Abstract
Quantitative neuroanatomical data are important for the study of many areas of neuroscience, and the complexity of problems associated with neuronal structure requires that research from multiple groups across many disciplines be combined. However, existing neuron-tracing systems, simulation environments, and tools for the visualization and analysis of neuronal morphology data use a variety of data formats, making it difficult to exchange data in a readily usable way. The NeuroML project was initiated to address these issues, and here we describe an extensible markup language standard, MorphML, which defines a common data format for neuronal morphology data and associated metadata to facilitate data and model exchange, database creation, model publication, and data archiving. We describe the elements of the standard in detail and outline the mappings between this format and those used by a number of popular applications for reconstruction, simulation, and visualization of neuronal morphology.
Keywords: Neuronal morphology, Neuronal digitization, MorphML, NeuroML, Neuroanatomical data, Neuroscience standards, Neurolucida
1. Introduction
Many computational models in neuroscience seek to elucidate how complex processes in the nervous system interact to produce emergent, higher-level behavior. To combine research from multiple groups, there must be an infrastructure for sharing data and exchanging model specifications (Gardner et al. 2003; Martone et al. 2004); however, the current use of multiple formats for encoding model information has hampered model exchange. The Neural Open Markup Language project (NeuroML) (http://www.neuroml.org) is an international, collaborative initiative that seeks to address this issue by encouraging the use of declarative specifications for models in neuroscience and by producing extensible markup language (XML) schemas for encoding information on the essential elements used by neuroscience applications. Some of the first efforts towards this goal dealt with how best to describe a model in a declarative format and how to restructure computational neuroscience applications to make full use of this conceptual framework (Goddard et al. 2001). However, the usage of modeling applications with proprietary formats has continued due to large existing user bases and the experimental data and models available in these formats.
The increasing enthusiasm in the community of neuroscience application developers for standards that will allow greater interoperability of models is driving the current NeuroML standards initiative. This initiative focuses on the key objects that need to be exchanged among existing applications and tries to anticipate those needed by future neuroscience applications. These objects include descriptions of neuronal morphologies, voltage-gated ion channels, synaptic mechanisms, and network structure. Enabling the exchange of these objects allows for the continued development of models using tried and trusted applications but also encourages export of the core elements of the models in a standard format for importation into another application or archival in a database, and thus facilitates the creation of composite models of independently developed components. The process of creating these common specifications also encourages discussion among users of independently developed applications, which leads to succinct, transparent descriptions of the essential elements of models. The openness of the standards and the encouragement of feedback from many sections of the community are some of the guiding principles of the NeuroML standards initiative, and the use of XML as a definition language helps to provide the transparency, portability, and extensibility required in these efforts.
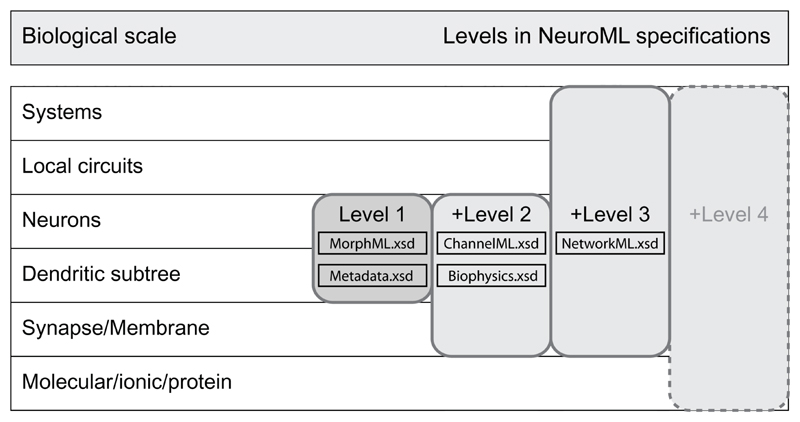
The declarative specifications produced by the NeuroML standards project are arranged into levels, with higher levels adding extra concepts. This approach is similar to the approach of the systems biology markup language (SBML, http://www.sbml.org) (Hucka et al. 2003), although for the NeuroML standards, each new level increases the scope of the language significantly. Level 1, which is the subject of this paper, deals with neuroanatomical information. MorphML forms the core of this level and describes the branching structures of neurons, along with other relevant reconstructed anatomical information. Level 1 also includes the specification for metadata, which are common across all levels. Level 2 allows for specification of cell models with realistic channel and synaptic mechanisms distributed on their membranes, and level 3 describes networks of these cells in three dimensions. These levels are under development as discussed below. Currently, neuroanatomical data in digitized format are used by (a) neuronal reconstruction and visualization software packages that focus on anatomical analysis and (b) neuronal simulation packages, which use detailed morphologies and cable theory to study topics such as dendritic computation. In this work, we provide a brief overview of how neuroanatomical data are handled by a number of these applications, followed by a description of how MorphML structures this information. The key elements in MorphML are discussed and the recommended mapping to several formats is given. We also discuss currently available tools that rely on the NeuroML standards, and we provide an outline of future plans for the NeuroML standards initiative.
