Significance
The molecular clock provides an anticipatory mechanism, allowing organisms to prepare and respond to daily changes in the external environment. The response of the innate immune system to pathogenic threats is dependent on time of day; however, the molecular mechanisms underlying this have yet to be fully uncovered. We observe that the core molecular clock component, BMAL1, is crucial in promoting an antioxidant response in myeloid cells. Deletion of Bmal1 in macrophages disrupts NRF2 activity, facilitating accumulation of reactive oxygen species and the proinflammatory cytokine, IL-1β. Thus the molecular clock directly controls NRF2 transcriptional activity and antioxidant capacity to regulate IL-1β in myeloid cells.
Keywords: circadian clock, BMAL1, oxidative stress, inflammation, macrophage
Abstract
A variety of innate immune responses and functions are dependent on time of day, and many inflammatory conditions are associated with dysfunctional molecular clocks within immune cells. However, the functional importance of these innate immune clocks has yet to be fully characterized. NRF2 plays a critical role in the innate immune system, limiting inflammation via reactive oxygen species (ROS) suppression and direct repression of the proinflammatory cytokines, IL-1β and IL-6. Here we reveal that the core molecular clock protein, BMAL1, controls the mRNA expression of Nrf2 via direct E-box binding to its promoter to regulate its activity. Deletion of Bmal1 decreased the response of NRF2 to LPS challenge, resulting in a blunted antioxidant response and reduced synthesis of glutathione. ROS accumulation was increased in Bmal1−/− macrophages, facilitating accumulation of the hypoxic response protein, HIF-1α. Increased ROS and HIF-1α levels, as well as decreased activity of NRF2 in cells lacking BMAL1, resulted in increased production of the proinflammatory cytokine, IL-1β. The excessive prooxidant and proinflammatory phenotype of Bmal1−/− macrophages was rescued by genetic and pharmacological activation of NRF2, or through addition of antioxidants. Our findings uncover a clear role for the molecular clock in regulating NRF2 in innate immune cells to control the inflammatory response. These findings provide insights into the pathology of inflammatory conditions, in which the molecular clock, oxidative stress, and IL-1β are known to play a role.
Life on Earth follows a 24-h rhythm that is largely entrained by daily oscillations in light, due to the earth’s axial rotation (1). Cellular molecular clocks dictate variations in physiology and behavior that peak and trough within this 24-h timescale, termed circadian rhythms. Circadian rhythms generated by the molecular clock are maintained by autoregulatory transcriptional and translational feedback loops. BMAL1 is the core orchestrator of the molecular clock, and the only single clock gene deletion that leads to complete ablation of all rhythms. BMAL1 forms a heterodimeric partnership with CLOCK and binds to E-box sites located across the genome, inducing rhythmic expression in clock-controlled genes (1). Circadian rhythms are orchestrated by the master clock, which resides in the suprachiasmatic nucleus (SCN) of the hypothalamus. The SCN clock keeps peripheral clocks in synchrony with the external environment (1). This system provides circadian rhythms across a range of biological processes, including hormone secretion (2), metabolism (1), and immune function (1).
There is now a growing body of evidence that cells of the innate immune system, such as macrophages, possess molecular clocks (1). These endogenous clocks impose temporal gating across a range of functions, including phagocytosis (3), cytokine production (4), and antibacterial (5) and antiviral activity (6). These daily oscillations in immune parameters are hypothesized to prepare an organism for pathogenic threats, optimizing pathogen clearance and recovery. For example, mice have a heightened response to Listeria monocytogenes, lipopolysaccharide (LPS), and induction of experimental autoimmune encephalomyelitis (EAE), a model of multiple sclerosis, ahead of their active phase, which is exacerbated with Bmal1 deletion (5, 7–9). Bmal1 was found to regulate inflammatory responses after Toll-like receptor 4 (TLR4) activation in macrophages by regulating the epigenetic state of enhancers (10). Increased acetylation of lysine 27 of histone 3 was increased globally with Bmal1 deletion, prolonging activation of NF-κB target genes (10). Therefore, BMAL1 in the myeloid lineage appears to be a potent regulator of the inflammatory state at the cellular and organismal levels.
Reactive oxygen species (ROS) are signaling molecules that are critical for the progression of an immune response (11). Evidence exists of a role for BMAL1 in regulating ROS in multiple tissue types. Global deletion of Bmal1 produces an advanced aging phenotype, which is a result of increased oxidative stress (12). Specific deletion of Bmal1 in the pancreas generates a diabetic phenotype due to oxidative stress-induced death of β-cells (13). Deletion of Bmal1 in the brain results in oxidative stress-induced neurodegeneration and astrogliosis (14). Circadian rhythms in CD45+ leukocyte trafficking and migration are dictated by endogenous rhythms in ROS levels, which stabilize HIF-1α (15). In macrophages, ROS promote a proinflammatory response following LPS-induced activation of TLR4, boosting production of the cytokine IL-1β via stabilization of HIF-1α (16). IL-1β is a well-established pyrogen, critical in producing symptoms of fever and driving inflammation by inducing the expression of downstream proinflammatory molecules, such as COX2 and nitric oxide (17), and promoting cellular proliferation and differentiation of immune cells (18). Intriguingly, IL-1β levels display time-of-day variation in mice following L. monocytogenes infection (5) and in the serum and joints of mice subjected to a model of rheumatoid arthritis (19).
Given the damaging potential of ROS, antioxidants are produced to inhibit the oxidation of biological molecules and balance the oxidative state of cells (20). NRF2 is a basic leucine zipper (bZIP) transcription factor that plays a major role in regulating the expression of antioxidant proteins, protecting cells against oxidative damage triggered by ROS-induced injury or inflammation (20). NRF2 is regulated by KEAP1, which sequesters NRF2 to the cytoplasm, facilitating its degradation. Oxidative stress promotes nuclear translocation of NRF2, facilitating binding to antioxidant response elements (AREs) in the promoters of antioxidant genes, such as Hmox1, Gsr, and Nqo1 (20). NRF2 expression and activity has emerged as being under the control of the molecular clock in lung tissue. The BMAL1:CLOCK heterodimer was demonstrated to bind directly to E-box sites within the Nrf2 promoter, inducing its expression (21). NRF2 is expressed in macrophages and has been demonstrated to inhibit IL-1β production via suppression of ROS and HIF-1α (22). NRF2 has also been demonstrated to limit IL-6 and IL-1β production through direct binding of NRF2 to promoter regulatory regions, preventing RNA Pol II recruitment (23). Whether clock-mediated regulation of NRF2 and oxidative stress exists in macrophages has yet to be examined. Circadian gating of NRF2 in macrophages would provide a mechanism by which BMAL1 may temporally regulate inflammation via modulation of ROS levels (16) and direct binding of NRF2 to the IL-1β promoter (23).
