Abstract
Homotypic cell-in-cell (CIC) structures forming between cancer cells were proposed to promote tumor evolution via entosis, a nonapoptotic cell death process. However, the mechanisms underlying their formation remained poorly understood. We performed a microarray analysis to identify genes associated with homotypic CIC formation. Cancer cells differing in their ability to form homotypic CIC structures were selected for the study. Association analysis identified 73 probe sets for 62 candidate genes potentially involved in CIC formation. Among them, twenty-one genes were downregulated while 41 genes were upregulated. Pathway analysis identified a gene interaction network centered on IL-8, which was upregulated in high CIC cells. Remarkably, CIC formation was significantly inhibited by IL-8 knockdown and enhanced upon recombinant IL-8 treatment, which correlated with altered cell-cell adhesion and expression of adhesive molecules such as P-cadherin and γ-catenin. Together, our work identified IL-8 as a positive regulator of homotypic CIC formation via enhancing intercellular adhesion.
Keywords: Cell-in-cell, Entosis, Expression profiling, IL-8, Intercellular adhesion
INTRODUCTION
Cell-in-cell (CIC) structures, characterized by fully enclosure of one or more viable cells inside another one, are prevalent in human tumors, where cell cannibalism was usually used for the description (1). In tumors, via internalizing and killing the less fit “loser” tumor cells, and endowing aneuploidy to the progenies of outer “winner” tumor cells (2, 3), CIC formation could serve as a mechanism of cell competition to promote clonal selection and therefore tumor evolution (4). Based on recent research on entosis, a CIC-mediated nonapoptotic cell death process (5), the mechanisms underlying formation of homotypic CICs between cells of epithelial origin are starting to be revealed (6). Current research fits a working model of two core elements: first, adherens junction, mediated by epithelial complexes formed by E- or P-cadherin and multiple essential catenins (7, 8), brings cells together and sets up asymmetric RhoA activity by junction localized-p190A RhoGAP. Then, activated RhoA/ROCKs signaling drives actomyosin contraction and the following cell internalization (6). Besides, glucose starvation and membrane cholesterol/lipids were found important regulators of entotic CIC formation probably by affecting the phosphorylation of myosin light chain (MLC) (9, 10), suggesting CIC formation is a complex and finely controlled process. Therefore, it warrants further investigation to decipher the mechanisms underlying its regulation.
In this study, by using microarray analysis of gene expression in different cancer cells, we attempt to screen for genes potentially involved in the formation of homotypic CIC structures. As a result, bioinformatic analysis, combined with expressional and functional validation, identified a gene network centered on IL-8 as a novel pathway participating in CIC regulation, and IL-8 promotes homotypic cell-in-cell formation probably via positive regulating P-cadherin mediated intercellular adhesion.
RESULTS
Formation of CIC structures in breast cancer cell lines
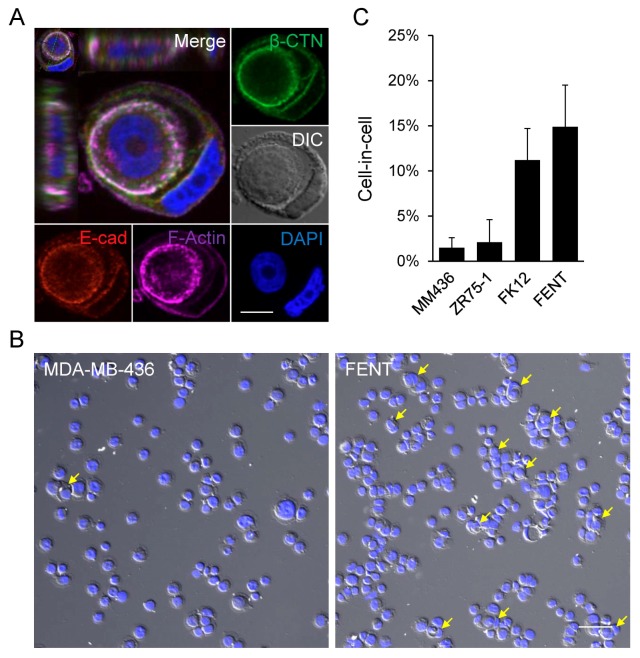
To make phenotype-based association analysis, we screened a group of breast cancer cell lines based on their ability to form homotypic CIC structures by entosis. Four cell lines were included in this study. As shown in Fig. 1, although typical CIC structures could be detected in all four cells (Fig. 1A), the frequency varied from cell to cell. MDA-MB-436 (MM436) and ZR75-1 cells could only form low frequency CIC structures upon induction (< 2% or so), whereas FK12 and FENT, two derivatives of ZR75-1 cells, demonstrate high levels of CIC formation (> 10%) under the same inducing conditions (Fig. 1B and 1C). Considering the similar genetic background, these four cells were chosen for expression profiling analysis by microarray.
