Abstract
The genomic complexity of profound copy-number aberration has prevented effective molecular stratification of ovarian cancers. To decode this complexity, we derived copy-number signatures from shallow whole genome sequencing of 117 high-grade serous ovarian cancer (HGSOC) cases, which were validated on 527 independent cases. We show that HGSOC comprises a continuum of genomes shaped by multiple mutational processes that result in known patterns of genomic aberration. Copy-number signature exposures at diagnosis predict both overall survival and the probability of platinum-resistant relapse. Measuring signature exposures provides a rational framework to choose combination treatments that target multiple mutational processes.
Introduction
The discrete mutational processes that drive copy-number change in human cancers are not readily identifiable from genome-wide sequence data. This presents a major challenge for the development of precision medicine for cancers that are strongly dominated by copy-number changes, including high-grade serous ovarian (HGSOC), esophageal, non-small-cell lung and triple negative breast cancers1. These tumors have low frequency of recurrent oncogenic mutations, few recurrent copy number alterations, and highly complex genomic profiles2.
HGSOCs are poor prognosis carcinomas with ubiquitous TP53 mutation3. Despite efforts to discover new molecular subtypes and targeted therapies, overall survival has not improved over two decades4. Current genomic stratification is limited to defining homologous recombination-deficient (HRD) tumors5–7 with approximately 20% HGSOC cases having a germline or somatic mutation in BRCA1/2 with smaller contributions from mutation or epigenetic silencing of other HR genes8. Classification using gene expression predominantly reflects the tumor microenvironment and is reliable in only a subset of patients9–11. Detailed genomic analysis using whole genome sequencing has shown frequent loss of RB1, NF1 and PTEN by gene breakage events12 and enrichment of amplification associated fold-back inversions in non-HRD tumors13. However, none of these approaches has provided a broad mechanistic understanding of HGSOC, reflecting the challenges of detecting classifiers in extreme genomic complexity.
Recent algorithmic advances have enabled interpretation of complex genomic changes by identifying mutational signatures — genomic patterns that are the imprint of mutagenic processes accumulated over the lifetime of a cancer cell14. For example, UV exposure or mismatch repair defects induce distinct, detectable single nucleotide variant (SNV) signatures14. The clinical utility of these signatures has recently been demonstrated through a combination of structural variant (SV) and SNV signatures to improve the prediction of HRD15. Importantly, these studies show that tumor genomes are shaped by multiple mutational processes and novel computational approaches are needed to identify coexistent signatures. We hypothesized that specific features of copy-number abnormalities could represent the imprints of distinct mutational processes, and developed methods to identify signatures from copy-number features in HGSOC.
Results
Experimental design and data collection
We generated absolute copy number profiles from 253 primary and relapsed HGSOC samples from 132 patients in the BriTROC-1 cohort16 using low-cost shallow whole-genome sequencing (sWGS; 0.1×) and targeted amplicon sequencing of TP53 (Supplementary Figure 1). These samples formed the basis of our copy-number signature identification. A subset of 56 of these cases had deep whole-genome sequencing (dWGS) performed for mutation analysis and comparison with sWGS data. Independent data sets for validation included 112 dWGS HGSOC cases from PCAWG17 and 415 HGSOC cases with SNP array and whole exome sequence from TCGA8. Supplementary Figure 1a shows the REMARK diagram for selection of BriTROC-1 patients. Supplementary Figure 1b outlines which samples were used in each analysis across the three cohorts. Clinical data for the BriTROC-1 cohort are summarized in Supplementary Table 1 and Supplementary Figure 2. Detailed information on experimental design is provided in the Life Sciences Reporting Summary.
Identification and validation of copy-number signatures
To identify copy-number (CN) signatures, we computed the genome-wide distributions of six fundamental CN features for each sample: the breakpoint count per 10MB, the copy-number of segments, the difference in CN between adjacent segments, the breakpoint count per chromosome arm, the lengths of oscillating CN segment chains and the size of segments. These features were selected as hallmarks of previously reported genomic aberrations, including breakage-fusion-bridge cycles18, chromothripsis19 and tandem duplication20,21.
We applied mixture modelling to separate the copy-number feature distributions from 91 BriTROC-1 samples with high quality CN profiles into mixtures of Poisson or Gaussian distributions. This resulted in a total of 36 mixture components (Figure 1a). For each sample, the posterior probability of copy-number events arising from these components was computed and summed. These sum-of-posterior vectors were then combined to form a sample-by-component sum-of-posteriors matrix. To identify copy-number signatures, this matrix was subjected to non-negative matrix factorization (NMF)22, a method previously used for deriving SNV signatures14.
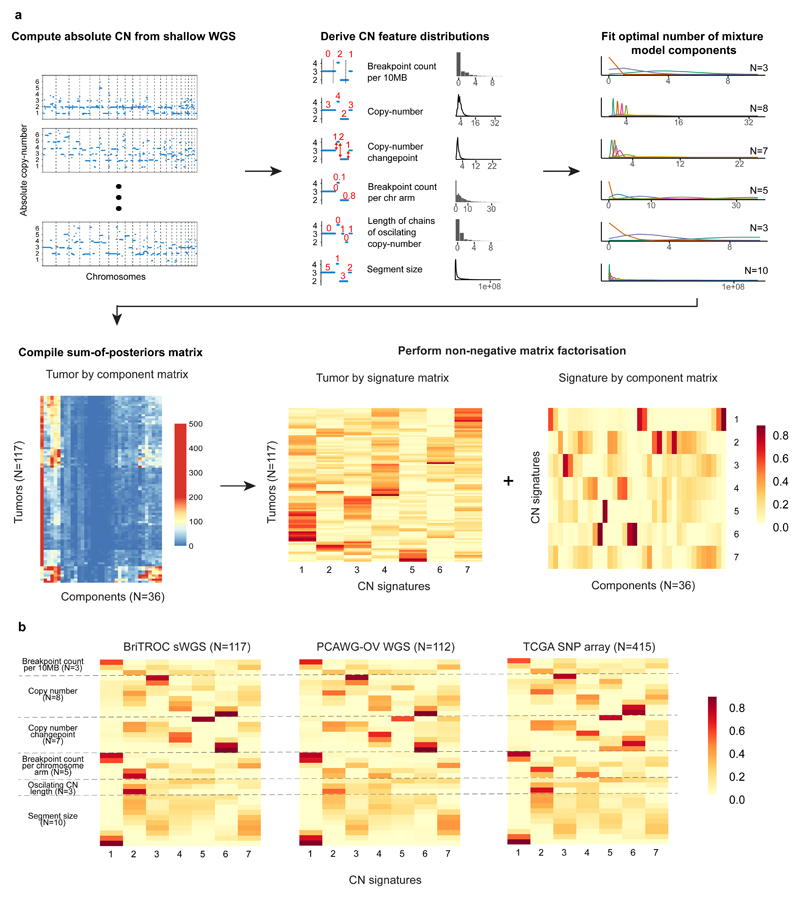
Figure 1. Copy-number signature identification from shallow whole genome sequence data and validation in independent cohorts.
a. Step 1: Absolute copy-numbers are derived from sWGS data; Step 2: genome-wide distributions of six fundamental copy-number features are computed; Step 3: Gaussian or Poisson mixture models (depending on data type) are fitted to each distribution and the optimal number of components is determined (ranging from 3–10) ; Step 4: the data are represented as a matrix with 36 mixture component counts per tumor. Step 5: Non-negative matrix factorization is applied to the components-by-tumor matrix to derive the tumor-by-signature matrix and the signature-by-components matrix.
b. Heat maps show component weights for copy number signatures in two independent cohorts of HGSOC samples profiled using WGS and SNP array. Correlation coefficients are provided in Supplementary Table 2.
