Significance
DNA lesions threaten cellular life and must be repaired to maintain genome integrity. During transcription, RNA polymerases (RNAPs) actively scan DNA to find bulky lesions and trigger their repair. In growing eukaryotic cells, most transcription involves synthesis of ribosomal RNA by RNAP I (Pol I), and Pol I activity thus influences survival upon DNA damage. We determined the high-resolution electron cryomicroscopy structure of Pol I stalled by a UV-induced lesion, cyclobutane pyrimidine dimer (CPD), to unveil how the enzyme manages this important DNA damage. We found that Pol I gets stalled when the lesion reaches the bridge helix, a structural element involved in enzyme advance along DNA. We identified Pol I-specific residues around the active site that contribute to CPD-induced arrest.
Keywords: transcription, UV damage, cyclobutane pyrimidine dimers, RNA polymerase I, DNA repair
Abstract
RNA polymerase I (Pol I) transcribes ribosomal DNA (rDNA) to produce the ribosomal RNA (rRNA) precursor, which accounts for up to 60% of the total transcriptional activity in growing cells. Pol I monitors rDNA integrity and influences cell survival, but little is known about how this enzyme processes UV-induced lesions. We report the electron cryomicroscopy structure of Pol I in an elongation complex containing a cyclobutane pyrimidine dimer (CPD) at a resolution of 3.6 Å. The structure shows that the lesion induces an early translocation intermediate exhibiting unique features. The bridge helix residue Arg1015 plays a major role in CPD-induced Pol I stalling, as confirmed by mutational analysis. These results, together with biochemical data presented here, reveal the molecular mechanism of Pol I stalling by CPD lesions, which is distinct from Pol II arrest by CPD lesions. Our findings open the avenue to unravel the molecular mechanisms underlying cell endurance to lesions on rDNA.
The maintenance of genomic DNA integrity is essential for the normal function of the cell. Consequently, chemical changes in the genetic material are connected to cellular dysfunction. One of the best-known environmental threats is UV light, which can generate bulky DNA lesions that are cytotoxic (1). The main UV light-induced DNA lesions are cis-syn cyclobutane pyrimidine dimers (CPDs) and 6-4 photodimers (2). These UV light-induced photoproducts can interfere with base pairing and introduce helix distortions that obstruct fundamental cell processes such as transcription (3). The nucleotide excision repair (NER) pathway is one of the major cellular mechanisms removing this class of helix-distorting lesions from DNA (4). Transcription-coupled repair (TCR) is a NER subpathway that activates when RNA polymerases (RNAPs) are blocked by bulky DNA lesions (5).
There are three classes of eukaryotic RNAPs, and each of them transcribes a different set of genes. Pol I transcribes ribosomal DNA (rDNA) to produce the ribosomal RNA (rRNA) precursor, which is matured to yield the rRNA molecules forming the ribosome scaffold. Pol II mainly produces mRNA, and Pol III synthesizes tRNA and other short RNAs. These three eukaryotic Pols share common structural features around the active site that are crucial for the transcription process (6). In the active site, three conserved aspartate residues coordinate a magnesium ion required for catalytic activity. The bridge helix is a conserved element involved in enzyme translocation along template DNA, likely through a fully folded/partially unfolded transition that allows displacement of the protein complex toward downstream DNA (7). During elongation, the trigger loop also alters its conformation to facilitate binding of the correct nucleoside triphosphate (NTP) at the nucleotide addition site (8). Conformational rearrangements of the bridge helix and the trigger loop need to be orchestrated to enable RNA synthesis (7).
Structural and biochemical studies of CPD recognition by Pol II suggested that the enzyme stalls due to misincorporation of uracil opposite the downstream thymine in the CPD lesion (9). While this can elicit TCR, Pol II has the capacity of bypassing CPD lesions by nontemplated addition of adenine opposite the downstream thymine in the CPD lesion (10). This property of Pol II critically depends on the trigger loop and surrounding regions, as point mutations in these structural elements can increase or decrease Pol II bypass and UV-light cell resistance. While the CPD damage recognition and bypass in Pol II were elucidated, whether such mechanisms also take place in other nuclear RNAPs remains unclear. Interestingly, it was shown that UV irradiation causes severe inhibition of rRNA transcripts that is compensated for by increased Pol I initiation, suggesting that this enzyme scans rDNA until lesions are repaired, thus increasing cell survival (11). The proteins involved in Pol II TCR have been reported to also participate in the repair of damage at Pol I genes (12, 13). This suggests that differences in TCR between the two transcription systems may take place at the level of damage recognition by each RNAP.