2. Neuroanatomical Data Formats
Quantitative neuroanatomical data that specify neuronal morphology are important for the study of many areas of neuroscience including neural development, aging, pathology, and neural computation (Cannon et al. 2002). Existing neuronal morphology data have been obtained using many different neuron tracing systems, and tools developed for reconstruction, visualization, analysis, and simulation of neurons are also based on their own formats for the representation of morphology data. There are a number of online archives where files in these various formats can be obtained (Ascoli 2006; Hines et al. 2004), and recent work using reconstructed morphologies from hippocampal neurons clearly demonstrates that the exchange of morphological data is beneficial for studies of the relationship between structure and function (Szilagyi and De Schutter 2004; Scorcioni et al. 2004; Holmes et al. 2006).
We discuss two broad categories of applications that use neuroanatomical data: neuronal reconstruction applications and neuronal simulators. For the first category, we focus on the Neurolucida (Glaser and Glaser 1990) system and for the second, we discuss the NEURON simulator (http://www.neuron.yale.edu) (Hines and Carnevale 1997; Hines and Carnevale 2001) and the general neural simulation system (GENESIS) (http://www.genesis-sim.org) (Bower and Beeman 1998; Beeman et al. 1997; Beeman and Bower 2004).
2.1. Neuronal Reconstruction Applications
Most neural tracing systems represent the axonal and dendritic arborizations of a neuron using a collection of points, diameters and connections in three dimensions. However, the various file formats such as the Nevin binary branch tree syntax (Jacobs and Nevin 1991), the SWC format (http://www.compneuro.org) (Cannon et al. 1998; Ascoli et al. 2001) used by NeuroMorpho (Brown et al. 2005) (http://neuromorpho.org), and the formats used by Eutectic’s neuron tracing system and Microbrightfield’s Neurolucida (Glaser and Glaser 1990) employ a variety of representations and vocabularies to describe the connectivity between points based on different conceptual models. Neurons are reconstructed for visualization and analysis using algorithms specific to each of these formats; often, there is no documentation available for the precise details of these tools. In contrast, the use of XML provides explicit structural information and also supplies a robust representation of data that is independent of file format because documents can be read and understood even if the corresponding schemas are lost. This additional functionality for the storage of morphological reconstructions, which are often the result of hours of tedious work, offsets the larger file size of the structured information provided by XML as compared to the compact, yet opaque, representations in current use.
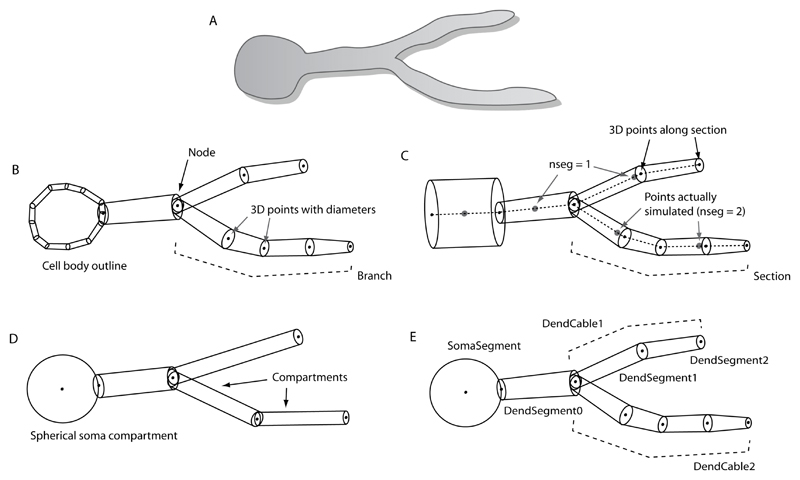
A schematic example of a simple cell (Fig. 1A) digitized in Neurolucida format is shown in Fig. 1B. An outline is used to represent the shape of the soma, but the remainder of the neuron is described using three-dimensional points with associated diameters along branches. Nodes are points at which new branches start. This information is encoded in a hierarchical tree in Neurolucida ASC files. Much more information can be present in a Neurolucida file, due to the fact that the format is designed to encode any interesting anatomical features found in the optical images being reconstructed. Items such as dendritic spines can be identified on the cell, and any points of interest, such as synaptic locations, can be highlighted. Free points and paths that are independent of any cell also can be included, which is useful for reconstructing contours or fixed points (fiducials) and aids in the registration of images to a common coordinate system.
