Abstract
Context: The Food and Agriculture Organization has estimated that every year considerable losses of the food crops occur due to plant diseases. Although fungicides are extensively used for management of plant diseases, they are expensive and hazardous to the environment and human health. Alternatively, biological control is the safe way to overcome the effects of plant diseases and to sustain agriculture. Since Monarda citriodora Cerv. ex Lag. (Lamiaceae/Labiatae) is known for its antifungal properties, it was chosen for the study.
Objective: The isolation of endophytic fungi from M. citriodora and assessing their biocontrol potential.
Material and methods: The isolated endophytes were characterized using ITS-5.8 S rDNA sequencing. Their biocontrol potential was assessed using different antagonistic assays against major plant pathogens.
Results: Twenty-eight endophytes representing 11 genera were isolated, of which, around 82% endophytes showed biocontrol potential against plant pathogens. MC-2 L (Fusarium oxysporum), MC-14 F (F. oxysporum), MC-22 F (F. oxysporum) and MC-25 F (F. redolens) displayed significant antagonistic activity against all the tested pathogens. Interestingly, MC-10 L (Muscodor yucatanensis) completely inhibited the growth of Sclerotinia sp., Colletotrichum capsici, Aspergillus flavus and A. fumigatus in dual culture assay, whereas MC-8 L (A. oryzae) and MC-9 L (Penicillium commune) completely inhibited the growth of the Sclerotinia sp. in fumigation assay.
Conclusions: Endophytes MC-2 L, MC-14 F, MC-22 F and MC-25 F could effectively be used to control broad range of phytopathogens, while MC-10 L, MC-8 L and MC-9 L could be used to control specific pathogens. Secondly, endophytes showing varying degrees of antagonism in different assays represented the chemo-diversity not only as promising biocontrol agents but also as a resource of defensive and bioactive metabolites.
Keywords: Phytopathogen, fumigation assay, dual culture assay, culture filtrate assay, antagonistic activity
Introduction
The Food and Agriculture Organization (FAO) has estimated that almost 25% of the world’s crops are affected by plant diseases each year (Schmale & Munkvold 2017). Traditionally, chemical fungicides are used to control the plant diseases caused by fungi, but they are expensive and hazardous for both the environment and human health. Alternatively, biological control is nowadays considered as safe option for plant disease management and to sustain agriculture (Bateman 2002). Biological control mainly relies on the use of antagonistic organisms that broadly/specifically target the pathogen. Antagonistic organisms act on pathogens by various mechanisms, which include mycoparasitism due to physical inter-hyphal interference, competition for nutrients and colonizing space, production of volatiles and nonvolatile metabolites or stimulation of host defence (Dennis & Webster 1971; Larkin et al. 1996; Benhamou et al. 2000; Walters et al. 2005; El-Hasan et al. 2009; Ting et al. 2010). To date, the genus Trichoderma Persoon (Hypocreaceae) remains the most studied and widely used economically efficient biological control agent (BCA) against various phytopathogens (Benitez et al. 2004).
Endophytes are the organisms, which colonize in the internal plant tissues without causing any apparent harm to them, and provide resistance against abiotic and/or biotic stresses (Walters et al. 2005). Taxonomically and biologically diverse endophytes have been reported from various wild as well as cultivated crops (Wilson 1995). In past, many endophytes have been evaluated for their biocontrol potential against various plant pathogens (Arnold et al. 2003; Holmes et al. 2004; Ting et al. 2010). Lahlali and Hijri (2010) reported Trichoderma atroviridae, an endophyte, as an efficient biocontrol agent for Rhizoctonia solani AG3, a causal agent of black scurf on potato.
In the last few decades, the market for microbial/endophytic inoculants with capability to be used as biological control has grown worldwide with an approximately annual growth rate of 10% (Lahlali & Hijri 2010). Thus, it is imperative to investigate biocontrol potential of fungal endophytes, which could be utilized to protect the crops from plant diseases and to increase the crop production.
Monarda citriodora Cerv. ex Lag. (Lamiaceae/Labiatae) is commonly called as lemon beebalm or lemon bergamot. It is an erect, widely distributed, medicinal, aromatic and ornamental annual herb (Bailey 1977; Duke 2007). The genus is reported to have origins in the USA, Canada and Mexico and is widely grown in Europe and Asia (Bailey 1977; Collins & Bishop 1993). It consists of roughly 30 species. In India, it has been grown in the Shivalik hills since 1990. There is no report of any fungal disease on M. citriodora. Primarily, it is used for variety of purposes such as a flavouring agent and gives delicious flavour to a variety of drinks, bakery and meat products. Due to the presence of citronellol, its leaves are pulverized and sprinkled on meat for its preservation as they repel insects. Its infusion is used to treat human ailments such as catarrh, cold, headache, gastric disorders, tooth ache, fever, flatulence, sore throat, nausea, insomnia and menstrual pain (Bee Balm c2006-2017). It is applied externally to treat skin eruption and infections. Duke (2007) reported its diaphoretic, antirheumatic, carminative, sedative, diuretic, stimulant antibacterial, anticoagulant and antiseptic properties, and its use as a tonic.
Monarda citriodora has a rich source of volatile compounds present in its essential oil (Collins et al. 1994; Dorman & Deans 2004; Zhan Guo et al. 2011). Its essential oil is reported to contain 30 constituents. Out of which, four were major ones: β-cymene 4.019%, α-phellandrene 4.815%, 1,8-cineole 23.613% and thymol 44.599% (Zhan Guo et al. 2011). However, Dorman and Deans (2004) reported terpinen-4-ol 1.2%, carvacrol 6.1%, p-cymene 10.6%, 7-octen-4-ol and thymol 70.6% as dominant components of its essential oil (95.9%). The essential oil of M. citriodora has antibacterial, antioxidant and antifungal properties. It was found to be active against various human pathogens such as Escherichia coli, Bacillus subtilis, Staphylococcus albus and many post-harvest pathogens infecting variety of crops (Collins et al. 1993; Bishop & Thornton 1997; Zhan Guo et al. 2011). Recently its essential oil was found to have anticancer property targeting PI3K pathway (Pathania et al. 2013). Thymol, an ingredient of its essential oil has antiseptic properties and is used in modern commercial mouthwash formulations. Thus, the plant has been exploited for its phytochemical and pharmacological activities.