In this study, we establish that NRF2 levels and activity are under circadian control in macrophages. Furthermore, we demonstrate that the lack of NRF2 activity in Bmal1-deficient macrophages results in higher ROS and an increase in the production of the proinflammatory cytokine, IL-1β. This provides a mechanism by which the molecular clock may regulate aspects of innate immunity, thus providing a potential mechanism for the rhythmicity in pathology observed in inflammatory conditions, including rheumatoid arthritis (19), osteoarthritis (24), asthma (25), multiple sclerosis (8), and sepsis (7).
Results
BMAL1 Is a Direct Positive Regulator of NRF2 Transcription and Activity.
In vivo studies were performed on Bmal1LoxP/LoxP Lys-MCre (Bmal1−/−) mice, in which Bmal1 is excised from cells of the myeloid lineage, versus control Bmal1wt/wt Lys-MCre (Bmal1+/+) mice. Bone marrow-derived macrophages (BMDMs) were prepared for in vitro studies. We compared unsynchronized wild-type BMDMs to macrophages with a dysfunctional clock by genetic deletion or knockdown of Bmal1. In certain experiments we synchronized BMDMs in vitro to observe basal rhythms in these cells and how they differed from Bmal1−/− BMDMs (26). Peritoneal myeloid cells were also utilized in this study to directly measure time-of-day differences.
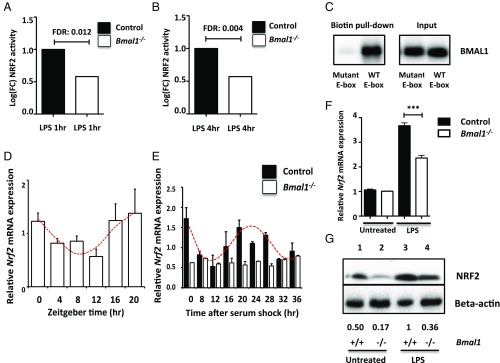
To understand the impact of Bmal1 deletion on the LPS response in macrophages, we performed an unbiased screening approach of transcription factor activity, termed transcription factor sequencing (TF-Seq) (27). The system utilizes lentiviral reporter vectors, which contain the response element for a transcription factor of interest. This response element was linked to a Luc2p promoter to allow detection. Ultimately this allows for unbiased measurement of the activity of numerous transcription factors in a cell type of interest. Relative activity of the NRF2 transcription factor was significantly decreased in Bmal1−/− macrophages following 1 (Fig. 1A) and 4 h (Fig. 1B) of LPS stimulation. The NRF2 pathway was the only pathway in the screen to be significantly altered with Bmal1 deletion (a full list of assayed transcription factors can be found in SI Appendix, Fig. S1).
Fig. 1.
Rhythms in NRF2 levels and activity are disrupted with Bmal1 deletion. Bmal1+/+ and Bmal1−/− BMDMs were analyzed by TF-Seq. Bar charts reveal the relative activity of NRF2 in Bmal1+/+ and Bmal1−/− BMDMs following (A) 1 or (B) 4 h of LPS stimulation (n = 3). (C) Wild-type BMDMs were lysed and the samples were exposed to biotinylated primer sequences for the E-box site located in the Nrf2 promoter (WT E-box) or a mutated control of the E-box. The sequences were then isolated using streptavidin beads before performing an immunoblot for BMAL1. (D) Peritoneal cells were isolated from wild-type mice at 4-h intervals (ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20) and Nrf2 mRNA was measured by qPCR. A cosinor regression model was used to test the null hypothesis that the amplitude of expression = 0. The presence of the red line indicates the presence of a significant rhythm (n= 3–6). (E) Bmal1+/+ and Bmal1−/− BMDMs were synchronized using 50% horse serum. RNA was extracted at 4-h intervals for 36 h and Nrf2 mRNA was measured by qPCR. A cosinor regression model was used to test the null hypothesis that the amplitude of expression = 0. The presence of the red line indicates the presence of a significant rhythm (n = 3). (F) Nrf2 mRNA was measured by qPCR in Bmal1+/+ and Bmal1−/− BMDMs following 24 h of LPS (100 ng/mL) (n = 4). (G) Immunoblot of NRF2 levels following 24 h of LPS (100 ng/mL) in Bmal1+/+ and Bmal1−/− BMDMs. Immunoblots are a representative of at least three independent experiments. Values provided below each lane indicate relative densitometry of each band. Statistical significance in graphs A and B was determined by false discovery rate (FDR); in D and E, by establishing a cosinor regression model; and in graph F, by one-way ANOVA. ***P ≤ 0.001.
To follow on from this result, we investigated the mechanisms by which BMAL1 might regulate NRF2 in macrophages. We performed an oligonucleotide pull-down experiment to demonstrate that BMAL1 binds to an E-box sequence in the Nrf2 promoter in macrophages, and binding is prevented when this E-box is mutated (Fig. 1C). We also performed an analysis of a previously published ChIP-Seq dataset, monitoring BMAL1 binding to DNA in peritoneal macrophages (10). We found a significant peak of BMAL1 binding to this same E-box in the Nrf2 promoter in this dataset (SI Appendix, Fig. S2). Binding of BMAL1 to this E-box in the Nrf2 promoter has previously been described in lung (21) and pancreatic tissue (13).
We next examined whether time of day impacted the transcription of Nrf2 in macrophages. Peritoneal cells were isolated from mice every 4 h over a 24-h period and RNA was analyzed. Zeitgeber time (ZT) is defined as the time in hours following the onset of light in the animal facility. ZT0 corresponds to lights on, whereas ZT12 refers to lights off. The expected rhythm in clock gene expression was observed for Bmal1 (SI Appendix, Fig. S3A) and a significant diurnal rhythm in Nrf2 expression was detected (Fig. 1D). Given the approximate trough of Bmal1 and Nrf2 at ZT8 and peak at ZT20, we extracted peritoneal cells from mice injected with PBS or LPS at these times and measured BMAL1 and NRF2 protein. In line with mRNA expression levels, BMAL1 and NRF2 protein were higher at ZT20 relative to ZT8 (SI Appendix, Fig. S3B). We next performed the serum shock protocol (26) in control and Bmal1−/− BMDMs to rule out any effects of light and/or cell extrinsic effect on the cycling of Nrf2 mRNA. Bmal1+/+ and Bmal1−/− BMDMs were synchronized with 50% horse serum and RNA was collected over a 36-h period. We observed clear rhythms in the expression of Bmal1 (SI Appendix, Fig. S3C) and Nrf2 (Fig. 1E) in Bmal1+/+ BMDMs. However, Nrf2 transcription was not rhythmic in Bmal1−/− BMDMs and expression was constitutively lower (Fig. 1E).
