Abstract
We describe convergent evidence from transcriptomics, morphology and physiology for a specialized GABAergic neuron subtype in human cortex. Using unbiased single nucleus RNA sequencing, we identify ten GABAergic interneuron subtypes with combinatorial gene signatures in human cortical layer 1 and characterize a novel group of human interneurons with anatomical features never described in rodents having large, “rosehip”-like axonal boutons and compact arborization. These rosehip cells show an immunohistochemical profile (GAD1/CCK-positive, CNR1/SST/CALB2/PVALB-negative) matching a single transcriptomically-defined cell type whose specific molecular marker signature is not seen in mouse cortex. Rosehip cells in layer 1 make homotypic gap junctions, predominantly target apical dendritic shafts of layer 3 pyramidal neurons and inhibit backpropagating pyramidal action potentials in microdomains of the dendritic tuft. These cells are therefore positioned for potent local control of distal dendritic computation in cortical pyramidal neurons.
Keywords: human, neocortex, interneuron, layer 1, cell type, transcriptomics, microcircuit
Understanding the cellular and circuit organization of the neocortex, the substrate for much of higher cognitive function, has been intensely studied since Ramón y Cajal 1. Morpho-physiological characterization using slice physiology has been the standard for decades2, but this approach suffers from undersampling, difficulties in quantitative classification of cell types3, and limited scalability to cover neuronal diversity. Single cell transcriptomics enables unbiased, high-throughput quantitative surveys of molecularly defined cell types4–6 that can be applied to any species including human. Initial application to mouse cortex has revealed approximately 50 transcriptomic types, demonstrating both the feasibility of the approach and the complexity of the cortex. There is now great promise in combining these morpho-electric and transcriptomic approaches for an unbiased molecular classification and characterization of these types.
Recent systematic efforts have provided insight into the cellular composition and organization of rodent neocortical circuits, suggesting the presence of several dozen inhibitory and excitatory cell types3–5,7. However, conservation of cellular and circuit principles in human cortex is assumed but largely untested to date. Indeed there is evidence for significant neuronal differences between rodents and human; for example, distinct membrane8,9 and synaptic10–14 properties and dendritic complexity15–17 of human neurons might contribute to human specific signal processing. With the mouse cortex as dominant model for understanding human cognition it is essential to establish whether the cellular architecture of human is conserved or whether there are specialized cell types and system properties that cannot be modeled in rodents. Here we combine single nucleus transcriptomics and slice physiology to study GABAergic neurons in layer 1 of human cortex and provide convergent lines of evidence for the identification of a cell type with human specialized features.
Results
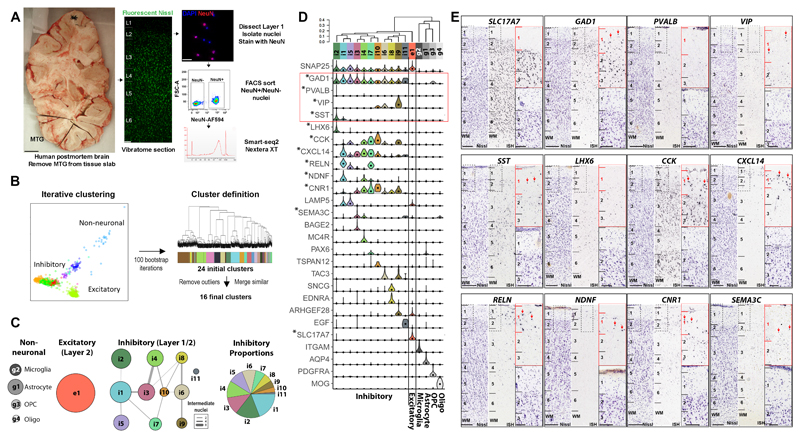
To allow an unbiased survey of transcriptionally-defined cell types in human cortical tissue we used single nucleus RNA sequencing18,19 to profile large numbers of nuclei from frozen postmortem brain specimens (Fig.1A). Briefly this method involved microdissection of regions of interest from fluorescent Nissl-stained vibratome sections of cortex, tissue homogenization to liberate nuclei, NeuN staining and FACS isolation, and Smart-seq2 based library preparation 20 (Fig.1, Suppl. Fig.1). We applied this strategy to profile n=769 quality control passed NeuN-positive neurons and n=102 NeuN-negative non-neuronal cells across 2 individuals from microdissected layer 1 of the middle temporal gyrus, expected to predominantly contain inhibitory neurons. Median gene detection (expression > 0) was 9937 in neurons and 6287 in glia. Iterative clustering was used to group nuclei with similar transcriptional profiles, thereby identifying a robust set of transcriptomic-defined cell types (Fig.1B). Based on expression of known marker genes (Suppl. Fig.2A), clusters corresponded to all major classes of neural cell types that were expected to be captured. These included major non-neuronal cell types (microglia, astrocytes, oligodendrocyte precursor cells (OPCs) and oligodendrocytes) and one excitatory neuron type sampled from upper cortical layer 2 incidentally included in the layer 1 dissection (Fig.1C). In addition, eleven distinct clusters corresponding to GABAergic neuron subtypes were identified (numbered by relative abundance).
Figure 1. Identification of transcriptomic cell types in layer 1 of human temporal cortex.
A, Isolation of single nuclei from post-mortem adult human cortex for RNA-sequencing. Scale bars, left 1 cm, right, 20 µm. B, Left: Nuclei were grouped based on similar gene expression profiles using an automated iterative clustering procedure. Clustering was repeated 100 times on random subsets of 80% of nuclei. Right: Hierarchical clustering of nuclei that were consistently co-clustered across iterations identified 24 clusters. 16 clusters remained after removal of clusters associated with quality control metrics and merging of clusters that lacked at least one binary marker gene. C, 4 non-neuronal, 1 excitatory and 11 inhibitory neuron clusters were identified, although the excitatory cluster and one inhibitory cluster were likely in Layer 2 due to incidental capture superficial layer 2 with Layer 1 dissection. For each cluster, the constellation diagram shows the cell type class (based on canonical marker gene expression), relative frequency (disc area), and discreteness (line thickness proportional to the number of nuclei with ambiguous cluster membership) of clusters. D, Clusters arranged by transcriptomic similarity based on hierarchical clustering, with the expression distributions of selective marker genes shown across clusters as violin plots. Expression is on a linear scale and dots indicate median expression. Cluster sample sizes: i2 (n=77); i1 (n=90); i5 (n=47); i3 (n=56); i4 (n=54); i7 (n=31); i10 (n=16); i6 (n=44); i8 (n=27); i9 (n=22); i11 (n=6); e1 (n=299); g2 (n=27); g1 (n=48); g3 (n=18); g4 (n=9). E, ISH of select marker genes in human temporal cortex at low magnification (left columns with near adjacent Nissl stain for cytoarchitectonic laminar identification) and high magnification in layers 1-3 (right column). Red arrows highlight cells expressing genes in layer 1. Note that LHX6 marks a single cluster (i2) that is not expressed in layer 1 and therefore nuclei in this cluster were likely sampled from upper layer 2. Other clusters are restricted to layer 1 (e.g. NDNF+) or may be distributed across layers 1 and 2. Scale bars=250 µm (low mag), 100 µm (high mag). ISH experiments were conducted on multiple tissue donors as follows: SLC17A7, LHX6, CNR1, SEMA3C (n=3); CXCL14 (n=5); GAD1, CCK, RELN, NDNF (n=6); SST (n=7); PVALB (n=8); VIP (n=10).
Transcriptomic cell types displayed highly selective gene expression (Fig.1D, Suppl. Fig.2A). For example, the pan-neuronal gene synaptosomal-associated protein 25 (SNAP25) clearly differentiated neuronal from non-neuronal types, which were in turn differentiated by highly specific marker genes. Glutamic acid decarboxylase 1 (GAD1) clearly delineated the GABAergic neurons. In cortical layers 2-6, most GABAergic neurons have mutually exclusive expression of parvalbumin (PVALB), somatostatin (SST) or vasoactive intestinal peptide (VIP)21. In contrast, Pvalb and Sst are not expressed in mouse layer 1 by in situ hybridization (ISH), while Vip labels only sparse cell populations (Suppl. Fig.2B). Interestingly, both SST and VIP (but not PVALB) are seen in human MTG layer 1 by ISH (Fig.1E). The layer 1 MTG transcriptomic clusters expressed either SST (i1,i2), VIP (i6, i9, i10), or neither marker, although cluster i2 represents a cell type restricted to layer 2 since it also expresses LHX6 which is not found in layer 1 (Fig.1D,E). Therefore, there appear to be ten inhibitory cell types within layer 1, although it is not clear whether any of these types are completely restricted to layer 1.We compared these layer 1 cell types to eight inhibitory clusters reported by Lake et al.22 and find increased diversity within several published clusters (In1-4) and decreased diversity of LHX6+ interneuron clusters (In5-8) that are enriched in deeper cortical layers and were not sampled in this study (Suppl. Fig.3A-C).
These clusters in layer 1 express different combinations of known markers of layer 1 interneurons, including cholecystokinin (CCK), reelin (RELN), neuron derived neurotrophic factor (NDNF), and lysosomal associated membrane protein family member 5 (LAMP), many of which were confirmed to have expression in layer 1 by ISH (Fig.1E). Furthermore, each cluster showed highly selective expression of known and previously uncharacterized individual marker genes. Interestingly, given the proximity of layer 1 to the overlaying pia, several of these markers appear to be related to interaction with endothelia, including endothelin receptor type A (EDNRA) and epidermal growth factor (EGF). Furthermore, voltage-gated ion channels and GABA and glutamate receptor subunits show diverse expression patterns among interneurons, including highly cell type-specific expression of CACNA2D1, GABRG1, KCNH5, and SCN5A (Suppl. Fig.4). To summarize, this unbiased transcriptomic approach identified ten GABAergic interneuron subtypes in layer 1 that have distinctive combinatorial and specific gene expression signatures suggestive of distinct morphological and functional properties.