Pol I synthesizes ∼60% of total transcripts in growing cells (14); thus, it is a key determinant for cell growth control (15). The crystal structure of yeast Pol I, comprising 14 subunits with an overall mass of about 600 kDa, revealed the enzyme architecture (16, 17). Subunits A190 and A135 are the largest and form the cleft that accommodates nucleic acids. These subunits are stabilized by the AC40/AC19 heterodimer shared with Pol III. Five additional subunits shared by all nuclear Pols, named Rpb5, Rpb6, Rpb8, Rpb10, and Rpb12, are located at the enzyme periphery. Subunit A12.2 completes the enzyme core and confers intrinsic RNA cleavage activity through its C-terminal zinc ribbon domain (A12-Ct), as truncation of A12-Ct abolishes this activity (18). Two additional heterodimers are positioned on opposite sides of the enzyme. The A49 N-terminal domain and subunit A34.5 (A49-Nt/A34.5) form a dimerization module on the enzyme lobe, while A43/A14 constitutes a stalk that is important for enzyme activation (19). Electron cryomicroscopy (cryo-EM) structures of the Pol I elongation complex (EC) with undamaged DNA scaffolds identified structural elements that are important for transcriptional activity, and also the presence of the A49 C-terminal tandem winged-helix domain (A49-Ct) next to upstream DNA (20, 21).
To understand the molecular basis of CPD lesion recognition by Pol I, we carried out structural and functional studies of the complex assembled on an RNA/DNA scaffold containing a chemical analog CPD lesion at the site i + 1/i + 2. Herein, we report the structure of Pol I EC containing a CPD lesion at a resolution of 3.6 Å. Our structural and biochemical study provides the molecular mechanism underlying persistent enzyme stalling that would subsequently trigger Pol I-dependent TCR.
Results
Pol I Stalls at CPD Lesions.
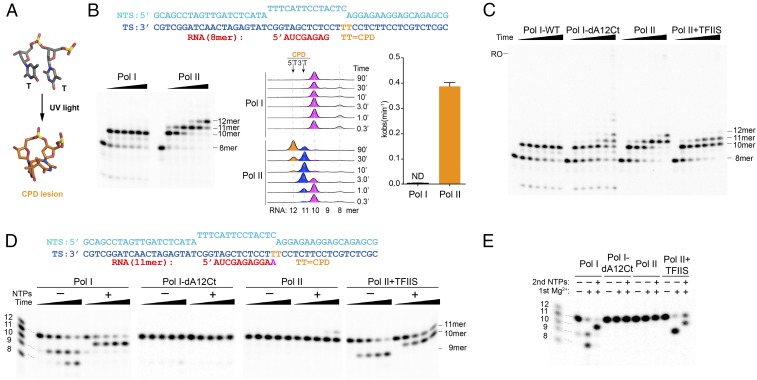
To investigate the behavior of Pol I in the presence of bulky CPD DNA lesions, we performed in vitro transcription tests using a nucleic acid scaffold containing a site-specific CPD lesion on the template strand (Fig. 1 A and B). The artificial transcription bubble includes an 8-mer RNA molecule whose 3′ end base-pairs with the template strand two nucleotides before the CPD lesion. Upon NTP addition, Pol I is able to elongate the RNA molecule from 8-mer to 10-mer and gets persistently stalled at this position, where the CPD lesion reaches the active site. No further transcript extension beyond the 10-mer is observed even after 90 min of incubation (Fig. 1B). In sharp contrast, we found that Pol II initially stalls at 10-mer but slowly extends to 11-mer and 12-mer after nucleotide incorporation opposite both thymines in the CPD lesion (Fig. 1B), in agreement with previous reports (9). We found that nucleotide addition opposite the CPD lesion, as measured by observed incorporation rate constant (kobs), by Pol II is significantly higher than that for Pol I (Fig. 1B). In particular, nucleotide addition opposite the upstream thymine (3′T) is efficiently achieved (within 3 min of incubation), while addition opposite the 5′T is much slower (90 min incubation).
Fig. 1.
CPD lesion has a distinct effect on Pol I and Pol II elongation. (A) Structure of the CPD lesion. (B) In vitro transcription assays of Pol I and Pol II on a scaffold containing a site-specific CPD lesion in the presence of 1 mM NTPs. Different RNA lengths are highlighted in different colors, with time points indicated on the graph. ND, not detectable. (C) Comparison of Pol I and Pol II processing of the CPD lesion in the presence (Pol I-wt and Pol II-TFIIS) and absence (Pol I-dA12Ct and Pol II) of strong RNA cleavage activity, using the same conditions as in B. RO indicates an expected position of the run-off product. (D) In vitro transcription assays using a scaffold with an 11-mer RNA that mimics transcript extension opposite the 3′T in the CPD lesion. The cleavage reaction was started by adding 5 mM MgCl2; incubated at room temperature; and stopped at 1 min, 3 min, 10 min, and 30 min. (E) RNAPs with cleaved RNA product remain active and can be chased by adding 1 mM NTPs, using the same scaffold as in D. The final concentration of TFIIS was 100 nM for those experiments that included TFIIS.
Since A12-Ct in Pol I confers intrinsic cleavage capacity to the enzyme, the final Pol I transcription outcome is determined by both its polymerization and intrinsic cleavage activities. In contrast, Pol II has much weaker intrinsic cleavage activity, which requires transcription factor IIS (TFIIS) to be stimulated to a comparable level as Pol I. To dissect the polymerization activity from intrinsic cleavage activity in Pol I, we purified a mutant harboring a truncation in A12-Ct (Pol I-dA12Ct) that abolishes its strong cleavage activity (SI Appendix, Fig. S1). We then compared transcription processing of a CPD lesion in the following scenarios: (i) Pol I wild type (Pol I-wt), (ii) Pol I-dA12Ct mutant, (iii) Pol II, and (iv) Pol II-TFIIS. Intriguingly, in sharp contrast to Pol I-wt, we found that cleavage-deficient Pol I-dA12Ct mutant is able to slowly incorporate nucleotides opposite CPD lesions to generate 11-mer and 12-mer in a manner similar to Pol II alone (Fig. 1C). Nevertheless, the nucleotide incorporation rate opposite the CPD lesion appears slower than that of Pol II. Interestingly, introduction of TFIIS in the Pol II system leads to a great reduction of long RNA transcript extension (11-mer) and very little 12-mer product was observed (Fig. 1C).