We next examined the effects of LPS challenge on NRF2 expression in unsynchronized Bmal1−/− BMDMs and found that both mRNA (Fig. 1F) and protein (Fig. 1G) were decreased in response to LPS. Interestingly, basal levels of NRF2 were also lower in Bmal1−/− BMDMs (Fig. 1G). To exclude the possibility that deletion of Bmal1 was impacting the differentiation of macrophages, we targeted Bmal1 by siRNA in fully differentiated macrophages (SI Appendix, Fig. S4A). We observed decreased expression of Nrf2 mRNA (SI Appendix, Fig. S3D) and protein (SI Appendix, Fig. S3E) in response to LPS stimulation with siRNA knockdown of Bmal1. Thus, we demonstrate circadian- and BMAL1-mediated regulation of NRF2.
NRF2 Antioxidant Pathway Is Down-Regulated upon Deletion of Bmal1.
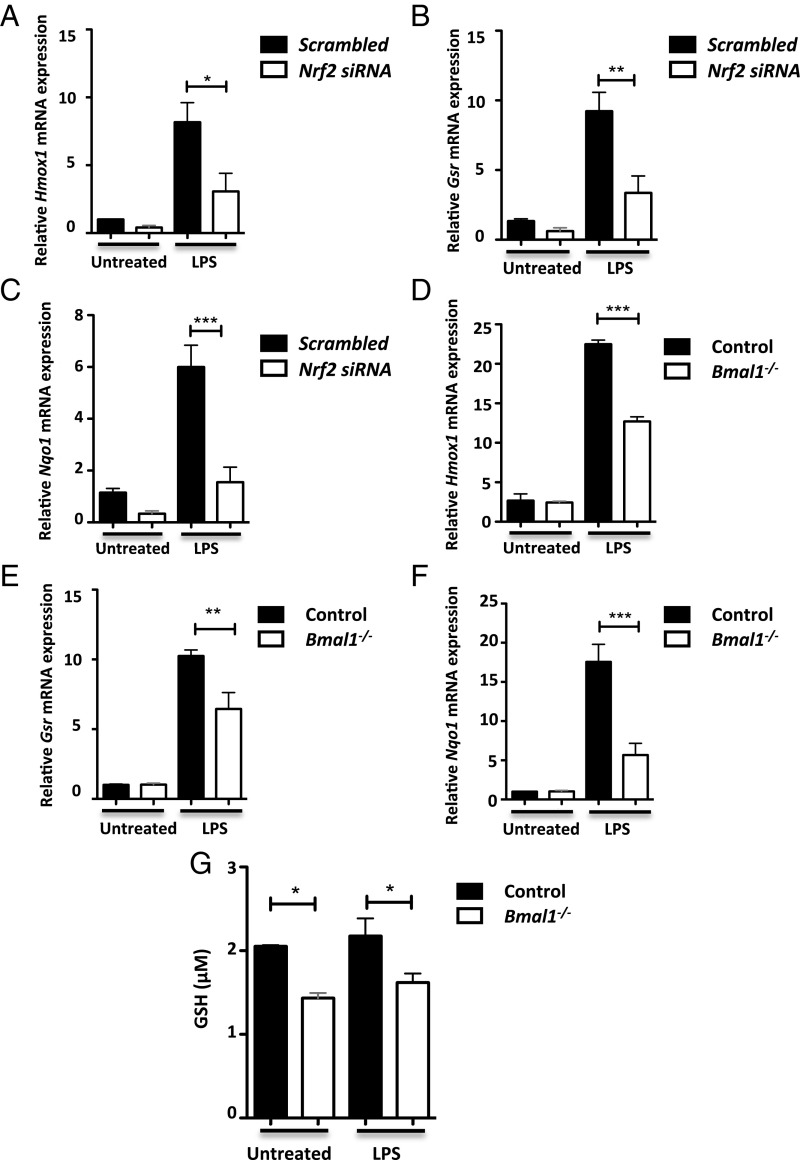
NRF2 mediates the induction of a panel of antioxidant genes that balance the oxidative state of the cell (20). Therefore, we next examined the impact of BMAL1 on NRF2-mediated antioxidant response genes. Knockdown of Nrf2 (SI Appendix, Fig. S4B) was shown to decrease induction of three core NRF2 target genes, Hmox1 (Fig. 2A), Gsr (Fig. 2B), and Nqo1 (Fig. 2C) in response to LPS. Hmox1 (Fig. 2D), Gsr (Fig. 2E), and Nqo1 (Fig. 2F) expression levels were also decreased in Bmal1−/− compared with Bmal1+/+ BMDMs following LPS stimulation. Induction of Hmox1 (SI Appendix, Fig. S5A) and Gsr (SI Appendix, Fig. S5B) were similarly decreased in Bmal1−/− BMDMs following hydrogen peroxide stimulation. siRNA-mediated knockdown of Bmal1 recapitulated the same effect on Hmox1 (SI Appendix, Fig. S5C), Gsr (SI Appendix, Fig. S5D), and Nqo1 (SI Appendix, Fig. S5E) expression following LPS stimulation. Reduced glutathione (GSH) synthesis is a downstream component of the NRF2-mediated antioxidant defense pathway. Glutathione reductase, the product of Gsr, which we determined was reduced with Bmal1 deletion, catalyzes the reduction of oxidized glutathione (GSSG) to the sulfhydryl GSH form (20). We found that total cell levels of GSH were decreased in Bmal1−/− BMDMs versus Bmal1+/+ controls (Fig. 2G). These results demonstrate that deletion or silencing of Bmal1 results in the same suppression of the antioxidant response as direct silencing of Nrf2, suggesting that BMAL1 activity in macrophages during inflammation is mediated in part by NRF2.
Fig. 2.
NRF2 antioxidant targets are decreased with Bmal1 deletion. (A) Hmox1, (B) Gsr, and (C) Nqo1 mRNA was measured by qPCR in wild-type BMDMs transfected with scrambled siRNA or siRNA targeting Nrf2 following 24 h of LPS (n = 4–6). (D) Hmox1, (E) Gsr, and (F) Nqo1 mRNA was measured by qPCR in Bmal1+/+ and Bmal1−/− BMDMs following 24 h of LPS (n = 4–6). (G) Bmal1+/+ and Bmal1−/− BMDMs were left untreated or treated with LPS for 24 h before measuring total cell levels of GSH by luminescence (n = 3). All graphs displayed were analyzed by one-way ANOVA. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Diurnal Variation in Oxidative Stress in Macrophages.
