Abstract
RNA viruses are a major threat to animals and plants. RNA interference (RNAi) and the interferon response provide innate antiviral defense against RNA viruses. Here we performed a large-scale screen using C. elegans and its natural pathogen, the Orsay virus (OrV), and identified cde-1 as important for antiviral defense. CDE-1 is a homologue of the mammalian TUT4 and TUT7 (collectively called TUT4(7)) terminal uridylyltransferases; its catalytic activity is required for its antiviral function. CDE-1 uridylates the 3′ end of the OrV RNA genome and promotes its degradation, independently of the RNAi pathway. Likewise, TUT4(7) uridylate influenza A virus (IAV) mRNAs in mammalian cells. Deletion of TUT4(7) leads to increased IAV mRNA and protein levels. We have defined 3′ terminal uridylation of viral RNAs as a conserved antiviral defense mechanism.
RNA viruses are a major threat to human health and food security. Understanding the fundamental mechanisms by which animals and plants combat viral infections might lead to new therapeutic antiviral approaches. RNA interference (RNAi) is an important antiviral pathway in most animals and plants: Dicer recognizes and cleaves the double-stranded viral RNA genome into virus-derived small interfering RNAs (viral siRNAs, viRNAs), which are loaded into Argonaute proteins to form the RNA-induced silencing complex (RISC) that in turn targets the viral RNA genome 1. Vertebrates have additionally evolved a cellular signaling-based pathway, the interferon response (IR): upon recognition of foreign RNAs (i.e. double-stranded or bearing a 5′ di/triphosphate), cytosolic receptors of the RIG-I family activate the IR which results in an antiviral state of the cell 2,3. In the evolutionary arms race between viruses and their hosts, however, animals must have evolved a diverse range of antiviral strategies, to not solely rely on the RNAi or IR pathways.
Here, we develop a system for antiviral gene discovery using the nematode Caenorhabditis elegans (C. elegans) and identify 3′ terminal uridylation of viral RNAs as a third antiviral mechanism in animals.
Results
A forward genetic screen identifies new genes required for antiviral defense in C. elegans
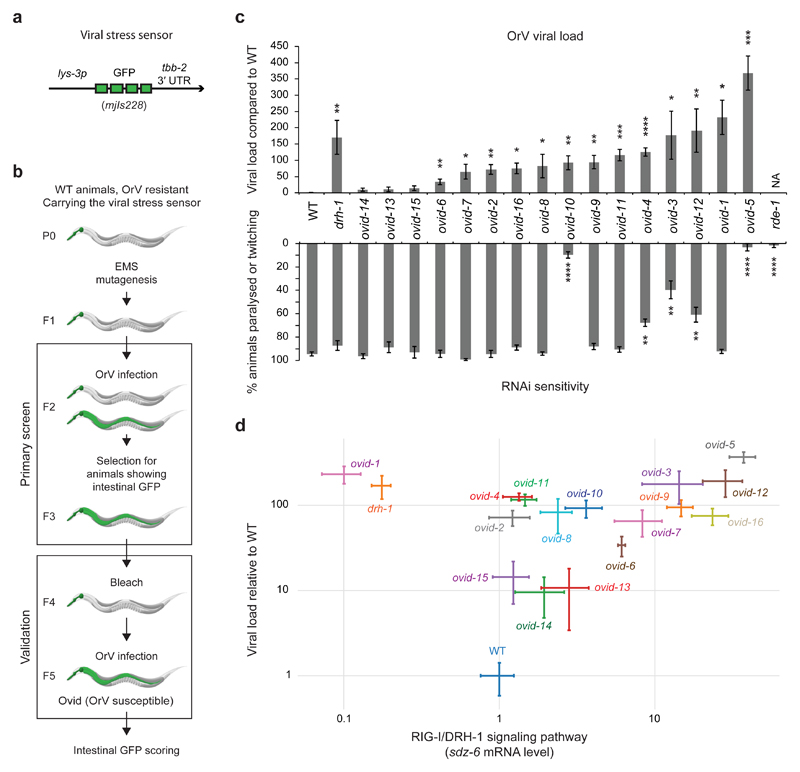
We carried out a forward genetic screen to discover antiviral pathways in animals using C. elegans and its natural intestinal pathogen, the Orsay virus (OrV) 4–12. OrV is a bipartite positive-strand RNA virus related to the Nodaviridae 4. As is typical for positive sense RNA viruses, the genomic strand of the OrV is a template for translation. The OrV spreads horizontally in populations of C. elegans: it is taken up orally, infects only intestinal cells and probably exits through defecation 4. While C. elegans lacks an interferon pathway, a RIG-I ortholog, DRH-1, acts in viral recognition. DRH-1 forms a Viral Recognition Complex (ViRC) with the C. elegans Dicer (DCR-1) and the RNA-binding protein RDE-4 to link viral recognition to a dedicated antiviral RNAi pathway, involving the Argonaute protein RDE-1 5,11,13,14. DRH-1 also induces a transcriptional immune response through a STAT-dependent signaling pathway (e.g. the gene sdz-6, as shown by qRT-PCR in Extended Data Fig. 1a) 10,15,16. However, the antiviral function of the DRH-1-mediated stress response remains to be elucidated. C. elegans also elicits a “biotic stress response” upon OrV infection that is independent of DRH-1 and partially overlaps with transcriptional responses induced by other types of pathogens, possibly as a result of perturbations in cell homeostasis and/or mechanical integrity (e.g. the gene lys-3, encoding an antibacterial enzyme, as shown by qRT-PCR in Extended Data Fig. 1a) 10. We generated a viral stress sensor transgene by placing the green fluorescent protein (GFP) under the control of the lys-3 promoter (allele mjIs228; Fig. 1a). Upon infection, the level of GFP expression in the intestine mirrored the viral load in wild type, drh-1 and rde-1 mutants (Extended Data Fig. 1b, c). We used chemical mutagenesis to screen ~50,000 haploid genomes (Fig. 1b) and identified 16 isolates we named Ovid (Orsay Virus Immune Deficient; Fig. 1c and Supplementary Table 1). 13 out of 16 ovid mutants showed increased viral loads (Fig. 1c). ovid-3,4,5,10,12 are compromised in somatic RNAi, as tested by RNAi knockdown of the gene unc-22, which normally results in impaired locomation (Fig. 1c), and ovid-3,4,10 carry new alleles of RNAi genes mut-16, rde-4 and rrf-1, respectively (Table 1). To further stratify our Ovid isolates, we assayed DRH-1 pathway activation using the expression of the downstream induced gene sdz-6 as readout (Fig. 1d). Only ovid-1 phenocopied drh-1 mutants and we subsequently demonstrated that ovid-1 defines a new allele of drh-1 (Fig. 1d). We identified a number of additional candidate genes (Table 1). ovid-9 and ovid-11 mutants are neither defective in canonical RNAi nor in the DRH-1 pathway and thus represent candidate genes for novel antiviral defense mechanisms.
Figure 1. A forward genetic screen identifies novel antiviral immunity genes.
a, Diagram of the lys-3p::gfp viral stress sensor.
b, Ovid screen workflow. Transgenic animals carrying the viral stress sensor were mutagenized using EMS and F2 progeny were assayed. OrV, Orsay virus. Ovid, Orsay virus immunodeficient.
c, Top panel: viral load of strains as indicated, measured by qRT-PCR of OrV RNA1, 4 dpi. Bars represent average value; error bars represent the standard error of the mean (SEM) of four independent infections. One-tailed student’s t-test: ****p<0.0001, *** p<0.001, **p<0.01, *p<0.05. Bottom panel: locomotion defects scored (paralyzed or twitching) after unc-22 RNAi feeding. Bars: average value; error: SEM; three independent RNAi treatments. Two-tailed student’s t-test: ****p<0.0001, **p<0.01.
d, Viral load compared to sdz-6 mRNA levels by qRT-PCR. Dots: average value; error: SEM; four independent infections. Samples as in c.