The results of Pol I-wt and Pol I-dA12Ct mutant suggest that for Pol I-wt, the intrinsic cleavage activity of 11-mer is faster than RNA extension over the CPD lesion (from 10-mer to 11-mer). To further test this, we assembled Pol I and Pol II ECs with a scaffold containing an RNA 11-mer (Fig. 1D), mimicking the insertion product opposite the 3′T in the CPD lesion. We found that the RNA 11-mer is readily cleaved to short transcripts (9-mer and even 8-mer at later time points) in Pol I-wt and Pol II-TFIIS systems, while it remains stable in Pol I-dA12Ct and Pol II complexes. The cleavage rates of Pol I-wt and Pol II-TFIIS (100 nM) are at comparable levels (Fig. 1D). The shortened transcripts can be chased by adding 1 mM NTP to the 10-mer in both the Pol I-wt and Pol II-TFIIS systems (Fig. 1E). We also observed that in the presence of 1 mM NTP, the RNA 11-mer can be slowly extended to 12-mer by Pol II after 30 min of incubation, but not by Pol I-dA12Ct (Fig. 1D). This suggests that Pol II is more prone to CPD lesion bypass than Pol I-dA12Ct. In systems with strong intrinsic cleavage activity (Pol I-wt or Pol II-TFIIS), the appearance of the 10-mer product is found to concur with the disappearance of 11-mer in the presence of NTP (Fig. 1D).
Taken together, our results revealed distinct behaviors between Pol I and Pol II upon CDP lesion encounter. While Pol II is able to insert additional nucleotides opposite the damage (11-mer and 12-mer), Pol I stalls right before the CPD lesion (10-mer). This difference is due to the combination of slower nucleotide incorporation opposite the CPD lesion and faster intrinsic cleavage activity in Pol I.
Structure Determination of CPD-Stalled Pol I.
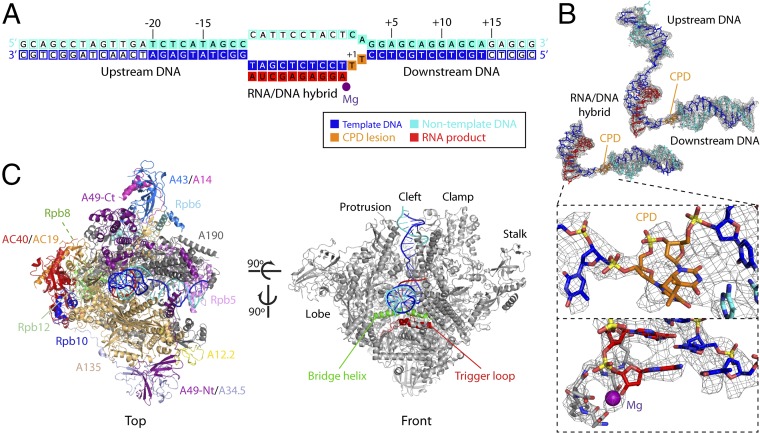
To investigate the structural basis of stable Pol I stalling at CPD lesions, we obtained the structure of Pol I in complex with a nucleic acid scaffold containing a CPD lesion located at positions i + 1/i + 2 of the template strand, where i + 1 is defined as the substrate addition site (Fig. 2A). The structure was determined by cryo-EM (SI Appendix, Fig. S2). Four independent runs of focused 3D classification were performed to isolate particles containing different areas of interest in our complex: (i) the nucleic acid scaffold within the cleft, (ii) the A49-Nt/A34.5 module, (iii) the stalk, and (iv) the upstream DNA plus A49-Ct. The first identified a subset of particles that yielded a cryo-EM map with an overall resolution of 3.6 Å, while local resolution showed detail up to 3.4 Å at the center of the enzyme and at scaffold regions close to the active site (SI Appendix, Figs. S2 and S3). This map allowed us to build an atomic model for the RNA/DNA hybrid and downstream DNA, including the CPD lesion, and for most of the Pol I enzyme (Fig. 2B and SI Appendix, Fig. S3B). Flexible regions in this map include the nontemplate strand at the mismatch, A12-Ct and the A190 DNA-mimicking loop. Regions corresponding to A49-Nt/A34.5, the stalk, A49-Ct, and upstream DNA were modeled using maps derived from focused 3D classifications around each of these regions (SI Appendix, Fig. S2C). This allowed us to complete the atomic model for the Pol I EC stalled at a CPD lesion (Fig. 2C).
Fig. 2.
