Abstract
Neandertals and Denisovans are extinct groups of hominins that separated from each other more than 390,000 years ago1,2. Here we present the genome of “Denisova 11”, a bone fragment from Denisova Cave (Russia)3, and show that it comes from an individual who had a Neandertal mother and a Denisovan father. The father, whose genome bears traces of Neandertal ancestry, came from a population related to a later Denisovan found in the cave4–6. The mother came from a population more closely related to Neandertals who lived later in Europe2,7 than to an older Neandertal found in Denisova Cave8, suggesting that migrations of Neandertals between eastern and western Eurasia occurred sometime after ~120,000 years ago. The finding of a first-generation Neandertal-Denisovan offspring among the small number of archaic specimens sequenced to date suggests that mixing between Late Pleistocene hominin groups was common when they met.
Neandertals and Denisovans inhabited Eurasia until they were replaced by modern humans around 40,000 years ago (40 kya)9. Neandertal remains are found in western Eurasia10; while physical remains of Denisovans have thus far been found only in Denisova Cave4–6,11,12, where Neandertal remains have also been recovered8. Although little is known about the morphology of Denisovans, their molars lack derived traits typical of Neandertals5,11.
DNA recovered from individuals of both groups suggests that they diverged from each other more than 390 kya1,2. The presence of small amounts of Neandertal DNA in the genome of “Denisova 3”, the first Denisovan individual identified4–6, indicates that the two groups mixed with each other at least once8. It has also been shown that Neandertals mixed with the ancestors of present-day non-Africans around 60 kya2,8,13, and possibly with earlier ancestors of modern humans1,14,15; and that Denisovans mixed with the ancestors of present-day Oceanians and Asians5,16,17. Denisovans may furthermore have received gene flow from an archaic hominin that diverged more than a million years ago from the ancestors of modern humans8.
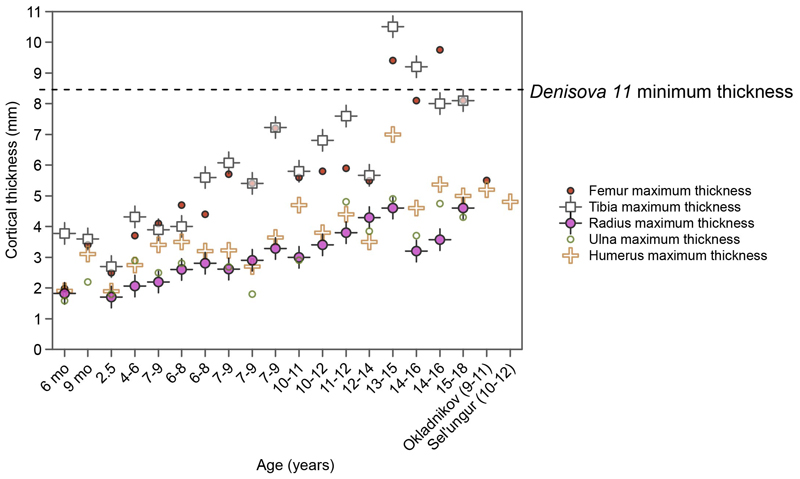
A fragment of a long bone, “Denisova 11” (Fig. 1), was identified among over 2,000 undiagnostic bone fragments excavated in Denisova Cave as being of hominin origin using collagen peptide mass fingerprinting3. Its mitochondrial (mt) DNA was found to be of the Neandertal type and direct radiocarbon dating showed it to be >50,000 years old3. From its cortical thickness, we infer that Denisova 11 was at least 13 years old at death (Extended Data Figure 1, Supplementary Information [SI] 1). We performed six DNA extractions18,19 from bone powder collected from the specimen, produced ten DNA libraries20 from the extracts (Extended Data Table 1, SI 2 and 3) and sequenced the Denisova 11 genome to an average coverage of 2.6-fold. The coverage of the X chromosome is similar to that of the autosomes, indicating that Denisova 11 was a female. Using three different methods, we estimate that contaminating present-day human DNA fragments constitute at most 1.7% of the data (SI 2).
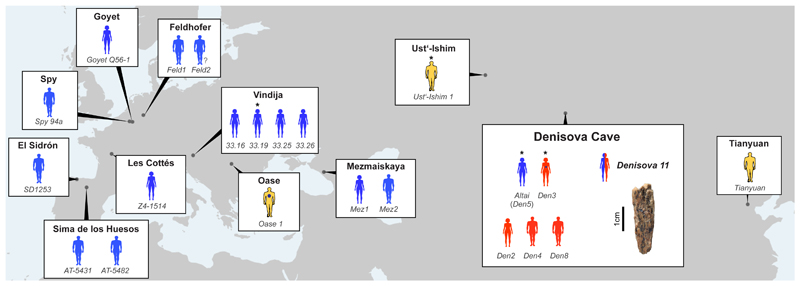
Figure 1. Location of Neandertals (blue), Denisovans (red) and ancient modern humans (yellow) dated to ~40 kya or older, from which sufficient nuclear DNA fragments have been recovered to enable their attribution to a hominin group.
