Abstract
Exposure to inorganic arsenic (iAs) during pregnancy is associated with adverse health outcomes present both at birth and later in life. A biological mechanism may include epigenetic and genomic alterations in fetal genes involved in immune functioning. To investigate the role of the maternal immune response to in utero iAs exposure, we conducted an analysis of the expression of immune-related genes in pregnant women from the New Hampshire Birth Cohort Study. A set of 31 genes was identified with altered expression in association with levels of urinary total arsenic, urinary iAs, urinary monomethylated arsenic and urinary dimethylated arsenic. Notably, maternal gene expression signatures differed when stratified on fetal sex, with a more robust inflammatory response observed in male pregnancies. Moreover, the differentially expressed genes were also related to birth outcomes. These findings highlight the sex-dependent nature of the maternal iAs-induced inflammatory response in relationship to fetal outcomes.
Keywords: Arsenic, In utero exposure, Maternal exposure, Inflammatory response
1. Introduction
Exposure to inorganic arsenic (iAs) in drinking water remains a significant public health problem [1]. Globally, it is estimated that more than 100 million people are exposed to iAs via drinking water at levels that exceed the Environmental Protection Agency and World Health Organization’s recommended limit of 10 μg/L [2,3]. Populations experiencing such exposures have been documented in Bangladesh, Chile, Mexico, Taiwan, and the United States [1,4]. Of concern, chronic exposure to iAs has been associated with numerous adverse health outcomes in adults, including cancers of the skin, lung, bladder, liver, and kidney; diabetes mellitus; cognitive impairment; and coronary heart disease [1,5,6].
There is great interest in understanding the biological mechanisms underlying the health effects related to iAs exposure that occurs during pregnancy and early childhood. iAs and its methylated metabolites cross the placental barrier and are observed in cord blood at levels similar to those found in the mother [7,8]. Moreover, exposure to iAs during this period may underlie diseases that emerge at birth and later in life [7,9,10]. Prenatal iAs exposure has been associated with outcomes including spontaneous abortion, stillbirth, and fetal growth, in a sex-dependent manner [11-13]. Children who experience such exposures are at risk for delayed neurodevelopment, increased risk of infection and immune dys-function, and face higher rates of cancer and non-cancer mortality later in life [9,14,15]. Taken together, these data suggest that the prenatal period represents a developmental window of susceptibility to the toxic effects of iAs.
Prenatal and early life exposure to iAs has been linked to dys-regulated immune system functioning in infants and children. At the molecular level, in utero iAs exposure is associated with an activation of inflammation-related gene expression, includeing the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB) signaling cascade, that may ultimately impact health status [16]. Additional research has demonstrated that epigenetic reprogramming is evident in cells from the fetus, with both CpG methylation and miRNA expression associated with maternal urinary iAs levels [17,18]. Notably, the genes that display altered expression are enriched for innate and adaptive immune processes [17]. At the functional level, both thymic and cell-mediated immune functioning have been inversely associated with increasing iAs exposure in school-aged children, indicating immunosuppression [19,20]. Moreover, in utero and early life exposure to iAs has been linked to increased susceptibility to infection [9,21]. In addition to immunosuppression, iAs exposure has been linked to the presence of pro-inflammatory markers in infants and children [22,23]. Such immunomodulatory effects on the immune system during early life have the potential to impact the maturation of the immune system and may play a role in the development of the diverse health effects associated with iAs exposure [10,24]. In spite of the large body of evidence for immune and inflammatory effects in children and infants with prenatal iAs exposure, there is a gap in research on the genomic effects of iAs exposure on pregnant women themselves.
Importantly, the impact of exposure to both iAs and its metabolites on health should be considered. Six major forms of arsenic have been identified in human urine. These include inorganic arsenicals (iAsIII and iAsV), which are metabolized to monomethylarsonic acid (MMAsV) and reduced to monomethylarsonous acid (MMAsIII). MMAsIII is then methylated to dimethylated arsenic species (DMAs) – dimethylarsinous acid (DMAsIII) and dimethyarsinic acid (DMAsV) [25,26]. There is growing evidence that levels of these individual arsenic species have differential toxico-logic effects in human populations. For instance, elevated levels of urinary MMAs have been specifically associated with cancers, cardiovascular disease, and birth outcomes in human populations [25,27,28].
Despite increasing research efforts into the effects of in utero iAs exposure, the effects of iAs on pregnant women themselves is understudied. Pregnancy represents a metabolically unique period of time for women during which iAs metabolism is altered [8,29]. To better understand the effects of iAs exposure during pregnancy on the mother, we conducted a targeted genomic analysis of changes in gene expression associated with urinary arsenic species in a cohort of pregnant women living in New Hampshire exposed to relatively low levels of iAs via well water. This analysis specifically targeted immune- and inflammatory-related gene expression because of strong evidence that immune functioning is altered in populations with prenatal iAs exposure [9,16-18,20,22,30-32]. This study is one of the first to analyze the relationship between iAs exposure, individual iAs metabolites, maternal gene expression and birth outcomes conducted in a pregnancy cohort.
2. Methods
2.1. Study population
The current study included samples from 48 pregnant women taking part in the New Hampshire Birth Cohort Study, a study of women using private wells during pregnancy, described previously [33]. Briefly, women, aged 18-45, were enrolled through prenatal clinics in New Hampshire between 24 and 28 weeks gestation if they used a private well in their home at enrollment and had no plans to move prior to delivery. The women selected for analysis in this study represented the range of drinking water iAs exposure in the larger New Hampshire Birth Cohort Study (Table 1). Questionnaires were given both at enrollment and at two weeks postpartum in order to assess changes in covariates. Information was collected regarding smoking status, drinking status, and medications taken during pregnancy. All participants provided informed consent prior to enrollment. All study procedures have been approved by the Internal Review Board at Dartmouth College.
Table 1.
Demographics for eligible births.
| StudySample N = 48 N (%) or mean, median [range] |
New Hampshire Birth Cohort Study N = 1033 N (%) or mean, median [range] |
|
|---|---|---|
| Race | ||
| White | 48 (100) | 1002 (98.4) |
| Non-White | 0 (0.0) | 16 (1.57) |
| Missing | 0 | 15 |
| Ever smoke | ||
| Yes | 4 (9.09) | 145 (15.7) |
| No | 40 (90.1) | 780 (94.3) |
| Missing | 4 | 108 |
| Drinking status during pregnancy | ||
| Yes | 7 (15.6) | 59 (6.38) |
| No | 38 (84.4) | 866 (93.6) |
| Missing | 3 | 108 |
| Fetal sex | ||
| Male | 24 (50.0) | 514 (49.8) |
| Female | 24 (50.0) | 519 (50.2) |
| Maternal age at enrollment (years) | 31.5, 31.5 [20.0-40.0] | 31.1, 30.9 [18.4-44.8] |
| DW-iAS μg/L) | 2.43, 0.500 [0.100-8.18] | 4.08, 0.440 [<0.01-189.34] |
| U-tAs μg/L) | 6.83, 5.56 [0.25-32.46] | 6.05, 3.78 [0.21-288.48] |
| U-iAs μg/L) | 1.95, 0.98 [0.11-12.27] | 0.75, 0.31 [0.02-37.29] |
| U-MMAsVμg/L) | 0.87, 0.90 [0.06-2.11] | 0.49, 0.31 [0.01-28.35] |
| U-DMAsV μg/L) | 4.02, 3.50 [0.07-23.26] | 4.80, 2.84 [0.07-281.07] |
| Gestational age (weeks) | 39.31, 39.74 [32.28-41.94] | 39.31, 39.57 [25.29-44.85] |
| Ponderal index | 2.61, 2.66 [1.90-3.28] | 2.62, 2.60 [1.47-4.26] |
| Birth weight (grams) | 3443.97, 3518.49 [1455.00-5194.00] | 3414.66, 3435.00 [357.20-5318.00] |
| Birth length (inches) | 19.98, 20.08 [16.14-22.00] | 19.96, 20.00 [11.00-24.50] |
| Head circumference (cm) | 34.55, 35.00 [28.50-38.00] | 34.6, 34.50 [28.00-53.34] |
Subject demographics for pregnant women in the sub cohort (n = 48) are shown in comparison to the overall New Hampshire Birth Cohort (n = 1033).