Figure 1.
Schematic comparing handling of morphological information for a simple cell by different applications. A) Original cell structure. B) Schematic of Neurolucida reconstruction where the soma is represented by an outline and three-dimensional points are specified along each branch. C) NEURON simulator format where cell structure is specified in sections. Only the center of the section is simulated unless the nseg parameter is greater than one. D) GENESIS simulator representation using compartments that are cylinders except for the soma, which can be spherical. The optimal length of each compartment is determined by the electrotonic length. E) MorphML representation where any of the information shown in panels B through D can be encoded. Example 1 gives the XML corresponding to this morphology. Ideally, the MorphML document will contain the same amount of detail as the original source document, and each application that uses the file can extract the relevant information. Note that these are simply representative examples of how morphology is handled by each application
2.2. Neuronal Simulators
Reconstruction and visualization applications need to store information on a wide range of anatomical details found in optical images. However, neuronal simulators have a different focus—that of reproducing the electrophysiological behavior of neurons through explicit modeling of voltage-dependent ionic conductances and synaptic inputs. The use of realistic cellular morphologies is essential for the study of many cell types due to the effect of complex dendritic trees and inhomogeneous distributions of channel densities on cell firing behavior (Mainen and Sejnowski 1996; Vetter et al. 2001). Morphologies are normally obtained via a reconstruction application like the one described above, and reconstructed data must be converted into a format that is acceptable for the chosen neuronal simulation package. Applications such as CVAPP (http://www.compneuro.org) (Cannon et al. 1998) have served this function for many years. We will outline how two of the most popular simulators handle morphological data.
NEURON’s method for encoding morphological information is based around the section object, which represents an unbranched cable in three-dimensional space. For a complex neuronal morphology, a number of three-dimensional points with associated diameters are specified along a section, as depicted in Fig. 1C. The membrane surface area and total axial resistance for a given length along the dendrite/axon are computed using these values. For simulation purposes, sections can be split into a number of evenly spaced segments, where the number of segments depends on the value of the parameter nseg. The appropriate spatial discretization for simulations depends on the consideration of electrotonic length. In practice, for a biologically realistic value for the specific axial resistance, a value of 1 for nseg is sufficient for many unbranched sections of dendrite in neuronal reconstructions; however, longer sections may require multiple segments. Because the membrane potential is only calculated at the center of these segments, NEURON simulates a neuron efficiently while maintaining morphological detail. It is important to note that the points along the section that are used for simulation purposes will generally not correspond to any of the points from the reconstruction. An additional point to note is that there is no standard format for a morphology-only NEURON script file (usually *.hoc or *.nrn), and the use of commands such as create and connect are often interspersed with other NEURON commands in published models, making translation into a file format used by another simulation application difficult. This is one of the factors that inhibits the exchange of morphologies in this format; however, the ModelView tool in NEURON allows analysis of the structure of neurons in a simulation and is being updated to allow export in MorphML format, as outlined in NeuroML-Related Software Tools.
In GENESIS, neurons are constructed from compartments, where each compartment is a cylinder or a sphere without internal divisions (Fig. 1D). There is no concept of a cable; thus, all compartments are modeled explicitly. For this reason, a model cell based on a morphology reconstructed using a system like Neurolucida, where one compartment is created between each two consecutive three-dimensional points along a branch, will normally contain too many compartments. Recompartmentalization of the cell morphology while retaining the cell’s overall structure, dendritic length, surface area, etc., can be carried out by applications such as CVAPP and neuroConstruct (NeuroML-Related Software Tools). The optimal length of each compartment is a compromise between choosing a sufficiently small maximum allowable value for the electrotonic length of a compartment and reducing the number of compartments to obtain shorter simulation time. GENESIS morphology files are often in readcell-compatible (*.p) format, which simplifies the creation of the model cell structure; compartments are automatically generated, messages between compartments are initiated, and the appropriate channels can be associated with compartments using the *.p file. This file format can be parsed easily by other applications.
3. NeuroML Level 1 Standards
The NeuroML standards have been designed to provide a markup for representing various types of neuroscience data where the chosen vocabulary is an abstract representation language that is independent of any particular software application. The chosen format for representing the standards is XML. Model description files, which are written as XML documents, can automatically be checked for validity and compliance to the various NeuroML schemas, which are written in the XML schema definition (XSD) format (Crook and Howell 2007). This functionality is present in the online NeuroMLValidator (http://www.morphml.org:8080/NeuroMLValidator) application as discussed in NeuroML-Related Software Tools. One of the key features of XML is the ease with which valid documents are converted into other formats. An extensible stylesheet language file contains instructions for converting the elements of an XML file into another structured format, which is not necessarily XML. This functionality has been used in the NeuroMLValidator application to convert submitted NeuroML files into NEURON or GENESIS scripts or HTML format.