NMF identified seven CN signatures (Figure 1a), as well as their defining features and exposures in each sample. The optimal number of signatures was chosen using a consensus from 1000 initializations of the algorithm and 1000 random permutations of the data combining four model selection measures (Supplementary Figure 3). We found highly similar component weights for the signatures in the two independent cohorts (PCAWG-OV and TCGA), demonstrating the robustness of both the methodology and the copy-number features (Figure 1b, P<9e-05, median r=0.86. Supplementary Table 2), despite a significant difference in exposures to CN signatures 2, 3, 4 and 5 between the cohorts (P<0.05, two-sided Wilcoxon rank sum test, Supplementary Figure 4).
Mutational processes underlying copy-number signatures
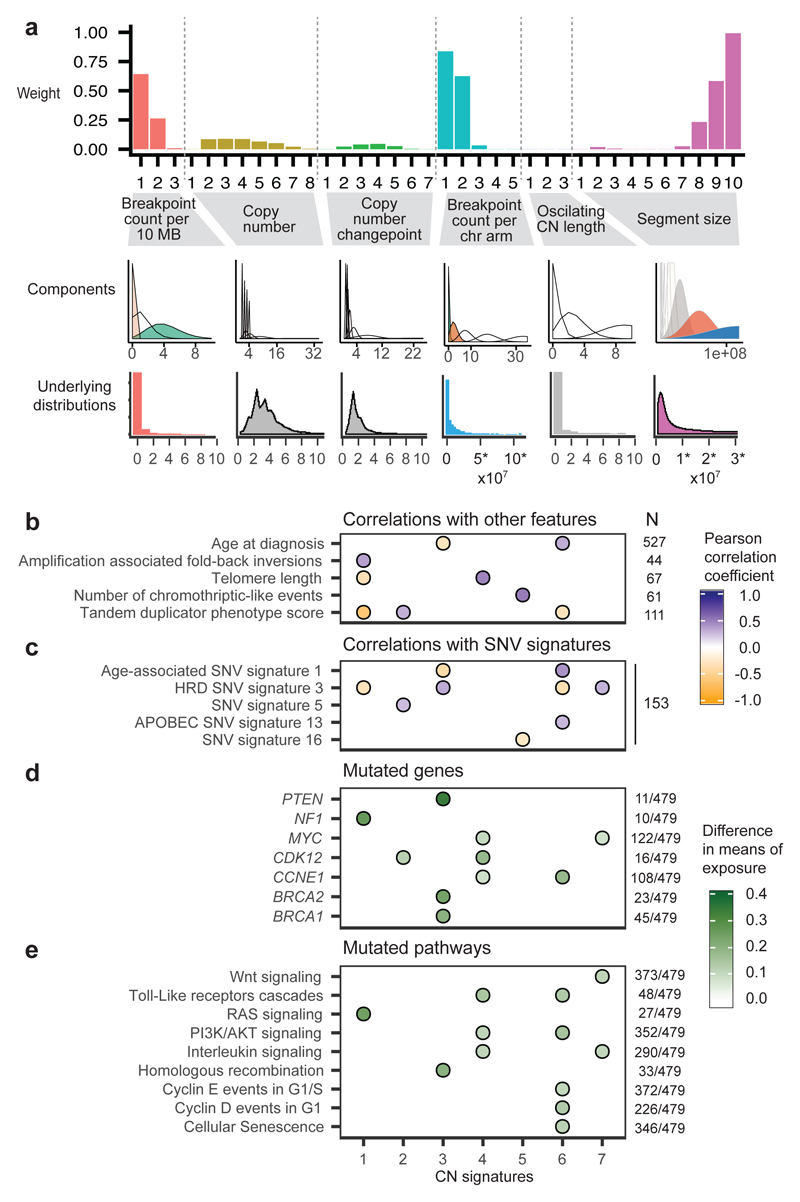
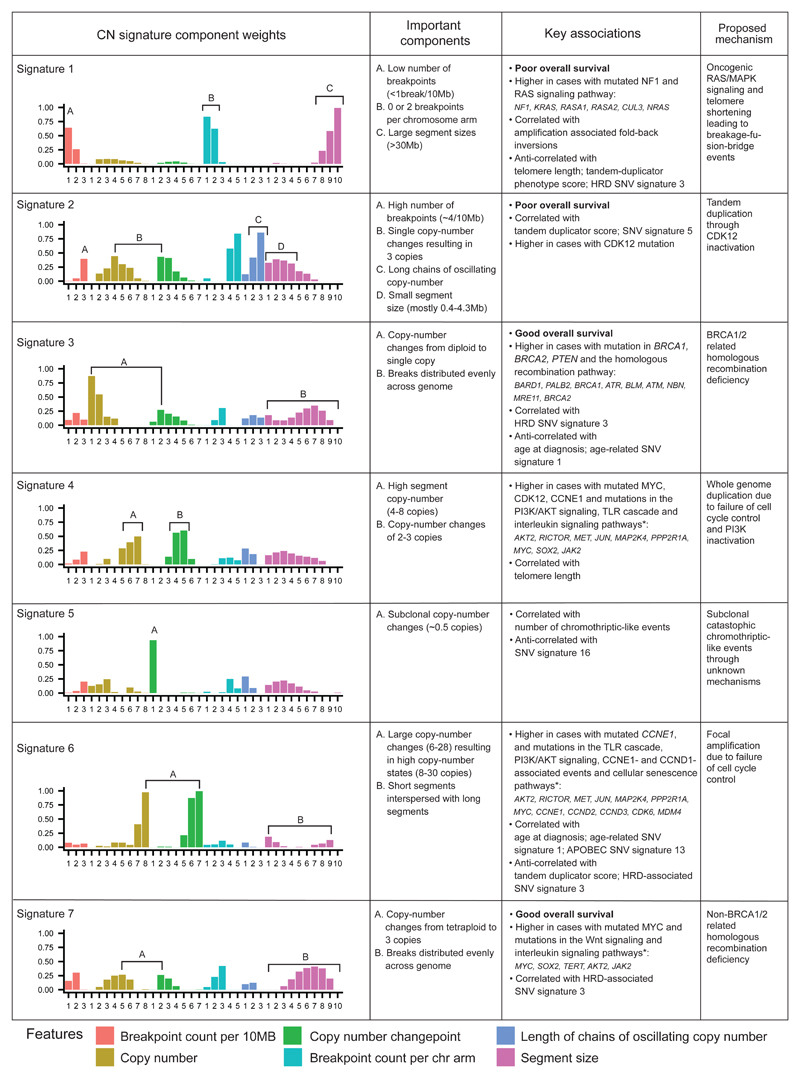
The majority of cases analysed exhibited multiple signature exposures suggesting that HGSOC genomes are shaped by more than one mutational process. As our signature analysis reduced this genomic complexity into its constituent components, we were able to link the individual copy-number signatures to their underlying mutational processes. To do this, we used the component weights identified by NMF to determine which pattern of global or local copy-number change defined each signature. For example, for CN signature 1, the highest weights were observed for components representing low numbers of breakpoints per 10MB, long genomic segments and two breaks occurring per chromosome arm (Figure 2a, Supplementary Figure 5). Two breaks per chromosome arm suggested that the mutational process underlying this signature might be breakage-fusion-bridge (BFB) events18.
