Significance
Earlier studies suggested that particles are released from the macrophage plasma membrane, but the mechanism has been unclear. We found that filopodia of macrophages release large numbers of vesicular particles. Nanoscale secondary ion mass spectrometry revealed that these particles are enriched in cholesterol, including the “accessible” pool of cholesterol detectable by the cholesterol-binding protein. The cholesterol content of macrophage particles increased when the cells were loaded with cholesterol and could be depleted by incubating the cells with high-density lipoproteins. Our studies suggest that the release of particles by macrophages could be one mechanism for cholesterol efflux and that particles could be an intermediate in the movement of cholesterol to high-density lipoproteins.
Keywords: cholesterol efflux, accessible cholesterol, nanoSIMS
Abstract
Macrophages are generally assumed to unload surplus cholesterol through direct interactions between ABC transporters on the plasma membrane and HDLs, but they have also been reported to release cholesterol-containing particles. How macrophage-derived particles are formed and released has not been clear. To understand the genesis of macrophage-derived particles, we imaged mouse macrophages by EM and nanoscale secondary ion mass spectrometry (nanoSIMS). By scanning EM, we found that large numbers of 20- to 120-nm particles are released from the fingerlike projections (filopodia) of macrophages. These particles attach to the substrate, forming a “lawn” of particles surrounding macrophages. By nanoSIMS imaging we showed that these particles are enriched in the mobile and metabolically active accessible pool of cholesterol (detectable by ALO-D4, a modified version of a cholesterol-binding cytolysin). The cholesterol content of macrophage-derived particles was increased by loading the cells with cholesterol or by adding LXR and RXR agonists to the cell-culture medium. Incubating macrophages with HDL reduced the cholesterol content of macrophage-derived particles. We propose that release of accessible cholesterol-rich particles from the macrophage plasma membrane could assist in disposing of surplus cholesterol and increase the efficiency of cholesterol movement to HDL.
Macrophages ingest senescent erythrocytes, remove cellular debris after an injury or infection, and ingest lipoproteins that enter the arterial intima. In carrying out these functions, macrophages internalize cholesterol. Some of the cholesterol is initially stored in cytosolic cholesterol ester droplets, but ultimately the cholesterol must be returned to the bloodstream for uptake and excretion by the liver. Cholesterol efflux from cells has been studied intensively and involves multiple mechanisms; the one that is generally highlighted involves movement of cellular cholesterol to HDL through direct interactions between HDL and ATP-binding cassette (ABC) transporters on the surface of the plasma membrane (1, 2). ABC transporters move phospholipids and cholesterol to the exofacial leaflet of the plasma membrane, facilitating uptake of the cholesterol by HDL. The expression of ABC transporters is regulated by liver X receptor (LXR) transcription factors, which are activated by sterols.
Another potential mechanism for macrophage cholesterol efflux is the release of particles containing cholesterol, variously described as “microparticles,” “cholesterol microdomains,” or “exosomes” (2–7). Phillips and coworkers (2) proposed in 2007 that a significant fraction of the cholesterol released by cultured macrophages is due to the release of microparticles. They proposed that the particles originated from the plasma membrane, but how the microparticles were released was not clear. A 2014 review of cholesterol efflux by Philips did not mention a role for microparticle release (8). Kruth and coworkers (3–6, 9) have published a series of articles on the release of cholesterol microdomains by cultured macrophages. The microdomains were detected by immunocytochemistry using a cholesterol-specific monoclonal antibody. In contrast to Philip’s work, they proposed that the cholesterol microdomains are not vesicles but are branching, irregularly shaped deposits that originate from the plasma membrane. How they were released was not defined. They suggested that the release of cholesterol microdomains could be important for reverse cholesterol transport.
Efforts to visualize cholesterol efflux by EM have been limited. The microparticles described by Phillips and coworkers (2) had a mean diameter of 24 nm, as judged by negative-staining EM. Transmission electron micrographs showing binding of apo-AI to poorly defined protrusions on the surface of ABCA1-expressing macrophages (10) and a budding vesicle on the plasma membrane of ABCA1-expressing baby hamster kidney (BHK) cells have been published (7), but the images were not optimal, and whether plasma membrane protrusions were enriched in cholesterol was unclear.
We suspected that a combination of EM and nanoscale secondary ion mass spectrometry (nanoSIMS) would make it possible to define the origin of macrophage particles and gain fresh insights into the cholesterol content of those particles. NanoSIMS uses a Cs+ beam to bombard a cell, releasing secondary ions (e.g., 12C−, 13C−, 12C14N−, and 12C15N−) that are collected and used to create high-resolution images of cells based solely on their isotopic content. When cultured cells are loaded with cholesterol containing a stable isotope (e.g., 13C), nanoSIMS images can define the distribution of cholesterol on the plasma membrane (11). NanoSIMS can also be used to assess the distribution of the “accessible” pool of cholesterol (12) on the plasma membrane. The accessible pool—a pool not sequestered by sphingomyelin or phospholipids—is present when the cholesterol content of the plasma membrane is high and can be detected because it binds specifically to modified versions of cholesterol-binding cytolysins (e.g., domain 4 of anthrolysin O; ALO-D4) (12, 13). The accessible pool of cholesterol on the plasma membrane is metabolically important because it is mobile, capable of moving to the endoplasmic reticulum and participating in the regulation of cholesterol biosynthetic enzymes (14). Recently, He et al. (15) used nanoSIMS, along with an 15N-labeled ALO-D4, to show that the accessible pool of cholesterol on the plasma membrane of CHO-K1 cells is concentrated on microvilli.
In the current study, we used SEM to visualize the formation and release of particles from the plasma membrane of macrophages. Using nanoSIMS imaging, we investigated the cholesterol content of particles, including the content of accessible cholesterol, under different cell-culture conditions. We also tested whether HDL is capable of removing cholesterol from particles after they have been released by macrophages.
Results
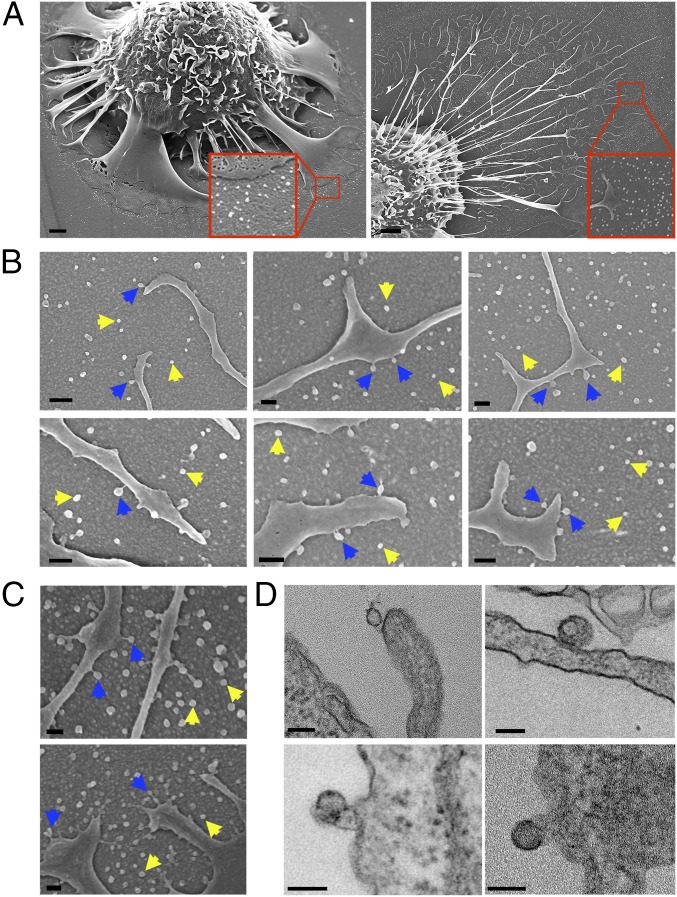
In our first experiments, we use SEM to visualize the release of particles from the plasma membrane of macrophages. Mouse peritoneal macrophages were plated on poly-d-lysine–coated silicon wafers, loaded with cholesterol by incubating the cells with acetylated LDLs (acetyl-LDLs) (16), and then visualized by SEM. SEM images revealed numerous particles on the substrate surrounding macrophages (Fig. 1A). The particles, ∼20–120 nm in diameter, were released from the filopodia of both primary macrophages (Fig. 1B) and RAW 264.7 cells (a mouse macrophage cell line) (Fig. 1C). The lawn of particles around macrophages was most often located preferentially on one or two sides of the cell rather than surrounding the entire circumference of the cell (Figs. 1A and 2A); the preferential localization of the lawn of particles on one side of the macrophage did not change when the cells were grown on an orbital shaker. The macrophage-derived particles were unilamellar and were surrounded by a lipid bilayer, as judged by transmission electron microscopy (TEM) (Fig. 1D). By TEM, we occasionally observed osmophilic material within particles.
Fig. 1.