2.2. Maternal urine collection and analysis
Women provided a spot urine sample at study enrollment (approximately 24-28 weeks gestational age). The specimen containers were pre-labeled, acid-washed, screw-top, 120-mL containers that had 30 uL of 10 mM diammonium diethyldithio-carbamate to stabilize arsenic species. Samples were sent to the University of Arizona Hazard Identification Core for analysis, as previously described [33]. In short, urine samples were analyzed for urinary arsenic levels using a high-performance liquid chromatography (HPLC) coupled to an inductively coupled plasma mass spectroscopy (ICP-MS) system capable of quantitatively determining levels of five arsenic species in urine: AsIII, AsV, MMAsV, DMAsV, and arsenobetaine [33]. Urinary iAs (U-iAs) was calculated by summing the levels of AsIII and AsV. Total urinary arsenic (U-tAs) was calculated by summing U-iAs and the metabolic products urinary MMAV (U-MMAs) and urinary DMAV (U-DMAs). Arsenobetaine was excluded from this analysis because it is not known to be absorbed or metabolized by humans [34].
2.3. Maternal blood collection and RNA extraction
Blood samples were collected at the visit during which the routine glucose challenge test was taken, approximately between 24 and 28 weeks gestation. Blood was drawn into a 2.5 mL Qiagen PAXgene RNA tube, containing an RNA stabilizing agent. Collection tubes were maintained at 4 °C and sent to the study laboratory at Dartmouth within 24 h and stored at −80 °C until they were processed for RNA isolation. Samples were thawed and processed in batches using the PAXgene Blood mRNA kit (PreAnalytix 763134), according to the manufacturer’s instructions, and quantified by Nanodrop. In order to remove any contaminating DNA, RNase-free DNase was added during the extraction process. RNA integrity of batch controls was assessed by BioAnalyzer fragment analysis.
2.4. Analysis of gene expression in maternal blood
Gene expression levels were measured using the Human Immunology v2 NanoString panel (NanoString Technologies), which assesses the levels of 594 inflammatory- and immune-related genes. The NanoString hybridizing technology provides direct quantification without requiring amplification of the sample [35]. Furthermore, this technology serves as a high-sensitivity alternative to RT-qPCR with established increased accuracy [36-38]. NanoString data from maternal blood were normalized using SAS v9.3 (SAS Institute, Cary, NC) according to the manufacturer’s recommendation. To perform normalization, gene expression values were adjusted by a sample normalization factor. This factor is calculated for each sample by dividing the geometric mean of housekeeping and control gene expression by the mean of the geometric means across all samples. These normalization steps control for batch effect and artifact error. A total of 15 housekeeping genes were included on the Human Immunology v2 NanoString panel, namely ATP Binding Cassette Subamily F Member 1 (ABCF1), 5-aminolevulinate synthase 1 (ALAS1), eukaryotic translocation elongation factor 1 gamma (EEF1G), glucose-6-phosphate dehydrogenase (GP6D), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glucuronidase beta (GUSB), hypoxanthine phosphoribo-syltransferase 1 (HPRT1), ornithine decarboxylate antizyme (OAZ1), RNA polymerase I subunit B (POLR1B), RNA polymerase II subunit A (POLR2A), peptidylprolyl isomerase A (PPIA) succinate dehydrogenase complex flavoprotein subunit A (SHDA), TATA-box binding protein (TBP), tubulin beta class 1 (TUBB), and ribosomal protein L19 (RPL19), were used for normalization. This suite of housekeeping genes is selected to span genes with both low and high expression levels. Genes that were expressed below the stated manufacturer threshold in more than 25% of subjects were excluded from analysis.
2.5. Statistical analyses
Differentially expressed genes were identified using multivariable linear regression models, adjusting for a history of smoking (ever smoked/never), drinking status during pregnancy (ever drank/never), maternal age at enrollment (years), the time point during pregnancy that blood samples were drawn (weeks gestation), and infant sex (male/female). Models including U-iAs, U-MMAs, or U-DMAs were analyzed in two ways. First, models were constructed including the proportional amount of each metabolite group (i.e.%U-iAs). Second, models were constructed using the levels of U-iAs, U-MMAs, or U-DMAs, with U-tAs included as a covariate in order to control for overall iAs exposure. Smoking status, drinking status, maternal age, time at blood draw, and the sex of the infant were selected as covariates a priori. No association was observed between any medications taken during pregnancy and the outcome, therefore, medications were not included in the model. Finally, stratified models analyzed based on fetal sex were run. Genes were considered differentially expressed if they had an overall p-value < 0.05 and a false discovery rate-corrected q-value < 0.1 in relation to urinary iAs measures. Analysis was carried out using Partek Genomic Suite (St. Louis, MO).
To determine whether differentially expressed genes were associated with each of the five birth outcomes, simple and multivariable linear regression models were used. The birth outcomes included gestational age, birth weight, birth length, head circumference, and ponderal index. No covariates were controlled for in the analysis with gestational age. For models with the remaining birth outcomes, gestational age was included as a covariate. Genes were considered significantly associated with the outcome if p-value < 0.05 and false discovery rate corrected q-value < 0.1. Analysis was done using SAS v9.3 (SAS Institute, Cary, NC).
2.6. Adjusting gene expression by white blood cell composition
Genes with expression associated with urinary arsenic levels were examined to determine whether the differential expression was detected due to differences in white blood cell composition of the samples. In order to take this into account, a list of 1135 genes associated with specific white blood cells was used [39]. These genes were previously identified as being associated with a white blood cell subset, including B-cells, CD4+ T-cells, CD8+ T-cells, granulocytes, and lymphocytes [39]. The white blood cell-associated genes were compared with genes identified as differentially expressed in association with urinary arsenic levels. This method of correction for white blood cell composition has previously been used where blood count data are unavailable [17,40-42].
2.7. Identification of enriched gene ontologies and biological pathways
Gene ontology and pathway enrichment were analyzed using the Functional Annotation Chart feature on the Database for Annotation, Visualization and Integrated Discovery v 6.7 (DAVID; david.ncifcrf.gov). Gene ontology terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were considered enriched in the gene sets if p-value < 0.05 and false discovery rate-corrected q-value < 0.1.
2.8. Comparison with previous published iAs-associated genes and proteins
The genes identified as differentially expressed in the non-stratified and stratified models were compared against genes previously identified in infants with in utero exposure to iAs. A previously published database of genes (n = 843 genes) associated with in utero iAs exposure [43] was updated with data from four additional studies [44-47]. The full list of genes and their source studies (n = 953 genes; n = 16 studies) included in the database can be found in Table S4 in the online version at DOI: 10.1016/j.reprotox.2017.07.023.
2.9. Validation of gene expression using maternal serum protein measures in the BEAR cohort
As data on maternal protein measures were unavailable in the New Hampshire Birth Cohort Study, the differential expression of maternal immune-related genes were partially validated by testing serum protein expression in a subset of 50 pregnant women from the Biomarkers of Exposure to ARsenic (BEAR) cohort, which is described elsewhere, in greater depth [17,32,43]. Briefly, pregnant women living in the Gómez Palacio region of Mexico were enrolled into BEAR cohort approximately 24 hours prior to delivery. They were recruited if they 1) lived in the Gómez Palacio region for at least one year, 2) had a confirmed singleton pregnancy without complications, and 3) had overall good health status with no sign of chronic or acute disease. Following written and informed consent, participants completed questionnaires to provide information on socioeconomic factors and potential sources of iAs exposure.
Maternal spot urine was collected before delivery, shipped to UNC-Chapel Hill on dry ice, and frozen at −80° C. Urinary levels of U-iAs, U-MMAs, and U-DMAs were measured using hydride generation-atomic absorption spectrometry in the UNC-Chapel Hill Nutrition Obesity Research Center, as previously described [27,32]. Urinary arsenic measures were adjusted for differences in hydration using specific gravity, which was measured using a handheld refractometer (Reichert TX 400 #13740000; Reichert Inc., Depew, NY). Urinary arsenic measures were adjusted according to the following equation: iAs x (mean SG-1)/(individual SG - 1).