Full or abbreviated names of specimens are shown near each figure. Asterisks indicate that the genome was sequenced to high-coverage, a question mark that the individual is of unknown sex. Note that Oase 1 has recent Neandertal ancestry (blue dot) that is beyond the amount seen in non-Africans. Denisova 3 has also been found to carry a small percentage of Neandertal ancestry. Data taken from 1,2,5–8,11–13,21–24.
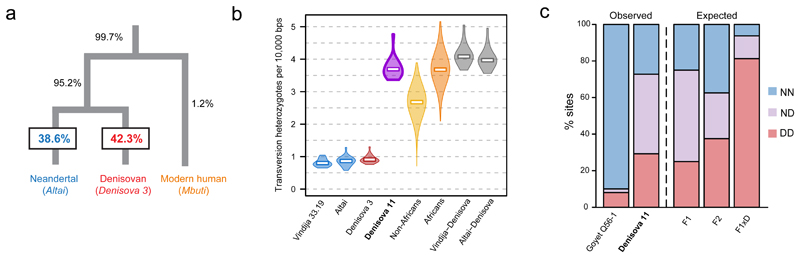
To determine from which hominin group Denisova 11 originated, we compared the proportions of DNA fragments that match derived alleles from a Neandertal genome (“Altai Neandertal”, also known as “Denisova 5”) or a Denisovan genome (Denisova 3), both determined from bones discovered in Denisova Cave6,8, as well as from a present-day African genome (Mbuti)6 (SI 4). At such informative sites1, 38.6% of fragments from Denisova 11 carried alleles matching the Neandertal genome and 42.3% carried alleles matching the Denisovan genome (Fig. 2a), suggesting that both archaic groups contributed to the ancestry of Denisova 11 to approximately equal extents (SI 4). An approximately equal proportion of such Neandertal-like and Denisovan-like alleles is found in each of the ten Denisova 11 libraries but not in libraries from other projects that were prepared, sequenced and processed in parallel with them, excluding an accidental mixing of DNA in the laboratory or a systematic error in data processing (SI 3).
Figure 2. Denisova 11 carries both Neandertal and Denisovan ancestry.
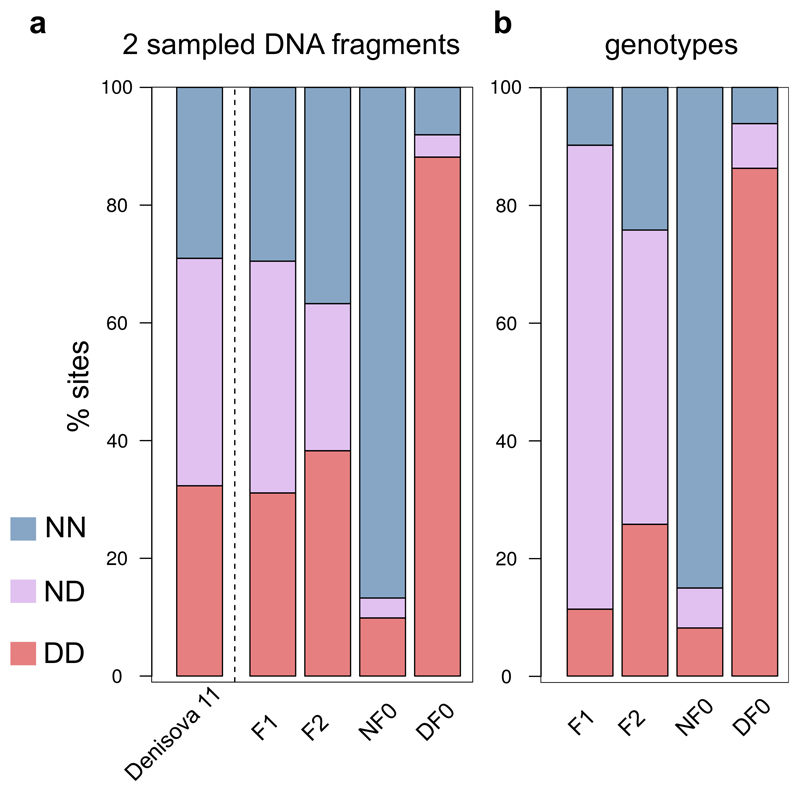
a. Percentage of DNA fragments in Denisova 11 carrying derived alleles seen on each branch of a tree relating a Neandertal, a Denisovan and a present-day human genome. b. Distribution of heterozygosity per chromosome in two Neandertals (blue), a Denisovan (red), Denisova 11 (purple) and present-day humans (N=235 non-African [yellow] and N=44 African individuals [orange] from 28), and the expectation for a Neandertal-Denisovan F1 offspring (grey). The violins represent the distribution from the minimum and maximum heterozygosity values for the autosomes of each archaic hominin and of present-day humans (n=5,170 pairs of chromosomes for non-Africans and n=968 for Africans). White squares represent autosome-wide estimates for the archaic hominins, and the average of estimates across individuals for present-day humans. c. Percentage of sites at which two sampled DNA fragments both carry “Neandertal alleles” (blue), “Denisovan alleles” (red), or one allele of each type (purple); and the expectations for an offspring of a Neandertal and a Denisovan (F1), of two F1 parents (F2), and of an F1 and a Denisovan (F1xD). The expected proportions for simulated Neandertal and Denisovan genomes are shown in Extended Data Figure 2.