Figure 2. Linking copy-number signatures with mutational processes.
a Component weights for copy number signature 1. Barplots (upper panel) are grouped by copy number feature and show weights for each of the 36 components. The middle panel shows the mixture model distributions which are shaded by the component weight - solid colours have a high weight and transparent have low weight (contrasting colours are randomly assigned). Lower panel shows genome-wide distribution (histogram or density) of each copy number feature, across the BriTROC-1 cohort, with coloured plots indicating important distributions (> 0.1 component weight). (Note: similar plots for other CN signatures are shown in Figure 3 and Supplementary Figure 5).
b Associations between CN signature exposures and other features. Purple indicates positive correlation and orange negative correlation (see also Supplementary Figure 6). Numbers at the right of the panel indicate cases included in each analysis. Only significant correlations are shown (P<0.05).
c Associations between CN signature exposures and SNV signatures. Purple indicates positive correlation and orange negative correlation (see also Supplementary Figure 6). The number at the right of the panel indicates cases included in the analysis.
d and e Difference in CN signature exposures between cases with mutations in specific genes (d) and mutated/wildtype reactome pathways (e). The absolute difference in mean signature exposures was calculated for cases with and without mutations. Colors in filled circles indicate extent of difference. Only differences with FDR P<0.05 (Mann-Whitney test) are shown (see also Supplementary Figure 7).
Numbers at the right of the panel indicate cases with mutations (SNVs, amplifications or deletions) in each gene/pathway.
To test this hypothesis, we correlated CN signature 1 exposures with mutation data, SNV signatures, and other measures derived from deep WGS and exome sequencing (Figure 2b-e, Supplementary Figures 6, 7, 8 and 9, Supplementary Tables 3, 4, 5, 6, 7 and 8). CN signature 1 was anti-correlated with sequencing estimates of telomere length (r=-0.32, P=0.009), consistent with BFB events. In addition, CN signature 1 was positively correlated with amplification-associated fold-back inversion structural variants (r=0.36, P=0.02), which have been strongly implicated in BFB events23 and have also been associated with inferior survival in HGSOC13. CN signature 1 was also enriched in cases with oncogenic RAS signaling, including NF1 loss and mutated KRAS (p=5e-06, Mann-Whitney test), which has previously been shown to induce chromosomal instability as a result of aberrant G2 and mitotic checkpoint controls and missegregation24,25. Taken together, these data provide independent evidence for BFB arising as a result of oncogenic RAS signaling and telomere shortening as the underlying mechanism for CN signature 1.
We applied these approaches to the remaining signatures to identify statistically significant genomic associations using a false discovery rate <0.05 (Figure 2b-e, Figure 3, Supplementary Figures 5, 6, 7, 8 and 9, Supplementary Tables 3, 4, 5, 6, 7 and 8).
Figure 3. The seven copy-number signatures in HGSOC.
Description of the defining component weights, key associations and proposed mechanisms for the seven copy number signatures.
*only the top three mutated genes for each of the pathways associated with CN signatures 4, 6 and 7 are shown (the list of all significant genes is provided in Supplementary Tables 7 and 8).
CN signature 2 showed frequent breakpoints per 10MB, single changes in copy-number (resulting in 3 copies), chains of oscillating copy-number, and was significantly correlated with tandem duplicator phenotype scores (r=0.3, P=0.004) and SNV signature 5 (r=0.26, P=0.02). In addition, this signature was enriched in patients with mutations in CDK12 (P=0.02, Mann-Whitney test, Supplementary Table 6), in keeping with previous studies that have demonstrated large tandem duplication in cases with inactivating CDK12 mutations26.
CN signature 4 was characterised by high copy-number states (4-8 copies) and predominant copy-number change-points of size 2. This pattern indicates a mutational process of late whole-genome duplication (WGD)27. Significantly increased signature 4 exposure in cases with aberrant PI3K/AKT signaling provided further support for late WGD as oncogenic PIK3CA induces tolerance to genome doubling28 (P=2e-22, Mann-Whitney test, mutation of PIK3CA or amplification of AKT, EGFR, MET, FGFR3 and ERBB2). Signature 4 was also seen at higher levels in cases with mutations in genes encoding proteins from Toll-like receptor signaling cascades (P=2e-07), interleukin signaling pathways (P=3e-24) and CDK12 (P=0.0009), as well as those with amplified CCNE1 (P=2e-10) and MYC (P=9e-12). It was also significantly correlated with telomere length (r=0.46, P=4e-05).
CN signature 6 showed extremely high copy-number states and high copy-number change-points for small segments interspersed among larger, lower-copy segments. This suggests a mutational process resulting in focal amplification. Increased signature 6 exposure was associated with mutations in genes encoding proteins across diverse pathways, including aberrant G1/S cell cycle checkpoint control (through either amplification of CCNE1, CCND1, CDK2, CDK4 or MYC, deletion/inactivation of RB1 or mutation in CDK12), Toll-like receptor signaling cascades and PI3K/AKT signaling (P<0.05). However, as many of these statistical associations are marked by gene amplification, it is difficult to determine whether the copy number states represent causal events or are simply a consequence of focal amplification. Exposure to CN signature 6 was also positively correlated with age at diagnosis (r=0.31, P=6e-12) and age-related SNV signature 114 (r=0.43, P=3e-06).
CN signature 5 was significantly associated with predicted chromothriptic-like events using the Shatterproof algorithm29 (r=0.44, P=2e-03). Chromothripsis is considered rare in HGSOC12,27,30. However, the key component of this signature—the presence of copy-number change points centered at 0.5 copies—suggests that the events are subclonal. This implies that chromothripsis may be an underestimated oncogenic mechanism in HGSOC that could reflect ongoing formation and rupture of micronuclei31.
CN signature 3 was characterized by an even distribution of breaks across all chromosomes, and copy number changes from diploid to single copy (LOH). CN signature 3 was significantly enriched in cases with mutations in BRCA1 and BRCA2, and other HR genes including BARD1, PALB2 and ATR (P=0.002, Mann-Whitney test). It was also correlated with the HRD-related SNV signature 3 (r=0.32, P=0.002) and anti-correlated with age at diagnosis and age-related SNV signature 1 (P<0.05). CN signature 3 was also enriched in cases with loss of function mutations in PTEN (P=0.002, Mann-Whitney test). Taken together, these data suggest that CN signature 3 is driven by BRCA1/2-related HRD mechanisms.
CN signature 7, like CN signature 3, also demonstrated an even distribution of breaks across all chromosomes. By contrast with CN signature 3, single copy-number changes were observed from a tetraploid rather than a diploid state (Figure 3). Although there was correlation with the HRD-related SNV signature 3, there was no enrichment with BRCA1/2 mutation, suggesting alternative HRD mechanisms as potential mutational processes.
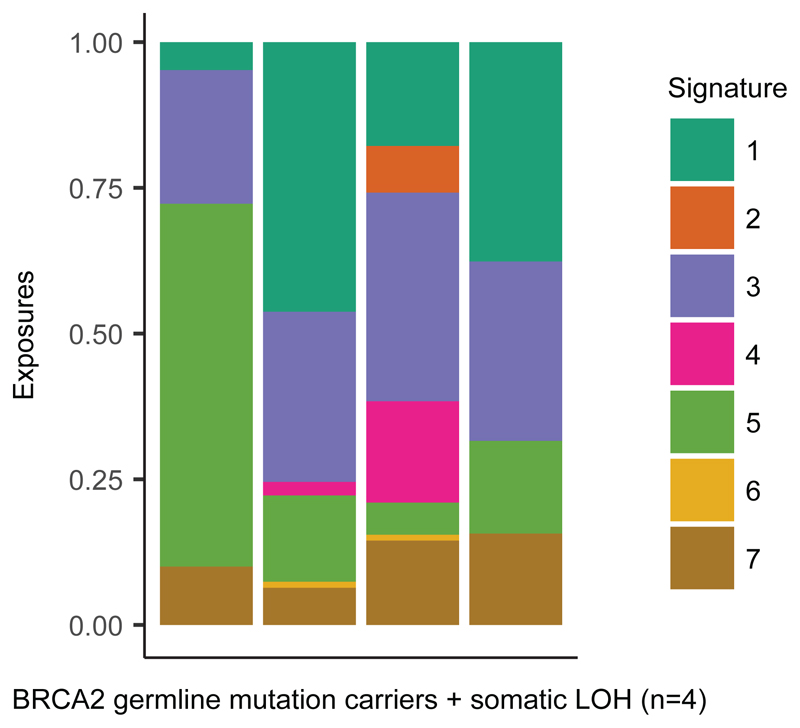
We also investigated relationships between CN signatures. BRCA1 dysfunction and CCNE1 amplification have been shown to be mutually exclusive in HGSOC32, and we observed that CN signature 3 (BRCA1/2 HRD) and CN signature 6 (marked by aberrant G1/S cell cycle checkpoint control) showed mutually exclusive associations (Figure 2b-e). Loss of BRCA1 and BRCA2 are early driver events in HGSOC, and to investigate acquisition of additional mutational processes, we studied four BriTROC-1 cases with deleterious germline BRCA2 mutations and confirmed somatic loss of heterozygosity at BRCA2 (Figure 4). A diverse and variable number of CN signatures was seen in these cases, including substantial exposures to CN signature 1 (RAS signaling) in three of the four cases.