Fig. 1.
Formation of CIC structures in breast cancer cell lines. (A) Representative image of CIC structures formed in MDA-MB-436 cells. Cytospins were immunostained with antibodies for E-cadherin and β-catenin, and phalloidin for F-actin. “E-cad” for E-cadherin, “β-CTN” for β-catenin. Nuclei were stained with DAPI. Scale bar: 10 μm. (B) Representative cytospin images of MDA-MB-436 and FENT cells. Yellow arrow heads indicate the entotic cell-in-cell structures. The nuclei were stained with DAPI. Scale bar: 100 μm. (C) Quantification of entotic CIC formation in breast cancer cell lines MDA-MB-436, ZR75-1, FK12 and FENT. “MM436” for MDAMB-436. FK12 and FENT were monoclonal cells isolated from ZR75-1. Data are mean ± SD of cells analyzed in triplicate and representative of three independent experiments.
Expression profiling of genes associated with CIC formation
Total 54,675 probe sets standing for genes were analyzed by Affymatrix microarray chip, and 48,460 probe sets, passing initially quality control with their signal intensity values satisfying the cutoff expression percentile (20–100), were used for further analysis. Given the limited sample number, we performed differential analysis sequentially to obtain a group of genes that were strong candidates involved in CIC regulation. Probe sets differentially expressing (fold change > 2) between FK12, FENT vs ZR75-1 (2536), or FK12, FENT vs MDA-MB-436 (398), or FK12, FENT vs ZR75-1 and MDA-MB-436 (634) were first captured, then plotted in Venn Diagram to identified probe sets shared by them as candidates (Fig. 2A). As a result, total 73 probe sets standing for 62 individual genes were identified. Among them, forty seven probes (41 genes in Table S1) were downregulated with 26 probe sets (21 genes in Table S2) upregulated in high CIC cells (Fig. 2B).
Fig. 2.
Expression profiling of genes associated with CIC formation. (A) A sequential screening for genes associated with CIC formation. The differentially expressed genes shared by three analysis sets were selected as the candidate genes. The number in brackets are probes differentially expressed in each comparison set. (B) A heat map of the 73 probes potentially involved in CIC formation. The up-regulated and down-regulated probes are marked as up (26) and down (47). The number in brackets are the differentially expressed probes. (C) A gene network centered on IL-8 constructed by the “Pathway” algorithm of GeneSpring software under the condition of “simple algorithm” and “direct interaction”. (D) Validation of candidate gene expression by quantitative RT-PCR. Five down-regulated genes NUPR1, STAT6, SERPINA1, DLC1 and PRSS23 and one up-regulated gene IL-8 were selected for qRT-PCR analysis. Data are mean ± SD of three independent experiments.
To find the potential signal pathways that coordinately worked in CIC formation, we employed the “Pathway Analysis” module in GeneSpring GX software to explore the potential interactions among the candidate genes. Interestingly, under the condition of “simple algorithm” and “direct interaction”, an IL-8-centered gene interaction network was identified. The network consists of 9 additional genes that make 7 direct links to IL-8 (Fig. 2C).
Validation of candidate gene expression by quantitative real-time PCR
To confirm gene expression from microarray analysis, six genes including NUPR1, STAT6, SERPINA1, DLC1, PRSS23 and IL-8 were selected for quantitative real-time PCR. As shown in Fig. 2D, despite slight variations such as NUPR1 and PRSS23, most of the gene expressions in microarray were consistently validated by quantitative real-time PCR. Therefore, the microarray data was likely to be reliable and most candidate genes identified might be genuinely differentially expressed.
