Abstract
Methuosis is a form of nonapoptotic cell death characterized by the accumulation of macropinosome-derived vacuoles. Herein, we identify PIKFYVE, a class III phosphoinositide (PI) kinase, as the protein target responsible for the methuosis-inducing activity of indolyl-pyridinyl-propenones (3-(5-methoxy-2-methyl-1H-indol-3-yl)-1-(4-pyridinyl)-2-propen-1-one). We further characterize the effects of chemical substitutions at the 2- and 5-indolyl positions on cytoplasmic vacuolization and PIKFYVE binding and inhibitory activity. Our study provides a better understanding of the mechanism of methuosis-inducing indolyl-pyridinyl-propenones.
Introduction
Methuosis is a form of nonapoptotic cell death triggered by alterations in the trafficking of clathrin-independent endosomes and is associated with vacuolization of macropinosome and endosome compartments.1 Methuosis was initially defined in glioblastoma cells expressing activated Ras,2 but several studies have reported the features of methuosis in a broad spectrum of cancer cells including gastric carcinoma cells and osteosarcoma cells.3,4 The vacuoles generated during methuosis are distinct from autophagosomes and show characteristics of enlarged macropinosomes. Vacuolated cells eventually undergo caspase-independent cell death initiated by the dysregulation of micropinocytosis.2
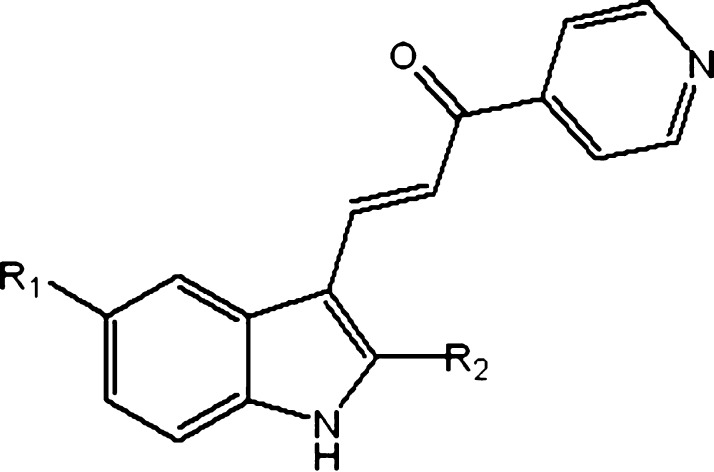
Previous investigations found that indolyl-pyridinyl-propenones (also referred to as indole-based chalcones) trigger methuosis.5 Specifically, 3-(5-methoxy-2-methyl-1H-indol-3-yl)-1-(4-pyridinyl)-2-propene-1-one (MOMIPP) was identified as an initial exemplar tool compound. Follow-up structure–activity relationship (SAR) studies defined key features associated with three phenotypic classifications, namely, the induction of methuosis, vacuolization without cell death,6 and disruption of microtubules leading to apoptosis.7 Selected substitutions on the indole at position 2 or moving the methoxy group from position 5 to 68 redirected the mode of cytotoxicity from distinctive methuosis to microtubule disruption with little observable effect on vacuoles. However, the protein target(s) of the compounds that prompt the methuosis phenotype have not been identified. In this study, we identified PIKFYVE, a class III phosphoinositide (PI) kinase, as at least one candidate protein that when inhibited triggers vacuolization, a hallmark of methuosis.
PI homeostasis is tightly regulated because the spatiotemporal restriction of different PI species is crucial for membrane trafficking within both the secretory and endocytic pathways.9,10 Disruption of endosome or macropinosome trafficking has been shown to induce cellular vacuolization.11 PI synthesis involves phosphorylating/dephosphorylating positions D3, D4, and/or D5 of their inositol ring by various lipid kinases and phosphatases.10 Accordingly, the inhibition of select lipid kinases or phosphatases by chemical or genetic perturbation can induce cellular vacuolization.12,13
Results and Discussion
To explore whether indolyl-pyridinyl-propenones disrupt PI homeostasis by inhibiting a specific inositol kinase or phosphatase, we used a competition binding assay to quantitatively measure interactions between the lead compound MOMIPP and 22 inositol kinases including clinically relevant mutants (Table 1). The competition binding assay showed that at 10 μM, MOMIPP competes away all binding of inositol kinases PIKFYVE and PIP5K1C as compared to the control.
Table 1. Lipid Kinase Competition Assay for 22 Inositol Kinases Including Clinically Relevant Mutantsa.
| target | % control |
|---|---|
| PIK3C2B | 76 |
| PIK3C2G | 39 |
| PIK3CA | 99 |
| PIK3CA(C420R) | 100 |
| PIK3CA(E542K) | 98 |
| PIK3CA(E545A) | 82 |
| PIK3CA(E545K) | 81 |
| PIK3CA(H1047L) | 72 |
| PIK3CA(H1047Y) | 92 |
| PIK3CA(I800L) | 72 |
| PIK3CA(M1043I) | 98 |
| PIK3CA(Q546K) | 82 |
| PIK3CB | 46 |
| PIK3CD | 66 |
| PIK3CG | 54 |
| PIK4CB | 87 |
| PIKFYVE | 0 |
| PIP5K1A | 61 |
| PIP5K1C | 0 |
| PIP5K2B | 96 |
| PIP5K2C | 66 |
| VPS34 | 50 |
MOMIPP (10 μM ) was used for the competition. Data are shown as % control, where lower numbers indicate more competition between the control compound and MOMIPP. % control was calculated as follows. (MOMIPP signal – positive control signal)/(negative control signal – positive control signal) × 100. Negative control = DMSO (100% control); positive control = control compound that binds specifically to each kinase (0% control).