Rosehip cells: novel morphological features in layer 1 of the human cerebral cortex
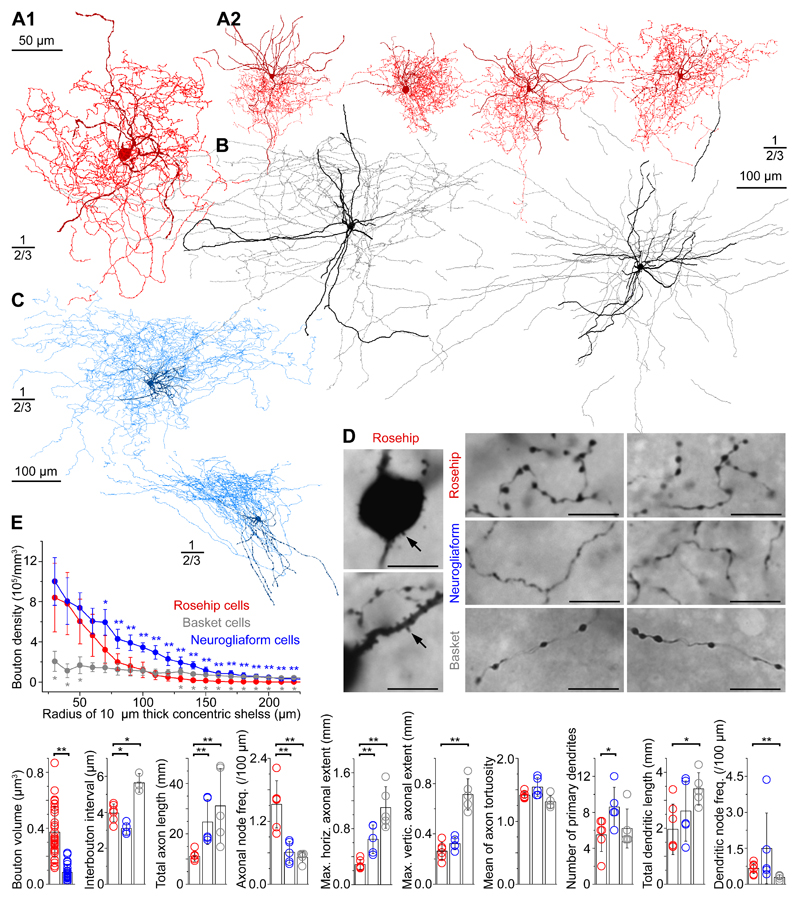
In parallel to the transcriptomic approach we developed a dataset of whole cell recorded, biocytin-filled interneurons in layer 1 of slices of nonpathological human samples of parietal, frontal and temporal cortices10,11,23. Unbiased recordings of layer 1 cell types yielded a set of interneurons with complete axo-somato-dendritic recovery (n=76). Light microscopic examination of these cells identified neurons with previously described morphological features, e.g. neurogliaform cells (NGFCs, n=16, 21%; Fig.2C,D)1,21,24 as well as a novel group of interneurons having large, rosehip-shaped axonal boutons forming very compact, bushy arborizations (rosehip cells, RCs, n=10, 13%; Fig.2A,D). To our knowledge, interneurons having the phenotype of RCs detailed below have not been identified previously in layer 1 of the cerebral cortex. Somata and dendrites of RCs were confined to layer 1 with only distal dendrites occasionally penetrating layer 2. Proximal dendrites and somata of RCs were decorated with stub-like spines. The axon of RCs usually emerged from the basal part of the soma and gave rise to very compact, dense axonal trees predominantly arborizing in layer 1 with tortuous collaterals having spindle-shaped boutons with diameters not seen in other types of human layer 1 interneurons in our sample. Targeted recordings increased the number of RCs in our database (n=120) and we quantitatively compared axo-dendritic parameters of randomly selected and three-dimensionally reconstructed RCs (n=6) to layer 1 neurogliaform (n=5) and layer 2/3 basket cells (BCs, n=5; Fig.2B,D)10,11,24,25.The number of primary dendrites of RCs (5.50±1.87) was similar to that of BCs (6.2±2.17, n=5) and was significantly fewer compared to NGFCs (8.6±2.19, n=5, p<0.04, Mann-Whitney (MW) U-test). Total dendritic length (1.96±0.90 mm) and dendritic node frequency per 100 µm (0.66±0.21) of RCs were significantly different from those of BCs (3.41±0.58 mm, p<0.031; 0.29±0.10, p<0.009, respectively, MW U-test) and were similar to those of NGFCs (2.62±1.08 mm, 1.50±1.47). Total length (11.13±1.99 mm) and maximal horizontal extent of axons (287.75±70.15 µm) of RCs were significantly smaller than those of NGFCs (24.74±8.90 mm, 648.68±202.60 µm, respectively; p<0.005 for both, MW U-test) and BCs (31.16±14.79 mm, p<0.009; 1102.76±296.99 µm, p<0.005, respectively, MW U-test). Maximal radial extent of axon of RCs (263.42±69.09 µm) was significantly smaller than that of BCs (713.22±124.87 µm, p<0.005, MW U-test), but were not different from those of NGFCs (323.18±49.60 µm). We measured axonal bouton densities of rosehip (n=6), neurogliaform (n=4) and basket (n=3) cells in 10 µm thick spherical shells of increasing diameter by Sholl analysis corrected with the portion of shells outside the brain slice. The bouton density of rosehip, neurogliaform and BCs almost monotonously decreased with increasing distances from the soma; however, bouton densities were lower in BCs 30-50 µm (p<0.04 for 30-50, MW U-test) and higher in NGFCs 70-220 µm (p<0.02, MW U-test) and in BCs 130-220 µm (p<0.03) from the soma. RCs had longer interbouton intervals compared to NGFCs (3.97±0.49 and 3.10±0.32 µm, respectively, p<0.038, MW U-test) and shorter compared to BCs (5.63±0.51 µm, p<0.024) measured as linear distances between neighboring boutons. RC axons branched more frequently, with RCs, NGFCs and BCs having 1.52±0.45, 0.61±0.21 and 0.52±0.10 nodes along 100 µm length of their axons (p<0.005 for both, MW U-test). Axon tortuosity (see Methods) of RCs (1.42±0.05) was similar to that of neurogliaform (1.54±0.15) and BCs (1.31±0.10). Measurements based on serial section electron microscopy and three-dimensional reconstructions revealed that the volume of boutons of RCs (0.37±0.18 µm3, n=31) was approximately four times larger (p<0.001; MW U-test) compared to that of NGFCs (0.08±0.06 µm3, n=24, Fig.2E). The size of active zones in RCs (0.11±0.03 µm2, n=11) was not correlated to bouton volumes (rho=0.34, p=0.29, Spearman correlation). All fully reconstructed boutons (n=31) formed single synapses targeting dendritic shafts.
Figure 2. Morphological phenotype of rosehip cells in layer 1 of the human cerebral cortex.
A1, A2, Anatomical reconstructions of RCs biocytin filled during whole cell recordings (somata and dendrites, burgundy; axons, red). B, Anatomical reconstructions of layer 2/3 BCs in the human cerebral cortex (somata and dendrites, black; axons, gray). C, Anatomical reconstructions of NGFCs in layer 1 of the human cerebral cortex (somata and dendrites, dark blue; axons, light blue). D, Left, Light micrographs of RCs (n=130) showing somata and proximal dendrites with stub-like spines (arrows). Right, Axons of RCs arborized densely with large, round boutons (top). Tortuous neurogliaform axons (n=16) posess very small boutons (middle). Axons of BCs (n=5) form longer segments with less convoluted branches with longer interbouton intervals (bottom). Scale bars: 10 µm. E, Quantitative comparison of axonal and dendritic parameters of rosehip (red, n=6), neurogliaform (blue, n=5) and basket (gray, n=5) cells. Top, bouton densities determined by Sholl analysis in 10 µm thick spherical shells were lower in BCs 30-50 µm and higher in NGFCs 70-220 µm and in BCs 130-220 µm from the soma compared to that of RCs. Bottom, Bouton volume (p<0.001)and the number of primary dendrites (p<0.04) of RCs were significantly different from that of NGFCs. Maximal vertical extent of axon (p<0.005), total dendritic length (p<0.031) and dendritic node frequency (/100 µm, p<0.009) of RCs differed significantly from that of BCs. Axonal tortuosity of RCs was similar in the two other cell types, however, the frequency of axonal branch points in RCs was 2.5 and 2.95 times that of neurogliaform (p<0.005) and BCs (p<0.005), respectively. Furthermore, interbouton interval, total axon length and maximal horizontal extent of the axon were also significantly different (two-sided Mann-Whitney U Test, * p ≤ 0.05; ** p ≤ 0.01; columns and error bars represent mean and standard deviation, respectively).
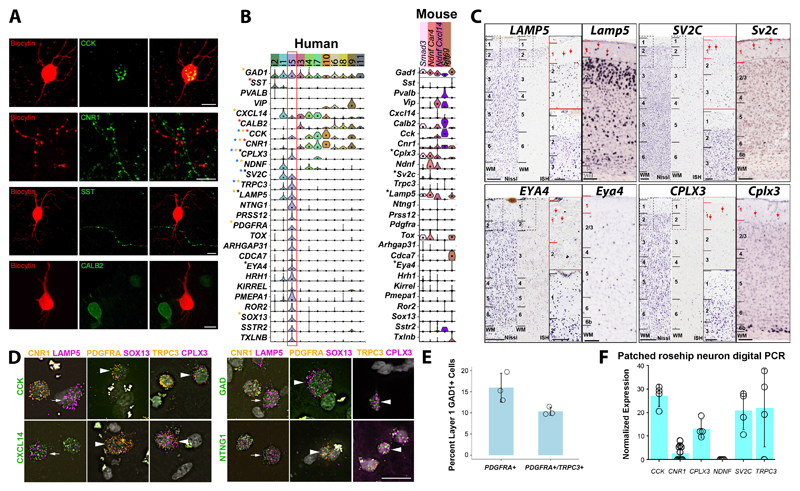
To understand the molecular identity of RCs and link them to the transcriptomic clusters, we performed immunohistochemistry (IHC) on electrophysiologically recorded and anatomically recovered cells for known markers of GABAergic cell types (see Methods for details)26. This revealed that RCs were immunopositive for CCK (n=10) but negative for CB1 cannabinoid receptor (CNR1, n=11), SST (n=9) and calretinin (CALB2; n=2; Fig.3A). Furthermore, RCs were immunopositive for gamma-aminobutyric acid (GABA; n=2), for chicken ovalbumin upstream promoter transcription factor II (NR2F2; n=2) and negative for parvalbumin (n=3), neuronal nitric oxide synthase (n=4), neuropeptide Y (n=2), calbindin (n=2), and choline acetyltransferase (n=3; Suppl. Fig.5A).
