Over the last few decades humanity has experienced a series of tremendous technological advances, especially in the realm of basic science. A plethora of new approaches has allowed us to appreciate life in previously unimaginable detail, prompting the realization that life in the microscopic world is not so different from what we can observe with our naked eyes. Microorganisms, for instance, like any other organism, compete with one another for resources and space. Bacteria often use simple mechanisms to occupy their niche such as rapid growth and biofilm formation. Bacteria also use ingenious strategies to maximize their success. Indeed, to engage in warfare, microorganisms often produce diffusible toxic antimicrobial compounds as well as other more complicated molecular weapons. Secretion systems are a particular kind of molecular weapon as they release or inject molecules and substrates that interact not only with hosts and predators but also with bacterial competitors. Given that the type VI secretion system (T6SS) has been recognized as a frequently used molecular weapon, it is not at all surprising that it is found widely distributed throughout diverse bacterial species (around 25% of all gram-negatives) (1). The T6SS system was discovered in Vibrio cholerae due to its toxicity against the social amoeba Dictyostelium discoideum (2). That same year, it was also suggested to contribute to the pathogenesis of Pseudomonas aeruginosa in cystic fibrosis patients (3). Follow-up studies showed that the T6SS of these two species could also be used against other bacteria (4, 5). Indeed, while being recognized for its involvement in host–pathogen and predator–prey interactions, the principal role of the T6SS is currently thought to be as an antibacterial weapon. In PNAS, Speare et al. (6) explore an accessory T6SS of the symbiotic bacterium Vibrio fischeri and describe how this T6SS contributes to light-organ occupancy within the squid host.
Since its discovery, knowledge about the T6SS has evolved considerably, especially concerning its structure, its molecular mechanism of action, and its possible function(s) under controlled laboratory conditions. The machinery is composed of two main parts: a membrane-spanning complex and a double tube structure that resembles a contractile bacteriophage tail (1, 7, 8). Upon contraction of the outer sheath structure, the inner tube and its membrane-puncturing spike are propelled out of the bacterium and into the neighboring cell (Fig. 1). During this process, effector proteins are delivered, which can intoxicate the target organism. These effectors have different functions but frequently target conserved bacterial or eukaryotic cellular structures such as the cell wall, the membrane compartment, nucleic acids, or the actin cytoskeleton (9). This suggests that the system is multipurpose and might have evolved to cope with a wide variety of both bacterial and eukaryotic competitors. Importantly, most effector proteins are coproduced with their cognate immunity protein, which prevents self-intoxication and undesired killing of sibling cells (10). This strategy also predicts that strains with matching sets of effector and immunity proteins could, in theory, coexist in the same environment, while those with incompatible effector–immunity sets will engage in a competitive relationship.
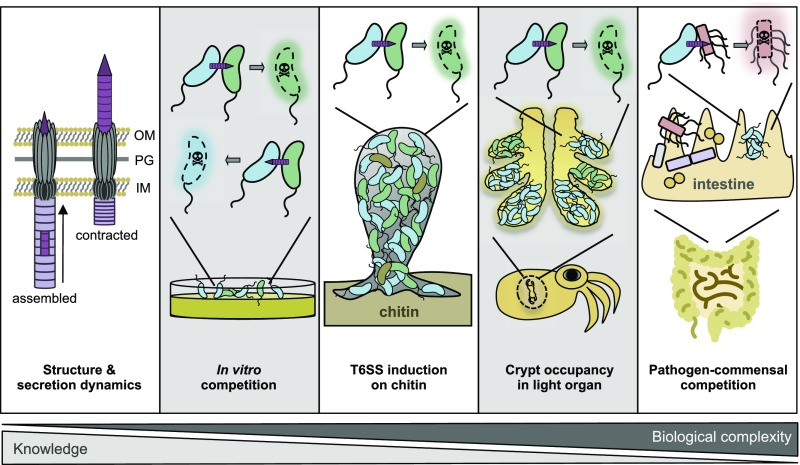
Fig. 1.
