Abstract
Neurodegeneration in Alzheimer’s disease (AD) is associated with amyloid-beta peptide accumulation into insoluble amyloid plaques. The five-familial AD (5XFAD) transgenic mouse model exhibits accelerated amyloid-beta deposition, neuronal dysfunction, and cognitive impairment. We aimed to determine whether connectome properties of these mice parallel those observed in patients with AD. We obtained diffusion tensor imaging and resting-state functional magnetic resonance imaging data for four transgenic and four nontransgenic male mice. We constructed both structural and functional connectomes and measured their topological properties by applying graph theoretical analysis. We compared connectome properties between groups using both binarized and weighted networks. Transgenic mice showed higher characteristic path length in weighted structural connectomes and functional connectomes at minimum density. Normalized clustering and modularity were lower in transgenic mice across the upper densities of the structural connectome. Transgenic mice also showed lower small-worldness index in higher structural connectome densities and in weighted structural networks. Hyper-correlation of structural and functional connectivity was observed in transgenic mice compared with nontransgenic controls. These preliminary findings suggest that 5XFAD mouse connectomes may provide useful models for investigating the molecular mechanisms of AD pathogenesis and testing the effectiveness of potential treatments.
Keywords: Alzheimer’s disease, Connectome, Neuroimaging, fMRI, Diffusion tensor imaging
Author Summary
Many connectome properties have been shown to be preserved across species, providing potentially novel insights regarding the mechanisms of various disease processes. In this study, we measured functional and structural connectomes in a transgenic mouse model of Alzheimer’s disease using resting-state functional MRI and diffusion tensor imaging. We showed that connectome organization was significantly altered in transgenic mice compared with nontransgenic controls in ways that parallel what has been observed in human patients. These findings suggest that transgenic mouse connectomes may be useful for studying the etiology and treatment of Alzheimer’s disease.
INTRODUCTION
Alzheimer’s disease (AD) is the most common form of age-related neurodegeneration and dementia (Risacher & Saykin, 2013). AD pathology initiates many years before diagnosis and develops slowly in some individuals and more rapidly in others. Patients with incipient AD initially are cognitively normal, but inevitably progress to severe dementia and death. Over 46 million people have Alzheimer’s dementia globally, and the prevalence is expected to double every 20 years (Prince et al., 2015). There currently are no effective treatments for reversing AD. Risk factors include age, first-degree family history, and the apolipoprotein E (APOE) e4 genotype (Green et al., 2002; Hebert et al., 2010; Saunders et al., 1993; ten Kate et al., 2016; Wolters et al., 2017). However, the only causative factors identified to date are mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1), or presenilin 2 (PSEN2) genes. These mutations are rare but tend to be associated with aggressive, early onset disease and therefore have provided unique information regarding the pathophysiology of AD (Bateman et al., 2011).
AD is associated with significant amyloid-beta peptide accumulation, which is produced from APP by PSEN1 and PSEN2, leading to the hypothesis that it is a primary mechanism of neurodegeneration (Hardy & Selkoe, 2002; Lloret, Fuchsberger, Giraldo, & Vina, 2015). Neuroimaging studies of patients with AD demonstrate significant abnormalities in brain structure and function. These abnormalities are abundant in frontal and temporal regions, including the hippocampus and prefrontal cortex, but tend to reflect widespread disruption of large-scale, distributed networks.
In vivo functional neuroimaging of transgenic mice may yield important insights regarding the mechanisms of AD and provide preclinical models for testing the effectiveness of candidate drugs on preventing or reversing AD-related neuropathology. Previous studies of APP/PS1 and ArcBeta transgenic mice have demonstrated deficits in the functional connectivity of multiple brain regions that is associated with amyloid deposition (Bero et al., 2012; Grandjean et al., 2014; Shah et al., 2013). The five-familial AD (5XFAD) transgenic mouse model expresses three APP and two PSEN1 mutations on a (C57BL/6 x SJL)F1 background. These mice demonstrate accelerated amyloid deposition and have an early onset, aggressive disease presentation. They are particularly useful for investigating the effects of amyloid-beta deposition on neuronal loss (Eimer & Vassar, 2013).
Few if any studies have examined connectome organization in transgenic AD mice or any disease group. Connectomics models brain networks as graphs with nodes (regions) and edges (connections; Bassett & Bullmore, 2006). These mathematical models of brain networks provide measurements of information-processing efficiency and resilience to pathology, among other topological properties that are highly relevant to AD (Contreras, Goni, Risacher, Sporns, & Saykin, 2015; Dai & He, 2014; Tijms, Wink et al., 2013). Connectome properties have been shown to be preserved across species and therefore provide a unique translational bridge between preclinical and clinical studies (Gorges et al., 2017; Oh et al., 2014; Stafford et al., 2014; van den Heuvel, Bullmore, & Sporns, 2016).
Connectome studies of patients with AD have most consistently demonstrated alterations in measures of network integration and efficiency (Daianu et al., 2013; Fischer, Wolf, Scheurich, Fellgiebel, & Alzheimer’s Disease Neuroimaging Initiative, 2015; Kim et al., 2016; Lo et al., 2010; Pereira et al., 2016; Reijmer et al., 2013; Stam, Jones, Nolte, Breakspear, & Scheltens, 2007; Wang et al., 2012; Zhao et al., 2012). Additionally, studies suggest that AD pathogenesis targets high-traffic hub regions in the brain, spreading from epicenters to secondary networks as the disease progresses (Buckner et al., 2009; Dai et al., 2015; Mallio et al., 2015; Stam et al., 2009; Zhou, Gennatas, Kramer, Miller, & Seeley, 2012; Zhou & Seeley, 2014). We compared functional and structural connectomes of 5XFAD transgenic mice with those of nontransgenic controls. The aim of this pilot study was to determine whether 5XFAD mice show alterations in brain networks that parallel those observed in patients with AD, including elevated characteristic path length, reduced network efficiency, and decreased hub presence.
METHODS
Subjects
5XFAD mice were purchased from the Jackson Laboratory and maintained on the B6SJLF1/J background. Mice were maintained on a 12-hour light/dark cycle at room temperature of 75°F with unrestricted access to food and water. In total, eight male mice, 23 weeks of age, were used in this experiment. Four mice were 5XFAD transgenic and four were nontransgenic littermate controls. 5XFAD mice at 23 weeks of age have been shown to have cognitive deficits that are prior to significant neuronal or synaptic loss (Eimer & Vassar, 2013; Oakley et al., 2006). Our protocols were approved by the University of Texas MD Anderson Institutional Animal Care and Use Committee.
Neuroimaging
We obtained in vivo resting-state functional magnetic resonance imaging (rsfMRI) data from mice using a 7 Tesla Bruker BioSpec (Bruker BioSpin, Billerica, MA) scanner while mice were anesthetized with isoflurane. Isoflurane was administered at 1% (mixed with O2) to keep the respiration rate between 80 and 120 beats per minute (Stafford et al., 2014). Mice were secured into the head coil with a bite bar and the head was taped down to minimize motion. We first acquired a single-shot gradient, axial echo planar imaging (EPI) functional sequence (slice thickness = 0.5 mm, gap = 0.0 mm, repetition time [TR] = 2,000 ms, echo time [TE] = 12 ms, matrix = 80 × 64 × 32, field of view [FOV] = 20 × 16 mm, flip angle = 75°, number of volumes = 450, averages = 1, scan time = 15 min) followed by a T2-weighted, turbo spin echo, rapid acquisition with refocused echoes (Turbo RARE) sequence (slice thickness: 0.5 mm, gap = 0.0 mm, TR = 4,000 ms, TE = 40.00 ms, matrix = 256 × 180, FOV = 26.600 × 18.000 mm, flip angle = 90°, number of images = 32, scan time = 4 min and 24 s).