Table 1. Ovid screen candidate genes.
| Genotype | High viral load? | RNAi intact? | High sdz-6 level? | Candidate gene | Candidate variation | Brief description |
|---|---|---|---|---|---|---|
| WT | No | Yes | Yes | |||
| rde-1 | Yes | No | Yes | RNAi factor | ||
| drh-1 | Yes | Yes | No | Viral RNA receptor | ||
| ovid-1 | Yes | Yes | No | drh-1 | Glu834Lys | Viral RNA receptor |
| ovid-2 | Yes | Yes | Yes | n.d. | ||
| ovid-3 | Yes | No | Yes | mut-16 | Gln861* | RNAi factor |
| ovid-4 | Yes | No | Yes | rde-4 | Ala220Thr | RNAi factor |
| ovid-5 | Yes | No | Yes | n.d. | ||
| ovid-6 | Yes | Yes | Yes | T09B4.2 | Pro330Leu | Putative rho guanine nucleotide exchange factor |
| ovid-7 | Yes | Yes | Yes | C41D11.6 | Gly596Ser | Putative RNA nuclease |
| ovid-8 | Yes | Yes | Yes | n.d. | ||
| ovid-9 | Yes | Yes | Yes | cde-1 | Gln910* | Terminal uridylyltransferase |
| ovid-10 | Yes | No | Yes | rrf-1 | Gly45Glu | RNAi factor |
| ovid-11 | Yes | Yes | Yes | C54D10.14 | Gly122Arg | Uncharacterized, DRH-1-dependent induction |
| ovid-12 | Yes | No | Yes | F27D4.6 | Arg717* | Uncharacterized |
| ovid-13 | n.s. | Yes | Yes | n.d. | ||
| ovid-14 | n.s. | Yes | Yes | n.d. | ||
| ovid-15 | n.s. | Yes | Yes | n.d. | ||
| ovid-16 | Yes | Yes | Yes | phi-32 ssl-1 | Pro75Ser Gly1119Glu | Ubiquitin gene SNF2-related |
n.s., not scored. n.d., not determined.
The terminal uridylyltransferase CDE-1 is required for antiviral defense in C. elegans
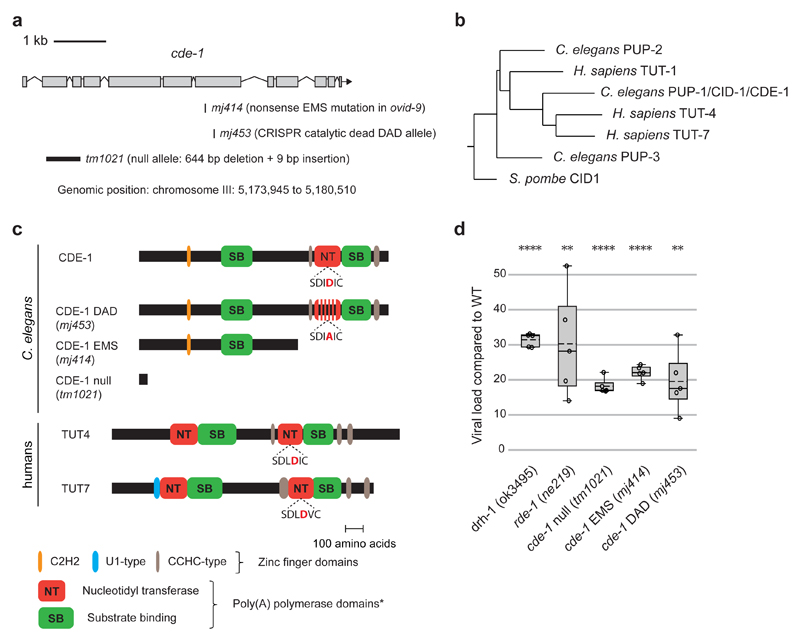
Whole-genome re-sequencing and genetic complementation tests revealed the causative mutation in ovid-9 to be a single-nucleotide nonsense mutation in the cde-1 gene (mj414, glutamine 910 to STOP) (Fig. 2a and Extended Data Fig. 2). cde-1 encodes a catalytically active 3′-terminal RNA uridylyltransferase (TUT), which is a homologue of mammalian TUT4 and TUT7 enzymes 17–19 (Fig. 2b, c). The independently derived cde-1 (tm1021) knockout strain also phenocopied viral stress sensor activation (Extended Data Fig. 3), high viral loads (Fig. 2d), and horizontal transmission of infection (Extended Data Fig. 3). RNA FISH revealed that viral infection is restricted to the intestine in cde-1 and in cde-1; drh-1 double mutants 4,9 (Extended Data Fig. 4a). We validated that CDE-1 is present in the intestine using a GFP fusion 18 (Extended Data Fig. 4b). To disentangle between the functions of CDE-1 in different tissues, cde-1 was exclusively expressed from an intestine-specific vha-6p promoter (Extended Data Fig. 4c). Animals with intestinal expression of cde-1 became resistant to viral infection (Extended Data Fig. 4d), but kept a defect in meiotic chromosome segregation (Extended Data Fig. 4e), probably caused by CDE-1 depletion in the germline 17. CDE-1 contains a conserved triad of acid aspartic residues (DDD) in its nucleotidyltransferase domain. Mutation of the corresponding DDD triad to DAD (D1011A) in human TUT4 resulted in loss of catalytic activity 20. A cde-1 DAD mutant strain (Fig. 2a,c) showed similar viral susceptibility as the cde-1 null mutants (Fig. 2d). In summary, we identify CDE-1-mediated 3′ terminal uridylation as an antiviral activity in the intestine of C. elegans.
Figure 2. The terminal uridylyltransferase CDE-1 restricts viral infection.
a, Diagram of cde-1 alleles. DAD, catalytic dead mutant.
b, Neighbor joining tree of the terminal uridylyl transferases (TUTs) of C. elegans and humans and S. pombe CID1.
c, Diagrams of C. elegans CDE-1 and human TUT4 and TUT7. Domains were predicted by Interpro. The central D of the conserved DDD catalytic triad is highlighted in red.
d, Viral load as measured by qRT-PCR of OrV RNA1 genome in adults two days after infection. Boxplots: whiskers from minimum to maximum; dots: independent infection; n=5. One-tailed student’s t-test: **** p<0.0001, **p<0.01
CDE-1 exert its antiviral function independently of antiviral RNAi
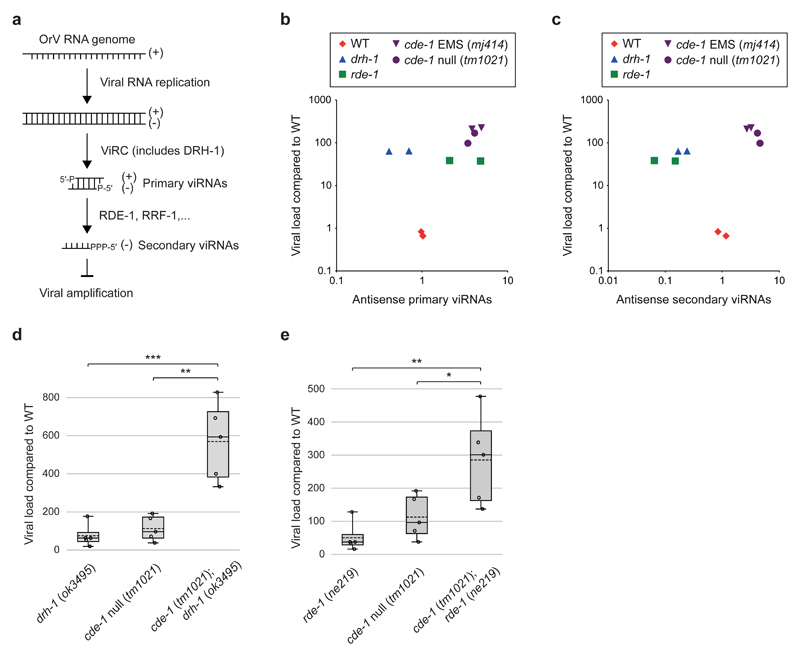
In eukaryotes, addition of 3′ uridyl-tails (U-tails) by TUTs is a degradation signal that can engage: (i) the XRN-family of exoribonucleases for 5′ to 3′ RNA decay; (ii) the 3′ to 5′ exoribonuclease DIS3L2; (iii) the 3′ to 5′ exosome complex 21–24. We sought to identify the RNA(s) targeted by CDE-1 in its antiviral role. CDE-1 is implicated in endogenous RNAi pathways that are restricted to the germline 17. Small RNA sequencing on whole animals revealed that siRNAs are targeted by CDE-1 for 3′ uridylation, miRNAs are occasionally targeted, and piRNAs are not targeted 17 (Extended Data Fig. 5a). The role of CDE-1 in small RNA function remains unclear as depletion of CDE-1 leads to only subtle changes in siRNA and miRNA steady state levels (Extended Data Fig. 5b, c). To understand if CDE-1 functions through modification of siRNAs in antiviral immunity, we tested cde-1 mutants directly for defects in antiviral RNAi. During an antiviral RNAi response in C. elegans, the ViRC complex recognizes the dsRNA of the replicating viral genome and dices it into sense and antisense ~23-nt long primary viRNAs, which are loaded into the RDE-1 Argonaute protein 5 (Fig. 3a). The RNAi response is further amplified by RNA-dependent RNA polymerase (RdRP, RRF-1) generated 22-nt long antisense secondary viRNAs, with a 5′ triphosphate guanine (22G-RNAs), which are incorporated into secondary Argonaute proteins to silence viral amplification 5 (Fig. 3a). Thus, in an animal with functional antiviral RNAi, a high viral load should correlate with a high level of viRNAs. We measured primary and secondary viRNAs in different genetic backgrounds (Fig. 3b,c). All the mutants tested (drh-1, rde-1, cde-1) accumulate high levels of the virus as compared to wild type. In drh-1 mutants, primary and secondary viRNAs are depleted when compared to wild type, despite the increase in viral load. In rde-1 mutants, primary viRNAs are abundant but secondary viRNAs are depleted, as in drh-1. In contrast, cde-1 mutants accumulate both primary and secondary viRNAs to a level that correlates with the high viral load. To determine if viRNAs can silence viral amplification in cde-1 mutants, we carried out epistasis analysis using null mutants of drh-1, rde-1 and cde-1 (Fig. 3d,e). Both cde-1;drh-1 and cde-1;rde-1 double mutants showed an increase in viral load as compared to drh-1 or rde-1 on its own. We conclude that CDE-1 does not exert its immune function through the antiviral RNAi pathway.