Maternal blood samples were collected at the time of delivery into a BD Vacutainer tube with a clot activator (Becton, Dickinson, and Company, Franklin Lakes, NJ). After clot formation, blood tubes were centrifuged and the supernatant was collected and stored at −70 °C until proteomic analyses were performed. Serum protein was analyzed using the Biotin Label-based Human Antibody Array I, L series 507 (RayBiotech, Norcross, GA), which measures the relative expression of 507 proteins that are involved in a range of cellular signaling functions, including cytokines, growth factors, and soluble receptors, among others [32].
We analyzed expression of the protein targets on the Human Antibody Array that overlapped with differentially expressed genes identified in mothers from the New Hampshire Birth Cohort Study using the NanoString Human Immunology v2 panel. Of the 507 proteins on the Human Antibody Array, 68 target proteins correspond to 65 differentially expressed genes identified in the overall and sex-stratified analyses performed above. The relationship between U-tAs, U-iAs, U-MMAs, and U-DMAs and serum protein levels were tested using multivariable linear regression models identical to those used for mRNA expression. As all samples were collected at the time of delivery, the time point during pregnancy that blood samples were drawn (weeks gestation) was not included as a covariate in the model.
3. Results
3.1. Study subject characteristics
Samples from a sub-cohort of 48 women from the New Hampshire Birth Cohort Study were analyzed. Study subject demographics are summarized in Table 1. The subjects ranged in age from 20 to 40, with an average of 31.5 years. The overall mean U-tAs in these subjects was 6.83 μg/L (median = 5.56 μg/L), with a range of 0.25 - 32.46 μg/L. The average urinary concentration for individual arsenic species was 1.95 μg/L, 0.87 μg/L, and 4.02 μg/L for U-iAs, U-MMAs, and U-DMAs, respectively. The number of women who reported that they had ever smoked was four (9.1%) and the number of women who reported that they had ever consumed alcohol during pregnancy was seven (15.6%). Women with incomplete urinary arsenic measures were excluded from further analysis, leaving samples from 47 women for the subsequent genomic analyses.
3.2. Urinary arsenic and gene expression in pregnant women
First, the association between gene expression levels for 594 genes and U-tAs was examined across all subjects. Three genes were positively associated with U-tAs, namely intracellular adhesion molecule 5 (ICAM5), C-C motif chemokine ligand 7 (CCL7), and major histocompatibility complex, class II, DQ, alpha 1 (HLA-DQA1) (Table 2).
Table 2.
Differentially expressed genes associated with U-tAs, U-iAs, U-MMAs, and U-DMAs in the non-stratified model (n = 31).
| Gene ID | Gene Name | U-tAs β (p-value) |
U-iAs β (p-value) |
U-MMAsβ(p-value) | U-DMAs β (p-value) |
|---|---|---|---|---|---|
| ICAM5 | Intracellular Adhesion Molecule 5 | 0.417 (0.002) |
-0.314 (0.01) |
- | 0.320 (0.01) |
| CCL7 | C-C Motif Chemokine Ligand 7 | 0.291 (0.03) |
- | - | - |
| HLA-DQAV | Major Histocompatability Complex, Class II, DQAlpha 1 |
0.298 (0.04) |
- | - | - |
| TNFSF11 | Tumor Necrosis Factor Superfamily 11 |
- | -0.373 (0.01) |
- | 0.358 (0.01) |
| IL18RAPa | Interleukin 18 ReceptorAccessory Protein |
- | -0.371 (0.01) |
- | 0.353 (0.02) |
| PLAU | Plasminogen Activator, Urokinase | - | -0.337 (0.02) |
- | 0.335 (0.02) |
| CCL26 | C-C MotifChemokine Ligand 26 | - | -0.335 (0.02) |
- | 0.344 (0.02) |
| AIRE | Autoimmune Regulator | - | -0.342 (0.02) |
- | 0.333 (0.02) |
| NOD2 | Nucleotide Binding Oligomerization Domain Containing 2 |
- | -0.312 (0.02) |
- | 0.315 (0.03) |
| SERPING1a | Serpin Family G Member1 | - | -0.318 (0.03) |
- | 0.291 (0.05) |
| THY1 | Thy-1 Cell Surface Antigen | - | -0.325 (0.03) |
- | 0.322 (0.03) |
| PPARG | Peroxisome ProliferatorActivated Receptor Gamma |
- | -0.329 (0.03) |
- | 0.309 (0.04) |
| HLA-DRB1a | Major Histocompatability Complex, Class II, DR Beta 1 |
- | -0.312 (0.03) |
- | - |
| C7 | Complement Component 7 | - | -0.297 (0.03) |
- | 0.290 (0.04) |
| EGR1a | Early Growth Response 1 | - | -0.302 (0.03) |
- | 0.290 (0.04) |
| C9 | Complement Component 9 | - | -0.289 (0.03) |
- | 0.285 (0.04) |
| CEACAM6 | Carcinoembryonic Antigen Related Cell Adhesion Molecule 6 |
- | -0.303 (0.03) |
- | 0.312 (0.03) |
| MUC1 | Mucin 1, Cell Surface Associated | -0.319 (0.03) |
- | - | |
| IL17A | Interleukin 17A | - | -0.306 (0.04) |
- | 0.289 (0.05) |
| PIGR | Polymeric Immunoglobulin Receptor |
- | -0.317 (0.04) |
- | - |
| GP1BB | Glycoprotein 1b Platelet Beta Subunit |
- | -0.308 (0.04) |
- | 0.311 (0.03) |
| CD1A | CD1a Molecule | - | -0.310 (0.04) |
- | 0.291 (0.05) |
| CTSG | Cathepsin G | - | -0.291 (0.04) |
- | 0.293 (0.04) |
| LAG3 | Lymphocyte Activating 3 | - | -0.291 (0.04) |
- | - |
| IGF2R | Insulin Like Growth Factor 2 Receptor |
- | -0.298 (0.04) |
- | - |
| MMEa | Membrane Metallo-Endopeptidase | - | -0.293 (0.04) |
- | - |
| ITLN2 | Intelectin 2 | - | -0.274 (0.05) |
- | - |
| GBP1a | Guanylate Binding Protein 1 | - | -0.285 (0.05) |
- | - |
| LILRA6 | Leukocyte Immunoglobulin Like Receptor A6 |
- | -0.295 (0.05) |
- | - |
| ICAM1 | Intracellular Adhesion Molecule 1 | - | -0.289 (0.05) |
- | - |
| MX1 | MX Dynamin Like GTPase 1 | - | - | 0.304 (0.04) | - |
Genes identified as differentially expressed are displayed with their associated β- and p-values in their respective multivariable linear regression models.
Indicates that differential gene expression is associated with shifts in white blood cell composition.