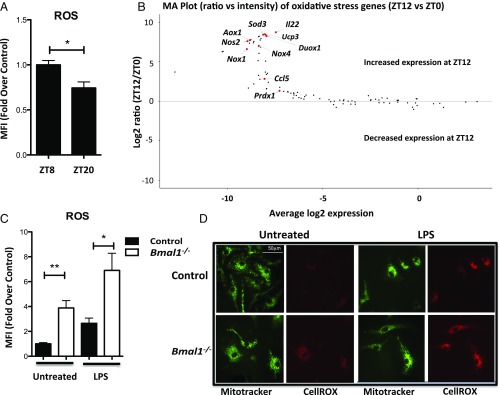
The observed diurnal rhythm of Nrf2 expression in peritoneal myeloid cells, in which Nrf2 expression is high before the onset of morning (ZT20) and low in the afternoon (ZT8/ZT12) (Fig. 1D), led us to investigate whether ROS levels showed similar diurnal variability. We discovered a significant increase in basal ROS levels in peritoneal myeloid cells at ZT8 versus ZT20 (Fig. 3A), correlating inversely with the diurnal rhythm in Bmal1 and Nrf2 expression. The diurnal pattern of reduced Nrf2 mRNA expression and increased ROS production at ZT8 may explain our previously reported increase in LPS-induced lethality at ZT12 (7). Therefore, we next investigated whether there was differential expression in genes of the oxidative stress pathway. We discovered an enrichment of genes involved in ROS production and regulation at ZT12 versus ZT0 (Fig. 3B), providing further evidence for a rhythm in ROS regulation in peritoneal myeloid cells. A full list of genes analyzed can be found in SI Appendix, Fig. S6. Given these results, we measured whole-cell ROS levels in macrophages lacking Bmal1. A substantial increase in the levels of ROS in Bmal1−/− BMDMs was detected both basally and with the addition of LPS in comparison with Bmal1+/+ BMDMs (Fig. 3C). This was further confirmed with siRNA-mediated knockdown of Bmal1 in wild-type BMDMs where increased ROS was also observed (SI Appendix, Fig. S7). We were also able to visualize this phenotype directly by utilizing fluorescent imaging (Fig. 3D), which once again highlighted greater ROS production with Bmal1 deletion.
Fig. 3.
Diurnal rhythms in ROS regulation. (A) Peritoneal cells were isolated from wild-type mice at ZT8 and ZT20. Cells were stained with a CD11b+ antibody and CellROX stain. ROS levels were then measured by mean fluoresence intensity (MFI) in the myeloid population by flow cytometry (n = 4–6). (B) Peritoneal cells were isolated from wild-type mice at ZT0 and ZT12. RNA was extracted and gene expression panels were used to measure fold change of oxidative stress pathway genes at ZT12 versus ZT0 (n = 3). (C) Bmal1+/+ and Bmal1−/− BMDMs were treated with LPS (100 ng/mL) for 24 h before staining with CellROX. ROS was then measured via flow cytometry (n = 4). (D) Bmal1+/+ and Bmal1−/− BMDMs were untreated or treated with LPS (100 ng/mL) for 24 h before staining with CellROX and MitoTracker green. The cells were imaged using confocal microscopy. Image shown is a representative of at least three independent experiments. Statistical significance of graph A was determined by unpaired Student’s t test. Statistical significance of C was determined by one-way ANOVA. *P ≤ 0.05 and **P ≤ 0.01.
Increased Production of IL-1β in Cells Lacking Bmal1.
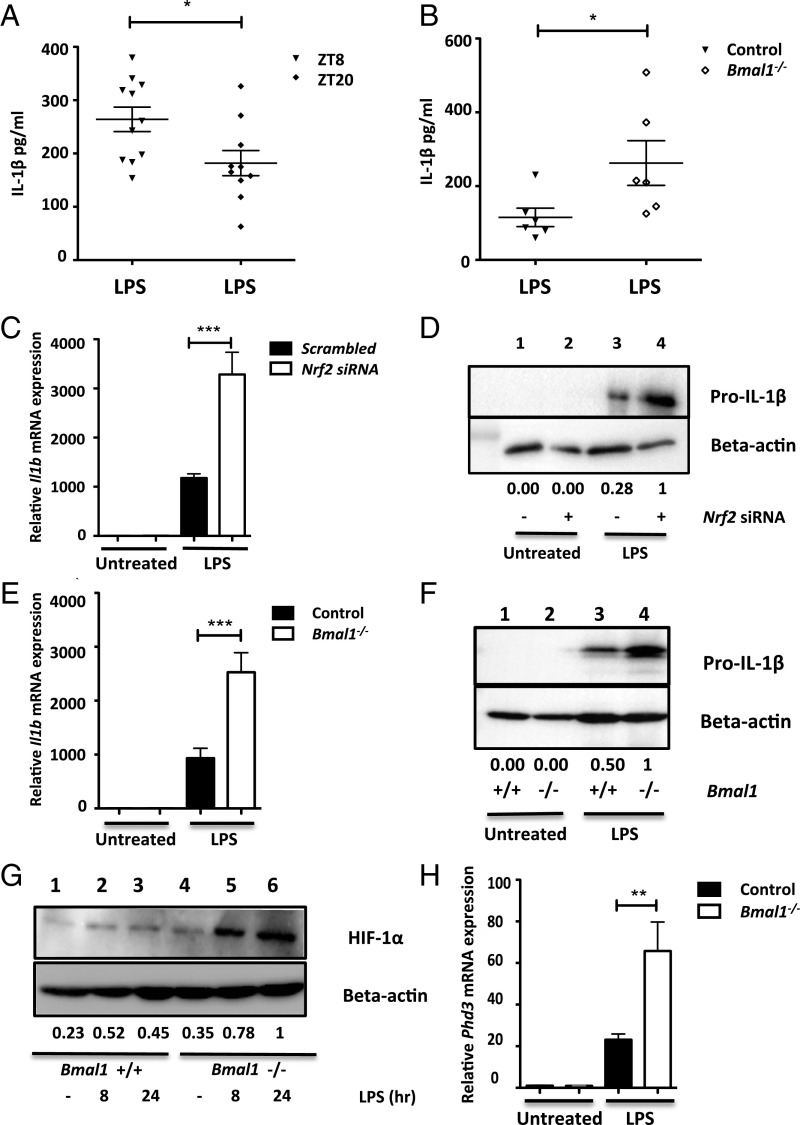
NRF2 and ROS have been linked to regulation of IL-1β (16, 22, 23). Therefore, we next investigated whether BMAL1-mediated regulation of Nrf2 could impact IL-1β production. To test for time-of-day variation in IL-1β production, we injected mice with LPS at ZT8 or ZT20 and measured levels of IL-1β in serum (Fig. 4A). We found a significant increase in IL-1β induction at ZT8 relative to ZT20, correlating with our previous results of diurnal variation in ROS (Fig. 3A), BMAL1, and NRF2 levels (SI Appendix, Fig. S3B). Furthermore, when myeloid Bmal1+/+ and myeloid Bmal1−/− mice were injected with LPS at ZT20, we saw increased levels of IL-1β in the serum of myeloid Bmal1−/− mice (Fig. 4B).