Figure 3. Molecular phenotype of rosehip cells in layer 1 of the human cerebral cortex.
A, Whole cell recorded and biocytin (red) filled RCs shows CCK (green; n=10) immunopositivity. All biocytin (red) labeled RCs tested for CB1 cannabinoid receptors (CNR1; n=11), somatostatin (SST; n=9), and calretinin (CALB2; n=2) were immuno-negative in spite of having labeled cells in the vicinity. Scale bars, 10 µm. B, Violin plots of gene expression for broad cell type and putative rosehip specific markers. Expression is on a linear scale and dots indicate median expression. Cluster sample sizes: i2 (n=77); i1 (n=90); i5 (n=47); i3 (n=56); i4 (n=54); i7 (n=31); i10 (n=16); i6 (n=44); i8 (n=27); i9 (n=22); i11 (n=6); Smad3 (n=12); Ndnf Car4 (n=24); Ndnf Cxcl14 (n=30); Igtp (n=10). Expression validated for select genes by immunohistochemistry (red stars), colorimetric ISH (black), multiplex FISH (orange), and single cell digital PCR (blue) in morphologically identified RCs. C, ISH of select marker genes in human temporal cortex (left) and mouse cortex (right). Red arrows highlight cells expressing genes in layer 1. Scale bars=250 µm (low mag), 100 um (high mag). ISH experiments were repeated on multiple human donors as follows: LAMP5 (n=2); EYA4, CPLX3 (n=3); SV2C (n=5). For mouse, ISH experiments were repeated on multiple specimens as follows: Lamp5, Sv2c, Cplx3 (n=2); Eya4 (n=3). D, Multiplex FISH validation of rosehip marker co-expression. Arrowheads and arrows show examples of RCs that are triple- and double-positive (i.e. CNR1-), respectively, for marker genes based on RNA-Seq expression data. Scale bar=25 µm. Multiplex FISH experiments were repeated on n=2 tissue donors. E, RCs comprise 10-15% of layer 1 interneurons based on multiplex FISH quantification of 408 GAD1+ cells in 2 subjects. 15% (+/- 3) of GAD1+ cells express the rosehip specific marker PDGFRA, although a small fraction of these cells may be oligodendrocyte precursor cells (see Suppl. Fig.5). 10% (+/-1) of GAD1+ cells express PDGFRA and a second rosehip marker TRPC3, although some RCs may lack TRPC3 expression based on RNA-seq. Error bars represent standard deviation. Cell counts were conducted on n=3 tissue sections from n=2 tissue donors. F, Expression of rosehip cluster markers in cytoplasm of whole cell recorded RCs. Quantified by single cell digital PCR and reported as a percentage of housekeeping gene (TBP) expression in n=9 cells (CNR1) or n=4 cells (CCK, CPLX3, NDNF, SV2C, TRPC3) per gene. Note that NDNF expression was not detected in any of the cells tested. Columns and error bars represent mean and standard deviation.
Remarkably, this immunohistochemical profile aligned closely with a single transcriptomic cell type, i5, which was similarly GAD1/CCK-positive but CNR1/SST/CALB2/PVALB-negative (Fig.3B, red box). This putative rosehip transcriptomic type, one of the most distinctive layer 1 GABAergic transcriptomic types, expresses many other genes either highly specifically or coexpressed in only one other layer 1 cell type. Intriguingly, given the rosehip synaptic phenotype, these markers include many genes with known associations to axon growth and synaptic structure and function, including synaptic vesicle glycoprotein 2c (SV2C), LAMP5, transient receptor potential cation channel subfamily C member 3 (TRPC3), complexin 3 (CPLX3), neurotrypsin (PRSS12), netrin G1 (NTNG1), histamine receptor H1 (HRH1), receptor tyrosine kinase like orphan receptor 2 (ROR2), somatostatin receptor 2 (SSTR2), and taxilin beta (TXLNB).
Since the rosehip anatomical phenotype has not been described in rodents, we asked whether a transcriptomic signature similar to the rosehip transcriptomic type had been observed in a recent large-scale analysis of mouse primary visual cortex using single cell RNA-seq analysis4. We attempted to find homologous cell types between species by correlating the median expression of 212 cell type-informative genes between all pairs of mouse and human clusters (Suppl. Fig.3D,E). Expression correlations were quite low (r < 0.5), and clusters could only be reliably grouped into broad classes of cell types. Human clusters i5, i7, i1, i4, and i3 matched mouse Smad3+ and Ndnf+ clusters. Clusters i6, i8, i9, i11, and i10 matched mouse Vip+ clusters, and cluster i2 matched mouse Sst+ clusters. The rosehip cluster i5 had a weak (r=0.34) reciprocal best match to mouse cluster Smad3, although several other human clusters (i7, i1, i4, i3) matched Smad3 almost as well (r>0.3).
Many rosehip marker genes are not expressed in Smad3 or other Pvalb/Sst/Vip-negative mouse cell types (Fig.3B, right panel) or the complete set of mouse GABAergic types (Suppl. Fig.5B, right panel). Importantly, it is the unique combinatorial expression of many marker genes that defines RCs. For example, expression of LAMP5, SV2C, EYA4 and CPLX3 is seen by ISH in human layer 1 (Fig.3C); similarly, as predicted by transcriptomics three of these four genes are also expressed in mouse layer 1 whereas cells expressing Eya4 are extremely rare. Many other rosehip-selective genes had no evidence of expression in layer 1 interneurons in mouse based on single cell transcriptomics (Fig.3B, Suppl. Fig.5B).
To demonstrate that layer 1 neurons with combinatorial expression patterns predicted by transcriptomics could be found in human layer 1, and to quantify their proportions, we systematically performed triple fluorescent ISH on human MTG tissue using discriminating positive and negative gene markers. For all combinations tested we observed cells with the predicted profiles. For example, we observed CCK+/CNR1-/LAMP5+, CCK+/PDGFRA+/SOX13+, and CCK+/TRPC3+/CPLX3+ cells, as well as cells where CCK was swapped with other positive rosehip markers (Fig.3D; additional gene combinations shown in Suppl. Fig 5C). Quantification of cell proportions using marker expression is complicated by two factors; first, markers for one cell type are often expressed in others, and second, individual markers are often not expressed in every cell in a cluster. We used the combination of GAD1, PDGFRA and TRPC3 to quantify the proportion of RCs among layer 1 GABAergic neurons (Fig.3E). PDGFRA is known to be expressed in OPCs at extremely high levels as well (which is why it appears to only be expressed in OPCs in Figure 1 but appears high in RCs in Figure 2 once levels are not normalized across all cell types including OPCs). PDGFRA+ cells represent ~15% of GAD1+ cells, therefore an upper bound. TRPC3 is not expressed in all cells in the rosehip cluster on the other hand. The proportion of GAD1+ cells that are PDGFRA+/TRPC3+ was ~10%, therefore a lower bound. The triple positive cells for this combination were sparsely distributed across layer 1, although not restricted to this layer (Suppl. Fig.5D). To determine if cells with the transcriptional signature of RCs could be found in cortical regions outside of MTG, we conducted triple fluorescent ISH on tissue sections from Brodmann Area (BA) 9 (frontal cortex) and BA40 (parietal cortex) using several combinatorial gene panels. We found that GAD1+ cells expressing rosehip marker genes with low or absent expression of CNR1 were present in layer 1 of both BA9 and BA40 (Suppl. Fig 6), suggesting that this cell type is found in the cortical areas sampled for morpho-electric profiling. Furthermore, Lake et al.22 identified cluster IN4 (the best match to rosehip cluster i5) in all six cortical areas sampled (frontal, temporal, and visual cortex).
Finally, to more concretely link morphologically and transcriptionally defined RCs, we performed digital PCR for additional marker genes on cellular content extracted from individual rosehip neurons. As predicted by the transcriptome data, RCs were positive for CCK, CPLX3, SV2C and TRPC3, and low (CNR1) or absent (NDNF) for genes not expressed by cells in that cluster (Fig.3F). Together, these data strongly link the anatomically-defined rosehip phenotype with a highly distinctive transcriptomic cell type signature that is found in human but apparently not in mouse layer 1.