Figure 3. CDE-1 acts in parallel to antiviral RNAi.
a, Schematic of antiviral RNAi in C. elegans. Viral Recognition Complex (ViRC) includes DCR-1; DRH-1; RDE-4.
b, Comparison between the viral load and primary viRNA populations. Primary viRNAs (23-nucleotide long, from 5′ monophosphate RNA sequencing). Only antisense RNAs were considered to exclude potential viral genome degradation products. Dots: independent infection.
c, Comparison between the viral load and secondary viRNA populations. Secondary viRNAs (22-nucleotide long, starting with a G, from 5′ tri/monophosphate RNA sequencing). Samples as in b.
d and e, Viral load as measured by qRT-PCR of OrV RNA1 genome in adults two days after infection. Boxplots: whiskers from minimum to maximum; dots: independent infection; n=5. One-tailed student’s t-test: *** p<0.001, **p<0.01, *p<0.05. Samples as in b.
CDE-1 defines a novel antiviral immunity pathway
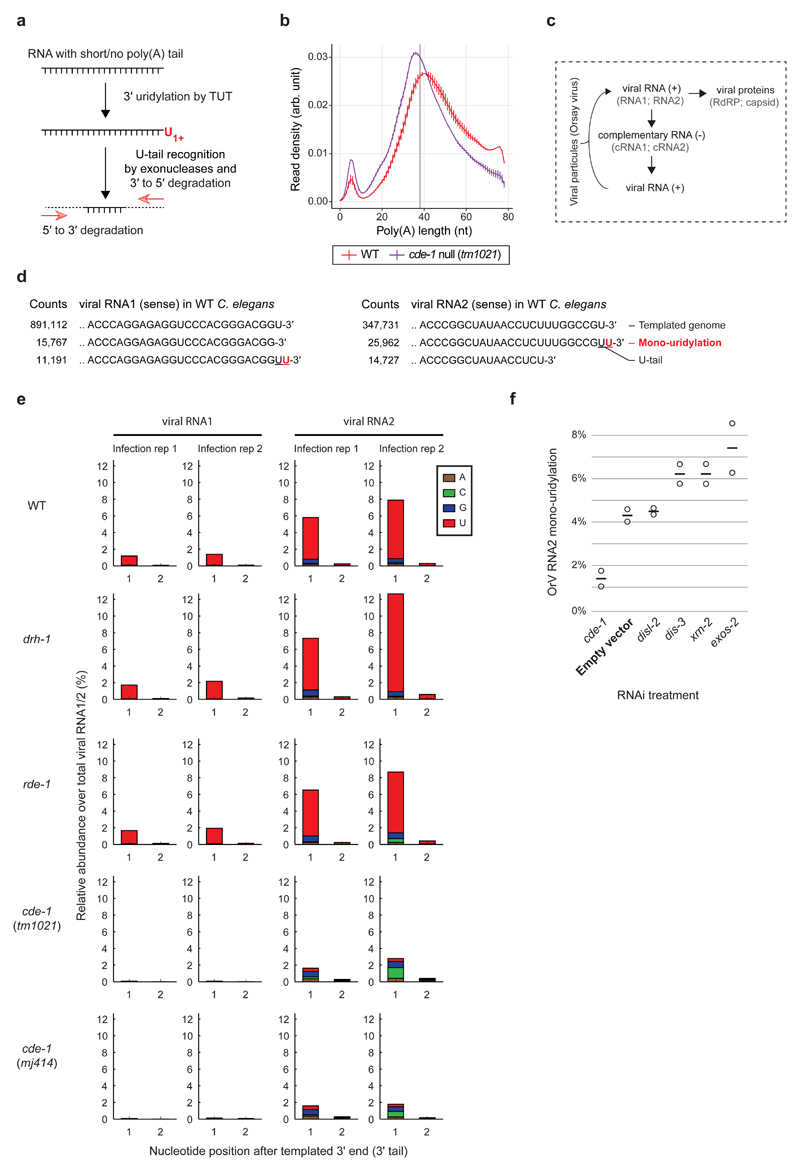
In mammals, uridylation is coupled to poly(A) tail length where TUT4 and TUT7 (collectively called TUT4(7)) preferentially uridylate mRNAs with short poly(A) tails (<25 nt) to facilitate their degradation 25,26 (Fig. 4a). We thus assessed the impact of CDE-1 on endogenous mRNA poly(A) tail lengths and terminal nucleotide addition in infected wild-type or cde-1 mutant animals using TAIL-seq 25,27. The C. elegans transcriptome revealed a bimodal distribution of poly(A) tail lengths, with a major peak of poly(A) tails of ~40 nt, and a second peak of poly(A) tails of ~10 nt (Fig. 4b; using our method we could not assess transcripts with poly(A) tails > 79 nt). In cde-1 mutants, there is a shift of the major ~40 nt peak to ~36 nt and an increase in transcripts with shorter poly(A) (Fig. 4b). We infer that CDE-1 promotes the degradation of transcripts with short poly(A) tails in C. elegans too. However, CDE-1 had no global effect on the poly(A) tail distribution of OrV-induced stress response genes (Extended Data Fig. 6a). Also, the OrV-induced stress response was stronger in cde-1 mutants than in wild-type upon infection (Extended Data Fig. 6b), reflecting the difference in viral load between these two strains. This indicates that CDE-1 is not required for the OrV-induced stress response. Although we cannot formally rule out that CDE-1 may regulate an endogenous target(s), the evidence indicates this is not CDE-1’s principal function in antiviral immunity.
Figure 4. CDE-1 directly targets the Orsay virus RNA genome for uridylation.
a, Schematic of TUT-mediated RNA degradation.
b, Poly(A) tail length distribution measured by TAIL-seq after two days of OrV infection. Vertical bars: range from minimum to maximum (two independent C. elegans culture plates). Vertical grey line represents the mean of cde-1 and wild type peaks (38 nt).
c, Schematic of Orsay virus replication
d, Most frequent collapsed reads after RACE-seq on OrV RNA1 and RNA2 (2 dpi), respectively. Non-templated residues (absent from the reference genome) are indicated in red.
e, Percentage reads with non-templated nucleotides detected at the 3′ end of OrV RNA1-2 in strains as indicated, two days post infection. Two independent infections per genotype.
f, Percentage reads with a non-templated mono-uridyl residue at the 3′ end of OrV RNA2, upon RNAi-mediated gene knockdown as indicated, one day post infection. Bars; average; dots: independent RNAi treatments and infections.
Instead, we postulated that the viral RNA genome itself may be uridylated by CDE-1. U-tails can only be observed on a small percentage of cellular RNAs as uridylated RNAs are prone to be degraded 26. To detect uridylated Orsay RNA degradation intermediates, we carried out 3′ rapid amplification of cDNA ends (RACE) followed by high-throughput sequencing of the OrV RNAs extracted from C. elegans two days postinfection (RACE-seq; Extended Data Fig. 7a). Mono(U) tails constituted the most abundant fraction of non-templated nucleotides detected at the 3′ end of both OrV RNA1 and OrV RNA2 (Fig. 4c-e). For both RNA1 and 2, U-tailing was lost in two independent cde-1 mutant alleles. In contrast, drh-1 and rde-1 mutants showed similar levels of viral RNA U-tails to wild-type, indicating that U-tailing is independent of viral load and that CDE-1 is not in limited quantities (Extended Data Fig. 7b,c). OrV RNA1 and RNA2 have a terminal uridylyl residue in their genome such that the addition of an extra non-templated uridine by CDE-1 forms a UU termination (Fig. 4d), which is a signal for uridylation-dependent RNA decay 21,23. The two XRN paralogs in C. elegans (XRN-1 and XRN-2) and the exosome components (e.g. DIS-3, EXOS-2) are essential 28,29, and these RNA degradation pathways normally act redundantly on uridylated RNAs 26. We therefore subjected C. elegans to a short (24 hours) RNAi treatment to effect a partial knockdown of cde-1, the exonuclease disl-2 (the C. elegans DIS3L2 homologue), the exosome components exos-2 and dis-3, and the exonuclease xrn-2. Treated animals, which appeared superficially wild type, were infected with OrV for 24 hours. The frequency of U-tails in OrV RNA2 was measured by RACE-seq (Fig. 4f). ~4% of OrV RNA2 were uridylated in animals exposed to the empty vector control RNAi, compared to ~1% in cde-1 knockdown. RNAi treatments against disl-2 did not affect the U-tail frequency. We measured a 1.4 to 1.7 fold increase in U-tail frequency upon RNAi treatment against exos-2, dis-3 and xrn-2, suggesting that these factors each contribute to the degradation of uridylated viral RNAs, in accordance with a study that shows that DIS3 and the exosome can degrade viral RNAs in Drosophila and human cells 30. We conclude that C. elegans uses uridylation of the OrV as an innate immune defense. This mechanism acts in parallel to antiviral RNAi to combat viral infection (Fig. 5).