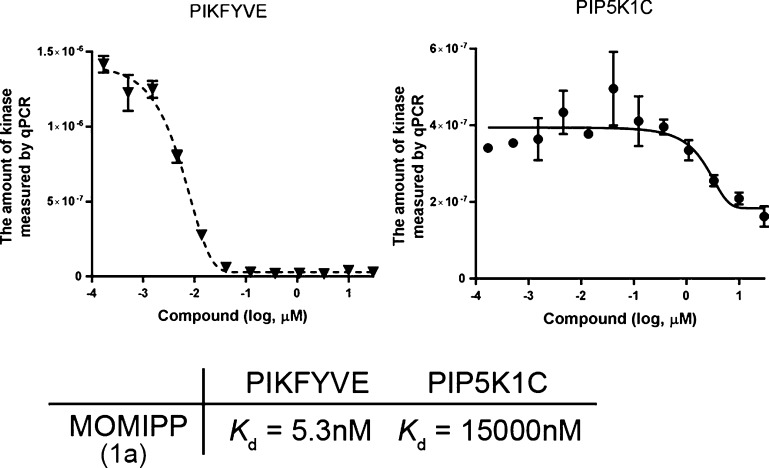
To further assess the binding potency and specificity, we calculated binding constants (Kd) against PIKFYVE and PIP5K1C with a standard dose–response curve (Figure 1). In this assay, MOMIPP exhibited a Kd of 5.3 nM against PIKFYVE, whereas the Kd against PIP5K1C was >15 000 nM, suggesting that MOMIPP specifically binds PIKFYVE with strong potency. Because previous SAR studies7 of methuosis-inducing activity resulted in three classes of indolyl-pyridinyl-propenones with distinct cellular phenotypes, we synthesized compounds from each class to confirm cellular phenotypes and characterize PIKFYVE inhibition (Table 2).
Figure 1.
Lipid kinase competition assay for PIKFYVE and PIP5K1C. Compound–kinase interactions were quantitated by quantitative PCR (qPCR) that detects the associated DNA label on each kinase as described in the Experimental Section. The amount of kinase measured by qPCR is plotted against the corresponding MOMIPP concentration. Binding constants (Kd) were calculated with a standard dose–response curve using the Hill equation12 as follows. Response = background + (signal – background)/(1 + (KdHill slope)/(doseHill slope)). The Hill slope was set to −1.
Table 2. Summary of Compound Activitya.
| compounds | class | R1 | R2 | vacuolization | cell death | Kd (PIKFYVE) (nM) |
|---|---|---|---|---|---|---|
| 2a | 1 | H3CH2CO | CH3 | yes | no | 23 |
| 2c | 1 | (H3C)2HCO | CH3 | yes | no | 51 |
| 1a (MOMIPP) | 2 | H3CO | CH3 | yes | yes | 6.9 |
| 2b | 2 | H3C(H2C)2O | CH3 | yes | yes | 6.5 |
| 2l | 3 | H3CO | COOCH2CH3 | no | yes | 540 |
| 2o | N/A | H3CO | COOH | no | no | 46 |
N/A: not assigned. Compound code numbers were assigned as previously reported.7
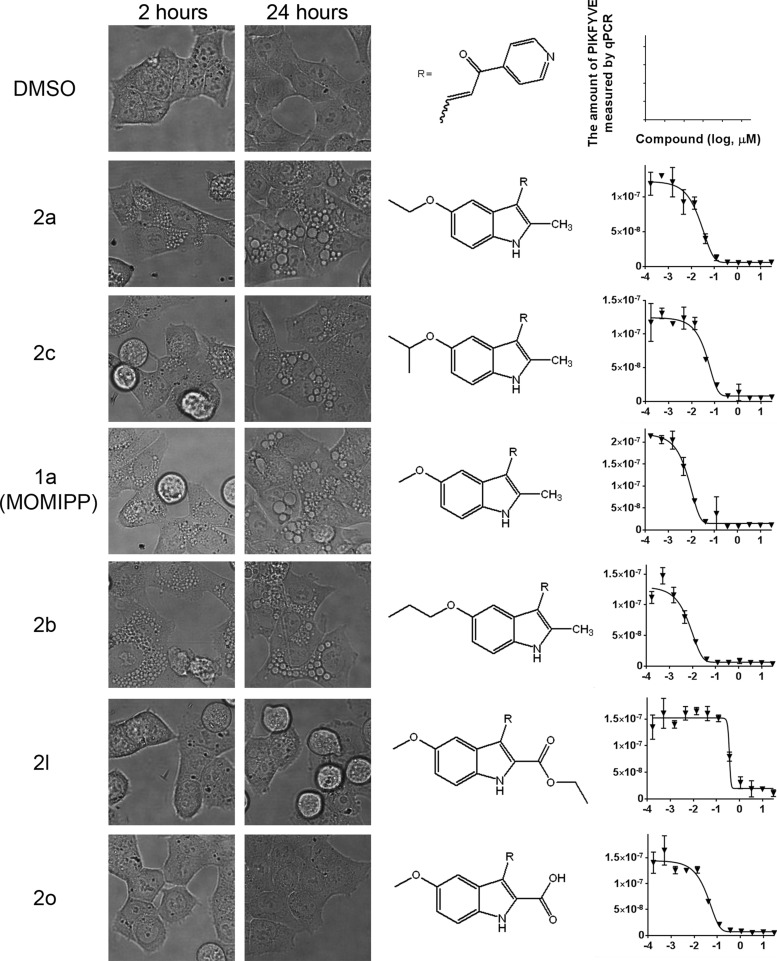
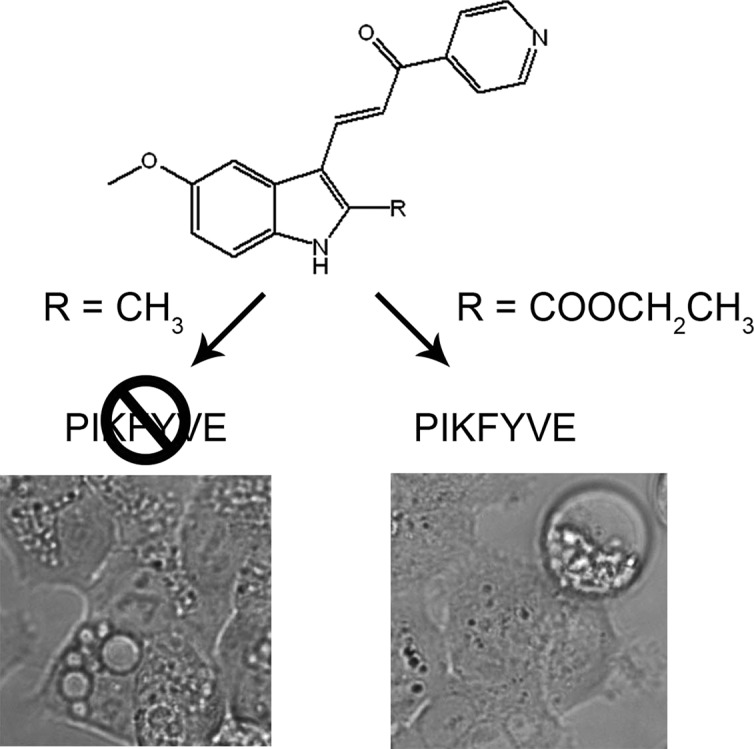