The NeuroML level 1 specifications consist of two XSD files: Metadata.xsd and MorphML.xsd. The metadata file specifies elements such as notes and properties, which are common to all levels of NeuroML. Elements for basic three-dimensional structures are also contained here, as are various elements for including references to external databases or publications. MorphML (Qi and Crook 2004; Crook et al. 2005) contains the elements needed to specify neuroanatomical data, as described in more detail below. The latest versions of these schemas are available at the NeuroML web site (http://www.neuroml.org) in both XML and more human-friendly formats, along with additional extensive documentation.
3.1. NeuroML Level 1 Element Structure
The hierarchical tree of the elements of the NeuroML schemas was designed to be compatible with any neuroanatomical data obtained using a three-dimensional digitization system, where the standards are meant to be as general as possible under this constraint. NeuroML level 1 documents can be used to represent anatomical structures such as dendritic branches, single neurons, groups of cells, and various histological features for use in database or visualization programs, or they can be used to specify neuroanatomical reconstructions for multicompartment conductance-based models using NEURON, GENESIS, or other model simulation software.
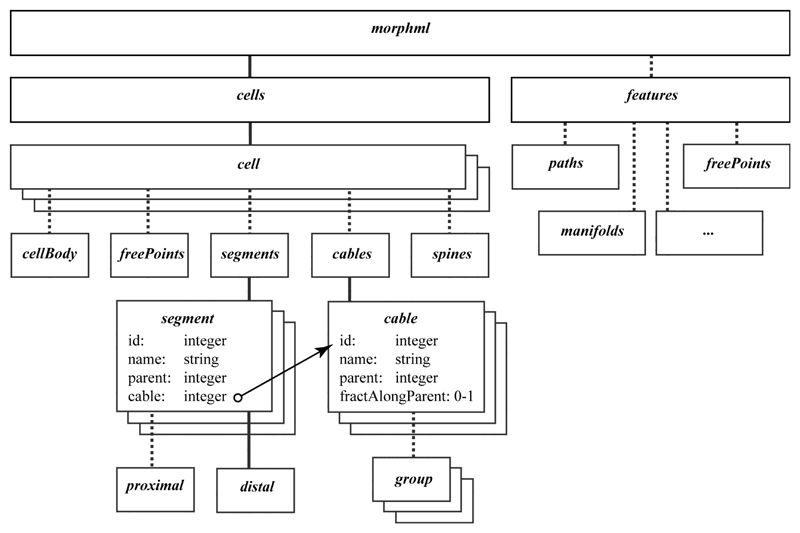
The morphml element provides a wrapper used to specify all of the information for a particular morphology document and serves as the root element of the hierarchical tree. Each morphml can contain a collection of cells where each cell groups together all of the information for a single neuron (see Fig. 2). The structures that make up a cell are constructed using point elements containing a three-dimensional location of a digitized point. These points may be associated with an optional diameter attribute to specify a cross-section at a point in space. Branching structures are created using a collection of successive points. Connectivity is described using segments and optionally cables. Each segment contains, in addition to a unique id and an optional name, a proximal point and a distal point, defined with respect to the distance from the soma. The parent attribute points to the id of the parent segment, thus defining electrical connectivity. The optional cable attribute of a segment points to one of the cable elements that, when present, define unbranched sections of dendrites, axons, etc. The inclusion of cables allows efficient specification of groups to which segments belong, e.g., basal dendrites, apical dendrites, etc. When segments are arranged in unbranched cables, the specification of a proximal point in a child segment is unnecessary, as this will be the same as the distal point of the parent. The fractAlongParent attribute in a cable is needed in the case that the first point of the initial segment of a cable does not coincide with the end of its parent segment in another cable. This is often the case in neuronal reconstructions where dendritic branches can “float in space,” although there may be a clear point on the parent cable where the electrical connection should be. The standard requires this representation to ensure lossless conversion from existing file formats, but applications that support MorphML should signal such issues if they could lead to problems (e.g., connecting disconnected cells). The cellBody element can be used to specify the shape and location of the soma. There is an additional element, freePoints, for describing the locations of varicosities or synaptic connections if one wants to maintain this information in a way that is independent of any part of the branching structure. There are also elements for describing the locations, lengths, volumes, and shapes of dendritic spines.
Figure 2.
Structure of the main elements in MorphML files. A dashed line indicates an optional child element. The root element morphml contains cells, with information on the cellular structures, and features, with information on other interesting anatomical items observed in the sample. Elements for adding notes, references, and other metadata are omitted for clarity. The core of the cell element is the specification of the segments, and the optional grouping of them (by a pointer to the id of the cable) into cables.