Figure 4. CN signature exposures of four BriTROC-1 patients with germline BRCA2 mutations and somatic loss of heterozygosity.
Stacked bar plots show copy-number signature exposures for four BriTROC-1 cases with pathogenic germline BRCA2 mutations and confirmed somatic loss of heterozygosity (LOH) at the BRCA2 locus.
Copy-number signatures predict overall survival
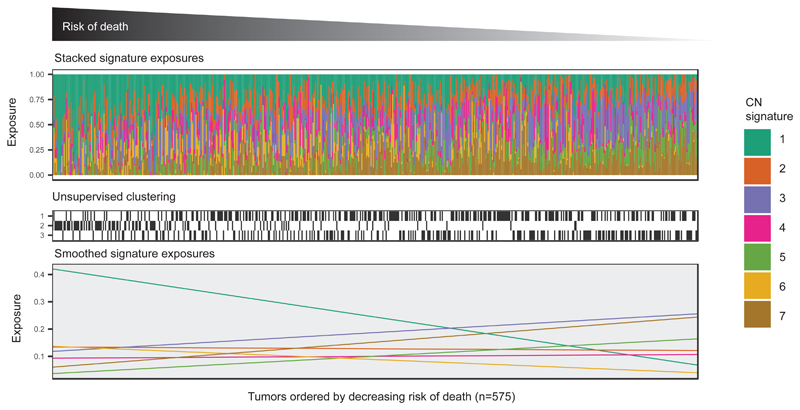
We next explored the association between individual CN signature exposures and overall survival using a combined dataset of 575 diagnostic samples with clinical outcomes. We trained a multivariate Cox proportional hazards model on 417 cases and tested this on the remaining 158 cases (Figure 5, Supplementary Table 9). CN signature exposure was significantly predictive of survival (Training: P=0.002, log-rank test; stratified by age and cohort; Test: P=0.05, C-index=0.56, 95% CI:0.50-0.62; Entire cohort: P=0.002, log-rank test; stratified by age and cohort). Across the entire cohort, poor outcome was significantly predicted by CN signature 1 (P=0.0008) and CN signature 2 exposures (P=0.03), whilst good outcome was significantly predicted by exposures to CN signatures 3 (P=0.05) and 7 (P=0.006).
Figure 5. Association of survival with copy-number signatures.
Upper panel: Stacked barplots show CN signature exposures for each patient. Patients were ranked by risk of death estimated by a multivariate Cox proportional hazards model stratified by age and cohort, with CN signature exposures as covariates.
Middle panel: The matrix indicates group for each patient assigned by unsupervised clustering of CN signature 1, 2, 3 and 7 exposures (see also Supplementary Figure 10).
Lower panel: Linear fit of signature exposures ordered by risk predicted by the Cox proportional hazards model.
Unsupervised hierarchical clustering of samples by signature exposures identified three clusters (Figure 5). Despite showing significant survival differences (P=0.004, log-rank test; stratified by age and cohort), these clusters did not provide any prognostic information in addition to that identified from the Cox proportional hazards model; cluster 2 was dominated by patients with high signature 1 exposures (poor prognosis), cluster 3 showed high signature 3 exposures (good prognosis) and cluster 1 had mixed signature exposures (Supplementary Figure 10).
Copy-number signatures indicate relapse following chemotherapy
Using a generalised linear model, we investigated whether copy-number signatures could be used to predict outcome following chemotherapy across 36 patients from the BriTROC-1 study with paired diagnostic and relapse samples16. The model showed CN signature 1 exposures at the time of diagnosis to be significantly predictive of platinum-resistant relapse (P=0.02, z-test, Supplementary Table 10).
Using the same 36 sample pairs, we also investigated whether chemotherapy treatment changed CN signature exposures. No significant effects on exposures were observed following chemotherapy treatment using a linear model that accounted for signature exposure at time of diagnosis, number of lines of chemotherapy and patient age (P>0.05, F-test, Supplementary Table 10). The only variable showing a significant association with exposure at relapse was signature exposure at diagnosis (P<0.01, F-test, Supplementary Table 11).
Discussion
Copy-number signatures provide a framework that is able to rederive the major defining elements of HGSOC genomes, including defective HR8, amplification of CCNE19 and amplification-associated fold-back inversions13. In addition, the CN signatures show significant associations with known driver gene mutations in HGSOC and provide the ability to detect novel associations with gene mutations. We derived signatures using inexpensive shallow whole genome sequencing of DNA from core biopsies. These approaches are rapid and cost effective, thus providing a clear path to clinical implementation. Copy-number signatures open new avenues for clinical trial design by highlighting contributions from underlying mutational processes that depend on oncogenic RAS and PI3K/AKT signaling.
We found that almost all patients with HGSOC demonstrated a mixture of signatures indicative of combinations of mutational processes. These results suggest that early TP53 mutation, the ubiquitous initiating event in HGSOC, may permit multiple mutational processes to co-evolve, potentially simultaneously. Although further work is needed to define the precise timing of signature exposures, early driver events such as BRCA2 mutation still permit a diverse and variable number of CN signatures in addition to an HRD signature (Figure 4). These additional signature exposures may alter the risk of developing therapeutic resistance, particularly when only a single mutational process such as HRD is targeted.
High exposure to CN signature 3, characterised by BRCA1/2-related HRD, is associated with improved overall survival, confirming prior data showing that BRCA1/2 mutation is associated with long survival in HGSOC33,34. Conversely, high exposure to signature 1, which is characterised by oncogenic RAS signaling (including NF1, KRAS and NRAS mutation), predicts subsequent platinum-resistant relapse and poor survival. This suggests that powerful intrinsic resistance mechanisms are present at the time of diagnosis and can be readily identified using CN signature analysis. This hypothesis is supported by the presence of exposure to CN signature 1 in germline BRCA2-mutated cases (Figure 4) as well as our previous work demonstrating the expansion of a resistant subclonal NF1-deleted population following chemotherapy treatment in HGSOC35 and poor outcomes in Nf1-deleted murine models of HGSOC36. Our CN signature analysis of BRCA2-mutated cases also concurs with PCAWG/ICGC data showing that over half (9/16) of NF1-mutated cases also harboured mutations in BRCA1 or BRCA212. These data suggest a complex interplay between RAS signaling and HRD. Thus, RAS signaling may be an important target, especially in first line treatment, to prevent emergence of platinum-resistant disease.
We found that CN signature exposures were not significantly altered between diagnosis and disease relapse in 36 sample pairs with a median interval of 30.6 months16. This suggests that the underlying mutational processes in HGSOC are relatively stable and that genome-wide patterns of copy-number change mainly reflect historic alterations to the genome acquired during tumorigenesis37. Relative invariant genomic changes were also observed in the ARIEL2 trial, where genome-wide loss-of-heterozygosity was used to predict HRD, and only 14.5% (17/117) cases changed LOH status between diagnosis and relapse7.