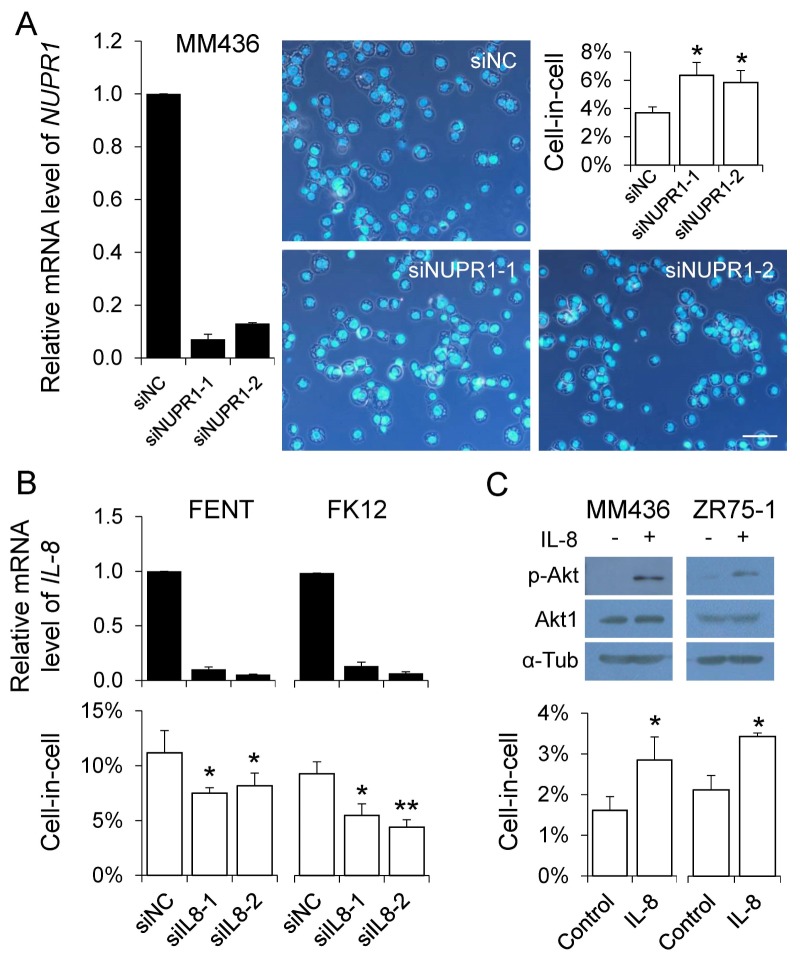
Regulation of CIC formation by NUPR1 and IL-8
To test whether the candidate genes we obtained above might participate in CIC regulation, several genes were subjected to RNA interference-mediated knockdown, most of them affected homotypic CIC formation upon knockdown (data not shown). Among them, NUPR1 was previously reported to suppress homotypic CIC formation in pancreatic cancer cells (11), we therefore test its role in our system. As shown in Fig. 3A, CIC formation in MDA-MB-436 cells, where NUPR1 was relatively highly expressed, was consistently enhanced upon NUPR1 knockdown by two individual siRNAs, confirming its negative role in homotypic CIC formation across different types of cancer cell lines (11). We also examined the effects of IL-8 knockdown on CIC formation. As shown in Fig. 3B, though slightly nevertheless significantly, CIC formation in FENT cells, where IL-8 expression is relatively high, was decreased by RNAi-mediated knockdown. Similarly, IL-8 depletion led to reduced CIC formation in FK12, another IL-8 high-expression cell line. To further confirm IL-8’s positive role, MDA-MB-436 and ZR75-1 cells were treated with recombinant IL-8. As shown in Fig. 3C, IL-8 treatment activated downstream signaling as indicated by increased Akt phosphorylation. And here also, CIC formation was significantly enhanced upon IL-8 treatment. Together, these results support the notion that IL-8 is a positive regulator of homotypic CIC formation.