Structure of CPD-stalled Pol I. (A) Schematic diagram of the CPD scaffold. Filled squares denote nucleotides with an interpretable map that were included in the model. (B) Cryo-EM maps and derived models of the scaffolds in Pol I-CPD at a resolution of 3.6 Å (Left), with zoomed-in views around the CPD lesion and the active site (Insets), and in Pol I-CPD + upstream DNA at a resolution of 4.6 Å (Right). (C) Two views of CPD-stalled Pol I, indicating the different subunits and structural elements in the enzyme.
The CPD Lesion Accommodates Above the Pol I Bridge Helix.
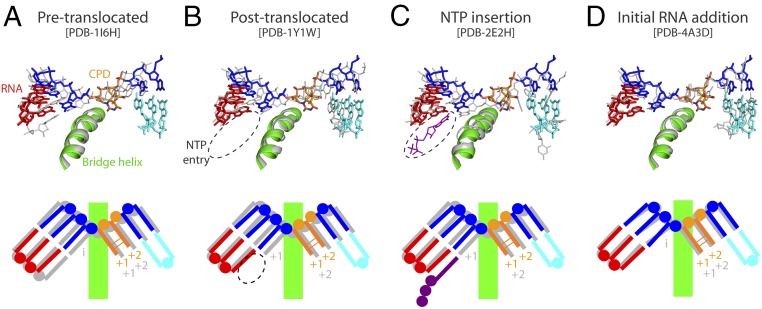
In agreement with our biochemical data, the two thymines in the CPD lesion at positions i + 1/i + 2 of the scaffold lie above the bridge helix (Fig. 3). This configuration is compatible with an intermediate in the translocation cycle, as shown by superposition with Pol II at different stages of elongation (8, 22, 23). The CPD thymines accommodate at a position that is similar to that occupied by nucleotides i + 1/i + 2 in the Pol II pretranslocated state, but they are slightly more advanced toward the active site (Fig. 3A). The base pair immediately upstream of the lesion occupies a midway location between canonical positions i and i−1 in the Pol II pretranslocated state. Consequently, the NTP entry site is partially occluded, as shown by comparison with Pol II in the posttranslocated state (Fig. 3B). Moreover, superposition with Pol II in the preinsertion stage shows that the incoming NTP would lie as close as 1.9 Å from the base at the 3′ end of RNA (Fig. 3C). A similar configuration of the RNA/DNA hybrid has been observed for Pol II initiation complexes with four- to six-nucleotide RNAs (24) and for paused bacterial RNAP (25, 26), with the RNA in the posttranslocated state and the DNA template halfway to that state. Nevertheless, template DNA in the hybrid of CPD-stalled Pol I is somewhat closer to the posttranslocated configuration than in the Pol II intermediate (Fig. 3 B and D). Overall, our results suggest that CPD-stalled Pol I presents an intermediate translocation state that is incompatible with nucleotide addition opposite the CPD lesion.
Fig. 3.
Translocation state of CPD-stalled Pol I. Superposition of CPD-stalled Pol I with Pol II in the pretranslocated (A), posttranslocated (B), and NTP insertion (C) stages of the nucleotide addition cycle, and with Pol II in an initial RNA addition intermediate (D). A schematic representation of each state, indicating the i + 1 and i + 2 positions, is shown below. PDB, Protein Data Bank.
The CPD Lesion Is Stabilized by Specific Contacts with Pol I.
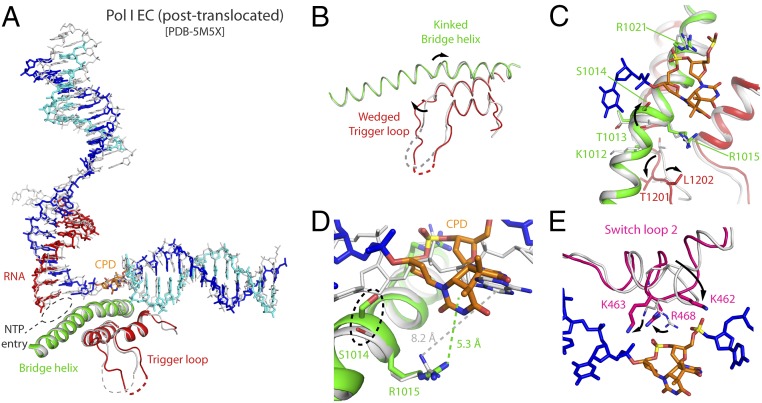
The configuration of CPD-stalled Pol I is overall similar to that found in lesion-free EC (20, 21). However, while the former represents an intermediate of translocation, the latter corresponds to the canonical posttranslocated state (Fig. 4A). In CPD-stalled Pol I, upstream DNA is minimally tilted toward A49-Ct, which is absent in the cryo-EM structure of lesion-free EC. The cleft in the CPD-stalled Pol I is closed, compatible with the recently defined conformation III of the enzyme (27). Noticeably, several Pol I structural elements inside the cleft undergo conformational changes that are likely relevant for enzyme stalling. In lesion-free EC, the bridge helix, comprising residues 992–1,028 in subunit A190, presents a fully regular helical configuration. In contrast, the bridge helix in CPD-stalled Pol I is kinked at its central region, including residues 1,009–1,015 in A190, by a maximum of 1.2 Å toward the template strand (Fig. 4 B and C). Within the kinked region, S1014 and R1015 are conserved in Pol I from yeasts to humans, while the same positions are almost invariably alanine and glutamate in Pol II and Pol III (SI Appendix, Fig. S4). Strikingly, R1015 lies at cation-π distance from the 3′T in the CPD lesion, while the distance from R1015 to the base at i + 1 in lesion-free EC is too far for such interaction (Fig. 4D). Moreover, S1014 lies at hydrogen bond distance from the template strand backbone next to the CPD lesion, while R1021 lies at hydrogen bond distance from the backbone phosphate within the thymine dimer (Fig. 4 C and D). The bridge helix kink associates with a wedged conformation in the trigger loop, where trigger loop residue T1201 lies next to bridge helix residue K1012. Additional rearrangements inside the Pol I cleft mainly affect switch loop 2 in A190, which is involved in clamp swinging in Pol II (6). In CPD-stalled Pol I, this loop approaches downstream DNA and contacts the backbone phosphate within the CPD lesion through R468 (Fig. 4E). Two additional basic residues in switch loop 2, K462 and K463, alter their configuration to approach the template strand, with K463 lying at hydrogen bond distance (Fig. 4E). In conclusion, structural rearrangements mainly affecting the central region of the bridge helix and switch loop 2 generate a network of interactions around the CPD lesion that likely contributes to enzyme stalling.