Release of particles from the plasma membrane of macrophages. (A) Scanning electron micrographs of mouse peritoneal macrophages surrounded by a lawn of small particles (attached to a poly-d-lysine–coated silicon wafer). The macrophages had been loaded with acetyl-LDL (50 μg/mL). (Scale bars, 2 μm.) (B and C) Scanning electron micrographs show particles surrounding filopodia of mouse peritoneal macrophages (B) and RAW 264.7 cells (C). Yellow arrows show particles attached to the poly-d-lysine–coated substrate. Blue arrows show the release of particles from the filopodia. (Scale bars, 100 nm.) (D) Transmission electron micrographs of particle release from filopodia of mouse peritoneal macrophages. (Scale bars, 100 nm.)
Fig. 2.
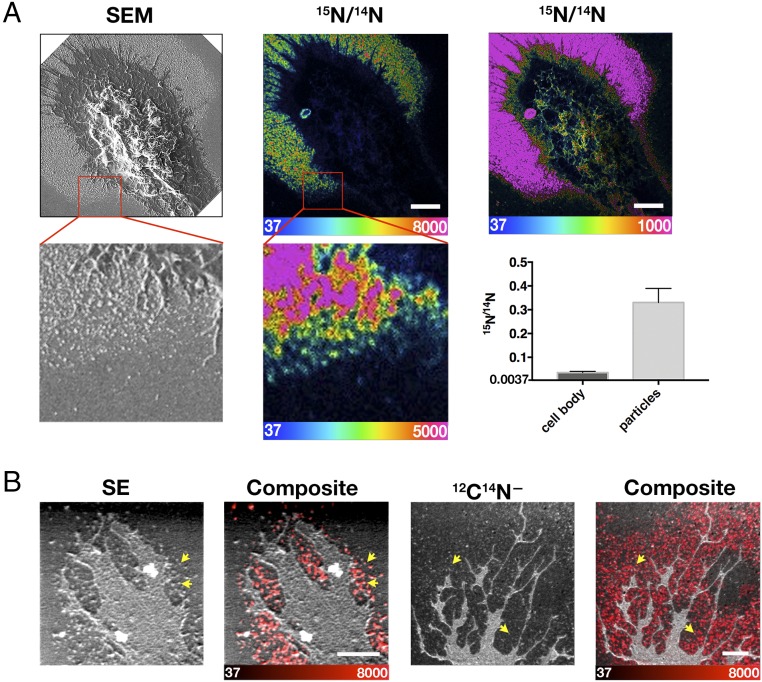
Macrophages release particles enriched in cholesterol. (A) Macrophages were loaded with cholesterol/MβCD. SEM and nanoSIMS images of the macrophage after a short incubation with [15N]ALO-D4. The SEM image shows a lawn of particles outside the macrophage; the nanoSIMS image (scaled at two different settings) reveals binding of [15N]ALO-D4 to the particles, indicating that they contain accessible cholesterol. The boxed regions are shown below at higher magnification, again showing binding of [15N]ALO-D4 to macrophage-derived particles. (Scale bars, 5 μm.) The bar graph shows 15N/14N levels for the cell body and particles of two cells (60 particles were quantified). The y axis starts at 0.0037, the natural abundance of 15N. Data are shown as mean ± SD. (B) Macrophages were loaded with acetyl-LDL. Particles released by macrophages that had been loaded with acetyl-LDL (50 μg/mL) (yellow arrows) are visible in secondary electron (SE) and 12C14N− nanoSIMS images. Composite 12C14N− or secondary electron (SE) (gray) and 15N/14N ratio (red) images show binding of [15N]ALO-D4 to particles. (Scale bars, 2 μm.)
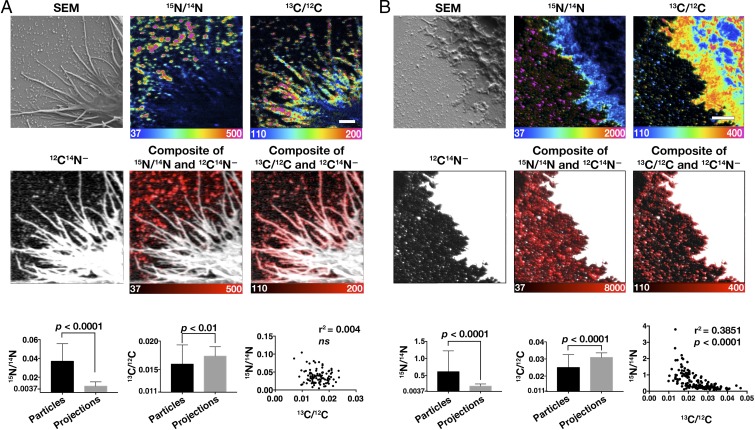
To determine if macrophage-derived particles contained accessible cholesterol, we loaded macrophages with acetyl-LDL (50 μg/mL) or cholesterol in complex with methyl-β-cyclodextrin (MβCD) and then incubated the macrophages with [15N]ALO-D4 for 2 h at 4 °C. After fixation, the cells were imaged by SEM and nanoSIMS. NanoSIMS revealed avid binding of [15N]ALO-D4 to the particles (Fig. 2), implying that the particles contained accessible cholesterol. Higher-magnification nanoSIMS images of macrophages showed that the particles are visible with secondary electrons, 12C14N− (Fig. 2B) ions, and other secondary ion signals and confirmed the binding of [15N]ALO-D4 to the macrophage-derived particles (Fig. 2B).
To further assess the cholesterol content of macrophage-derived particles, MβCD was used to load mouse peritoneal macrophages with [13C]cholesterol. We then lifted the cells with EDTA and replated them onto fresh poly-d-lysine–coated silicon wafers. After the cells were allowed to recover for 24 h, they were incubated with [15N]ALO-D4 for 2 h and processed for nanoSIMS imaging. 13C/12C nanoSIMS images revealed that the particles on the substrate surrounding macrophages were enriched in [13C]cholesterol (Fig. 3). The 15N/14N image of the same macrophage revealed that the particles contained accessible cholesterol, as judged by [15N]ALO-D4 binding (Fig. 3). The 13C/12C and 15N/14N ratios in 150 particles were positively correlated (P < 0.0001), and the r2 value was somewhat lower than we had expected (0.3663) (Fig. 3B). In hindsight, however, the low r2 value is probably not surprising. The primary Cs+ beam vaporizes cells and tissues to a depth of several nanometers, releasing secondary ions that are collected for analysis. We suspect that the depth of [15N]ALO-D4 overlying the [13C]cholesterol was variable in different particles, depending on particle size and geometry, and that this variability may have contributed, at least in part, to the moderate correlation between the 13C/12C and 15N/14N ratios. The negligible binding of ALO-D4 to particles with low levels of cholesterol probably contributed to the lower-than-expected correlation between 15N and 13C enrichments.
Fig. 3.
NanoSIMS images of macrophages that were loaded with [13C]cholesterol and then incubated with [15N]ALO-D4 revealing that macrophage-derived particles are rich in cholesterol (high 15N/14N and 13C/12C ratios). (A) Low-magnification secondary electron (SE) and 15N/14N nanoSIMS images. The lawn of particles surrounding the macrophages is enriched in 15N, reflecting the binding of [15N]ALO-D4 to accessible cholesterol. (Scale bar, 10 μm.) (B) The boxed region in A is shown at higher magnification. 13C/12C ratio and 15N/14N ratio images reveal that the particles surrounding the macrophages are enriched in both 13C and 15N. (Scale bar, 2 μm.) The scatter plot shows the 15N/14N and 13C/12C ratios for 150 particles. A linear regression analysis revealed a positive correlation between 15N and 13C enrichments (P < 0.0001). 15N/14N and 13C/12C scales are multiplied by 10,000.