Fig. 3.
Regulation of CIC formation by NUPR1 and IL-8. (A) CIC formation in MDA-MB-436 cells upon NUPR1 knockdown. Representative images show the cytospins of MDA-MB-436 cells transfected with control and NUPR1 siRNAs. Nuclei were stained with DAPI. Scale bar: 100 μm. (B) CIC formation in FENT and FK12 cells with IL-8 knockdown. (C) CIC formation in MDA-MB-436 and ZR75-1 cells treated with recombinant IL-8. IL-8 activity was determined by Akt phosphorylation. The black bar graphs show relative mRNA level examined by qRT-PCR. Data are mean ± SD of three independent experiments. The white bar graphs show the quantification of CIC formation. Data are mean ± SD of cells analyzed in triplicate and are representative of three independent experiments. “*” for P < 0.05. “**” for P < 0.01. “MM436” for MDA-MB-436. “si” for siRNA. “NC” for negative control.
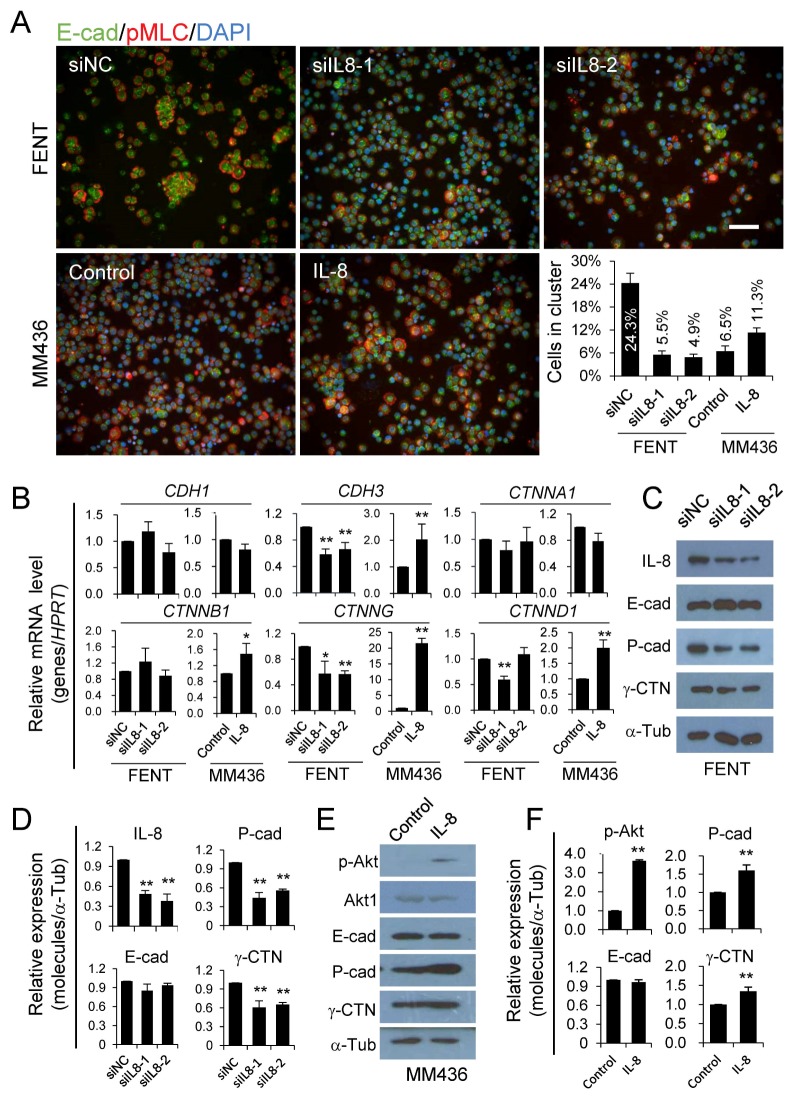
Regulation of cell-cell adhesion by IL-8
To explore the underlying mechanisms whereby IL-8 regulates the formation of homotypic CIC structures, we examined intercellular adhesion, the essential mediator of CIC formation (6, 8), by cluster assay. As shown in Fig. 4A, cells with IL-8 depletion formed much fewer clusters than did control cells, while those treated with human IL-8 protein formed much more clusters as compared with control cells. These results suggest that altered cell-cell adhesion may directly affect CIC formation. In light of this, we examined expression of key adhesive molecules that mediate adherens junction, whose defects would strikingly impair homotypic CIC formation (7, 8). As shown in Fig. 4, IL-8 depletion caused significant reduction in the expression of CDH3 and CTNNG genes at both mRNA (Fig. 4B) and protein levels (Fig. 4C, 4D, 4E and 4F), and IL-8 treatment significantly increased their expression. These results are in agreement with altered cell-cell adhesion and therefore homotypic CIC formation.