Intrinsic electrophysiological properties of rosehip cells
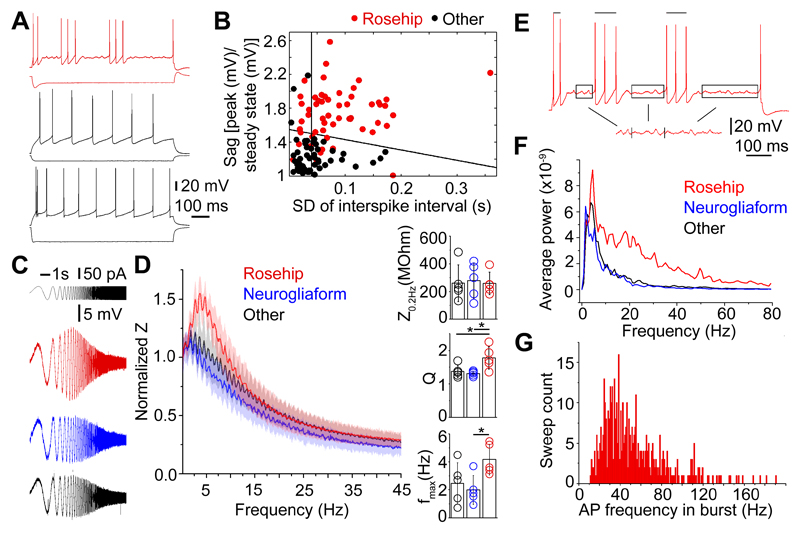
Anatomically identified RCs responded to long (800 ms) suprathreshold current injections with stuttering or irregular spiking firing patterns2 when activated from resting membrane potential (-61.34±5.8 mV, Fig.4A). Analysis of silent and suprathreshold periods during rheobasic firing of RCs indicated that membrane oscillations and firing of RCs were tuned to beta and gamma frequencies (Fig.4E-G). The power of averaged fast Fourier transforms (FFT) of subthreshold membrane potential oscillations27 was higher between 3.8 and 80 Hz in RCs compared to neurogliaform and other interneurons (Fig.4F) and intraburst frequency of stuttering firing also peaked in the beta-gamma range (Fig.4G). The standard deviation of interspike intervals was higher in RCs (87±64 ms, n=55) compared to neurogliaform (41±34 ms, n=16, p<0.001, Wilcoxon-test) or unclassified (47±41 ms, n=36, p<0.001, Wilcoxon-test) interneurons, indicating alternating silent and active periods during rheobasic stimulation. As described previously23, human interneurons recorded in layer 1 had a characteristic sag when responding to hyperpolarizing current pulses. However, the amplitude of the sag measured in anatomically classified RCs (1.73±0.30, n=55) exceeded that of interneurons morphologically identified as NGFCs (1.19±0.12, n=16, p<0.001, Wilcoxon-test) or unclassified interneurons (1.29±0.28, n=36, p<0.001, Wilcoxon-test). Input resistances of RCs (139.6±54.1 MΩ) were similar to those of NGFCs (160.1±55.9 MΩ) and lower compared to other interneurons (216.3±84.4 MΩ, p<0.001, Wilcoxon-test); however, time constants of RCs (7.3±3.7 ms) were similar to neurogliaform (8.9±2.4 ms, p<0.001) and faster compared to other cells (11.1±12.5 ms, p<0.001). Anatomically identified RCs showed distinct impedance profiles relative to other layer 1 interneurons in response to current injections with an exponential chirp (0.2-200 Hz, Fig.4 C-D). Impedance at the lowest frequency (Z0.2Hz) was similar in layer 1 interneurons (rosehip, 258±81 MΩ, neurogliaform, 279±128 MΩ, unclassified, 261±133 MΩ, Lillefors test followed by one-way ANOVA with Bonferroni correction). Resonance magnitude (Q, see methods) of RCs (1.77±0.34) was significantly higher compared to NGFCs (1.31±0.07; p<0.021, Lillefors test followed by one way ANOVA with Bonferroni correction) and unclassified interneurons (1.37±0.19; p<0.049). In addition, frequencies of maximal impedance (fmax) in RCs (4.17±1.1 Hz) were significantly higher than in NGFCs (1.98±1.04 Hz; p<0.045) but the difference was not significant compared to unclassified interneurons (2.47±1.47 Hz, p<0.142). We did not find significant differences between NGFCs and unclassified interneurons in impedance parameters. Support vector machine (SVM)-based wrapper feature selection of electrophysiological parameters ranked the amplitude of the sag and the standard deviation of interspike intervals as the two best delineators out of n=200 measured electrophysiological parameters for separating anatomically identified rosehip, neurogliaform and unclassified interneurons in layer 1 (Fig.4B). Indeed, the best hyperplane separating RCs from other interneuron types according to SVM analysis had a false positive rate of 0% for identifying RCs (n=37) in the total population of anatomically recovered layer 1 interneurons (n=107). Thus, we included cells defined by the hyperplanes of SVM analysis referred to as SVM identified RCs in case anatomical recovery was lacking in some experiments as indicated below.
Figure 4. Intrinsic electrophysiological properties of rosehip cells.
A, Examples of different firing patterns induced by current injections in layer 1 interneurons. Firing pattern of a RC (top), a NGFC (middle) and an unidentified layer 1 interneuron (bottom). B, Support vector machine (SVM) based wrapper feature selection of electrophysiological parameters for the identification of RCs. Anatomically identified RCs (red dots) and other types of interneurons with known morphology (black dots) are mapped to the distribution of electrophysiological features ranked as the two best delineators by SVM. Black lines show the best hyperplane separating RCs from other interneuron types. C-D, RCs exhibit distinct impedance profile relative to neurogliaform and other human interneurons in layer 1. C, Individual responses of anatomically identified rosehip (red), neurogliaform (blue) and other (black) interneurons to current injections with an exponential chirp (0.2-200 Hz, top). Traces were normalized to the amplitude of the rosehip response at 200 Hz. D, Left, Normalized impedance (Z) profiles of distinct groups of interneurons. RCs (n=5) had higher impedance in the range of 0.9 - 12.4 Hz compared to neurogliaform (n=5) and other (n=5) interneurons. Shaded regions represent standard deviation. Right, Impedances were similar at the lowest frequency (Z0.2 Hz, left), but resonance magnitude (Q) calculated as maximal impedance value divided by the impedance at lowest frequency (middle) and frequencies of maximal impedance (fmax, right) showed significant differences (p<0.05, ANOVA with and Bonferroni post hoc correction). E, Automatized selection of recording periods for the assessment of subthreshold membrane potential oscillations (boxed segments) and detection of bursts (bars) for measuring intraburst spiking frequency demonstrated on a RC response to near rheobasic stimulation showing stuttering firing behavior. F, Averaged fast Fourier transforms (FFT) of membrane potential oscillations had higher power between 3.8 and 80 Hz in RCs compared to neurogliaform and other interneurons. G, Intraburst frequency of RCs peaked in the gamma range.
Function of rosehip cells in local microcircuits
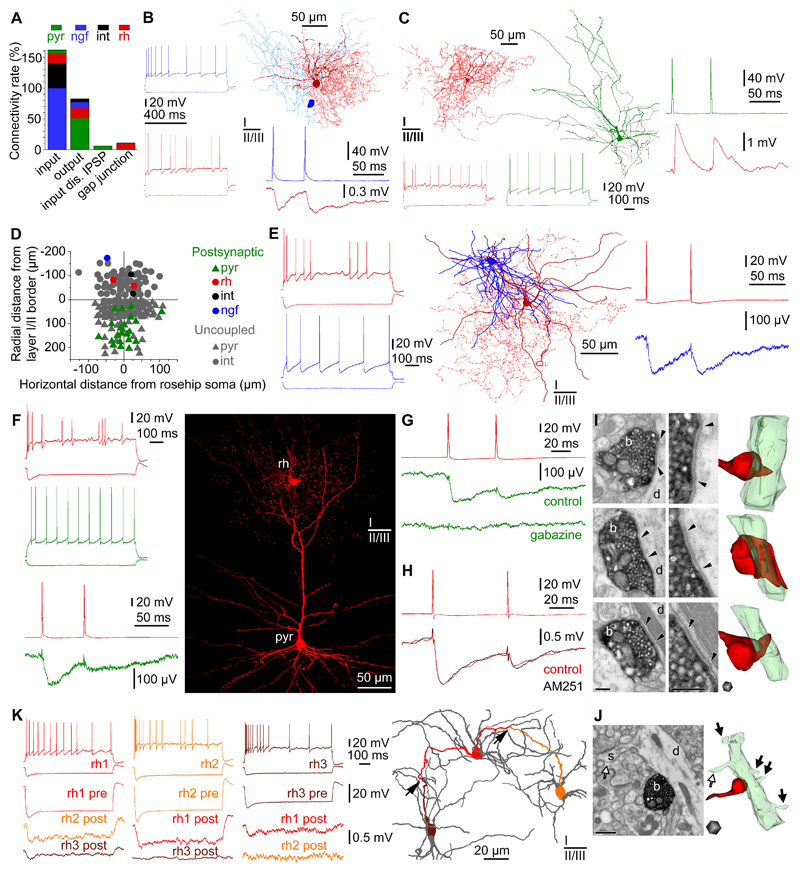
To assess functional connectivity of RCs in the local microcircuit, we established recordings from RCs and then searched for potential pre- and postsynaptic partners without any cell type preference in an area of the brain slices within a horizontal and vertical radius of ~100 µm and ~200 µm, respectively (Fig.5A-F). Monosynaptic input connections (n=226) were tested on anatomically (n=43) and SVM (n=24) identified RCs. Presynaptic layer 1 interneurons were connected to RCs with an overall coupling ratio (CR) of 45%. GABAergic cells evoking IPSPs on RCs included layer 1 NGFCs (n=10, CR 100 %), RCs (n=2, CR 17%) and unclassified interneurons (n=14, CR 40%); however, none of the tested interneurons (n=9) having somata in layer 2 (defined as <70 µm below the layer 1/2 border) were connected to RCs. Fast components of IPSPs arriving to RCs evoked by different presynaptic interneurons had similar amplitudes (0.982±0.705, 0.915±0.594 and 1.504±1.308 mV, respectively) and showed paired pulse depression with paired pulse ratios of (0.42±0.48, 0.27±0.04 and 0.71±0.26, respectively). RCs received local excitatory inputs from layer 2-3 pyramidal cells sporadically (n=8, CR 5%) with monosynaptic EPSP amplitudes of 3.357±1.458 mV and paired pulse ratios of 0.68±0.12. Very large unitary EPSPs, described to drive human basket and axo-axonic cells to suprathreshold postsynaptic responses10,11,14, were not encountered on RCs. Thus, local inputs to RCs appear to be predominantly GABAergic, acknowledging that some axon collaterals of pyramidal cells were cut during the slicing procedure (Fig.5C) leading to a potential underrepresentation of pyramidal cell triggered EPSPs.
Figure 5. Connections of rosehip cells in the local microcircuit.