Six days following rsfMRI mice were euthanized using carbon dioxide and transcardially perfused with 20 ml of 10 U/ml heparin (Sagent Pharmaceuticals, Schaumburg, IL) in PBS pH 7.4 (Invitrogen, Carlsbad, CA) at room temperature followed by 20 ml 4% paraformaldehyde in PBS pH 7.4 (Electron Microcopy Sciences, Hatfield, PA) at room temperature. Following perfusion, the heads were removed and the skin, muscle, eyes, ears, nose tip, and lower jaw were removed to expose the skull. The skulls were then immersed in 20 ml of 4% paraformaldehyde in PBS pH 7.4 overnight at 4°C with continuous mixing. The skulls were then transferred to 50 ml of 0.01% sodium azide (Teknova, Hollister, CA) in PBS pH 7.4 at 4°C for seven days with continuous mixing. At the end of the seven days the skulls were transferred to 50 ml of 5 mM Magnevist (Gadopentetate Dimeglumine; Bayer Healthcare Pharmaceuticals, Indianola, PA), 0.01% sodium azide in PBS pH 7.4 at 4°C for 24 days with continuous mixing. Following the Magnevist treatment the skulls were transferred to 50 ml of 0.01% sodium azide in PBS pH 7.4 at 4°C and maintained in this solution with continuous mixing until the day of imaging, when the skulls were transferred to Fomblin Y (Sigma-Aldrich, Saint Louis, MO). We then acquired ex vivo diffusion tensor imaging (DTI) data using a 9.4 Tesla Bruker Avance BioSpec scanner (fMRI was not available on this scanner at the time of this study). The following parameters were used: spin echo, b-value = 0 and 1,000 s/mm2, 20 diffusion directions with one non–diffusion weighted image, TR = 500 ms, TE = 14.8 ms, FOV = 17 × 12.5 × 15 mm, matrix = 180 × 133 × 160, NEX = 1, δ = 3 ms, Δ = 7 ms, scan time = ∼35 hr.
A brain mask was manually delineated in 3D for the T2 and rsfMRI volumes in FMRIB Software Library (FSL) View v3.2.0 to remove the skull. RsfMRI data were preprocessed in Statistical Parametric Mapping v8 including realignment and warping of the EPI volume via the co-registered T2-weighted volume to a male C57BL/6 mouse brain template (Ma et al., 2005). CONN Toolbox v13 software was then used to filter data to the <0.1 Hz range of spontaneous activity (Raichle, 2011; Whitfield-Gabrieli & Ford, 2012). CONN implements the CompCor method to remove motion and physiologic/nonneuronal artifacts. This method involves extracting signal from white matter and cerebrospinal fluid regions using principal component analysis and then regressing these signals out of the total fMRI signal (Behzadi, Restom, Liau, & Liu, 2007). Functional time series were extracted from each of 32 bilateral cortical and subcortical gray matter regions of interest to cover the entire brain (Supplementary Figure 1; Kesler, Acton, Rao, & Ray, 2018), cross-correlated and normalized using Fisher r-to-z transformation.
DTI preprocessing was performed in FSL v5.0 (Smith et al., 2004) including eddy current correction and tensor reconstruction. Deterministic tractography was performed in TrackVis v0.6.1 (Wang, Benner, Sorensen, & Wedden, 2007) using an FA threshold of 0.1 and a curvature threshold of 40°, based on the study by Chen et al. (2015). The 32 regions of interest described above were warped into DTI native space via inverse transformation of the b0 volume to the mouse brain template. We determined the number of DTI streamlines connecting each pair of regions, and regions were considered connected if one streamline endpoint terminated within one region and the other endpoint terminated within the other region. A threshold of three streamlines was applied to minimize false-positive streamlines, and each valid edge was weighted by the average streamline fractional anisotropy (Kesler, Watson, & Blayney, 2015).
Functional and structural connectomes were constructed for each participant with N = 32 nodes, network degree of E = number of edges, and a network density of D = E/[(N × (N − 1))/2] representing the fraction of present connections to all possible connections. Negative functional edges were zeroed given evidence that properties of negative correlation networks are different than those of positive correlation networks (Hosseini & Kesler, 2013; Schwarz & McGonigle, 2011). Structural connectomes were scaled to the range of 0 to 1 (Wang, Ghumare, Vandenberghe, & Dupont, 2017).
Statistical Analysis
Connectome properties were calculated using graph theoretical analysis. Specifically, we measured characteristic path length and global/local efficiencies to test our hypothesis that these properties would be altered in transgenic mice consistent with studies of patients with AD. We additionally measured normalized clustering coefficient, small-worldness index, and modularity as these have also been reported in human studies of AD (Dai & He, 2014; Tijms, Wink et al., 2013). Connectome properties were defined as previously described (Bassett & Bullmore, 2006; Rubinov & Sporns, 2010; Sporns & Betzel, 2016). Briefly, characteristic path length is the average shortest path length between all pairs of nodes normalized by the characteristic path length of random networks. Normalized clustering coefficient is the proportion of actual connections to possible connections between a node’s neighbors normalized by the clustering coefficient of random networks. Small-worldness index is defined as normalized clustering coefficient/characteristic path length. Path length and clustering coefficient were normalized using 20 benchmark random networks (Zalesky, Fornito, & Bullmore, 2012). Global efficiency is the inverse average shortest path length of the network, while local efficiency is the inverse of the average shortest path connecting all neighbors of a node, or in other words, the average efficiency of the local subgraphs. Modularity analysis involves decomposing the network into nonoverlapping groups of regions (modules) that have maximal within-group connections and minimal between-group connections. Connectome measurement was conducted using Brain Connectivity Toolbox (Rubinov & Sporns, 2010).
Thresholding connectomes is necessary for removing false-positive edges and facilitating between-group comparisons but can remove potentially valid information regarding differences in network topology (Fornito, Zalesky, & Breakspear, 2013; van Wijk, Stam, & Daffertshofer, 2010). Further, there tends to be a large difference in network densities between rsfMRI- and DTI-derived connectomes. Therefore, we compared connectome properties across multiple densities using the area under the curve (AUC; Bassett, Meyer-Lindenberg, Achard, Duke, & Bullmore, 2006; Bassett, Nelson, Mueller, Camchong, & Lim, 2012). Specifically, we measured connectome properties at each density from minimum connection density to the last density associated with a small-world organization (Basset et al., 2008; Humphries & Gurney, 2008) up to a maximum density of 0.5 (Kaiser & Hilgetag, 2006). We then measured the AUC across this entire range as well as in a windowed manner where target windows were determined from visual inspection of the small-worldness index curves. This approach was based loosely on the clustering method introduced by Drakesmith et al. (2015). AUCs were compared between groups using nonparametric permutation testing (Basset et al., 2008) using 2,000 iterations and two-tailed p values.
We also evaluated weighted networks without any thresholding. Connectome properties from weighted networks were compared between groups using the general linear model with network density as a covariate (Brown et al., 2011). The weighted network data are provided in the Supplementary Information (Kesler et al., 2018).
To examine hub profiles, we determined whether the cumulative degree distribution of the networks followed an exponentially truncated power-law indicating the presence of hub regions (Achard, Salvador, Whitcher, Suckling, & Bullmore, 2006). This analysis was performed with weighted networks and at minimum density. Power-law fitting and comparison was conducted in the R statistical package v3.3.2 (R Foundation) using the “poweRlaw” library.
We supplemented hypothesis testing with exploratory analysis of regional effects using the Network-Based Statistic Toolbox v1.2 (Zalesky, Fornito, & Bullmore, 2010). This method identifies connected substructures, or components, within the larger network, similar to the cluster-based thresholding approach used in traditional voxel-wise neuroimaging analyses (Zalesky et al., 2010). Permutation testing with 2,000 permutations was then used to determine group differences in components controlling for multiple comparisons using family-wise error (FWE). Because the network-based statistic (NBS) can be less sensitive to focal effects, we also examined regional effects using false discovery rate (FDR; Zalesky et al., 2010). We examined NBS using both extent and intensity; the latter improves the sensitivity of NBS to focal effects (Zalesky et al., 2010).
We also explored the relationship between structure and function. First, network communication measures (e.g., search information of shortest paths, path transitivity) were computed for each pair of nodes in the structural connectivity matrix for each subject. The structural communication measures were then entered into a multiple linear regression model to generate a predicted functional connectivity matrix for each subject (Goñi et al., 2014). In other words, the communication measures for each structural node pair were used as the predictors, and the functional connectivity between that same node pair was used the response. The fitted responses from the linear regression were used to construct the predicted functional connectivity matrix. Finally, a Pearson correlation was computed between the predicted and observed functional matrices for each participant (Goñi et al., 2014). It was unknown how data collected from two different field strengths and/or rodent neurobiology would affect structure-function relationships, so we tested the default communication measures (shortest path length and search information of shortest paths; Goñi et al., 2014) as well as all available measures in the Brain Connectivity Toolbox. These included the default measures plus path transitivity, column-wise z-scored mean first passage time, neighborhood overlap, and matching index (Goñi et al., 2014). These are measures of information flow and community structure that do not require global knowledge of the network’s topology (Goñi et al., 2014; Meghanathan, 2016). Between-group difference in these correlations was measured using two-tailed t test.