These features element contains a collection of all of the histological features that are separate from the reconstructed neurons. These anatomical features can be specified using collections of additional elements such as paths, freePoints, manifolds, polygons, polyhedrons, and spheres. For example, a path may be used to represent an unclosed fiducial contour, or a group of freePoints might be used to record the locations of a series of fiducial points used for image registration. Each of these elements is specified by a collection of points but with different conceptual interpretations.
Additional elements provide functionality for metadata such as textual notes, references, directives for downstream applications, user-defined tag-value pairs, and ways to group anatomical elements, such as segments or cables, and provide group names and attributes. One of the requirements of NeuroML is clarity in the description language (Goddard et al. 2001). In the schemas, collections of elements are identified by pluralization. Qualifiers such as Set, SetOf, or ListOf have not been used because they have precise definitions regarding element duplication and order, which these collections may not satisfy.
The mapping of these elements to and from the formats described in Neuroanatomical Data Formats requires some discussion. Branching structures in Neurolucida can be encoded by specifying a segment for each pair of successive points along an unbroken branch where each segment belongs to the same cable. While branching structures can be specified without cables, their use allows for easier grouping of segments, e.g., reflecting the properties often found in Neurolucida files such as “apical dendrite” or “color red.” A comment is often present after a point in a Neurolucida file, which gives textual information about the point, e.g., “R-1, 2” for the second point along the first branch off the root. This information can be stored in the name attribute of the segment. The soma outline information can be encoded in the cellBody element, or an application may use some algorithm to generate appropriate segments to fill the outline. Further information present in these files about histological features of interest can be mapped on to the features element, and the metadata allowing notes, grouping information, and tag-value properties is flexible enough to encode any additional information in the Neurolucida files.
The mapping between NEURON and MorphML is straightforward. Sections are mapped to cables, with the section’s name being used for the name attribute in MorphML. Lists of sections are often present in cells in NEURON, and these can be specified with a group in the cable element. The three-dimensional points along a cable are given by the end points of segments. Only the first segment will have both a proximal point and distal point; each subsequent segment in the cable will only have a distal point. Cables can be connected at arbitrary points (0 for proximal and 1 for distal) along their parent cables, and this can be specified with the fractAlongParent attribute. This corresponds closely with the usage of the connect statement to define electrical connectivity in NEURON. The value nseg can be specified for a cable by adding a property tag-value pair. The soma in a NEURON cell consists of a section like any other, and so should be encoded as a cable/ segment set in MorphML, as opposed to using cellBody. Cable groups such as soma_group, dendrite_group, and axon_group can be used to specify the type of cable so that the soma cable can be identified.
The mapping to and from GENESIS is complicated by the fact that there is no cable construct in that simulator. One possible approach is a one-to-one mapping between a compartment and a segment. This is sufficient for converting a GENESIS cell to a MorphML representation; however, as discussed above, translating a MorphML file originally obtained from a reconstruction application such as Neurolucida into GENESIS would most likely lead to a GENESIS model cell with too many compartments. Specifying the correct way to convert morphologies to specific simulation formats is beyond the scope of this paper, but any application for viewing and editing MorphML files for use in a simulator should have the ability to perform the appropriate recompartmentalization, as discussed below for the application neuroConstruct.
3.2. Example
The following is an example of a MorphML file showing the usage of segments and corresponding cables as shown in Fig. 1E. Each of the segments is part of a cable, and the soma and the dendrites can be distinguished by their grouping.
Example 1.
<?xml version="1.0" encoding="UTF-8"?>
<morphml xmlns= "http://morphml.org/morphml/schema" xmlns:meta= "http://morphml.org/metadata/schema" xmlns:xsi= "http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation = "http://morphml.org/morphml/schema MorphML.xsd" lengthUnits = "micron">
<cells>
<cell name = "SampleCell">
<meta:notes>A Simple cell</meta:notes>
<segments>
<segment id = "0" name = "SomaSegment" cable = "0">
<proximal x = "0.0" y = "0.0" z = "0.0" diameter = "16.0"/>
<distal x = "0.0" y = "0.0" z = "0.0" diameter = "16.0"/>
</segment>
<segment id = "1" name = "DendSegment0" parent = "0" cable = "1">
<proximal x = "8.0" y = "0.0" z = "0.0" diameter = "5.0"/>
<distal x = "28.0" y = "2.0" z = "0.0" diameter = "6.0"/>
</segment>
<segment id = "2" name = "DendSegment1" parent = "1" cable = "2">
<proximal x = "28.0" y = "2.0" z = "0.0" diameter = "3.6"/>
<distal x = "48.0" y = "12.0" z = "0.0" diameter = "4.0"/>
</segment>
<segment id = "3" name = "DendSegment2" parent = "2" cable = "2">
<distal x = "65.0" y = "14.0" z = "0.0" diameter = "3.2"/>
</segment>
<!-- The 4 segments associated with DendCable2 have been omitted -->
</segments>
<cables>
<cable id = "0" name = "SomaCable">
<meta:group>soma_group</meta:group>
</cable>
<cable id = "1" name = "DendCable0">
<meta:group>dendrite_group</meta:group>
</cable>
<cable id = "2" name = "DendCable1">
<meta:group>dendrite_group</meta:group>
</cable>
<cable id = "3" name = "DendCable2">
<meta:group>dendrite_group</meta:group>
</cable>
</cables>
</cell>
</cells>
</morphml>
More detailed examples of MorphML files are available on the NeuroMLValidator website under Samples.