As previously characterized,7 class 1 compounds (2a, 2c) induced cellular vacuolization without causing cell death, class 2 compounds (1a; MOMIPP, 2b) induced cellular vacuolization and cell death via methuosis, and class 3 compounds (2l) induced cell death without showing distinct cellular vacuolization, consistent with prior reports of tubulin modulation (Figures 2, 3, and summarized in Table 2). To explore the correlation between the cellular vacuolization phenotype and the binding activity against PIKFYVE, we calculated binding constants (Kd) against PIKFYVE with a standard dose–response curve.
Figure 2.
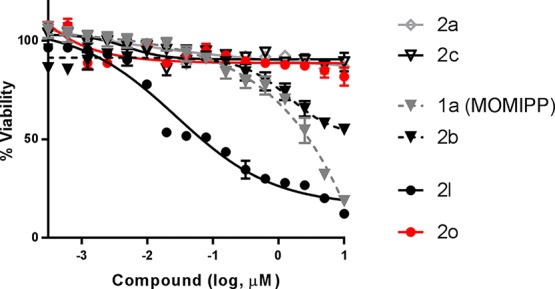
Effects of selected indolyl-pyridinyl-propenones on cell viability. Percent viability relative to vehicle [dimethyl sulfoxide (DMSO)] control after treatment for 72 h with indicated concentrations of compounds. Error bars indicate mean ± SD of at least three replicates.
Figure 3.
Vacuolization phenotype of compounds and its correlation to the PIKFYVE binding activity. Phase contrast images of live cells were obtained at 2 and 24 h after compound treatment as described in the Experimental Section at 40× magnification. The PIKFYVE binding assay with each compound was performed as described.
In this assay, class 1 compounds (2a, 2c) exhibited Kd of 23 and 51 nM, respectively, and class 2 compounds (1a; MOMIPP, 2b) exhibited Kd of 6.9 and 6.5 nM, respectively, whereas the Kd of class 3 compound (2l) was >500 nM. The Kd of free acid compound (2o) was 46 nM (Figure 3 and summarized in Table 2). These results show that compounds with the cellular vacuolization phenotype (class 1 and 2) strongly bind PIKFYVE. Compounds with carboxylic acid substitutions at R2 (class 3) do not show the cellular vacuolization phenotype. Compound 2l, a tubulin modulator,7 displays a weak binding activity against PIKFYVE but triggers cell death. The related compound (2o) binds PIKFYVE with strong in vitro potency but likely loses the cellular activity because of the limited cellular permeability.