Fig. 4.
Increased IL-1β production with Bmal1 deletion. (A) Wild-type C57Bl6 mice were injected with PBS or LPS (5 mg/kg) for 90 min at ZT8 or ZT20. Serum was isolated from whole blood and levels of IL-1β were measured by ELISA (n = 10–11). (B) Bmal1myeloid+/+ and Bmal1myeloid−/− mice were injected with PBS or LPS (5 mg/kg) for 90 min at ZT20. Serum was isolated from whole blood and levels of IL-1β were measured by ELISA (n = 6). (C and D) Wild-type BMDMs were transfected with scrambled siRNA or siRNA targeting Nrf2 and treated with LPS (100 ng/mL) for 24 h (n = 3), (C) Il1b mRNA was measured by qPCR, and (D) pro–IL-1β was measured by immunoblot. (E and F) Bmal1+/+ and Bmal1−/− BMDMs were treated with LPS (100 ng/mL) for 24 h, (E) Il1b mRNA was measured by qPCR (n = 8), and (F) pro–IL-1β was measured by immunoblot. (G and H) Bmal1+/+ and Bmal1−/− BMDMs were treated with LPS (100 ng/mL), (G) HIF-1α levels were measured by immunoblot, and (H) Phd3 mRNA was measured by qPCR (n = 4). Immunoblots presented are a representative of at least three independent experiments. Values provided below each lane indicate relative densitometry of each band. Statistical significance of A–C, E, and H was determined by one-way ANOVA. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
We next decided to investigate the effect of NRF2 on IL-1β production in vitro. Knockdown of Nrf2 resulted in an increase in IL-1β mRNA (Fig. 4C) and protein (Fig. 4D) following LPS stimulation. A similar phenotype was seen with LPS treatment of Bmal1−/− BMDMs, which revealed a boost in IL-1β mRNA (Fig. 4E) and protein (Fig. 4F) relative to Bmal1+/+ BMDMs. This increased pro–IL-1β in cells lacking Bmal1 was also evident when peritoneal cells were extracted from Bmal1+/+ or Bmal1−/− mice and treated with LPS ex vivo (SI Appendix, Fig. S8A). We also observed an increase in IL-6 secretion in Bmal1−/− BMDMs (SI Appendix, Fig. S8B); however, TNFα levels remained unchanged (SI Appendix, Fig. S8C). This phenotype of IL-1β was confirmed with siRNA-mediated knockdown of Bmal1 in wild-type BMDMs at the RNA (SI Appendix, Fig. S8D) and protein (SI Appendix, Fig. S8E) levels. HIF-1α is a hypoxic response protein that is activated in response to increased ROS levels and directly binds to the IL-1β promoter to induce its expression (28). We observed increased HIF-1α accumulation following LPS stimulation in Bmal1−/− BMDMs (Fig. 4G) and increased HIF-1α activity, as measured indirectly by increased Phd3 expression, a target of HIF-1α (16) (Fig. 4H).
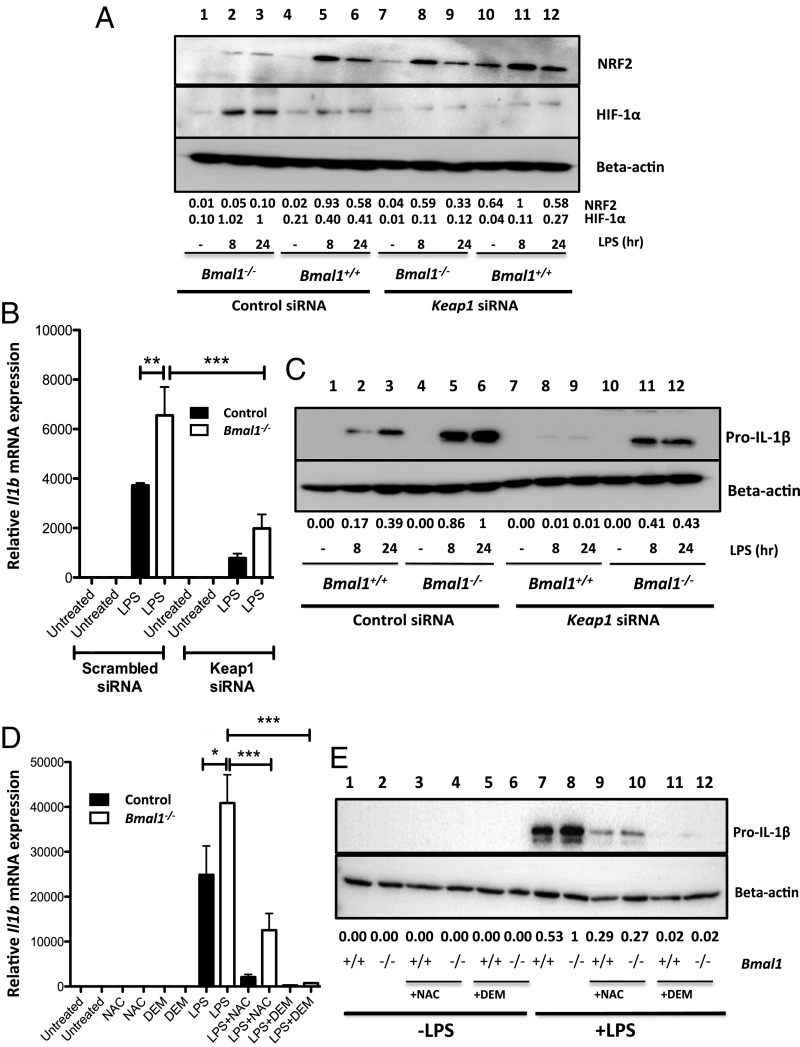
Boosting NRF2 Activity and Limiting ROS Rescues the IL-1β Phenotype of Bmal1-Deficient Macrophages.