A, Distribution of local connections mapped in layers 1-3 between RCs (rh, red), pyramidal cells (pyr, green), NGFCs (ngf, blue) and other types of layer 1 interneuron (int, black) based on unbiased targeting of postsynaptic cells. RCs predominantly innervate pyramidal cells, receive monosynaptic EPSPs from layer 2-3 pyramidal cells, monosynaptic IPSPs from neurogliaform and other types of interneurons, however, IPSPs arriving from RCs were not encountered. In addition, RCs are interconnected by homologous electrical synapses (gap junctions). B, Example of a NGFC to RC connection. Left, Firing patterns of the presynaptic NGFC (blue) and the postsynaptic RC (red). Right, Anatomical reconstruction of the recorded NGFC (soma, dark blue; axon, light blue) and RC (soma and dendrites, burgundy; axon: red). Action potentials in the NGFC (blue) elicited slow IPSPs in the RC (red). C, Example of a pyramidal cell to RC connection. Left, Anatomical reconstruction and firing pattern of the presynaptic pyramidal cell (firing, soma and dendrites, green; axon, black) and the postsynaptic RC (firing, soma and dendrites, burgundy; axon, red). Right, action potentials in the pyramidal cell (green) elicited EPSPs in the RC (burgundy). D, Spatial distribution of coupled and uncoupled neurons tested as postsynaptic targets of RCs. Note the relative dominance of layer 2-3 pyramidal cells among neurons receiving input from RCs. E, The only RC to NGFC connection successfully tested for synaptic coupling. Left, Firing patterns of the presynaptic RC (burgundy) and the postsynaptic NGFC (blue). Middle, Anatomical reconstruction of the RC (soma and dendrites, burgundy; axon, red) and the NGFC (soma and dendrites, blue; axon not shown). Right, Action potentials in the RC (red) elicited slow IPSPs in the NGFC (blue). F, Example of RC to layer 3 pyramidal cell connections (n=16). Left, Firing patterns of the presynaptic RC (red) and the postsynaptic pyramidal cell (green). Action potentials in the RC (burgundy) elicited IPSPs in the pyramidal cell (green). Right, Confocal fluorescence image showing the recorded RC (rh) forming its axonal cloud in the tuft of the apical dendrite of the layer 2-3 pyramidal cell (pyr). G, Pharmacological characterization of a rosehip-to-pyramidal cell connection. Presynaptic spikes in the RC (red) elicited IPSPs in the layer 2-3 pyramidal cell (green) which could be blocked by application of gabazine (n=4, 10 µM). H, Functional test of presynaptic CNR1expression in RCs show the absence of modulation by the CNR1antagonist AM251 (n=4). Presynaptic spikes in the RC 1 (red, top) elicited IPSPs in the RC 2 (red, bottom). Application of AM251 (5 µM) had no effect on IPSPs (black). I, Representative electron microscopic images (left) and three-dimensional reconstructions (right, n=31) showing axon terminals (b, red) of biocytin filled RCs (n=3) targeting exclusively dendritic shafts (d, green) (100%, n=31). Synaptic clefts are indicated between arrowheads. Scale bars: 200 nm. J, Representative electron microscopic image (left) and three-dimensional reconstruction (right) of a biocytin filled RC bouton (b, red) targeting a pyramidal dendritic shaft (d, green) identified based on emerging dendritic spines (s, arrows). Scale bars: 500 nm. K, RCs form a network of electrical synapses. Top left, firing patterns of three RCs (red, rh1; orange, rh2; burgundy, rh3). Bottom left, Hyperpolarization of RC rh1 was reciprocally transmitted to RCs rh2 and rh3 confirming electrical coupling. Right, Route of the hyperpolarizing signals through putative dendro-dendritic gap junctions (arrows) between RCs rh1, rh2 and rh3 is shown by corresponding colors in the dendritic network of the three cells (gray).
In turn, monosynaptic output connections triggered by anatomically (n=49) and SVM (n=13) identified RCs rarely innervated postsynaptic interneurons (overall CR 8%). Even though a NGFC (n=1, CR 10%), RCs (n=2, CR 17%), unclassified layer 1 interneurons (n=2, CR 5%) and superficial layer 2 pyramidal cells (n=5, CR 5%) were targeted when testing a total number of n=197 connections, the output of RCs were predominantly directed towards layer 3 pyramidal cells (n=16, CR 46%) having somata >70 µm below the layer 1/2 border. IPSPs elicited by RCs were mediated by GABAA receptors based on experiments showing blockade of IPSPs by application of the GABAA receptor antagonist gabazine (n=4, 10 µM, Fig.5G). Amplitudes of RC triggered IPSPs arriving to interneurons (0.428±0.370 mV) were larger compared to those targeting layer 3 pyramidal cells (0.087±0.059 mV, p<0.05, MW U-test), in agreement with dendritic filtering of distally elicited IPSPs during signal propagation along the apical dendrite to the somatically placed electrode. The results above indicate that RCs in layer 1 might preferentially target pyramidal cells sending terminal branches of their apical dendrites to layer 1. Indeed, when randomly sampling the output formed by RCs (n=6) using serial electron microscopic sections, we found that axon terminals (n=64) exclusively targeted dendritic shafts (Fig.5I). Moreover, further ultrastructural analysis of postsynaptic dendrites (n=46) revealed dendritic spines and sparse innervation by symmetrical synapses on the shaft, suggesting that these dendrites belonged to pyramidal cells (n=41, 86%, Fig.5J). The remaining n=5 (11 %) dendrites had no spines and received asymmetric synapses on the shaft were likely to be formed by interneurons.
Previous studies on rodent cortical interneurons containing CCK show functional presynaptic expression of the CB1 cannabinoid receptor28, however, application of the CB1 receptor antagonist AM251 was ineffective in modulating RC evoked IPSPs (n=4, Fig.5H), supporting our results of single cell digital PCR, IHC and ISH data (Fig.3). Earlier reports on human microcircuits identified single cell triggered polysynaptic network events10,11,14. We found that RCs were involved in single cell activated ensembles detected through disynaptic IPSPs triggered by layer 2 (n=1) and layer 3 (n=2) pyramidal cells and polysynaptic EPSPs triggered by an axo-axonic cell, respectively (data not shown). In addition to mono- and polysynaptic chemical synaptic communication, human interneurons are also involved in gap junctional signaling23. RCs also formed homologous electrical synapses (n=5, CR 57%) between each other (Fig.5K) and established convergent heterologous electrical synapses (n=2, CR 11%) with an unclassified layer 1 interneuron. When applying hyperpolarizing current steps in the first neuron to elicit response in the second neuron, coupling coefficients for gap junctions (0.05±0.05) were similar to those found earlier between human and rodent interneurons23.
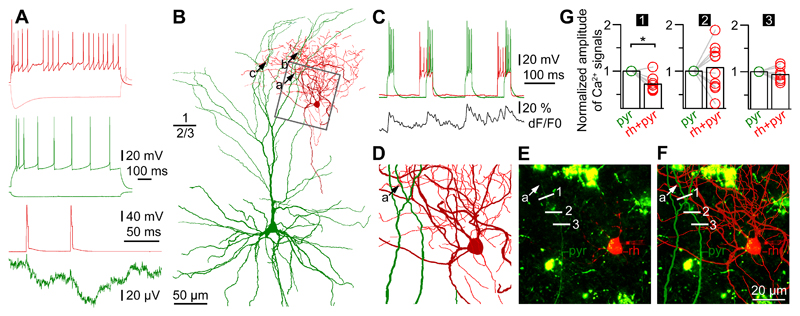
Preferential placement of output synapses on distal dendritic shafts of pyramidal cells reaching layer 1 suggest that RCs might specialize in the control of dendritic signal processing. RCs established 2.6±1.5 (range, 1-4) close appositions on dendrites of layer 3 pyramidal cells at distances 290±98 µm (range, 94-455 µm) from the somata of postsynaptic pyramidal cells (n=5). We found correlation between the rise times of IPSPs arriving to the postsynaptic pyramidal cells (n=5, 7.3±2.4 ms, range 3.8-10.1 ms) and the distances of close axo-dendritic appositions from the somata (ρ=0.90, P= 0.037, Spearman correlation). In dual recordings of synaptically connected RCs to pyramidal cell pairs (n=6), we loaded RCs with Alexa Fluor 594 to label presynaptic axons and filled the postsynaptic pyramidal cells with Oregon Green BAPTA 1 in order to structurally map the course of dendrites and to measure dendritic Ca2+ dynamics (Fig.6). The amplitude of the IPSPs triggered by the first action potential of RCs and evoked on distal dendrites of the postsynaptic pyramidal was (35.6±24.7 µV) at the soma (Fig.6A,B). Backpropagation of action potentials to dendrites of human neurons has been shown in previous studies13,29 and we confirmed these results by detecting dendritic Ca2+ responses following somatically elicited burst firing (100 ms current injections, 4 spikes/burst) in layer 3 pyramidal cells. Changes in ΔF/F (17.2±7.3%) in distal branches of the apical dendrites in layer 1 were consistently detected at multiple (17±8) locations on the postsynaptic neurons confirming action potential backpropagation into distal apical dendritic branches of human pyramidal cells (Fig.6C). We chose regions of interest on Oregon Green BAPTA 1-filled branches of the postsynaptic apical dendrites overlapping with the Alexa Fluor 594 labeled axonal arborization of presynaptic RCs and triggered somatically evoked bursts in the pyramidal cells alone for control and together with bursts in the RC in an alternating fashion (Fig.6B-F). Inputs from RCs simultaneous with backpropagating action potentials were effective in suppressing the amplitude of Ca2+ signals relative to control (n=6, 12.8±4.6% vs. 18.8±5.7% ΔF/F, p<0.02, Wilcoxon-test, Fig.6C) in one or two locations heuristically line scanned on dendrites of postsynaptic cells (Fig.6G). The anatomical arrangement of presynaptic axons and imaged segments of postsynaptic dendrites was recovered in n=4 pairs. Rosehip inputs simultaneous with backpropagating pyramidal action potentials were effective in suppressing Ca2+ signals only at sites that were neighboring (8±5 µm) to the putative synapses between the two cells. No effect of RCs was detected at dendritic sites one step further in distance (21±14 µm, Fig.6D-F). This suggests that RCs specialize in providing tightly compartmentalized control of dendritic Ca2+ electrogenesis of human pyramidal cells, thereby enforcing inhibitory microdomains in dendritic computation.
Figure 6. Human rosehip interneurons perform segment specific regulation of action potential backpropagation to apical dendritic tufts of pyramidal cells.
