
To identify potential fatty acylation site(s) of RFX3, we aligned the sequences of RFX3 protein across different species including mouse, Xenopus, zebrafish, Drosophila (RFX), and C. elegans (DAF-19) and found one evolutionarily conserved cysteine residue (Cys544) at the dimerization domain of RFX3 (Fig. 1D). Cys544 is also conserved in RFX1, RFX2, and RFX3 (SI Appendix, Fig. S3). According to the CSS-Palm software (csspalm.biocuckoo.org/online.php) for the prediction of protein palmitoylation sites, cysteine residues including Cys429, Cys431, Cys542, Cys544, and Cys739 are potential palmitoylation sites with relatively high scores. To confirm the fatty acylated cysteine residues, we constructed a series of RFX3 mutants, in which the cysteine residues of RFX3 were mutated to serines. HEK293A cells were transfected with Flag-tagged wild-type (WT) RFX3 or the mutant constructs (C429S, C431S, C542S, C544S, and C739S) and metabolically labeled with Alk-C16. Streptavidin blot revealed that only the mutation of Cys544 could completely abolish RFX3 fatty acylation, whereas the mutation of other cysteine residues had little effect (Fig. 1E). Notably, Cys542 is very close to Cys544 and relatively conserved across different species and different members including RFX1–3, but it has no effect on fatty acylation of RFX3, suggesting that Cys544 is specific for RFX3 fatty acylation. To confirm the modification site, we also performed aycl-PEG exchange (APE) assay, which separates the acylated protein from nonacylated protein by adding a mass tag (54). Our APE results showed that there is only one slower migrating PEGylated polypeptide band that corresponds to mono–S-fatty acylated WT RFX3, whereas the slower migrating band disappears in the C544S mutant (Fig. 1F).
To further characterize RFX3 fatty acylation, we performed more LC-MS/MS analysis by employing chemical tagging methods that were modified from a recent study (55). Briefly, Flag-RFX3 was overexpressed in HEK293A cells and immunoprecipitated using anti-Flag beads, followed by sequential treatment with N-ethylmaleimide (NEM), NH2OH, and iodoacetamide (IAA) (SI Appendix, Fig. S4A). After washing, RFX3 was either eluted from beads followed by in-solution digestion or separated by SDS/PAGE (SI Appendix, Fig. S4B) and subjected to in-gel digestion before LC-MS/MS analysis. The peptide containing IAA-modified Cys544 was identified from both in-solution and in-gel tryptic digestion sample (Fig. 1G). We next prepared the samples with or without NH2OH treatment side-by-side. Compared with the sample not treated with NH2OH, the peptide containing IAA-modified Cys544 was identified from an NH2OH-treated sample, and its abundance increased significantly (SI Appendix, Fig. S4C), suggesting that RFX3 could be S-fatty acylated at Cys544. In addition, the peptides containing IAA-modified Cys542 and Cys739 were also identified from in-gel digestion sample (SI Appendix, Table S1), suggesting that Cys542 and Cys739 might be additional potential sites of RFX3 S-fatty acylation. Taken together, our results suggest that Cys544 is the primary site of RFX3 fatty acylation. However, we cannot completely exclude the possibility that Cys542 and Cys739 are minor sites of RFX3 fatty acylation.
Fatty Acid Preferences and Dynamics of RFX3 Fatty Acylation.
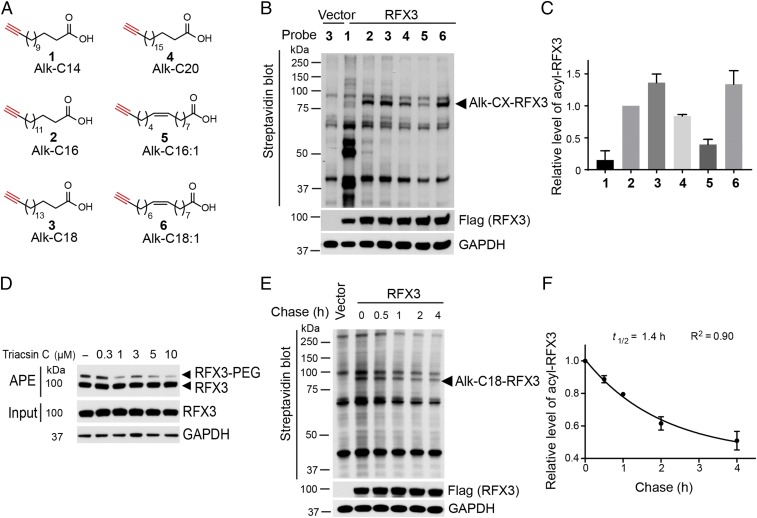
Our streptavidin beads pull-down assay suggested that saturated fatty acid Alk-C16 displays higher labeling efficiency for RFX3 than that of the monounsaturated fatty acid Alk-C16:1 (Fig. 1C). We asked whether fatty acylation of RFX3 has preferences for different types of fatty acids. To answer this question, we utilized a series of fatty acid chemical probes including different chain length and unsaturation levels (Fig. 2A). Flag-tagged RFX3 was transfected into the HEK293A cells, labeled cells with 50 µM of probes, followed by Click reaction with biotin-azide. The streptavidin blot results suggested that 18-carbon stearoylation probe (Alk-C18) displays the highest labeling efficiency, whereas 14-carbon myristoylation probe (Alk-C14) shows the lowest labeling efficiency (Fig. 2B). In addition, 16-carbon palmitoylation probe (Alk-C16) shows slightly lower labeling efficiency than that of 18-carbon stearic acid probe but higher than that of 20-carbon arachidic acid (Alk-C20). Notably, the labeling efficiency of 18-carbon monounsaturated oleic acid probe (Alk-C18:1) appears to be substantially higher than that of 16-carbon monounsaturated palmitoleic acid probe (Alk-C16:1), similar to 18-carbon stearic acid probe. We quantified RFX3 fatty acylation level in the HEK293A cells treated with different fatty acid probes in four independent experiments. The quantification results confirmed that RFX3 fatty acylation has preferences for 18-carbon saturated stearic acid and monounsaturated oleic acid in HEK293A cells (Fig. 2C), suggesting that RFX3 fatty acylation might be regulated by fatty acid metabolism. In addition, we treated HEK293A cells with long-chain fatty acyl-CoA synthetase (ACSL) inhibitor Triacsin C and analyzed the fatty acylation level of RFX3 using APE assay. The APE results showed that inhibition of ACSL by Triacsin C substantially decreased the fatty acylated RFX3 level in HEK293A cells (Fig. 2D), suggesting that fatty acylation of RFX3 might respond to changes in intracellular long-chain fatty acyl-CoA levels. Taken together, our results suggested that RFX3 fatty acylation might be regulated by intracellular metabolites.