Our results demonstrate that depletion of the NRF2 pathway in Bmal1−/− BMDMs results in a prooxidant and proinflammatory phenotype. We performed knockdown of Keap1 in macrophages to facilitate NRF2 accumulation. Keap1 knockdown (SI Appendix, Fig. S4C) increased protein levels of NRF2 in Bmal1−/− BMDMs (Fig. 5A). The increased accumulation of NRF2 in Bmal1−/− BMDMs resulted in a decrease in HIF-1α protein levels. Keap1 knockdown was demonstrated to recover NRF2 activity in Bmal1−/− BMDMs as Nqo1 expression levels were enhanced (SI Appendix, Fig. S9A). We anticipated that increased NRF2 activity would rescue the proinflammatory phenotype of Bmal1−/− BMDMs. Keap1 knockdown decreased the levels of Il1b mRNA (Fig. 5B) and protein (Fig. 5C) in Bmal1−/− BMDMs. To determine whether the reduction in IL-1β levels was due to direct inhibition of Il1b expression by NRF2, or if ROS and HIF-1α decrease, mediated by the antioxidant defense pathway, also played a role, we treated Bmal1+/+ and Bmal1−/− BMDMs with diethylmaleate (DEM) and N-acetyl-l-cysteine (NAC). DEM activates NRF2 through direct modification of KEAP1. NAC suppresses ROS by increasing intracellular GSH levels. Treatment of Bmal1+/+ and Bmal1−/− BMDMs with DEM significantly enhanced expression of Nqo1, validating enhanced activity of NRF2 with this compound (SI Appendix, Fig. S9B). Treatment with both NAC and DEM decreased IL-1β mRNA (Fig. 5D) and protein levels (Fig. 5E) in both Bmal1+/+ and Bmal1−/− BMDMs. A greater decrease in IL-1β was seen with DEM treatment. This compound not only decreases ROS levels by activating NRF2, but would also increase levels of NRF2 binding directly to the IL-1β promoter (23). By freeing NRF2 from regulation of KEAP1 in Bmal1 knockout macrophages, we were able to rescue the proinflammatory phenotype of these cells. Therefore, these results confirm that deletion of Bmal1 facilitates increased production of IL-1β due to suppression of NRF2 activity.
Fig. 5.
Boosting NRF2 activity and limiting ROS rescues the IL-1β phenotype of Bmal1-deficient macrophages. Bmal1+/+ and Bmal1−/− BMDMs were transfected with scrambled siRNA or siRNA targeting Keap1 and treated with LPS (100 ng/mL) and (A) NRF2 and HIF-1α were detected by immunoblot following 8 and 24 h of LPS stimulation. (B) Il1b mRNA was measured by qPCR (n = 4). (C) Protein levels of pro–IL-1β were measured by immunoblot. Bmal1+/+ and Bmal1−/− BMDMs were pretreated with either 100 μM DEM or 10 mM NAC before LPS (100 ng/mL) stimulation. (D) Il1b mRNA was measured by qPCR (n = 3) and (E) pro–IL-1β protein levels were measured by immunoblot. Immunoblots presented are a representative of at least three independent experiments. Values provided below each lane indicate relative densitometry of each band. Statistical significance of all graphs was determined by one-way ANOVA. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
Discussion
The significant finding of this study is that BMAL1 regulates oxidative stress pathways in macrophages to limit the production of the proinflammatory cytokine IL-1β. We build upon previous findings of circadian-mediated regulation of NRF2 (13, 21). Specifically, we uncover that in macrophages the activity of NRF2 over time of day and under LPS induction is directly mediated by BMAL1. Furthermore, we have shown that reduction of NRF2 activity leads to a loss of redox homeostasis and aberrant production of IL-1β, providing an explanation as to why macrophages lacking BMAL1 are highly proinflammatory in response to LPS. This mechanism may underlie the observed time-of-day differences in LPS-induced lethality between mice lacking myeloid Bmal1 versus controls (7).
Previous studies have determined the role of circadian rhythms in modulating Nrf2 expression levels in various tissue types (13, 21). However, the molecular clock does not regulate NRF2 in all tissues. The BMAL1:CLOCK heterodimer was found to directly induce Nqo1 and Aldh2 in the cerebral cortex, and not through the intermediate of NRF2 (14). In our study, however, we observed clear binding of BMAL1 to the Nrf2 promoter, and 24-h rhythms in the expression of Nrf2 in myeloid cells and macrophages were lost with Bmal1 deletion. We also discovered that activity of NRF2 was decreased in Bmal1−/− macrophages. Intriguingly, suppression of NRF2 activity has previously been linked with pathogenesis of inflammatory conditions that are at least partly mediated by both myeloid cells and the circadian clock, such as EAE (29), sepsis (30), and asthma (31). A surprising but intriguing outcome of our TF-Seq screen was that the only significant pathway altered between macrophages lacking Bmal1 and controls was that of the NRF2 pathway. This may have been due to having limited LPS timepoints (1 and 4 h), and not an extended timecourse for pathway analysis. However, the fact that a significant decrease in NRF2 activity was detected in Bmal1-deficient macrophages highlights the importance of BMAL1 in its regulation.
The molecular clock has previously been associated with regulation of ROS in multiple tissues. Complete Bmal1 deletion has been shown to induce an advanced aging phenotype via ROS-induced tissue atrophy (12). Our results demonstrate an increase in ROS when Nrf2 and Bmal1 mRNA expression and protein are at their lowest point, and vice versa, in peritoneal myeloid cells. This also correlates with increased levels of IL-1β in the serum of mice upon LPS stimulation. However, Bmal1−/− mice produced increased IL-1β when injected with LPS at ZT20 relative to wild-type controls. This suggests that rhythms in Nrf2 may facilitate rhythms in ROS production that impact IL-1β production in vivo. This was further substantiated by an increase in the production of ROS observed with Bmal1 deletion in BMDMs. ROS are clearly linked with driving inflammation and tissue damage (11). We have previously reported that BMAL1 expression in myeloid cells is crucial to providing mice with time-of-day protection from LPS at ZT0 relative to ZT12 in a mouse model of LPS lethality (7). Here we discovered greater basal expression in genes involved in ROS regulation at ZT12 relative to ZT0. For example, Nox1, which forms a catalytic subunit of NADPH oxidase, a major producer of ROS that is crucial to macrophage differentiation (32), was significantly up-regulated at ZT12. Similarly, Nos2, an initiator of nitric oxide production and a marker of proinflammatory macrophage differentiation (33), was also increased. We hypothesize that diurnal variation of ROS could be contributing to circadian gating of LPS-induced endotoxic shock and that deletion of Bmal1 results in constitutively elevated ROS in macrophages.
NRF2 and oxidative stress are regulators of IL-1β. NRF2 has been found to bind specific inhibitory sites in the promoter of Il1b, suppressing its transcription, independently of ROS (23). However, in later stages of macrophage activation with LPS, ROS levels accumulate, stabilizing HIF-1α to drive IL-1β production (16, 28). NRF2 has been demonstrated to limit IL-1β through this mechanism (22). We discovered that Nrf2 silencing leads to a clear boost in IL-1β production, which is phenocopied when Bmal1 is silenced or deleted. We also show that HIF-1α levels are significantly enhanced with deletion of Bmal1 following LPS stimulation. Deletion of Bmal1 has previously been demonstrated to lead to increased Hif1a mRNA in macrophages, and ChIP-Seq revealed BMAL1 binding to the regulatory regions of the Hif1α promoter (10). It is possible that we observe increased HIF-1α with deletion of Bmal1 through a lack of BMAL1-mediated suppression of Hif1α mRNA transcription, rather than through increased oxidative stress-induced stabilization of the protein. However, Keap1 knockdown led to increased accumulation of NRF2, boosted antioxidant response, and decreased HIF-1α in Bmal1 knockout macrophages. This resulted in decreased levels of IL-1β being produced in Bmal1−/− BMDMs, indicating that oxidative stress plays a key role in this pathway.