Fig. 4.
Regulation of cell-cell adhesion by IL-8. (A) RNAi-mediated IL-8 knockdown and IL-8 treatment regulates cluster formation. Scale bar: 100 μm. The graph shows the percentage of cells forming clusters. Cells in cluster = cells forming cluster / total cells counted. Cell cluster was defined as a cell colony that contains more than 6 cells. Data are mean ± SD of cells in three fields of view, n > 200 for each field. (B) Relative mRNA level of CDH1, CDH3, CTNNA1, CTNNB1, CTNNG and CTNND1 by quantitative real-time PCR. Data are mean ± SD of three independent experiments. (C and E) Expression of adhesion molecules as detected by western blot. (D and F) Quantification of the blots of C and E. Data are mean ± SD of triplicate quantification. “*” for P < 0.05. “**” for P < 0.01. “MM436” for MDA-MB-436. “si” for siRNA. “NC” for negative control.
DISCUSSION
It’s now clear that cell death programs, such as apoptosis, necroptosis and autosis, were genetically controlled (12). As a non-autonomous cell death mechanism that involves at least two cells (13), CIC-mediated death was conceivably subjected to more complex regulation. Though due to technical limitations, a systemic screening for CIC-associated genes remains to be made. In this study, we identified a group of candidate genes by phenotype-expression association analysis. These genes were likely participating into homotypic CIC formation for two reasons. First, NUPR1, a gene known to negatively regulate homotypic CIC formation in pancreatic cancer cells (11), was included in the gene list. Second, functional analysis by RNA interference-mediated knockdown confirmed that candidate genes such as NUPR1 and IL-8 affect homotypic CIC formation in the cells used for expression profiling. Thus, the way we employed in this study might be feasible method to systemically identify genes related to homotypic, and presumably heterotypic, CIC formation.
It is interesting that our work identifies IL-8, an inflammatory cytokine that recruits leukocytes such as neutrophil (14), as a regulator of homotypic CIC formation. Although IL-8 may affect CIC formation via regulating intercellular adhesion as proposed here, it does not necessarily rule out other mechanisms. Actually, IL-8 played a critical role in epithelial-mesenchymal transition (EMT) of human carcinoma cells by promoting both acquisition and maintenance of mesenchymal features (15). Previous work proposed that EMT induced by TGF-β was associated with homotypic CIC formation in pancreatic cancer cells. TGF-β could potentiate homotypic CIC formation in the background of NUPR1 depletion. Blocking TGF-β signaling could effectively reverse the phenotype (11). In our study, IL-8 was highly expressed in high CIC cancer cells (FK12 and FENT) where NUPR1 was expressed at relatively low level, suggesting that IL-8, probably via promoting EMT, worked together with low NUPR1 expression to promote homotypic CIC formation in FK12 and FENT cells. While it was argued that the EMT-associated CIC formation was mechanistically different from entotic CIC formation (11), our data supports a working model with shared molecular mechanism for these two types of CIC formation. Since NUPR1 was an effector gene of and could be induced by TGF-β, it would be intriguing to know whether IL-8 could also regulate the expression of NUPR1. Moreover, we identified a gene network centered on IL-8, future works are warranted to address whether and how this network may regulate homotypic CIC formation.