RESULTS
Small-World Organization
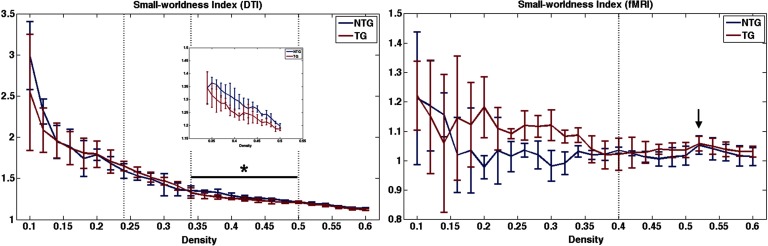
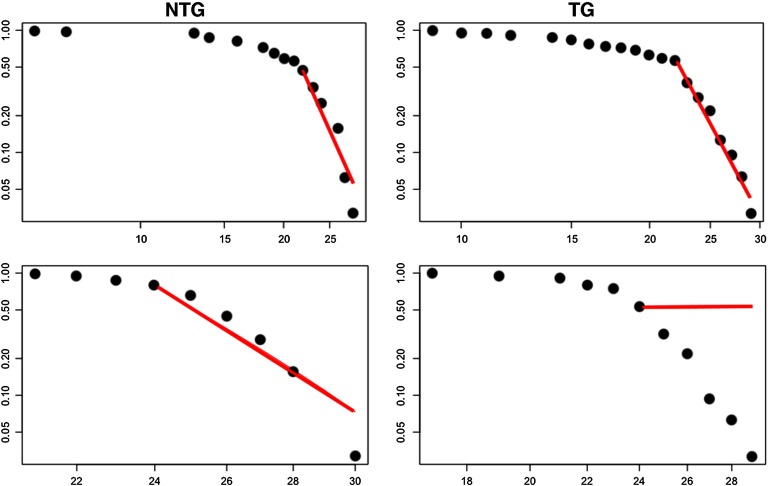
As shown in Figure 1, structural networks demonstrated expected small-world organization defined as a small-worldness index greater than 1 (Humphries & Gurney, 2008) across multiple densities. However, functional networks were small-world for all subjects at only one density (0.52), which was one step above our upper density boundary. Minimum connection density for structural networks occurred at 0.24 and at 0.4 for functional networks.
Figure 1. . Small-worldness index across network densities. For structural connectomes (DTI), vertical lines indicate area under the curve (AUC) windows. Minimum connection density was 0.24 (first dotted vertical line). At a density of 0.34 (second dotted vertical line), the group curves appear to cross over, and therefore this is where we defined the first AUC window. Maximum density was set at 0.5 (third dotted vertical line) for both modalities based on previous research. For functional connectomes (fMRI), the dotted vertical line indicates minimum connection density (0.4). The black bar and asterisk indicate the significant AUC window, and the inset figure shows the curve on a smaller scale for easier viewing. The arrow indicates the only density where all mice showed small-worldness greater than 1. NTG = nontransgenic; TG = transgenic.
AUC Across Densities
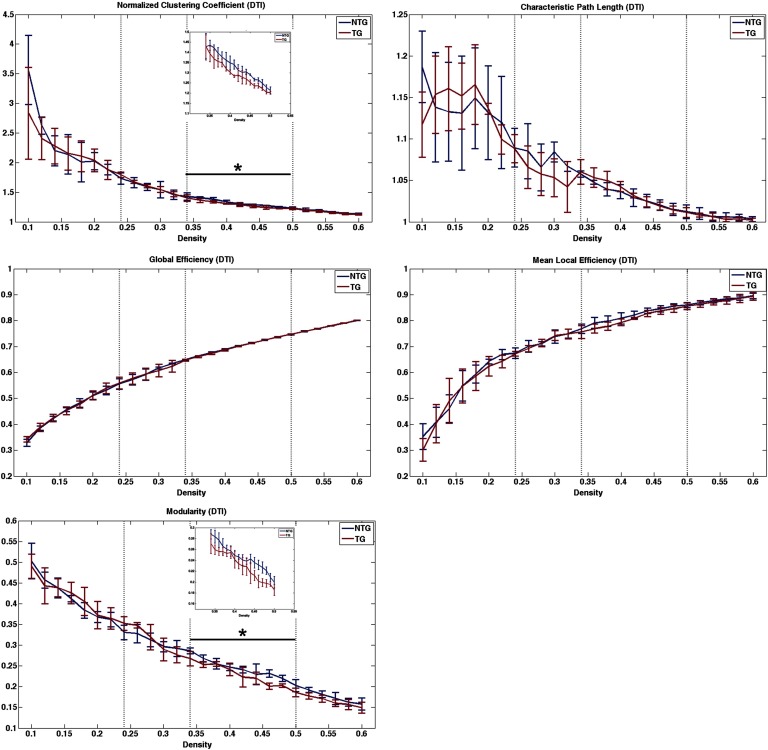
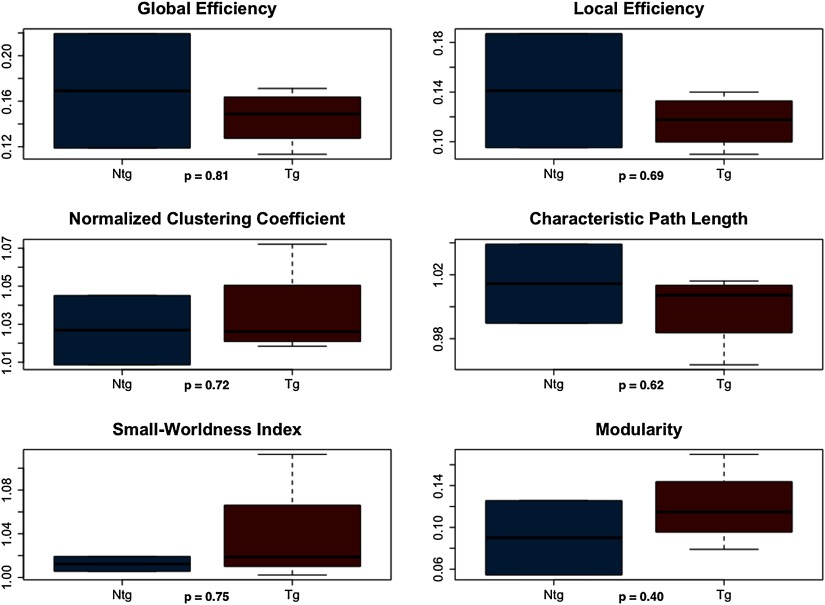
For structural connectomes, permutation testing indicated no significant differences between groups (p > 0.19, Figure 2) across the entire range of densities measured (0.24 to 0.5) or across the first density window (p > 0.17, Figure 2), which was defined from minimum density to 0.34 where the group curves crossed over. However, transgenic mice demonstrated significantly lower normalized clustering coefficient (p = 0.01), small-worldness index (p = 0.02), and modularity (p = 0.03) compared with nontransgenic mice across the second density window from 0.34 to maximum density (Figures 1 and 2). Module regions are presented in Table 1.
Figure 2. . Structural connectome properties. Dotted vertical lines indicate area under the curve (AUC) windows. The black bar and asterisk indicate the significant AUC window, and the inset figure shows the curve on a smaller scale for easier viewing. NTG = nontransgenic; TG = transgenic.
Table 1. . Module regions.