Fig. 4.
Comparison of CPD-stalled Pol I with undamaged Pol I EC. (A) Side view of the superposition with undamaged Pol I EC shown in gray. PDB, Protein Data Bank. (B) Front view where only the bridge helix and trigger loop are shown. (C) Close-up view around the bridge helix and trigger loop. (D) Close-up view around the CPD lesion, with distances between the side chain of R1015 and the closest base in the downstream template strand. (E) Close-up view around switch loop 2.
CPD-Mediated Stalling Is Different in Pol I and Pol II.
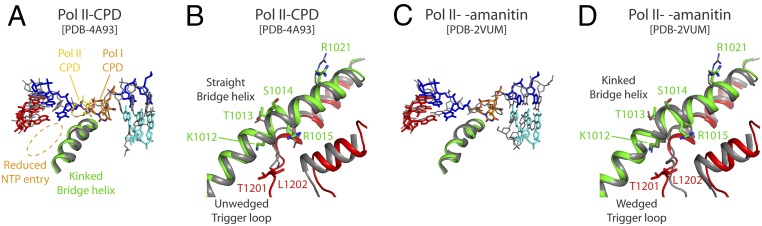
To obtain further insights into CPD-mediated Pol I stalling, we compared our structure with that of CPD-stalled Pol II (10). In the Pol II complex, the CPD thymines at positions i + 1/i + 2 are disengaged from downstream DNA but the 3′T is not able to template at the active site (Fig. 5A), thus leaving an enlarged NTP binding site that allows for nontemplated addition (i.e., A-rule) (10). In contrast, the NTP entry site in CPD-stalled Pol I is reduced by the nucleotide at the 3′ end of RNA (Fig. 5A). In agreement, the CPD lesion and the immediately upstream base pair at the i position in our structure lie about 7 Å backward, compared with the equivalent Pol II structure. This argues toward an early blockage of translocation in Pol I upon encounter of the CPD lesion in comparison to Pol II. Moreover, the bridge helix kink and the trigger loop wedge are absent in the Pol II–CPD complex (Fig. 5B). These results suggest that the mechanism of Pol I stalling at CPD lesions is caused by blockage of translocation rather than by nucleotide misincorporation opposite the lesion, as described for Pol II (9).
Fig. 5.
Comparison of CPD-stalled Pol I and Pol II. (A) Side view of the superposition with Pol II-CPD, with Pol II shown in gray and the CPD lesion shown in yellow. PDB, Protein Data Bank. (B) Close-up view where only the bridge helix and trigger loop are shown. (C) Side view of the superposition with α-amanitin–inhibited Pol II shown in gray. (D) Close-up view, where only the bridge helix and trigger loop are shown.
We further compared our CPD-stalled Pol I structure with that of Pol II inhibited by α-amanitin (28, 29). Interestingly, we found that the scaffold in α-amanitin–arrested Pol II adopts a configuration where downstream nucleotides at positions i + 1/i + 2 are located similarly as in CPD-stalled Pol I (Fig. 5C). However, the i + 1 base in α-amanitin–arrested Pol II has essentially crossed over the bridge helix and reached the canonical i + 1 template position, while the 3′T in the CPD lesion in CPD-stalled Pol I essentially lies on the downstream side of the bridge helix (Fig. 5C), which is likely due to the covalent bond between the thymines in the CPD lesion. Moreover, in the case of CPD-stalled Pol I, the RNA base pair at the i position is tilted toward the NTP site and its DNA template counterpart is significantly retarded and tilted with respect to that observed in the α-amanitin–inhibited Pol II. Furthermore, while the bridge helix is kinked in both cases, the bridge helix residues that establish contacts with template DNA in CPD-stalled Pol I are either not conserved or differently oriented in Pol II inhibited by α-amanitin (Fig. 5D). This further confirms that CPD stalls Pol I at an early stage of translocation.