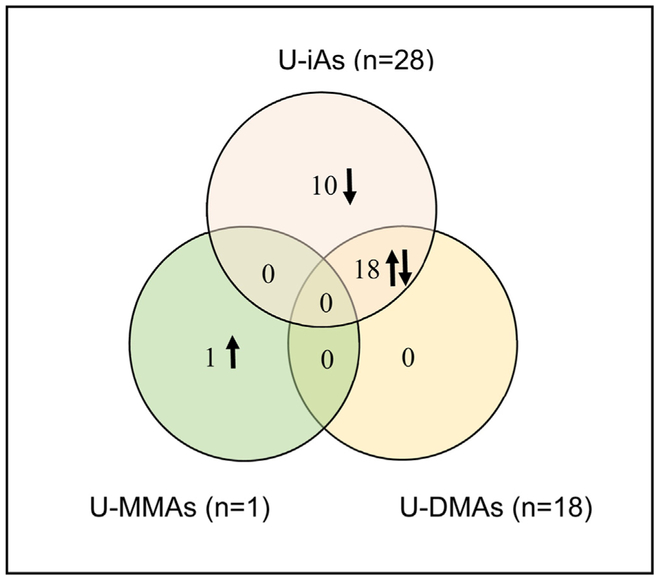
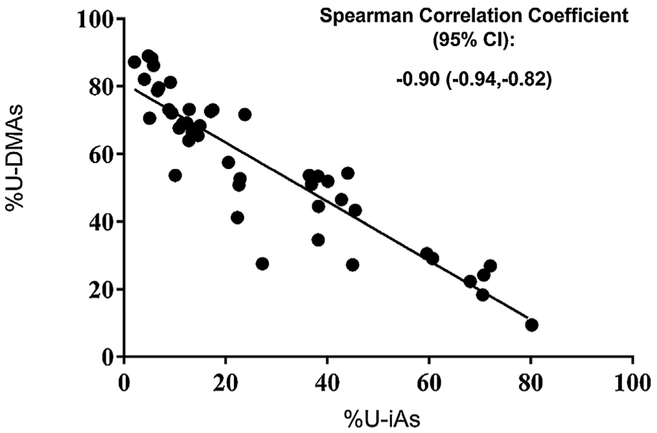
Next, associations between U-iAs, U-MMAs, and U-DMAs and maternal gene expression were tested using two different models. The first model, where exposure was measured as the proportion of the urinary arsenic metabolites, resulted in the identification of no genes with expression associated with any urinary arsenic metabolites. The second model, examining gene expression in relation to levels of urinary arsenic metabolites while adjusting for U-tAs was tested. Here, a total of 29 genes were identified as differentially expressed in association with U-iAs or its metabolites (Table 2). Of these, 28 genes had expression levels that were negatively associated with U-iAs, while one gene displayed expression that was positively associated with U-MMAs and 18 with U-DMAs. Across U-tAs, U-iAs, U-MMAs, and U-DMAs, a total of 31 genes were identified as differentially expressed. Overlaps between genes associated with U-iAs, U-MMAs, and U-DMAs are shown in Fig. 1. Only seven of the 31 genes are known to be associated with shifts in white blood cell composition [39]. Notably, every gene identified as positively associated with U-DMAs was also negatively associated with U-iAs. This is likely due to the fact that the proportions of U-iAs and U-DMAs are significantly negatively associated with one another because as arsenic is metabolized, U-iAs levels decrease as U-DMAs levels increase (Fig. 2).
Fig. 1.
Venn-diagram depicting relationships between genes with expression associated with U-iA, U-MMAs, and U-DMAs. Complete overlap between genes with expression associated with U-DMAs and U-iAs was observed while no overlap was observed between the other genes.
Fig. 2.
Correlation between proportions of U-iAs and U-DMAs across all subjects (n = 47). The results indicate that there is a strong negative relationship between the two urinary species. The Pearson correlation coefficient is −0.909 and is significant at a 95% significance level.
3.3. Maternal immune gene signatures differ by fetal sex
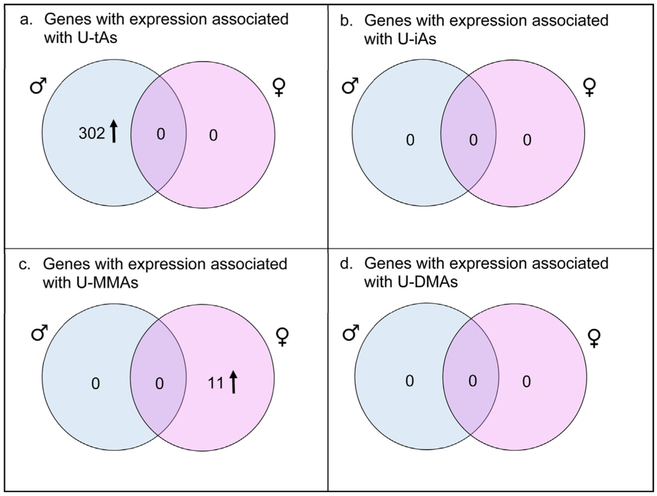
After analysis across all subjects, stratified models were run based on infant sex. A total of 302 genes were identified to have expression levels that were positively associated with U-tAs in the lymphocytes of women pregnant with a male infant. No genes had expression that was significantly associated with U-iAs, U-MMAs, or U-DMAs in women pregnant with a male infant. Of the 302 differentially expressed genes in women pregnant with a male infant, 53 (~17%) of them are known to be associated with shifts in white blood cell composition [39]. For women pregnant with a female infant, there were 11 genes with expression that was positively associated with U-MMAs, but none with U-tAs, U-iAs, and U-DMAs (Fig.3; seeTable S1 in the online version at DOI: 10.1016/j.reprotox2017.07.023. When compared with genes known to be associated with white blood cell composition, only two of the 11 genes were identified as being associated with white blood cell composition (see Table S1 in the online version at DOI: 10.1016/j.reprotox2017.07.023. [39].
Fig. 3.
Venn-diagrams depicting relationships between genes with expression associated with U-tAs and U-MMAs in the sex-stratified models. The results indicate that there is no overlap between the genes associated with arsenical species in women pregnant with a male infant (shown in blue) and women pregnant with a female infant (shown in pink). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Gene enrichment for innate and adaptive immune-response functions
Genes identified as differentially expressed in association with U-tAs and the individual arsenic species were combined into three gene sets representing the non-stratified model (n = 31 genes) and two sex-stratified models (male infants n = 302 genes; female infants n = 11 genes). These three gene sets were then tested for enrichment of gene ontology and KEGG pathways. In the genes representing the non-stratified model, several gene ontologies were enriched, including innate and adaptive immune response and leukocyte-mediated immunity (Tables 3 and 4; see Table S2 in the online version at DOI: 10.1016/j.reprotox.2017.07.023).
Table 3.
Top ten enriched gene ontology terms in differentially expressed genes in the non-stratified and sex-stratified models.
| Genes Differentially Expressed in the Non-Stratified Model | ||||
|---|---|---|---|---|
| GOTERM | Description | # of Genes | p-value | FDR q-value |
| GO:0006955 | Immune response | 18 | 1.08E-15 | 1.65E-12 |
| GO:0006952 | Defense response | 11 | 2.19E-07 | 3.26E-04 |
| GO:0002684 | Positive regulation of immune system process |
7 | 7.58E-06 | 0.011295 |
| GO:0009611 | Response to wounding | 9 | 8.18E-06 | 0.012178 |
| GO:0045087 | Innate immune response | 6 | 8.36E-06 | 0.012451 |
| GO:0050778 | Positive regulation of immune response |
6 | 1.06E-05 | 0.015844 |
| GO:0002449 | Lymphocyte mediated immunity |
5 | 1.23E-05 | 0.018253 |
| GO:0002460 | Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin domains |
5 | 1.79E-05 | 0.026673 |
| GO:0002250 | Adaptive immune response | 5 | 1.79E-05 | 0.026673 |
| GO:0002443 | Leukocyte mediated immunity | 5 | 2.77E-05 | 0.04132 |
| Genes Differentially Expressed in Women Pregnant with a Male Infant | ||||
| GOTERM | Description | # of Genes | p-value | FDR q-value |
| GO:0006955 | Immune response | 160 | 5.19E-131 | 9.13E-128 |
| GO:0006952 | Defense response | 116 | 2.83E-79 | 4.98E-76 |
| GO:0006954 | Inflammatory response | 83 | 4.50E-66 | 7.92E-63 |
| GO:0009611 | Response to wounding | 96 | 1.93E-62 | 3.40E-59 |
| GO:0002684 | Positive regulation of immune system process |
65 | 5.57E-53 | 9.81E-50 |
| GO:0048584 | Positive regulation of response to stimulus |
56 | 1.03E-41 | 1.81E-38 |
| GO:0050778 | Positive regulation of immune response |
46 | 3.14E-40 | 5.53E-37 |
| GO:0045087 | Innate immune response | 41 | 1.83E-34 | 3.22E-31 |
| GO:0001775 | Cell activation | 53 | 1.43E-33 | 2.52E-30 |
| GO:0001817 | Regulation of cytokine production |
44 | 5.45E-33 | 9.59E-30 |
| Genes Differentially Expressed in Women Pregnant with a Female Infant | ||||
| GOTERM | Description | # of Genes | p-value | FDR q-value |
| GO:0006955 | Immune response | 6 | 6.92E-05 | 0.09 |
Displays the top ten enriched gene ontologies in the differentially expressed genes in the non-stratified model (n = 31) and sex-stratified models (n = 302, n = 11).
Table 4.