However, the endophytic fungi associated with M. citriodora and their role in plant is still unclear and needs to be explored and exploited. The aim of the present study isolates and assesses the biocontrol potential of endophytic fungi associated with different tissues of M. citriodora against major fungal phytopathogens using three in vitro assays: dual culture, culture filtrate and fumigation assays. The results could be further exploited for the management of plant pathogens and thus reducing the losses caused by plant diseases.
Materials and methods
Isolation, identification and phylogenetic characterization of endophytes
Fully mature disease-free healthy plants of M. citriodora were collected during March-April, 2013 from the Shivalik hills of Jammu and Kashmir (32.73°N 74.87°E), India. The species was identified by taxonomist (Dr. Bikarma Singh) via leaf and flower morphology and preserved in the Janaki Ammal Herbarium (IIIM) (accession no. 18554) and in the farm of IIIM as genetic resource.
The endophytic fungi were isolated from M. citriodora (Katoch et al. 2014). Different tissues (leaves, roots and flowers) of the plants were carefully excised with a sterile scalpel. The tissues were thoroughly cleaned by washing in running tap water, followed by deionized (DI) water. Clean tissue pieces were sterilized by keeping them in a series of solution: 70% ethanol; 1.0% sodium hypochlorite (v/v); again 70% ethanol for 1 min in each solution. Finally the tissues were rinsed twice with sterile distilled water to remove extra surfactant. After surface sterilization, tissues were dried on blotting sheets and cut into small pieces of 1 cm2. These sterile tissue pieces were placed on Petri plates containing water/potato dextrose agar (WA, PDA) supplemented with streptomycin (250 μg mL−1) to inhibit the bacterial growth. The plates were incubated at 25 ± 2 °C and regularly observed for emergence of any fungal growth. The mycelium originating from the tissue in the plates was subcultured on fresh PDA plate. The endophytic fungal isolates so obtained, were coded as per their tissue origin (MC-1 L, MC-2 L, MC-3 L, etc. from leaves, MC-13 R, MC-20 R from roots and MC-7 F, MC-14 F, MC-21 F from flowers). Regular sub culturing was performed after every two months to maintain the cultures. Bits of endophytes were stored in paraffin oil at 4 °C and were deposited in RN Chopra, Microbial Repository, IIIM, Jammu.
ITS-based rDNA sequencing was used to identify the endophytes. Genomic DNA of the endophytes was extracted from the in vitro grown biomass of endophytes using the protocol described by Raeder and Broda (1985). Approximately 1 g of dried mycelia was kept in liquid nitrogen and crushed into a fine powder. It was transferred to 10 mL of extraction buffer and vortexed thoroughly. The samples were incubated in water bath set at 65 °C for 30 min with intermittent mixing. The tubes were centrifuged at 10,000 g for 5–10 min followed by extraction of aqueous layer with chloroform:isoamyl alcohol (24:1). Aqueous layer was collected and DNA was precipitated with 2.5–3 volume of absolute ethanol in the presence of 1/10th volume of sodium acetate. Tubes were inverted slowly to mix the contents and centrifuged at 8000 g for 20 min at 4 °C. So obtained white/transparent pellets were washed with ice-cold 70% ethanol followed by air drying. Dried pellets were dissolved in 20 μL of water (molecular biology grade). ITS sequences containing ITS1-5.8 S-ITS2 region spanning 500–600 bp were amplified with the universal primers ITS1 and ITS4 (White et al. 1990). PCR reaction was set up in 50 μL containing DNA (1–10 ng), 1× PCR buffer (with 15 mM MgCl2), each dNTP (200 mM), each primer (10 pmol, Sigma) and 1 U Taq DNA polymerase (Bangalore Genei, India). Cycling parameters were 5 min at 94 °C followed by 30 cycles at 94 °C for 30 s, 55 °C for 1 min, 72 °C for 1 min and a final extension for 10 min at 72 °C. The PCR product (10 μL) was resolved using agarose gel electrophoresis at 100 V. The amplified product was purified using a Gel Extraction Kit (Qiagen) and sequencing reaction was set up in a 10 μL: 40–60 ng of purified PCR product, 3.2 pmol forward/reverse primer, Big Dye Terminator sequencing mix 8 μL (v. 3.1, Applied Biosystems, US). Samples were sequenced on an automated sequencing system (Applied Biosystems). Resultant sequences (KU527781-KU527806, KU680345) were submitted to the gene bank. These sequences were blasted against the nucleotide database using BLASTn Tool of the National Centre for Biotechnology Information (NCBI), US to identify the endophytes (Altschul et al. 1997).
Biocontrol potential of endophytes
Fungal pathogens used
Antagonistic activity of all the fungal endophytes isolated from M. citriodora was evaluated against common phytopathogens namely, Aspergillus flavus (accession number MTCC 1783), F. solani (MTCC 350), Sclerotinia sp. (MTCC 7114), Colletotrichum capsici (MTCC 2071), A. fumigatus (MTCC 343), procured from The Microbial Type Culture Collection and Gene Bank (MTCC), India. The fungal cultures were revived and regularly subcultured on potato dextrose agar (PDA) under aseptic conditions as per the guidelines given by MTCC, India.
Antagonistic studies
Antagonistic potential of the endophytic fungal isolates was assessed through dual culture assay, culture filtrate assay and fumigation assay (Lahlali et al. 2007; Miles et al. 2012). All the experiments were performed in triplicate and mean values were taken.
Dual culture assay
Dual culture assay was used for comparative evaluation of the endophytes for their ability to inhibit the fungal pathogen’s growth on potato dextrose agar (PDA) (Lahlali et al. 2007). Petri plates containing 15–20 mL potato dextrose agar (PDA) were prepared. Bits of isolated endophyte and pathogen (0.5 cm each) were co-cultured at the two opposite ends of the plates. The plates were incubated at 25 ± 2 °C after sealing them with parafilm. The pathogens alone (at one end of plate) without endophyte were served as control. After 7 days, the radial growth of pathogen in the presence/absence of the fungal endophyte was measured. The percent antagonism was calculated using the formula A (%) = [(CDC-CDT)/CDC] × 100, where CDC represents the colony radial growth of pathogen (measured in mm) on the control plates i.e., in the absence of endophyte and CDT is the radial growth of pathogen in test plate i.e., in the presence of endophyte (Ezra et al. 2004; Chamberlain & Crawford 1999).