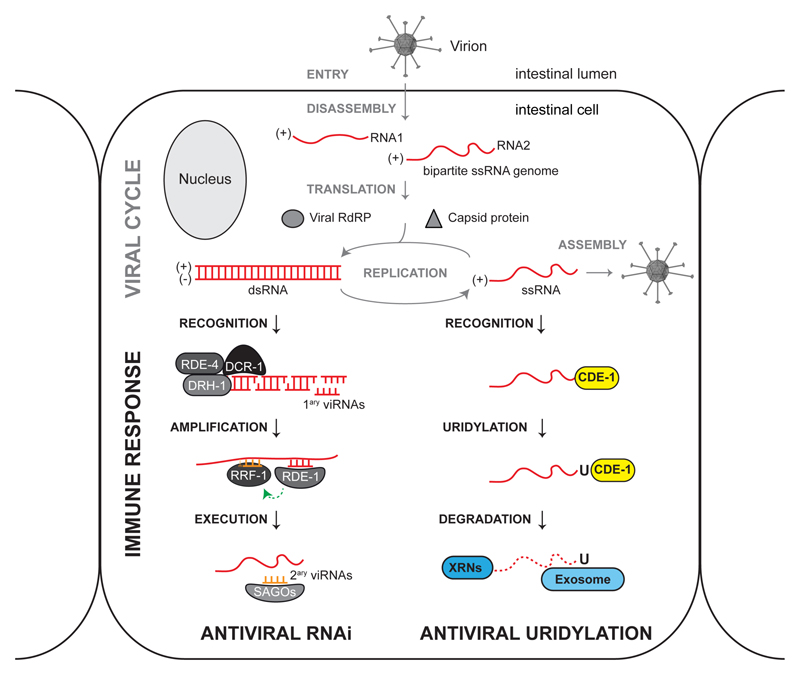
Figure 5. Antiviral RNAi and virus terminal uridylation are parallel immune defense pathways in C. elegans.
Virion cartoon adapted from 12. The Orsay virus primarily infects intestinal cells in C. elegans9. Once entered in the host cell, the virus disassembles in the cytoplasm and exposes its RNA genome. The Orsay virus has a bipartite positive-stand RNA genome that can be directly translated by the cellular machinery. The Orsay RNA1 molecule encodes a viral RNA dependent RNA polymerase (RdRP) and the Orsay RNA2 molecule encodes the capsid protein4. Virus amplification occurs in the cytoplasm in two steps: (i) the positive-strand genome serves as a template for the synthesis of a negative strand antigenome by the viral RdRP; (ii) the antigenome serves, in turn, as a template for further synthesis of genomic RNAs. The C. elegans antiviral RNAi response is initiated by DCR-1–DRH-1–RDE-4 (Viral RNA Recognition Complex) that recognize replicating double-stranded viral RNAs and process them into virus-derived small interfering RNAs (primary viRNAs). Primary viRNAs are loaded into the RDE-1 Argonaute protein and trigger the synthesis of secondary viRNAs by the host RdRP RRF-1, that uses the viral RNA as a template. Secondary viRNAs, loaded into somatic Argonaute proteins (SAGOs), target viral RNAs by base complementarity and potently reduce virus replication5,11. Viral RNAs are also targeted by the terminal uridylyltransferase CDE-1. CDE-1 marks viral RNAs with a 3′ U-tail to recruit 5′ to 3′ exonucleases of the XRN family, and 3′ to 5′ exonucleases of the exosome complex. C. elegans may have additional antiviral pathways, for example a set of stress response genes is induced by a STAT signaling pathway upon virus infection, that could promote virus clearance (not shown in this model) 15,16.
Terminal uridylyltransferases target viral RNAs in mammalian cells
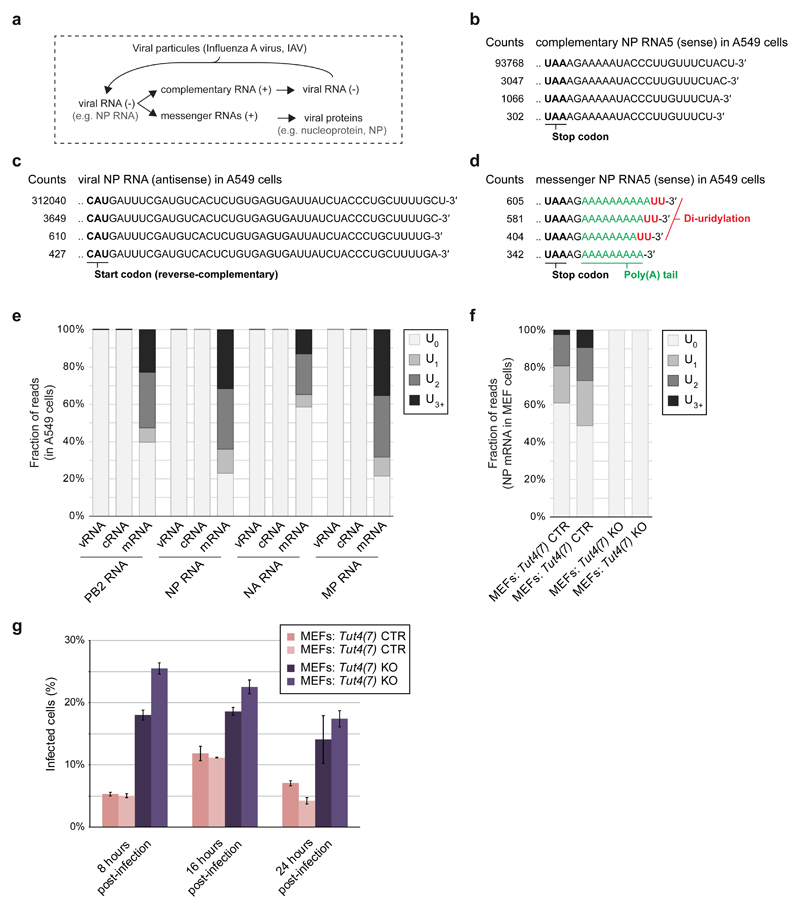
The U-tail modification is conserved in eukaryotes and could impact a broad range of viruses in a variety of hosts 31. We tested if U-tailing affects the replication of Influenza A virus (IAV), which can infect human and murine cells. The IAV genome consists of eight antisense RNA segments (viral RNAs, vRNAs) from which the viral RdRP produces: (i) the sense complementary RNAs (cRNAs), which serve as templates to produce more vRNAs; and (ii) the mRNAs that are 3′ polyadenylated and exported to the cytosol for translation into viral proteins 32 (Fig. 6a). We examined the 3′ end of a set of IAV RNAs, at 8 hours post-infection (hpi), in A549 human lung cells by RACE-seq. We could not detect U-tails at the ends of vRNAs or cRNAs. In contrast, viral mRNAs were highly uridylated at their 3′ end, with ~77% of the IAV Nucleoprotein (NP) mRNA containing a U-tail, and a di(U)-tail being the most common type of 3′ end (~32%) (Fig. 6b-e). The IAV NP mRNA is also uridylated (~40-50%) at 8 hpi in mouse embryonic fibroblasts (MEFs), but uridylation was lost in MEFs deficient in both Tut4 and Tut7 25 (Fig. 6f). Thus TUT4(7) can uridylate the 3′ end of viral RNAs in mammalian cells. The RACE-seq can only detect IAV mRNAs with poly(A) tails of <70 nt; it is possible that some IAV mRNAs with very long poly(A) tails are less prone to be uridylated. To test the impact of TUT4(7) on IAV, we measured the quantity of NP mRNA by qRT-PCR in infected MEFs (Extended Data Fig. 8a). The IAV NP mRNA accumulated more rapidly and to a higher level at the peak in MEFs Tut4(7) KO cells (peak at 8 hpi) compared to WT cells (peak at 16 hpi) before decreasing later in infection (24 hpi). Consistent with the difference in mRNA levels, the NP mRNA-encoded viral nucleoprotein (NP) accumulated more rapidly in MEFs Tut4(7) KO cells compared to WT during the first eight hours of infection (Extended Data Fig. 8b). Accordingly, more infected cells overall were observed in MEFs Tut4(7) KO compared to WT (Fig. 6g). In conclusion, TUT4(7) could act as an early barrier against IAV infection in mammalian cells. Although we cannot rule out that TUT4(7) may impact other steps of the IAV viral cycle, such as entry, our data strongly supports a model where TUT4(7) act by reducing the expression levels of IAV mRNAs during the early stages of IAV infection in MEFs, leading to a decrease in viral protein levels and rates of infection. Future studies will need to address the antiviral function of TUT4(7) in a variety of relevant host-virus models.