Fig. 2.
Fatty acid preferences and dynamics of RFX3 fatty acylation. (A) Fatty acylation chemical reporters used in the study. (B) Fatty acid preference of RFX3 fatty acylation by metabolic labeling using chemical reporters of fatty acylation and streptavidin blot. (C) Quantification of RFX3 fatty acylation level normalized by the expression level of Flag-RFX3. (D) APE assay showed the effect of long-chain fatty ACSL inhibitor Triacsin C on RFX3 S-fatty acylation. (E) Pulse-chase analysis showed dynamic S-stearoylation of RFX3. (F) Determination of the half-life for the stearic acid turnover on RFX3 from pulse-chase experiments. Values represent the average ± SEM of three or four experiments.
We next asked whether RFX3 undergoes cycles of fatty acylation and de-fatty acylation. To determine the rates of fatty acylation cycling of RFX3, pulse-chase experiments were performed. Results of multiple pulse-chase experiments showed that the turnover rate of stearoylation of RFX3 is approximately 1.4 h, while the protein remained stable during the assay period (Fig. 2 E and F), suggesting that fatty acylation of RFX3 is a dynamic process. In addition, we measured the turnover rates of palmitic acid and oleic acid on RFX3, with a half-life of about 1.5 h and 1.2 h, respectively (SI Appendix, Fig. S5).
Recent studies have shown that palmostatin B can inhibit depalmitoylases APT1/2 and α/β-hydrolase domain-containing protein 17 (ABHD17) (56, 57). We asked whether palmostatin B could affect RFX3 fatty acylation. Our results revealed that 10 µM of palmostatin B could not increase RFX3 fatty acylation level (SI Appendix, Fig. S6A). In addition, overexpression of APT1/2 and ABHD17 could not decrease the RFX3 fatty acylation level (SI Appendix, Fig. S6B), consistent with palmostatin B treatment results. Recent studies suggested that ABHD family proteins are potential depalmitoylases (57, 58). We thus screened other ABHDs and found that ABHD2 and ABHD6 might be potential candidate proteins regulating RFX3 defatty acylation (SI Appendix, Fig. S7).
RFX3 Undergoes Auto-Fatty Acylation That Is Required for Its Dimerization.
To identify potential S-fatty acylation enzymes of RFX3, we screened the known palmitoyl acyltransferases (PATs). We cotransfected each of the 23 HA-tagged mouse ZDHHC family members of S-acyltransferases (ZDHHC-PATs) and Flag-tagged RFX3 into HEK293A cells and metabolically labeled RFX3 using Alk-C16. Overexpression of ZDHHC-PATs failed to significantly enhance fatty acylation levels of RFX3 (SI Appendix, Fig. S8), suggesting that ZDHHC-PATs might not be involved in RFX3 fatty acylation and RFX3 might be fatty acylated through a different mechanism. However, we cannot exclude the possibility that an unknown enzyme contributes to the RFX3 fatty acylation. We previously have reported that TEA domain transcription factors (TEADs) are autopalmitoylated proteins (59). TEADs possess intrinsic “enzyme-like” activities and bind to palmitoyl-CoA directly. The cysteine residues located near the acyl-CoA binding site could undergo self-catalyzed, “proximity”-mediated acylation, resulting in nonenzymatic palmitoylation. We speculate that RFX3, like TEADs, might possess such enzyme-like activities and undergo auto-fatty acylation.
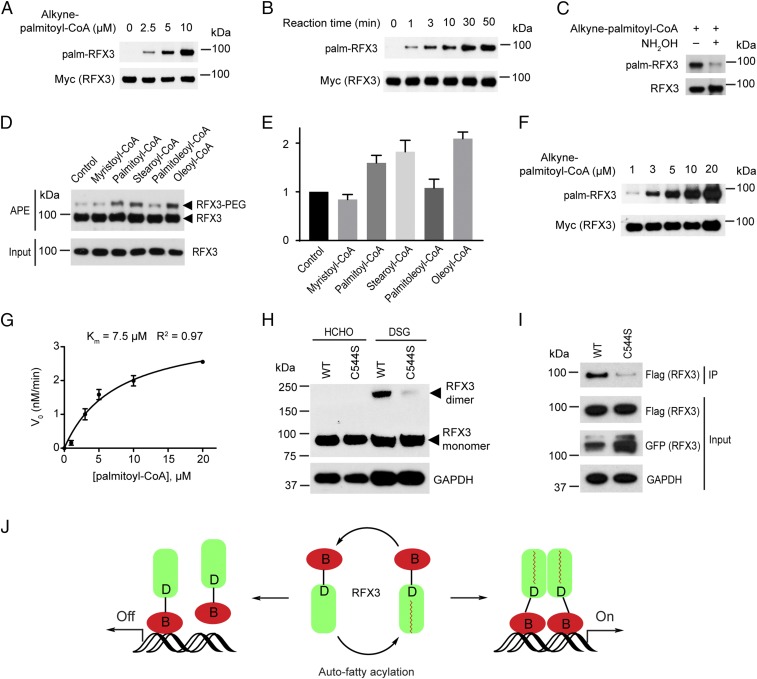
To test this hypothesis, we used the recombinant human RFX3 protein in biochemical assays in vitro. RFX3 protein was incubated with physiologically relevant concentrations (2.5 μM, 5 μM, or 10 μM) of a clickable analog of palmitoyl-CoA (15-hexadecynoic-CoA) for 30 min at neutral pH, followed by Click reaction with biotin-azide. The streptavidin blot showed that RFX3 is strongly fatty acylated in vitro in the absence of ZDHHC-PATs in a palmitoyl-CoA concentration-dependent manner (Fig. 3A), confirming that RFX3 fatty acylation is an “autoacylation” and nonenzymatic process. Next, we fixed the concentration of 15-hexadecynoic-CoA at 5 μM and tested the autoacylation of RFX3 at different time points. Our results suggested that the fatty acylation level of RFX3 increases in a time-dependent manner (Fig. 3B). Treating the Click reaction products with 2.5% (vol/vol) NH2OH substantially decreased RFX3 fatty acylation levels, suggesting the in vitro fatty acylation of recombinant RFX3 is sensitive to NH2OH (Fig. 3C). To measure the stoichiometry of fatty acyl-CoA modification of the recombinant RFX3 in vitro, we performed in vitro APE assay. Briefly, we incubated different fatty acyl-CoA with the recombinant RFX3 for 45 min followed by APE analysis in vitro. The in vitro APE results revealed that there is only one slower migrating PEGylated polypeptide band corresponding to the fatty acylated RFX3 (Fig. 3D), suggesting only one cysteine residue of RFX3 is modified by fatty acyl-CoA in vitro. As expected, we found that recombinant RFX3 contains a certain amount of fatty acylated form. In addition, the fatty acylated RFX3 level was quantified in three independent experiments. Consistent with the cell-based APE results, the recombinant RFX3 has preferences for 18-carbon saturated stearoyl-CoA and unsaturated oleoyl-CoA (Fig. 3E). Next, we fixed the concentrations of RFX3 protein at 0.1 μM and reaction time for 10 min and determined initial velocity (V0) of RFX3 autoacylation reactions under different concentrations of palmitoyl-CoA. The apparent Km of palmitoyl-CoA on RFX3 palmitoylation is around 7.5 μM (Fig. 3 F and G), which is comparable to the Km of ZDHHC-PATs (60). Taken together, we found RFX3 possesses enzyme-like activities and could bind to fatty acyl-CoA and undergo autoacylation.