Role of Pol I-Specific Bridge Helix Residues on Transcription Processing of CPD Lesions.
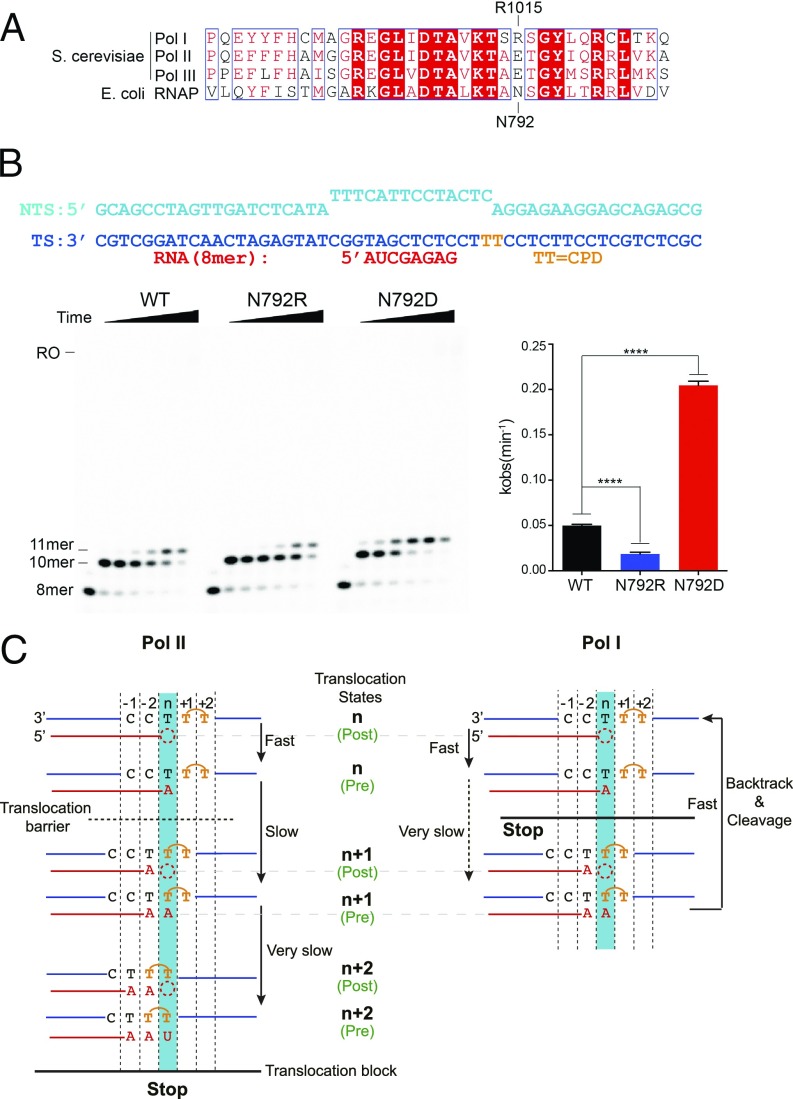
Our structure reveals that Pol I-specific bridge helix residues S1014 and R1015 may play important roles in CPD-induced Pol I stalling. We were intrigued by the fact that these residues, especially R1015, do not exist in other types of RNAPs, including bacterial RNAP, archaeal RNAP, and eukaryotic Pol II and Pol III (Fig. 6A and SI Appendix, Fig. S4). This prompted us to further investigate the role of these residues in CPD lesion processing from an evolutionary perspective. To this end, we used Escherichia coli RNAP as a model system and generated Pol I-like (N792R, A791S, and A791S/N792R) and Pol II-like (N792D) mutants in RNAP subunit β′. We found that substitution of N792 (equivalent to R1015 in Pol I) has a very strong effect, whereas substitution of A791 has almost no effect, on transcription processing over a CPD lesion. Quite remarkably, nucleotide insertion opposite the CPD lesion (11-mer) is greatly reduced for Pol I-like mutants (N792R and A791S/N792R) in comparison to a Pol II-like mutant (N792D) or wt E. coli RNAP (Fig. 6B and SI Appendix, Fig. S5). These biochemical results are fully consistent with our structural observations that R1015 in Pol I may establish a cation-π interaction with the 3′T in CPD, and therefore stabilize this lesion in the stalled position, thus reducing forward translocation. Therefore, we can recapitulate the distinct behaviors of Pol I and Pol II processing of CPD lesions using this E. coli RNAP model system by introducing single-point mutations on β′-N792 (functional equivalent of A190-R1015 in Pol I).
Fig. 6.
Mechanism of CPD lesion-induced Pol I stalling. (A) Pol I-specific residues revealed by sequence alignment of the bridge helix region for the three RNAPs in yeast and E. coli RNAP. Fully and partially conserved residues are boxed in red and white. A190-R1015 in Pol I and β′-N792 in RNAP are labeled on the top and bottom, respectively. (B, Left) Comparison between N792 mutants in the E. coli RNAP system using in vitro transcription assays. The NTP concentration is 1 mM, with time points as in Fig. 1B. (B, Right) Quantification of the 10-mer RNA extension. Data are mean and SD (n = 3). ****P < 0.0001, two-tailed Student’s t test. (C) Scheme of distinct mechanisms of CPD lesion recognition by transcribing Pol I (Right) and Pol II (Left). The translocation barrier and the translocation block are indicated with a dashed line and a solid horizontal line, respectively.