Studies on T6SSs range from the atomic over the molecular to the organismal scale. Little over a decade since its discovery, extensive knowledge has been gained on the structural aspects of T6SSs, as well as their assembly–contraction dynamics within individual bacterial cells. T6SS functionality is typically tested using in vitro assays in which the killing of prey (often E. coli as a surrogate) is enumerated. Recently, more complex systems have been studied, such as chitin-mediated T6SS induction, which fosters horizontal gene transfer in V. cholerae, light-organ occupancy among competing V. fischeri strains (6), and T6SS-mediated commensal invasion by human pathogens. Gray shaded boxes refer to the study by Speare et al. (6). IM, inner membrane; OM, outer membrane; PG, peptidoglycan.
The majority of studies on T6SS have focused on in vitro settings, for instance by competing bacterial strains with laboratory lines of Escherichia coli or with immunity-depleted variants of the same bacterial species (Fig. 1). While such experiments are of prime importance for advancing our understanding of how the T6SS works at the molecular scale, they neither provide insight into laboratory-silent T6SS systems (e.g., those that are only expressed under certain conditions) nor do they recapitulate the population dynamics of natural bacterial communities. Moving toward more natural but still tractable settings is often challenging but can provide crucial and pertinent insights into the impact that T6SSs have on competing bacteria. For example, Ma et al. (11) demonstrated that by using its T6SS, the soil bacterium Agrobacterium tumefaciens could kill E. coli in vitro but failed to eliminate P. aeruginosa under the same experimental conditions due to the competitor’s counterattack. Repeating the same experiments in planta, the natural niche of the tumor-inducing pathogen A. tumefaciens, resulted in a different outcome. Under such conditions, the T6SS-transported effector toxin provided A. tumefaciens with a colonization advantage over P. aeruginosa (11). A second example is related to the T6SS of V. cholerae, which, in pandemic isolates of this species, has long been recognized as being silent under laboratory conditions (2). Borgeaud et al. (12) discovered that chitin is a natural inducer of the T6SS in these strains (Fig. 1), consistent with the fact that many Vibrio species interact with and feed on natural chitinous surfaces. The authors further deciphered a regulatory link between T6SS induction and natural competence for transformation and demonstrated that T6SS-mediated interbacterial competition can be used by V. cholerae to acquire DNA from killed prey, thereby fostering horizontal gene transfer (12, 13). These examples highlight the need to move research toward more realistic conditions that mimic the natural habitats of the bacteria under investigation. Indeed, the V. cholerae example clearly highlights the danger that important interbacterial strategies might often be hidden under more artificial laboratory conditions.
Another appealing example of how bacteria use their T6SS for interbacterial competition under natural conditions comes from the study by Speare et al. (6) in PNAS. After colonizing the light organ of the bobtail squid Euprymna scolopes (which contains six spatially separated crypts), symbiotic V. fischeri multiply rapidly and upon reaching a high-cell-density state produce light. The squid uses this symbiont-produced light as an antipredatory camouflage strategy called counterillumination. The light emitted from the ventral light organ thereby mimics the down-welling moonlight and thus, by disguising its presence, protects the squid from predators during the night. As a consequence, the symbiosis is essential for the host, but not for the bacterium, as V. fischeri can also interact with other animals or adopt a planktonic lifestyle. Speare et al. (6) investigate the early stages of squid colonization, when the hatchling acquires its symbiotic partner from the surrounding seawater. While several mechanisms have been elucidated that contribute to the unique selection of V. fischeri as the sole symbiont of the animal’s light organ (14), the underlying molecular mechanism of a recent discovery, namely that crypts are rarely cocolonized by more than one strain of V. fischeri (15), remained elusive. This finding suggests that either a bottleneck restricts access to more than a single bacterium, or, as shown here, that interbacterial competition occurs inside the light organ (6). To address the monocolonization phenotype, the authors used six recent V. fischeri isolates derived from the light organs of two separate squids and competed those strains in vitro against the reference strain ES114. The results suggest the occurrence of two classes of V. fischeri isolates, namely those that eliminated ES114 from the coculture (hereafter called “lethal” strains) and those that did not impact the survival of the reference strain (“nonlethal” strains). Notably, the lethal attacks required direct cell-to-cell contact, suggesting the involvement of a molecular weapon. Indeed, comparative genomics revealed that the lethal strains harbor a genomic island encoding a bona fide T6SS. As all V. fischeri strains contain a primary T6SS (T6SS1), the function of which remains unknown, the lethal strain-specific T6SS was named T6SS2. Experimental validation of the in silico prediction showed a clear correlation between the presence of the T6SS2 and lethality. Furthermore, deletion of genes encoding an essential core component of either the T6SS1 or the T6SS2 confirmed the specific involvement of the T6SS2 in interbacterial killing in vitro. The authors also provide evidence that nonlethal strains could cooccur in vitro, while lethal strains outcompeted each other, especially after physical mixing and dilution (6). The latter process is expected to occur frequently inside the spatially separated crypts, as the animal vents its light organ each morning at dawn.