Fig. 3.
Auto-fatty acylation of RFX3 is required for its dimerization. (A) RFX3 is fatty acylated in a fatty acyl-CoA concentration-dependent manner in vitro. (B) RFX3 is fatty acylated in a time-dependent manner in vitro. (C) Hydroxylamine treatment substantially decreased in vitro fatty acylated RFX3. (D) Fatty acyl-CoA preferences of RFX3 in vitro by APE assay. (E) Quantification of in vitro fatty acylated RFX3 normalized by RFX3 protein level. (F) Representative streptavidin blot for measuring Km of in vitro fatty acylation of RFX3. (G) Plot of Km of in vitro fatty acylation of RFX3. Values represent the average ± SEM of three experiments. (H) Protein cross-linking results show stronger homodimerization of WT RFX3 than that of the fatty acylation-deficient C544S mutant. (I) Co-IP experiment shows the dimerization level decreased in the fatty acylation-deficient C544S mutant. (J) Cartoon showing fatty acylation of RFX3 regulates its dimerization and transcriptional activities.
To test whether RFX3 undergoes spontaneous de-fatty acylation in vitro, we have performed auto-defatty acylation assay in vitro. Briefly, we first incubated alkyne-palmitoyl-CoA with the recombinant RFX3 for 2 h at room temperature and then removed the excess unreacted alkyne-palmitoyl-CoA by buffer exchange using the Amicon Ultra-0.5 centrifugal filter unit. RFX3 fatty acylation level was detected every hour for 5 times. After removal of alkyne-palmitoyl-CoA, there was no substantial decrease of RFX3 fatty acylation level in a time-dependent manner (SI Appendix, Fig. S9), suggesting RFX3 might not undergo spontaneous de-fatty acylation in vitro.
As the RFX3 fatty acylation site is located in the dimerization domain of RFX3, we speculate that fatty acylation of RFX3 might affect its dimerization. We performed in vivo cross-linking assay to analyze RFX3 protein dimerization. We transfected WT RFX3 or the C544S mutant into the HEK293A cells. After 36 h, cells were treated with 1% formaldehyde (HCHO) as a negative control or 1 mM disuccinimidyl glutarate (DSG), which is a homobifunctional cross-linker with the amine-reactive N-hydroxysuccinimide ester group. After 10 min of incubation in cold PBS, HCHO- and DSG-treated cells were quenched with 100 mM glycine, pH 3.0, and 500 mM Tris⋅HCl, pH 8.0, respectively, for an additional 15 min. We observed a strong dimerization band of WT RFX3, whereas the dimerization band of the fatty acylation-deficient mutant C544S decreased substantially in Western blotting analysis (Fig. 3H). To further confirm that fatty acylation could regulate RFX3 dimerization, we carried out a coimmunoprecipitation (co-IP) experiment. Flag-tagged RFX3 (WT or C544S mutant) and GFP-tagged RFX3 (WT or C544S mutant) were cotransfected into HEK293A cells, respectively. The expression levels of Flag-tagged RFX3 in WT and the C544S mutant are at a comparable level. Although the expression level of the GFP-tagged C544S mutant is slightly higher than that of GFP-tagged WT RFX3, we have observed markedly decreased dimerization with fatty acylation-deficient C544S mutant (Fig. 3I), confirming that fatty acylation of RFX3 promotes its dimerization.
Previously, fatty acylation of proteins has been linked to the regulation of protein membrane localization. We carried out immunofluorescence (IF) assay to analyze the localization of WT or C544S mutant of RFX3. We found that the fatty acylation-deficient C544S mutant of RFX3, similar to the WT RFX3, mainly localizes to the nucleus in NIH 3T3 cells (SI Appendix, Fig. S10), suggesting that fatty acylation does not affect RFX3 localization. Taken together, we speculate that fatty acylation of RFX3 is required for its dimerization, thereby affecting its transcriptional activity (Fig. 3J)
Fatty Acylation Regulates RFX3-Mediated Ciliary Gene Expression and Ciliogenesis.