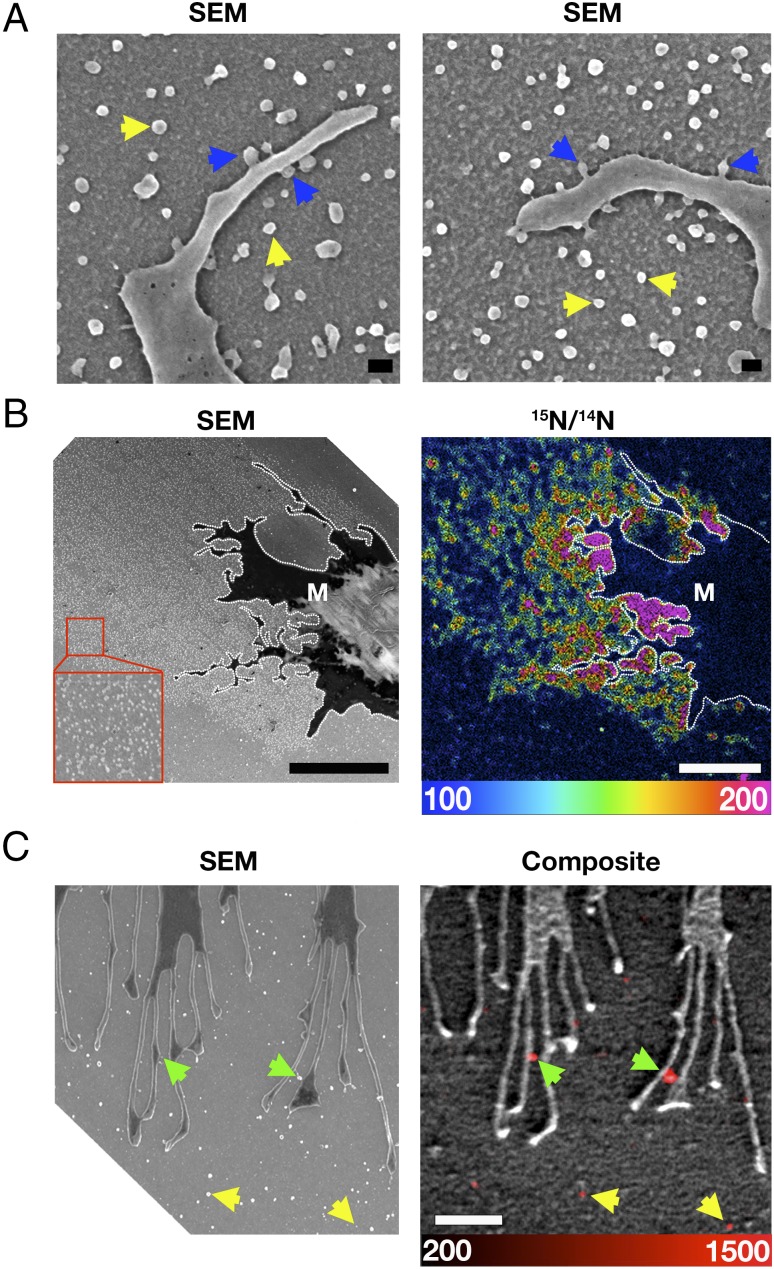
The release of particles and their attachment to the substrate was also observed in macrophages that had been incubated in medium containing 1% lipoprotein-deficient serum (LPDS) and not loaded with cholesterol (Fig. 4A). By nanoSIMS, the particles released from these macrophages contained accessible cholesterol, as judged by the binding of [15N]ALO-D4 (Fig. 4 B and C), even though accessible cholesterol was nearly absent from the plasma membrane of adjacent filopodia. In these studies, the 15N enrichment of particles was lower than in the studies shown in Fig. 2, in which cells had been loaded with cholesterol (note the different 15N/14N scales).
Fig. 4.
Macrophages release cholesterol-enriched particles when incubated in medium containing 1% LPDS. (A) Scanning electron micrographs show the release of particles from the filopodia of macrophages. Blue arrows indicate particle formation on filopodia; yellow arrows show particles on the substrate. (Scale bars, 100 nm.) (B) SEM and nanoSIMS images after a short incubation with [15N]ALO-D4. The scanning electron micrograph shows a lawn of particles on the substrate; the 15N/14N image shows [15N]ALO-D4 binding (enrichment of the particles with accessible cholesterol). The filopodia of the macrophage (M) are outlined by a dotted white line. The 15N/14N ratio scale is multiplied by 10,000 and is between two and five times the natural abundance of 15N. (Scale bars, 5 μm.) (C) Higher-magnification SEM and nanoSIMS images of the filopodia of macrophages in the absence of cholesterol loading. Note the binding of [15N]ALO-D4 to particles on the surface of filopodia (green arrows) and to particles on the surrounding substrate (yellow arrows). The 15N/14N ratio scale is multiplied by 10,000 and is between 5 and 40 times the natural abundance of 15N. (Scale bar, 1 μm.) Note that the scale in this figure differs from that in Fig. 2B, making it possible to visualize 15N enrichment that is lower than in cholesterol-loaded cells.
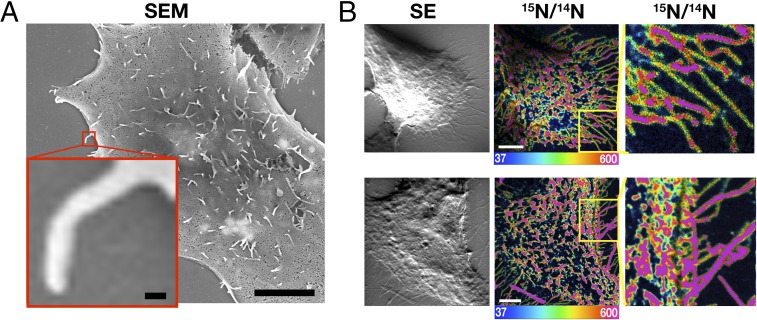
Our SEM and nanoSIMS findings in cultured CHO-K1 cells were different from the findings in macrophages (Fig. 5). The microvilli of CHO cells, both those over the cell body and those projecting from the perimeter of the cell, were enriched in accessible cholesterol, as judged by nanoSIMS images of [15N]ALO-D4 binding. Unlike the observations in macrophages, we did not observe particle release from the plasma membrane of CHO cells by SEM, and there were very few structures on the substrate resembling macrophage particles. By nanoSIMS, there was very little [15N]ALO-D4 binding to the substrate between the microvilli that projected from the perimeter of the cell (Fig. 5).
Fig. 5.
CHO-K1 cells do not produce cholesterol-rich particles. (A) SEM image of CHO-K1 cells grown in medium containing 10% FBS reveals very few particles on the substrate surrounding the cell. (Scale bar in the low-magnification image, 5 μm; scale bar in the boxed image, 100 nm.) (B) nanoSIMS secondary electron (SE) images show a lack of particles on the substrate surrounding the cell. NanoSIMS 15N/14N images reveal binding of [15N]ALO-D4 to the microvilli on the cell body and the microvilli that extend from the perimeter of the cell. (Scale bars, 5 μm.) The boxed regions are shown at higher magnification on the right.
When a 13C atom in one molecule and a 15N atom in a second molecule are located within 3 nm (as is the case when [15N]ALO-D4 is bound to [13C]cholesterol), 13C and 15N atoms released from the two molecules can combine to create a cluster secondary ion, 13C15N− (17). Thus, in experiments involving binding of [15N]ALO-D4 to [13C]cholesterol-loaded macrophages, we expected to observe a robust 13C15N− signal. Indeed, that was the case; we could image both macrophages and the surrounding particles based solely on 13C15N− secondary ions (SI Appendix, Fig. S1). In our experiments, the number of 13C15N− ions in particles surrounding macrophages was more than 2,000 times greater than expected from the natural abundance of the two stable isotopes.
Because the particles originate from the plasma membrane, we suspected that they would contain sphingomyelin. To test this idea, we incubated cholesterol-loaded RAW 264.7 macrophages and primary macrophages with [15N]ALO-D4 and [13C]lysenin [a sphingomyelin-binding cytolysin (15)]. 15N/14N and 13C/12C nanoSIMS images revealed that macrophage-derived particles were enriched in both 15N and 13C (Fig. 6). Interestingly, [13C]lysenin bound more to filopodia than to particles (i.e., the 13C/12C ratio was higher in filopodia than in particles). In contrast, the 15N/14N ratio, reflecting [15N]ALO-D4 binding to accessible cholesterol, was greater in particles than in the filopodia (Fig. 6). We found a significant inverse correlation between 13C/12C and 15N/14N ratios in the primary macrophages (Fig. 6B).
Fig. 6.
Particles released by macrophages contain cholesterol and sphingomyelin, as judged by ALO-D4 and lysenin binding. (A) RAW 267.4 murine macrophages were loaded with cholesterol/MβCD for 2 h. The cells were incubated for 2 h with [15N]ALO-D4 and for 1 h with [13C]lysenin and then were prepared for SEM and nanoSIMS imaging. A scanning electron micrograph showed particle formation on the filopodia of macrophages and particles attached to the poly-d-lysine–coated substrate. 15N/14N and 13C/12C images reveal [15N]ALO-D4 and [13C]lysenin, respectively, on macrophage-derived particles. 15N/14N ratio and 13C/12C ratio scales are multiplied by 10,000. Composite 12C14N− (gray) and 15N/14N ratio (red) or 13C/12C (red) images show binding of [15N]ALO-D4 and [13C]lysenin to particles and projections. (Scale bar, 1 μm.) ns, not significant. (B) Primary macrophages were loaded with acetyl-LDL (50 μg/mL), incubated with [15N]ALO-D4 and [13C]lysenin, and were prepared for SEM and nanoSIMS imaging. 15N/14N ratio and 13C/12C ratio scales are multiplied by 10,000. Composite 12C14N− (gray) and 15N/14N ratio (red) or 13C/12C ratio (red) images show binding of [15N]ALO-D4 and [13C]lysenin to particles and the filopodial projections. (Scale bar, 2 μm.) For quantification of 15N/14N ratios in particles released by macrophages and in the filopodia (projections) of macrophages, ratios in 150 particles and 20 projections were measured. The y axis in the 15N/14N bar graphs starts at 0.0037 (the natural abundance of 15N). The y axis in the 13C/12C bar graph starts at 0.011 (the natural abundance of 13C). Data are presented as mean ± SD. Differences were assessed with a Student’s t test with Welch’s correction. The scatter plot shows the correlation between 15N/14N and 13C/12C ratios for 150 particles. A linear regression analysis revealed a significant inverse correlation between 15N and 13C enrichments in primary macrophages (P < 0.0001).