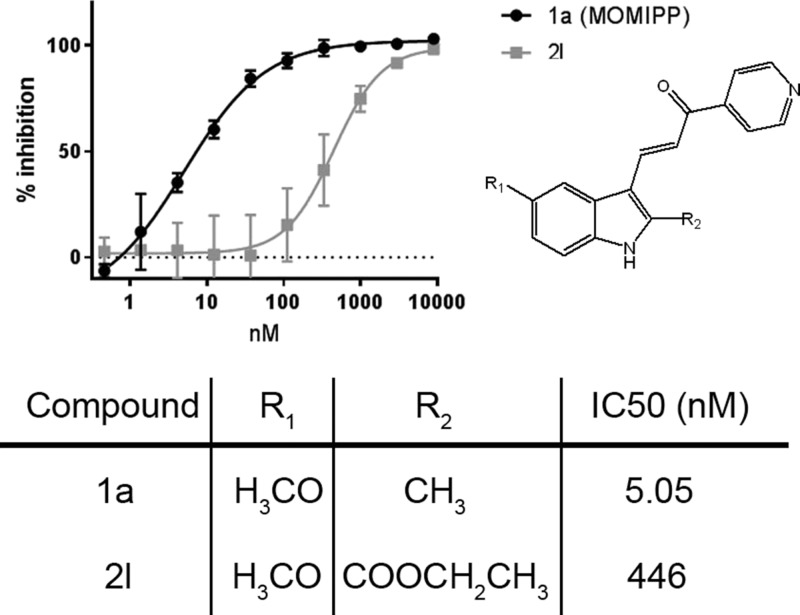
PIKFYVE is a 240 kD lipid kinase that phosphorylates the D-5 position in endosomal phosphatidylinositol-3-phosphate (PI3P) to yield 3,5-bisphosphate (PI(3,5)P2).14 To determine whether MOMIPP directly inhibits the PIKFYVE kinase activity, we performed an in vitro kinase assay with the purified PIKFYVE full-length protein. MOMIPP potently inhibited the PIKFYVE kinase activity (IC50 = 5.05 nM), whereas the class 3 compound (2l) had a reduced activity (IC50 = 446 nM) toward PIKFYVE (Figure 4). This result is consistent with the PIKFYVE binding assay where MOMIPP showed a stronger binding activity (Kd = 6.9 nM) in comparison to the class 3 compound (2l) (Kd = 540 nM) (Figure 3 and Table 2).
Figure 4.
PIKFYVE inhibition by MOMIPP. The PIKFYVE inhibitory activity by compounds 1a (MOMIPP) and 2l was measured as described in the Experimental Section. Data are shown as percent inhibition of kinase activity calculated from the percent conversion of adenosine triphosphate (ATP) to adenosine diphosphate (ADP).
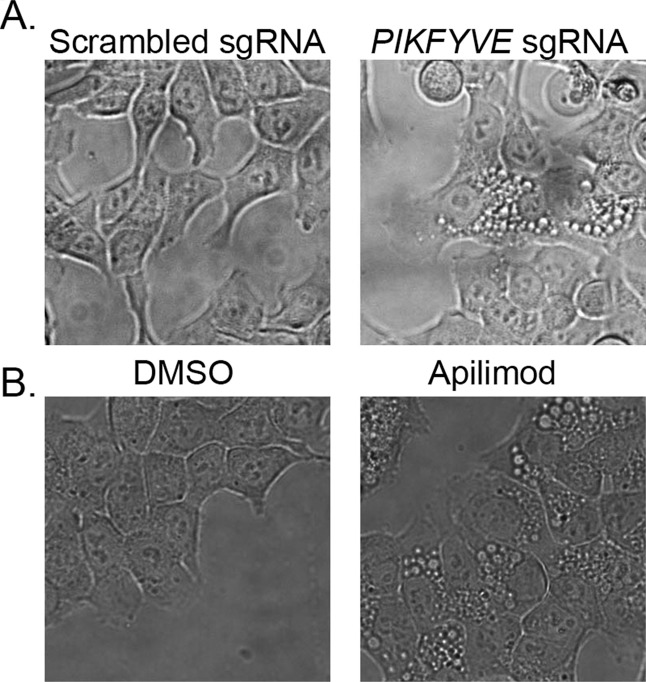
The genetic manipulation of PIKFYVE with CRISPR (clustered regularly interspaced short palindromic repeats)-mediated genome editing (Figure 5A) and chemical perturbation of PIKFYVE activity using apilimod (Figure 5B) also showed similar vacuolization. These results effectively demonstrate that MOMIPP and its analogues (class 1 and class 2 compounds) trigger cellular vacuolization by inhibiting the PIKFYVE kinase activity.
Figure 5.
Genetic and chemical perturbations of PIKFYVE phenocopies the MOMIPP treatment. (A) Phase contrast images of HCT116 cells stably expressing scrambled single-guide RNA (sgRNA) or sgRNA targeting PIKFYVE. (B) Phase contrast images of HCT116 cells treated with DMSO or 5 nM PIKFYVE inhibitor apilimod for 24 h.
In conclusion, we showed that MOMIPP binds PIKFYVE and inhibits its kinase activity. The preliminary SAR study effectively demonstrates that the binding and inhibitory activity of MOMIPP analogues against PIKFYVE determines their biological activity in cellular vacuolization, which is a hallmark of methuosis.1 The cytotoxic activity of MOMIPP (1a) and 2b may have additional target(s) and has unique pleiotropic effects on cells that lead to metabolic collapse and cell death.6 We showed that the inhibition of the PIKFYVE kinase activity results in cellular vacuolization, which is a contributing factor to methuosis.