As RFX3 plays a well-defined role in ciliogenesis in vertebrates (17, 18, 24), we ask whether fatty acylation of RFX3 is required for ciliary gene expression and cilia formation. Mouse embryonic fibroblast NIH 3T3 cells are commonly used for the study of ciliogenesis. To further explore the functions of RFX3 fatty acylation, we generated NIH 3T3 cell lines stably expressing Flag-tagged WT RFX3 or the fatty acylation-deficient C544S mutant. Western blot analysis confirmed the comparable expression levels of WT and the mutant in NIH 3T3 stable cell lines (SI Appendix, Fig. S11A). We then performed quantitative RT-PCR (qRT-PCR) assay to analyze expression levels of RFX3 target genes including Dync2li1, Dnaic1, and Dnali1. Dync2li1 is a well-defined target gene of RFX3, encoding a protein of the dynein-2 complex that is essential for the intraflagellar transport system as well as generation and maintenance of cilia (17, 18, 22, 27, 61–63). Defects in this gene lead to dysfunction of the dynein-2 complex, abnormal cilia function, and Hh pathway impairment that causes several types of ciliopathy including short-rib polydactyly syndrome (SRPS) (64–66). Dnaic1/DNAI1 encodes a dynein complex protein in respiratory cilia, and defects in this gene lead to abnormal ciliary structure and function involved in the first identified human ciliopathy called primary ciliary dyskinesia (PCD) or Kartagener syndrome (67–70). Dnali1/DNALI1 encodes a flagella protein and is a potential candidate gene associated with PCD (71, 72). DNAI1 and DNALI1 have been identified as RFX3-associated ciliary genes in human airway epithelium (24). Our qRT-PCR results suggested that overexpression of WT RFX3, but not the fatty acylation-deficient mutant C544S, significantly increased the expression of Dync2li1, Dnaic1, and Dnali1 in NIH 3T3 cells (SI Appendix, Fig. S11B). To minimize the confounding effects from endogenous RFX3, we generated RFX3-knockout cell lines (KO-1 and KO-2) by using the CRISPR/Cas9 genome-editing system, and the knockout efficiency was evaluated by Western blot analysis (SI Appendix, Fig. S12A). We then performed qRT-PCR assay to analyze the expression of three RFX3 target genes (Dync2li1, Dnaic1, and Dnali1) and found loss of RFX3 led to decreased expression of these ciliary genes (SI Appendix, Fig. S12B), which is consistent with previous reports (17, 22, 27). RFX3 and other RFX family members, such as RFX1, may cooperatively regulate an overlapping set of target genes and might have functional redundancy (10), which might cause the relatively low-fold decrease of the ciliary genes in RFX3-knockout cells. We next performed an IF assay to analyze cilia formation in the two knockout cell lines. Interestingly, compared with control cell lines, the ciliated cells were statistically significantly decreased in the two RFX3 knockout cell lines (SI Appendix, Fig. S12 C and D), consistent with the previous reports that RFX3 plays a regulatory role in cilia formation in NIH 3T3 cells.
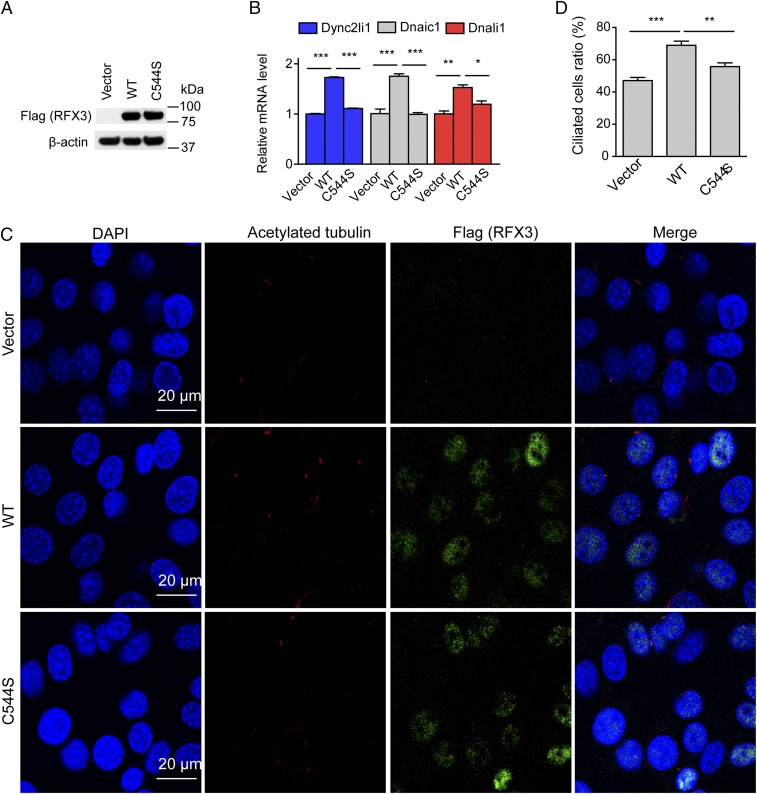
To further evaluate the effect of RFX3 fatty acylation on its target gene expression and cilia formation, we next re-expressed WT RFX3 and the fatty acylation-deficient C544S mutant in the established RFX3-knockout NIH 3T3 cells. Western blot confirmed that the RFX3 protein expression levels are comparable in cells expressing WT RFX3 and the C544S mutant (Fig. 4A). Our qRT-PCR results suggested that re-expression of WT RFX3, but not the fatty acylation-deficient C544S mutant, markedly increased the expression of the RFX3 target genes, including Dync2li1, Dnaic1, and Dnali1 (Fig. 4B). We next performed IF assay to analyze the cilia formation in the established stable cells, and the percentage of ciliated cells was quantified. Compared with the RFX3-knockout cells, reintroducing WT RFX3, but not the C544S mutant, significantly increased the percentage of ciliated cells (Fig. 4 C and D). Taken together, our results suggested that fatty acylation of RFX3 plays a regulatory role in ciliary gene expression and ciliogenesis.
Fig. 4.
Fatty acylation of RFX3 regulates ciliary gene expression and cilia formation. (A) Western blot analysis shows comparable expression levels of WT RFX3 and the C544S mutant in the established stable cells. (B) qRT-PCR results show expression of ciliary genes in different stable cell lines expressing vector control, WT RFX3, and C544S mutant. (C) Representative images of cilia in different NIH 3T3 stable cell lines expressing vector control, WT RFX3, and C544S mutant. Cells were stained with anti-acetylated tubulin antibody (red), anti-RFX3 antibody (green), and DAPI (blue). (Scale bars: 20 μm.) (D) Quantification of the percentage of ciliated cells in different stable cell lines. At least 200 cells were analyzed for each cell line in three independent experiments. Values represent the average ± SEM of three experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (different from control; Student’s t test).
Fatty Acylation of RFX3 Regulates Cilia Elongation and Hh Signaling.