Next, we compared the binding of [15N]ALO-D4 to particles from macrophages that had been incubated in medium containing 10% FBS and to particles from macrophages that had been incubated in medium containing 1% LPDS. Mouse peritoneal macrophages were incubated overnight in medium containing 10% FBS and then were incubated for two more days in medium containing either 10% FBS or 1% LPDS. The cells were then lifted, replated, and incubated in either 10% FBS or 1% LPDS for 12 h (allowing cells to release particles). Finally, the cells were incubated with [15N]ALO-D4 and processed for nanoSIMS. The 15N/14N ratio in particles was approximately sixfold higher in the cells incubated in 10% FBS than in the cells incubated in 1% LPDS (SI Appendix, Fig. S2). The 15N/14N ratio in projections (or filopodia) incubated in 10% FBS was only approximately twofold higher.
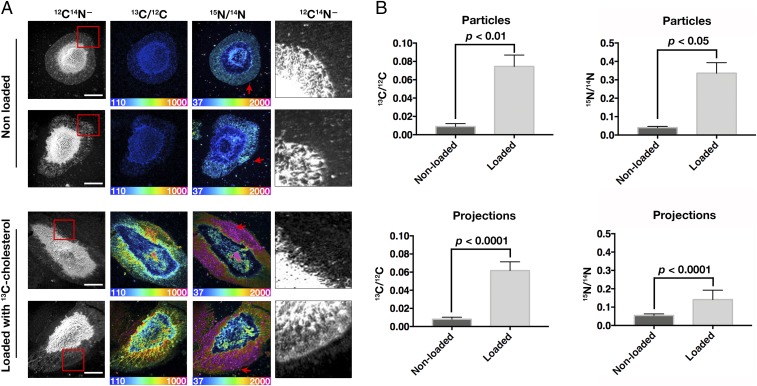
To determine whether cholesterol loading increases the binding of [15N]ALO-D4 to the particles, peritoneal macrophages were collected, and one-half of the cells were loaded with [13C]cholesterol. After incubation in medium containing 1% LPDS for 24 h, the cells were lifted with EDTA, replated, and incubated in 1% LPDS medium for 12 h. Finally, the cells were incubated with [15N]ALO-D4 for 2 h and processed for nanoSIMS (Fig. 7A). The 13C/12C ratios in the particles and projections of the [13C]cholesterol-loaded macrophages were similar (Fig. 7B). In contrast, the 15N/14N ratio was higher in the particles than in the filopodia, implying that accessible cholesterol is more abundant in particles than in filopodia (Fig. 7B).
Fig. 7.
Binding of [15N]ALO-D4 to macrophage-derived particles depends on the cholesterol content in macrophages. (A) NanoSIMS images assessing the binding of [15N]ALO-D4 to macrophages that were not loaded with cholesterol (Upper) and macrophages that had been loaded with [13C]cholesterol (Lower). Cell morphology was visualized with 12C14N− images. The 13C/12C and 15N/14N images depict [13C]cholesterol and [15N]ALO-D4, respectively. Boxed areas in the 12C14N− images are shown at higher magnification in the images on the right, revealing particles on the substrate (red arrows). 15N/14N and 13C/12C ratio scales are multiplied by 10,000. (Scale bar, 10 μm.) (B) Bar graphs show the 13C/12C (Left) and 15N/14N (Right) ratios in particles (Upper) and macrophage projections (Lower). Three 40 × 40-μm images, each containing one or two macrophages, were quantified. For particle quantification, a minimum of 50 particles per image was selected on the basis of the 12C14N− image, and the mean 15N/14N ratio in 15–30 projections per cell was calculated. Data are presented as mean ± SD. Differences were assessed with a Student’s t test with Welch’s correction.
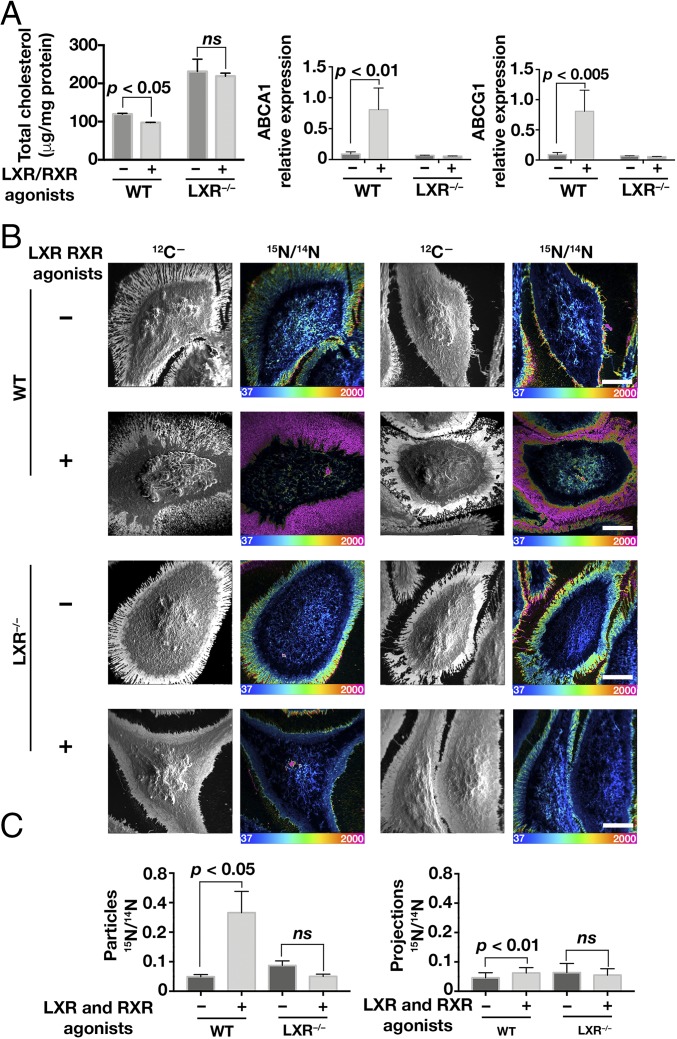
To determine whether the cholesterol content of particles is influenced by the LXR signaling pathway, we tested the effects of LXR/retinoid X receptor (RXR) agonists on the cholesterol content of particles from peritoneal macrophages of wild-type and Lxrα/Lxrβ double-knockout (LXR−/−) mice. Macrophages were loaded with acetyl-LDL, lifted with EDTA, and replated. The cells were then treated with the LXR/RXR agonists or vehicle (DMSO) alone for 12 h. As expected, cholesterol levels in wild-type macrophages fell during treatment with LXR/RXR agonists; cholesterol levels in LXR−/− macrophages were somewhat higher and did not fall when the cells were treated with LXR/RXR agonists (Fig. 8A). The LXR/RXR agonists increased ABCA1 and ABCG1 expression levels in wild-type but not in LXR−/− macrophages (Fig. 8A).
Fig. 8.
NanoSIMS analysis of [15N]ALO-D4 binding to cholesterol-loaded wild-type macrophages or Lxrα/Lxrβ double-knockout macrophages (LXR−/−) in the presence of LXR and RXR agonists or vehicle (DMSO) alone. Macrophages were loaded with acetyl-LDL before being lifted with EDTA, replated, and incubated with LXR/RXR agonists or DMSO alone for 12 h. (A) Bar graphs depicting cellular cholesterol levels and ABC transporter expression levels in wild-type and LXR−/− macrophages treated with LXR/RXR agonists or DMSO alone. Cholesterol was measured in two wells of macrophages in a six-well plate; each data point in the bar graph represents cholesterol content in one well of macrophages. RNA was isolated from macrophages in a six-well plate, and levels of gene expression were measured (n = 5 for WT and n = 3 for LXR−/−). Data are presented as mean ± SD. Differences were assessed with a Student’s t test with Welch’s correction. (B) NanoSIMS images of WT and LXR−/− macrophages after incubation with [15N]ALO-D4. Cell morphology was visualized with 12C− images; 15N/14N images show binding of [15N]ALO-D4 to macrophages and macrophage-derived particles. 15N/14N ratio scales are multiplied by 10,000. (Scale bars, 10 μm.) (C) Quantification of 15N/14N ratios in particles and filopodia (projections) of wild-type and LXR−/− macrophages. Two or three 40 × 40-μm images, each containing one or two macrophages, were quantified. A minimum of 50 particles per image were selected from the 12C− image (particles were visible after adjusting the contrast of the images), and the mean 15N/14N ratio was calculated. For the filopodial projections, the 15N/14N ratio in 15–30 projections per cell was calculated. The 13C/12C ratios in cells that did not receive [13C]cholesterol reflected the natural abundance of 13C. Data are presented as mean ± SD. Differences were assessed with a Student’s t test with Welch’s correction; ns, not significant.