3.3. NeuroML Related Software Tools
We have developed tools for online validation of NeuroML files and their conversion into several useful data formats. After a NeuroML document is generated using an application such as neuroConstruct as described below, it can be validated against the NeuroML schemas using the NeuroMLValidator, which provides the version and level of compliance for the given document. The NeuroMLValidator web site also provides software for translating valid NeuroML level 1 documents into scripts for running simulations using GENESIS or NEURON. Detailed examples of valid documents that describe components of published models are also provided.
One example of the down-stream use of NeuroML level 1 for neuroanatomical data exchange and analysis is the Virtual Ratbrain Project (http://www.ratbrain.org), which includes a database for peer-reviewed, three-dimensional, cellular anatomical data from the rat brain, as well as visualization and analysis tools. In this project, data are stored and manipulated in the MorphML format, and one of the available tools is a MorphML viewer.
For modelers who want to use NeuroML level 1 to specify morphology for a multicompartment conductance-based neuronal model, the modeling-related application neuroConstruct (http://www.neuroConstruct.org) (Gleeson et al. 2007) will import, visualize, and write NeuroML documents. neuroConstruct has been designed to simplify the specification of single-cell models or models of complex three-dimensional networks of morphologically realistic neurons. It allows a user to import morphology files in Neurolucida, MorphML, GENESIS, NEURON, or SWC formats, to specify the three-dimensional positioning of groups of neurons and to define complex connectivity for a network of cells. It then generates output for simulating network activity using either NEURON or GENESIS as a simulation environment and provides tools for the visualization and analysis of simulation-generated data. The application also includes functionality to optimize large neuronal morphologies for particular simulators. For example, a more efficient compartmentalization can be created from a Neurolucida reconstruction for use in a GENESIS simulation.
The NeuroML standards will be used extensively in future implementations of neuronal simulation applications.
The GENESIS simulator is being updated to a faster, easier-to-use, and backwards-compatible version with a modern Java-based user interface. The model specification in Moose-GENESIS 3.0 (http://sourceforge.net/projects/moose-g3) will utilize the NeuroML standards for cell morphology, membrane properties, subcellular processes, and network connectivity. The NEURON simulator group is working on exporting information in NeuroML format, including cellular morphology, channel density information, and channels created with the ChannelBuilder tool. Due to compatibility with NEURON and GENESIS and the descriptive nature of NeuroML documents for model specification, NeuroML provides an ideal format for archiving published neuroscience models in databases such as ModelDB (http://senselab.med.yale.edu/senselab/modeldb).
4. Future Work and Relation to Other Projects
MorphML, which forms the core of the level 1 standard, is quite stable and no major changes to the specification are planned. Level 2 of NeuroML builds on level 1 to include standards for describing passive membrane properties such as specific capacitance and axial resistance, the distribution of ionic conductance densities on the neuronal membrane, and the dynamics of voltage and concentration-gated ion channels and synaptic mechanisms. A beta version of the NeuroML level 2 standards is available on the NeuroML web site, and these standards will be described in detail in a subsequent publication. Specification of three-dimensional positioning and network connectivity for populations of these biologically realistic cells is included in level 3, which is also available in an initial version. Subcellular processes such as detailed calcium dynamics and signaling pathways are the target of level 4. The level 4 standards will rely heavily on existing standards for biochemical networks provided by SBML, and this work is at an early stage of development. With the gradual expansion of the scope of NeuroML in this way, it is hoped that models covering the full range of biological scales at which information is processed in the nervous system can be expressed and exchanged in declarative specifications (Fig. 3). Additional information on current development activities is available at the NeuroML Project Development web site (http://sourceforge.net/projects/neuroml).
Figure 3.