| Nontransgenic | |||
|---|---|---|---|
| Module 1 | Module 2 | Module 3 | Module 4 |
| Right amygdala | Left external capsule | Left amygdala | Right brainstem |
| Right striatum | Right external capsule | Left brainstem | Right central gray |
| Right cerebellum | Left hypothalamus | Left striatum | Right hypothalamus |
| Right pallidum | Left superior colliculi | Left central gray | Right midbrain |
| Right hippocampus | Right superior colliculi | Left cerebellum | |
| Right inferior colliculi | Left pallidum | ||
| Right neocortex | Left hippocampus | ||
| Left olfactory | Left inferior colliculi | ||
| Right olfactory | Left neocortex | ||
| Right basal forebrain/septum | Left midbrain | ||
| Right thalamus | Left basal forebrain/septum | ||
| Left thalamus | |||
| Transgenic | |||
| Module 1 | Module 2 | Module 3 | |
| Left external capsule | Right amygdala | Left amygdala | |
| Right external capsule | Right brainstem | Left brainstem | |
| Left hypothalamus | Right striatum | Left striatum | |
| Right hypothalamus | Right central gray | Left central gray | |
| Left superior colliculi | Right cerebellum | Left cerebellum | |
| Right superior colliculi | Right pallidum | Left pallidum | |
| Right hippocampus | Left hippocampus | ||
| Right inferior colliculi | Left inferior colliculi | ||
| Right neocortex | Left neocortex | ||
| Right olfactory | Left olfactory | ||
| Right midbrain | Left midbrain | ||
| Right basal forebrain/septum | Left basal forebrain/septum | ||
| Right thalamus | Left thalamus | ||
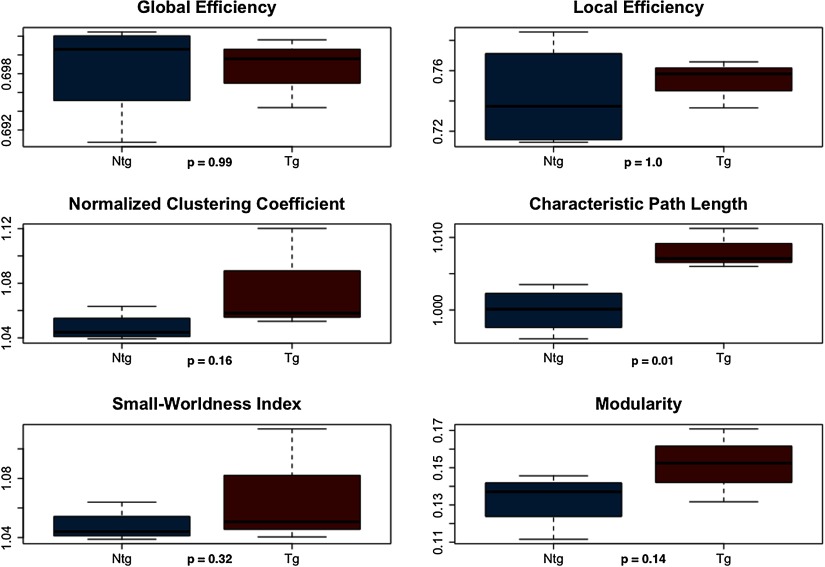
Given the above small-worldness results, we did not compare the AUCs of functional connectomes between groups. It was not possible to simply exclude data since the lack of small-worldness affected different mice at different densities. However, at minimum connection density, connectomes of all subjects but one in the transgenic group showed small-world organization, and therefore we compared connectome metrics at this specific density after excluding the transgenic subject. T test indicated significantly higher characteristic path length in transgenic mice (t = 3.64 p = 0.01, Figure 3).
Figure 3. . Functional connectome properties at minimum connection density. NTG = nontransgenic; TG = transgenic.
Weighted Networks
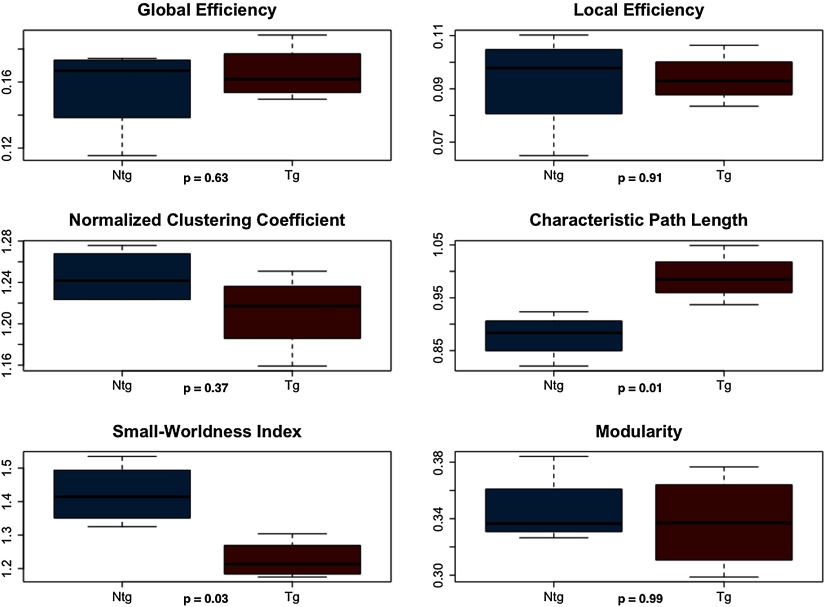
All weighted structural networks demonstrated small-world organization. As shown in Figure 4, general linear models covaried for density indicated that structural connectomes of transgenic mice showed significantly higher characteristic path length (F = 15.2, p = 0.01) and lower small-worldness index (F = 9.73, p = 0.03) compared with controls.
Figure 4. . Weighted structural connectome properties. NTG = nontransgenic; TG = transgenic.
Weighted functional connectomes for two nontransgenic mice did not demonstrate small-worldness, so these were excluded. There were no significant group differences (Figure 5).
Figure 5. . Weighted functional connectome properties. NTG = nontransgenic; TG = transgenic.
Hubs: Degree Distribution
Both groups showed goodness of fit with the power-law with no between-group difference (p = 0.68). There was no significant group difference in power-law fit for either modality at minimum density (p > 0.343). For weighted functional networks, the transgenic group showed poor fit with the power-law (Figure 6), and this was significantly lower than that of the control group (p < 0.001).
Figure 6. . Power-law fit of cumulative degree distributions. Power-law fit is shown as a plot of log degree (x-axis) by log cumulative degree distribution (y-axis). Left column = nontransgenic (NTG), right column = transgenic (TG), top row = weighted structural connectomes, bottom row = weighted functional connectomes.
Regional Connectivity
There were no significant regional effects for structural or functional connectomes weighted or at minimum density via NBS or FDR comparison.
Relationship Between Structure and Function
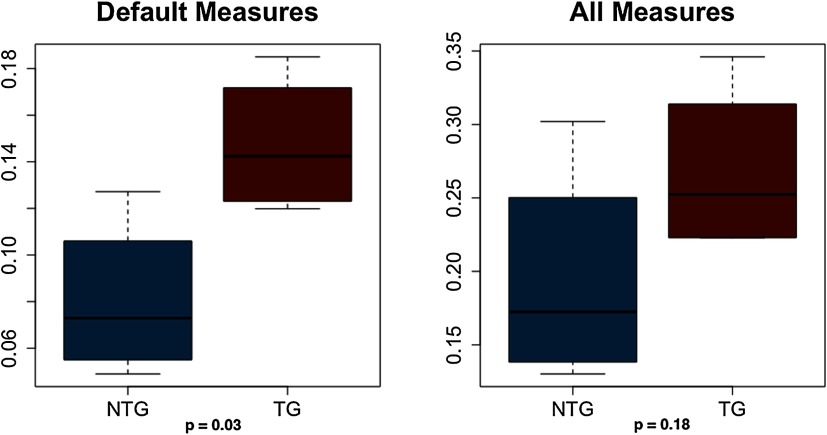
This analysis was conducted only on weighted networks without excluding any subjects. Using default communication measures resulted in significant correlations between structure and function for five out of eight subjects (p < 0.008). Using all communication measures resulted in significant correlations for all subjects (p < 0.004). T tests indicated that correlation coefficients based on the default model were significantly higher in the transgenic group (t = 2.92, p = 0.03), but there was no difference in the coefficients from the all-measures model (t = 1.53, p = 0.18, Figure 7).
Figure 7. . Correlation coefficients for structural and functional connectivity. Default measures = shortest path length, search information of shortest path length; all measures = default measures plus path transitivity, column-wise z-scored mean first passage time, neighborhood overlap, and matching index. NTG = nontransgenic; TG = transgenic.
DISCUSSION
To our knowledge, this is the first study to evaluate connectome organization in an AD mouse model. Few if any studies have compared connectomes in rodent models of disease groups. Using in vivo resting-state fMRI and ex vivo DTI, we constructed and measured functional and structural connectomes for 5XFAD transgenic mice. This mouse model is characterized by aggressive amyloid pathology. We evaluated connectome properties in these mice across multiple network densities (i.e., thresholds) and also for weighted, unthresholded networks and compared them with the connectomes of nontransgenic control mice.
Weighted DTI-based structural networks demonstrated significantly higher path length and lower small-worldness index in transgenic mice after controlling for network density. Weighted functional networks also demonstrated higher characteristic path length at minimum connection density. Clinical studies of patients with AD have demonstrated higher characteristic path lengths of DTI- or fMRI-based connectomes (Lo et al., 2010; Wang et al., 2012; Zhao et al., 2012). Increased characteristic path length has also been observed in patients with AD using gray matter structural connectomes (Kim et al., 2016; Pereira et al., 2016) and electroencephalography (EEG)-based networks (Stam et al., 2007). In a small-world network, clustering coefficient is greater than that of random networks, while the path length is comparable to that of random networks (Bassett & Bullmore, 2006; Humphries & Gurney, 2008; Watts & Strogatz, 1998). Characteristic path length is defined as the average shortest path length between all pairs of network nodes divided by the mean path length of benchmark random networks (Bassett & Bullmore, 2006; Humphries & Gurney, 2008; Watts & Strogatz, 1998; Zalesky et al., 2012). Therefore, higher characteristic path length suggests disconnection within the network such that longer, less efficient routes of information exchange must be taken. It also suggests that the network is less random in terms of path length.