Particle release from the plasma membrane was observed in both wild-type and LXR−/− macrophages (SI Appendix, Fig. S3). LXR/RXR agonist treatment of wild-type macrophages increased accessible cholesterol in both particles and the filopodia, as judged by [15N]ALO-D4 binding (Fig. 8B). Interestingly, the LXR/RXR agonist-induced increase in [15N]ALO-D4 binding was greater in particles than in filopodia (Fig. 8 B and C). In LXR−/− macrophages, LXR/RXR agonists resulted in little or no change in [15N]ALO-D4 binding to either particles or filopodia (Fig. 8 B and C). In an independent experiment, LXR/RXR agonists had little or no effect on [15N]ALO-D4 binding to LXR−/− macrophages, but they increased [15N]ALO-D4 binding to particles and projections of wild-type macrophages in both the presence and absence of cholesterol loading (SI Appendix, Fig. S4). Again, the LXR/RXR agonist-induced increase in [15N]ALO-D4 binding was greater in particles than in filopodia (SI Appendix, Fig. S4).
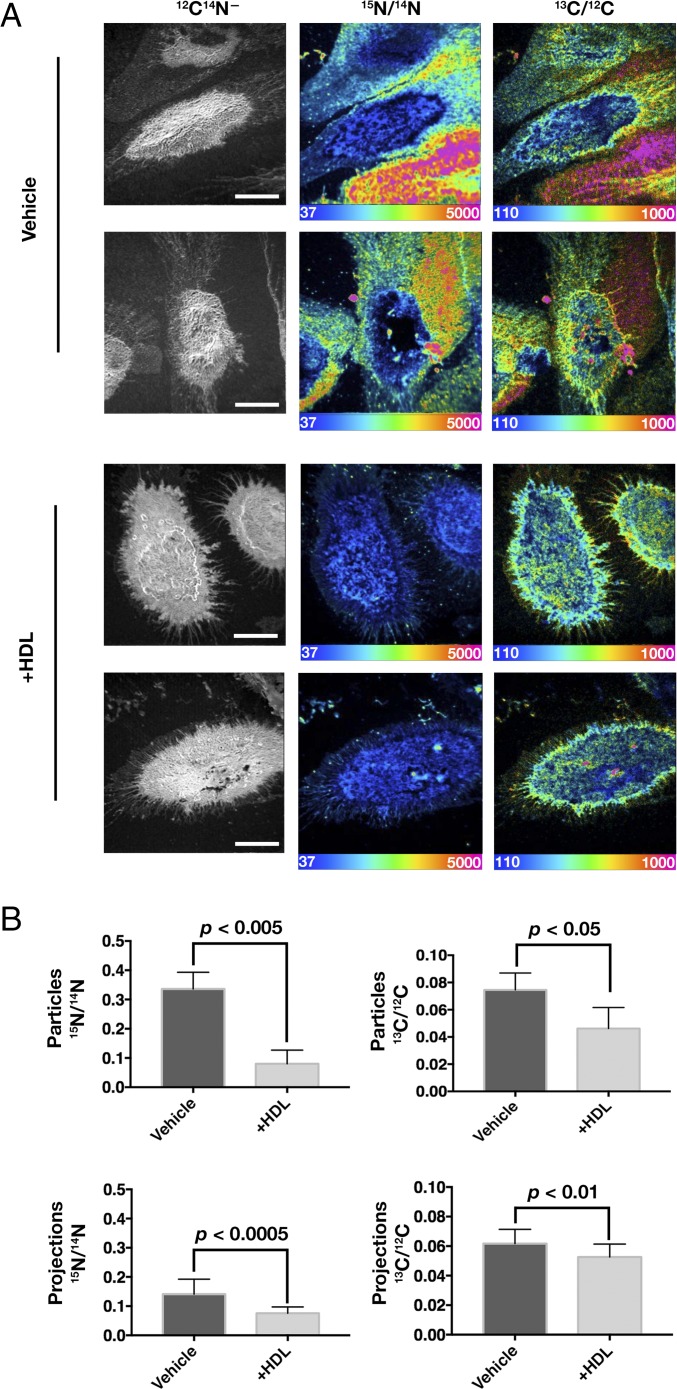
To determine if HDL is capable of unloading cholesterol from macrophage-derived particles, we loaded macrophages with [13C]cholesterol/MβCD for 24 h, lifted the cells with EDTA, and replated the cells in medium in the presence or absence of HDL for 24 h. Next, the cells were incubated with [15N]ALO-D4 for 2 h and processed for nanoSIMS. The HDL incubation did not have an obvious effect on particle production, as judged by SEM (SI Appendix, Fig. S3). However, the incubation with HDL reduced both the [13C]cholesterol content of particles and the binding of [15N]ALO-D4 to the particles (Fig. 9). Even after the HDL incubation, it was possible to visualize particles by adjusting the scale of the nanoSIMS images (SI Appendix, Fig. S5). Interestingly, the 15N/14N ratio in particles fell by ∼75% with the HDL incubation, while the 13C/12C ratio fell by only ∼40% (Fig. 9B). HDL reduced [13C]cholesterol content and [15N]ALO-D4 binding to filopodia of macrophages but to a lesser extent than with particles. A 15-min incubation of macrophages with MβCD also reduced the binding of [15N]ALO-D4 to the particles and projections of macrophages (SI Appendix, Fig. S6). Interestingly, the decrease in [15N]ALO-D4 binding to particles with MβCD treatment was proportionately greater than the decrease in [15N]ALO-D4 binding to the filopodia (SI Appendix, Fig. S6).
Fig. 9.
HDL removes cholesterol from macrophage-derived particles. Macrophages were loaded with [13C]cholesterol for 24 h before being lifted, replated, and incubated in the presence or absence of HDL for 24 h. After cells were incubated with [15N]ALO-D4 for 2 h, nanoSIMS imaging was performed. (A) Cell morphology was visualized with 12C14N− images; the 15N/14N image reflects [15N]ALO-D4 binding (accessible cholesterol); the 13C/12C ratio reflects total cholesterol. 15N/14N ratio and 13C/12C ratio scales are multiplied by 10,000. (Scale bars, 10 μm.) (B) Quantification of 15N/14N and 13C/12C ratios revealed reduced [13C]cholesterol and [15N]ALO-D4 binding in particles after incubation with HDL. Three or four 40 × 40-μm images, each with one or two macrophages, were quantified. For particle quantification, a minimum of 50 particles per image were selected from the 12C14N− image (particles were visible after the contrast was adjusted), and the mean 15N/14N ratio for the particles was calculated. For the filopodial projections, the 15N/14N ratio of 15–30 projections per cell was calculated. Data are presented as mean ± SD. Differences were assessed with a Student’s t test with Welch’s correction.
Discussion
We found that filopodia of mouse peritoneal macrophages and RAW 264.7 cell macrophages release ∼20- to 120-nm unilamellar vesicular particles. In the past, macrophages have been reported to release ∼24-nm particles into the cell-culture medium (2) or to deposit irregularly shaped cholesterol microdomains on the substrate (3), but mechanisms for the genesis and release of the particles and microdomains have never been clear. In the current study, we demonstrated by SEM and TEM that particles appear to be released from the plasma membrane of filopodia, attaching to the surrounding substrate. By nanoSIMS analyses, these particles are enriched in cholesterol. When macrophages were loaded with [13C]cholesterol, the particles on the surrounding substrate were enriched in 13C. The particles also bound [15N]ALO-D4 avidly. Because macrophage-derived particles originate from the plasma membrane, we expected that they would contain sphingomyelin. Indeed, nanoSIMS studies showed that lysenin, a sphingomyelin-specific cytolysin, bound to macrophage-derived particles.
The appearance and release of particles from the macrophage plasma membrane was evident under multiple conditions—whether the cells were incubated in LPDS- or FBS-containing medium, whether or not the macrophages were loaded with cholesterol, whether or not they were deficient in LXRs, whether or not they were treated with LXR/RXR agonists, and whether or not the medium contained HDL. While we could not discern clear differences in particle size or numbers under these different conditions, we certainly cannot exclude the possibility that such differences exist, given that we do not have protocols for accurately quantifying particle numbers or measuring the dynamics of particle formation and release. On the other hand, we are confident that the cholesterol content of particles changes with different conditions. Loading macrophages with cholesterol, incubating the cells in FBS rather than LPDS, or adding LXR/RXR agonists to the cell-culture medium increases the content of cholesterol in particles, as judged by nanoSIMS analyses.