To estimate the heterozygosity of Denisova 11, we restrict the analysis to transversion polymorphisms to prevent deamination-derived substitutions from inflating the estimates, and find 3.7 transversions per 10,000 autosomal base pairs. This is over four times higher than the heterozygosity of the two Neandertal (Altai Neandertal and “Vindija 33.19”) and one Denisovan (Denisova 3) genomes sequenced to date, and similar to the heterozygosity seen in present-day Africans. In fact, the heterozygosity of Denisova 11 is similar to what would be expected if this individual carried one set of chromosomes of Neandertal origin and one of Denisovan origin, as estimated from the number of differences between randomly sampled DNA fragments from either the Vindija 33.19 or the Altai Neandertal genome and the Denisova 3 genome (Fig. 2b, SI 5).
Denisova 11 could have had approximately equal amounts of Neandertal and Denisovan ancestry because she belonged to a population with mixed Neandertal and Denisovan ancestry, or because her parents were each from one of these two groups. To determine which of these two scenarios fits the data best, we considered sites where the genomes of the Altai Neandertal and Denisova 3 carry a transversion difference in a homozygous form. At each of these sites, we recorded the alleles carried by two randomly drawn DNA fragments from Denisova 11. Note that in 50% of cases, both fragments will come from the same chromosome, making 50% of heterozygous sites appear homozygous. As a consequence, the expected proportion of apparent heterozygous sites is 50% for a first-generation (F1) offspring, while it is 25% in a population at Hardy-Weinberg equilibrium with mixed ancestry in equal proportions (SI 6). We find that in 43.5% of cases, one fragment from Denisova 11 matches the Neandertal genome and the other matches the Denisovan genome, while in 27.3% and 29.2% of cases both fragments match the state seen in the Neandertal or the Denisovan genome, respectively (Fig. 2c). For comparison, when a low-coverage Neandertal genome (“Goyet Q56-1”)7 is analysed in the same way, the two fragments match different states in 2.1% of cases, while they both match the Neandertal state in 90.3% of cases and the Denisovan state in 7.5% of cases (Fig. 2c).
Obviously, the Altai Neandertal and Denisova 3 are unlikely to be identical to the genomes of the individuals that contributed ancestry to Denisova 11. To take this into account, we used coalescent simulations to estimate the expected proportions of DNA fragments matching a Neandertal or a Denisovan genome in populations with demographic histories similar to those of the Altai Neandertal and Denisova 3 (SI 6). The