Figure 6. The terminal uridylyltransferases TUT4(7) attenuate Influenza A mRNAs in mammalian cells.
a, Schematic of Influenza A virus replication
b-d, Most frequent collapsed reads after RACE-seq on IAV NP cRNA, NP vRNA and NP mRNA, respectively in A549 cells at 8 hpi.
e, Percentage of reads with a non-templated U-tail (no U-tail; 1 U; 2 Us or ≥ 3 Us) in different RNAs as indicated measured by RACE-seq in A549 cells 8 hpi.
f, Percentage of reads with a non-templated U-tail (as in e) in MEF cells of different genotypes as indicated (with two independently created cell lines per genotype).
g-h, Percentage of infected cells measured by immunofluorescence against NP (FACS). Bars: average; Error: SEM; three independent infections. MEFs Tut4(7) KO are full null independent lines.
Discussion
Previously, we have shown that the antiviral RNAi pathway and DRH-1 are central to the innate immune response of C. elegans 5. Here, we demonstrate that the terminal uridylyltransferases also play a critical role in antiviral immunity, uridylating viral RNAs (with 1-2 Us) to mark them for degradation. It is unclear how terminal uridylyltransferase recognize viral RNAs as bona fide targets. Receptors of the RIG-I family commonly recognize pathogen-associated patterns at the 5′ termini of viral RNAs. In contrast, terminal uridylyltransferases interact with the 3′ termini of cytosolic RNAs with no poly(A)-tail or a short poly(A)-tail. As many RNA viruses, like OrV, lack a poly(A) tail at the 3′ termini of their RNA genomes, this may be a pathogen-associated pattern-recognition feature. We speculate that the IAV mRNAs and a fraction of the OrV RNAs are vulnerable to TUTs when exposed in the cytosol for translation. In conclusion, we find that terminal uridylyltransferases are potent antiviral factors during the early stages of RNA virus infections in C. elegans and in mammalian cells. This finding supports a scenario where eukaryotic mRNA decay pathways originally evolved as intrinsic cellular defenses against pathogens 33,34. Vertebrates also benefit from the interferon response and adaptive immune system, serving as potent lines of defense against pathogenic viruses; future studies will thus need to address the relative importance of antiviral uridylation in whole organisms. Terminal uridylyltransferases are widely conserved in eukaryotes and could potentially target a wide range of RNA viruses 31. Perhaps as a response to this threat, some viruses evolved to protect their RNA termini, such as single-stranded RNA viruses of the Flaviviridae family, which have highly structured 3′ ends resistant to degradation by cellular exonucleases 35. Our study illustrates that the 3′ termini of viral RNAs are key in the evolutionary arms race between viruses and their hosts.
Methods
Genetics
Animals were grown on agar plates, at 20°C, and fed with E. coli strain HB101 (obtained from the Caenorhabditis Genetics Center, University of Minnesota, USA). Standard C. elegans procedures were used for maintenance and genetic crosses 36. The wild-type strain refers to Bristol N2 unless stated otherwise. All strains used in this study are listed in the Supplementary Table 2.
PCR primers
All PCR primers used in this study are listed in the Supplementary Table 3.
Viral filtrate preparation
Viral filtrate was prepared as in 8. Briefly, JU1580 animals were first stably infected by the Orsay virus (OrV) in solid culture and then transferred in a liquid culture containing OP50 bacteria for seven days. The liquid culture with infected JU1580 was then centrifuged at 16,000 g for 30 min and the supernatant was filtered (0.22 µm filter) to produce the viral filtrate (stored at -80°C).
Transgenesis of C. elegans with the lys-3p::GFP viral stress sensor
The 452 bp region upstream of the lys-3 start codon and the first 57 bp of the coding region of lys-3 were used as a promoter and cloned into an entry clone using Multi-Site Gateway cloning (Invitrogen) according to manufacturer's instructions. The lys-3 donor plasmid was validated by sequencing. Gateway technology was then used to clone the lys-3 fragment in frame with a GFP cDNA. The 3′ UTR of the tbb-2 (tubulin, beta) gene was used. The lys-3p::GFP:tbb-2-3′UTR plasmid was amplified and purified according to Invitrogen's instruction. The C. elegans microinjection mix was: 5 ng/µl plasmid lys-3p::GFP:tbb-2-3′UTR; 5 ng/µl co-injection marker (myo-2::mcherry::unc-54-3′UTR, pharynx expression) and 85 ng/µl 1 kb Invitrogen ladder in 1× injection buffer (20 mM potassium phosphate, 3 mM potassium citrate, pH 7.5). This mix was microinjected into the gonads of rde-1 (ne219) mutants to generate a multicopy extrachromosomal array (allele mjEx547). X-ray integration of the transgene into the C. elegans genome was performed as described previously 37. Animals carrying an integrated transgene (allele mjIs228) were outcrossed three times to generate SX2635 (lacking ne219), referred to as wild-type viral stress sensor strain in this study.
Confocal images of the biostress reporter
A 2% agar pad was used on top of a glass slide and a drop of 10 µM tetramisol in M9 medium was placed on this agar pad. Animals were picked into the tetramisol solution. Imaging was performed with an Olympus Upright FV1000 microscope at 10× or 20× magnification, as specified, using the FluoView image software (Olympus). Identical microscope settings were used for all images within a figure.
Forward genetic screen for Ovid screen isolates
Approximately 4,000 viral stress sensor transgenic animals were mutagenized using ethyl methanesulfonate (EMS) as described in 36 and 38. Approximately 50,000 F2 animals were infected for 3-4 days and ~2,000 animals showing intestinal GFP were picked individually for re-testing. 16 F2 families showed transmission of the viral stress sensor activation. Bleach treatment confirmed that removing OrV lead to a loss of intestinal GFP signal.
C. elegans infection by the Orsay virus
Animals were either infected for four days as asynchronous populations or for two days as synchronous populations. Infections of asynchronous populations were performed as in 5. Briefly, two L4 hermaphrodites were distributed in each 50 mm plates and, on the next day, 20 µl of viral filtrate was spread on the plates. Animals were harvested (for viral load measurement) or observed under a Leica M165 FC fluorescent microscope (for scoring of the viral stress sensor) four days post-infection (4 dpi). This method was typically used for the characterization of the Ovid screen isolates. For the infection of synchronous populations, 200 animals at the larval stage L1 were deposited on each 50 mm plate. On the next day, L2 animals were infected with 20 µl of viral filtrate homogeneously spread on the plate. Plates were kept up-side-up for 24 hrs. Animals were harvested for viral load measurement at 2 dpi. This method was used to measure the viral load in cde-1 mutants, as indicated in the figure legends.
RNA level measurement by qRT-PCR
Harvested animals were washed three times by pelleting-resuspension in M9 solution. Lysis and qRT-PCR was then performed from 5 µl of animal pellet using the Power SYBR Green Cells-to-Ct kit (Ambion, Austin, TX) as described in 5. The primers M1835 and M1836 13, and M4410 and M4411 4, were used to measure RNA levels of gapdh and OrV gRNA1, respectively.
RNAi-mediated knockdown of unc-22
All the bacterial feeding clones used in this study were a kind gift from the laboratory of Julie Ahringer. Bacteria were grown in LB-Ampicillin (50 μg/ml) for 6 hrs, then seeded onto 50 mm NGM agar plates containing 1 mM IPTG and 25 μg/ml Carbenicillin at a volume of 300 μl bacterial culture per plate and left to dry at room temperature, protected from the light, for 48 hrs. Two L4 animals were picked onto each RNAi plates and the young adult progeny were scored for the phenotype of interest after five days.