Discussion
In this work, we provide biochemical and structural evidence for the mechanism of Pol I stalling by CPD lesions, which are commonly formed on DNA as a result of UV light irradiation. We show that Pol I firmly stalls when the cross-linked thymines in the CPD lesion reach positions i + 1/i + 2. Unlike Pol II, Pol I is not able to bypass the CPD lesion even after long incubation at a high NTP concentration. We further reveal that this difference is due to the combination of slower nucleotide incorporation opposite the CPD lesion and faster intrinsic cleavage activity in Pol I in comparison to Pol II.
The structure of CPD-stalled Pol I provides further structural insights as to why Pol I is less prone to nucleotide addition opposite the CPD lesion. Upon CPD stalling, Pol I is trapped at an intermediate configuration between the pre- and posttranslocation states that is characterized by unforeseen structural features. First, the lesion locates above the bridge helix, in a position that is only slightly advanced from the pretranslocated state, with the 5′T in the lesion partially base-paired with its nontemplate strand counterpart. Second, the base pair at the i position adopts an intermediate position between the pre- and posttranslocated states, with the template base strongly tilted toward the bridge helix, which partly occludes the NTP entry site. Third, the bridge helix is kinked at its central region, where the Pol I-specific residue R1015 forms a cation-π interaction with the 3′T in the CPD lesion, while S1014 lies at hydrogen bond distance from the DNA backbone next to the lesion. We confirmed the central role of R1015 in CPD lesion recognition by mutational analysis, using bacterial RNAP as a model system. Finally, the unique position of the CPD lesion allows contacts with two conserved arginine residues in the bridge helix and switch loop 2.
Molecular dynamics has identified two translocation intermediates between the pre- and posttranslocated states in Pol II (30). In the first intermediate, the DNA/RNA hybrid has been translocated, while the base at i + 1 lies above the bridge helix. In the second intermediate, the template base at i + 1 has passed the bridge helix but not yet occupied the canonical templating position. CPD-stalled Pol I represents an early intermediate that is only slightly advanced from the pretranslocation state. As the base pair at the i position presents a unique midway configuration between the pre- and posttranslocation states, CPD stalls Pol I earlier than the first intermediate identified by molecular dynamics. While certain similarities can be observed with Pol II inhibition by α-amanitin (28, 29), our intermediate is secured by a unique set of specific interactions between the template DNA strand and the bridge helix.
According to our structural and biochemical data, the mechanism of Pol I stalling by CPD lesions significantly differs from that observed in Pol II (Fig. 6C). The latter stalls by nucleotide misincorporation opposite the 5′T in the lesion and can be slowly bypassed through untemplated addition of adenine (9, 10). In striking contrast, the same lesion blocks Pol I at a much more upstream position right before the 3′T of the CPD lesion, leading to an intermediate of translocation that is stabilized by a unique set of contacts (Fig. 6C).
CPD lesions trigger TCR both in Pol I and Pol II genes. In the latter case, it was proposed that Cockayne Syndrome Group B (CSB)-induced advance counteracts TFIIS-induced backtracking, which triggers TFIIH recruitment to extend the transcription bubble, followed by removal of the damaged DNA fragment together with the RNA transcript and Pol II (9, 31). While a similar molecular mechanism likely operates in Pol I, as shown for the human system (12, 13), the firm Pol I blockage revealed by our biochemical and structural data indicates that CPD lesions are distinctly handled by different RNAPs. This may have an impact on how cells react to DNA damage. While transcription of protein-coding genes may accept certain mutations through lesion bypass, rDNA transcription may be less prone to accept equivalent DNA modifications. In agreement, rRNA transcripts are strongly inhibited after UV irradiation, which, in turn, activates Pol I initiation to scan rDNA until lesions are repaired (11). Our results open the avenue to unravel the molecular mechanisms underlying cell endurance to lesions on rDNA.
Methods
Purification of yeast Pol I and Pol I-dA12Ct, the latter of which lacks the 47 C-terminal residues in subunit A12.2, was performed as described (32). Purification of yeast Pol II was performed as reported (31). Yeast TFIIS was expressed essentially as described (33). E. coli RNAP variants were purified as reported (34). Standard or chemically modified oligos (TriLink) were all purified by HPLC. Transcription elongation assays were performed based on reported methods with slight modifications (35). For cryo-EM studies, equimolar amounts of the CPD-containing scaffold and Pol I were mixed, deposited on grids with empty holes, and imaged on an FEI Titan Krios microscope using a K2 summit electron detector (Gatan). Cryo-EM data processing, model building, and refinement were performed following reported strategies (19), with minor modifications. Cryo-EM maps were deposited in the Electron Microscopy Database under accession codes EMD-0146 (Pol I-CPD) and EMD-0147 (Pol I-CPD + upstream DNA). The derived atomic models were deposited in the Protein Data Bank under ID codes 6H67 and 6H68, respectively.
Extended method information is included in SI Appendix.