MATERIALS AND METHODS
Cell culture
The two breast cancer cells MDA-MB-436 and ZR75-1 used in this study were from our lab. FK12 and FENT were two monoclonal cell lines isolated from ZR75-1. All cells used in this study were cultured in DMEM (M&C GENE Technology Ltd, Beijing, China) supplemented with 10% fetal bovine serum (PAN-Biotech, Aidenbach, Germany).
Proteins and Antibodies
Human IL-8 / CXCL8 protein (aa 23–99) was purchased from Sino Biological (Beijing, China). Antibodies used in the study are listed as follows: primary antibodies include anti-E-cadherin, anti-β-catenin, and anti-γ-catenin (BD Biosciences, Franklin Lakes, NJ, USA), anti-P-cadherin, anti-Akt1, anti-p-Akt (Ser473), and anti-pMLC (Cell Signaling Technology, Danvers, MA, USA), anti-α-tubulin (Proteintech, Wuhan, Hubei, China), anti-IL-8 (Abcam, Cambridge, UK). Secondary antibodies include AlexaFluor 568 anti-rabbit and Alexa Fluor 488 anti-mouse (Invitrogen, California, CA, USA) for immunostaining, HRP labeled anti-rabbit and anti-mouse (CWBIO, Beijing, China) for immunoblotting. Fluorescent dyes including DAPI and Phalloidin 647 were purchased from Thermo Fisher Scientific (Waltham, MA, USA).
CIC formation assays
To investigate the CIC formation frequency of each cell line, cells were seeded at the concentration of 2.5 × 105 cells/well in six well plates and incubated at 37°C for 24 hours, then cultured in suspension in the complete medium for 6 hours to induce CIC formation, followed by cytospin. For the cells into which siRNA had been delivered, they were incubated at 37°C for 36 hours post transfection, followed by suspension and cytospin. CIC quantification was performed as described in (7).
Immunostaining and immunoblotting
Cytospins were fixed in 10% trichloroacetic acid or 4% paraformaldehyde for 10 minutes and then washed with PBS three times, followed by immunostaining with the indicated antibodies. Nuclei were counterstained with DAPI when slides were mounted with coverslips by Prolong Gold Antifade reagent for image capture after 24 hours of air-drying. Images were taken at five channels (FITC, TRITC, Cy5, DAPI and DIC) by confocal microscopy. Western blot was performed as described (7). Briefly, protein samples were subjected to SDS-PAGE and then transferred onto nitrocellulose membrane, after which appropriate antibodies were used to probe the specified protein.
Microarray and expression profiling
Total RNA was extracted from the four breast cancer cell samples using TRIzol reagent according to the manufacturer’s protocol (Invitrogen, California, CA, USA). Then sent for microarray detection (CapitalBio Technology, Beijing, China). Affymetrix Human Genome U133 Plus 2.0 (Affymetric, Inc., Santa Clara, CA, USA) was used to detect the gene expression profile. Detection Method Number was LSS-SOP-AE-03. The Data Analysis Method Number was LSS-SOP-DLAE-01. Cluster analysis was performed on differentially expressing genes. The molecular interaction networks were constructed by using the “Pathway Analysis” module. Both cluster analysis and molecular interaction networks construction were performed with the GeneSpring GX software (Agilent Technologies, Santa Clara, CA, USA).
RNA interference mediated gene knockdown
Cells were seeded at the concentration of 1.2 × 105 cells/well in 12 well plates and incubated at 37°C for 24 hours, then transfected with siRNAs targeting NUPR1 (1#: sense-5′-GGCAGCAACAAUAAAUAGATT-3′, antisense-5′-UCUAUU UAUUGUUGCUGCCTT-3′; 2#: sense-5′-GGAUGAAUCUGA CCUCUAUTT-3′, antisense-5′-AUAGAGGUCAGAUUCAUCC TT-3′) and IL-8 (1#: sense-5′-GCAUAAAGACAUACUCCA ATT-3′, antisense-5′-UUGGAGUAUGUCUUUAUGCTT-3′; 2#: sense-5′-GCACGGGAGAAUAUACAAATT-3′, antisense-5′-UU UGUAUAUUCUCCCGUGCTT-3′) and negative control siRNA using Lipofectamine RNAiMAX (Invitrogen, California, CA, USA) according to the manufacturer’s recommendations. The siRNA was purchased from GenePharma Co., Ltd (Suzhou, China). Cells were incubated at 37°C for 36 hours for gene knockdown. Quantitative real-time PCR was applied to detect mRNA expression level.