Transgenesis of C. elegans with the CDE-1::GFP fosmid and imaging
The modified fosmid WRM064A_D06 where the GFP sequence is added at the N-terminal end of cde-1 was provided by the TransgeneOme Project (Max Planck Institute of Molecular Cell Biology and Genetics, TransgeneOme Unit, Pfotenhauerstr. 108, 01307 Dresden, Germany; construct 09318202437763223 H08) 39. The construct was injected into the gonad of N2 animals to produce an extrachromosomal array (as described for the biostress reporter), using a myo-3p::mCherry::unc-54-3′UTR construct as a co-injection reporter. Transgenic animals (strain SX3123; allele mjEx594) were imaged with an Olympus Upright FV1000 microscope at 10x magnification.
Fluorescence in situ hybridization of the Orsay virus RNA2
Animals were harvested in 15 ml of nanopure water and washed three times by pelleting-resuspension in nanopure water. Animals were then transferred to 1.5 ml tubes with a glass pipette. 1 ml of fixative solution (4% formaldehyde in 1X PBS) was added and samples were incubating at room temperature, on a rotating wheel, for 45 min. Nematodes were then washed twice by pelleting-resuspension in 1 ml of 1x PBS. Pellet of animals was resuspended in 1 ml 70% ethanol and stored at 4°C. After removal of the ethanol, fixed nematodes were washed once in 1 ml of wash solution (10% formamide, 2X SSC). The animal pellet was resuspended in 100 µl of hybridization solution (10% dextran sulfate, 2X SSC, 10% formamide) with 1 µl 1:50 of the probe v1580-RNA2-TexRed (ACCATGCGAGCATTCTGAACGTCA), a kind gift of Marie-Anne Félix, and incubated overnight at 30°C protected from the light. The next day, animals were washed three times in wash solution by pelleting-resuspension. Eventually, animals were resuspended in 1 ml wash solution with DAPI and incubated at 30°C for 30 min. Samples were centrifuged and supernatant was discarded. The animal pellet was resuspended in 1 ml of 2X SSC solution and stored at 4°C protected from light. Animals were then placed on a glass slide, in a drop of Vectashield anti-fade solution (Vector). Imaging was performed on an Olympus Upright FV1000 at 40x magnification, using the FluoView image software (Olympus). Same settings of fluorescence were used for all images compared.
Transgenesis of C. elegans with the vha-6p::gfp plasmid and viral load measurement
The 878 bp region upstream of the vha-6 start codon was used as a promoter and cloned into an entry clone using Multi-Site Gateway cloning (Invitrogen) according to manufacturer's instructions. The vha-6p donor plasmid was validated by sequencing. Gateway technology was then used to clone the vha-6p upstream of (i) the GFP cDNA, or (ii) the full length cde-1 gene (from ATG to STOP with endogenous introns). The 3′ UTR of the tbb-2 (tubulin, beta) gene was used. The vha-6p::GFP::tbb-2-3′UTR and vha-6p::cde-1::tbb-2-3′UTR plasmids were amplified and purified according to Invitrogen's instruction. The C. elegans microinjection mix was: 10 ng/µl plasmid vha-6p::GFP:tbb-2-3′UTR; 10 ng/µl plasmid vha-6p::cde-1::tbb-2-3′UTR; 5 ng/µl co-injection marker (myo-2::mcherry::unc-54-3′UTR, pharynx expression) and 75 ng/µl 1 kb Invitrogen ladder in 1× injection buffer (20 mM potassium phosphate, 3 mM potassium citrate, pH 7.5). This mix was microinjected into the gonads of cde-1 (tm1021) mutants to generate a multicopy extrachromosomal array (allele mjEx595). vha-6p driven GFP expression was only observed in the intestine. 100 animals carrying the extrachromosomal array were manually selected for infection (from the L2 larval stage to young adult).
Small RNA sequencing
Small RNA libraries were prepared from infected animals as previously described in 5. We used pellets of animals, washed three times in M9 solution and resuspended in 1 ml of TriSure (Bioline) as a starting material. RNA extraction was performed according to manufacturer’s instructions. Some populations of siRNAs (including secondary viRNAs) contain a characteristic 5′ triphosphate group that has to be replaced by a 5′ monophosphate to allow the 5′ ligation step of the library preparation. For this purpose, 1 µg of RNA was put in solution with 1X 5′p polyphophatase buffer and 1 µl of 5′ polyphophatase (Epicentre) for a total volume of 20 µl, incubated for 30 min at 37°C and then submitted to phenol purification and resuspended in 5 µl of nuclease-free water. Treated RNA sample was entirely used as starting material for the TruSeq Small RNA kit (Illumina), following the manufacturer’s instructions, to make the so-called 5′ independent libraries. So-called 5′ dependent libraries were made by a similar procedure but without polyphophatase treatment, so that only 5′ monophosphate siRNAs (such as primary viRNAs) could be cloned. Libraries were submitted to the Gurdon Institute sequencing facility for Illumina HiSeq sequencing (SR36). Small RNA sequencing data was aligned to the Ensemble WBcel235 release of the C. elegans genome using STAR 40 (v2.5.1b). Briefly, the aligner will allow untemplated residues at the ends of an aligned sequence when run in local mode. Untemplated 3′ sequences were extracted and analysed using custom Python scripts. Details of the analyses for each small RNA subtype can be found in the source code. For miRNA differential expression, reads were counted against the miRBase miRNA annotations (miRBase21 hairpins, WBcel235 genome) using featureCounts 41 (v1.5.0-p1). Differential expression analysis was performed on the counts using DESeq2 42 (v1.10.1).
CRISPR/Cas9 for cde-1 catalytic dead mutant
A CRISPR/Cas9-mediated mutation of cde-1 was generated as previously described 43. Guide RNA: UUUGCUGUCAAAUCCUUUGG. Homologous recombination template: TCAGCTATTGCTATTTGTTTGAGATTCGGAGATGGAGATGTTCCGCCTAAAGACTTGACAGCAAAAGAAGTTATTCAGAAAACTGAATCCGTTCTCAGAAAATGTCATTT. Only the D1069A missense mutation was introduced, as verified by sequencing.
TAIL-seq
The TAIL-seq was performed as previously described in 27. Tail-seq libraries were processed using Tailseeker 2 27. The 5′ and 3′ libraries were subsequently adapter trimmed using cutadapt 1.10 44 with Illumina small RNA-seq adapters and filtered to a minimum length of 5bp. Trimmed 5′ reads were mapped with STAR 2.5.2a 40 against a combined meta-genome consisting of the C. elegans reference genome WBcel235 45 and the OrV genome 4. Mapping was performed in end-to-end mode allowing no mismatches and a gap opening and extension penalty of 10,000. Reads were assigned to genes with bedtools 2.26.0 46. Subsequently, 3′ reads without poly(A) tail or too many dark cycles were removed from the data. For the subsequent analysis, all C. elegans tags with poly(A) tail length equal to zero were discarded. Average poly(A) tail lengths and uridylation lengths for each sample were calculated as the arithmetic mean weighted by the support for each tag, reported by Tailseeker 2. The complete code is at https://github.com/klmr/poly-u/tree/submitted.
mRNA libraries for deep sequencing
mRNA libraries were prepared from three independent infections, using the NEBNext Ultra RNA non-directional Library kit with poly(A) selection (NEB), according to manufacturer’s instructions. Libraries were submitted to the Gurdon Institute sequencing facility for Illumina HiSeq sequencing (SR30). Differentially expressed genes were then called using EdgeR 47.
3′ RACE-seq on the Orsay virus RNAs
The 3′ RACE was performed on the same RNA input than that used for small RNA libraries, without polyphosphatase treatment. 200 ng of RNA were submitted to 3′ ligation using the TruSeq Small RNA kit (Illumina), following the manufacturer’s instructions. 3′ ligated RNA was used for reverse-transcription, still using the TruSeq Small RNA kit whilst bypassing the 5′ ligation step. The 3′ end of OrV RNA1 (or RNA2) genome was amplified by PCR (“PCR1”) from 2 µl of cDNA, using the primers M7454 and M7456 (or M7455 and M7456) and the Phusion High-Fidelity Taq Polymerase (NEB) with CG buffer, according to manufacturer's instructions. The thermocyler was programmed to 30 seconds at 98°C; 15 cycles of 5 seconds at 98 °C followed by 20 seconds at 60°C and 10 seconds at 72°C. The 5′ adapter sequence from the TruSeq Small RNA kit was then introduced at the 5′ end of the amplicons by PCR (“PCR2”) using the primers M7456 and M7601 for OrV RNA1 (or M7456 and M7602 for the OrV RNA2), using 2 µl of 1/10 diluted amplicon from PCR1 as a template and the same PCR conditions than that used in PCR1. The amplicons from PCR2 were purified using the DNA Clean & Concentrator-5 kit (Zymo Research) and resuspended in 10 µl of water. Resulting DNA was used as an input for the PCR amplification step of the TruSeq Small RNA kit, following the manufacturer’s instructions. Libraries were submitted to the Gurdon Institute sequencing facility for Illumina HiSeq sequencing (PE100). The libraries were run on a 10% polyacrylamide gel for size selection (the amplicons could be visualized under UV light and the bands were cut at the same distance of migration for all samples). Paired-end reads obtained from the 3′ RACE experiment on the viral genome show overlap. The PEAR software 48 was used to merge the paired reads into a single read (v0.9.6, default parameters). Merged reads not starting with the targeted 3′ viral genome sequence fragment were discarded. The targeted viral genome sequence was removed from the remaining reads using custom python scripts (https://github.com/tdido/cde-1_analysis). The resulting sequences representing the untemplated tails were analyzed using custom python scripts.