proportion of cases where one of the two DNA fragments sampled from Denisova 11 matches the Neandertal state and the other the Denisovan state fits the expectation for an F1 Neandertal-Denisovan offspring, but not an offspring of two F1 individuals, an offspring of an F1 parent and a Neandertal or a Denisovan parent, nor an individual from a population of mixed ancestry at Hardy-Weinberg equilibrium (Extended Data Figure 2, SI 6). We conclude that Denisova 11 did not originate from a population carrying equal proportions of Neandertal and Denisovan ancestry. Rather, she was the offspring of a Neandertal mother, who contributed her mtDNA, and a Denisovan father.
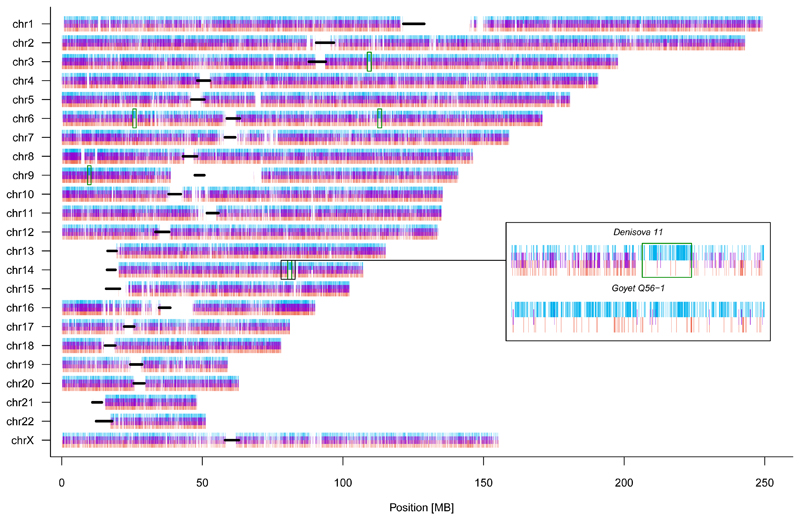
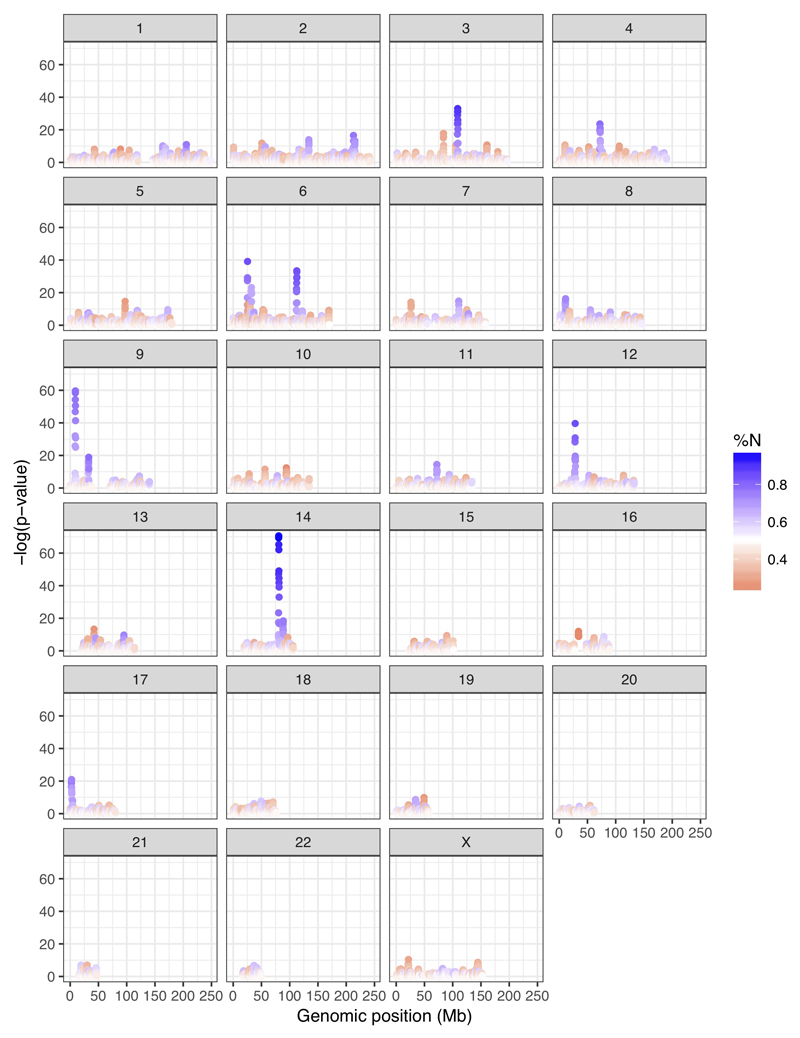
We next plotted the distribution of sites across the genome, where Denisova 11 carries an allele matching the Altai Neandertal genome and a different allele matching the Denisova 3 genome. Such sites are distributed largely uniformly (Fig. 3), as would be expected for an F1 offspring of Neandertal and Denisovan parents. To explore the ancestry of the parents of Denisova 11, we looked for regions in the genome that deviate from a pattern consistent with Denisova 11 being an F1 offspring (Extended Data Figure 3). Using four tests for enrichment of Denisovan or Neandertal ancestry, we identify at least five ~1 Mb long (0.72-0.95 Mb) regions, all of which are homozygous for Neandertal ancestry. This suggests that the Denisovan father of Denisova 11 had some Neandertal ancestry. Given conservative estimates of the size and number of these regions, it is likely that there was more than one Neandertal ancestor in his genealogy, possibly as far back as 300-600 generations beforehand (SI 7). Interestingly, the heterozygosity in the regions of Neandertal ancestry in Denisova 11 is higher than in the same regions in the genomes of Vindija 33.19 or the Altai Neandertal, suggesting that the Neandertals that contributed to the ancestry of Denisova 11’s father were from a different population than her mother (SI 5).
Figure 3. The distribution of Neandertal-like and Denisovan-like alleles across the Denisova 11 genome.
Positions where one randomly drawn DNA fragment matches the Neandertal genome and another matches the Denisovan genome are marked in purple. Positions are marked in blue if both DNA fragments match the Neandertal genome, and in red if both match the Denisovan genome. Black lines indicate centromeres. The inset shows one region out of five (green boxes) where both chromosomes carry predominantly Neandertal-like alleles. For comparison, the distribution of alleles in this region is shown for a Neandertal genome (Goyet Q56-1).