To understand whether the decreased IL-1β with Keap1 silencing was due to the transcriptional repression of Il1b by NRF2, or suppression of ROS, we utilized DEM to pharmacologically enhance NRF2 activity, and NAC, to limit ROS independent of NRF2. Both DEM and NAC decrease IL-1β in Bmal1−/− BMDMs. However, there was greater inhibition of IL-1β with DEM than NAC. Our findings suggest that this further decrease is possibly due to the dual role of NRF2, as an antioxidant transcription factor and as a direct transcriptional repressor of IL-1β (Fig. 6).
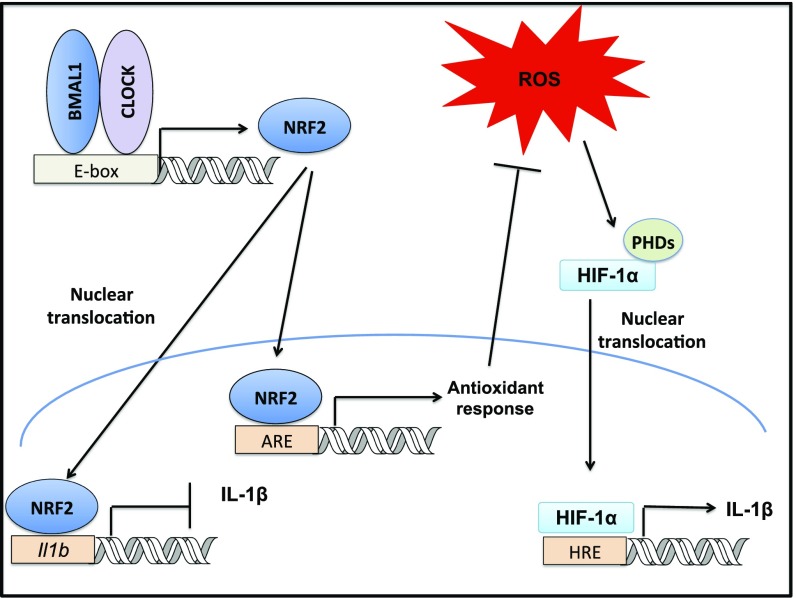
Fig. 6.
Schematic of proposed model. We observe that the core molecular clock component, BMAL1, is crucial in promoting Nrf2 transcription in myeloid cells. NRF2 can repress IL-1β through two possible mechanisms: direct repression of Il1b transcription or induction of antioxidant response elements. This second pathway results in ROS suppression, which reduces levels of HIF-1α binding to hypoxia response elements (HRE), preventing induction of Il1b transcription. Thus, the molecular clock directly controls NRF2 transcriptional activity and antioxidant capacity to regulate IL-1β in myeloid cells.
Rhythmic regulation of ROS and inflammatory mechanisms by NRF2 activity could provide an explanation for circadian rhythms exhibited in a variety of inflammatory processes and diseases involving oxidative stress and IL-1β. The molecular clock acts as a timed regulator of Nrf2 activity in mouse macrophages, which has a large effect on the inflammatory output of the cell through oxidative stress modulation. Given the importance of ROS in innate immunity, it is distinctly possible that many aspects of the innate immune system mediated by oxidative stress could be under circadian regulation. Recent reports have demonstrated that HIF-1α levels regulated by circadian variation in ROS dictates increased immune cell trafficking into tissues at ZT7 (15). Ly6Chi monocytes show higher accumulation in tissues at ZT8 (5), the time at which we showed increased ROS levels in myeloid cells and enhanced IL-1β in serum. It is plausible that rhythms in immune cell trafficking dictated by ROS could be maintained by rhythms in Nrf2. Severity of circadian inflammatory conditions involving oxidative stress and IL-1β, such as asthma (25), sepsis (7), rheumatoid arthritis (19), osteoarthritis (24), and multiple sclerosis (8, 9), could very well be mediated at the molecular level by this BMAL1:NRF2:IL-1β axis. Further understanding of how the clock intersects with NRF2 function and activity may reveal opportunities for chronotherapies in the treatment of circadian gated inflammatory disorders.
Experimental Procedures
Transgenic Animals.
Mice with the gene Bmal1 containing LoxP sites were provided by Christropher Bradfield. Bmal1LoxP/LoxP were crossed with Lyz2Cre, which express Cre recombinase under the control of the Lyz2 promoter to produce progeny that have Bmal1 excised in the myeloid lineage. Bmal1LoxP/LoxPLyz2Cre (Bmal1−/−) mice where compared with control Lyz2Cre (Bmal1+/+). Offspring were genotyped to confirm the presence of LoxP sites and Cre recombinase.
All mice were maintained according to European Union regulations and the Irish Health Products Regulatory Authority. Experiments were performed under Health Products Regulatory Authority license with approval from the Trinity College Dublin BioResources Ethics Committee.
In Vivo Studies.
Mice were injected intraperitoneally with PBS or LPS (5 mg/kg) at the relevant time of day. After 90 min, mice were humanely killed and serum was isolated from whole blood and peritoneal cells were harvested.
Affinity Purification with Biotinylated Nucleotide (Oligo Pulldown).
BMDMs were seeded at 1 × 106 cells per milliliter and incubated overnight. Cells were lysed in 100 μL of oligonucleotide buffer (25 mM Tris, 50 mM EDTA, 5% glycerol, 5 mM NaF, 1% Nonidet P-40, 1 mM DTT, 150 mM NaCl, and protease and phosphatase inhibitors). Samples were kept on ice and 4.5 mL of oligonucleotide buffer without NaCl was added. A 50-μL sample was isolated and the remainder was split between two tubes and incubated for 2 h with streptavidin-agarose beads, which conjugated to the biotinylated promoter regions (either WT E-box or mutant E-box). The lysates were centrifuged to pellet the beads before addition of 50 μL of sample lysis buffer. Presence of BMAL1 binding was detected by Western blot.
Transcription Factor Sequencing.