After a series of additional in vitro experiments to better understand the compatibility of strains isolated from the same or different animals, the authors tested whether T6SS2-mediated interstrain competition was required for the crypt monocolonization phenotype (15). To address this question, animals were inoculated with a mixture of bacterial strains that were distinguishable through their production of two different fluorescent proteins. The tested strains were the nonlethal strain ES114 and the lethal isolate FQ-A001, as well as its T6SS2-negative derivative. These in vivo data show that T6SS-incompatible strains did not cooccur inside individual crypts but were spatially separated within the same light organ (6) (Fig. 1). Importantly, standard isolation methods that are based on the enumeration of bacteria from animal tissues would not have captured the spatial separation between incompatible strains and might have led to the erroneous conclusion that the isolates do not engage in interbacterial warfare.
Besides the beauty of the concept, the results of this study bring up many additional questions. For instance, why do so many strains (∼50% in this study) lack the T6SS2 despite the striking fitness advantage it conveys in vivo? Is the primary T6SS1 also involved in interbacterial competition? While the authors tested T6SS1-deficient mutants in vitro, no such data were presented under in vivo conditions. It is well established that chitin-degradation products play a key role in the establishment of the V. fischeri–squid symbiosis (16). As chitin-degradation products induce the T6SS in V. cholerae (12, 13), it is conceivable that the same holds true for the T6SS1 of V. fischeri. Indeed, chitin-induced T6SS1 production could occur within the chitin-rich mucus matrix that coats the ciliated surface of the squid’s light organ, which might foster competitor exclusion upon initial association with the host.
In summary, the evidence provided by Speare et al. (6) suggests that the T6SS of a symbiont can be relevant in shaping microbial communities inside an animal host (6). Recent studies have addressed similar questions in the context of the mammalian microbiota (17–19) (Fig. 1). In these studies, the authors demonstrated that pathogens such as Shigella sonnei, Salmonella enterica serovar Typhimurium, and V. cholerae use their T6SS for interbacterial competition in vivo (17–19) and for niche occupancy (18). However, due to the complexity of the mammalian microbiota, which often complicates interpretation or in some cases even prevents the initial colonization by the species under test, most of these studies used animal models in which the microbiota had been artificially depleted. Future studies will therefore be required to better understand the contribution of T6SSs to pathogen invasion of otherwise healthy microbiota.
Acknowledgments
This work was supported by Swiss National Science Foundation Grant 31003A_162551, National Research Programme “Antimicrobial Resistance” (NRP72) Grant 407240_167061, and Consolidator Grant 724630-CholeraIndex from the European Research Council. M.B. is a Howard Hughes Medical Institute International Research Scholar (Grant 55008726).
Footnotes
The authors declare no conflict of interest.
See companion article on page E8528.
References
- 1.Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016;24:51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Pukatzki S, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mougous JD, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hood RD, et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci USA. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speare L, et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci USA. 2018;115:E8528–E8537. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand E, et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 9.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: Poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unterweger D, et al. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014;16:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]
- 13.Metzger LC, et al. Independent regulation of type VI secretion in Vibrio cholerae by TfoX and TfoY. Cell Reports. 2016;15:951–958. doi: 10.1016/j.celrep.2016.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFall-Ngai MJ. The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, et al. Intraspecific competition impacts Vibrio fischeri strain diversity during initial colonization of the squid light organ. Appl Environ Microbiol. 2016;82:3082–3091. doi: 10.1128/AEM.04143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kremer N, et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe. 2013;14:183–194. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sana TG, et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci USA. 2016;113:E5044–E5051. doi: 10.1073/pnas.1608858113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson MC, Vonaesch P, Saffarian A, Marteyn BS, Sansonetti PJ. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe. 2017;21:769–776. doi: 10.1016/j.chom.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhao W, Caro F, Robins W, Mekalanos JJ. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science. 2018;359:210–213. doi: 10.1126/science.aap8775. [DOI] [PMC free article] [PubMed] [Google Scholar]