Treatment with human IL-8
The recombinant human IL-8 protein was dissolved as a 200 μg/ml solution in sterile water. Cells of 2.5 × 105 /well were seeded in a 6-well plate and grown overnight. The next day, the medium was replaced with the one containing 200 ng/ml of IL-8 protein. After 12 hours the cells were collected for cell-in-cell formation assays, immunostaining, and immunoblotting.
Quantitative real-time PCR
Total RNA was extracted from the cell samples. First strand cDNA was reverse transcribed from 1 to 2 μg total RNA using cDNA synthesis kit ReverTra Ace® qPCR RT Master Mix with gDNA Remover (Toyoba Co., Ltd., Dalian, China) following the procedure of 37°C for 15 minutes and 98°C for 5 minutes. The resulting cDNA was diluted 1:5 for all genes. The mRNA levels of the candidate genes were quantified by real-time PCR using the SYBR® Green Realtime PCR Master Mix (Toyoba Co., Ltd., Dalian, China) according to the manufacturer’s instructions. The Applied BiosystemsTM 7500 Fast Dx Real-Time PCR Instrument (Applied Biosystems, Foster City, Calif., USA) were used to perform the experiments. Gene specific primers for real-time PCR were designed using OLIGO Primer Analysis Software Version 7 (Molecular Biology Insights, Inc., Cascade, CO, USA), and primer-target specificity was checked using the nucleotide BLAST program (http://blast.ncbi.nlm.nih.gov). PCR products were analyzed by thermal dissociation curve to verify specific PCR product amplification. Data were normalized against the reference gene HPRT. The primers used in real-time PCR analysis were as follow: HPRT (F 5′-AGGCCATCA CATTGTAGCCCTCTGT-3′, R 5′-TACTGCCTGACCAAGGAAA GCAAAGT-3′), NUPR1 (F 5′-CCTGGCCCATTCCTACCTC-3′, R 5′-GCAGTCCCGTCTCTATTGCT-3′), STAT6 (F 5′-CAACCA AGACAACAATGCCAA-3′, R 5′-TCCTTGTTGAACTGCGACC A-3′), SERPINA1 (F 5′-AGCCTTCACTGTCAACTTCGG-3′, R 5′-TCTTCCTCGGTGTCCTTGACT-3′), DLC1 (F 5′-CAGAGGA AATCTTAAAGCGCCTA-3′, R 5′-TGGACAAGAGCACATTAAC CC-3′), PRSS23 (F 5′-TTCATCTGAACAGTGCTCGG-3′, R 5′-G ATATTGCTTGGCCTCTTCGT-3′), IL-8 (F 5′-CCTTTCCACCC CAAATTTATCA-3′, R 5′-TTTCTGTGTTGGCGCAGTGT-3′), CDH1 (F 5′-GCCGCCGGCGTCTGTAGGAA-3′, R 5′-GTCACTTCAG GCCGAGCGTCCAG-3′), CDH3 (F 5′-GAGGTGGTTCTCCGC AATG-3′, R 5′-TGTTAGCCGCCTTCAGGT-3′), CTNNA1 (F 5′-CTCGGCGGGGAGCTTGTTGTCTCT-3′, R 5′-GAGGGCCT GCACCGGGTTCAC-3′), CTNNB1 (F 5′-GACAAGCCACAAG ATTACAAG-3′, R 5′-TCAACTGGATAGTCAGCACC-3′), CTNNG (F 5′-GCTGCCGTCCTGTTCCGCATCT-3′, R 5′-TGAGGCCGT CGCTGTAGGTGTCG-3′) and CTNND1 (F 5′-ACCACGGT CAAGAAAGTAG-3′, R 5′-GAAATCACGACCCAAAGT-3′). All fold changes were calculated using the 2−ΔΔCt method and were expressed as means ± SD of triplicate quantification.