Using biochemical approaches, Radhakrishnan and coworkers (12) demonstrated that ALO-D4 binds to an accessible pool of cholesterol on the plasma membrane. The accessible pool is not sequestered by sphingomyelin or phospholipids and appears when the cholesterol content of the plasma membrane exceeds ∼30 mol% (12, 14). More recently, the same group showed that the accessible pool in the plasma membrane is mobile, readily moving to the endoplasmic reticulum and participating in the regulation of genes involved in cholesterol synthesis (14). Our studies revealed that the particles released from macrophage filopodia are enriched in accessible cholesterol. (i) [15N]ALO-D4 binds avidly to macrophage particles even when the macrophages were incubated in medium containing 1% LPDS and the binding of the [15N]ALO-D4 to the surrounding filopodia was extremely low (Fig. 4). (ii) When macrophages were incubated in medium containing 10% FBS (rather than 1% LPDS), the amount of accessible cholesterol in particles increased substantially while the increase in filopodia was modest (SI Appendix, Fig. S2). (iii) When macrophages were loaded with [13C]cholesterol, [15N]ALO-D4 binding to particles increased substantially more than the increase in binding to the macrophage plasma membrane (Fig. 7). (iv) When cells were loaded with cholesterol, adding LXR/RXR agonists to the cell-culture medium increased [15N]ALO-D4 binding to both particles and filopodia, but the increase in the binding of [15N]ALO-D4 to particles was greater (Fig. 8). (v) [13C]lysenin bound preferentially to the filopodial projections of macrophages, whereas [15N]ALO-D4 bound preferentially to particles (Fig. 6). Each of these observations suggests that the particles that are formed and then released from the plasma membrane are enriched in accessible cholesterol.
The combination of LXR and RXR agonists increased the cholesterol content of macrophage particles, as judged by nanoSIMS analyses. These drugs induce the production of many proteins involved in cholesterol metabolism and cholesterol efflux. We suspect that increased expression of ABC transporters is important for enriching the macrophage plasma membrane and plasma membrane-derived particles with cholesterol. By nanoSIMS analyses, we also found that HDL unloads cholesterol from macrophage-derived particles. It is noteworthy that the decrease in [15N]ALO-D4 binding to particles with the HDL incubation was greater than the decrease in the [13C]cholesterol content of the particles. We suspect that this difference relates to the greater mobility of accessible cholesterol.
We do not yet have a firm understanding of the mechanisms for the formation and release of particles from the plasma membrane of macrophages. By SEM, the particles appear to bud from the plasma membrane. One possibility is that the particles form by ballooning or outpouching of a segment of the plasma membrane that is not firmly attached to the cytoskeleton. However, a second possibility is that discrete segments of the macrophage plasma membrane are strongly affixed to the substrate by substrate adhesion proteins and that those segments are left behind in the form of vesicular particles as the cell “pulls away” during locomotion. This second possibility is attractive because it would help explain why the lawn of particles is generally located on one or two sides of the macrophage rather than around the entire circumference of the cell. (Note that the two possibilities are not mutually exclusive.) However, if we assume that cell mobility is largely responsible for the genesis of particles, the remaining mystery is why the particles are more enriched in accessible cholesterol than the adjacent filopodia and lamellipodia. Cholesterol affects many properties of lipid bilayers, including viscosity, elasticity, permeability, and protein association. Perhaps the accessible cholesterol within the plasma membrane is less firmly attached to the cytoskeleton and is more prone to being “left behind” on vesicular particles as macrophages move across the substrate.
One limitation of our study is that all our studies dealt with cultured macrophages. Whether particles are released from macrophages in vivo is unknown, but we suspect that particle release will prove to be a feature of resident macrophages in mammalian tissues. Following the phagocytosis of senescent erythrocytes, splenic macrophages need to unload substantial amounts of cholesterol, and particle release could assist in that function. We further suspect that a failure to enrich particles in cholesterol in the setting of ABCA1 deficiency could help to explain the hallmark histopathology of Tangier disease (cholesterol-laden macrophages in the spleen and lymph nodes). Particle release from macrophages in atherosclerotic plaques could also be a mechanism for unloading cholesterol and promoting reverse cholesterol transport.
Whether the macrophage microparticles described by the laboratory of Phillips and coworkers (2) were similar to those in this study is unclear. The microparticles in the Phillips study averaged 24 nm in diameter. By SEM, many of the vesicular particles that we observed were in a similar size range (30 ± 10 nm in cholesterol-loaded macrophages vs. 34 ± 17 nm in nonloaded macrophages; n = 30 particles for each). Kruth and coworkers (3–6, 9) have published a series of papers on the release of cholesterol microdomains from macrophages, but how they were released from cells was not defined. We believe it is likely that the cholesterol microdomains in the studies by Kruth and coworkers are the same as the particles that we document in the current studies, namely vesicular particles that are released from the plasma membrane of macrophage filopodia. The studies by Kruth relied largely on immunofluorescence microscopy with a cholesterol-specific antibody. In our studies, cholesterol enrichment of the macrophage particles was documented by nanoSIMS with [13C]cholesterol and [15N]ALO-D4. In the studies by Kruth and coworkers (4–6), microdomain production virtually disappeared in the absence of cholesterol loading. In our studies, we found particle release under a variety of cell-culture conditions, including in cells in the absence of cholesterol loading and cells incubated in 1% LPDS. However, the different cell-culture conditions (e.g., cholesterol loading, LXR/RXR agonists) in our studies clearly influenced the cholesterol content of the particles. We do not understand why particle release disappeared in the studies by Kruth and coworkers (4–6), but one possibility is that they used human monocyte-derived macrophages rather than mouse peritoneal macrophages.
The release of vesicular particles that we documented in macrophages is morphologically similar to the budding of HIV-1 virions from macrophages (18–21). HIV-1 virions are released by a budding process from cholesterol- and sphingolipid-rich regions of the plasma membrane (18). HIV-1 budding may be driven by the formation of a latticework of Gag proteins along budding sites. In the case of our studies, the identity of proteins important for particle release is unknown. Another process that closely resembles particle release from macrophages is the budding of vesicles from the canalicular membrane of hepatocytes (22, 23). Like macrophages, hepatocytes need to dispose of large amounts of cholesterol, and that involves releasing cholesterol-rich vesicular particles from the canalicular microvilli. Another physiological event resembling macrophage particle release is the release of vesicles from the tips of the microvilli of intestinal enterocytes (24, 25). In that case, the size of the vesicles released from the tips of microvilli was altered in mice lacking myosin 1a, implicating cytoplasmic motors in particle formation and/or release (25). Myosins are required for the formation of macrophage filopodia and their movement (26–29), but their relevance to particle release by macrophages is not yet known. However, we suspect that the application of motor-driven forces to the plasma membrane is important for the formation and release of particles.
Materials and Methods
Mouse Strains.
Wild-type mice (C57BL/6) were purchased from the Jackson Laboratory. Lxrα/Lxrβ double-knockout mice were originally obtained from David Mangelsdorf (University of Texas Southwestern Medical Center, Dallas) and have been backcrossed to strain C57BL/6 for more than 10 generations (30).
Cell Lines.
RAW 264.7 cell line murine macrophages (purchased from ATCC) and hamster CHO-K1 cells were grown in monolayer cultures at 37 °C with 8–9% CO2. RAW cells were maintained in macrophage medium (DMEM) (Thermo Fisher Scientific) containing 1 mM sodium pyruvate, 2 mM glutamine, and 10% (vol/vol) FBS (HyClone). CHO-K1 cells were maintained in Ham’s F-12 Nutrient Mixture (Thermo Fisher Scientific) containing 10% (vol/vol) FBS (HyClone) and 2 mM glutamine.
Mouse Peritoneal Macrophages.
Wild-type and Lxrα/Lxrβ double-knockout mice were injected i.p. with 1 mL of 3% (wt/vol) Difco Fluid Thioglycollate Medium (Becton, Dickinson and Co.). Three days later, peritoneal macrophages were harvested with 10 mL of cold Dulbecco’s PBS without Ca2+ and Mg2+. Cells were centrifuged at 400 × g for 5 min at 4 °C, treated with 5 mL of red blood cell lysing buffer Hybri-Max (Sigma) for 5 min, and washed two times with cold PBS. Macrophages were plated on FBS-coated Petri dishes (8 × 106 cells per dish) and were cultured overnight in macrophage medium. Macrophages were then lifted by incubation in cold PBS without Ca2+ and Mg2+ containing 5% FBS and 5 mM EDTA for 45 min at 4 °C. For nanoSIMS and SEM, cells were replated on 0.5-cm2 silicon wafers coated with 0.1 mg/mL poly-d-lysine hydrobromide (Sigma) in 24-well plates.
Preparation of [13C]cholesterol.
[13C]cholesterol was produced as described (31) using a Saccharomyces cerevisiae strain (RH6829) engineered to produce cholesterol rather than ergosterol (32, 33). RH6829 yeast cells were grown in medium containing 0.7% yeast nitrogen base (US Biological), 0.5% yeast extract (BD), 1.5% glucose (>98% 13C; >99% glucose) (Martek Isotopes LLC), and 0.4 mg/L uracil and leucine. Precultures were diluted 1:2,000 in this medium and were grown for 3 d at 30 °C. The [13C]cholesterol was purified from harvested cells and analyzed by GC-MS and by NMR (32). The cholesterol contained 94% 13C, and the yield was ∼10 mg/L of medium.
Cholesterol Loading of Macrophages.