A, Top, Firing patterns of a presynaptic RC (burgundy) and a postsynaptic pyramidal cell (green). Bottom, Action potentials in the RC (burgundy) elicited IPSPs in the pyramidal cell (green). B, Anatomical reconstruction of the RC (soma and dendrites: burgundy; axon, red) and the layer 2-3 pyramidal cell (soma and dendrites, green; axon not shown). Presynaptic axonal boutons of the RC formed close appositions (a, b, and c) with three separate branches on the tuft of the pyramidal apical dendrite. C, Repetitive burst firing was triggered to initiate backpropagating Ca2+ signals in the pyramidal cell (green) while the output of the RC (red) was switched on and off timed prior and during every second pyramidal burst. Simultaneously, Ca2+ dynamics of the pyramidal apical dendritic tuft was measured at several locations and signals detected at location no.1 shown on panels E and F are shown in black. D, The area boxed in panel B shows the dendritic branch of the apical tuft of the pyramidal cell (green) with a putative synaptic contact (a) arriving from the RC. E, Confocal Z-stack image of the same area shown on panel D taken during paired whole cell recordings. The soma of the RC (rh, red), the dendrite of the pyramidal cell (pyr, green), the putative synaptic contact (a) arriving from the RC to the pyramidal cell and sites of line scans performed across the dendrite (1, 2 and 3) are indicated. Cytoplasmic lipofuscin autofluorescence characteristic to human tissue is seen as green patches. The experiment was repeated independently with similar results in n=4 cell pairs. F, Superimposition of the anatomical reconstruction of panel D and the confocal image of panel E. G, Normalized amplitudes of Ca2+ signals during pyramidal cell firing with and without coactivation of the RC detected at the three sites of line scans (1, 2 and 3) on the pyramidal dendrite. Rosehip input simultaneous with the backpropagating pyramidal action potentials was significant (p=0.02) in suppressing Ca2+ signals only at site 1 which was closest (8 µm) to the putative synapse between the two cells, no effect (n=10 trials, p=1 and p=0.27, respectively, two-sided Wilcoxon-test) of the RC was detected at sites 2 and 3 located at distances of 21 and 28 µm, respectively from the putative synaptic contact.
Discussion
Understanding the cellular makeup of the cortex and its conservation across species represent twin challenges difficult to address in human tissue. Historically, forming a representative overview of cell type diversity in a particular brain region has been achieved based on molecular marker expression cross referenced to axonal and dendritic morphology3,4,21,30. Conserved patterns of molecular and morpho-physiological features for particular cell classes have been reported2,23,24, but inter-species variation26,31–34 and cell types potentially characteristic to several species have also been described35–37. Recent studies have overcome some of the difficulties associated with the scarcity of human tissue of sufficient quality9,10,38,11–14,22,24,25,31 propelling understanding of human circuits. Here we demonstrate the strength of a modern version of this approach that can be applied to human postmortem and neurosurgical tissues. Single nucleus transcriptomics provides the scale for an unbiased survey of molecular expression, while human slice physiology characterizes the functional properties of those types. Together these approaches provide convergent evidence for robust cell type identities and concomitant evidence for species conservation or specialization.
The targeted application of single nucleus sequencing reported here has demonstrated a significantly higher degree of GABAergic neuron complexity in just one layer of the human cerebral cortex (10 types) than what has previously been described in all of the cortex (8 types22). This difference is likely due to a combination of improved sequencing technique and increased sampling in a targeted anatomical domain enriched in GABAergic neurons. This diversity also appears to be higher than described for layer 1 in mouse4, although by covering all layers that study likely underrepresented layer 1. Indeed, a recent characterization of rat cortex3 described 6 morphological and 17 morpho-electric types in layer 1, so our results are consistent with the neuronal diversity described using other methods. The RC represents a type with highly distinctive transcriptomic signature, a highly distinctive morphological, physiological and connectional phenotype, and a strong correspondence between these properties. In this respect, it appears similar to other highly specialized and distinctive cortical cell types such as chandelier cells39. To our knowledge a similar anatomical cell type has not been described in rodent. While we cannot prove the negative, given the extent of cellular studies of rodent cortex such cells would have to be either extremely rare or experimentally difficult to study to have escaped detection to date. Similarly, the rosehip molecular marker signature appears highly distinctive from any published data from rodent. Although the transcriptomic comparison is between human temporal cortex and mouse visual cortex, regional differences seem unlikely to account for this difference as we found the anatomically defined rosehip type in multiple human regions. A complete comparison of all cortical cell types and assessment of relative similarities between cell types should be possible in the future as more comprehensive transcriptome data become available and linked to other cellular phenotypes in multiple species. Our study is based on a relatively limited number of multimodally characterized cells due to the scarcity of high quality human samples required and further systematic analysis of human cell types in well-defined cytoarchitectonic areas using increased sample sizes are needed to substantiate further interpretations.
It is widely accepted28 that CCK-positive cells in the rodent show selectively high expression of cannabinoid receptors and are involved in perisomatic inhibition. The bouton morphology and/or the compact axonal field of RCs resembles that of cell types described in deeper layers of the cat cortex that innervate relatively proximal dendrites (dendrite targeting and clutch cells)40,41. In contrast, RCs are CCK-positive but cannabinoid receptor-negative, and appear to selectively target distal dendrites of pyramidal neurons. Moreover, when assessing layer 1 canonical inhibitory pathways in rodent with high throughput electrophysiology capable of sampling all cell types in layer 1, Lee at al.42 found two interneuron types and two canonical pathways involving feed forward interneuron-to-interneuron connections. Thus, the monosynaptic pyramidal cell-preferring pathway initiated by RCs does not appear to have a homologue in the rodent layer 1 circuit. Furthermore, focal intralayer inhibition restricted by the compact axonal arbor of RCs to distal dendrites of a column of pyramidal cells is also missing from the rodent; rather, mouse feedforward inhibitory connections are vertically spread to all somatodendritic domains42.
Addition of new human cell types, or specialization of existing types through major modification of cellular features, would be expected to alter circuit function3,43,44, and therefore cannot be studied in rodents. Dissimilarities of RCs and other dendrite-targeting interneurons are not fully understood without further experiments testing differences directly. RCs may be of particular importance in compartmental control of backpropagating action potentials and their pairing with incoming excitatory inputs. The uniquely small membrane capacitance (Cm) found in human pyramidal cells8 promotes backpropagation of action potentials and increases excitability in human dendrites13,29 relative to rodent dendrites having larger Cm. Action potentials backpropagate to distal dendrites of human pyramidal cells and can be attenuated by RC activation. Thus, RCs may provide supplementary inhibitory control required to balance the potentially higher excitability in human dendrites8 and modulate interactions between long range excitatory connections arriving to layer 1 and backpropagating action potentials suggested to participate in interhemispheric modulation45. The sharp resonance in the theta-range detected in individual RCs and its potential spread through gap junctions to a rosehip network could phase-selectively interact with long range inputs similarly to mechanisms suggested for example in oscillation dependent memory consolidation30,46. The function of neuron types specific to the human circuit could be important in understanding pathological alterations of network functions. For example, several highly selective markers for RCs have been implicated as risk factors for neuropsychiatric disease, including netrin G1 (NTNG1) for Rett syndrome47 and neurotrypsin (PRSS12) for mental retardation48. A better understanding of human cellular and circuit organization may help counteract the current lack of success in translating promising rodent results to effective treatment against human neuropsychiatric disorders49,50.
Methods
Postmortem human brain specimens
After obtaining permission from decedent next-of-kin, postmortem adult human brain tissue was collected by the San Diego Medical Examiner’s office and provided to the Allen Institute for Brain Science. All tissue collection was performed in accordance with the provisions of the Uniform Anatomical Gift Act described in Health and Safety Code §§ 7150, et seq., and other applicable state and federal laws and regulations. The Western Institutional Review Board reviewed tissue collection processes and determined that they did not constitute human subjects research requiring IRB review. The tissue specimens used in this study were pre-screened for known neuropsychiatric and neuropathological history, and underwent routine serological testing and toxicology screening. Specimens were further screened for RNA quality and had an RNA integrity number (RIN) ≥7. The specimens used for RNA-sequencing in this study were from two individual control Caucasian male donors, aged 50 and 54 years. Postmortem interval was 24 hours for both specimens. For multiplex fluorescent in situ hybridization (FISH) on frontal and parietal brain regions, postmortem tissue was obtained from two different male Caucasian donors, aged 60 (24 hr PMI) and 66 (22 hr PMI) years.
Tissue processing for nuclei isolation
Whole postmortem brain specimens were bisected through the midline and individual hemispheres were embedded in alginate for slabbing. Coronal brain slabs were cut at 0.5-1cm intervals through each hemisphere and the slabs were frozen in a bath of dry ice and isopentane and stored at -80°C. For RNA-sequencing experiments, middle temporal gyrus (MTG) was identified on slabs of interest and removed for further sectioning. MTG tissue was then thawed in a buffer containing PBS supplemented with 10mM DL-Dithiothreitol (DTT, Sigma Aldrich), mounted on a vibratome (Leica), and sectioned at 500µm in the coronal plane. Sections were transferred to a fluorescent Nissl staining solution (Neurotrace 500/525, ThermoFisher Scientific) prepared in PBS with 10mM DTT and 0.5% RNasin Plus RNase inhibitor (Promega). After staining for 5 min, sections were visualized on a fluorescence dissecting microscope (Leica) and layer 1 was microdissected using a needle blade micro-knife (Fine Science Tools).
Nuclei isolation and FACS
Microdissected sections of layer 1 from MTG were transferred into nuclei isolation medium containing 10mM Tris pH 8.0 (Ambion), 250mM sucrose, 25mM KCl (Ambion), 5mM MgCl2 (Ambion) 0.1% Triton-X 100 (Sigma Aldrich), 1% RNasin Plus, 1X protease inhibitor (Promega), and 0.1mM DTT and placed into a 1ml dounce homogenizer (Wheaton). Tissue was homogenized to liberate nuclei using 10 strokes of the loose dounce pestle followed by 10 strokes of the tight pestle. Homogenate was strained through a 30µm cell strainer (Miltenyi Biotech) and centrifuged at 900xg for 10 min to pellet nuclei. Nuclei were then resuspended in staining buffer containing 1X PBS (Ambion), 0.8% nuclease-free BSA (Omni-Pur, EMD Millipore), and 0.5% RNasin Plus. Mouse monoclonal anti-NeuN antibody (EMD Millipore) was applied to nuclei preparations at a concentration of 1:1000 and samples were incubated for 30 min at 4°C. Control samples were incubated with mouse IgG1,k isotype control (BD Pharmingen). Samples were then centrifuged for 5 min at 500xg to pellet nuclei and pellets were resuspended in staining buffer as described above. Nuclei samples were incubated with secondary antibody (goat anti-mouse IgG, Alexa Fluor 594, ThermoFisher Scientific) for 30 min at 4°C, centrifuged for 5 min at 500xg, and resuspended in staining buffer. DAPI (4′, 6-diamidino-2-phenylindole, ThermoFisher Scientific) was applied to nuclei samples at a concentration of 1µg/ml.