Diagram showing different levels of biological scale at which information is processed and stored in the nervous system (left) and the related levels in NeuroML. The corresponding XSD files are indicated. This paper deals only with level 1, which describes neuronal morphology. Level 2 allows specification of mechanisms at the membrane and synapse level, allowing the creation of biologically realistic models of spiking neurons. Level 3 adds network connectivity so that neuronal circuits and systems can be described. Future work includes level 4, which will allow specification of interactions with subcellular signaling pathways.
NeuroML is complementary to other efforts to develop markup languages for model and data specification. CellML (http://www.cellml.org) (Hedley et al. 2001) is a language for describing cellular and subcellular level models that is built around systems of equations and is mostly procedural rather than using a descriptive, objectoriented approach. SBML and NeuroML have many common applications, and as mentioned above, future levels of the NeuroML standards for specifying models at the subcellular level will interface with SBML; however, SBML currently lacks functionality for representing the spatial aspects of models. BrainML (http://www.brainml.org) provides a language for neuroscience data that is centered around interoperability and exchange in the area of experimental neuroscience but can also be used for data that are used by neural simulators. The functional imaging research science and technology group of the Biomedical Informatics Research Network (http://www.nbirn.net) provides an XML schema and tools for storage of neuroimaging results and anatomical labels (Keator et al. 2006).
By engaging the wider neuroscience community in discussions about what forms the essential and reusable core of published computational models, we hope to remove the barriers caused by multiple formats that have hindered closer collaboration on the fundamental problems of brain research.
Acknowledgements
We would like to thank David Beeman, Robert Cannon, Hugo Cornelis, Laszlo Zaborszky, Zoltan Nadasdy, Michael Hines, Sushil Kambampati, and Nicola McDonnell for helpful discussions that ultimately led to the design of the NeuroML schemas. We also would like to thank Weihong Qi for her thoughtful early contributions to this project. SC gratefully acknowledges the support of the National Science Foundation through IBN 0091117 for some of the early part of this work. PG is in receipt of a Special Research Training Fellowship in Bioinformatics from the Medical Research Council and RAS is in receipt of a Wellcome Trust Senior Research Fellowship.
Contributor Information
Sharon Crook, Department of Mathematics and Statistics and School of Life Sciences, Arizona State University, Tempe, Arizona, USA.
Padraig Gleeson, Department of Physiology, University College London, London, UK.
Fred Howell, Institute of Adaptive and Neural Computation, University of Edinburgh, Edinburgh, UK.
Joseph Svitak, Research Imaging Center, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
R. Angus Silver, Department of Physiology, University College London, London, UK.
References
- Ascoli GA. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nature Reviews Neuroscience. 2006;7:318–324. doi: 10.1038/nrn1885. [DOI] [PubMed] [Google Scholar]
- Ascoli GA, Krichmar JL, Nasuto SJ, Senft SL. Generation, description and storage of dendritic morphology data. Philosophical transactions of the Royal Society of London. Series B, Biological Sciences. 2001;356:1131–1145. doi: 10.1098/rstb.2001.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeman D, Bower JM. Simulator-independent representation of ionic conductance models with ChannelDB. Neurocomputers. 2004;58–60:1085–1090. [Google Scholar]
- Beeman D, Bower JM, De Schutter E, Efthimiadis EN, Goddard N, Leigh J. The GENESIS simulator-based neuronal database. In: Koslow SH, Huerta MF, editors. Neuroinformatics: An overview of the human brain project. Mahwah: Lawrence Erlbaum Associates; 1997. pp. 57–80. [Google Scholar]
- Bower JM, Beeman D. The book of GENESIS: Exploring realistic neural models with the general neural simulation system. 2nd ed. New York: Springer; 1998. [Google Scholar]
- Brown KM, Donohue DE, D’Alessandro G, Ascoli GA. A cross-platform freeware tool for digital reconstruction of neuronal arborizations from image stacks. Neuroinformatics. 2005;3:343–360. doi: 10.1385/NI:3:4:343. [DOI] [PubMed] [Google Scholar]
- Cannon RC, Howell FW, Goddard NH, De Shutter E. Non-curated distributed databases for experimental data and models in neuroscience. Network: Computation in Neural Systems. 2002;13:415–428. [PubMed] [Google Scholar]