For some experiments, macrophages were loaded with 50 μg/mL of acetyl-LDL (Alfa Aesar) in macrophage medium containing 1% LPDS (15) instead of 10% FBS for 24 h at 37 °C. In other experiments, macrophages were loaded with [13C]cholesterol/MβCD (34). Briefly, [13C]cholesterol in 100% ethanol (7.5 mg/mL) was added in 50-μL aliquots to a stirring solution of 5% (wt/vol) methyl-β-cyclodextrin (Sigma) in double-distilled water (ddH2O) in an 80 °C water bath to achieve an MβCD:cholesterol ratio of 10:1. The solution was lyophilized and then reconstituted in ddH2O to a cholesterol concentration of 2.5 mM and an MβCD concentration of 25 mM. The [13C]cholesterol/MβCD solution was filtered through a 0.22-μm filter and stored at 4 °C. Macrophages were loaded with 20 μL/mL of [13C]cholesterol/MβCD (final cholesterol concentration, 50 μM) in macrophage medium containing 0.1% LPDS, 50 μM mevastatin (Calbiochem), and 50 μM mevalonolactone (Sigma) for 24 h at 37 °C. RAW 267.4 cells were loaded with unlabeled cholesterol/MβCD complexes (Sigma).
Treatment of Macrophages with LXR/RXR Agonists, HDL, and MβCD.
In some experiments, macrophages were treated with GW3965 (35), an LXR agonist, and LG268 (Ligand Pharmaceuticals), an RXR agonist. Cells were treated with 1 μM LXR ligand and 100 nM RXR ligand in DMSO or with DMSO alone in macrophage medium containing 1% LPDS for 12 h at 37 °C. In other experiments, cells were incubated with 200 μg/mL of human HDL (Alfa Aesar) in macrophage medium containing 1% LPDS for 24 h or were incubated with 10 mM MβCD in DMEM for 15 min at 37 °C.
Preparation of 15N-Labeled His-Tagged ALO-D4.
A plasmid for ALO-D4 (ALO amino acids 404–512 with C472A and S404C substitutions) was obtained from Arum Radhakrishnan (University of Texas Southwestern Medical Center, Dallas), and 15N-labeled ALO-D4 was prepared (15, 36). Briefly, ALO-D4 was expressed in BL21(DE3) pLysS Escherichia coli (Invitrogen) and induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) in 1 L of minimal medium containing 20.2 mM 15NH4Cl at 25 °C for 16 h. Cells were pelleted and lysed by sonication, and the lysate was centrifuged at 4 °C. The supernatant was mixed with 4 mL of HisPur Cobalt resin (50% bed volume; Thermo Fisher Scientific). The mixture was loaded into a column and allowed to flow through by gravity. The column was washed, and [15N]ALO-D4 was eluted with a buffer containing 300 mM imidazole. The eluates were pooled and concentrated to 1 mL with an Amicon 10-kDa cut off concentrator (Millipore). The purified [15N]ALO-D4 was stored at 4 °C.
Preparation of 13C-Labeled His-mCherry–Tagged Lysenin.
To produce [13C]lysenin, E. coli BL21(ED3) (Invitrogen) was transformed with the plasmid encoding lysenin (15) and grown in 1 L of minimal medium containing 95.5 mM KH2PO4, 57.4 mM K2HPO4, 63.4 mM Na2HPO4, 13.8 mM K2SO4, 20.2 mM NH4Cl, 5 MgCl2, 0.2% (wt/vol) 13C6 glucose (Cambridge Isotope Laboratories), and 100 μg/mL carbenicillin. The expression of lysenin was induced with 0.2% (wt/vol) arabinose at 25 °C for 16 h; [13C]lysenin was purified as described (15).
Binding of ALO-D4 and Lysenin to Cells.
Primary macrophages were washed three times for 10 min in PBS/Ca/Mg containing 0.2% (wt/vol) BSA. Cells were then incubated with 20 μg/mL of [15N]ALO-D4 in PBS/Ca/Mg containing 0.2% (wt/vol) BSA for 2 h at 4 °C. RAW cells were incubated in macrophage medium containing 0.1% LPDS, unlabeled cholesterol/MβCD (final cholesterol concentration of 50 μM), 50 μM mevastatin (Calbiochem), and 50 μM mevalonolactone (Sigma) for 2 h at 37 °C. Cells were lifted, washed, and replated onto fresh poly-d-lysine–coated silicon wafers for 20 h. Next, cells were incubated with 20 μg/mL of [13C]lysenin for 1 h and 20 μg/mL of [15N]ALO-D4 in PBS/Ca/Mg containing 0.2% (wt/vol) BSA for 2 h at 4 °C. Unbound [15N]ALO-D4 and [13C]lysenin were removed by washing with PBS/Ca/Mg containing 0.2% (wt/vol) BSA three times for 2 min each.
Preparing Samples for NanoSIMS and SEM.
Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) and 2.5% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M phosphate buffer (1.14 g NaH2PO4, 1.69 g Na2HPO4 in a 100-mL final volume of ddH2O, pH 7.4) for 20 min at 4 °C followed by 1 h at room temperature. The samples were washed three times for 7 min each in 0.1 M phosphate buffer, postfixed with 1% osmium tetroxide (Electron Microscopy Sciences) in 0.1 M phosphate buffer for 45 min, and washed three times for 7 min each in ice-cold ddH2O. For nanoSIMS, cells were air-dried. For SEM, cells were dehydrated with a graded series of ethanol concentrations (50, 70, 85, 95, and 100% ×3 for 7 min each) and then were critical-point dried with a Tousimis Autosamdri 810 critical point dryer (Tousimis). Samples were then coated with 2 nm of platinum (Pelco) with an ion-beam sputtering system (South Bay Technologies). Cells were imaged with a Zeiss Supra 40VP scanning electron microscope with a 3-KeV incident beam.
Preparing Samples for TEM.
Primary macrophages were cholesterol-loaded and plated onto poly-d-lysine–coated Thermanox plastic coverslips (Thermo Scientific) in a 24-well plate. After 24 h, the cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer for 2 h at 4 °C. The samples were then washed five times for 2 min each in 0.1 M phosphate buffer, postfixed with ice-cold 2% osmium tetroxide in 0.1 M phosphate buffer for 45 min, and washed five times for 2 min each in ice-cold ddH2O. The cells were then incubated with 2% (vol/vol) aqueous uranyl acetate overnight at 4 °C, washed five times with ddH2O, and dehydrated with a graded series of ethanol concentrations (30, 50, 70, 85, 95, and 100% ×3, 7 min each) before infiltration with increasing concentrations (33, 66, and 100%) of Epon (Ted Pella) (15). Next, the coverslips were embedded by inverting on a BEEM capsule (Ted Pella) filled with fresh resin and were polymerized for 48 h at 60 °C. The coverslips removed, and 65-nm-thick sections were cut en face with a Diatome diamond knife. Sections were placed on formvar-coated 100-mesh copper grids that had been glow-discharged. The sections on grids were stained with Reynold’s lead citrate for 9 min. Next, samples were imaged at 200 kV with an FEI T12 iCorr microscope equipped with an Eagle 2K CCD camera or were imaged at 60 kV with a JEOL 100CX transmission electron microscope.
NanoSIMS Analyses.
Platinum-coated (5-nm) cells were analyzed with a nanoSIMS 50L instrument (CAMECA) as described (15, 37) with some modifications. Briefly, samples were bombarded with a focused 133Cs+ primary beam, and secondary ions (e.g., 12C−, 13C−, 16O−, 12C14N−, 12C15N−) and secondary electrons were collected. Before imaging, a high 133Cs+ primary beam (1-nA beam current; primary aperture D1 = 1) was used to presputter an area of 50 × 50 μm for 25 s to remove the platinum coating and implant 133Cs+. In the same region, low-magnification images (∼40 × 40 μm) were obtained with an ∼2.5-pA beam current (primary aperture D1 = 2), a dwell time of 2.5 ms per pixel, and scans of 512 × 512 pixels. High-magnification images (∼10 × 10 μm) were obtained with an ∼0.8-pA beam current (primary aperture D1 = 3), a dwell time of ∼10 ms per pixel, and scans of 512 × 512 pixels.
To quantify 13C/12C and 15N/14N ratios in particles, we identified particles by SEM and/or 12C−, 12C14N−, 16O−, or secondary electron nanoSIMS images, and regions of interest in the middle of the particles were defined with the OpenMIMS plugin in ImageJ (NIH). The region in the middle of each particle was quantified rather than the whole particle because we wanted to avoid pixels overlapping the perimeter of the particle and/or the substrate immediately adjacent to the particle. For each image, 50–100 particles were selected. To quantify 13C/12C and 15N/14N ratios in macrophage filopodia, straight lines were drawn, pixel by pixel, in the middle of the filopodial projections, and line scan analyses were performed. The average 13C/12C and 15N/14N ratios of line scan analyses were calculated. For each cell, 15–30 projections were assessed. The mean 15N/14N and 13C/12C ratios of the regions of interest were measured and processed by Prism 7.0. Differences were assessed by a Student’s t test with Welch’s correction.
Gene-Expression Analyses.