Transcription factor sequencing was performed as previously described by O’Connell et al. (27). TF-Seq lentiviral vectors were transfected into HEK293T cells. After 48 h, the media containing a pool of lentiviral particles, which represented the TF-Seq reporter library, were transfected into Bmal1+/+ and Bmal1−/− BMDMs. After 72 h, the BMDMs were replated into 96-well plates and left untreated or treated with LPS for 1 or 4 h before assay collection and assessment of transcription factor activity was carried out.
Quantification of Oxidative Stress Genes.
RT2 Profiler Plates were used to screen for differences in mouse oxidative stress pathways (PAMM-065ZC, Qiagen) between peritoneal cells isolated at ZT0 and ZT12, based on SYBR Green qPCR array. cDNA was generated from RNA samples using an RT2 First Strand Kit (330401, Qiagen) and mixed with SYBR Green qPCR Mastermix (330520, Qiagen) and distributed to RT2 Profiler plates which were preloaded with primer sets for pathway relevant genes. Amplification was then initiated in a real-time cycler and data were analyzed by the ΔΔCT method. Ratios between ZT12 and ZT0 were plotted against the log2 mean average expression values, which are based on ZT12 and ZT0 2^(–Avg(Delta(Ct)) values. The MA plot was generated using the statistical software R with the packages ggplot2 and ggrepel (for labeling).
Statistical Analyses.
Data were evaluated on Prism 5 or Stata 14 statistical software (Stata Corp). Differences were compared by using analysis of variance (ANOVA) followed by Tukey’s posttest analysis for comparison of multiple independent groups or unpaired Student’s t test for direct analysis of two independent groups. Circadian rhythms were detected and analyzed using a cosinor regression model. Results are presented as mean ± SEM from at least three independent experiments. Differences were considered significant at the values of *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. Anne McGettrick for critical reading of this manuscript and Dr. Moritz Haneklaus for bioinformatics assistance. This work was supported by Science Foundation Ireland (SFI) Starting Investigator Research Award 13/SIRG/2130 (to A.M.C.), SFI Career Development Award 17/CDA/4688 (to A.M.C.), and Wellcome Trust Metabolic Grant 205455 (to L.A.J.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800431115/-/DCSupplemental.
References
- 1.Early JO, Curtis AM. Immunometabolism: Is it under the eye of the clock? Semin Immunol. 2016;28:478–490. doi: 10.1016/j.smim.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Hastings M, O’Neill JS, Maywood ES. Circadian clocks: Regulators of endocrine and metabolic rhythms. J Endocrinol. 2007;195:187–198. doi: 10.1677/JOE-07-0378. [DOI] [PubMed] [Google Scholar]
- 3.Oliva-Ramírez J, Moreno-Altamirano MM, Pineda-Olvera B, Cauich-Sánchez P, Sánchez-García FJ. Crosstalk between circadian rhythmicity, mitochondrial dynamics and macrophage bactericidal activity. Immunology. 2014;143:490–497. doi: 10.1111/imm.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs JE, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen KD, et al. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagnidze K, et al. Nuclear receptor REV-ERBα mediates circadian sensitivity to mortality in murine vesicular stomatitis virus-induced encephalitis. Proc Natl Acad Sci USA. 2016;113:5730–5735. doi: 10.1073/pnas.1520489113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis AM, et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci USA. 2015;112:7231–7236. doi: 10.1073/pnas.1501327112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton CE, et al. Loss of the molecular clock in myeloid cells exacerbates T cell-mediated CNS autoimmune disease. Nat Commun. 2017;8:1923. doi: 10.1038/s41467-017-02111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Druzd D, et al. Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity. 2017;46:120–132. doi: 10.1016/j.immuni.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oishi Y, et al. Bmal1 regulates inflammatory responses in macrophages by modulating enhancer RNA transcription. Sci Rep. 2017;7:7086. doi: 10.1038/s41598-017-07100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mittal M, Siddiqui MR, Tran K, Reddy SP, Malik AB. Reactive oxygen species in inflammation and tissue injury. Antioxid Redox Signal. 2014;20:1126–1167. doi: 10.1089/ars.2012.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondratov RV, Vykhovanets O, Kondratova AA, Antoch MP. Antioxidant N-acetyl-L-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein BMAL1. Aging (Albany NY) 2009;1:979–987. doi: 10.18632/aging.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, et al. Bmal1 and β-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced β-cell failure in mice. Mol Cell Biol. 2013;33:2327–2338. doi: 10.1128/MCB.01421-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musiek ES, et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J Clin Invest. 2013;123:5389–5400. doi: 10.1172/JCI70317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Uncovering the mystery of opposite circadian rhythms between mouse and human leukocytes in humanized mice. Blood. 2017;130:1995–2005. doi: 10.1182/blood-2017-04-778779. [DOI] [PubMed] [Google Scholar]
- 16.Mills EL, et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470.e413. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinarello CA. The history of fever, leukocytic pyrogen and interleukin-1. Temperature (Austin) 2015;2:8–16. doi: 10.1080/23328940.2015.1017086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burger D, Molnarfi N, Gruaz L, Dayer JM. Differential induction of IL-1beta and TNF by CD40 ligand or cellular contact with stimulated T cells depends on the maturation stage of human monocytes. J Immunol. 2004;173:1292–1297. doi: 10.4049/jimmunol.173.2.1292. [DOI] [PubMed] [Google Scholar]
- 19.Hand LE, et al. The circadian clock regulates inflammatory arthritis. FASEB J. 2016;30:3759–3770. doi: 10.1096/fj.201600353R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pekovic-Vaughan V, et al. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev. 2014;28:548–560. doi: 10.1101/gad.237081.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi EH, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gossan N, Boot-Handford R, Meng QJ. Ageing and osteoarthritis: A circadian rhythm connection. Biogerontology. 2015;16:209–219. doi: 10.1007/s10522-014-9522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaslona Z, et al. The circadian protein BMAL1 in myeloid cells is a negative regulator of allergic asthma. Am J Physiol Lung Cell Mol Physiol. 2017;312:L855–L860. doi: 10.1152/ajplung.00072.2017. [DOI] [PubMed] [Google Scholar]
- 26.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 27.O’Connell DJ, et al. Simultaneous pathway activity inference and gene expression analysis using RNA sequencing. Cell Syst. 2016;2:323–334. doi: 10.1016/j.cels.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linker RA, et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 30.Thimmulappa RK, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sussan TE, et al. Nrf2 reduces allergic asthma in mice through enhanced airway epithelial cytoprotective function. Am J Physiol Lung Cell Mol Physiol. 2015;309:L27–L36. doi: 10.1152/ajplung.00398.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Q, et al. NADPH oxidases are essential for macrophage differentiation. J Biol Chem. 2016;291:20030–20041. doi: 10.1074/jbc.M116.731216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jablonski KA, et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS One. 2015;10:e0145342. doi: 10.1371/journal.pone.0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.