Larger association studies will be required to further refine CN signature definitions and interpretation. The application of our approach to other tumour types is likely to extend the set of signatures beyond the robust core set identified here. Basal-like breast cancers, squamous cell and small cell lung carcinoma, which all have high rates of TP53 mutation and genomic instability2, are promising next targets. Although it is likely that the strong associations have identified the driver mutational processes for CN signatures 1 and 3, functional studies will be required to establish causal links for the remaining signatures. For example, CN signature 6 was significantly associated with multiple mutated pathways, and this association was primarily driven by amplification of target genes. As this signature represented focal amplification events, it is difficult to determine whether amplification of specific genes drives the underlying mutational process or the amplifications emerge as a consequence of strong selection of advantageous phenotypes. Our data does not provide timing information for exposures and there is the real possibility that one mutational process may well drive the emergence of other mutational processes. For example, the association between signature 6 and PI3K signalling is also shared with signature 4.
Other limitations of this work are technical: we integrated data from three sources, using three different pre-processing pipelines, and the ploidy determined by different pipelines can have a significant effect on the derived signatures. For example, high-ploidy CN signature 4 was predominantly found in the sequenced samples that underwent careful manual curation to identify whole-genome duplication events. When extending to larger sample sets, a unified processing strategy with correct ploidy determination is likely to produce improved signature definitions. Another technical limitation is the resolution of copy-number calling from sWGS (limited to 30kb bins) and future application to large cohorts of deeply sequenced samples will be needed to improve the resolution of the CN signatures.
Efforts to identify discrete, clinically relevant subtypes of disease have been successful in many cancer types38–40. However, HGSOC lacks clinically-relevant patient stratification, which is reflected in continued poor survival. We show that HGSOC genomes are shaped by multiple mutational processes that preclude simple subtyping. Thus, our results suggest that HGSOC is a continuum of genomes. By dissecting the mutational forces shaping HGSOC genomes, our study paves the way to understanding extreme genomic complexity, as well as revealing the evolution of tumors as they relapse and acquire resistance to chemotherapy.
Online Methods
Patients and samples
The BriTROC-1 study has been described previously16. Characteristics of the 142 patients included in this study are given in Supplementary Table 1. The study is sponsored by NHS Greater Glasgow and Clyde and ethics/IRB approval was given by Cambridge Central Research Ethics Committee (Reference 12/EE/0349). The study enrolled patients with recurrent ovarian high-grade serous or grade 3 endometrioid carcinoma who had relapsed following at least one line of platinum-based chemotherapy and whose disease was amenable either to image-guided biopsy or secondary debulking surgery. At study entry, patients were classified as having either platinum-sensitive relapse (i.e. relapse six months or more following last platinum chemotherapy) or platinum-resistant relapse (i.e. relapse less than six months following prior platinum chemotherapy) (Supplementary Figure 2). All patients provided written informed consent. Access to archival diagnostic formalin-fixed tumor was also required. Survival was calculated from the date of enrolment to the date of death or the last clinical assessment, with data cutoff at 1 December 2016. At subsequent relapse or progression after chemotherapy following study entry, patients could optionally have a second biopsy under separate consent.
DNA was extracted from 300 samples of 142 patients - 158 methanol-fixed relapse biopsies and 142 FFPE archival diagnostic tissues. Germline DNA was extracted from blood samples of 137 patients.
Tagged-amplicon sequencing
Mutation screening of TP53, PTEN, EGFR, PIK3CA, KRAS and BRAF was performed on all 300 samples using tagged-amplicon sequencing as previously described16. DNA extracted from blood was analyzed by tagged-amplicon sequencing for BRCA1 and BRCA2 germline mutations.
Shallow whole genome sequencing (sWGS)
Libraries for sWGS were prepared from 100ng DNA using modified TruSeq Nano DNA LT Sample Prep Kit (Illumina) protocol41. Quality and quantity of the libraries were assessed with DNA-7500 kit on 2100 Bioanalyzer (Agilent Technologies) and with Kapa Library Quantification kit (Kapa Biosystems) according to the manufacturer's protocols. Sixteen to twenty barcoded libraries were pooled together in equimolar amounts and each pool was sequenced on HiSeq4000 in SE-50bp mode.
Prior to sequencing we estimated the required sequencing depth by adapting calculations made in previous work that explored the relationship between sequencing depth (reads per sample) and copy number calling accuracy42. Based on these analyses, we devised a power calculator for sWGS copy number analysis (see URL 1, described in 43). We estimated that with an average ploidy of 3 and purity of 0.65, a sequencing depth of at least 2.7 million reads is required to detect single, clonal copy-number changes (minimum 60kb) at 90% power and alpha 0.05. After analysis we determined that BritROC 3-star samples had an average purity of 0.66, ploidy of 2.7, and were sequenced to an average depth of 8.6 million reads. This allowed us to detect single copy-number changes with 90% power, and alpha 0.05 down to subclonal frequencies of 55%.
Deep whole genome sequencing
Deep whole-genome sequencing was performed on 56 tumors with confirmed TP53 mutations and matched normal samples, of which 48 passed quality control. Libraries were constructed with ~350-bp insert length using the TruSeq Nano DNA Library prep kit (Illumina) and sequenced on an Illumina HiSeq X Ten System in paired-end 150-bp reads mode. The average depth was 60× (range 40-101×) in tumors and 40× (range 24-73×) in matched blood samples.
Variant calling
Read alignment and variant calling of tagged-amplicon sequencing data were processed as described41. Deep WGS samples were processed with bcbio-nextgen44 using Ensemble somatic variants called by two methods out of VarDict45, Varscan46 and FreeBayes47. Somatic SNV calls were further filtered based on mapping quality, base quality, position in read, and strand bias as described40. In addition, the blacklisted SNVs from the Sanger Cancer Genomics Project pipeline derived from a panel of unmatched normal samples were used for filtering48.
Data download
PCAWG-OV
Consensus SNVs and INDELs (October 2016 release), consensus structural variants (v 1.6), consensus copy-number calls (January 2017 release), donor clinical (August 2016 v7-2) and donor histology information (August 2016 v7) for 112 ovarian cancer samples were downloaded from the PCAWG data portal. ABSOLUTE49 copy-number calls were used for analysis.
TCGA
ABSOLUTE49 copy-number profiles from Zack et al27 for 415 ovarian cancer TCGA samples were downloaded from Synapse50. SNVs for these samples were downloaded from the Broad Institute TCGA Genome Data Analysis Center (Broad Institute TCGA Genome Data Analysis Center: Firehose stddata__2016_01_28 run. doi:10.7908/C11G0KM9, Broad Institute of MIT and Harvard). Donor clinical data were downloaded from the TCGA data portal.
Absolute copy-number calling from sWGS
Segmentation
sWGS reads were aligned and relative copy-number called as described41. After inspection of the TP53 mutation status and relative copy-number profiles of the 300 sequenced BriTROC-1 samples, 47 were excluded from downstream analysis for the following reasons: low purity (24), mislabeled (7), pathology re-review revealed sample was not HGSOC (3), no detectable TP53 mutation (13). Of the 253 BriTROC-1 samples analysed, 111 were FFPE-fixed. Fifty seven out of 253 showed an over segmentation artefact (likely due to fixation). A more strict segmentation was subsequently applied to these samples to yield a usable copy-number profile.