- Cannon RC, Turner DA, Pyapali G, Wheal HV. An on-line archive of reconstructed hippocampal neurons. Journal of Neuroscience Methods. 1998;84:49–54. doi: 10.1016/s0165-0270(98)00091-0. [DOI] [PubMed] [Google Scholar]
- Crook S, Beeman D, Gleeson P, Howell F. XML for model specification in neuroscience: An introduction & workshop summary. Brains, Minds, and Media. 2005;1 bmm228 (urn:nbn: de:0009-3-2282) [Google Scholar]
- Crook S, Howell F. XML for data representation & model specification in neuroscience. In: Crasto C, editor. Neuroinformatics, methods in molecular biology book series. Totowa: Humana; 2007. [DOI] [PubMed] [Google Scholar]
- Gardner D, Toga AW, Ascoli GA, Beatty JT, Brinkley JF, Dale AM, et al. Towards effective & rewarding data sharing. Neuroinformatics. 2003;1:289–295. doi: 10.1385/NI:1:3:289. [DOI] [PubMed] [Google Scholar]
- Glaser JR, Glaser EM. Neuron imaging with Neurolucida—a PC-based system for image combining microscopy. Computerized Medical Imaging and Graphics. 1990;14:307–317. doi: 10.1016/0895-6111(90)90105-k. [DOI] [PubMed] [Google Scholar]
- Gleeson P, Steuber V, Silver RA. neuroConstruct: A tool for modeling networks of neurons in 3D space. Neuron. 2007;54:219–235. doi: 10.1016/j.neuron.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard NH, Hucka M, Howell F, Cornelis H, Shankar K, Beeman D. Towards NeuroML: Model description methods for collaborative modelling in neuroscience. Philosophical Transactions of the Royal Society of London B. 2001;356:1209–1228. doi: 10.1098/rstb.2001.0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley WJ, Nelson MR, Bullivant DP, Nielsen PF. A short introduction to CellML. Philosophical Transactions of the Royal Society of London A. 2001;359:1073–1089. [Google Scholar]
- Hines ML, Carnevale NT. The NEURON simulation environment. Neural Computation. 1997;9:1179–1209. doi: 10.1162/neco.1997.9.6.1179. [DOI] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. Neuron: a tool for neuroscientists. The Neuroscientist. 2001;7:123–135. doi: 10.1177/107385840100700207. [DOI] [PubMed] [Google Scholar]
- Hines ML, Morse T, Migliore M, Carnevale NT, Shepherd GM. ModelDB: A database to support computational neuroscience. Journal of Computational Neuroscience. 2004;17:7–11. doi: 10.1023/B:JCNS.0000023869.22017.2e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WR, Ambros-Ingerson J, Grover LM. Fitting experimental data to models that use morphological data from public databases. Journal of Computational Neuroscience. 2006;20:349–365. doi: 10.1007/s10827-006-7189-8. [DOI] [PubMed] [Google Scholar]
- Hucka M, et al. The system biology markup language (SBML): A medium for representation & exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Jacobs GA, Nevin R. Anatomical relationships between sensory afferent arborizations in the cricket cercal system. Anatomical Record. 1991;231:563–572. doi: 10.1002/ar.1092310418. [DOI] [PubMed] [Google Scholar]
- Keator DB, Gadde S, Grethe JS, Taylor DV, Potkin SG, FIRST BIRN A general XML schema & SPM toolbox for storage of neuro-imaging results & anatomical labels. Neuroinformatics. 2006;4:199–212. doi: 10.1385/ni:4:2:199. [DOI] [PubMed] [Google Scholar]
- Mainen ZF, Sejnowski TJ. Influence of dendritic structure on firing pattern in model neocortical neurons. Nature. 1996;382:363–366. doi: 10.1038/382363a0. [DOI] [PubMed] [Google Scholar]
- Martone ME, Gupta A, Ellisman MH. e-Neuroscience: Challenges & triumphs in integrating distributed data from molecules to brains. Nature Neuroscience. 2004;7:467–472. doi: 10.1038/nn1229. [DOI] [PubMed] [Google Scholar]
- Qi W, Crook S. Tools for neuroinformatic data exchange: An XML application for neuronal morphology data. Neurocomputing. 2004;58C–60C:1091–1095. [Google Scholar]
- Scorcioni R, Lazarewicz MT, Ascoli GA. Quantitative morphometry of hippocampal pyramidal cells: Differences between anatomical classes and reconstructing laboratories. Journal of Comparative Neurology. 2004;473:177–193. doi: 10.1002/cne.20067. [DOI] [PubMed] [Google Scholar]
- Szilagyi T, De Schutter E. Effects of variability in anatomical reconstruction techniques on models of synaptic integration by dendrites: a comparison of three Internet archives. European Journal of Neuroscience. 2004;19:1257–1266. doi: 10.1111/j.1460-9568.2004.03222.x. [DOI] [PubMed] [Google Scholar]
- Vetter P, Roth A, Hausser M. Propagation of action potentials in dendrites depends on dendritic morphology. Journal of Neurophysiology. 2001;85:926–937. doi: 10.1152/jn.2001.85.2.926. [DOI] [PubMed] [Google Scholar]