We also observed lower normalized clustering coefficient, small-worldness index, and modularity, indicating that the connectomes of transgenic mice were more similar to random networks in these properties. Lower clustering coefficient suggests lower network segregation or specialization, while lower small-worldness index reflects the overall similarity to random networks in that segregation and integration (i.e., path length) are imbalanced (Bassett & Bullmore, 2006; Watts & Strogatz, 1998). Previous DTI and fMRI connectome studies of patients with AD have also demonstrated lower normalized clustering, small-worldness, and modularity (Brier et al., 2014; Sun et al., 2014; Supekar, Menon, Rubin, Musen, & Greicius, 2008).
However, some studies have observed conflicting results, including lower characteristic path length in DTI (Daianu et al., 2013), gray matter (Tijms, Moller et al., 2013), fMRI (Sanz-Arigita et al., 2010), EEG (de Haan et al., 2009), and magneto-encephalogram (MEG; Stam et al., 2009) connectomes in patients with AD. Inconsistencies in connectome findings are a well-known issue in the literature (for reviews, see Dai & He, 2014; Tijms, Wink et al., 2013) and often reflect differences in methodology such as imaging modality and/or choice of thresholding method. This was part of our rationale for a multimodal study including different thresholding methods. Our most consistent findings included higher characteristic path length in transgenic mice, which was noted in weighted, unthresholded DTI connectomes, and fMRI connectomes thresholded to minimum connection density. We also observed lower small-worldness index in transgenic mice in DTI connectomes thresholded to higher densities and in weighted, unthresholded DTI graphs. Small-worldness index is the ratio of clustering coefficient to path length (Bassett & Bullmore, 2006; Watts & Strogatz, 1998) and therefore, lower values can reflect lower normalized clustering and/or higher characteristic path length.
Clinical studies have also noted decreased network efficiency in patients with AD (Daianu et al., 2013; Fischer et al., 2015; Lo et al., 2010; Reijmer et al., 2013), which we did not observe. Efficiency and path length are related measures (Achard & Bullmore, 2007). Since this was a preliminary study, we may have lacked power to detect differences in efficiencies. The 5XFAD mouse model used in this study is associated with accelerated amyloid-beta pathology. Both amyloid-beta and tau are believed to synergistically drive neurodegenerative processes involved in AD (Bloom, 2014; Lloret et al., 2015; Stancu, Vasconcelos, Terwel, & Dewachter, 2014). 5XFAD do not have significant tau pathology. It is possible that greater tau burden and/or some other AD-related neuropathology is more associated with impairments in connectome efficiency. We evaluated the 5XFAD mice at an age prior to significant neuronal and synaptic loss, which may have preserved network efficiency.
Lower cerebrospinal fluid (CSF) levels of amyloid-beta have been associated with higher path length and lower clustering of the gray matter structural connectome in human participants; low levels of amyloid-beta in the CSF indicate higher amyloid plaque burden in the brain (Tijms et al., 2015). Gray matter and DTI-based connectome properties show moderate convergence (Gong, He, Chen, & Evans, 2012). Accordingly, our findings provide indirect support for amyloid-beta effects on connectome properties of the 5XFAD transgenic mouse connectome. We did not have molecular assays available for analysis in this study. Future AD transgenic mouse connectome studies could provide unique insights regarding the molecular mechanisms underlying impairments in various connectome properties associated with AD. For example, Golgi staining and longitudinal fluorescent microscopy could be used to examine neuronal morphology and survival rates in impaired connectome regions. Evaluating the role of mitochondrial dysfunction (Lin & Beal, 2006) via Seahorse flux technology (Brand & Nicholls, 2011) is another potential application.
Previous research has demonstrated lower connectivity among regions involved in specific brain networks of patients with AD, including the default mode network (Bai et al., 2011; Damoiseaux, Prater, Miller, & Greicius, 2012; Lehmann et al., 2013; Simic, Babic, Borovecki, & Hof, 2014). Relevant subnetworks have been shown to be present in mice both structurally and functionally (Liska, Galbusera, Schwarz, & Gozzi, 2015; Stafford et al., 2014), but we did not find any significant regional connectome differences between transgenic mice and controls. This may again reflect limited statistical power and/or may indicate diffuse regional effects. Modularity was lower at higher densities in structural connectomes of transgenic mice, suggesting fewer dissociable networks. Transgenic mice appeared to lack separation between certain sensorimotor/homeostatic regions and other networks.
Both functional and structural connectome topologies showed the expected goodness of fit with a power-law distribution. The power-law fit is believed to reflect the brain network’s hub organization wherein the majority of information processing is handled by a small number of core regions (Achard et al., 2006). This is consistent with other studies showing presence of hubs in the mouse brain (Liska et al., 2015; Rubinov, Ypma, Watson, & Bullmore, 2015). There was no difference in power-law fit between the groups for structural connectomes, but transgenic mice showed a significantly poorer power-law fit in weighted functional connectomes. Previous studies have suggested that AD pathogenesis may selectively target certain hub regions (Dai et al., 2015; Stam et al., 2009; Xie & He, 2011; Yao et al., 2010; Zhou et al., 2012).
Despite problematic functional connectomes that did not show adequate small-world characteristics, functional connectivity was predicted from structural connectivity. Transgenic mice tended to show hyper-correlation of structural and functional networks compared with controls. Such hyper-correlation has been noted in patients with neurologic disorders (Kesler et al., 2017; Rudie et al., 2012; Wirsich et al., 2016). However, Sun et al. (2014) observed lower structure-function coupling in connectomes of patients with AD. Few studies have examined both structural and functional connectomes in AD, and therefore further investigation regarding the relationship between structure and function is required.
There are several limitations to consider for this preliminary, pilot study. It is unclear why functional connectomes failed to demonstrate expected small-worldness. This could be the results of anesthesia, which has been shown to attenuate intrinsic functional networks (Boveroux et al., 2010; Peltier et al., 2005). The effects of anesthesia on connectome organization are currently unknown. MRI field strength could play a role, although a previous study was also conducted at 7 Tesla (Liska et al., 2015). Another drawback is that fMRI and DTI were acquired at different MRI field strengths. This study is also limited by the small sample, which may have reduced our power to detect certain effects. However, we used a conservative statistical approach, including permutation analysis with a large number of permutations and correction for multiple comparisons where appropriate. Currently there is no standard regarding the parcellation scheme for connectome analyses, and therefore a different approach may yield alternate results. DTI-based connectomes have particular limitations, as they have been shown to correspond poorly with neuron tracer data (Calabrese, Badea, Cofer, Qi, & Johnson, 2015). However, as noted above, DTI connectome properties have been shown to differentiate patient groups and therefore seem to provide valuable insights regarding the effects of AD on brain networks.
In conclusion, we demonstrated preliminary evidence that connectome properties of 5XFAD transgenic mice show some correspondence with results observed in patients with AD. There were several innovative aspects of this study, including connectome measurement in an AD mouse model, multimodal connectome measurement, and the use of different network thresholding methods. Future studies in mice could allow us to better understand the molecular mechanisms underlying connectome disruption in AD. These models could also aid in drug discovery and preclinical trials for AD by providing outcome measurements of connectome organization.
ACKNOWLEDGMENTS
The authors would like to thank the faculty and staff of the MD Anderson Small Animal Imaging Facility as well as Robia Pautler, PhD, and others at the Baylor College of Medicine Small Animal MRI.
AUTHOR CONTRIBUTIONS
Shelli R. Kesler: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Software; Supervision; Validation; Visualization; Writing – original draft; Writing – review & editing. Paul Acton: Data curation; Methodology; Resources; Writing – review & editing. Vikram Rao: Formal analysis; Methodology; Writing – review & editing. William J. Ray: Conceptualization; Data curation; Funding acquisition; Investigation; Methodology; Project administration; Resources; Supervision; Validation; Visualization; Writing – review & editing.
FUNDING INFORMATION
This research was funded by the Neurodegeneration Consortium, the MD Anderson Foundation, and the National Institutes of Health (1R03CA191559, 1R01NR014195, 1R01CA172145:SK). The sponsors had no role in the design, implementation, analysis, or interpretation of the study.
TECHNICAL TERMS
- Apolipoprotein E (APOE):
A gene that encodes a protein important for the metabolism and transport of fats.