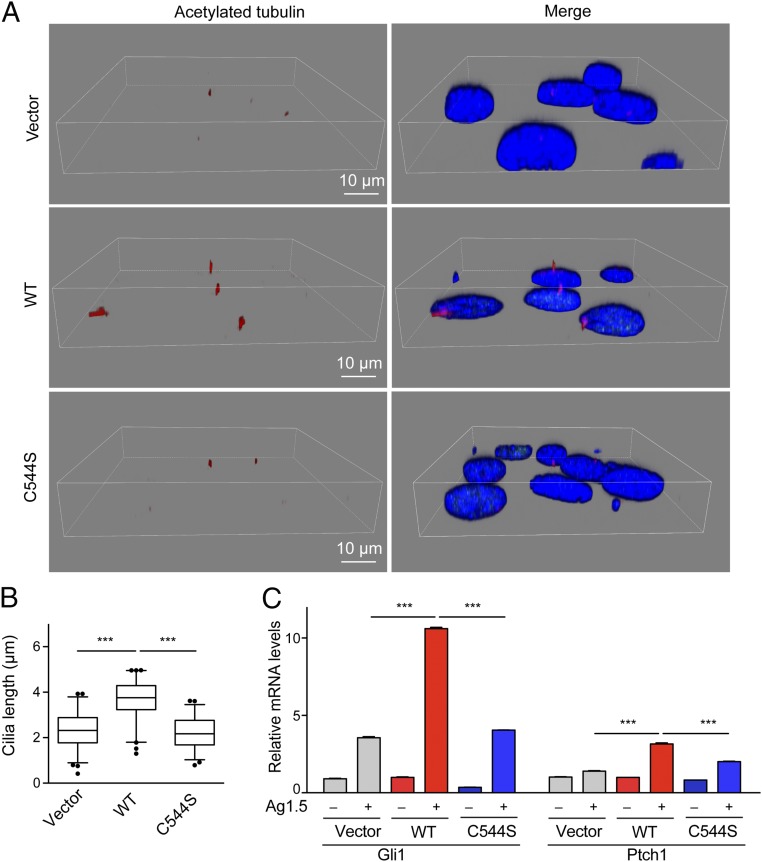
RFX3 has been reported as a key transcription factor associated with cilia growth (18, 22). We noticed that cilia are shorter in the RFX3-knockout NIH 3T3 cells than in control cells. To analyze the cilia length more quantitatively and accurately, we performed three-dimensional image constructions using confocal microscopy to visualize and measure the cilia length in cells. Reintroducing WT RFX3 into the RFX3-knockout NIH 3T3 cells, but not the fatty acylation-deficient mutant C544S, significantly increased the cilia length (Fig. 5 A and B), suggesting that fatty acylation of RFX3 is required for cilia elongation in NIH 3T3 cells.
Fig. 5.
Fatty acylation of RFX3 regulates cilia growth and Hh signaling. (A) 3D reconstructions of confocal imaging of cilia show the length of cilia from the indicated cell lines. Cells were stained with anti-acetylated tubulin antibody (red), anti-RFX3 antibody (green) and DAPI (blue). (Scale bars: 10 μm.) (B) The box plot shows the mean length of cilia from different cell lines measured in confocal-microscopy stacks. At least 200 cilia were measured for each cell line. Statistical significance of the difference in cilia length was analyzed using Student’s t test (***P < 0.0001). (C) Reintroducing WT RFX3 into the RFX3 knockout cells, but not the fatty acylation-deficient mutant C544S, increased the expression of Hh target genes Gli and Ptch1 upon stimulation with Hh agonist Hh-Ag1.5. Values represent the average ± SEM of three experiments. ***P < 0.001 (different from control; Student’s t test).
Cilia are crucial for Hh signaling, and multiple Hh pathway components are required to localize to cilia for proper signal transduction (2). Loss of cilia is linked to defective Hh signaling. Therefore, we asked whether fatty acylation of RFX3 could regulate Hh signaling, through regulating ciliogenesis. After treatment with 100 nM of Smoothened agonist Hh-Ag-1.5 overnight, qRT-PCR assay was performed to evaluate Hh target genes (Gli1 and Ptch1) in the stable RFX3-knockout NIH 3T3 cell lines. Compared with vector control, the expression levels of Gli and Ptch1 are significantly decreased in the RFX3-knockout cells (SI Appendix, Fig. S13), suggesting that RFX3 is required for Hh signaling. Reintroducing WT RFX3, but not the fatty acylation-deficient C544S mutant, significantly restored the expression levels of Gli1 and Ptch1 (Fig. 5C), suggesting that fatty acylation of RFX3 could indeed regulate Hh signaling, possibly through regulating ciliogenesis.
Discussion
Protein S-fatty acylation is a reversible lipid modification, playing important roles in regulating localization and functions of many proteins. Here we show that the transcription factor RFX3 is S-fatty acylated on an evolutionarily conserved cysteine residue in the dimerization domain (Fig. 1). Using different chemical reporters of fatty acylation, we found that fatty acylation of RFX3 exhibits preferences for 18-carbon stearic acid and oleic acid (Fig. 2 B and C). Future 3D structural studies of the RFX3 dimerization domain may provide direct evidences for the fatty acid selectivity profiles of RFX3. Pharmacological inhibition of ACSL reduced the RFX3 fatty acylation level in cells (Fig. 2D), indicating that intracellular metabolites, such as the long-chain fatty acyl-CoA level, might regulate RFX3 fatty acylation. A recent study found that the metabolite stearic acid can function as a regulator of mitochondrial function in response to diet by stearoylation of human transferrin receptor 1 (TFR1) (73), suggesting an important role of metabolite stearic acid as a signaling molecule. Our study showed that RFX3 undergoes cycles of fatty acylation and de-fatty acylation, and the stearoylation turnover rate on RFX3 is about 1.4 h (Fig. 2 E and F). The 18-carbon stearic acid oleic acid (t1/2 = 1.2 h) exhibits a slightly shorter half-life than 16-carbon palmitic acid (t1/2 = 1.5 h) (SI Appendix, Fig. S5). Our ABHD screening results revealed that ABHD2/6 could decrease RFX3 fatty acylation (SI Appendix, Fig. S7), implying that de-fatty acylation of RFX3 might be an enzyme-mediated process. It is also possible that other unknown enzymes are de-fatty acylases of RFX3. Future follow-up studies will uncover the detailed regulation mechanism of RFX3 de-fatty acylation. We further demonstrated that RFX3, like TEAD, undergoes an auto-fatty acylation process (Fig. 3 A and B). Surprisingly, the recombinant RFX3 displays preferences for 18-carbon stearoyl-CoA and oleoyl-CoA in vitro (Fig. 3 D and E), which is consistent with the cell-based results. We calculated the apparent Km value (around 7.5 μM) of palmitoyl-CoA on RFX3 palmitoylation (Fig. 3 F and G), which is within the physiological range of intracellular fatty acyl-CoA concentrations. It has been reported that the total cellular concentrations of long-chain (14–20 carbon) fatty acyl-CoA range from 5 μM to 160 μM, depending on the tissue and its metabolic state (74–76). Our in vitro de-fatty acylation results indicated that RFX3 might not undergo spontaneous de-fatty acylation (SI Appendix, Fig. S9), which is in line with cell-based results supporting that RFX3 de-fatty acylation might be an enzymatic process. Another possibility is that RFX3 might exhibit enzyme-like properties and need a “substrate-like” protein to accept its fatty acyl group. We further found that fatty acylation of RFX3 is required for its dimerization (Fig. 3 H and I) and transcriptional activities (Fig. 4B), but not for its nuclear localization (SI Appendix, Fig. S10). Intracellular metabolites may affect the availability of long-chain fatty acyl-CoA and thereby regulate RFX3 auto-fatty acylation and its transcriptional activities. Taken together, our study provided a potential connection between fatty acylation of RFX3 and long-chain fatty acyl-CoA in signal transduction. However, we cannot completely exclude that Cys544 may also contribute to RFX3 dimerization via disulfide bond formation, which could be an alternative or complementary mechanism regulating RFX3.