PIKFYVE has also been reported to regulate TLR (toll-like receptor)-induced IL-12/23 expression by modulating PI(3)P and PI(3,5)P2 levels, and dysregulated TLR signaling can lead to autoimmune diseases such as Crohn’s disease, rheumatoid arthritis, and psoriasis.13 Our study suggests that MOMIPP might be an important tool to study the function of the PIKFYVE lipid kinase activity in various biological processes including the TLR pathway regulation in IL-12/23-mediated diseases as well as methuosis.
Experimental Section
General Chemistry
All commercially available solvents (VWR and Fisher) and compounds (Fluka, Lancaster, Aldrich) were used as received. Compounds were synthesized according to the reported experimental procedures (Supporting Information).7 NMR spectra were recorded with a Bruker AV 400 spectrometer in the solvents indicated. Chemical shifts (δ) are given in ppm relative to trimethylsilane. All coupling constants (J) are reported in hertz. The data are reported as follows: chemical shift, intergration, and multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, and b = broad peak). High-resolution mass spectrometry (HRMS) was performed using electrospray ionization (ESI) in positive ion mode after separation by liquid chromatography (Nexera from Shimadzu). The elemental composition was derived from the mass spectra acquired at the high resolution of about 30 000 on an LTQ Orbitrap XL mass spectrometer (Thermo Scientific). The high mass accuracy below 1 ppm was obtained by using a lock mass. Preparatory silica gel chromatography was performed on a CombiFlash Rf 200 (Teledyne ISCO) flash chromatography system with RediSep Rf normal phase silica gel (25–70 μm) columns and 254 nm UV detection.
To determine the chemical purity of final compounds, reverse-phase high-performance liquid chromatography (HPLC) analysis was performed. An ACQUITY UPLC HSS T3 column (1.8 μm; 2.1 × 100 mm) and a mobile phase (0.8 mL/min flow rate) were used with the following composition: eluent A (water + 0.05% formic acid + 3.75 mM acetic acid), eluent B (acetonitrile + 0.04% formic acid), with gradient elution from 5 to 98% eluent B. HPLC was performed at 60 °C. As determined by HPLC, all compounds were of 95% or of a greater purity. Mass spectra were obtained with a Finnigan MAT 8200 (70 eV) spectrometer with an ESI-MS source: Finnigan MAT 95 for accurate mass determination. The mass spectra (m/z) were recorded using ESI.
Synthetic Protocols
5-Methoxy-2-methyl-[1H]-indole-3-carbaldehyde (4)
To a dried two-neck round-bottom flask, flushed under argon at 0 °C, POCl3 (0.25 mL, 2.68 mmol) was added to a solution of anhydrous dimethylformamide (DMF) (1 mL) and stirred for 10 min before the dropwise addition of a solution of 5-methoxy-2-methyl-[1H]-indole (100 mg, 0.620 mmol) dissolved in anhydrous DMF (2 mL). The reaction mixture was then warmed to room temperature and stirred for an additional 30 min. NaOH (1 N, 5 mL) was added slowly, and the aqueous layer was extracted with dichloromethane (DCM) (2 × 25 mL). The organic phases were combined, dried over Na2SO4, and concentrated under a reduced pressure to yield the corresponding compound (4) as a colorless oil (107 mg, 82%). LC/MS (ESI) m/z: (M + H)+ 190.2. The compound was taken to the next step without further purification.
(E)-3-(5-Methoxy-2-methyl-[1H]-indol-3-yl)-1-(pyridin-4-yl)prop-2-en-1-one (1a)
To a solution of 4 (105 mg, 0.555 mmol) in anhydrous MeOH (3 mL), pyrrolidine (0.101 mL, 1.221 mmol) and 4-acetylpyridine (168 mg, 1.387 mmol) were added. The mixture was then heated at 65 °C for 24 h. As the reaction progressed, a precipitate was formed. When the reaction was complete, the precipitate was collected, washed with ice-cold MeOH (3 × 10 mL), and dried at room temperature for 24 h to yield 1a as a red solid (111.3 mg, 68%): 1H NMR (400 MHz, DMSO-d6): δ 11.90 (s, 1H), 8.85–8.78 (m, 2H), 8.09 (d, J = 15.3 Hz, 1H), 7.98–7.91 (m, 2H), 7.44 (d, J = 2.3 Hz, 1H), 7.41–7.28 (m, 2H), 6.85 (dd, J = 8.7, 2.3 Hz, 1H), 3.87 (s, 3H), 2.58 (s, 3H). 13C NMR (101 MHz, DMSO-d6): δ 188.01, 155.13, 150.56, 145.73, 145.06, 139.54, 130.98, 126.55, 121.37, 112.77, 112.23, 110.86, 109.26, 103.47, 12.13. HPLC (10 min): tR = 3.28 min, HPLC purity 100%. LC/MS (ESI) m/z: (M + H)+ 293.2; HRMS (EI) m/z: 293.12845 [M + H]+ calcd for C18H17O2N2, 293.12845; found, 293.12845.