Single nucleus sorting was carried out on a BD FACS Aria Fusion instrument (BD Biosciences) using a 130µm nozzle. Nuclei were first gated on DAPI and then passed through doublet discrimination gates prior to being gated on NeuN (Alexa Fluor 594) signal. Approximately 10% of nuclei were intentionally sorted as NeuN-negative to allow for the collection of non-neuronal nuclei. Single nuclei were sorted into 96-well PCR plates (ThermoFisher Scientific) containing 2µl of lysis buffer (0.2% Triton-X 100, 0.2% NP-40 (Sigma Aldrich), 1U/µl RNaseOut (ThermoFisher Scientific), PCR-grade water (Ambion) and ERCC spike-in synthetic RNAs [Ambion]). 96-well plates containing sorted nuclei were then snap frozen and stored at -80°C. Positive controls (10 nuclei pools and/or 10 pg and 1 pg total RNA) were included on every 96-well plate of sorted nuclei.
cDNA and sequencing library preparation
cDNA libraries from single nuclei were prepared using Smart-seq2 20 with minor modifications. Briefly, Protoscript II (New England Biolabs) was used for reverse transcription, the final dilution of ERCCs in the reverse transcription reaction was 1:55 million, and the template switching oligonucleotide was 5’-biotinylated. Additionally, the number of PCR cycles used for cDNA amplification was increased to 21 to compensate for lower RNA content in single nuclei. cDNA yield was quantified using PicoGreen (ThermoFisher Scientific) and a subset of single nuclei libraries were screened for quality on a Bioanalyzer (High Sensitivity DNA Chip, Agilent Technologies). cDNA library quality was further assessed using qPCR for a housekeeping gene (ACTB) and an ERCC spike-in control RNA (ERCC-00009) 51.
Sequencing libraries were prepared using Nextera® XT (Illumina) with minor modifications. Briefly, the input amount of cDNA was 250pg per reaction, reactions were carried out a 1/4X the volume recommended by the manufacturer, and the tagmentation step was extended to 10 min. Sequencing library concentration was determined using PicoGreen and 53-57 samples were pooled per sequencing lane. Pooled libraries were purified using Ampure XP beads and eluted to a concentration of 5nM. Following purification, the pooled library size using a Bioanalyzer and Kapa Library QC was used to determine nM concentrations. Final library pools were then diluted to 3nM final concentration. Pooled samples were sequenced on a HiSeq® 4000 instrument (Illumina) using 150 base paired end reads at a mean untrimmed read depth of ~19 million reads/sample and a mean trimmed read depth of ~16 million reads/sample.
RNA-Seq processing
The RNASeq data obtained from single nuclei is processed and analyzed according to the procedure described in detail previously51. Briefly, following the demultiplexing of the barcoded reads generated on the Illumina HiSeq platform, the amplification (cDNA & PCR) and sequencing primers (Illumina) and the low quality bases were removed using the Trimmomatic 0.35 software package52. The trimmed reads were mapped to the human reference genome version, GRCh38 (Ensembl) guided by the version 21 annotations obtained from the GENCODE repository. RSEM 1.2.3153, TOPHAT 2.1.154 and CUFFLINKS 2.2.154 were used to quantify transcript expression at the transcriptome (exon) and the whole genome (exon plus intron) level, respectively. The fastQC 0.10.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), FASTX 0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/download.html), RSeQC 2.6.1 55, RNA-seq-QC 1.1.8 56 programs were used to generate various sequence and alignment quality metrics used for classification of the sample quality. A novel pipeline (“SCavenger”, unpublished) was created to automate execution across statistical analysis tools, integrate pre-formatted laboratory and clustering metrics, and calculate new statistics specific to biases identified in the single nuclei lab and sequence preparation protocol. The normalized expression counts (FPKM/TPM) generated at both gene and isoform level by RSEM and TOPHAT-CUFFLINKS analyses and the raw counts generated from the RSEM/TOPHAT alignment (BAM) files by the HTSeq-count program 57 were used for differential expression analysis.
RNA-Seq quality control
To remove data from low quality nuclei samples prior to downstream analysis, we implemented a Random Forest machine learning classification approach as described in detail in Aevermann et al. 58. The overall workflow for sample quality classification and filtering was to i) establish a training set using a representative subset of samples, ii) collect a series of 108 quality control metrics (e.g. percent unique reads, percent reads surviving trimming, transcript isoform counts) spanning both the laboratory and data analysis workflows as model features, iii) use these training data and quality control metrics to build a classification model using the Random Forest method, and iv) apply the model to the entire data set for quality classification and data filtering.
A training set of 196 samples, including 169 single nuclei samples, was selected and a set of high confidence pass/fail calls for individual samples determined based on the qualitative assessment of data produced by fastQC, which includes quality Phred scores, GC content, Kmer distributions, and sequence over-representation information. Pass samples (152 samples, including single nuclei and purified bulk RNA positive controls) were identified as having high average quality per read across the entire length of the sequenced fragment and a unimodal average GC content around 40%, reflecting the GC content of the expressed human transcriptome. In contrast, two types of Fail samples were identified. One type of Fail samples (29 samples) exhibited a significant number of reads with low mean Phred quality, and average Phred quality scores that fall off down the length of the sequence read. A second type of Fail samples (15 samples) showed a second peak in the GC content distribution with a mean around 48% GC; this peak appears to be generated from ERCC reads, which are derived from bacterial genome sequences.
The quality control metrics for these training data were then used as features to construct a Random Forest model to distinguish these three quality classes (Pass, Fail-Phred, and Fail-ERCC) comprised of one hundred thousand decision trees generated by standard bagging methods as implemented in KNIME v3.1.2. Using this Random Forest classification model, all 196 samples in the training set were classified correctly with high confidence scores. To test the classification accuracy of the resulting random forest model, we used an independent test set of 185 single nuclei samples classified using the same fastQC evaluation criteria applied to the training data, with 135 determined to be Pass samples, 29 determined to be Fails and 21 determined to be Marginals. Application of the random forest model to these test Pass and Fail samples resulted in only 8 misclassifications (4.9%), for a classification accuracy of 95%. The Random Forest model was then applied to the remaining data and final classification determined. A Pass confidence cutoff of 0.6 or greater was used to select single nuclei data for downstream analysis. Using this Random Forest model applied to the entire layer 1 dataset including contaminating layer 2 excitatory and inhibitory nuclei, 79% of 1154 single nuclei samples passed quality control. For these Pass samples, the average number of reads after trimming was 16,383,881 (±19,810,661), the number of ERCC transcripts detected was 43.76 (± 3.77), and the number of genes detected at a level of >1 FPKM was 6337 (± 1659), giving an average coverage of 879 reads per human gene detected.
Gene expression calculation
For each nucleus, expression levels were estimated based on the scaled coverage across each gene. Specifically, bam files were read into R 59 using the “readGAlignmentPairs” function in the “GenomicAlignments” library, and genomic coverage was calculated using the “coverage” function in “GenomicRanges”60. All genes in GENCODE human genome GRCh38, version 21 (Ensembl 77; 09-29-2014) were included, with gene bounds defined as the start and end locations of each unique gene specified in the gtf file (https://www.gencodegenes.org/releases/21.html). Total counts for each gene (including reads from both introns and exons) were estimated by dividing total coverage by twice the read length (150bp, paired end). Expression levels were normalized across nuclei by calculating counts per million (CPM) using the “cpm” function in “edgeR” 61.
Clustering nuclei
Nuclei that passed quality control were grouped into transcriptomic cell types based on an iterative clustering procedure. For each gene, log2(CPM + 1) expression was centered and scaled across nuclei. Gene expression dropout was more likely to occur in nuclei with lower quality cDNA libraries and for genes with lower average expression in nuclei isolated from the same cell type. Expression noise models were estimated for each nucleus based on the 8 most similar nuclei using the “knn.error.models” function of the “scde” R package as described in 62. These noise models were used to select significantly variable genes (adjusted variance > 1.25) and to estimate a zero-weight matrix that represented the likelihood of dropouts based on average gene expression levels. Dimensionality reduction was performed with principal components analysis (PCA) on variable genes, and the covariance matrix was adjusted by the zero-weight matrix to account for gene dropouts. Principal components (PCs) were retained for which more variance was explained than the broken stick null distribution or PCs based on permuted data. If more than 2 PCs were retained, then dimensionality was further reduced to 2-dimensions using t-distributed stochastic neighbor embedding (tSNE)63 with a perplexity parameter of 80.
After dimensionality reduction, nuclei were clustered using a conservative procedure that attempted to split them into the fewest number of clusters possible. Nearest-neighbor distances between all nuclei were calculated and sorted, and segmented linear regression (R package “segmented”) was applied to estimate the distribution breakpoint to help define the distance scale for density clustering. Next, density clustering (R package “dbscan” 64) was applied to nuclei, and the number of clusters calculated for a range of 10 nearest-neighbor distances (parameter epsilon), starting from the maximum distance between nuclei to the distance breakpoint identified in the last step. If only one cluster was found using all values of epsilon, then the above procedure was repeated using a perplexity parameter of 50, 30, and 20 for tSNE, and stopping when more than one cluster was detected. Finally, if no cluster splitting was possible using tSNE, then a final density clustering was applied to the first two significant PCs. If more than one cluster was identified, then the statistical significance of each cluster pair was evaluated with the R package “sigclust” 65, which compares the distribution of nuclei to the null hypothesis that nuclei are drawn from a single multivariate Gaussian. Iterative clustering was used to split nuclei into sub-clusters until the occurrence of one of four stop criteria: 1) <6 nuclei in a cluster (because it cannot be split due a minimum cluster size of 3), 2) no significantly variable genes, 3) no significantly variable PCs, 4) no significant sub-clusters.