RFX3 is ubiquitously expressed in various tissues with ciliated cells, such as embryonic node, brain, epidermis, and pancreas, and plays a pivotal role in formation, growth, and function of cilia (17, 18, 22, 23, 25). The cilia can be substantially affected in multiple ways due to the defects in RFX3, such as shortened cilia and a reduced number of cilia (17, 23, 27). Our results suggested that fatty acylation of RFX3 is required for the expression of ciliary genes (Fig. 4B), cilia formation (Fig. 4 C and D), and growth (Fig. 5 A and B). The three detected ciliary genes (Dync2li1, Dnaic1, and Dnali1) are involved in assembly and function of cilia and are associated with several types of ciliopathies (17, 18, 22, 27, 61–72), suggesting fatty acylation of RFX3 may play a regulatory role in the related ciliopathies. Cilia are essential for Hh signaling pathways (2). Our results indicated that fatty acylation of RFX3 regulates the expression of Hh target genes Gli1 and Ptch1 (Fig. 5C), probably through regulating cilia formation. In addition to ciliogensis, RFX3 regulates pancreatic β-cell differentiation and glucokinase gene Gck expression, which is crucial for the mature β-cell functions (22, 27, 28). We found that knockout RFX3 in the β-cell line (MIN6) leads to significantly reduced expression of the Gck gene (SI Appendix, Fig. S14 A and B), consistent with the previous study (27). Reintroducing WT RFX3, but not the fatty acylation-deficient C544S mutant, significantly restored the expression levels of Gck (SI Appendix, Fig. S14 C and D), suggesting that fatty acylation of RFX3 is required for Gck expression in the MIN6 pancreatic β-cell line. Previous studies showed that RFX6 is a master regulator of the pancreatic program and is involved in progenitor specification, endocrine cell differentiation, β-cell function, and glucose homeostasis (28, 77–79). RFX3 and RFX6 have been proposed to cooperatively regulate common genes required for the pancreatic program (28). Fatty acylation of RFX3 may be involved in this process and regulate the interactions between RFX3 and RFX6 by forming the RFX3–RFX6 heterodimer.
RFX1, RFX2, and RFX3 are evolutionarily related proteins that share a highly conserved dimerization domain (SI Appendix, Fig. S15) (13, 14, 33). Our MS data as well as previously reported chemoproteomic profiling data suggested that RFX1 and RFX3 are potential fatty acylated proteins (SI Appendix, Fig. S1) (50–53). Our biochemical study confirmed that RFX1 and RFX3 are S-fatty acylated proteins (Fig. 1 and SI Appendix, Fig. S2). It is likely that RFX2 is also fatty acylated (SI Appendix, Figs. S3 and S15). Further studies may provide detailed information on how fatty acylation affects the function of RFX1 and RFX2. A recent study found that RFX2 can recruit and stabilize distal enhancers to promoters via a chromatin loop created by RFX2 dimerization, enabling distantly bound factor FOXJ1 to promote cilia gene expression during multiciliated cell differentiation (80). RFX3 may activate target genes via dimerization in a manner similar to RFX2. Another recent study reported that transcription factors including RFX1–6 are highly associated with the risk of T2D (29). RFX3 has been demonstrated to interact directly with RFX1, RFX2, RFX4, RFX6, and another key ciliary transcription factor, FOXJ1 (SI Appendix, Fig. S16) (14, 24, 28, 40, 43). Overall, our study suggested that fatty acylation of RFX3 may play a regulatory role in the protein interaction network by affecting homo- or heterodimerization, thereby regulating ciliogenesis as well as tissue specification. Deregulation of fatty acid metabolism might affect fatty acylation of RFX3 and thereby the RFX3-mediated protein interaction network, leading to ciliopathies and metabolic disorders such as diabetes.
Materials and Methods
Cell Labeling and Click Reaction.
Cells were treated with chemical probes or DMSO for 4 h or overnight. Labeled cells were washed three times with cold PBS and lysed with lysis buffer containing 50 mM TEA-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM PMSF, and 1× EDTA-free cOmplete protease inhibitor mixture (Roche). Click reaction was performed in 100 μL lysis buffer containing 50 μg protein, 100 μM biotin-azide, 1 mM TCEP, 100 μM TBTA, and 1 mM CuSO4 for 1 h at room temperature. The reactions were terminated by the addition of 20 μL of 6× SDS sample loading buffer. The samples were heated for 5 min at 95 °C before being subjected to SDS/PAGE analysis.
MS Analysis.
Cells were harvested, followed by Click reaction. Click reaction products were precipitated using methanol. The protein pellets were dissolved in resuspension buffer containing 50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 10 mM EDTA, and 2% SDS. Then samples were diluted with an equal volume of buffer containing 50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 10 mM EDTA, and 1% Nonidet P-40 and incubated with prewashed streptavidin-agarose beads overnight at room temperature. The beads were washed three times with buffer containing 50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 10 mM EDTA, and 0.2% SDS. Then samples were washed an additional 15 times with buffer containing 50 mM Tris⋅HCl, pH 7.4, and 150 mM NaCl. The prepared samples were directly digested on beads, and supernatants were collected for LC-MS/MS analysis.