Top ten enriched KEGG Pathways in differentially expressed genes in the non-stratified and sex-stratified models.
| Genes Differentially Expressed in the Non-Stratified Model | ||||
|---|---|---|---|---|
| Pathway ID | Description | # of Genes | p-value | FDR q-value |
| - | - | - | - | - |
| Genes Differentially Expressed in Women Pregnant with a Male Infant | ||||
| Pathway ID | Description | # of Genes | p-value | FDR q-value |
| hsa04060 | Cytokine-cytokine receptor interaction | 64 | 1.16E-31 | 1.30E-28 |
| hsa04640 | Hematopoietic cell lineage | 31 | 3.69E-20 | 4.15E-17 |
| hsa04620 | Toll-like receptor signaling pathway | 33 | 5.37E-20 | 6.04E-17 |
| hsa04630 | Jak-STAT signaling pathway | 33 | 5.42E-14 | 6.09E-11 |
| hsa04672 | Intestinal immune network for IgA production | 16 | 1.12E-09 | 1.26E-06 |
| hsa04210 | Apoptosis | 20 | 3.63E-09 | 4.08E-06 |
| hsa05332 | Graft-versus-host disease | 14 | 4.63E-09 | 5.20E-06 |
| hsa04514 | Cell adhesion molecules (CAMs) | 24 | 8.24E-09 | 9.26E-06 |
| hsa05330 | Allograft rejection | 13 | 1.86E-08 | 2.09E-05 |
| hsa04610 | Complement and coagulation cascades | 17 | 2.64E-08 | 2.97E-05 |
| Genes Differentially Expressed in Women Pregnant with a Female Infant | ||||
| Pathway ID | Description | # of Genes | p-value | FDR q-value |
| - | - | - | - | - |
Displays the top ten enriched KEGG pathways in the differentially expressed genes in the non-stratified model (n = 31) and sex-stratified models (n = 302, n=11).
The 302 genes upregulated in women pregnant with a male infant were enriched for their role in cytokine-mediated signaling, Toll-like receptor signaling, JAK-STAT cascade, and positive regulation of tumor necrosis factor production, among others (Table 3; see Table S2 in the online version at DOI: 10.1016/j.reprotox.2017.07.023. The genes were enriched for KEGG pathways involved in auto-immune functioning, as well as diseases and signaling pathways that have been previously associated with arsenic exposure (Table 4; see Table S2 in the online version at DOI: 10.1016/j.reprotox.2017.07.023). These pathways include Toll-like signaling pathway, type I diabetes mellitus, and asthma. For women pregnant with a female infant, the eleven genes with differential expression were enriched for only immune response and no KEGG pathways were enriched (Table 3; see Table S2 in the online version at DOI: 10.1016/j.reprotox.2017.07.023).
3.5. Gene expression associations with birth outcomes
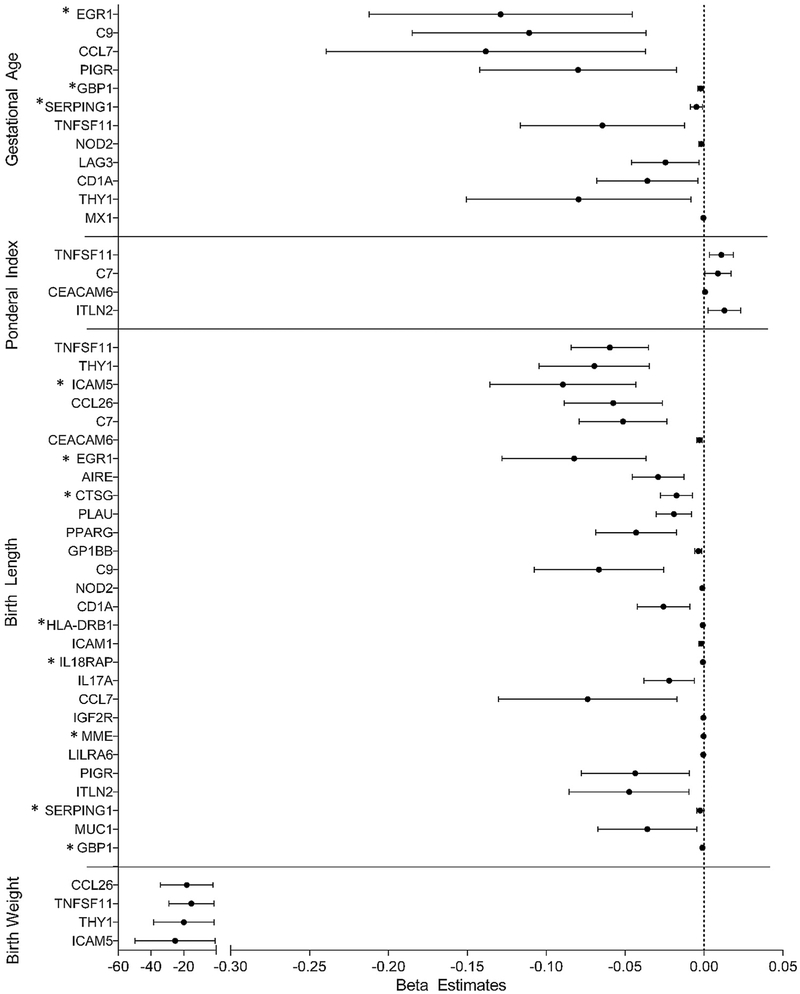
Having identified differentially expressed genes in circulating lymphocytes of pregnant women associated with iAs exposure in the non-stratified and sex-stratified models, we next set out to assess these genes in relation to birth outcomes. Each of the five birth outcomes, namely gestational age, birth weight, birth length, ponderal index, and head circumference, was regressed onto each differentially expressed gene. For the 31 genes identified in the non-stratified model, expression levels for 12 were significantly negatively associated with gestational age, four with birth weight, and 28 with birth length. The expression levels of four genes were significantly positively associated with the ponderal index. No genes displayed expression that was associated with head circumference (Fig. 4; Table 5). Expression of early growth response 1 (EGR1) was most strongly associated with gestational age. Tumor necrosis factor superfamily 11 (TNFSF11) was most strongly associated with both the ponderal index and birth length. C-C motif chemokine ligand 26 (CCL26) was most strongly associated with birth weight.
Fig. 4.
Forest plot displaying beta coefficients and 95% confidence intervals for arsenic-associated genes with expression that are associated with birth outcomes in the non-stratified cohort. *indicates that differential gene expression is associated with differences in white blood cell composition.
Table 5.
Differentially expressed genes in the non-stratified model (n = 31) are associated with birth outcomes.