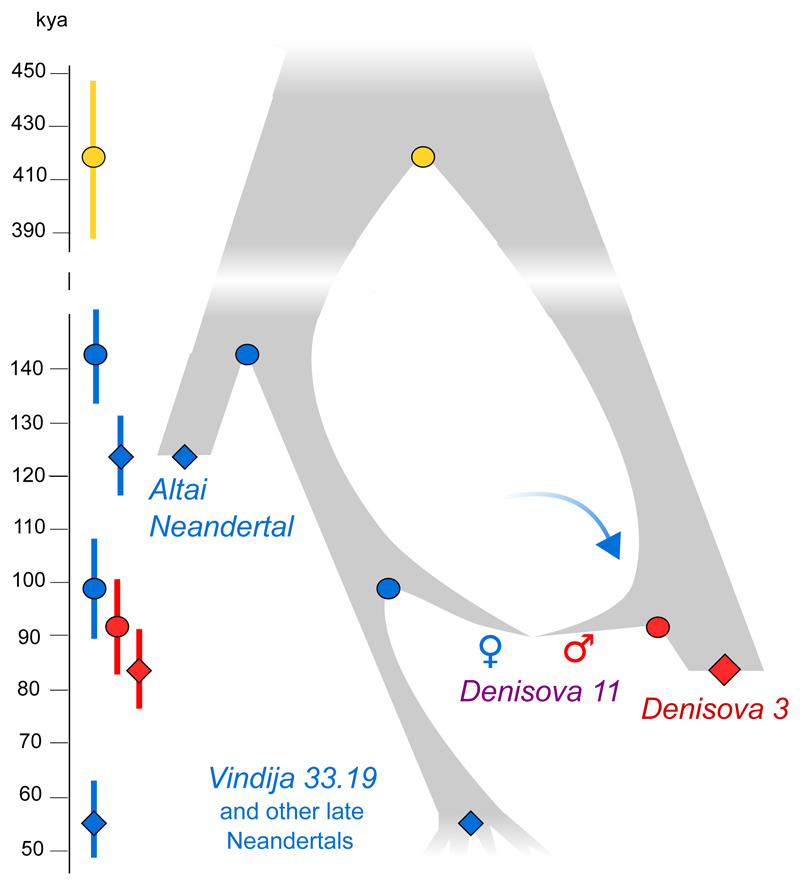
To explore how the mother of Denisova 11 was related to the two Neandertals that have been sequenced to high coverage to date, we evaluated the proportions of fragments from Denisova 11 that match derived alleles from either of these two Neandertal genomes. Denisova 11 shares derived alleles seen in the Altai Neandertal genome in 12.4% of cases and those present in the Vindija 33.19 genome in 19.6% of cases, showing that the Neandertal mother of Denisova 11 came from a population that was more closely related to Vindija 33.19 than to the Altai Neandertal (SI 8). We estimate the population split times of Denisova 11’s Neandertal mother from the ancestors of the Altai Neandertal to ~20,000 years (20 ky) prior to the time when the Altai Neandertal lived, and her split time from the ancestors of Vindija 33.19 to ~40 ky prior to Vindija 33.19. The population split between the Denisovan father of Denisova 11 and Denisova 3 is estimated to ~7 ky prior to the latter individual (SI 8). In Fig. 4 we present a population scenario that is compatible with these observations as well as with the population split times and molecular estimates of the ages of the three high-coverage archaic genomes2. We caution that the age estimates are associated with uncertainties, e.g., regarding demography, mutation rates and generation times, and that additional gene flow events are likely to have affected the population split times. Nevertheless, that a Neandertal in Siberia ~90 kya shared more alleles with Neandertals that lived at least ~20 ky later in Europe2,7 than with an earlier Neandertal from the same cave8 suggests that eastern Neandertals spread into Western Europe sometime after ~90 kya or that western Neandertals spread to Siberia before that time and partially replaced the local population. These two non-mutually exclusive hypotheses could be tested by sequencing the genomes of early Neandertals from Western Europe.
Figure 4. Relationships and gene flow events between Neandertal and Denisovan populations inferred from genome sequences.
Diamonds indicate ages of specimens estimated via branch shortening2; circles indicate population split times estimated from allele sharing between Denisova 11 and the high-coverage genomes (blue and red) and among the three high-coverage genomes (yellow, from 2); the arrow indicates Neandertal gene flow into Denisovans. Markers indicate the means of these estimates, error bars indicate 95% confidence intervals (CIs) based on block jackknife resampling across the genome (n=523 blocks). Note that the CIs do not take the uncertainty with respect to population size, mutation rates or generation times into account. Ages before present are based on a human-chimpanzee divergence of 13 million years22,29.
In conclusion, the genome of Denisova 11 provides direct evidence for genetic mixture between Neandertals and Denisovans on at least two occasions: once between her Neandertal mother and her Denisovan father, and at least once in the ancestry of her Denisovan father. Therefore, of the six individuals from Denisova Cave from whom nuclear DNA is available5,6,8,11,12, two (Denisova 3 and Denisova 11) show evidence of gene flow between Neandertals and Denisovans. We note that of the three genomes21–24 retrieved from modern humans who lived at a time when Neandertals were present in Eurasia (i.e., at least ~40 kya)9, one individual, “Oase 1”, had a Neandertal ancestor four to six generations back in his family tree23.