Supplementary Material
Acknowledgments
We thank Srdja Drakulic for experimental assistance and for critically reading the manuscript. We acknowledge Daniel Clare at the Diamond Light Source for support on cryo-EM data collection, Rafael Núñez at the Centro de Investigaciones Biológicas (CIB) Electron Microsopy Facility and Carlos F. Rodríguez for help, and the Centro de Supercomputación de Galicia (CESGA) for computing resources. C.F.-T. and M.S.-M. were supported by the Spanish Ministry of Science (Grant BFU2017-87397-P) and the Ramón Areces Foundation. D.W. and J.X. were supported by NIH Grant GM102362. This study was also supported by Infrastructure for NMR, EM and X-rays for Translational Research (iNEXT) funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic models reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 6H67 and 6H68), and the cryo-EM maps have been deposited in the Electron Microscopy Database (accession codes EMD-0146 and EMD-0147).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802626115/-/DCSupplemental.
References
- 1.Mitchell JR, Hoeijmakers JH, Niedernhofer LJ. Divide and conquer: Nucleotide excision repair battles cancer and ageing. Curr Opin Cell Biol. 2003;15:232–240. doi: 10.1016/s0955-0674(03)00018-8. [DOI] [PubMed] [Google Scholar]
- 2.Ravanat JL, Douki T, Cadet J. Direct and indirect effects of UV radiation on DNA and its components. J Photochem Photobiol B. 2001;63:88–102. doi: 10.1016/s1011-1344(01)00206-8. [DOI] [PubMed] [Google Scholar]
- 3.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 4.Peyresaubes F, et al. RNA polymerase-I-dependent transcription-coupled nucleotide excision repair of UV-induced DNA lesions at transcription termination sites, in Saccharomyces cerevisiae. Photochem Photobiol. 2017;93:363–374. doi: 10.1111/php.12690. [DOI] [PubMed] [Google Scholar]
- 5.Tornaletti S, Hanawalt PC. Effect of DNA lesions on transcription elongation. Biochimie. 1999;81:139–146. doi: 10.1016/s0300-9084(99)80046-7. [DOI] [PubMed] [Google Scholar]
- 6.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 7.Cheung AC, Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: Role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brueckner F, Hennecke U, Carell T, Cramer P. CPD damage recognition by transcribing RNA polymerase II. Science. 2007;315:859–862. doi: 10.1126/science.1135400. [DOI] [PubMed] [Google Scholar]
- 10.Walmacq C, et al. Mechanism of translesion transcription by RNA polymerase II and its role in cellular resistance to DNA damage. Mol Cell. 2012;46:18–29. doi: 10.1016/j.molcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assfalg R, et al. Cellular sensitivity to UV-irradiation is mediated by RNA polymerase I transcription. PLoS One. 2017;12:e0179843. doi: 10.1371/journal.pone.0179843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iben S, et al. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 2002;109:297–306. doi: 10.1016/s0092-8674(02)00729-8. [DOI] [PubMed] [Google Scholar]
- 13.Bradsher J, et al. CSB is a component of RNA pol I transcription. Mol Cell. 2002;10:819–829. doi: 10.1016/s1097-2765(02)00678-0. [DOI] [PubMed] [Google Scholar]
- 14.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 15.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell Mol Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Tornero C, et al. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 17.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn CD, et al. Functional architecture of RNA polymerase I. Cell. 2007;131:1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 19.Torreira E, et al. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. eLife. 2017;6:e20832. doi: 10.7554/eLife.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neyer S, et al. Structure of RNA polymerase I transcribing ribosomal DNA genes. Nature. 2016;540:607–610. doi: 10.1038/nature20561. [DOI] [PubMed] [Google Scholar]
- 21.Tafur L, et al. Molecular structures of transcribing RNA polymerase I. Mol Cell. 2016;64:1135–1143. doi: 10.1016/j.molcel.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 23.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 24.Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J. 2011;30:4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, et al. Structural basis for NusA stabilized transcriptional pausing. Mol Cell. 2018;69:816–827.e4. doi: 10.1016/j.molcel.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang JY, et al. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol Cell. 2018;69:802–815.e1. doi: 10.1016/j.molcel.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernández-Tornero C. RNA polymerase I activation and hibernation: Unique mechanisms for unique genes. Transcription. January 26, 2018 doi: 10.1080/21541264.2017.1416267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva DA, et al. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc Natl Acad Sci USA. 2014;111:7665–7670. doi: 10.1073/pnas.1315751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017;551:653–657. doi: 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno-Morcillo M, et al. Solving the RNA polymerase I structural puzzle. Acta Crystallogr D Biol Crystallogr. 2014;70:2570–2582. doi: 10.1107/S1399004714015788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Awrey DE, et al. Yeast transcript elongation factor (TFIIS), structure and function. II: RNA polymerase binding, transcript cleavage, and read-through. J Biol Chem. 1998;273:22595–22605. doi: 10.1074/jbc.273.35.22595. [DOI] [PubMed] [Google Scholar]
- 34.Svetlov V, Artsimovitch I. Purification of bacterial RNA polymerase: Tools and protocols. Methods Mol Biol. 2015;1276:13–29. doi: 10.1007/978-1-4939-2392-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kellinger MW, et al. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat Struct Mol Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.