(E)-3-(5-Ethoxy-2-methyl-1H-indol-3-yl)-1-(pyridin-4-yl)prop-2-en-1-one (2a)
In a 20 mL pressure-release vial, 5-ethoxy-2-methyl-1H-indole-3-carbaldehyde 4a (50 mg, 0.25 mmol) was dissolved in anhydrous MeOH (2 mL), followed by the addition of 4-acetylpyridine (74.5 mg, 0.62 mmol) and piperidine (0.054 mL, 0.54 mmol). The solution was then heated at 65 °C for 24 h. The reaction mixture was diluted with DCM and purified using silica gel flash chromatography (0–100% EtOAc in heptane) to yield an orange solid (9.7 mg, 13%): 1H NMR (400 MHz, DMSO-d6): δ 11.88 (s, 1H), 8.88–8.73 (m, 2H), 8.07 (d, J = 15.3 Hz, 1H), 7.99–7.88 (m, 2H), 7.41 (d, J = 2.3 Hz, 1H), 7.37–7.26 (m, 2H), 6.83 (dd, J = 8.7, 2.3 Hz, 1H), 4.13 (q, J = 7.0 Hz, 2H), 2.57 (s, 3H), 1.37 (t, J = 6.9 Hz, 3H). 13C NMR (101 MHz, DMSO-d6): δ 188.16, 154.40, 145.77, 145.18, 139.63, 131.00, 126.61, 121.43, 112.24, 111.37, 109.29, 104.40, 63.58, 40.15, 39.94, 39.73, 39.52, 39.31, 38.89, 14.91, 12.15. HPLC (5 min): tR = 1.59 min, HPLC purity 98%. LC/MS (ESI) m/z: (M + H)+ 307.1; HRMS (EI) m/z: 306.1368 [M + H]+ calcd for C19H18N2O2, 307.1447; found, 307.1448.
(E)-3-(2-Methyl-5-propoxy-1H-indol-3-yl)-1-(pyridin-4-yl)prop-2-en-1-one (2b)
2-Methyl-5-propoxy-[1H]-indole-3-carbaldehyde (80 mg, 0.37 mmol) was reacted following the protocol described for 2a to yield the corresponding compound as a yellow solid (46.7 mg, 39%): 1H NMR (400 MHz, DMSO-d6): δ 11.89 (s, 1H), 8.85–8.78 (m, 2H), 8.08 (d, J = 15.3 Hz, 1H), 7.96–7.90 (m, 2H), 7.41 (d, J = 2.3 Hz, 1H), 7.37–7.27 (m, 2H), 6.85 (dd, J = 8.7, 2.3 Hz, 1H), 4.04 (t, J = 6.5 Hz, 2H), 2.58 (s, 3H), 1.78 (q, J = 6.8 Hz, 2H), 1.03 (t, J = 7.4 Hz, 3H). 13C NMR (101 MHz, DMSO-d6): δ 188.15, 154.52, 150.54, 145.66, 145.14, 139.59, 130.94, 126.60, 121.37, 112.89, 112.20, 111.38, 109.23, 104.35, 69.59, 22.23, 12.14, 10.53. HPLC (10 min): tR = 4.27 min, HPLC purity 100%. LC/MS (ESI) m/z: (M + H)+ 321.3; HRMS (EI) m/z: 321.15973 [M + H]+ calcd for C20H21O2N2, 321.15975; found, 321.15973.
(E)-3-(5-Isopropoxy-2-methyl-1H-indol-3-yl)-1-(pyridin-4-yl)prop-2-en-1-one (2c)
2-Methyl-5-propoxy-[1H]-indole-3-carbaldehyde (80 mg, 0.37 mmol) was reacted following the protocol described for 2a to yield the corresponding compound as an orange solid (2.0 mg, 3%): 1H NMR (400 MHz, DMSO-d6): δ 11.87 (s, 1H), 8.84–8.79 (m, 2H), 8.06 (d, J = 15.3 Hz, 1H), 7.94–7.89 (m, 2H), 7.41 (d, J = 2.3 Hz, 1H), 7.34–7.25 (m, 2H), 6.83 (dd, J = 8.7, 2.3 Hz, 1H), 4.68 (p, J = 6.1 Hz, 1H), 3.30 (s, 4H), 2.56 (s, 3H), 1.30 (d, J = 6.0 Hz, 6H). HPLC (5 min): tR = 1.74 min; HPLC purity 100%. LC/MS (ESI) m/z: [M + H]+; HRMS (EI) m/z: 321.2 [M + H]+ calcd for C20H20N2O2, 321.1643; found, 321.1635.
(E)-Ethyl 5-Methoxy-3-(3-oxo-3-(pyridin-4-yl)prop-1-en-1-yl)-[1H]-indole-2-carboxylate (2l)
Ethyl 3-formyl-5-methoxy-[1H]-indole-2-carboxylate (50 mg, 0.202 mmol) was reacted following the protocol described for 1a using piperidine (2 equiv) instead of pyrrolidine to yield the desired product as a yellow solid (61.9 mg, 86%): 1H NMR (400 MHz, DMSO-d6): δ 12.5 (s, 1H), 8.84 (d, J = 5.9 Hz, 2H), 8.74 (d, J = 16.0 Hz, 1H), 7.93 (d, J = 5.9 Hz, 2H), 7.65 (d, J = 15.8 Hz, 1H), 7.57–7.47 (m, 2H), 7.09 (d, J = 8.9 Hz, 1H), 4.40 (q, J = 7.1 Hz, 2H), 3.91 (s, 3H), 1.35 (t, J = 7.1 Hz, 3H). 13C NMR (101 MHz, DMSO-d6): δ 189.84, 160.68, 155.73, 150.61, 144.44, 139.46, 125.44, 121.58, 116.56, 115.49, 114.50, 103.09, 61.12, 55.55, 14.09. HPLC (10 min): tR = 4.45 min, HPLC purity 100%. LC/MS (ESI) m/z: 351.3 (M + H)+; HRMS (EI) m/z: 351.13393 [M + H]+ calcd for C20H19O4N2, 351.13393; found, 351.13391.