| Gene ID | Gene Name | B (p-value) | FDR q-value |
|---|---|---|---|
| Gestational Age (Weeks) | |||
| EGR1a | Early growth response 1 | −0.13 (0.003) | 0.003 |
| C9 | Complement component 9 | −0.11 (0.004) | 0.003 |
| CCL7 | C-C motif chemokine ligand 7 | −0.14 (0.008) | 0.003 |
| PIGR | Polymeric immunoglobulin receptor | −0.080 (0.01) | 0.003 |
| GBP1a | Glycoprotein 1b platelet beta subunit | −0.0022 (0.01) | 0.003 |
| SERPING1a | Serpin family G member 1 | −0.0049 (0.01) | 0.003 |
| TNFSF11 | Tumor necrosis factor superfamily 11 | −0.064 (0.02) | 0.003 |
| NOD2 | Nucleotide binding oligomerization domain containing 2 |
−0.0018 (0.02) | 0.003 |
| LAG3 | Lymphocyte activating 3 | −0.025 (0.03) | 0.003 |
| CD1A | CD1a molecule | −0.036 (0.03) | 0.003 |
| THY1 | Thy-1 cell surface antigen | −0.080 (0.03) | 0.003 |
| MX1 | MX Dynamin like GTPase 1 | −0.00046 (0.03) | 0.003 |
| Ponderal Index | |||
| TNFSF11 | Tumor necrosis factor superfamily 11 | 0.011 (0.006) | 0.02 |
| ITLN2 | Intelectin 2 | 0.013 (0.02) | 0.02 |
| CEACAM6 | Carcinoembryonic antigen related cell adhesion molecule 6 |
0.00058 (0.02) | 0.02 |
| C7 | Complement component 7 | 0.0087(0.04) | 0.02 |
| Birth Length (Inches) | |||
| TNFSF11 | Tumor necrosis factor superfamily 11 | −0.060 (0.0001) | 0.0001 |
| THY1 | Thy-1 cell surface antigen | −0.070 (0.0002) | 0.0001 |
| ICAM5a | Intracellular adhesion molecule 5 | −0.089 (0.0003) | 0.0001 |
| CCL26 | C-C motif chemokine ligand 26 | −0.058 (0.0005) | 0.0001 |
| C7 | Complement component 7 | −0.051 (0.0005) | 0.0001 |
| EGR1a | Early growth response 1 | −0.082 (0.0007) | 0.0001 |
| CEACAM6 | Carcinoembryonic antigen related cell adhesion molecule 6 |
−0.0030 (0.0007) | 0.0001 |
| AIRE | Autoimmune regulator | −0.029 (0.0008) | 0.0001 |
| CTSGa | Cathepsin G | −0.018(0.001) | 0.0001 |
| PLAU | Plasmonigen activator, urokinase | −0.019(0.001) | 0.0001 |
| PPARG | Peroxisome proliferator activated receptor gamma |
−0.043 (0.002) | 0.0001 |
| GP1BB | Glycoprotein 1b platelet beta subunit | −0.0036 (0.002) | 0.0001 |
| C9 | Complement component 9 | −0.067 (0.002) | 0.0002 |
| NOD2 | Nucleotide binding oligoerization domain containing 2 |
−0.0012(0.003) | 0.0002 |
| CD1A | CD1a molecule | −0.026 (0.003) | 0.0002 |
| HLA-DRB1a | Major histocompatibility complex, class II, DQ alpha 1 |
−0.00086 (0.005) | 0.0004 |
| ICAM1 | Intracellular adhesion molecule 1 | −0.0019(0.006) | 0.0004 |
| IL18RAPa | Interleukin 18 receptor accessory protein | −0.00071 (0.006) | 0.0004 |
| IL17A | Interleukin 17A | −0.022 (0.008) | 0.0004 |
| CCL7 | C-C motif chemokine ligand 7 | −0.074 (0.01) | 0.0006 |
| IGF2R | Insulin like growth factor 2 recceptor | −0.00044 (0.01) | 0.0006 |
| MMEa | Membrane metallo-endopeptidase | −0.00039 (0.01) | 0.0006 |
| LILRA6 | Leukocyte immunoglobulin like receptor A6 | −0.00056 (0.01) | 0.0006 |
| PIGR | Polymeric immunoglobulin receptor | −0.044 (0.01) | 0.0006 |
| ITLN2 | Intelectin 2 | −0.047 (0.01) | 0.0007 |
| SERPING1a | Serpin family G member 1 | −0.0025 (0.02) | 0.0009 |
| MUC1 | Mucin 1, cell surface associated | −0.036 (0.03) | 0.001 |
| GBP1 a | Guanylate binding protein 1 | −0.0011 (0.03) | 0.001 |
| Birth Weight (Grams) | |||
| CCL26 | C-C motif chemokine ligand 26 | −18.1918(0.03) | 0.01 |
| TNFSF11 | Tumor necrosis factor superfamily 11 | −15.3846 (0.03) | 0.01 |
| THY1 | Thy-1 cell surface antigen | −20.0278 (0.03) | 0.01 |
| ICAM5 | Intracellular adhesion molecule 5 | −25.2839 (0.04) | 0.01 |
The beta estimates, p-values, and FDR q-values for arsenic-associated genes that have expression associated with birth outcomes are displayed.
Indicates that differential gene expression is associated with shifts in white blood cell composition.
For the 302 genes with increased expression levels in women pregnant with a male infant, expression of 21 was negatively associated with gestational age, three with birth weight, and 240 with birth length. The expression of 23 genes was positively associated with ponderal index. For women pregnant with a female infant, six of the 11 genes had expression that was positively associated with gestational age, while five were significantly associated with birth length. Additionally, three of the genes had expression that was positively associated with ponderal index. No genes displayed expression levels that were significantly associated with head circumference in women pregnant with either a male or female infant (see Table S3 in the online version at DOI: 10.1016/j.reprotox.2017.07.23
3.6. Comparison with genes previously identified in iAs-exposed infants
A database of 953 genes with known differential expression and/or regulation in infants with in utero iAs exposure was developed (see Table S4 in the online version at DOI: 10.1016/j.reprotox.2017.07.023). A total of 47 of the genes in this database overlap with the genes that were identified as differentially expressed in the circulating lymphocytes of pregnant women with iAs exposure (see Table S5 in the online version at DOI: 10.1016/j.reprotox.2017.07.23). Of these 47 genes, 45 were previously identified as differentially expressed at the transcript or protein level, while the remaining two genes displayed differential CpG methylation in infants with in utero iAs exposure. Interestingly, over half of these genes (n = 28) were shared with those identified in a population of iAs-exposed infants in Thailand [16]. Moreover, several of these genes were also proposed biomarkers of in utero iAs exposure, namely chemokine C-X-C motif ligand 1 (CXCL1), EGR1, prostaglandin-endoperoxide synthase 2 (PTSG2) and suppressor of cytokine signaling 3 (SOCS3) [16]. However, EGR1 and SOCS3 are also both differentially expressed based on white blood cell composition, and changes in the expression of these genes may be due to the effects of iAs on white blood cell composition [39,46].
3.7. Differentially expressed genes correspond to differentially expressed proteins in the BEAR cohort
Using maternal serum protein measures available in the BEAR cohort, we sought to validate differential gene expression findings in the New Hampshire Birth Cohort Study. The 507 proteins included on the Human Antibody Array overlapped with seven genes differentially expressed in the overall New Hampshire Birth Cohort Study, and 61 and three genes differentially expressed in pregnant women carrying male and female infants, respectively (Table S6 in the online version at DOI: 10.1016/j.reprotox.2017.07.023). Of the overlapping proteins identified, we were not able to confirm differential expression of proteins corresponding to genes in the overall cohort or among women carrying female infants. However, in pregnant women carrying male infants, 20 (~33%) of the 61 overlapping proteins were significantly differentially expressed in association with U-tAs. Interestingly, all 20 of the differentially expressed proteins were inversely related to U-tAs levels (Table S7 in the online version at DOI: 10.1016/j.reprotox.2017.07.023).
4. Discussion
We used a high-sensitivity assay to assess differential gene expression of immune- and inflammatory-related genes in the serum of pregnant women living in New Hampshire exposed to relatively low levels of iAs via drinking water. Based upon the strong evidence from infant populations experiencing prenatal iAs exposure, our hypothesis was that the expression of immune- and inflammatory-genes would be altered in pregnant women with iAs exposure [9,16-18,20,22,30-32]. Maternal gene expression was examined in association with indicators of exposure to total arsenic and individual arsenic species, including U-iAs, U-MMAs, and U-DMAs, across all subjects, as well as stratified by the sex of the fetus. A total of 31 genes was identified as differentially expressed in relation to levels of urinary arsenic species in pregnant women. Using sex-stratified models, gene expression patterns differed in relation to individual arsenic species and between women pregnant with a male or a female. In support of links to children’s health, expression of many of these genes was associated with birth outcomes, namely, gestational age, birth weight, birth length, and ponderal index. The results provide further evidence of the molecular basis for the fetal sex-specific effects of iAs exposure on gene expression.
Within this cohort of pregnant women, gene expression patterns in lymphocytes differed in relation to the levels of U-iAs, U-MMAs, and U-DMAs. Specifically, the expression levels of 28 genes were negatively associated with U-iAs; one was positively associated with U-MMAs, and 18 were positively associated with U-DMAs. These genes are involved in innate and adaptive immune response, as well as cell-mediated immunity. The differential gene expression was established to be largely independent of white blood cell composition [48]. Interestingly, while maternal genomic responses have not been investigated in relation to iAs exposure during pregnancy, these results are supported by previous research on infants with in utero iAs exposure. For instance, it has been demonstrated that differentially expressed miRNAs and mRNAs are enriched for innate and adaptive immune processes in the cord blood of infants prenatally exposed to iAs [17]. Research in children with in utero and early life exposure has also found that iAs exposure is associated with impaired cell-mediated immunity [19]. The direction of the associations between individual arsenicals and gene expression indicate that increased levels of U-iAs may serve as an immunosuppressant, while increased levels of U-MMAs and U-DMAs may induce a pro-inflammatory response. These differences warrant further research into the effects, both at the molecular and phenotypic level, of exposure to the individual methylated metabolites of iAs.