Peritoneal macrophages were isolated from wild-type and Lxrα/Lxrβ double-knockout mice as described earlier. Cells were plated in macrophage medium containing 10% FBS. On the next day, cells were washed and were incubated in macrophage medium containing 1% LPDS and 50 μg/mL acetyl-LDL for 24 h. Cells were treated with 1 mM LXR ligand (GW3965) and 100 nM RXR ligand (LG268) for 12 h, and then RNA and lipids were extracted. Total RNA was isolated with TRIzol reagent (Invitrogen) and reverse-transcribed with the iScript cDNA synthesis kit (Bio-Rad). cDNA was quantified by real-time PCR with SYBR Green Master Mix (Diagenode) on an ABI 7900 instrument. Gene-expression levels were determined with a standard curve. Each gene was normalized to the housekeeping gene 36B4 and analyzed in duplicate. Primers for real-time PCR were 5′-CGTTTCCGGGAAGTGTCCTA-3′ (mABCA1 forward primer), 5′-GCTAGAGATGACAAGGAGGAT-3′ (mABCA1 reverse primer), 5′-TCACCCAGTTCTGCATCCTCT-3′ (mABCG1 forward primer), and 5′-GCAGATGTGTCAGGACCGAGT-3′ (mABCG1 reverse primer). In these experiments, cellular cholesterol was extracted with hexane/isopropanol (3:2). The organic phase was collected and dried under nitrogen and resuspended with Tag Replication Buffer (TRB) buffer [100 mM KH2PO4, 100 mM K2HPO4, 50 mM NaCl, 5 mM sodium chlorate, and 0.1% Triton X-100 (pH 7.4)]. Cholesterol was quantified with the Amplex Red Cholesterol kit (Thermo Fisher Scientific). The values were normalized to cellular protein content determined with a BCA protein assay kit (Thermo Fisher Scientific).
Study Approval.
Mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited vivarium at the University of California, Los Angeles (UCLA). All animal procedures were approved by the UCLA Chancellor’s Animal Research Committee.
Supplementary Material
Acknowledgments
We thank the nanoSIMS facilities at the California Institute of Technology and the University of Western Australia; Dr. Kevin Williams and Dylan Zhou for assistance in measuring [13C]cholesterol by mass spectrometry; and Dr. Marianne Cilluffo at the UCLA Brain Research Institute for help in obtaining TEM images. This work was supported by Fondation Leducq Transatlantic Network Grant 12CVD04 (to S.G.Y.), NIH Grants P01 HL090553, R01 HL087228, R35 HL139725-01, and HL125335 (all to S.G.Y.), Ruth L. Kirschstein National Research Service Award F32 HL132471 (to C.H.), the Swiss National Science Foundation (H.R.), and the National Centre of Competence in Research in Chemical Biology (H.R.). H.J. is supported by Australian Research Council Discovery Early Career Researcher Award DE180100080 and a Healy Research Collaboration Award. Support was also received from the Australian Microscopy and Microanalysis Research Facility and the Science and Industry Endowment Fund for supporting the Ion Probe Facility at the Centre for Microscopy, Characterisation and Analysis at the University of Western Australia.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810724115/-/DCSupplemental.
References
- 1.Tall AR, Costet P, Wang N. Regulation and mechanisms of macrophage cholesterol efflux. J Clin Invest. 2002;110:899–904. doi: 10.1172/JCI16391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duong PT, et al. Characterization of nascent HDL particles and microparticles formed by ABCA1-mediated efflux of cellular lipids to apoA-I. J Lipid Res. 2006;47:832–843. doi: 10.1194/jlr.M500531-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Jin X, et al. ABCA1 (ATP-binding cassette transporter A1) mediates ApoA-I (apolipoprotein A-I) and ApoA-I mimetic peptide mobilization of extracellular cholesterol microdomains deposited by macrophages. Arterioscler Thromb Vasc Biol. 2016;36:2283–2291. doi: 10.1161/ATVBAHA.116.308334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin X, et al. ABCA1 contributes to macrophage deposition of extracellular cholesterol. J Lipid Res. 2015;56:1720–1726. doi: 10.1194/jlr.M060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman SR, et al. ABCG1-mediated generation of extracellular cholesterol microdomains. J Lipid Res. 2014;55:115–127. doi: 10.1194/jlr.M044552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong DS, et al. Extracellular cholesterol-rich microdomains generated by human macrophages and their potential function in reverse cholesterol transport. J Lipid Res. 2010;51:2303–2313. doi: 10.1194/jlr.M005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hafiane A, Genest J. ATP binding cassette A1 (ABCA1) mediates microparticle formation during high-density lipoprotein (HDL) biogenesis. Atherosclerosis. 2017;257:90–99. doi: 10.1016/j.atherosclerosis.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Phillips MC. Molecular mechanisms of cellular cholesterol efflux. J Biol Chem. 2014;289:24020–24029. doi: 10.1074/jbc.R114.583658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin X, et al. Macrophages shed excess cholesterol in unique extracellular structures containing cholesterol microdomains. Arterioscler Thromb Vasc Biol. 2018;38:1504–1518. doi: 10.1161/ATVBAHA.118.311269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vedhachalam C, et al. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J Biol Chem. 2007;282:25123–25130. doi: 10.1074/jbc.M704590200. [DOI] [PubMed] [Google Scholar]
- 11.Frisz JF, et al. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J Biol Chem. 2013;288:16855–16861. doi: 10.1074/jbc.M113.473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A, Brown MS, Anderson DD, Goldstein JL, Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 2014;3:02882. doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das A, Goldstein JL, Anderson DD, Brown MS, Radhakrishnan A. Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc Natl Acad Sci USA. 2013;110:10580–10585. doi: 10.1073/pnas.1309273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Infante RE, Radhakrishnan A. Continuous transport of a small fraction of plasma membrane cholesterol to endoplasmic reticulum regulates total cellular cholesterol. eLife. 2017;6:25466. doi: 10.7554/eLife.25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He C, et al. High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proc Natl Acad Sci USA. 2017;114:2000–2005. doi: 10.1073/pnas.1621432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein JL, Ho YK, Basu SK, Brown MS. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc Natl Acad Sci USA. 1979;76:333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss FR, 3rd, Boxer SG. Atomic recombination in dynamic secondary ion mass spectrometry probes distance in lipid assemblies: A nanometer chemical ruler. J Am Chem Soc. 2016;138:16737–16744. doi: 10.1021/jacs.6b10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundquist WI, Kräusslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abbas W, Herbein G. Plasma membrane signaling in HIV-1 infection. Biochim Biophys Acta. 2014;1838:1132–1142. doi: 10.1016/j.bbamem.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 21.Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell. 2010;102:335–350. doi: 10.1042/BC20090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crawford AR, et al. Hepatic secretion of phospholipid vesicles in the mouse critically depends on mdr2 or MDR3 P-glycoprotein expression. Visualization by electron microscopy. J Clin Invest. 1997;100:2562–2567. doi: 10.1172/JCI119799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford JM, et al. Imaging biliary lipid secretion in the rat: Ultrastructural evidence for vesiculation of the hepatocyte canalicular membrane. J Lipid Res. 1995;36:2147–2163. [PubMed] [Google Scholar]
- 24.McConnell RE, et al. The enterocyte microvillus is a vesicle-generating organelle. J Cell Biol. 2009;185:1285–1298. doi: 10.1083/jcb.200902147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tyska MJ, et al. Myosin-1a is critical for normal brush border structure and composition. Mol Biol Cell. 2005;16:2443–2457. doi: 10.1091/mbc.E04-12-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stendahl OI, Hartwig JH, Brotschi EA, Stossel TP. Distribution of actin-binding protein and myosin in macrophages during spreading and phagocytosis. J Cell Biol. 1980;84:215–224. doi: 10.1083/jcb.84.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang FS, Wolenski JS, Cheney RE, Mooseker MS, Jay DG. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 1996;273:660–663. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 28.Tuxworth RI, et al. A role for myosin VII in dynamic cell adhesion. Curr Biol. 2001;11:318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- 29.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 30.Bensinger SJ, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alfonso-García A, Pfisterer SG, Riezman H, Ikonen E, Potma EO. D38-cholesterol as a Raman active probe for imaging intracellular cholesterol storage. J Biomed Opt. 2016;21:61003. doi: 10.1117/1.JBO.21.6.061003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Souza CM, et al. A stable yeast strain efficiently producing cholesterol instead of ergosterol is functional for tryptophan uptake, but not weak organic acid resistance. Metab Eng. 2011;13:555–569. doi: 10.1016/j.ymben.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Shivapurkar R, Souza CM, Jeannerat D, Riezman H. An efficient method for the production of isotopically enriched cholesterol for NMR. J Lipid Res. 2011;52:1062–1065. doi: 10.1194/jlr.D014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 35.Collins JL, et al. Identification of a nonsteroidal liver X receptor agonist through parallel array synthesis of tertiary amines. J Med Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 36.Gay A, Rye D, Radhakrishnan A. Switch-like responses of two cholesterol sensors do not require protein oligomerization in membranes. Biophys J. 2015;108:1459–1469. doi: 10.1016/j.bpj.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He C, Fong LG, Young SG, Jiang H. NanoSIMS imaging: An approach for visualizing and quantifying lipids in cells and tissues. J Investig Med. 2017;65:669–672. doi: 10.1136/jim-2016-000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.