Culture filtrate assay
Each endophyte was cultured in 50 mL of potato dextrose broth (PDB) in 250 mL Erlenmeyer flasks by inoculating two plugs (0.5 cm) of actively growing endophytic fungus. The flasks were incubated for 10 days in rotatory shaker (150 rpm) at 25 ± 2 °C. The cultures were centrifuged to remove biomass so as to collect the broth containing antagonist metabolites (Dennis & Webster 1971). Endophytic culture filtrate (200 μL) was spread on PDA plates to test their antagonistic activity. On drying of filtrate, respective pathogenic fungus (0.5 cm plug) was inoculated at the centre of PDA plate. Simultaneously PDA plates containing pathogenic fungi without endophytic culture filtrate served as control. The plates were incubated at 25 ± 2 °C. After 7 days, the radial growth of pathogen in the presence/absence of endophytic culture filtrate was monitored and percent antagonism was calculated for respective culture filtrate.
Fumigation assay
The assay was conducted in PDA Petri plates, which were inoculated separately with endophytic and pathogenic cultures (0.5 cm plug from actively growing fungus). The plates were incubated at 25 ± 2 °C after sealing with parafilm. After 2–4 days, lids were removed and Petri dish inoculated with endophytic culture was inverted on the petri dish inoculated with pathogen under aseptic conditions and were tightened with double layer of parafilm. This creates no physical contact between the agar containing pathogen and endophyte. Plates were incubated at 25 ± 2 °C for 7 days. Petri dish containing PDA without any endophyte inverted on the Petri dish inoculated with pathogen served as control. Radial growth of pathogen in the presence/absence of endophyte culture was monitored and percent antagonism was calculated for each antagonist-pathogen combination.
Clustering of endophytes with biocontrol potential
For clustering the endophytes with biocontrol potential, a tree was generated using NTSYS program. Results of biocontrol potential were converted into binary form of data. Endophytes inhibiting more than 50% growth of pathogen were designated as 1, whereas those inhibiting less than 50% growth of pathogen were given as 0. Data were compiled for all the isolated endophytes against all the tested plant pathogens in three different assays. These data sets were used to generate the tree.
Results
Identification and characterization of the endophytic fungi
Twenty-eight endophytic fungi were isolated from healthy and symptomless tissues (leaves, roots and flowers) of M. citriodora to determine their biocontrol potential. Their morphology and growth characteristics on potato dextrose agar were recorded (Supplementary file Figure S1; Table S1). Their molecular identification was carried out on the basis of rDNA sequences. The ITS sequences of each endophyte were blasted against the nucleotide database of NCBI for the most homologous sequence. The endophytes were assigned as particular species only if minimum threshold similarity was ≥99% compared to the most closely related strain (Yuan et al. 2010). In the present study, only two sequences (for isolates MC-8 L and MC-20 L) showed <99% similarity to the known sequences. Details of the fungal endophytes, GenBank accession numbers and closest sequence homologs are given in Table 1.
Table 1.
Fungal endophytes isolated from various tissues of M. citriodora with their respective strain codes, Gen-Bank accession numbers and closest affiliations of the representative isolates in the GenBank according to rDNA ITS analysis.
| S No. | Endophyte | EMBL-Bankaccession number | Most closely related strain(accession number) | Maximumidentity (%) |
|---|---|---|---|---|
| 1 | MC-1L | KU527781 | C. boninense JQ676184 | 100 |
| 2 | MC-2L | KU527782 | F. chlamydosporum KP641161 | 99 |
| 3 | MC-3L | KU527783 | C. aeria KP131939 | 99 |
| 4 | MC-4L | KU527784 | A. alternata GQ121322 | 100 |
| 5 | MC-5L | KU527785 | A. flavus KM285408 | 99 |
| 6 | MC-6L | KU527786 | N. hiratsukae GQ461906 | 98 |
| 7 | MC-8La | KU680345 | A. oryzae KT964480 | 93 |
| 8 | MC-9L | KU527787 | P. commune KF938402 | 99 |
| 9 | MC-10L | KU527788 | M. yucatanensis KJ572191 | 99 |
| 10 | MC-12L | KU527789 | F. solani FJ426390 | 99 |
| 11 | MC-14L | KU527790 | F. oxysporum JX406507 | 99 |
| 12 | MC-15L | KU527791 | Aspergillus sp. GQ352493 | 99 |
| 13 | MC-16L | KU527792 | Neurospora sp. KJ676544 | 99 |
| 14 | MC-17L | KU527793 | A. waksmanii EF669934 | 99 |
| 15 | MC-18L | KU527794 | A. fumigatus KM207771 | 99 |
| 16 | MC-20La | KU680346 | Cladosporium sp. KP050606 | 89 |
| 17 | MC-24L | KU527795 | C. tenuissimum KJ589554 | 100 |
| 18 | MC-25L | KU527796 | Fusarium sp. KC007281 | 100 |
| 19 | MC-13R | KU527797 | A. carthami JF710542 | 99 |
| 20 | MC-20R | KU527798 | Cladosporium sp. JQ388271 | 99 |
| 21 | MC-7F | KU527799 | Fusarium sp. KJ567458 | 99 |
| 22 | MC-14F | KU527800 | F. oxysporum KT876658 | 99 |
| 23 | MC-17F | KU527801 | C. gloeosporioides KM520010 | 99 |
| 24 | MC-21F | KU527802 | C. cladosporioides KP900248 | 99 |
| 25 | MC-22F | KU527803 | F. oxysporum KF264963 | 100 |
| 26 | MC-23F | KU527804 | G. intermedia JQ846048 | 99 |
| 27 | MC-25F | KU527805 | F. redolens KJ540090 | 99 |
| 28 | MC-26F | KU527806 | F. oxysporum KF998987 | 99 |
Homology is less but it was supported by morphology.