An extended section is provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Masaki Fukata (National Institute for Physiological Sciences, Japan) for the expression vectors of ZDHHC and ABHD proteins, the Confocal Imaging Core at Cutaneous Biology Research Center of Massachusetts General Hospital and the Shared Instrumentation Grant that covered the purchase of the microscope (1S10RR027673-01), and the Taplin Mass Spectrometry Core Facility at Harvard Medical School for proteomic studies. This work was supported by a Samuel M. Fisher Memorial–MRA (Melanoma Research Alliance) Established Investigator Award and National Institutes of Health Grants R01CA181537 and R01DK107651-01 (to X.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.D.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800949115/-/DCSupplemental.
References
- 1.Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ezratty EJ, et al. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145:1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phua SC, et al. Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell. 2017;168:264–279.e15. doi: 10.1016/j.cell.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novarino G, Akizu N, Gleeson JG. Modeling human disease in humans: The ciliopathies. Cell. 2011;147:70–79. doi: 10.1016/j.cell.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waters AM, Beales PL. Ciliopathies: An expanding disease spectrum. Pediatr Nephrol. 2011;26:1039–1056. doi: 10.1007/s00467-010-1731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piasecki BP, Burghoorn J, Swoboda P. Regulatory factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc Natl Acad Sci USA. 2010;107:12969–12974. doi: 10.1073/pnas.0914241107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: Transcriptional networks regulating ciliogenesis. Development. 2014;141:1427–1441. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- 11.Thomas J, et al. Transcriptional control of genes involved in ciliogenesis: A first step in making cilia. Biol Cell. 2010;102:499–513. doi: 10.1042/BC20100035. [DOI] [PubMed] [Google Scholar]
- 12.Reith W, et al. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 13.Chu JS, Baillie DL, Chen N. Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC Evol Biol. 2010;10:130. doi: 10.1186/1471-2148-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aftab S, Semenec L, Chu JS, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 2008;8:226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- 16.Chung MI, et al. RFX2 is broadly required for ciliogenesis during vertebrate development. Dev Biol. 2012;363:155–165. doi: 10.1016/j.ydbio.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnafe E, et al. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol Cell Biol. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Zein L, et al. RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. J Cell Sci. 2009;122:3180–3189. doi: 10.1242/jcs.048348. [DOI] [PubMed] [Google Scholar]
- 19.Ashique AM, et al. The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Sci Signal. 2009;2:ra70. doi: 10.1126/scisignal.2000602. [DOI] [PubMed] [Google Scholar]
- 20.Manojlovic Z, Earwood R, Kato A, Stefanovic B, Kato Y. RFX7 is required for the formation of cilia in the neural tube. Mech Dev. 2014;132:28–37. doi: 10.1016/j.mod.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurençon A, et al. Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 2007;8:R195. doi: 10.1186/gb-2007-8-9-r195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ait-Lounis A, et al. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007;56:950–959. doi: 10.2337/db06-1187. [DOI] [PubMed] [Google Scholar]
- 23.Baas D, et al. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur J Neurosci. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- 24.Didon L, et al. RFX3 modulation of FOXJ1 regulation of cilia genes in the human airway epithelium. Respir Res. 2013;14:70. doi: 10.1186/1465-9921-14-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benadiba C, et al. The ciliogenic transcription factor RFX3 regulates early midline distribution of guidepost neurons required for corpus callosum development. PLoS Genet. 2012;8:e1002606. doi: 10.1371/journal.pgen.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnani D, et al. The ciliogenic transcription factor Rfx3 is required for the formation of the thalamocortical tract by regulating the patterning of prethalamus and ventral telencephalon. Hum Mol Genet. 2015;24:2578–2593. doi: 10.1093/hmg/ddv021. [DOI] [PubMed] [Google Scholar]
- 27.Ait-Lounis A, et al. The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression. Diabetes. 2010;59:1674–1685. doi: 10.2337/db09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith SB, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varshney A, et al. Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc Natl Acad Sci USA. 2017;114:2301–2306. doi: 10.1073/pnas.1621192114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, et al. Genome-wide association analysis identifies three new risk loci for gout arthritis in Han Chinese. Nat Commun. 2015;6:7041. doi: 10.1038/ncomms8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkon R, et al. RFX transcription factors are essential for hearing in mice. Nat Commun. 2015;6:8549. doi: 10.1038/ncomms9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reith W, et al. MHC class II regulatory factor RFX has a novel DNA-binding domain and a functionally independent dimerization domain. Genes Dev. 1990;4:1528–1540. doi: 10.1101/gad.4.9.1528. [DOI] [PubMed] [Google Scholar]
- 33.Emery P, Durand B, Mach B, Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24:803–807. doi: 10.1093/nar/24.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reith W, et al. Cooperative binding between factors RFX and X2bp to the X and X2 boxes of MHC class II promoters. J Biol Chem. 1994;269:20020–20025. [PubMed] [Google Scholar]
- 35.Moreno CS, et al. Purified X2 binding protein (X2BP) cooperatively binds the class II MHC X box region in the presence of purified RFX, the X box factor deficient in the bare lymphocyte syndrome. J Immunol. 1995;155:4313–4321. [PubMed] [Google Scholar]
- 36.Linhoff MW, Wright KL, Ting JP. CCAAT-binding factor NF-Y and RFX are required for in vivo assembly of a nucleoprotein complex that spans 250 base pairs: The invariant chain promoter as a model. Mol Cell Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwama A, et al. Dimeric RFX proteins contribute to the activity and lineage specificity of the interleukin-5 receptor alpha promoter through activation and repression domains. Mol Cell Biol. 1999;19:3940–3950. doi: 10.1128/mcb.19.6.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caretti G, et al. Dissection of functional NF-Y-RFX cooperative interactions on the MHC class II Ea promoter. J Mol Biol. 2000;302:539–552. doi: 10.1006/jmbi.2000.4028. [DOI] [PubMed] [Google Scholar]
- 39.Zhu XS, et al. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morotomi-Yano K, et al. Human regulatory factor X 4 (RFX4) is a testis-specific dimeric DNA-binding protein that cooperates with other human RFX members. J Biol Chem. 2002;277:836–842. doi: 10.1074/jbc.M108638200. [DOI] [PubMed] [Google Scholar]