- Amyloid precursor protein (APP):
A protein believed to be involved in neural development and degeneration.
- Presenilin 1 (PSEN1):
A gene that encodes a protein involved in processing APP.
- Presenilin 2 (PSEN2):
A gene that encodes another protein involved in processing APP.
- 5XFAD transgenic mouse model:
A mouse whose DNA has been altered to express five genes that are known to be associated with Alzheimer’s disease.
- Connectome:
The brain network.
- Global efficiency:
A measure of network efficiency of information exchange based on path lengths between regions.
- Small-world:
A network organization or topology associated with high local connectivity and economical large-range connectivity; that is, a balance between segregation and integration.
- Modularity:
A measure of how well a network can be decomposed into nonoverlapping subnetworks or modules.
Contributor Information
Shelli R. Kesler, Department of Neuro-oncology, University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Paul Acton, Neurodegeneration Consortium, Institute for Applied Cancer Science, University of Texas MD Anderson Cancer Center, Houston, TX, USA.
Vikram Rao, Department of Neuro-oncology, University of Texas MD Anderson Cancer Center, Houston, TX, USA.
William J. Ray, Neurodegeneration Consortium, Institute for Applied Cancer Science, University of Texas MD Anderson Cancer Center, Houston, TX, USA
REFERENCES
- Achard S., & Bullmore E. (2007). Efficiency and cost of economical brain functional networks. PLoS Computational Biology, 3(2), e17 10.1371/journal.pcbi.0030017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard S., Salvador R., Whitcher B., Suckling J., & Bullmore E. (2006). A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. Journal of Neuroscience, 26(1), 63–72. 10.1523/JNEUROSCI.3874-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F., Watson D. R., Shi Y., Wang Y., Yue C., YuhuanTeng, … Zhang Z. (2011). Specifically progressive deficits of brain functional marker in amnestic type mild cognitive impairment. PLoS ONE, 6(9), e24271 10.1371/journal.pone.0024271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett D. S., & Bullmore E. (2006). Small-world brain networks. The Neuroscientist, 12(6), 512–523. 10.1177/1073858406293182 [DOI] [PubMed] [Google Scholar]
- Bassett D. S., Bullmore E., Verchinski B. A., Mattay V. S., Weinberger D. R., & Meyer-Lindenberg A. (2008). Hierarchical organization of human cortical networks in health and schizophrenia. Journal of Neuroscience, 28(37), 9239–9248. 28/37/9239 10.1523/JNEUROSCI.1929-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett D. S., Meyer-Lindenberg A., Achard S., Duke T., & Bullmore E. (2006). Adaptive reconfiguration of fractal small-world human brain functional networks. Proceedings of the National Academy of Sciences, 103(51), 19518–219523. 10.1073/pnas.0606005103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett D. S., Nelson B. G., Mueller B. A., Camchong J., & Lim K. O. (2012). Altered resting state complexity in schizophrenia. NeuroImage, 59(3), 2196–2207. 10.1016/j.neuroimage.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman R. J., Aisen P. S., De Strooper B., Fox N. C., Lemere C. A., Ringman J. M., … Xiong C. (2011). Autosomal-dominant Alzheimer’s disease: A review and proposal for the prevention of Alzheimer’s disease. Alzheimer’s Research and Therapy, 3(1), 1–1. 10.1186/alzrt59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y., Restom K., Liau J., & Liu T. T. (2007). A component based noise correction method (compcor) for BOLD and perfusion based fMRI. NeuroImage, 37(1), 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bero A. W., Bauer A. Q., Stewart F. R., White B. R., Cirrito J. R., Raichle M. E., … Holtzman D. M. (2012). Bidirectional relationship between functional connectivity and amyloid-beta deposition in mouse brain. Journal of Neuroscience, 32(13), 4334–4340. 10.1523/JNEUROSCI.5845-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom G. S. (2014). Amyloid-beta and tau: The trigger and bullet in Alzheimer disease pathogenesis. JAMA Neurology, 71(4), 505–508. 10.1001/jamaneurol.2013.5847 [DOI] [PubMed] [Google Scholar]
- Boveroux P., Vanhaudenhuyse A., Bruno M.-A., Noirhomme Q., Lauwick S., Luxen A., … Phillips C. (2010). Breakdown of within- and between-network resting state functional magnetic resonance imaging connectivity during propofol-induced loss of consciousness. Journal of the American Society of Anesthesiologists, 113(5), 1038–1053. [DOI] [PubMed] [Google Scholar]
- Brand Martin D., & Nicholls David G. (2011). Assessing mitochondrial dysfunction in cells. Biochemical Journal, 435(Pt. 2), 297–312. 10.1042/BJ20110162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier M. R., Thomas J. B., Fagan A. M., Hassenstab J., Holtzman D. M., Benzinger T. L., … Ances B. M. (2014). Functional connectivity and graph theory in preclinical Alzheimer’s disease. Neurobiology of Aging, 35(4), 757–768 . 10.1016/j.neurobiolaging.2013.10.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. A., Terashima K. H., Burggren A. C., Ercoli L. M., Miller K. J., Small G. W., & Bookheimer S. Y. (2011). Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proceedings of the National Academy of Sciences, 108(51), 20760–20765. 10.1073/pnas.1109038108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner R. L., Sepulcre J., Talukdar T., Krienen F. M., Liu H., Hedden T., … Johnson K. A. (2009). Cortical hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer’s disease. Journal of Neuroscience, 29(6), 1860–1873. 10.1523/JNEUROSCI.5062-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E., Badea A., Cofer G., Qi Y., & Johnson G. A. (2015). A diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cerebral Cortex, 25(11), 4628–4637. 10.1093/cercor/bhv121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Liu T., Zhao Y., Zhang T., Li Y., Li M., … Liu T. (2015). Optimization of large-scale mouse brain connectome via joint evaluation of DTI and neuron tracing data. NeuroImage, 115, 202–213. 10.1016/j.neuroimage.2015.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J. A., Goni J., Risacher S. L., Sporns O., & Saykin A. J. (2015). The structural and functional connectome and prediction of risk for cognitive impairment in older adults. Current Behavioral Neuroscience Reports, 2(4), 234–245. 10.1007/s40473-015-0056-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., & He Y. (2014). Disrupted structural and functional brain connectomes in mild cognitive impairment and Alzheimer’s disease. Neuroscience Bulletin, 30(2), 217–232. 10.1007/s12264-013-1421-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Yan C., Li K., Wang Z., Wang J., Cao M., … He Y. (2015). Identifying and mapping connectivity patterns of brain network hubs in Alzheimer’s disease. Cerebral Cortex, 25(10), 3723–3742. 10.1093/cercor/bhu246 [DOI] [PubMed] [Google Scholar]
- Daianu M., Jahanshad N., Nir T. M., Toga A. W., Jack C. R. Jr., Weiner M. W., & Thompson P. M., for the Alzheimer’s Disease Neuroimaging Initiative. (2013). Breakdown of brain connectivity between normal aging and Alzheimer’s disease: A structural k-core network analysis. Brain Connectivity, 3(4), 407–422. 10.1089/brain.2012.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux J. S., Prater K. E., Miller B. L., & Greicius M. D. (2012). Functional connectivity tracks clinical deterioration in Alzheimer’s disease. Neurobiology of Aging, 33(4), 828.e19–828.e30. 10.1016/j.neurobiolaging.2011.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan W., Pijnenburg Y. A. L., Strijers R. L. M., van der Made Y., van der Flier W. M., Scheltens P., & Stam C. J. (2009). Functional neural network analysis in frontotemporal dementia and Alzheimer’s disease using EEG and graph theory. BMC Neuroscience, 10, 101 10.1186/1471-2202-10-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakesmith M., Caeyenberghs K., Dutt A., Lewis G., David A. S., & Jones D. K. (2015). Overcoming the effects of false positives and threshold bias in graph theoretical analyses of neuroimaging data. NeuroImage, 118, 313–333. 10.1016/j.neuroimage.2015.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimer W. A., & Vassar R. (2013). Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Aβ42 accumulation and Caspase-3 activation. Molecular Neurodegeneration, 8(1), 2 10.1186/1750-1326-8-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F. U., Wolf D., Scheurich A., Fellgiebel A., & Alzheimer’s Disease Neuroimaging Initiative. (2015). Altered whole-brain white matter networks in preclinical Alzheimer’s disease. NeuroImage: Clinical, 8, 660–666. 10.1016/j.nicl.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A., Zalesky A., & Breakspear M. (2013). Graph analysis of the human connectome: Promise, progress, and pitfalls. NeuroImage, 80, 426–444. 10.1016/j.neuroimage.2013.04.087 [DOI] [PubMed] [Google Scholar]