The results of blast analysis revealed that the endophytic fungi belonged to the phylum Ascomycota. Out of these, 46.4% of endophytic fungi belonged to the class Sordariomycetes followed by Eurotiomycetes (28.5%), and Dothideomycetes (25%). Thus, Sordariomycetes was the most abundant class of fungi, which was represented by the orders Hypocreales, Xylariales, Glomerellales and Sordariales. They are mostly distributed in leaves and flowers. Eurotiomycetes were isolated only from leaves and represented by the order Eurotiales, whereas Dothideomycetes were isolated from all the tissues and are represented by the order Pleosporales and Capnodiales. The isolated endophytic fungi belonged to 11 different genera. The dominant fungi observed were Fusarium spp., Aspergillus spp. Cladosporium spp. In leaves, Aspergillus spp. (27%) showed the highest isolation frequency followed by Fusarium spp. (22%) and Cladosporium spp. (11%), while in flowers, Fusarium spp. (62.5%) showed the highest isolation frequency. Fusarium spp. were common in leaves and flowers, whereas Cladosporium sp. was common in leaves and roots. Endophytes isolated only from the leaves tissues were Curvularia aeria, Neosartorya hiratsukae, M. yucatanensis, Neurospora sp., C. boninense, Alternaria alternata, Penicillium commune, Fusarium spp., Aspergillus spp. and Cladosporium spp., whereas Gibberella intermedia, Fusarium spp., C. cladosporioides, and C. gloeosporioides were restricted to flowers and roots were observed to harbour only A. carthami and Cladosporium spp.
In vitro antagonistic activity of endophytes
All the endophytic strains were assayed for antagonistic activity against major phytopathogens using three different assays. The percentage antagonism (growth inhibition percentage) was calculated for each endophyte and summarized in Table 2 (dual culture assay), Table 3 (culture filtrate assay) and Table 4 (fumigation assay). Percentage antagonism varied significantly among endophytes investigated through different assays, which supported the OSMAC hypothesis (Bode et al. 2002). These endophyte–pathogen interaction studies gave an insight to understand the correlation between diversity of micro-flora associated with the plant tissues and their chemo-diversity.
Table 2.
Biocontrol potential of endophytes against plant pathogens using dual culture assay in terms of percent growth inhibition.
| S No. | Endophyte | F. solani | Sclerotinia sp. | Colletotricum capsici | A. flavus | A. fumigatus |
|---|---|---|---|---|---|---|
| 1 | MC-1-L | – | 47.6 | – | – | – |
| 2 | MC-2-L | 59.6 | 69.9 | 78.4 | 73.4 | 74 |
| 3 | MC-3-L | – | 22.8 | 53.2 | – | – |
| 4 | MC-4-L | – | 43 | – | – | 66.2 |
| 5 | MC-5-L | 39.1 | – | 73.2 | – | – |
| 6 | MC-6-L | – | – | – | – | – |
| 7 | MC-8-L | – | – | – | – | – |
| 8 | MC-9-L | – | – | – | – | – |
| 9 | MC-10-L | – | 100 | 100 | 98 | 100 |
| 10 | MC-12-L | – | – | 78.2 | – | – |
| 11 | MC-14-L | 9.5 | 17.3 | – | 40.2 | 28.2 |
| 12 | MC-15-L | 11.5 | 11.3 | – | 17.07 | 4.8 |
| 13 | MC-16-L | 15.07 | 41 | 78.1 | 29.7 | 41.5 |
| 14 | MC-17-L | – | – | - | – | – |
| 15 | MC-18-L | – | – | 78.7 | – | – |
| 16 | MC-20-L | – | 42 | 20 | – | 35 |
| 17 | MC-24-L | 26.9 | 50 | – | 3.5 | 36.8 |
| 18 | MC-25-L | 31.2 | – | – | 28.6 | 29.4 |
| 19 | MC-13-R | – | 78.9 | 51 | 62.7 | 43.1 |
| 20 | MC-20-R | 41.6 | 39.8 | – | – | – |
| 21 | MC-7-F | 5.2 | – | – | 0.61 | 44.15 |
| 22 | MC-14-F | 56.2 | 34 | 52.6 | – | – |
| 23 | MC-17-F | – | 40 | 15.09 | 62.4 | – |
| 24 | MC-21-F | 24.6 | 40 | 22 | 46.8 | 0.6 |
| 25 | MC-22-F | – | – | 59.4 | – | – |
| 26 | MC-23-F | – | 3.4 | – | – | 38.4 |
| 27 | MC-25-F | 52.3 | 64.1 | 72.3 | – | – |
| 28 | MC-26-F | 6.3 | 24.05 | 54.12 | 10.08 | 42.9 |
>50% inhibition is in bold.
Table 3.
Biocontrol potential of endophytes against plant pathogens using culture filtrate assay in terms of percent growth inhibition.