RNAi-mediated knockdown of exonucleases
Synchronized animals were grown on normal HB101 food until the L2 larval stage and then transferred RNAi food. Animals were left on RNAi plate (24 hours prior to infection) and infected for 24 hours, from the old L3/young L4 larval stages to adult. RACEseq was performed as described above.
Cell culture
MEF cells were cultured with DMEM (GIBECO) supplemented with 12.5% FBS, 2mM L-glutamine, non-essential amino acid,100 units/ml penicillin/streptomycin, 100 uM β-mercaptoethanol (Sigma). Cells were splitted 1:4 and passaged every three days. A549 cells were cultured with DMEM (GIBECO) supplemented with 10% FBS, 2mM L-glutamine, non-essential amino acid,100 units/ml penicillin/streptomycin and 25mM HEPES.
Cell lines used in this study
All cell lines were tested for negative for mycoplasma. Tut4(7) CTR and KO MEFs were derived from E13.5 embryos from crosses of Tut4+/fl;Tut7+/fl;R26+/+ and Tut4+/fl;Tut7+/fl;R26ERT-cre/ERT-cre mice by standard procedures and immortalized at passage 2 by two consecutive infections with pBabeSV40LT. Cre-mediated deletion to obtain Tut4(7) null alleles was induced with 600 nM 4-hydroxytamoxifen for three days 25. All mice used in this study were bred and maintained in EMBL Mouse Biology Unit, Monterotondo, and subsequently in the Centre for Regenerative Medicine, Edinburgh. All procedures were done in accordance to the current Italian legislation (Art. 9, 27. Jan 1992, nu116) under license from the Italian health ministry or the UK Home Office regulations, respectively.
A549 and MEF cells infection by Influenza A virus and RACE-seq
Influenza A virus (A/WSN/1933, H1N1) used in this study was titrated on MDCK cells. All the inoculation MOI of influenza A virus described here and below was calculated as an equivalent MOI on the originally titrated MDCK cells.
A549 or MEF cells were trypsinized and seeded as 2X10^6 cells per T25 flask one day before infection. 16 hours after seeding, culture media were removed and cells were washed once with pre-warmed DMEM. Influenza A virus (A/WSN/1933, H1N1) were inoculated at MOI 3 diluted with 1000 µl DMEM supplemented with 0.1% BSA (D0.1B). Cells were trypsinized and collected 8 hours post infection. 750 µl TRIzol were added into each infected sample and were then freezed at -80 °C. RNA extraction was performed according to the standard TRIzol procedure.
For the A549 RACE-seq, 2 µg of RNA were submitted to 3′ ligation using the TruSeq Small RNA kit (Illumina), following the manufacturer’s instructions. 3′ ligated RNA was used for reverse-transcription, still using the TruSeq Small RNA kit (except that the Invitrogen Suprescript III was used instead of the Superscript II) whilst bypassing the 5′ ligation step. The RT final volume was 12.5 µl. After the RT, water was added to the samples to reach 18.5 µl, final volume. The 3′ end of IAV RNAs were amplified by PCR (“PCR1”) from 2 µl of cDNA, using the left primers M8443, M8444, M8451, M8452, M8453, M8454, M8455, M8456 (depending on the target, see the Supplementary Table 3) with the right primer M7456 and the NEB Q5 polymerase, according to manufacturer's instructions (25 µl reaction). The thermocyler was programmed to 30 seconds at 98°C; 5 cycles of 5 seconds at 98 °C followed by 20 seconds at 60°C and 20 seconds at 72°C. Each PCR product was purified using the DNA Clean & Concentrator-5 kit (Zymo Research) and eluted in 11 µl of water. The 5′ adapter sequence from the TruSeq Small RNA kit was then introduced at the 5′ end of the amplicons by PCR (“PCR2”) using the left primers M8459, M8460, M8467, M8468, M8469, M8470, M8471, M8472 (depending on the target, see the Supplementary Table 3) with the right primer M7601, using 10 µl of purified PCR1 amplicon as a template and the same PCR conditions that used in PCR1. Again, the amplicons from PCR2 were purified using the Zymo columns and eluted in 11 µl of water. Resulting DNA was used as an input for the PCR amplification step of the TruSeq Small RNA kit, following the manufacturer’s instructions. Libraries were submitted to the Gurdon Institute sequencing facility for Illumina HiSeq sequencing (PE100). The libraries were run on a 10% polyacrylamide gel for size selection (the amplicons could be visualized under UV light and the bands were cut at the same distance of migration for all samples). Paired-end reads obtained from the 3′ RACE experiment on the viral genome show overlap. The PEAR software 48 was used to merge the paired reads into a single read (v0.9.6, default parameters). Merged reads not starting with the targeted 3′ viral RNA sequence fragment were discarded. The targeted viral genome sequence was removed from the remaining reads using custom python scripts (https://github.com/tdido/cde-1_analysis). The resulting sequences representing the untemplated tails were analyzed using custom python scripts. The MEFs RACE-seq was identical to the A549 cells RACE-seq, except: (i) the starting material was 1 µg, (ii) the Invitrogen Superscript II was used for the RT, (iii) PCR1 and PCR2 had 10 cycles each.
MEFs infection by Influenza A virus and qRT-PCR
MEF cells were trypsinized and seeded as 8X10^4 cells per well of 24-well plate one day before infection. 16 hours after seeding, culture media were removed and cells were washed once with pre-warmed DMEM. Influenza A virus (A/WSN/1933, H1N1) were inoculated at MOI 3 diluted with 250 ul DMEM supplemented with 0.1% BSA (D0.1B). Cells were trypsinized and collected 8, 16 and 24 hours post infection. 350 ul TRIzol were added into each infected sample. RNA was extracted using Direct-zol™ RNA MiniPrep (Zymo Research) purification according to the manufacture’s protocol and was finally eluted into 60 ul RNase/DNase free water. The extracted RNA was subjected to strand specific qRT-PCR to quantify influenza virus replication as described in 49.
MEFs infection by Influenza A virus and FACS assay
MEF cells were trypsinized and seeded as 1X10^4 cells per well of 96-well plate one day before infection. 16 hours after seeding, culture media were removed and cells were washed once with pre-warmed DMEM. Influenza A virus (A/WSN/1933, H1N1) were inoculated at MOI 3 diluted with 50 µl DMEM supplemented with 0.1% BSA (D0.1B). Inoculum was removed after 1 hour of incubation at 37 °C. The infected cells were cultured with MEF cell culture medium with 2.5% FBS. 8 hours post inoculation, culture media were removed and cells were trypsinized through incubation with 30 µl 0.05% trypsin for 3 minutes at 37 °C. Trypsinized cells were resuspended with 70ul of P2F (PBS with 2% FBS) and then fixed with 100 µl 4% PFA for 15 minutes. Fixed cells were centrifuged at 300g for 5 minutes and then washed once with 100 µl P2F. Cells were then permeablized with buffer (0.1% Saponin, 10mM HEPES, 0.025% Sodium Azide in 1XHBSS) for 15 minutes at room temperature and then spinned at 500g for 2 minutes to remove buffer. Primary anti-influenza A virus nucleoprotein antibodies were purchased from Millipore (MAB8258B | clone A3, biotin-conjugated). The primary antibodies were diluted 1:2000 in permeable buffer and 50 µl diluted antibodies were added into each well of 96-well plate. Primary antibodies were incubated with infected cells at room temperature for 1 hour. The cells were then washed 3 times with permeable buffer. FITC conjugated goat anti-mouse secondary antibodies were purchased from Invitrogen and diluted at 1:1000 in permeable buffer. Secondary antibodies were incubated for 1 hour at room temperature and washed as described before. The stained cells were finally resuspended in 70 µl P2F. The cell suspension was run on a high throughput FACS machine (MACSQuant® analyzer 10 - Miltenyi Biotec). Uninfected cells were stained the same as infected cells and were used as negative staining cell populations. Any cells/events that had fluorescence intensity higher than all the negative staining cell population were gated as virus infection positive. Data were analyzed using flowjo software (version 10).