- Gong G., He Y., Chen Z. J., & Evans A. C. (2012). Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. NeuroImage, 59(2), 1239–1248. 10.1016/j.neuroimage.2011.08.017 [DOI] [PubMed] [Google Scholar]
- Goñi J., van den Heuvel M. P., Avena-Koenigsberger A., Velez de Mendizabal N., Betzel R. F., Griffa A., … Sporns O.(2014). Resting-brain functional connectivity predicted by analytic measures of network communication. Proceedings of the National Academy of Sciences, 111(2), 833–838. 10.1073/pnas.1315529111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorges M., Roselli F., Müller H.-P., Ludolph A. C., Rasche V., & Kassubek J. (2017). Functional connectivity mapping in the animal model: Principles and applications of resting-state fMRI. Frontiers in Neurology, 8(200). 10.3389/fneur.2017.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean J., Schroeter A., He P., Tanadini M., Keist R., Krstic D., … Rudin M. (2014). Early alterations in functional connectivity and white matter structure in a transgenic mouse model of cerebral amyloidosis. Journal of Neuroscience, 34(41), 13780–13789. 10.1523/JNEUROSCI.4762-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. C., Cupples L., Go R., Benke K. S., Edeki T., Griffith P. A., … Farrer L. A. (2002). Risk of dementia among White and African American relatives of patients with Alzheimer disease. JAMA, 287(3), 329–336. 10.1001/jama.287.3.329 [DOI] [PubMed] [Google Scholar]
- Hardy J., & Selkoe D. J. (2002). The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science, 297(5580), 353–356. 10.1126/science.1072994 [DOI] [PubMed] [Google Scholar]
- Hebert L. E., Bienias J. L., Aggarwal N. T., Wilson R. S., Bennett D. A., Shah R. C., & Evans D. A. (2010). Change in risk of Alzheimer disease over time. Neurology, 75(9), 786–791. 10.1212/WNL.0b013e3181f0754f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S. M., & Kesler S. R. (2013). Comparing connectivity pattern and small-world organization between structural correlation and resting-state networks in healthy adults. NeuroImage, 78, 402–414. 10.1016/j.neuroimage.2013.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries M. D., & Gurney K. (2008). Network “small-world-ness”: A quantitative method for determining canonical network equivalence. PLoS One, 3(4), e0002051 10.1371/journal.pone.0002051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M., & Hilgetag C. C. (2006). Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Computational Biology, 2(7), e95 10.1371/journal.pcbi.0020095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler S. R., Acton P., Rao V., & Ray W. J. (2018). Supplemental material for “Functional and structural connectome properties in the 5XFAD transgenic mouse model of Alzheimer’s disease.” Network Neuroscience, 2(2), 241–258. 10.1162/netn_a_00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler S. R., Adams M., Packer M., Rao V., Henneghan A. M., Blayney D. W., & Palesh O. (2017). Disrupted brain network functional dynamics and hyper-correlation of structural and functional connectome topology in patients with breast cancer prior to treatment. Brain and Behavior, 7(3), e00643 . 10.1002/brb3.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesler S. R., Watson C. L., & Blayney D. W. (2015). Brain network alterations and vulnerability to simulated neurodegeneration in breast cancer. Neurobiology of Aging, 36(8), 2429–2442. 10.1016/j.neurobiolaging.2015.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Shin J. H., Han C. E., Kim H. J., Na D. L., Seo S. W., … Alzheimer’s Disease Neuroimaging Initiative. (2016). Using individualized brain network for analyzing structural covariance of the cerebral cortex in Alzheimer’s patients. Frontiers in Neuroscience, 10, 394 10.3389/fnins.2016.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., Madison C. M., Ghosh P. M., Seeley W. W., Mormino E., Greicius M. D., … Rabinovici G. D. (2013). Intrinsic connectivity networks in healthy subjects explain clinical variability in Alzheimer’s disease. Proceedings of the National Academy of Sciences, 110(28), 11606–11611. 10.1073/pnas.1221536110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. T., & Beal M. F. (2006). Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature, 443(7113), 787–795. 10.1038/nature05292 [DOI] [PubMed] [Google Scholar]
- Liska A., Galbusera A., Schwarz A. J., & Gozzi A. (2015). Functional connectivity hubs of the mouse brain. NeuroImage. 10.1016/j.neuroimage.2015.04.033 [DOI] [PubMed] [Google Scholar]
- Lloret A., Fuchsberger T., Giraldo E., & Vina J. (2015). Molecular mechanisms linking amyloid beta toxicity and Tau hyperphosphorylation in Alzheimer’s disease. Free Radical Biology and Medicine, 83, 186–191. 10.1016/j.freeradbiomed.2015.02.028 [DOI] [PubMed] [Google Scholar]
- Lo C. Y., Wang P. N., Chou K. H., Wang J., He Y., & Lin C. P. (2010). Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer’s disease. Journal of Neuroscience, 30(50), 16876–16885. 10.1523/JNEUROSCI.4136-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Hof P. R., Grant S. C., Blackband S. J., Bennett R., Slatest L., … Benveniste H. (2005). A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience, 135(4), 1203–1215. 10.1016/j.neuroscience.2005.07.014 [DOI] [PubMed] [Google Scholar]
- Mallio C. A., Schmidt R., de Reus M. A., Vernieri F., Quintiliani L., Curcio G., … van den Heuvel M. P. (2015). Epicentral disruption of structural connectivity in Alzheimer’s disease. CNS Neuroscience and Therapeutics, 21(10), 837–8845. 10.1111/cns.12397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meghanathan N. (2016). A greedy algorithm for neighborhood overlap-based community detection. Algorithms, 9(1). 10.3390/a9010008 [DOI] [Google Scholar]
- Oakley H., Cole S. L., Logan S., Maus E., Shao P., Craft J., … Vassar R. (2006). Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. Journal of Neuroscience, 26(40), 10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. W., Harris J. A., Ng L., Winslow B., Cain N., Mihalas S., … Zeng H. (2014). A mesoscale connectome of the mouse brain. Nature, 508(7495), 207–214. 10.1038/nature13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier S. J., Kerssens C., Hamann S. B., Sebel P. S., Byas-Smith M., & Hu X. (2005). Functional connectivity changes with concentration of sevoflurane anesthesia. NeuroReport, 16( 3), 285–288. [DOI] [PubMed] [Google Scholar]
- Pereira J. B., Mijalkov M., Kakaei E., Mecocci P., Vellas B., Tsolaki M., … Westman E. (2016). Disrupted network topology in patients with stable and progressive mild cognitive impairment and Alzheimer’s disease. Cerebral Cortex, 26(8), 3476–3493. 10.1093/cercor/bhw128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince M., Wimo A., Guerchet M., Ali G.-C., Wu Y.-T., & Prina M. (2015). World Alzheimer report 2015: The global impact of dementia. Retrieved from http://www.worldalzreport2015.org/ [Google Scholar]
- Raichle M. E. (2011). The restless brain. Brain Connectivity, 1(1), 3–12. 10.1089/brain.2011.0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmer Y. D., Leemans A., Caeyenberghs K., Heringa S. M., Koek H. L., & Biessels G. J. (2013). Disruption of cerebral networks and cognitive impairment in Alzheimer disease. Neurology, 80(15), 1370–1377. 10.1212/WNL.0b013e31828c2ee5 [DOI] [PubMed] [Google Scholar]
- Risacher S. L., & Saykin A. J. (2013). Neuroimaging and other biomarkers for Alzheimer’s disease: The changing landscape of early detection. Annual Review of Clinical Psychology, 9, 621–648. 10.1146/annurev-clinpsy-050212-185535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M., & Sporns O. (2010). Complex network measures of brain connectivity: Uses and interpretations. NeuroImage, 52(3), 1059–1069. 10.1016/j.neuroimage.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Rubinov M., Ypma R. J., Watson C., & Bullmore E. T. (2015). Wiring cost and topological participation of the mouse brain connectome. Proceedings of the National Academy of Sciences, 112(32), 10032–10037. 10.1073/pnas.1420315112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudie J. D., Brown J. A., Beck-Pancer D., Hernandez L. M., Dennis E. L., Thompson P. M., … Dapretto M. (2012). Altered functional and structural brain network organization in autism. NeuroImage: Clinical, 2, 79–94. 10.1016/j.nicl.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Arigita E. J., Schoonheim M. M., Damoiseaux J. S., Rombouts S. A., Maris E., Barkhof F., … Stam C. J. (2010). Loss of “small-world” networks in Alzheimer’s disease: Graph analysis of fMRI resting-state functional connectivity. PLoS One, 5(11), e13788 10.1371/journal.pone.0013788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A. M., Strittmatter W. J., Schmechel D., St. George-Hyslop P. H., Pericak-Vance M. A., Joo S. H., … Roses A. D. (1993). Association of apolipoprotein E allele ε 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology, 43(8), 1467–1472. [DOI] [PubMed] [Google Scholar]