| S No. | Endophyte | F. solani | Sclerotinia sp. | C. capsici | A. flavus | A. fumigatus |
|---|---|---|---|---|---|---|
| 1 | MC-1-L | 25 | 15 | 14.28 | 10 | 35 |
| 2 | MC-2-L | 4 | 20 | 50 | 50 | 50 |
| 3 | MC-3-L | 25 | 20 | 21.42 | 15 | 30 |
| 4 | MC-4-L | 50 | 69.56 | 63.15 | 80 | 44 |
| 5 | MC-5-L | 33.33 | 65.21 | 68.42 | 65 | 44 |
| 6 | MC-6-L | 50 | 56.52 | 73.68 | 60 | 22.22 |
| 7 | MC-8-L | 50 | 60.87 | 21.05 | 50 | 22.22 |
| 8 | MC-9-L | 56.66 | 56.52 | 47.36 | 30 | 55 |
| 9 | MC-10-L | 25 | 20 | 14.28 | 15 | 40 |
| 10 | MC-12-L | 29.16 | 10 | 50 | 20 | 10 |
| 11 | MC-14-L | 66 | 75 | 28.57 | 75 | 25 |
| 12 | MC-15-L | 56.66 | 34.78 | 21.05 | 35 | 50 |
| 13 | MC-16-L | 16 | 25 | 21.42 | 55 | 20 |
| 14 | MC-17-L | 33.33 | 47.82 | 57.89 | 10 | 44 |
| 15 | MC-18-L | 56.66 | 65.21 | 5.2 | 75 | 5 |
| 16 | MC-20-L | 25 | 10 | 7.1 | 5 | 5 |
| 17 | MC-24-L | 53.33 | 43.47 | 52.63 | 55 | 22.22 |
| 18 | MC-25-L | 46.66 | 65.21 | 47.36 | 50 | 11 |
| 19 | MC-13-R | 16 | 40 | 21.42 | 90 | 20 |
| 20 | MC-20-R | 16 | 0 | 28.57 | 25 | 35 |
| 21 | MC-7-F | 29.16 | 50 | 42.85 | 50 | 40 |
| 22 | MC-14-F | 66.66 | 69.56 | 63.15 | 65 | 72.2 |
| 23 | MC-17-F | 25 | 15 | 42.85 | 60 | 10 |
| 24 | MC-21-F | 25 | 20 | 0 | 10 | 50 |
| 25 | MC-22-F | 56.66 | 65.21 | 57.89 | 65 | 61.1 |
| 26 | MC-23-F | 25 | 40 | 7.1 | 20 | 20 |
| 27 | MC-25-F | 56.66 | 60.87 | 57.89 | 60 | 55 |
| 28 | MC-26-F | 50 | 73.91 | 10.52 | 65 | 27 |
>50% inhibition is in bold.
Table 4.
Biocontrol potential of endophytes against plant pathogens using fumigation assay in terms of percent growth inhibition.
| S No. | Endophyte | F. solani | Sclerotinia sp. | C. capsici | A. flavus | A. fumigatus |
|---|---|---|---|---|---|---|
| 1 | MC-1-L | 2 | 47.91 | 33.33 | 71.05 | 71.11 |
| 2 | MC-2-L | 0 | 16.67 | 27.77 | 26.82 | 37.77 |
| 3 | MC-3-L | 33.33 | 39.58 | 27.77 | 22.22 | 39.74 |
| 4 | MC-4-L | 16 | 20.83 | 33.33 | 4.87 | 68.88 |
| 5 | MC-5-L | 23.33 | 21.05 | 0 | 0 | 48.88 |
| 6 | MC-6-L | 6 | 0 | 16.66 | 46.34 | 55.55 |
| 7 | MC-8-L | 10 | 100 | 60 | 75.61 | 66.66 |
| 8 | MC-9-L | 33.33 | 100 | 72.22 | 51.22 | 55.55 |
| 9 | MC-10-L | 40 | 0 | 83.33 | 88 | 68.88 |
| 10 | MC-12-L | 16 | 68.75 | 34.14 | 51.11 | |
| 11 | MC-14-L | 27.08 | 73.68 | 22.22 | 36.58 | 40 |
| 12 | MC-15-L | 40 | 41.67 | 33.3 | 56.09 | 33.33 |
| 13 | MC-16-L | 6 | 43.75 | 72.22 | 26.82 | 46.66 |
| 14 | MC-17-L | 0 | 52.08 | 27.77 | 0 | 33.33 |
| 15 | MC-18-L | 20.8 | 26.31 | 72.22 | 39.02 | 51.11 |
| 16 | MC-20-L | 23.33 | 31.15 | 44.44 | 7.31 | 0 |
| 17 | MC-24-L | 47.91 | 36.84 | 44.44 | 37.5 | 40 |
| 18 | MC-25-L | 36.6 | 50 | 0 | 52 | 24.44 |
| 19 | MC-13-R | 10.41 | 8.33 | 27.77 | 43.9 | 11.11 |
| 20 | MC-20-R | 0 | 35.41 | 44.44 | 21.95 | 55.55 |
| 21 | MC-7-F | 10 | 10.52 | 33.33 | 26.82 | 48.88 |
| 22 | MC-14-F | 10 | 36.84 | 38.88 | 7.3 | 40 |
| 23 | MC-17-F | 10 | 0 | 55.55 | 39.02 | 26.66 |
| 24 | MC-21-F | 3 | 58.33 | 61.11 | 39.02 | 0 |
| 25 | MC-22-F | 25 | 0 | 27.77 | 60 | 28.88 |
| 26 | MC-23-F | 0 | 35.41 | 44.44 | 25 | 33.33 |
| 27 | MC-25-F | 6 | 47.91 | 88.88 | 26.82 | 40 |
| 28 | MC-26-F | 75 | 33.33 | 16.66 | 39.02 | 22.22 |
>50% inhibition is in bold.
Growth inhibition ≥50% against one or more pathogens was recorded by 61% of the endophytes using dual culture assay followed by 71% and 82% endophytes using fumigation assay and culture filtrate assay, respectively (Figure S2). In dual culture and fumigation assay, percentage of growth inhibition by different endophytes was ranged between 50–100%, whereas in culture filtrate assay, it was ranged between 50–90%. The eight endophytes (MC-4 L, MC-12 L, MC-16 L, MC-18 L, MC-17 F, MC-22 F, MC-25 F and MC-26 F) inhibited the growth (≥50%) of one or more plant pathogens in all the three assays. The endophytes, which showed growth inhibition (≥50%) in particular assay against one or more plant pathogens were MC-3 L (Dual culture assay), MC-7 F (culture filtrate assay) and MC-1 L and MC-20 R (fumigation assay). Endophytes MC-2 L, MC-5 L, MC-24 L, MC-13 R and MC-14 F inhibited the growth (≥50%) of one or more plant pathogens in both dual culture and culture filtrate assay, whereas endophytes MC-6 L, MC-8 L, MC-9 L, MC-14 L, MC-15 L, MC-17 L, MC-25 L and MC-21 F inhibited the growth (≥50%) of one or more plant pathogens in both culture filtrate and fumigation assay. Endophyte MC-10 L inhibited the growth (≥50%) of one or more plant pathogens in both dual culture and fumigation assay.