- 41.Briggs L, Laird K, Boss JM, Garvie CW. Formation of the RFX gene regulatory complex induces folding of the interaction domain of RFXAP. Proteins. 2009;76:655–664. doi: 10.1002/prot.22379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newton FG, et al. Forkhead transcription factor Fd3F cooperates with Rfx to regulate a gene expression program for mechanosensory cilia specialization. Dev Cell. 2012;22:1221–1233. doi: 10.1016/j.devcel.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 44.Linder ME, Deschenes RJ. Palmitoylation: Policing protein stability and traffic. Nat Rev Mol Cell Biol. 2007;8:74–84. doi: 10.1038/nrm2084. [DOI] [PubMed] [Google Scholar]
- 45.Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol. 2010;6:498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 46.Hang HC, Linder ME. Exploring protein lipidation with chemical biology. Chem Rev. 2011;111:6341–6358. doi: 10.1021/cr2001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng B, et al. 2-Bromopalmitate analogues as activity-based probes to explore palmitoyl acyltransferases. J Am Chem Soc. 2013;135:7082–7085. doi: 10.1021/ja311416v. [DOI] [PubMed] [Google Scholar]
- 48.Zheng B, Jarugumilli GK, Chen B, Wu X. Chemical probes to directly profile palmitoleoylation of proteins. ChemBioChem. 2016;17:2022–2027. doi: 10.1002/cbic.201600403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen B, et al. ZDHHC7-mediated S-palmitoylation of Scribble regulates cell polarity. Nat Chem Biol. 2016;12:686–693. doi: 10.1038/nchembio.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2011;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang R, et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Martin BR, Cravatt BF, Hofmann SL. DHHC5 protein palmitoylates flotillin-2 and is rapidly degraded on induction of neuronal differentiation in cultured cells. J Biol Chem. 2012;287:523–530. doi: 10.1074/jbc.M111.306183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Percher A, et al. Mass-tag labeling reveals site-specific and endogenous levels of protein S-fatty acylation. Proc Natl Acad Sci USA. 2016;113:4302–4307. doi: 10.1073/pnas.1602244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thinon E, Fernandez JP, Molina H, Hang HC. Selective enrichment and direct analysis of protein S-palmitoylation sites. J Proteome Res. 2018;17:1907–1922. doi: 10.1021/acs.jproteome.8b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dekker FJ, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- 57.Lin DT, Conibear E. ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife. 2015;4:e11306. doi: 10.7554/eLife.11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokoi N, et al. Identification of PSD-95 depalmitoylating enzymes. J Neurosci. 2016;36:6431–6444. doi: 10.1523/JNEUROSCI.0419-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan P, et al. Autopalmitoylation of TEAD proteins regulates transcriptional output of the Hippo pathway. Nat Chem Biol. 2016;12:282–289. doi: 10.1038/nchembio.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jennings BC, Linder ME. DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J Biol Chem. 2012;287:7236–7245. doi: 10.1074/jbc.M111.337246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grissom PM, Vaisberg EA, McIntosh JR. Identification of a novel light intermediate chain (D2LIC) for mammalian cytoplasmic dynein 2. Mol Biol Cell. 2002;13:817–829. doi: 10.1091/mbc.01-08-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mikami A, et al. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J Cell Sci. 2002;115:4801–4808. doi: 10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- 63.Perrone CA, et al. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in chlamydomonas and mammalian cells. Mol Biol Cell. 2003;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor SP, et al. University of Washington Center for Mendelian Genomics Consortium Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome. Nat Commun. 2015;6:7092. doi: 10.1038/ncomms8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kessler K, et al. DYNC2LI1 mutations broaden the clinical spectrum of dynein-2 defects. Sci Rep. 2015;5:11649. doi: 10.1038/srep11649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schmidts M, et al. UK10K Exome sequencing identifies DYNC2H1 mutations as a common cause of asphyxiating thoracic dystrophy (Jeune syndrome) without major polydactyly, renal or retinal involvement. J Med Genet. 2013;50:309–323. doi: 10.1136/jmedgenet-2012-101284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zariwala MA, et al. Mutations of DNAI1 in primary ciliary dyskinesia: Evidence of founder effect in a common mutation. Am J Respir Crit Care Med. 2006;174:858–866. doi: 10.1164/rccm.200603-370OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leigh MW, Zariwala MA, Knowles MR. Primary ciliary dyskinesia: Improving the diagnostic approach. Curr Opin Pediatr. 2009;21:320–325. doi: 10.1097/MOP.0b013e328329cddb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noone PG, et al. Mutations in DNAI1 (IC78) cause primary ciliary dyskinesia. Chest. 2002;121(3) Suppl:97S. [PubMed] [Google Scholar]
- 70.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423–450. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 71.Loges NT, et al. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am J Hum Genet. 2009;85:883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rashid S, et al. The murine Dnali1 gene encodes a flagellar protein that interacts with the cytoplasmic dynein heavy chain 1. Mol Reprod Dev. 2006;73:784–794. doi: 10.1002/mrd.20475. [DOI] [PubMed] [Google Scholar]
- 73.Senyilmaz D, et al. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature. 2015;525:124–128. doi: 10.1038/nature14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tardi PG, Mukherjee JJ, Choy PC. The quantitation of long-chain acyl-CoA in mammalian tissue. Lipids. 1992;27:65–67. doi: 10.1007/BF02537062. [DOI] [PubMed] [Google Scholar]
- 76.Rosendal J, Knudsen J. A fast and versatile method for extraction and quantitation of long-chain acyl-CoA esters from tissue: Content of individual long-chain acyl-CoA esters in various tissues from fed rat. Anal Biochem. 1992;207:63–67. doi: 10.1016/0003-2697(92)90500-7. [DOI] [PubMed] [Google Scholar]
- 77.Zhu Z, et al. Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Cell Stem Cell. 2016;18:755–768. doi: 10.1016/j.stem.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chandra V, et al. RFX6 regulates insulin secretion by modulating Ca2+ homeostasis in human β cells. Cell Rep. 2014;9:2206–2218. doi: 10.1016/j.celrep.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 79.Piccand J, et al. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep. 2014;9:2219–2232. doi: 10.1016/j.celrep.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quigley IK, Kintner C. Rfx2 stabilizes Foxj1 binding at chromatin loops to enable multiciliated cell gene expression. PLoS Genet. 2017;13:e1006538. doi: 10.1371/journal.pgen.1006538. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.