Sodium (E)-5-Methoxy-3-(3-oxo-3-(pyridin-4-yl)prop-1-en-1-yl)-[1H]-indole-2-carboxylate (2o)
To a solution of 2l (20 mg, 0.057 mmol) in tetrahydrofuran/MeOH (2:1, 3 mL), 4 N NaOH (0.071 mL, 0.285 mmol) was added at room temperature and further stirred for 4 h at 50 °C until completion of the reaction. The volatiles were then evaporated under a reduced pressure and cold diethyl ether (10 mL) was added. The resulting precipitate was collected and dried at room temperature for 24 h to yield the desired compound as a red solid sodium salt (15.9 mg, 77%). 1H NMR (400 MHz, DMSO-d6): δ 9.32 (d, J = 13.7 Hz, 1H), 8.69 (d, J = 5.9 Hz, 2H), 7.77 (d, J = 5.7 Hz, 2H), 7.31 (d, J = 8.5 Hz, 1H), 7.25 (s, 1H), 6.88 (s, 1H), 6.61 (d, J = 7.1 Hz, 1H), 3.81 (s, 3H). 13C NMR (126 MHz, DMSO-d6): δ 189.63, 163.52, 155.44, 150.95, 146.13, 145.58, 130.58, 127.05, 122.02, 115.01, 114.01, 112.69, 111.00, 104.30, 55.97. HPLC (10 min): tR = 3.10 min, HPLC purity >98%. LC/MS (ESI) m/z: 323.2 (M + H)+; HRMS (EI) m/z: 323.10272 [M + H]+ calcd for C18H15O4N2, 323.10263; found, 323.10272.
Lipid Kinase Competition Assay
Lipid kinase competition assay for 22 inositol kinases including clinically relevant mutants were performed using DiscoveRX KINOMEscan profiling. Briefly, streptavidin-coated magnetic beads were treated with biotinylated positive control compounds (compounds that specifically bind each kinase) to generate affinity resins. Binding reactions were performed by combining DNA-tagged kinases, liganded affinity beads, and test compounds. After binding, DNA-tagged kinases were eluted and each kinase concentration in the eluates was measured by qPCR of DNA tag on each kinase.
Cell Viability Assay
Cells were plated into each well of Greiner 384-well plates at a density of 1500 cells/well. The cells were incubated with increasing concentration of compounds (0.1% v/v DMSO) for 72 h, and cell viability was measured by an EnVison plate reader (PerkinElmer) with CellTiter-Glo (Promega).
Cell Morphology
For acquisition of phase contrast images of live cells, HCT116 cells were seeded on a 384-well high-content imaging microplate. Next day, the compounds were added and images were obtained at the indicated time after the addition of compounds using an EVOS FL Cell Imaging System (Life Technologies, Paisley, UK).
PIKFYVE Inhibition Assay
The kinase inhibition assay was performed using an ADP-Glo system (Promega) according to the manufacturer protocol. Briefly, the kinase reaction was performed by adding the active PIKFYVE protein (SignalChem), lipid substrate PI(3)P (SignalChem), protein kinase C lipid activator (SignalChem), and 50 μM ATP (SignalChem) in kinase buffer (25 mM MOPS (pH 7.2), 12.5 mM β-glycerol-phosphate, 25 mM MgCl2, 5 mM egtazic acid, 2 mM ethylenediaminetetraacetic acid, and 0.25 mM dithiothreitol). The reaction was terminated by adding the ADP-Glo reagent (Promega). The ATP–ADP conversion was quantitated by adding the kinase detection reagent (Promega) and reading the luminescence signal with the plate reader (EnVision).
PIKFYVE KnockOut
Pooled PIKFYVE-/- cells were generated using CRISPR-Cas9-mediated knockout. HCT116 cells stably expressing Cas9 were transduced with lentivirus containing sgRNA against PIKFYVE. sgRNA with a scrambled sequence was used as a control.
Acknowledgments
We thank the teams of High-Throughput Biology and Screening Informatics (Novartis, Cambridge, USA) for the collaboration and support.
Glossary
Abbreviations
- SAR
structure–activity relationship
- MOMIPP
3-(5-methoxy-2-methyl-1H-indol-3-yl)-1-(4-pyridinyl)-2-propene-1-one
- CRISPR
clustered regularly interspaced short palindromic repeats
- sgRNA
single-guide RNA
- TLR
toll-like receptor
- qPCR
quantitative PCR
- CD
Crohn’s disease
- RA
rheumatoid arthritis
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00202.
Synthetic protocol scheme, 1H NMR spectra, HPLC spectra, and HRMS spectra of compounds 1a, 2a, 2b, 2c, 2l, and 2o (PDF)
Author Present Address
§ Department of Pharmacology, Merck Research Laboratories, 33 Avenue Louis Pasteur, Boston, Massachusetts 02115, USA (H.C.).
Author Present Address
∥ Department of Scientific Computing, Vertex Pharmaceuticals, 50 Northern Avenue, Boston, Massachusetts 02210, USA (E.G.).