It is striking that one direct offspring of a Neandertal and a Denisovan (Denisova 11) and one modern human with a close Neandertal relative (Oase 1) have been identified among the few individuals from whom DNA has been retrieved and who lived at the time of overlap of these groups (Fig. 1). In conjunction with the presence of Neandertal and Denisovan DNA in ancient and present-day people2,5,8,13,16,17,25–27, this suggests that mixing among archaic and modern hominin groups may have been frequent when they met. However, Neandertals inhabited western Eurasia10 while Denisovans inhabited yet unknown parts of eastern Eurasia5,17. Thus, their zones of overlap may have been restricted in space and time. This, as well as possibly reduced fitness of individuals of mixed ancestry, may explain why Neandertals and Denisovans remained genetically distinct. In contrast, the spread of modern humans across Eurasia after ~60,000 years ago may have allowed repeated interactions with archaic groups over a wider spatial range. Admixture between them may have resulted in archaic populations becoming partly absorbed into what were probably larger modern human populations6,8.
Methods
Sampling and pre-treatment of bone powder
An overview of the laboratory experiments is in Extended Data Table 1. Bone powder was removed from the specimen using disposable sterile dentistry drills after the removal of a thin layer of surface material. Six samples were collected, each consisting of ~30mg of bone powder. Because a previous analysis of the bone revealed that it is contaminated with present-day human DNA3, each sample of bone powder was incubated with 1ml 0.5% sodium hypochlorite solution as described19 and as indicated in Extended Data Table 1, to reduce the amounts of present-day human and microbial DNA7,19. Residual sodium hypochlorite was removed by three consecutive 3-minute washes with 1ml water19. One extraction negative control (no powder) was included in each set of extractions.
DNA extraction and DNA library preparation
DNA was extracted using silica columns18 as described19, and eluted in 50µl 10mM Tris-HCl, 1mM EDTA, 0.05% Tween-20, pH 8.0. Ten µl of each DNA extract were used to prepare single-stranded DNA libraries as described19,20. Extraction negative controls were carried along, and a library preparation negative control was included in every experiment. Two additional 5µl aliquots from the extracts E3652 and E3655 were used to generate additional libraries (library preparation setup C in Extended Data Table 1), resulting in a total of ten DNA libraries. The number of DNA molecules in the libraries was estimated by digital droplet PCR30 or quantitative PCR20. Each library was amplified into plateau while incorporating a pair of unique indexes31 using 1µM primers19,31 and AccuPrime Pfx DNA polymerase (Life Technologies)32. Amplification products were purified using the MinElute PCR purification kit (Qiagen) or SPRI technology33 on a Bravo NGS workstation (Agilent Technologies) as described34. Indexed DNA libraries were pooled with libraries from other projects. Heteroduplices, which confound DNA separation and concentration measurements in chromatography, were removed from the pools by a single cycle amplification using Herculase II Fusion DNA polymerase (Agilent Technologies)32 with primers IS5 and IS6 (ref. 35). Prior to deeper sequencing of libraries R5507, R5509, R9880, R9881, R9882, R9883 and R9873, heteroduplices were removed from each library separately. The concentration of DNA in each pool or each individual library, respectively, was determined using the electrophoresis system implemented on the DNA-1000 chip (Agilent Technologies).
Sequencing and data processing
Sequencing was performed on Illumina platforms (MiSeq or HiSeq 2500) using 76-cycle paired-end runs adapted to double-indexed libraries31. Bases were called using Bustard (Illumina). Adapter sequences were trimmed and overlapping paired-end reads were merged into single sequences using leeHom36. Demultiplexing was carried out using jivebunny7. Sequences generated from a given library were merged using SAMtools37 and aligned to the human reference genome (hg19/GRCh37) with the decoy sequences as in 2 using BWA38 with parameters adjusted to ancient DNA6. PCR duplicates were collapsed using bam-rmdup (https://bitbucket.org/ustenzel/biohazard) and DNA fragments of length ≥35 bases that map within regions of unique mappability (Map35_100% from ref. 8) with a mapping quality of 25 or higher7 were used for analyses. Further filtering criteria used for certain analyses are detailed in the supplementary sections.
Extended Data
Extended Data Figure 1. Maximum cortical thickness of femora, tibiae, humeri, radii and ulnae among humans from the Bronze Age and two Neandertals, compared to the minimum thickness of Denisova 11 (dashed line).
Extended Data Figure 2. Percentage of sites at which Denisova 11 and genomes simulated under the demographic model detailed in SI 6 carry two Neandertal alleles (NN, blue), two Denisovan alleles (DD, red), or one allele of each type (ND, purple).
a. Percentages calculated for two random DNA fragments from Denisova 11 (leftmost column) and from simulated F1, F2, Neandertal (NF0) or Denisovan (DF0) genomes (columns 2-5). b. Proportions of sites for the simulated genotypes, prior to sampling two fragments.
Extended Data Figure 3. Neandertal and Denisovan allele proportions from Denisova 11 in 1 Mb windows (100 kb step, n=26,414 windows).
Y-axis shows –log(p-value) of the deviation of Neandertal and Denisovan allele counts from the genome-wide average (chi-square test of goodness-of-fit; see SI 7); color shows the proportion of alleles matching the Neandertal state (%N) within each 1 Mb window.
Extended Data Table 1. DNA extracts and DNA libraries prepared from the Denisova 11 specimen.