Endophytes MC-2 L (F. chlamydosporum), MC-14 F (F. oxysporum), MC-22 F (F. oxysporum) and MC-25 F (F. redolens) displayed broad range of antagonistic activity i.e., active against all the plant pathogens tested. Interestingly, the endophytic fungal strain MC-10 L completely inhibit the growth of the Sclerotinia sp., C. capsici, A. flavus, and A. fumigatus in dual culture assay (Figure S2), whereas the endophytic fungal strain MC-8 L, and MC-9 L completely inhibited the growth of the Sclerotinia sp., in fumigation assay. In culture filtrate assay, no endophyte could completely inhibit the growth of phytopathogens.
In dual culture assay, the most susceptible pathogen was found to be C. capsici, whose growth (≥50%) was inhibited by most of the endophytic fungi MC-2 L, MC-3 L, MC-5 L, MC-10 L, MC-12 L, MC-16 L, MC-18 L, MC-13 R, MC-14 F, MC-22 F, MC-25 F and MC-26 F, whereas in culture filtrate and fumigation assays, the most susceptible pathogen was found to be A. flavus and A. fumigatus, respectively. The most resistant pathogen was found to be A. fumigatus, A. fumigatus and Fusarium sp. in dual culture, culture filtrate and fumigation assay, respectively.
Clustering analysis
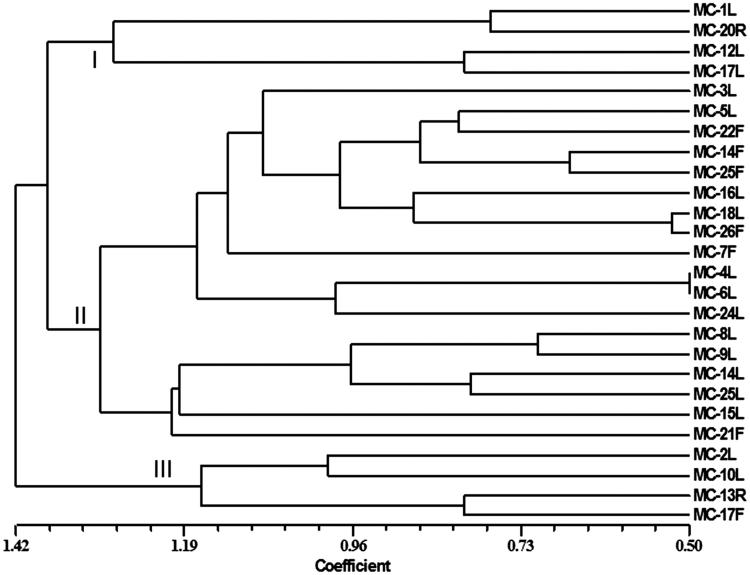
For clustering the endophytes with biocontrol potential, a tree was generated, which resulted into three major groups (Figure 1). First and third group contained four endophytes each while the second group contained rest of the endophytes.
Figure 1.
Phylogenetic tree generated using NTSYS program showing the clustering of endophytes with varying degree of antagonism.
Discussion
Endophytes have been characterized and preserved from diverse plants but still huge part of this natural treasure remains unexplored. Ecosystem plays a vital role in the process of endophytism or vice versa through host disease resistance, adaptation to unique niches, plant secondary metabolism etc. (Porras & Bayman 2011; Singh et al. 2011). Although, there is no genuine model available to study its various aspects including host specificity, geographical distribution and phylogenetic affinities, still different strategies have been adopted to study the endophytes. One of the model emphasized detailed investigation of endophytes from randomly collected plants followed by the study of ecosystem. Another model includes establishment of evolutionary relationships between plants at a particular site through morphological and molecular data, followed by characterization of endophytes from the desired plants. Pharmacologically useful plants could also be the legitimate target to explore the endophytes (Debbab et al. 2012). Many of the aromatic plants have medicinal potential but they are under studied in terms of the presence of endophytes. This present study is therefore the first report of endophytes associated with M. citriodora and their biocontrol potential.
There is no report of fungal disease infecting M. citriodora. This suggests that either plant’s defence system is quite strong or its endophytes manage to control the infection of plant pathogen. To understand the involvement of endophytes in biocontrol, their antagonistic behaviour was studied against major phytopathogens using different assay techniques. Consecutively in two assays, endophytes were grown on solid and liquid potato dextrose medium, whereas in third assay only volatiles metabolites of endophytes were evaluated against plant pathogens. In our experiments, varying growth inhibitory percentages were observed suggesting the chemo-diversity of endophytic fungi under three different assay conditions. These results were found in agreement with the concept of OSMAC in different culturing conditions; different molecules are produced by microbes (Bode et al. 2002). Kusari et al. (2013) also observed varying degree of antagonistic behaviour on five different growth media (Sabouraud agar, malt extract, Potato dextrose, water agar, nutrient agar) against two host plant pathogens. Our results are in concurrence with Paranagama et al. (2007), who reported that Chaetomium chiversii (J.C. Cooke) A. Carter (Chaetomiceae) produces the radicicol instead of chaetochromin A while changing the solid medium to liquid medium. The present study revealed that change in media, growth conditions and/or challenging it with plant pathogen might have elicited the endophytes for the production of ‘cryptic’ metabolites.
In the present study, endophytes MC-2 L (F. chlamydosporum), MC-14 F (F. oxysporum), MC-22 F (F. oxysporum) and MC-25 F (F. redolens) displayed broad range of antagonistic activity. Interestingly, the endophytic fungal strain MC-10 L completely inhibit the growth of the Sclerotinia sp., C. capsici, A. flavus and A. fumigatus in dual culture assay, whereas the endophytic fungal strain MC-8 L, and MC-9 L completely inhibited the growth of the Sclerotinia sp. Six endophytes 4 L (A. alternata), 8 L (A. oryzae), 9 L (P. commune), 10 L (M. yucatanensis), 13 R (A. carthami) and 25 F (F. redolens) of the present study showing more than 80% growth inhibition against the plant pathogenic fungi were also considered promising (Table 1).