Statistics and reproducibility
Statistics as shown in the figure legends. Table 1: ovid-9 data was reproduced in an independent experiment, data on other Ovid isolates only produced in the shown experiment. ovid-9 GFP and RNAi scoring not blinded but reproduced by different authors. Fig. 1: ovid-9 data was reproduced in an independent experiment, data on other Ovid isolates only produced in the shown experiment. Paralyzed animals were not moving even after tapping the plate. Twitching animals were moving but distinctively twitching laterally. Fig. 2: reproduced in an independent experiment (except for the DAD catalityc mutant, only produced in the shown experiment). Fig. 3: reproduced in an independent experiment. Fig. 4b: only produced in the shown experiment. Fig. 4d,e: reproduced in an independent experiment. Fig. 4f: only produced in the shown experiment. Fig. 6b,f: only produced in the shown experiment. Figure 6g: reproduced in an independent experiment. Supp. Fig. 1a: only produced in the shown experiment, corroborates published data as cited in the text. Supp. Fig. 1b: at least two pictures taken by condition, except for the male for which the only picture available is shown in the manuscript, all phenotypes (including in males) were observed multiple times. Supp. Fig. 2: reproduced in an independent experiment but with 28 F4 families instead of 64 F8 families. Not blinded but reproduced by different authors. Supp. Fig. 3: only produced in the shown experiment. Not blinded. Supp. Fig. 4a: only produced in the shown experiment, at least two pictures per condition. Supp. Fig. 4b: only produced in the shown experiment, at least two pictures per condition, corroborates published data as cited in the text. Supp. Fig. 4d,e: only produced in the shown experiment. Supp. Fig. 5a-c: only produced in the shown experiment, corroborates published data as cited in the text. Supp. Fig. 6a,b: only produced in the shown experiment. Supp. Fig. 7: reproduced in an independent experiment. Supp. Fig. 8: reproduced in an independent experiment.
Supplementary Material
Acknowledgments
We thank Mélanie Tanguy for OrV viral filtrates, Lise Frézal for help with the OrV RNA FISH, Isabel Wilkinson for support with the genetic screen, Nicolas J. Lehrbach for help with microinjections, and Marc Ridyard for lab management. We thank Kay Harnish, Fabian Braukmann and Sylviane Moss for high-throughput sequencing support. We are grateful to V. Narry Kim and Hyeshik Chang for sharing information on TAIL-seq and Adrianus C.M. Boon for providing IAV. We thank Alyson Ashe and Peter Sarkies for their theoretical input on the screen design. We thank the International C. elegans gene knockout consortium and the TransgeneOme project for providing reagents. We thank Vladimir Benes and the EMBL genome core for sequencing support. We thank George Allen and Charles Bradshaw for core bioinformatics support. We thank Ragini Medhi and Dick Zijlmans for help with TUTs Western blots. This work was supported by Cancer Research UK (C13474/A18583, C6946/A14492), the Wellcome Trust (104640/Z/14/Z, 092096/Z/10/Z) and The European Research Council (ERC, grant 260688). DW holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Data availability statement
All raw sequencing data are deposited in GEO (small RNA sequencing: GSE80169; mRNA sequencing: GSE76901; TAILseq: GSE85893). All C. elegans strains created in this study will be freely available on a non-collaborative basis.
Author Information
The authors have made the following declarations about their contributions: Conceived and designed the experiments: J.L.P., H.J., E.A.M. Performed the experiments: J.L.P., H.J., E.K., J.K., C.L., M.M., C.M. Analyzed the data: J.L.P., H.J., T.D.D., K.L.M.R., A.J.E, D.O.C., D.W., EAM. Wrote the manuscript: J.L.P., E.A.M.
References
- 1.Ding S-W, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic Sensing of Viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Félix M-A, et al. Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 2011;9:e1000586. doi: 10.1371/journal.pbio.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashe A, et al. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. Elife. 2013;2:e00994. doi: 10.7554/eLife.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashe A, Sarkies P, Le Pen J, Tanguy M, Miska EA. Antiviral RNAi against Orsay virus is neither systemic nor transgenerational in Caenorhabditis elegans. J Virol. 2015;89 doi: 10.1128/JVI.03664-14. JVI.03664–14–12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo YR, et al. Crystal structure of a nematode-infecting virus. Proc Natl Acad Sci U S A. 2014;111:12781–12786. doi: 10.1073/pnas.1407122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, et al. Orsay virus utilizes ribosomal frameshifting to express a novel protein that is incorporated into virions. Virology. 2014;450-451:213–221. doi: 10.1016/j.virol.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franz CJ, et al. Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology. 2014;448:255–264. doi: 10.1016/j.virol.2013.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Sarkies P, Ashe A, Le Pen J, McKie MA, Miska EA. Competition between virus-derived and endogenous small RNAs regulates gene expression in Caenorhabditis elegans. Genome Res. 2013;23:1258–1270. doi: 10.1101/gr.153296.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, Zhang R, Wang J, Ding S-W, Lu R. Homologous RIG-I-like helicase proteins direct RNAi-mediated antiviral immunity in C. elegans by distinct mechanisms. Proc Natl Acad Sci U S A. 2013;110:16085–16090. doi: 10.1073/pnas.1307453110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y, et al. Structure of a pentameric virion-associated fiber with a potential role in Orsay virus entry to host cells. PLoS Pathog. 2017;13:e1006231. doi: 10.1371/journal.ppat.1006231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 14.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Chen K, Sandoval LE, Leung C, Wang D. An Evolutionarily Conserved Pathway Essential for Orsay Virus Infection ofCaenorhabditis elegans. MBio. 2017;8:e00940–17. doi: 10.1128/mBio.00940-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanguy M, et al. An Alternative STAT Signaling Pathway Acts in Viral Immunity inCaenorhabditis elegans. MBio. 2017;8:e00924–17. doi: 10.1128/mBio.00924-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Wolfswinkel JC, et al. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Norbury CJ. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol. 2013;14:643–653. doi: 10.1038/nrm3645. [DOI] [PubMed] [Google Scholar]
- 22.Wickens M, Kwak JE. Molecular biology. A tail tale for U. Science. 2008;319:1344–1345. doi: 10.1126/science.1154946. [DOI] [PubMed] [Google Scholar]
- 23.Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6)A and U-tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Rissland OS, Norbury CJ. Decapping is preceded by 3' uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan M, et al. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature. 2017;548:347–351. doi: 10.1038/nature23318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim J, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159:1365–1376. doi: 10.1016/j.cell.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang H, Lim J, Ha M, Kim VN. TAIL-seq: Genome-wide Determination of Poly(A) Tail Length and 3′ End Modifications. Mol Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 28.Miki TS, Rüegger S, Gaidatzis D, Stadler MB, Großhans H. Engineering of a conditional allele reveals multiple roles of XRN2 in Caenorhabditis elegans development and substrate specificity in microRNA turnover. Nucleic Acids Res. 2014;42:4056–4067. doi: 10.1093/nar/gkt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 30.Molleston JM, et al. A conserved virus-induced cytoplasmic TRAMP-like complex recruits the exosome to target viral RNA for degradation. Genes Dev. 2016;30:1658–1670. doi: 10.1101/gad.284604.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huo Y, et al. Widespread 3'-end uridylation in eukaryotic RNA viruses. Sci Rep. 2016;6:25454. doi: 10.1038/srep25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samji T. Influenza A: understanding the viral life cycle. Yale J Biol Med. 2009;82:153–159. [PMC free article] [PubMed] [Google Scholar]
- 33.Rehwinkel J. Is anti-viral defence the evolutionary origin of mRNA turnover? (Comment on DOI 10.1002/bies.201600100) Bioessays. 2016;38:817–817. doi: 10.1002/bies.201600140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamid FM, Makeyev EV. Exaptive origins of regulated mRNA decay in eukaryotes. Bioessays. 2016;38:830–838. doi: 10.1002/bies.201600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manokaran G, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015;350:217–221. doi: 10.1126/science.aab3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fire A. Integrative transformation of Caenorhabditis elegans. EMBO J. 1986;5:2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen EM, Mango SE. The art and design of genetic screens: caenorhabditis elegans. Nat Rev Genet. 2002;3:356–369. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- 39.Sarov M, et al. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell. 2012;150:855–866. doi: 10.1016/j.cell.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 42.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paix A, Folkmann A, Rasoloson D, Seydoux G. High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics. 2015;201:47–54. doi: 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10. [Google Scholar]
- 45.Harris TW, et al. WormBase 2014: new views of curated biology. Nucleic Acids Res. 2014;42:D789–93. doi: 10.1093/nar/gkt1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawakami E, et al. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods. 2011;173:1–6. doi: 10.1016/j.jviromet.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.