Data are shown by DNA extraction set, and libraries prepared in the same setup are denoted by the same letter (A, B or C). Relevant negative controls are marked in grey. The number of molecules in each library was quantified by digital droplet PCR or quantitative PCR (denoted by *). The number of DNA fragments sequenced per library are for the combined data from all sequencing runs. Mapped fragments were counted if they were at least 35 bases long and mapped to the human reference genome with a mapping quality of 25 or higher; and their percentage was calculated out of sequenced fragments of length 35 bases or more. Following the removal of PCR duplicates, unique DNA fragments were retained if they mapped to the reference genome within the mappability track used. Such fragments were considered to contain a terminal C to T substitution relative to the human reference genome if a putative cytosine deamination was within the first three or last three bases of the strand. Extr. – extraction; Prep. – preparation; L – length; MQ – Mapping quality; Map35_100% - mappability track from 8; bp – base pairs; C – Cytosine; T – Thymine; ENC – Extraction negative control; LNC – Library preparation negative control.
| Extr. set | Bone powder [mg] | Pre-treatment [minutes] | Extract ID | Library prep.setup | Input in library [µI] | Molecules in library | Indexed library ID | DNA fragments sequenced | DNA fragments sequenced (L ≥35) | Mapped fragments (L ≥35, MQ ≥ 25) | Mapped fragments (%) | Unique fragments (L ≥35, MQ ≥ 25. Map35_100%) | Average fragment length [bp] | Fragments with C to T substitution | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 27.4 | 15 | E3259 | A | 10 | 2.27E+08 | R5507 | 133,898,498 | 89,793,496 | 2,139,377 | 2.4 | 1,656,500 | 52.7 | 404,188 | |
| 27.8 | 15 | E3261 | A | 10 | 2.00E+08 | R5509 | 145,234,847 | 94,543,170 | 1,712,750 | 1.8 | 1,201,280 | 48.8 | 329,411 | ||
| Denisova 11 | B | 10 | 4.33E+08 * | R5780 | 2,391,986 | 1,565,197 | 171,150 | 10.9 | 152,336 | 56.4 | 31,758 | ||||
| 29.0 | 15 | E3652 | C | 5 | 4.63E+08 | R9880 | 379,368,999 | 228,704,750 | 22,501,299 | 9.8 | 14,767,988 | 56.5 | 3,028,792 | ||
| C | 5 | 4.03E+08 | R9881 | 333,009,774 | 203,041,282 | 20,747,195 | 10.2 | 13,805,425 | 56.6 | 2,850,253 | |||||
| 2 | 29.7 | 15 | E3654 | B | 10 | 4.16E+08 * | R5782 | 2,671,910 | 1,669,048 | 81,750 | 4.9 | 72,730 | 54.5 | 15,618 | |
| B | 10 | 3.49E+08 * | R5783 | 2,348,997 | 1,510,249 | 199,762 | 13.2 | 177,860 | 59.3 | 31,952 | |||||
| 33.5 | 15 | E3655 | C | 5 | 4.19E+08 | R9882 | 368,237,790 | 225,412,495 | 27,395,573 | 12.2 | 17,849,890 | 59.8 | 3,173,678 | ||
| C | 5 | 3.69E+08 | R9883 | 343,471,978 | 224,455,462 | 28,522,051 | 12.7 | 18,048,369 | 60.4 | 3,215,383 | |||||
| 3 | 27.1 | 30 | E3922 | C | 10 | 7.43E+07 | R9873 | 348,156,224 | 222,947,600 | 62,160,161 | 27.9 | 17,009,638 | 53.3 | 4,282,134 | |
| Controls | 1 | ENC | 15 | E3262 | A | 10 | 2.55E+07 | R5510 | 12,220 | 4,123 | 38 | 0.9 | 35 | 55.2 | 2 |
| LNC | - | - | A | - | 7.60E+06 | R5521 | 12,444 | 2,170 | 11 | 0.5 | 10 | 47.2 | 1 | ||
| 2 | ENC | 15 | E3663 | B | 10 | 1.83E+06 * | R5791 | 32,008 | 4,183 | 473 | 11.3 | 412 | 51.2 | 8 | |
| LNC | - | - | B | - | 2.54E+06 * | R5792 | 31,455 | 2,908 | 70 | 2.4 | 58 | 49.0 | 3 | ||
| 3 | ENC | 30 | E3926 | C | 10 | 2.73E+07 | R9877 | 61,825 | 13,861 | 2,472 | 17.8 | 2,145 | 57.0 | 9 | |
| LNC | - | - | C | - | 1.30E+07 | R9888 | 68,130 | 5,275 | 100 | 1.9 | 67 | 46.2 | 6 | ||
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank Barbara Schellbach and Antje Weihmann for DNA sequencing; Gabriel Renaud and Udo Stenzel for data processing; Fiona Brock for the CT scans; Ronny Barr, Petra Korlević and Cristina Zickert for graphics; and Montgomery Slatkin and Linda Vigilant for comments on the manuscript. This work was funded by the Max Planck Society; the Max Planck Foundation (grant 31-12LMP Pääbo to S.Pä.); the European Research Council (grant agreements no. 694707 to S.Pä., no. 324139 (PalaeoChron) to T.H. and no. 715069 (FINDER) to K.D.); and the Russian Science Foundation (project No. 14-50-00036 to M.B.K., M.V.S. and A.P.D.).