A sex-dependent pattern in the gene expression signatures of immune-related genes was observed in pregnant women. In those pregnant with a male infant, a total of 302 genes displayed increased expression in relation to U-tAs levels, while only 11 genes were positively associated with U-MMAs in women pregnant with a female infant. These results were partially validated using protein expression from a separate cohort of iAs-exposed pregnant women, with women pregnant with male infants showing the altered serum expression of 20 of these proteins related to U-tAs levels. This highlights that women pregnant with male infants may experience a more robust inflammatory response to iAs exposure. This is supported by research demonstrating that males may experience greater risk of certain diseases associated with prenatal iAs exposure compared to females [14,23,49]. These differentially expressed genes are involved in Toll-like receptor signaling and tumor necrosis factor (TNF) production. In prior research, both Toll-like receptor and TNF signaling pathways have been shown to be dysregulated in children with in utero iAs exposure [17,23]. These results suggest that signaling pathways disrupted in pregnant women with iAs exposure are similar to those observed in infants and children with early life iAs exposure, in a sex-dependent manner.
Interestingly, we identified differentially expressed genes in the lymphocytes of pregnant women exposed to iAs that have previously been found to be differentially regulated in infants with in utero iAs exposure. Specifically, 47 (~15%) of the differentially expressed genes in pregnant women have also been previously identified in infants with in utero iAs exposure. Prenatal iAs exposure is associated with a robust immune response in infants. In prior research, 11 genes were identified as biomarkers of prenatal iAs exposure [16]. Interestingly, four of the genes that were dys-regulated in this study are members of this set of 11 biomarkers, namely, CXCL1, EGR1, PTSG2, and SOCS3 [16]. Of interest is EGR1, a transcription factor that is induced by growth factors and involved in cytokine signaling. EGR1 mRNA expression has also been previously shown to increase following in vitro iAs exposure [50]. Data from human populations and mouse models also suggests that early life EGR1 expression is critical in neurogenesis and may influence later life metabolic dysfunction, although it is unclear whether maternal expression of EGR1 during pregnancy may have similar effects [51,52]. However, as noted above, both EGR1 and SOCS3 are differentially expressed based on white blood cell composition and changes in the expression of these genes may be the result of iAs exposure on leukocyte subpopulations rather than the direct effect of iAs on gene expression [39,46,53]. Aside from these four biomarkers, the set of genes that overlap between iAs-exposed pregnant women and infants with in utero iAs exposure includes several TNF superfamily members and Toll-like receptors. This provides further evidence that iAs exposure disrupts these critical immune signaling pathways, and suggests that the maternal inflammatory response to iAs exposure is similar to that of infants with prenatal iAs exposure.
Genes differentially expressed in relation to levels of urinary arsenicals also were associated with birth outcomes, namely gestational age, birth length, birth weight, and the ponderal index. The same trends between arsenic-associated gene expression and birth outcomes was noted in the non-stratified and sex-stratified models. These genes were negatively associated with gestational age, birth weight, and birth length, while positively associated with the ponderal index. A proposed biomarker of in utero iAs exposure, EGR1, was the top gene associated with gestational age in the non-stratified model. Providing further support for these results, the 302 genes that were differentially expressed in women pregnant with a male infant are enriched for the Jak-STAT cascade, which is involved in growth hormone signaling, indicating that growth functions may be dysregulated [54]. Importantly, these results suggest that the maternal response to iAs exposure may have functional consequences in their infants, perhaps via the transfer of immune factors across the placenta. This is supported by additional evidence from the New Hampshire Birth Cohort Study indicating that levels of infant inflammatory markers may, in part, be mediated by maternal levels of inflammatory markers during pregnancy [55].
Several factors should be considered in the interpretation of our results. First, the study sample from New Hampshire Birth Cohort Study is relatively small and is racially and ethnically homogenous. This may limit both the statistical power and generalizability of these results. However, this sample homogeneity may also be regarded as a potential strength as it minimizes the inter-individual variability of the sample. Second, this study did not take into account contaminants other than iAs that may be present in well water, including other toxic metals that may similarly alter the expression of immune-related genes. Still, according to the United States Geological Survey, the New Hampshire population at risk of elevated drinking water iAs far outnumbers those at risk of other contaminants of concern, therefore, it is unlikely to significantly confound these results [56]. Third, because of the formal hypothesis on immune- and inflammatory-related gene expression in maternal samples, this study presents a targeted analysis of inflammatory genes, rather than an unbiased global analysis of gene expression. Therefore, future studies could examine genome-wide gene expression in pregnant women exposed to low levels of iAs to validate these findings and capture non-inflammatory and immune-related signaling pathways that may also be altered by iAs exposure during pregnancy. Lastly, this study examined gene expression in circulating lymphocytes. Further studies additionally may want to examine gene expression changes occurring in other tissues that may be more directly related to reproductive outcomes, such as the placenta.
In summary, this study identified the novel findings that immune-related gene expression signatures in pregnant women may depend on fetal sex. The inflammatory response in women pregnant with a male infant was much more robust than that observed in women pregnant with a female infant. These results provide molecular support for previous research demonstrating that iAs-associated diseases may be more severe in male infants and children that experience in utero iAs exposure [10,14,49]. The genes identified as differentially expressed in association with urinary arsenic species were also significantly associated with birth outcomes, namely gestational age, birth weight, birth length, and ponderal index. These results suggest that the maternal response to iAs exposure may be an important route to consider in determining the mechanism by which iAs exposure yields adverse birth outcomes in children.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institute of Environmental Health Sciences (T32ES007018, T32ES007126-34, P42ES005948, P42ES007373, P01ES022832), the National Institute of General Medical Sciences (P20GM104416) and the U.S. Environmental Protection Agency (RD-83544201).
No funding bodies had a role in the design, data collection, or analysis of this study, in the decision to publish, or the preparation of this manuscript.
Footnotes
Conflict of interests
The authors declare no conflict of interests.