Absolute copy number
We combined relative copy-number profiles generated by QDNAseq42 with mutant allele frequency identified using tagged amplicon sequencing in a probabilistic graphical modelling approach to infer absolute copy-number profiles. Using Expectation-Maximisation, the model generated a posterior over a range of TP53 copy-number states, using the TP53 mutant allele frequency to estimate purity for each state. The TP53 copy-number state that provided the highest likelihood of generating a clonal absolute copy-number profile was used to determine the final absolute copy-number profile. To test the validity of this approach, we compared purity and ploidy estimates derived from sWGS to those derived from 60× WGS using the Battenberg algorithm for copy-number calling51. Pearson correlation coefficients were computed for both ploidy and purity estimates using 34 3-star (see Quality rating) BriTROC-1 samples with matched sWGS and WGS (Supplementary Figure 11).
Quality rating
Following absolute copy-number fitting, samples were rated using a 1-3 star system. 1-star samples (n=54) showed a noisy copy-number profile and were considered likely to have incorrect segments and missing calls. These were excluded from further analysis. 2-star samples (n=52) showed a reasonable copy-number profile with only a small number of miscalled segments. These samples were used (with caution) for some subsequent analyses. 3-star samples (n=147) showed a high-quality copy-number profile that was used in all downstream analyses. The maximum star rating observed per patient was 1-star in 15 patients, 2-star in 26, and 3-star in 91 patients. Seventy-two out of 111 FFPE-fixed samples (64%) were amenable to signature analysis. This is consistent with typical sequencing success rates for archival material52.
Copy-number signature identification
Preprocessing
91 3-star BriTROC-1 absolute copy-number profiles were summarized using the genome-wide distribution of six different features (outlined in Figure 1):
Segment size - the length of each genome segment;
Breakpoint count per 10MB - the number of genome breaks appearing in 10MB sliding windows across the genome;
Change-point copy-number - the absolute difference in CN between adjacent segments across the genome;
Segment copy-number - the observed absolute copy-number state of each segment;
Breakpoint count per chromosome arm - the number of breaks occurring per chromosome arm;
Length of segments with oscillating copy-number - a traversal of the genome counting the number of contiguous CN segments alternating between two copy-number states, rounded to the nearest integer copy-number state.
Mixture modelling
For each of the feature density distributions, we applied mixture modelling to identify its distinct components. For distributions representing segment-size, change-point copy-number, and segment copy-number we employed mixtures of Gaussians. For distributions representing breakpoint count per 10MB, length of segments with oscillating copy-number, and breakpoint count per chromosome arm we employed mixtures of Poissons. Mixture modelling was performed using the FlexMix V2 package in R53. The algorithm was run for each distribution with the number of components ranging from 2-10. The optimal number of components was selected as the run showing the lowest Bayesian Information Criterion, resulting in a total of 36 components (see Figure 1 and Supplementary Table 3 for breakdown). Next, for each copy-number event, we computed the posterior probability of belonging to a component. For each sample, these posterior event vectors were summed resulting in a sum-of-posterior probabilities vector. All sum-of-posterior vectors were combined in a patient-by-component sum-of-posterior probabilities matrix.
Signature identification
The NMF Package in R54, with the Brunet algorithm specification55 was used to deconvolute the patient-by-component sum-of-posteriors matrix into a patient-by-signature matrix and a signature-by-component matrix. A signature search interval of 3-12 was used, running the NMF 1000 times with different random seeds for each signature number. As provided by the NMF Package54, the cophenetic, dispersion, silhouette, and sparseness coefficients were computed for the signature-by-component matrix (basis), patient-by-signature matrix (coefficients) and connectivity matrix (consensus, representing patients clustered by their dominant signature across the 1000 runs). 1000 random shuffles of the input matrix were performed to get a null estimate of each of the scores (Supplementary Figure 3). We sought the minimum signature number that yielded stability in the cophenetic, dispersion and silhouette coefficients, and that yielded the maximum sparsity which could be achieved without exceeding that which was observed in the randomly permuted matrices. As a result, 7 signatures were deemed optimal under these constraints and were chosen for the remaining analysis.
Signature assignment
For the remaining 26 2-star patient samples, and the 82 secondary patient samples (from patients with 2- or 3-star profiles from additional tumor samples), the LCD function in the YAPSA package in Bioconductor56 was used to assign signature exposures.
Copy-number signature validation
The signature identification procedure described above was applied to copy-number profiles from two independent datasets: 112 whole-genome sequenced (approximately 40×) HGSOC samples processed as part of ICGC Pan-Cancer Analysis of Whole Genomes Project17, (denoted here as PCAWG-OV) and 415 SNParray profiling of HGSOC cases as part of TCGA27. The number of signatures was fixed at 7 for matrix decomposition with NMF. Pearson correlation was computed between the BriTROC-1 signature-by-component weight matrix and each of the PCAWG-OV and TCGA signature-by-component matrices, signature by signature (Supplementary Table 2).
Association of copy-number signature exposures with other features
Association of signature exposures with other features was performed using one of two procedures: for a continuous association variable, correlation was performed; for a binary association variable, patients were divided into two groups and a Mann-Whitney test was performed to test for differences in signature exposure medians between the two groups. A more detailed explanation of each of these association calculations is given below. (Note: of the 48 deep WGS BriTROC-1 samples that passed QC, only 44 had matched 2- and 3-star sWGS copy-number profiles. As signature exposures from sWGS were used for BriTROC-1 sample associations, only these 44 samples could be used).
Age at diagnosis
Patient age at diagnosis for 112 PCAWG-OV samples and 415 TCGA samples was used to compute Pearson correlation with signature exposures.
Amplification associated fold-back inversions
For 111 PCAWG-OV samples, the fraction of amplification associated fold-back inversion events per sample was calculated as the proportion of head-to-head inversions (h2hINVs) within a 100kb window amplified region (copy number ≥5) relative to the total number of SV calls per sample. 94 samples had at least 1 h2hINV event out of which 58 had h2hINV events in amplified regions. On average they accounted for 4% of SV calls. As these are rare events, only samples showing a non-zero fraction of fold-back inversions (n=67) were used to compute Pearson correlation with signature exposures.
Telomere length
Telomere lengths of 44 deep WGS tumor samples from the BriTROC-1 cohort were estimated using the Telomerecat algorithm57. Telomere length estimates ranged from 1.5kb - 11kb with an average of 4kb. Correlation between telomere length and copy-number signature exposures was calculated with age and tumor purity as covariates using the ppcor package in R58.
Chromothripsis
Copy-number and translocation information from 111 PCAWG-OV samples were used to detect chromothripsis-like events using the Shatterproof software with default parameters29. Shatterproof, a state-of-the-art software, incorporates a wide range of hallmarks of chromothripsis in its detection algorithm as a precise definition of chromothripsis remains elusive. Govind et al. recommend a threshold of 0.37 based on their observations that normal samples produced a low number of calls with low scores (maximum 0.37) while prostate, colorectal and small cell lung cancer samples that were known to have chromothriptic events, produced the highest scores 29. Previous studies have reported a low incidence of chromothriptic events in HGSOC 12,27,30. The number of calls per sample in the PCAWG-OV samples ranged from 5 to 47 with an average of 23. The score per call ranged from 0.15-0.62 with a median of 0.38. Therefore, a conservative threshold was set at the 95th percentile of our distribution of scores to minimise false positives and calls with scores greater than 0.48 were used to obtain a count of chromothriptic events per sample. As chromothriptic events are rare in HGSOC, only samples showing a non-zero number of events (n=61) were used to compute Pearson correlation with signature exposures. Of 61 samples with scores above the threshold, 49 (80.3%) had 1-2 events, 11 samples (18%) had 3-6 events and 1 sample (1.6%) had 10 events.
Tandem duplicator phenotypes
Tandem duplicator phenotype (TDP) scores were calculated for 111 PCAWG-OV samples using the method described in Menghi et al21. The number of duplication events per chromosome normalized by chromosome length per sample was used to calculate a score relative to the expected number of duplication events per chromosome per sample. The scores ranged from -1.11 to 0.53 with an average score of 0.02.