Footnotes
Contributions: V.S. and S.N. performed the laboratory work; V.S., F.M., B.Ve., C.d.F., S.G., M.H., S.Pe., J.K., M.M., K.P. and S.Pä. analyzed the genetic data; B.Vi. carried out the morphological analysis; S.B., K.D., T.H., M.B.K., M.V.S. and A.P.D. discovered Denisova 11 and provided archaeological data; V.S., K.P. and S.Pä. wrote the manuscript with input from all authors.
Author Information: Reprints and permissions information are available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
The authors declare no competing financial interests.
Data availability statement: Sequences generated from Denisova 11 have been deposited in the European Nucleotide Archive under study accession number PRJEB24663.
Code availability: The computer code used for simulations is presented in SI 6.
References
- 1.Meyer M, et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature. 2016;531:504–507. doi: 10.1038/nature17405. [DOI] [PubMed] [Google Scholar]
- 2.Prüfer K, et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 2017;358:655–658. doi: 10.1126/science.aao1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown S, et al. Identification of a new hominin bone from Denisova Cave, Siberia using collagen fingerprinting and mitochondrial DNA analysis. Scientific Reports. 2016;6:23559. doi: 10.1038/srep23559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krause J, et al. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hajdinjak M, et al. Reconstructing the Genetic History of Late Neandertals. Nature. 2018;555:652–656. doi: 10.1038/nature26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higham T, et al. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature. 2014;512:306–309. doi: 10.1038/nature13621. [DOI] [PubMed] [Google Scholar]
- 10.Vandermeersch B, Garralda MD. In: Continuity and Discontinuity in the Peopling of Europe: One Hundred Fifty Years of Neanderthal Study. Condemi S, Weniger G-C, editors. Springer; Dordrecht: 2011. pp. 113–125. [Google Scholar]
- 11.Sawyer S, et al. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc Natl Acad Sci U S A. 2015;112:15696–15700. doi: 10.1073/pnas.1519905112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slon V, et al. A fourth Denisovan individual. Science Advances. 2017;3:e1700186. doi: 10.1126/sciadv.1700186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhlwilm M, et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 2016;530:429–433. doi: 10.1038/nature16544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Posth C, et al. Deeply divergent archaic mitochondrial genome provides lower time boundary for African gene flow into Neanderthals. Nature Communications. 2017;8 doi: 10.1038/ncomms16046. 16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skoglund P, Jakobsson M. Archaic human ancestry in East Asia. Proc Natl Acad Sci U S A. 2011;108:18301–18306. doi: 10.1073/pnas.1108181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reich D, et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dabney J, et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci U S A. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korlević P, et al. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques. 2015;59:87–93. doi: 10.2144/000114320. [DOI] [PubMed] [Google Scholar]
- 20.Gansauge MT, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc. 2013;8:737–748. doi: 10.1038/nprot.2013.038. [DOI] [PubMed] [Google Scholar]
- 21.Fu Q, et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc Natl Acad Sci U S A. 2013;110:2223–2227. doi: 10.1073/pnas.1221359110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Q, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang MA, et al. 40,000-Year-Old Individual from Asia Provides Insight into Early Population Structure in Eurasia. Curr Biol. 2017;27:3202–3208. doi: 10.1016/j.cub.2017.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seguin-Orlando A, et al. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346:1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 26.Fu Q, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sikora M, et al. Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science. 2017;358:659–662. doi: 10.1126/science.aao1807. [DOI] [PubMed] [Google Scholar]
- 28.Mallick S, et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nature Reviews Genetics. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 30.Slon V, et al. Mammalian mitochondrial capture, a tool for rapid screening of DNA preservation in faunal and undiagnostic remains, and its application to Middle Pleistocene specimens from Qesem Cave (Israel) Quatern Int. 2016;398:210–218. [Google Scholar]
- 31.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dabney J, Meyer M. Length and GC-biases during sequencing library amplification: a comparison of various polymerase-buffer systems with ancient and modern DNA sequencing libraries. Biotechniques. 2012;52:87–94. doi: 10.2144/000113809. [DOI] [PubMed] [Google Scholar]
- 33.Deangelis MM, Wang DG, Hawkins TL. Solid-Phase Reversible Immobilization for the Isolation of PCR Products. Nucleic Acids Research. 1995;23:4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slon V, et al. Neandertal and Denisovan DNA from Pleistocene sediments. Science. 2017;356:605–608. doi: 10.1126/science.aam9695. [DOI] [PubMed] [Google Scholar]
- 35.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb prot5448. [DOI] [PubMed] [Google Scholar]
- 36.Renaud G, Stenzel U, Kelso J. leeHom: adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 2014;42:e141. doi: 10.1093/nar/gku699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.