References
- [1].Naujokas MF, et al. , The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem, Environ. Health Perspect. 121 (3) (2013) 295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].WHO, Guidelines for Drinking-water Quality, WHO Press, 2011. [Google Scholar]
- [3].USEPA, Technical Fact Sheet: Final Rule for Arsenic in Drinking Water, O.o. Water, 2001. [Google Scholar]
- [4].Huang L, Wu H, van der Kuijp TJ, The health effects of exposure to arsenic-contaminated drinking water: a review by global geographical distribution, Int. J. Environ. Health Res. (2014) 1–21 (ahead-of-print). [DOI] [PubMed] [Google Scholar]
- [5].McClintock TR, et al. , Arsenic exposure in latin america: biomarkers, risk assessments and related health effects, Sci. Total Environ. 429 (2012) 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bardach AE, et al. , Epidemiology of chronic disease related to arsenic in Argentina: a systematic review, Sci. Total Environ. 538 (2015) 802–816. [DOI] [PubMed] [Google Scholar]
- [7].Vahter M, Effects of arsenic on maternal and fetal health, Annu. Rev. Nutr. 29 (2009) 381–399. [DOI] [PubMed] [Google Scholar]
- [8].Concha G, et al. , Exposure to inorganic arsenic metabolites during early human development, Toxicol. Sci. 44 (2) (1998) 185–190. [DOI] [PubMed] [Google Scholar]
- [9].Farzan SF, et al. , In utero arsenic exposure and infant infection in a United States cohort: a prospective study, Environ. Res. 126 (2013) 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bailey KA, et al. , Mechanisms underlying latent disease risk associated with early-life arsenic exposure: current research trends and scientific gaps, Environ. Health Perspect. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rahman A, et al. , Arsenic exposure and risk of spontaneous abortion: stillbirth, and infant mortality, Epidemiology 21 (6) (2010) 797–804. [DOI] [PubMed] [Google Scholar]
- [12].Rahman A, et al. , Arsenic exposure during pregnancy and size at birth: a prospective cohort study in Bangladesh, Am.J. Epidemiol. 169 (3) (2009) 304–312. [DOI] [PubMed] [Google Scholar]
- [13].Gilbert-Diamond D, et al. , Relation between in utero arsenic exposure and birth outcomes in a cohort of mothers and their newborns from new hampshire, Environ. Health Perspect. 124 (8) (2016) 1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Smith AH, et al. , Mortality in young adults following in utero and childhood exposure to arsenic in drinking water, Environ. Health Perspect. 120 (11) (2012) 1527–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hamadani JD, et al. , Critical windows of exposure for arsenic-associated impairment of cognitive function in pre-school girls and boys: a population-based cohort study, Int. J. Epidemiol. 40 (6) (2011) 1593–1604. [DOI] [PubMed] [Google Scholar]
- [16].Fry RC, et al. , Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers, PLoS Genet. 3 (11) (2007) e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rager JE, et al. , Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood, Environ. Mol. Mutagen. 55 (3) (2014) 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rojas D, et al. , Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes, Toxicol. Sci. 143 (1) (2015) 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ahmed S, et al. , Arsenic exposure and cell-mediated immunity in pre-school children in rural Bangladesh, Toxicol. Sci. 141 (1) (2014) 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ahmed S, et al. , In utero arsenic exposure is associated with impaired thymic function in newborns possibly via oxidative stress and apoptosis, Toxicol. Sci. 129 (2) (2012) 305–314. [DOI] [PubMed] [Google Scholar]
- [21].Raqib R, et al. , Effects of in utero arsenic exposure on child immunity and morbidity in rural Bangladesh, Toxicol. Lett. 185 (3) (2009) 197–202. [DOI] [PubMed] [Google Scholar]
- [22].Ahmed S, et al. , Arsenic-associated oxidative stress: inflammation, and immune disruption in human placenta and cord blood, Environ. Health Perspect. 119(2) (2011) 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bailey KA, et al. , Prenatal arsenic exposure and shifts in the newborn proteome: interindividual differences in tumor necrosis factor (TNF)-responsive signaling, Toxicol. Sci. 139 (2) (2014) 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Luna AL, et al. , Arsenic alters monocyte superoxide anion and nitric oxide production in environmentally exposed children, Toxicol. Appl. Pharmacol. 245 (2) (2010) 244–251. [DOI] [PubMed] [Google Scholar]
- [25].Tseng C-H, Arsenic methylation, urinary arsenic metabolites and human diseases: current perspective, J. Environ. Sci. Health Part C 25 (1) (2007) 1–22. [DOI] [PubMed] [Google Scholar]
- [26].Drobna Z, Styblo M, Thomas DJ, An overview of arsenic metabolism and toxicity, Curr. Protoc.Toxicol. 42 (431) (2009) 4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Laine JE, et al. , Maternal arsenic exposure: arsenic methylation efficiency, and birth outcomes in the biomarkers of exposure to ARsenic (BEAR) pregnancy cohort in Mexico, Environ. Health Perspect. 123 (2) (2015) 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang Y-K, et al. , Arsenic methylation capability and hypertension risk in subjects living in arseniasis-hyperendemic areas in southwestern Taiwan, Toxicol. Appl. Pharmacol. 218 (2) (2007) 135–142. [DOI] [PubMed] [Google Scholar]
- [29].Hopenhayn C, et al. , Profile of urinary arsenic metabolites during pregnancy, Environ. Health Perspect. 111 (16) (2003) 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ahmed S, et al. , Arsenic exposure and cell-mediated immunity in pre-school children in rural Bangladesh, Toxicol. Sci. 141 (1) (2014) 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Raqib R, et al. , Effects of in utero arsenic exposure on child immunity and morbidity in rural Bangladesh, Toxicol. Lett. 185 (3) (2009) 197–202. [DOI] [PubMed] [Google Scholar]
- [32].Bailey KA, et al. , Prenatal arsenic exposure and shifts in the newborn proteome: interindividual differences in tumor necrosis factor (TNF)-responsive signaling, Toxicol. Sci. 139 (2) (2014) 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gilbert-Diamond D, et al. , Rice consumption contributes to arsenic exposure in US women, Proc. Natl. Acad. Sci. 108 (51) (2011) 20656–20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tseng C-H, A review on environmental factors regulating arsenic methylation in humans, Toxicol. Appl. Pharmacol. 235 (3) (2009) 338–350. [DOI] [PubMed] [Google Scholar]
- [35].Kulkarni MM, Digital multiplexed gene expression analysis using the NanoString nCounter system, Curr. Protoc. Mol. Biol. (2011) 10 (Chapter 25, Unit25 B). [DOI] [PubMed] [Google Scholar]
- [36].Bentley-Hewitt KL, et al. , Comparison of quantitative real-time polymerase chain reaction with NanoString(R) methodology using adipose and liver tissues from rats fed seaweed, N. Biotechnol. 33 (3) (2016) 380–386. [DOI] [PubMed] [Google Scholar]
- [37].Prokopec SD, et al. , Systematic evaluation of medium-throughput mRNA abundance platforms, RNA 19 (1) (2013) 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hu D, et al. , Development of a NanoString assay to detect leukemogenic fusion transcripts in acute myeloid leukemia, Int.J. Lab. Hematol. 38 (6) (2016) 663–673. [DOI] [PubMed] [Google Scholar]
- [39].Palmer C, et al. , Cell-type specific gene expression profiles of leukocytes in human peripheral blood, BMC Genomics 7 (2006), 115–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bolen CR, Uduman M, Kleinstein SH, Cell subset prediction for blood genomic studies, BMC Bioinf. 12 (2011), 258–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rager JE, et al. , Formaldehyde-associated changes in microRNAs: tissue and temporal specificity in the rat nose, white blood cells, and bone marrow, Toxicol. Sci. 138 (1) (2014) 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Adkins RM, et al. , Racial differences in gene-Specific DNA methylation levels are present at birth. Birth defects research, Part A Clin. Mol. Teratol. 91 (8) (2011) 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Laine JE, Fry RC, A systems toxicology-based approach reveals biological pathways dysregulated by prenatal arsenic exposure, Ann. Glob. Health 82 (1) (2016) 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Green BB, et al. , Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the new hampshire birth cohort study (USA), Environ. Health Perspect. 124 (8) (2016) 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Remy S, et al. , Expression of the sFLT1 gene in cord blood cells is associated to maternal arsenic exposure and decreased birth weight, PLoS One 9 (3) (2014) e92677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kile ML, et al. , Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood, Epigenetics 9 (5) (2014) 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cardenas A, et al. , In utero arsenic exposure and epigenome-wide associations in placenta: umbilical artery, and human umbilical vein endothelial cells, Epigenetics 10 (11) (2015) 1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Allantaz F, et al. , Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression, PLoS One 7 (1) (2012) e29979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Drobna Z, et al. , Analysis of maternal polymorphisms in arsenic (+3 oxidation state)-methyltransferase AS3MT and fetal sex in relation to arsenic metabolism and infant birth outcomes: implications for risk analysis, Reprod. Toxicol. 61 (2016) 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shi Q, et al. , Sequential activation of Elk-1/Egr-1/GADD45α by arsenic, Oncotarget 5 (11) (2014) 3862–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Thakali KM, et al. , Maternal pregravid obesity changes gene expression profiles toward greater inflammation and reduced insulin sensitivity in umbilical cord, Pediatr. Res. 76 (2) (2014) 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Veyrac A, et al. , Zif268/egr1 gene controls the selection: maturation and functional integration of adult hippocampal newborn neurons by learning, Proc. Natl. Acad. Sci. U. S. A. 110 (17) (2013) 7062–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nygaard UC, et al. , Cord blood T cell subpopulations and associations with maternal cadmium and arsenic exposures, PLoS One 12 (6) (2017) e0179606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Huang Y, et al. , Impaired growth hormone receptor signaling during non-catch-up growth in rats born small for gestational age, Horm. Res. Paediatr. 74 (2) (2010) 106–113. [DOI] [PubMed] [Google Scholar]
- [55].Farzan SF, et al. , Maternal and infant inflammatory markers in relation to prenatal arsenic exposure in a U.S. pregnancy cohort, Environ. Res. 156 (2017) 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Flanagan S, Beleval M, Ayotte J, Arsenic, Iron, Lead, Manganese, and Uranium Concentrations in Private Bedrock Wells in Southeastern New Hampshire, 2012–2013, U.S.G. Survey, 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.