Author Contributions
H.C. designed and led the study under the supervision of J.L.J. and M.H. M.P. and A.R. synthesized the compounds under the supervision of R.M. R.M. and E.G. contributed to the study design. H.C. wrote the manuscript with input from all authors. All authors gave approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): The authors (H.C., E.G., R.M., M.H., and J.L.J.) were employees of Novartis during the time of this study. The research was fully funded by the Novartis Institutes for Biomedical Research.
Supplementary Material
References
- Maltese W. A.; Overmeyer J. H. Methuosis: Nonapoptotic Cell Death Associated with Vacuolization of Macropinosome and Endosome Compartments. Am. J. Pathol. 2014, 184, 1630–1642. 10.1016/j.ajpath.2014.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmeyer J. H.; Kaul A.; Johnson E. E.; Maltese W. A. Active Ras Triggers Death in Glioblastoma Cells through Hyperstimulation of Macropinocytosis. Mol. Cancer Res. 2008, 6, 965–977. 10.1158/1541-7786.mcr-07-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S.; Kitanaka C.; Noguchi K.; Mochizuki T.; Nagashima Y.; Shirouzu M.; Fujita H.; Yoshida M.; Chen W.; Asai A.; Himeno M.; Yokoyama S.; Kuchino Y. Oncogenic Ras Triggers Cell Suicide through the Activation of a Caspase-Independent Cell Death Program in Human Cancer Cells. Oncogene 1999, 18, 2281–2290. 10.1038/sj.onc.1202538. [DOI] [PubMed] [Google Scholar]
- Bhanot H.; Young A. M.; Overmeyer J. H.; Maltese W. A. Induction of Nonapoptotic Cell Death by Activated Ras Requires Inverse Regulation of Rac1 and Arf6. Mol. Cancer Res. 2010, 8, 1358–1374. 10.1158/1541-7786.mcr-10-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. W.; Overmeyer J. H.; Young A. M.; Erhardt P. W.; Maltese W. A. Synthesis and Evaluation of Indole-Based Chalcones as Inducers of Methuosis, a Novel Type of Nonapoptotic Cell Death. J. Med. Chem. 2012, 55, 1940–1956. 10.1021/jm201006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabbic C. J.; Dietsch H. M.; Alexander E. M.; Nagy P. I.; Robinson M. W.; Overmeyer J. H.; Maltese W. A.; Erhardt P. W. Differential Induction of Cytoplasmic Vacuolization and Methuosis by Novel 2-Indolyl-Substituted Pyridinylpropenones. ACS Med. Chem. Lett. 2014, 5, 73–77. 10.1021/ml4003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabbic C. J.; Overmeyer J. H.; Alexander E. M.; Crissman E. J.; Kvale H. M.; Smith M. A.; Erhardt P. W.; Maltese W. A. Synthesis and Biological Evaluation of Indolyl-Pyridinyl-Propenones Having Either Methuosis or Microtubule Disruption Activity. J. Med. Chem. 2015, 58, 2489–2512. 10.1021/jm501997q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabbic C. J.; George S. M.; Alexander E. M.; Du S.; Offenbacher J. M.; Crissman E. J.; Overmeyer J. H.; Maltese W. A.; Erhardt P. W. Synthesis and Biological Evaluation of Isomeric Methoxy Substitutions on Anti-Cancer Indolyl-Pyridinyl-Propenones: Effects on Potency and Mode of Activity. Eur. J. Med. Chem. 2016, 122, 79–91. 10.1016/j.ejmech.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billcliff P. G.; Lowe M. Inositol Lipid Phosphatases in Membrane Trafficking and Human Disease. Biochem. J. 2014, 461, 159–175. 10.1042/bj20140361. [DOI] [PubMed] [Google Scholar]
- De Craene J.-O.; Bertazzi D. L.; Bär S.; Friant S. Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 634. 10.3390/ijms18030634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltese W. A.; Overmeyer J. H. Non-Apoptotic Cell Death Associated with Perturbations of Macropinocytosis. Front. Physiol. 2015, 6, 38. 10.3389/fphys.2015.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton L. M.; Ikonomov O. C.; Sbrissa D.; Garg P.; Shisheva A. Active Vacuolar H + ATPase and Functional Cycle of Rab5 Are Required for the Vacuolation Defect Triggered by PtdIns(3,5)P 2 Loss under PIKfyve or Vps34 Deficiency. Am. J. Physiol.: Cell Physiol. 2016, 311, C366–C377. 10.1152/ajpcell.00104.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X.; Xu Y.; Cheung A. K.; Tomlinson R. C.; Alcázar-Román A.; Murphy L.; Billich A.; Zhang B.; Feng Y.; Klumpp M.; Rondeau J.-M.; Fazal A. N.; Wilson C. J.; Myer V.; Joberty G.; Bouwmeester T.; Labow M. A.; Finan P. M.; Porter J. A.; Ploegh H. L.; Baird D.; De Camilli P.; Tallarico J. A.; Huang Q. PIKfyve, a Class III PI Kinase, Is the Target of the Small Molecular IL-12/IL-23 Inhibitor Apilimod and a Player in Toll-like Receptor Signaling. Chem. Biol. 2013, 20, 912–921. 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisheva A. PIKfyve: Partners, Significance, Debates and Paradoxes. Cell Biol. Int. 2008, 32, 591–604. 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.