Mutational signatures
Motif matrices were extracted using the SomaticSignatures R package59 and the weights of all known COSMIC signatures were determined using the deconstructSigs R package60 for 44 deep WGS BriTROC-1 samples and 109 PCAWG-OV samples. SNV signatures showing an exposure >0 for at least one sample were retained. The rcorr function in the Hmisc R package61 was used to calculate the correlation matrix between the remaining SNV and CN signature exposures.
The significance of all observed correlations was estimated from a t-distribution where the null hypothesis was that the true correlation was 0. All reported p-values have been adjusted for multiple testing with Benjamini & Hochberg (BH) method62. Comparison plots can be found in Supplementary Figure 6.
Mutated pathways
A combined set of 479 samples (44 deep WGS BriTROC-1, 112 PCAWG-OV and 323 TCGA) showing at least one driver mutation was used for mutated pathway enrichment analysis. We focused on 765 driver genes reported by Cancer Genome Interpreter (CGI)63. SNVs, INDELs, amplifications (CN>5) or deletions (CN<0.4) affecting these genes were considered bona fide driver mutations if CGI predicted them as TIER1 or TIER2 (Supplementary Tables 4 and 5, see URL 2, run date: 2018-01-13). 320 of the 765 genes were mutated in a least one case. These genes were used to test for enriched pathways in the Reactome database using the ReactomePA R package64 with a p-value cutoff of 0.05 and q-value cutoff of 0.05. Pathways mutated in at least 5% of the cohort (n≥24) were retained. For each pathway, patients were split into two groups: those with mutated genes in the pathways, and those with wild-type genes in the pathways. A one-sided Mann-Whitney was carried out for each signature to determine if the exposure was significantly higher in mutated cases versus wild-type cases. After multiple testing correction using the Benjamini & Hochberg method (thresholding the p-value <0.005 and the median difference in exposures ≥0.1), 186 pathways were significantly enriched. Visual inspection revealed significant redundancy in the list and 9 representative pathways were manually selected as a final output (Supplementary Table 6).
Mutated genes
A combined set of 479 samples (44 deep WGS BriTROC-1, 112 PCAWG-OV and 323 TCGA) was used test if signature exposures were significantly higher in cases with mutated driver genes, including NF1, PTEN, BRCA1, BRCA2, PIK3CA, MYC and CDK12. Patients were split into two groups: those with the mutated gene and those with wild-type genes. A one-sided Mann-Whitney was carried out for each signature to determine if the exposure was significantly higher in mutated cases versus wild-type cases. After multiple testing correction using the Benjamini & Hochberg method (thresholding the p-value <0.05 and the median difference in exposures ≥0.0.08), 10 gene/signature combinations were significantly enriched (Supplementary Table 6).
Survival analysis
Censoring and truncation
Overall survival in BriTROC-1 patients was calculated from the date of enrolment to the date of death or the last documented clinical assessment, with data cutoff at 1 December 2016. As the BriTROC-1 study only enrolled patients with relapsed disease, left truncation was used in the survival analysis. In addition, cases where the patient was not deceased were right censored. Survival data for the PCAWG-OV and TCGA cohorts were right censored as required (left truncation was not necessary). The combined samples were split into training (100% BriTROC-1, 70% PCAWG-OV and 70% TCGA = 417) and test (30% PCAWG-OV and 30% TCGA = 158) cohorts. All of the BriTROC-1 samples were used in the training set to avoid issues calculating prediction performance on left-truncated data.
Cox regression
As the signature exposures for a given sample summed to 1, it was necessary to select one normalizing signature to perform regression. Signature 5 was chosen as it showed the lowest variability across the cohorts. To avoid division errors all 0 signature exposures were converted to 0.02. The remaining signature exposures were normalized taking the log ratio of their exposure to signature 5’s exposure. A Cox proportional hazards model was fitted on the training set, with the signature exposures as covariates, stratified by cohort (BriTROC-1, PCAWG-OV:AU, PCAWG-OV:US, TCGA) and age (<39; 40:44; 45:49; 50:54; 55:59; 60:64; 65:69; 70:74; 75:79; >80), using the survival package in Bioconductor65. After fitting, the model was used to predict risk in the test set and performance was assessed using the concordance index calculation in the survcomp package in Bioconductor47. A final Cox regression was performed using all data for reporting of hazard ratios and p-values.
Unsupervised clustering of patients using signature exposures
Hierarchical clustering of the exposure vectors of the 575 samples used in the survival analysis was performed using the NbClust66 package in R. The optimal number of clusters was 3 as determined by a consensus voting approach across 23 metrics for choosing the optimal numbers of clusters. 12/23 metrics reported 3 clusters as the optimal number. A Cox proportional hazards model was fitted using the cluster labels as covariates, stratified by cohort (BriTROC-1, PCAWG-OV:AU, PCAWG-OV:US, TCGA) and age (<39; 40:44; 45:49; 50:54; 55:59; 60:64; 65:69; 70:74; 75:79; >80), using the survival package in Bioconductor65.
Analysis of copy-number signature changes during treatment
Thirty-six BriTROC-1 cases with matched diagnosis and relapse samples were used to investigate the effects of treatment on signature exposures. A linear model was fitted to test for treatment effects with exposure at relapse as the dependent variable and exposure at diagnosis, age at diagnosis, number of lines of chemotherapy, and days between diagnosis and relapse as independent variables. Prior to fitting, age at diagnosis was centered and exposures transformed by log(x+0.1) to ensure normality. Fitting was done using the lm() function in R.
To test whether signature exposures at diagnosis were predictive of platinum sensitivity, a generalized linear model with Binomial error was fitted using type of relapse (platinum-sensitive or platinum-resistant) as the dependent variable and exposure at diagnosis and age at diagnosis as independent variables.
Supplementary Material
Acknowledgements
The BriTROC-1 study was funded by Ovarian Cancer Action (to IMcN and JDB, grant number 006). We would like to acknowledge funding and support from Cancer Research UK (grant numbers A15973, A15601, A18072, A17197, A19274 and A19694), the Universities of Cambridge and Glasgow, National Institute for Health Research Cambridge and Imperial Biomedical Research Centres, National Cancer Research Network, the Experimental Cancer Medicine Centres at participating sites, the Beatson Endowment Fund and Hutchison Whampoa Limited. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank the Biorepository, Bioinformatics, Histopathology and Genomics Core Facilities of the Cancer Research UK Cambridge Institute and the Pathology Core at the Cancer Research UK Beatson Institute for technical support. We would like to thank members of PCAWG Evolution and Heterogeneity Working Group for the consensus copy-number analysis, PCAWG Structural Variation Working Group for the consensus structural variants and PCAWG Technical Working Group for annotating driver mutations in the 112 PCAWG-OV samples.
Footnotes
Data Availability
Sequence data that support the findings of this study have been deposited in the European Genome-phenome Archive with the accession code EGAS00001002557. All code required to reproduce the analysis outlined in this manuscript can be found in the following repository (see URL 3).
URLs
Accession Codes
EGAS00001002557
Author contributions
G.M., T.E.G., F.M., I.McN., J.D.B. conceptualized the study; S.D., R.M.G., M.L., E.B., A.M., A.W., S.S., R.E., G.D.H., A.C., C.G., M.H., C.F., H.G., D.M., A.Ho., G.B., I.McN., J.D.B. conducted sample collection; T.E.G., D.E., A.M.P., L.A.L., A.Ha., C.W., C.N., L.Mi., L.N.S., M.J.L., L.Mo., A.S., J.P. performed experiments; G.M., T.E.G., D.D.S., M.E., D.S., B.Y., O.H., F.M. performed data analysis; G.M., D.D.S., F.M. developed the methodology and software; G.M., T.E.G., D.D.S., F.M., I.McN., J.D.B. wrote the manuscript.