Statistics
All the experiments were carried out at least three times. Results were displayed as mean ± SD, and the difference between groups was calculated by a two-tailed student’s t-test using SPSS 20.0 software (IBM, Chicago, IL, USA), with statistical significance assumed at P < 0.05.
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by the National Key Research & Development Program of China (2016YFC1303303, Q.S.), the National Natural Science Foundation of China (81572799, H.H., 81472588, 31671432, Q.S., 81471578, 31770975, X.W.), the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2015ZX 09501-009, Q.S.), Beijing Natural Science Foundation (7162091, H.H.), the National Basic Research Program of China (2015CB553704, Q.S.), the Beijing Health System High-level Health Technology Personnel Training Program (2015-3-058, H.H.), the open fund of State Key Laboratory of cancer biology (CBSKL201704, Y.Z.), the Youth Innovation Fund (2017CXJJ31, Y.Z.).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Wang S, Guo Z, Xia P, et al. Internalization of NK cells into tumor cells requires ezrin and leads to programmed cell-in-cell death. Cell Res. 2009;19:1350–1362. doi: 10.1038/cr.2009.114. [DOI] [PubMed] [Google Scholar]
- 2.Huang H, Chen Z, Sun Q. Mammalian cell competitions, cell-in-cell phenomena and their biomedical implications. Curr Mol Med. 2015;15:852–860. doi: 10.2174/1566524015666151026101101. [DOI] [PubMed] [Google Scholar]
- 3.Sun Q, Huang H, Overholtzer M. Cell-in-cell structures are involved in the competition between cells in human tumors. Mol Cell Oncol. 2015;2:e1002707. doi: 10.1080/23723556.2014.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q, Luo T, Ren Y, et al. Competition between human cells by entosis. Cell Res. 2014;24:1299–1310. doi: 10.1038/cr.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overholtzer M, Mailleux AA, Mouneimne G, et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell. 2007;131:966–979. doi: 10.1016/j.cell.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 6.Ning X, Luo T, Chen Z, Sun Q. The physics for the formation of cell-in-cell structures. Curr Mol Med. 2015;15:867–872. doi: 10.2174/1566524015666151026105123. [DOI] [PubMed] [Google Scholar]
- 7.Wang M, Ning X, Chen A, et al. Impaired formation of homotypic cell-in-cell structures in human tumor cells lacking alpha-catenin expression. Sci Rep. 2015;5:12223. doi: 10.1038/srep12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Q, Cibas ES, Huang H, Hodgson L, Overholtzer M. Induction of entosis by epithelial cadherin expression. Cell Res. 2014;24:1288–1298. doi: 10.1038/cr.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan B, Zhang B, Chen A, et al. Cholesterol inhibits entotic cell-in-cell formation and actomyosin contraction. Biochem Biophys Res Commun. 2018;495:1440–1446. doi: 10.1016/j.bbrc.2017.11.197. [DOI] [PubMed] [Google Scholar]
- 10.Hamann JC, Surcel A, Chen R, et al. Entosis Is Induced by Glucose Starvation. Cell Rep. 2017;20:201–210. doi: 10.1016/j.celrep.2017.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cano CE, Sandi MJ, Hamidi T, et al. Homotypic cell cannibalism, a cell-death process regulated by the nuclear protein 1, opposes to metastasis in pancreatic cancer. EMBO Mol Med. 2012;4:964–979. doi: 10.1002/emmm.201201255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martins I, Raza SQ, Voisin L, et al. Entosis: The emerging face of non-cell-autonomous type IV programmed death. Biomed J. 2017;40:133–140. doi: 10.1016/j.bj.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David JM, Dominguez C, Hamilton DH, Palena C. The IL-8/IL-8R axis: a double agent in tumor immune resistance. Vaccines (Basel) 2016;4 doi: 10.3390/vaccines4030022. pii: E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernando RI, Castillo MD, Litzinger M, Hamilton DH, Palena C. IL-8 signaling plays a critical role in the epithelial-mesenchymal transition of human carcinoma cells. Cancer Res. 2011;71:5296–5306. doi: 10.1158/0008-5472.CAN-11-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.