For clustering the endophytes with biocontrol potential, a tree was generated, which resulted into three major groups. Four endophytes with lowest biocontrol potential were categorized in the first group, while another four endophytes with broad spectrum biocontrol potential were placed in the third group. The second group contained rest of the endophytes with medium range of biocontrol potential against one/two plant pathogens. It is a well-known fact that endophytes produce variety of natural products, which are basically responsible for the biocontrol potential. These grouping also indicate about the molecules active against plant pathogens. It can be speculated that group III produced highest number of bioactive molecules or molecules active against many plant pathogens, while group I produced minimum bioactive molecules or molecules with no biocontrol potential.
Endophytes typically protects the plants by various mechanisms such as production of antifungal compounds, outcompete or displace pathogens, and induces cell death in the pathogen by disrupting pathogen’s cellular membranes (Ganley et al. 2008; Shittu et al. 2009). Endophytic fungi have been source of various bioactive secondary metabolites (Gao et al. 2010). These metabolites might have involved in controlling the pathogenic incidences. Ownley et al. (2010) also observed that Beauveria spp. produces a battery of bioactive metabolites and restrict the growth of fungal plant pathogens under in vitro conditions, which was in agreement with our results. In this study, Fusarium spp., A. oryzae, P. commune, Alternaria spp. and M. yucatanensis endophytes showed promising biocontrol potential ability. These fungi produce variety of natural products (Firakova et al. 2007; Brakhage 2013). These natural products either triggers the cell death in the pathogen or lowers the metabolism of pathogens required for its further growth. Use of these bioactive ingredients as biocontrol agent would be cumbersome and costlier because it requires purification of active ingredient from the complex mixture of bio-molecules present in fermented culture of endophyte. Earlier researches also reported the inoculation of fungal endophytes to plants for mitigation of pathogenic diseases (Arnold et al. 2003; Mejia et al. 2008; Lee et al. 2009).
Thus, these endophytes could directly be used to manage plant diseases under field conditions. To understand the biocontrol potential scientifically and pin pointing the active principle, we have started suitable devised fermentation of endophytes with challenging pathogens. Concerted efforts in this direction would in turn elucidate the mechanism behind it.
Supplementary Material
Funding Statement
Project GAP1182 (BT/PR4669/PBD/17/784/2012), Department of Biotechnology, Govt. of India.
Acknowledgements
This article bears the Institutional Publication No. IIIM/1991/2016 GAP1182. We acknowledge the Council of Scientific and Industrial Research (CSIR) for proving the platform for research.
Disclosure statement
The authors declare that they have no conflict of interest.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. . 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AE, Mejia LC, Kyllo D, Rojas E, Maynard Z, Robbins N, Herre EA.. 2003. Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci USA. 100:15649–15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey LH.1977. Manual of cultivated plants. New York: MacMillan. [Google Scholar]
- Bateman R.2002. Best-bet solutions for cocoa diseases. GRO-Cocoa Newsletter.1:4–5 [Google Scholar]
- Bee Balm: c2006-2017. Available from: https://altnature.com/gallery/beebalm.htm. [Google Scholar]
- Benhamou N, Gagne S, Lequere D, Dehbi L.. 2000. Bacterial-mediated induced resistance in cucumber: beneficial effect of the endophytic bacterium Serratia plymuthica on the protection against infection by pythium ultimum. Phytopathology. 90:45–56. [DOI] [PubMed] [Google Scholar]
- Benitez T, Rincon AM, Limon MC, Codon A.. 2004. Biocontrol mechanisms of Trichoderma strains. Int Microbiol. 7:249–260. [PubMed] [Google Scholar]
- Bishop CD, Thornton IB.. 1997. Evaluation of the antifungal activity of the essential oils of Monarda citriodora var. citriodora and Melaleuca alternifolia on post harvest pathogens. J Essent Oil Res. 9:77–82. [Google Scholar]
- Bode HB, Bethe B, Hofs R, Zeeck A.. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 3:619–627. [DOI] [PubMed] [Google Scholar]
- Brakhage AA.2013. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 11:21–32. [DOI] [PubMed] [Google Scholar]
- Chamberlain K, Crawford DL.. 1999. In vitro and in vivo antagonism of pathogenic turfgrass fungi by Streptomyces hygroscopicus strains YCED9 and WYE53. J Ind Microbiol Biotechnol. 23:641–646. [DOI] [PubMed] [Google Scholar]
- Collins JE, Bishop CD.. 1993. Evaluation of Monarda citriodora var citriodora as an alternative Western European non-food oil crop. In: Proceedings of the Second European symposium on Industrial Crops and Products Pisa, Italy. (Abstract). [Google Scholar]
- Collins JE, Bishop CD, Deans SG, Svoboda KP.. 1993. Antibacterial and antioxidant properties of the volatile oil of Monarda citriodora var citriodora. In: Proceedings of the 23rd International symposium on Essential oils SAC: Auchincruive Scotland. [Google Scholar]
- Collins JE, Bishop CD, Deans SG, Svoboda KP.. 1994. Composition of the essential oil from the leaves and flowers of Monarda citriodora var citriodora grown in the United Kingdom. J Essent Oil Res. 6:27–29. [Google Scholar]
- Debbab A, Aly AH, Proksch P.. 2012. Endophytes and associated marine derived fungi-ecological and chemical perspectives. Fungal Divers. 57:45–83. [Google Scholar]
- Dennis C, Webster J.. 1971. Antagonistic properties of species-groups of Trichoderma I. Production of volatile antibiotics. Trans Br Mycol Soc. 57:25–39. [Google Scholar]