- Schwarz A. J., & McGonigle J. (2011). Negative edges and soft thresholding in complex network analysis of resting state functional connectivity data. NeuroImage, 55(3), 1132–1146. 10.1016/j.neuroimage.2010.12.047 [DOI] [PubMed] [Google Scholar]
- Shah D., Jonckers E., Praet J., Vanhoutte G., Delgado y Palacios R., Bigot C., et al. (2013). Resting state fMRI reveals diminished functional connectivity in a mouse model of amyloidosis. PLoS ONE, 8(12), e84241 10.1371/journal.pone.0084241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic G., Babic M., Borovecki F., & Hof P. R. (2014). Early failure of the default-mode network and the pathogenesis of Alzheimer’s disease. CNS Neuroscience and Therapeutics, 20(7), 692–7698. 10.1111/cns.12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. M., Jenkinson M., Woolrich M. W., Beckmann C. F., Behrens T. E., Johansen-Berg H., … Matthews P. M. (2004). Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 23(Suppl. 1), S208–S219. 10.1016/j.neuroimage.2004.07.051 [DOI] [PubMed] [Google Scholar]
- Sporns O., & Betzel R. F. (2016). Modular brain networks. Annual Review of Psychology, 67, 613–640. 10.1146/annurev-psych-122414-033634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford J. M., Jarrett B. R., Miranda-Dominguez O., Mills B. D., Cain N., Mihalas S., … Fair D. A. (2014). Large-scale topology and the default mode network in the mouse connectome. Proceedings of the National Academy of Sciences, 111(52), 18745–18750. 10.1073/pnas.1404346111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam C. J., de Haan W., Daffertshofer A., Jones B. F., Manshanden I., van Cappellen van Walsum A. M., … Scheltens P. (2009). Graph theoretical analysis of magnetoencephalographic functional connectivity in Alzheimer’s disease. Brain, 132(Pt. 1), 213–224. 10.1093/brain/awn262 [DOI] [PubMed] [Google Scholar]
- Stam C. J., Jones B. F., Nolte G., Breakspear M., & Scheltens P. (2007). Small-world networks and functional connectivity in Alzheimer’s disease. Cerebral Cortex, 17(1), 92–99. 10.1093/cercor/bhj127 [DOI] [PubMed] [Google Scholar]
- Stancu I.-C., Vasconcelos B., Terwel D., & Dewachter I. (2014). Models of β-amyloid induced Tau-pathology: The long and “folded” road to understand the mechanism. Molecular Neurodegeneration, 9(1), 51 10.1186/1750-1326-9-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Yin Q., Fang R., Yan X., Wang Y., Bezerianos A., … Sun J. (2014). Disrupted functional brain connectivity and its association to structural connectivity in amnestic mild cognitive impairment and Alzheimer’s disease. PLoS One, 9(5), e96505 10.1371/journal.pone.0096505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K., Menon V., Rubin D., Musen M., & Greicius M. D. (2008). Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Computational Biology, 4(6), e1000100 10.1371/journal.pcbi.1000100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Kate M., Sanz-Arigita E. J., Tijms B. M., Wink A. M., Clerigue M., Garcia-Sebastian M., … Villanua J. (2016). Impact of APOE-ε4 and family history of dementia on gray matter atrophy in cognitively healthy middle-aged adults. Neurobiology of Aging, 38, 14–20. [DOI] [PubMed] [Google Scholar]
- Tijms B. M., Kate M. T., Wink A. M., Visser P. J., Ecay M., Clerigue M., … Barkhof F. (2015). Gray matter network disruptions and amyloid beta in cognitively normal adults. Neurobiology of Aging. 10.1016/j.neurobiolaging.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Tijms B. M., Moller C., Vrenken H., Wink A. M., de Haan W., van der Flier W. M., … Barkhof F. (2013). Single-subject grey matter graphs in Alzheimer’s disease. PLoS One, 8(3), e58921 10.1371/journal.pone.0058921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijms B. M., Wink A. M., de Haan W., van der Flier W. M., Stam C. J., Scheltens P., & Barkhof F. (2013). Alzheimer’s disease: Connecting findings from graph theoretical studies of brain networks. Neurobiology of Aging, 34(8), 2023–2036. 10.1016/j.neurobiolaging.2013.02.020 [DOI] [PubMed] [Google Scholar]
- van den Heuvel M. P., Bullmore E. T., & Sporns O. (2016). Comparative connectomics. Trends in Cognitive Sciences, 20(5), 345–361. 10.1016/j.tics.2016.03.001 [DOI] [PubMed] [Google Scholar]
- van Wijk B. C. M., Stam C. J., & Daffertshofer A. (2010). Comparing brain networks of different size and connectivity density using graph theory. PLoS ONE, 5(10), e13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zuo X., Dai Z., Xia M., Zhao Z., Zhao X., et al. (2012). Disrupted functional brain connectome in individuals at risk for Alzheimer’s disease. Biological Psychiatry. 10.1016/j.biopsych.2012.03.026 [DOI] [PubMed] [Google Scholar]
- Wang R., Benner T., Sorensen A. G., & Wedden V. J. (2007). Diffusion Toolkit: A software package for diffusion imaging data processing and tractography. Proceedings of the International Society for Magnetic Resonance in Medicine, 15, 3720. [Google Scholar]
- Wang Y., Ghumare E., Vandenberghe R., & Dupont P. (2017). Comparison of different generalizations of clustering coefficient and local efficiency for weighted undirected graphs. Neural Computation, 29(2), 313–331. 10.1162/NECO_a_00914 [DOI] [PubMed] [Google Scholar]
- Watts D. J., & Strogatz S. H. (1998). Collective dynamics of “small-world” networks. Nature, 393(6684), 440–442. 10.1038/30918 [DOI] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S., & Ford J. M. (2012). Default mode network activity and connectivity in psychopathology. Annual Review of Clinical Psychology, 8, 49–76. 10.1146/annurev-clinpsy-032511-143049 [DOI] [PubMed] [Google Scholar]
- Wirsich J., Perry A., Ridley B., Proix T., Golos M., Benar C., … Guye M. (2016). Whole-brain analytic measures of network communication reveal increased structure-function correlation in right temporal lobe epilepsy. NeuroImage: Clinical, 11, 707–718. 10.1016/j.nicl.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters F. J., van der Lee S. J., Koudstaal P. J., van Duijn C. M., Hofman A., Ikram M. K., … Ikram M. A. (2017). Parental family history of dementia in relation to subclinical brain disease and dementia risk. Neurology, 88(17), 1642–1649. [DOI] [PubMed] [Google Scholar]
- Xie T., & He Y. (2011). Mapping the Alzheimer’s brain with connectomics. Frontiers in Psychiatry, 2, 77 10.3389/fpsyt.2011.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Zhang Y., Lin L., Zhou Y., Xu C., & Jiang T. (2010). Abnormal cortical networks in mild cognitive impairment and Alzheimer’s disease. PLoS Computational Biology, 6(11), e1001006 10.1371/journal.pcbi.1001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A., Fornito A., & Bullmore E. (2012). On the use of correlation as a measure of network connectivity. NeuroImage, 60(4), 2096–2106. 10.1016/j.neuroimage.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Zalesky A., Fornito A., & Bullmore E. T. (2010). Network-based statistic: Identifying differences in brain networks. NeuroImage, 53(4), 1197–1207. 10.1016/j.neuroimage.2010.06.041 [DOI] [PubMed] [Google Scholar]
- Zhao X., Liu Y., Wang X., Liu B., Xi Q., Guo Q., … Wang P. (2012). Disrupted small-world brain networks in moderate Alzheimer’s disease: A resting-state fMRI study. PLoS One, 7(3), e33540 10.1371/journal.pone.0033540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Gennatas E. D., Kramer J. H., Miller B. L., & Seeley W. W. (2012). Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron, 73(6), 1216–1227. 10.1016/j.neuron.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., & Seeley W. W. (2014). Network dysfunction in Alzheimer’s disease and frontotemporal dementia: Implications for psychiatry. Biological Psychiatry, 75(7), 565–573. 10.1016/j.biopsych.2014.01.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.