Competing Financial Interests Statement
The following authors the authors have a competing interest as defined by Nature Research:
C.G. Personal interest: Roche, AstraZeneca, Tesaro, Clovis, Foundation One, Nucana. Research funding: AstraZeneca, Novartis, Aprea, Nucana, Tesaro. Named co-inventor on five patents (issued: PCT/US2012/040805; pending: PCT/GB2013/053202, 1409479.1, 1409476.7 and 1409478.3)
H.G. Employment: AstraZeneca
I.McN. Personal interest: Clovis Oncology.
J.D.B. Cofounder and shareholder of Inivata Ltd (a cancer genomics company that commercializes ctDNA analysis)
All other authors declare that they have no competing financial or non-financial interests as defined by Nature Research.
References
- 1.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–33. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed AA, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan S, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–725. doi: 10.1038/nrc3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fong PC, et al. Poly(ADP)-Ribose Polymerase Inhibition: Frequent Durable Responses in BRCA Carrier Ovarian Cancer Correlating With Platinum-Free Interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 6.Gelmon KA, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 7.Swisher EM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 8.TCGA. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etemadmoghadam D, et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin Cancer Res. 2009;15:1417–1427. doi: 10.1158/1078-0432.CCR-08-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhaak RG, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–25. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen GM, et al. Consensus on Molecular Subtypes of Ovarian Cancer. bioRxiv. 2017 [Google Scholar]
- 12.Patch A-M, et al. Whole–genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 13.Wang YK, et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet. 2017;49:856–865. doi: 10.1038/ng.3849. [DOI] [PubMed] [Google Scholar]
- 14.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goranova T, et al. Safety and utility of image-guided research biopsies in relapsed high-grade serous ovarian carcinoma-experience of the BriTROC consortium. Br J Cancer. 2017;116:1294–1301. doi: 10.1038/bjc.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell PJ, et al. Pan-cancer analysis of whole genomes. bioRxiv. 2017 [Google Scholar]
- 18.Murnane JP. Telomere dysfunction and chromosome instability. Mutat Res. 2012;730:28–36. doi: 10.1016/j.mrfmmm.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–36. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Ng CK, et al. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. J Pathol. 2012;226:703–12. doi: 10.1002/path.3980. [DOI] [PubMed] [Google Scholar]
- 21.Menghi F, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci U S A. 2016;113:E2373–82. doi: 10.1073/pnas.1520010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M, et al. Comparative analysis of whole genome sequencing-based telomere length measurement techniques. Methods. 2017;114:4–15. doi: 10.1016/j.ymeth.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Zakov S, Kinsella M, Bafna V. An algorithmic approach for breakage-fusion-bridge detection in tumor genomes. Proc Natl Acad Sci U S A. 2013;110:5546–51. doi: 10.1073/pnas.1220977110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knauf JA, et al. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J Biol Chem. 2006;281:3800–9. doi: 10.1074/jbc.M511690200. [DOI] [PubMed] [Google Scholar]
- 25.Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. MAPK mediates RAS-induced chromosome instability. J Biol Chem. 1999;274:38083–90. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- 26.Popova T, et al. Ovarian Cancers Harboring Inactivating Mutations in CDK12 Display a Distinct Genomic Instability Pattern Characterized by Large Tandem Duplications. Cancer Res. 2016;76:1882–91. doi: 10.1158/0008-5472.CAN-15-2128. [DOI] [PubMed] [Google Scholar]
- 27.Zack TI, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berenjeno IM, et al. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat Commun. 2017;8:1773. doi: 10.1038/s41467-017-02002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Govind SK, et al. ShatterProof: operational detection and quantification of chromothripsis. BMC Bioinformatics. 2014;15:78. doi: 10.1186/1471-2105-15-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malhotra A, et al. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;23:762–76. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakhoum SF, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–472. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etemadmoghadam D, et al. Synthetic lethality between CCNE1 amplification and loss of BRCA1. Proc Natl Acad Sci U S A. 2013;110:19489–94. doi: 10.1073/pnas.1314302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Candido Dos Reis FJ, et al. Germline mutation in BRCA1 or BRCA2 and ten-year survival for women diagnosed with epithelial ovarian cancer. Clin Cancer Res. 2015;21:652–7. doi: 10.1158/1078-0432.CCR-14-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norquist BM, et al. Mutations in Homologous Recombination Genes and Outcomes in Ovarian Carcinoma Patients in GOG 218: An NRG Oncology/Gynecologic Oncology Group Study. Clin Cancer Res. 2018;24:777–783. doi: 10.1158/1078-0432.CCR-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarz RF, et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 2015;12:e1001789. doi: 10.1371/journal.pmed.1001789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walton JB, et al. CRISPR/Cas9-derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Scientific Reports. 2017;7:16827. doi: 10.1038/s41598-017-17119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerstung M, et al. The evolutionary history of 2,658 cancers. bioRxiv. 2017 [Google Scholar]
- 38.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kandoth C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Secrier M, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48:1131–41. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piskorz AM, et al. Methanol-based fixation is superior to buffered formalin for next-generation sequencing of DNA from clinical cancer samples. Ann Oncol. 2016;27:532–539. doi: 10.1093/annonc/mdv613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheinin I, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–32. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macintyre G, Ylstra B, Brenton JD. Sequencing Structural Variants in Cancer for Precision Therapeutics. Trends Genet. 2016;32:530–42. doi: 10.1016/j.tig.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 44.bcbio-nextgen. (2017).
- 45.Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koboldt DC, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–5. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrison E, M G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012 [Google Scholar]
- 48.Jones D, et al. cgpCaVEManWrapper: Simple Execution of CaVEMan in Order to Detect Somatic Single Nucleotide Variants in NGS Data. Curr Protoc Bioinformatics. 2016;56:15 10 1–15 10 18. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schumacher S. pancan12_absolute.segtab.txt. 2015 [Google Scholar]
- 51.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–5. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Al-Kateb H, Nguyen TT, Steger-May K, Pfeifer JD. Identification of major factors associated with failed clinical molecular oncology testing performed by next generation sequencing (NGS) Mol Oncol. 2015;9:1737–43. doi: 10.1016/j.molonc.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grün B, Leisch F. FlexMix Version 2: Finite Mixtures with Concomitant Variables and Varying and Constant Parameters. J Stat Soft. 2008;28:35. [Google Scholar]
- 54.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brunet J-P, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huebschmann D, Gu Z, Schlesner M. YAPSA: Yet Another Package for Signature Analysis. R package version 1.2.0. 2015 [Google Scholar]
- 57.Farmery JHS, Mike L, Lynch Andy G. Telomerecat: A Ploidy-Agnostic Method For Estimating Telomere Length From Whole Genome Sequencing Data. bioRxiv. 2017 doi: 10.1038/s41598-017-14403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S. ppcor: Partial and Semi-Partial (Part) Correlation. 2015 [Google Scholar]
- 59.Gehring JS, Fischer B, Lawrence M, Huber W. SomaticSignatures: inferring mutational signatures from single-nucleotide variants. Bioinformatics. 2015;31:3673–5. doi: 10.1093/bioinformatics/btv408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrell FE. Hmisc: Harrell Miscellaneous. R package version 4.0-0. 2016 [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 63.Tamborero D, et al. Cancer Genome Interpreter Annotates The Biological And Clinical Relevance Of Tumor Alterations. bioRxiv. 2017 doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 65.Therneau TM, Grambsch Patricia M. Modeling Survival Data: Extending the Cox Model. Springer; New York: 2000. [Google Scholar]
- 66.Charrad M, Ghazzali N, Boiteau V, Niknafs A. NbClust: An R Package for Determining the Relevant Number of Clusters in a Data Set. J Stat Soft. 2014;61:36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.