- Dorman HJD, Deans SG.. 2004. Chemical composition, antimicrobial and in vitro antioxidant properties of Monarda citriodora var. citriodora, Myristica fragrans, composition and activity of Thymus fontanesii, Origanum vulgar ssp. hirtum, Pelargonium sp. and Thymus zygis oils. J Essent Oil Res. 16:145–150. [Google Scholar]
- Duke JA.2007. Farmacia verde, Editura All. [Google Scholar]
- El-Hasan A, Walker J, Schone J, Buchenaurer H.. 2009. Detection of viridiofungin A and other antifungal metabolites excreted by Trichoderma harzanium active against different plant pathogens. Eur J Plant Pathol. 124:457–470. [Google Scholar]
- Ezra D, Hess WH, Strobel GA.. 2004. New endophytic isolates of Muscodor albus, a volatile-antibiotic-producing fungus. Microbiology (Reading, Engl.). 150:4023–4031. [DOI] [PubMed] [Google Scholar]
- Firakova S, Sturdikova M, Muckova M.. 2007. Bioactive secondary metabolites produced by microorganisms associated with plants. Biologia 62:251–257. [Google Scholar]
- Ganley RJ, Sniezko RA, Newcombe G.. 2008. Endophyte mediated resistance against white pine blister rust in Pinus monticola. For Ecol Manage. 255:2751–2760. [Google Scholar]
- Gao FK, Dai CC, Liu XZ.. 2010. Mechanisms of fungal endophytes in plant protection against pathogens. Afr J Microbiol Res. 4:1346–1351. [Google Scholar]
- Holmes KA, Schroers HJ, Thomas SE, Evans HC, Samuels GJ.. 2004. Taxonomy and biocontrol potential of a new species of Trichoderma from the Amazon basin of South America. Mycol Prog. 3:199–210. [Google Scholar]
- Katoch M, Singh G, Sharma S, Gupta N, Sangwan PL, Saxena AK.. 2014. Cytotoxic and antimicrobial activities of endophytic fungi isolated from Bacopa monnieri (L.) Pennell (Scrophulariaceae). BMC Complem Altern Med. 14:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusari P, Kusari S, Spiteller M, Kayser O.. 2013. Endophytic fungi harbored in Cannabis sativa L.: diversity and potential as biocontrol agents against host plant specific phytopathogens. Fungal Divers. 60:137–151. [Google Scholar]
- Lahlali R, Bajii M, Jijakli MH.. 2007. Isolation and evaluation of bacteria and fungi as biological control agents against Rhizoctonia solani. Commun Agric Appl Biol Sci. 72:973–982. [PubMed] [Google Scholar]
- Lahlali R, Hijri M.. 2010. Screening, identification and evaluation of potential biocontrol fungal endophytes against Rhizoctonia solani AG3 on potato plants. FEMS Microbiol Lett. 311:152–159. [DOI] [PubMed] [Google Scholar]
- Larkin RP, Hopkins D, Martin FN.. 1996. Suppression of Fusarium wilt of watermelon by nonpathogenic Fusarium oxysporum and other microorganisms recovered from a disease- suppressive soil. Phytopathology. 86:812–819. [Google Scholar]
- Lee K, Pan JJ, May G.. 2009. Endophytic Fusarium verticillioides reduces disease severity caused by Ustilago maydis on maize. FEMS Microbiol Lett. 299:31–37. [DOI] [PubMed] [Google Scholar]
- Mejia LC, Rojas EI, Maynard Z, Bael SV, Arnold AE, Hebbar P, Samuels GJ, Robbins N, Herre EA.. 2008. Endophytic fungi as biocontrol agents of Theobroma cacao pathogens. Biol Control. 46:4–14. [Google Scholar]
- Miles LA, Lopera CA, Gonzalez S, Cepero de Garcia MC, Franco AE, Restrepo S.. 2012. Exploring the biocontrol potential of fungal endophytes from an Andean Colombian Paramo ecosystem. BioControl. 57:697–710. [Google Scholar]
- Ownley BH, Gwinn KD, Vega FE.. 2010. Endophytic fungal entomopathogens with activity against plant pathogens: ecology and evolution. BioControl. 55:113–128. [Google Scholar]
- Paranagama PA, Wijeratne EMK, Gunatilaka AAL.. 2007. Uncovering biosynthetic potential of plant associated fungi: effect of culture conditions on metabolite production by Paraphaeosphaeria quadriseptata and Chaetomium chiversii. J Nat Prod. 70:1939–1945. [DOI] [PubMed] [Google Scholar]
- Pathania AS, Guru SK, Verma MK, Sharma C, Abdullah ST, Malik F, Chandra S, Katoch M, Bhushan S.. 2013. Disruption of the PI3K/AKT/mTOR signaling cascade and induction of apoptosis in HL-60 cells by an essential oil from Monarda citriodora. Food Chem Toxicol. 62:246–254. [DOI] [PubMed] [Google Scholar]
- Porras AA, Bayman P.. 2011. Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol. 49:291–235. [DOI] [PubMed] [Google Scholar]
- Raeder U, Broda P.. 1985. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1:17–20. [Google Scholar]
- Schmale IIIDG, Munkvold GP.. 2017. Mycotoxins in Crops: a threat to human and domestic animal health- economic impact. [Internet]. Available from: http://www.apsnet.org/EDCENTER/INTROPP/TOPICS/MYCOTOXINS/Pages/EconomicImpact.aspx. [Google Scholar]
- Shittu HO, Castroverde DC, Nazar RN, Robb J.. 2009. Plant-endophyte interplay protects tomato against a virulent Verticillium. Planta. 229:415–426. [DOI] [PubMed] [Google Scholar]
- Singh LP, Gill SG, Tuteja N.. 2011. Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal Behav. 6:175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting ASY, Mah SW, Tee CS.. 2010. Identification of volatile metabolites from fungal endophytes with biocontrol potential towards Fusarium oxysporum F. sp. cubense Race 4. Am J Agric Biol Sci. 5:177–182. [Google Scholar]
- Walters D, Walsh D, Newton A, Lyon G.. 2005. Induced resistance for plant disease control: maximizing the efficacy of resistance elicitors. Phytopathology. 95:1368–1373. [DOI] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J.. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis M, Gelfand D, Sninsky J, White T editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press, pp 315–322. [Google Scholar]
- Wilson D.1995. Endophytes- The evolution of a term and clarification of its use and definition. Oikos. 73:274–276. [Google Scholar]
- Yuan ZL, Zhang CL, Lin FC, Kubicek CP.. 2010. Identity, diversity, and molecular phylogeny of the endophytic mycobiota in the roots of rare wild rice (Oryza granulate) from a nature reserve in Yunnan, China. Appl Environ Microbiol. 76:1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Guo L, Xiu Hui L, Wei L.. 2011. Chemical composition of antibacterial activity of essential oil from M. citriodora flowers. Adv Mat Res. 183-185:920–923. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.