To assess the robustness of clusters, the iterative clustering procedure described above was repeated 100 times for random subsamples of 80% of nuclei. A co-clustering matrix was generated that represented the proportion of clustering iterations that each pair of nuclei were assigned to the same cluster. Average-linkage hierarchical clustering was applied to this matrix followed by dynamic branch cutting (R package “WGCNA”) with cut height ranging from 0.01 to 0.99 in steps of 0.01. A cut height resulting in 25 clusters was selected to balance cohesion (average within cluster co-clustering) and discreteness (average between cluster co-clustering) across clusters. Finally, gene markers were identified for all cluster pairs, and clusters were merged if they lacked binary markers (gene expressed in >50% nuclei in first cluster and <10% in second cluster) with average CPM > 1 (see also Marker gene selection below).
Cluster visualization
The relationships between cell type clusters were represented as a constellation diagram where the area of each disc is proportional to the number of nuclei in each cluster and the width of the lines connecting clusters is proportional to the number of “intermediate nuclei” between these clusters, as described below and in 66. To define core and intermediate nuclei we used a nearest-centroid classifier, which assigns a nucleus to the cluster whose median is most highly correlated based on expression of the 1200 best marker genes, as described below. We performed 5-fold cross-validation 100 times: in each round, the nuclei were randomly partitioned into 5 equally-sized groups where the nuclei in each group were classified by a nearest centroid classifier trained using the remaining nuclei. Nuclei classified to the same cluster fewer than 90 times or classified to a cluster different from the originally assigned cluster were defined as core, while the others were designated as intermediate. In total, 443/470 (94.3%) of nuclei were defined as core.
Next, clusters were arranged by transcriptomic similarity based on hierarchical clustering. First, the average expression level of each gene was calculated for each cluster. Genes were then sorted based on variance and the top 2000 genes were used to calculate a correlation-based distance matrix, Dxy=1-(cor(x,y))/2, between each cluster average. A cluster tree was generated by performing hierarchical clustering on this distance matrix (using “hclust” with default parameters), and then reordered to show inhibitory clusters first, followed by excitatory clusters and glia, with larger clusters first, while respecting the tree structure. Note that this measure of cluster similarity is complementary to the co-clustering similarity described above. For example, two clusters with high transcriptomic similarity but a few distinct marker genes may have low co-clustering similarity.
Marker gene selection
Initial sets of marker genes for each pair of clusters were selected by assessing significance of differential expression using the “limma” 67 R package, and then filtering these sets of significant genes to include only those expressed in more than 50% of nuclei in the “on” cluster and fewer than 20% of nuclei in the “off” cluster. Potential marker genes for individual clusters were chosen by ranking the significance of pairwise marker genes, summing the ranks across all possible pairs for a given cluster, and sorting the resulting gene list ascending by summed rank. The final set of marker genes was selected by comparing the gene expression distribution for the top ranked marker genes for each cluster using the visualization described below.
Scoring marker genes based on cluster specificity
Many genes were expressed in the majority of nuclei in a subset of clusters. A marker score (beta) was defined for all genes to measure how binary expression was among clusters, independent of the number of clusters labeled. First, the proportion (xi) of samples in each cluster that expressed a gene above background level (CPM > 1) was calculated. Then, scores were defined as the squared differences in proportions normalized by the sum of absolute differences plus a small constant (ε) to avoid division by zero. Scores ranged from 0 to 1, and a perfectly binary marker had a score equal to 1.
Matching clusters based on marker gene expression
Human MTG layer 1 clusters were compared to published cell types from human cortex 22 and mouse primary visual cortex 66. The proportion of nuclei or cells expressing each gene with CPM > 1 was calculated for all clusters. Genes were selected that cluster-specific (beta score > 0.3) in this study and the published human and mouse studies. Weighted correlations were calculated between all pairs of clusters across these genes and weighted by beta scores to increase the influence of more informative genes. Heatmaps were generated to visualize all cluster correlations and pairs of clusters that were reciprocal best matches were labeled. Finally, scatter plots were generated to compare the expression detection of marker genes in these labeled cluster pairs.
Gene expression visualization
Gene expression (CPM) was visualized using heat maps and violin plots, which both show genes as rows and nuclei as columns, sorted by cluster. Heat maps display each nucleus as a short vertical bar, color-coded by expression level (blue=low; red=high), and clusters ordered as described above. The distribution of marker gene expression across nuclei in each cluster were represented as violin plots, which are density plots turned 90 degrees and reflected on the Y-axis. Black dots indicate the median gene expression in nuclei of a given cluster; dots above Y=0 indicate that a gene is expressed in more than half of the nuclei in that cluster.
Colorimetric in situ hybridization
In situ hybridization data for human temporal cortex and mouse cortex was from the Allen Mouse Brain Atlas 32 and a comparable study in human temporal cortex 68. All data is publicly accessible through www.brain-map.org. Data was generated using a semiautomated technology platform as described 32. with modifications to work with postmortem human tissues as described in 68. Digoxigenin-labeled riboprobes were generated for each human gene such that they would have >50% overlap with the orthologous mouse gene in the Allen Mouse Brain Atlas 32. Mouse ISH data shown is from the region most closely corresponding to human temporal cortex, corresponding to the medial portion of TeA in Paxinos Atlas 69.
Multiplex fluorescent in situ hybridization (FISH)
Tissue specimens used for multiplex FISH came from either neurosurgical resections (all MTG tissue) or postmortem brain specimens (frontal and parietal regions). Tissue procurement from donors undergoing surgery was performed at hospitals, fully outside of the supervision of the Allen Institute. Tissue was provided to researchers under the supervision and authority of the Internal Review Board (IRB) of each participating hospital. All surgical tissue donors met with a hospital-appointed surgical case coordinator to review the option of tissue donation and voluntarily signed an IRB-approved Informed Consent Form. Tissue donors for these experiments ranged in age from 28-37 years old. Tissue from surgical resections was transported in chilled, oxygenated ACSF and then mounted for slice preparation on a Compresstome VF-200 or VF-300 vibrating microtome (Precisionary Instruments) to be sliced perpendicular to pial surface. Slices (350 µm) were embedded in OCT (optimal cutting temperature medium), rapidly frozen, and sub-sectioned at 20 µm on a Leica cryostat.
For experiments using postmortem tissue, coronal brain slabs containing Brodmann Area 9 (rostrodorsal portion of dorsolateral prefrontal cortex) and Brodmann Area 40 (rostral division of posteroventral parietal cortex) were identified and regions of interest were removed from frozen slabs and subdivided into small blocks. Blocks were embedded in OCT and sectioned at 16 µm using a Leica cryostat.
The RNAscope multiplex fluorescent kit was used according to the manufacturer’s instructions for fresh frozen tissue sections (ACD Bio), with the exception that fixation at 4 °C with 4% PFA was performed for 60 minutes on 16-20 µm human brain sections, and the protease treatment step was shortened to 15 min. Probes used to identify specific cell types in layer 1 were designed antisense to the following human genes: CCK (hs-539041, NM_000729.4), CNR1 (hs-591521, NM_001160226.1), CPLX3 (hs-487681-C3, NM_001030005.2), GAD1 (hs-404031 and hs-404031-C3, NM_000817.2), LAMP5 (hs487691-C3, NM_012261.3), SV2C (hs448361-C3, NM_014979.3), PRSS12 (hs-493931-C3 NM_003619.3), SOX13 (hs-493941-C3, NM_005686.2), TRPC3 (hs-427641-C2, NM_001130698.1), NTNG1 (hs-446101, NM_001113226.1), CXCL14 (hs-425291, NM_004887.4), PDGFRA (hs-604481-C2, NM_006206.4), SOX9 (hs-404221-C2, NM_000346.3). Positive controls (POLR2A, UBC and PPIB) were used on each tissue sample to ensure RNA quality (ACD Bio, 320861). Following hybridization and amplification, FISH sections were imaged using a 40X oil immersion lens on a Nikon TiE fluorescent microscope. RNA spots in each channel were quantified manually using the ImageJ 1.51 cell counting plug- in. To count the percentage of RCs in layer 1, GAD1+ cells were first identified, followed by the PDGFRA+ cells within that population, followed by the TRPC3+ cells in that population. These counts were used to calculate the percentage of the GAD1+ cells expressing PDGFRA and TRPC3. A total of 408 GAD1+ cells were identified from two individuals for this quantification.
Electrophysiological recordings
All procedures were performed according to the Declaration of Helsinki with the approval of the University of Szeged Ethical Committee. We used neocortical tissue surgically removed from patients (n=42, n=22 female and n=20 male, aged 49±18 years) in a course of five years as part of the treatment protocol for aneurysms (n=9 and brain tumors (n=33). Patients with a history of epilepsy were excluded from this study. Anesthesia was induced with intravenous midazolam and fentanyl (0.03 mg/kg, 1– 2 lg/kg, respectively). A bolus dose of propofol (1–2 mg/kg) was administered intravenously. To facilitate endotracheal intubation, the patient received 0.5 mg/kg rocuronium. After 120 seconds, the trachea was intubated and the patient was ventilated with a mixture of O2 -N2O at a ratio of 1:2. Anesthesia was maintained with sevoflurane at monitored anesthesia care (MAC) volume of 1.2–1.5. Tissue blocks were removed from prefrontal (n=16), temporal (n=6) and parietal (n=10) areas. Blocks of tissue were immersed in ice-cold solution containing (in mM) 130 NaCl, 3.5 KCl, 1 NaH2PO4, 24 NaHCO3, 1 CaCl2, 3 MgSO4, 10 d(+)-glucose, saturated with 95% O2 and 5% CO2 in the operating theatre. Slices were cut perpendicular to cortical layers at a thickness of 350 µm with a vibrating blade microtome (Microm HM 650 V) and were incubated at room temperature for 1 h in the same solution. The solution used during recordings differed only in that it contained 2 mM CaCl2 and 1.5 mM MgSO4. Somatic whole-cell recordings were obtained at approximately 36 ºC from up to four concomitantly recorded cells visualized by infrared differential interference contrast videomicroscopy at depths 60–130 µm from the surface of the slice. Signals were filtered at 8 kHz, digitized at 16 kHz, and acquired with Patchmaster software. Micropipettes (5–7 MΩ) were filled with a low [Cl]i solution containing (in mM) 126 K-gluconate, 4 KCl, 4 ATP-Mg, 0.3 GTP-NA2, 10 HEPES, 10 phosphocreatine, and 8 biocytin (pH 7.20; 300 mOsm). Presynaptic cells were stimulated with brief (2–10 ms) suprathreshold pulses delivered at >7-s intervals, to minimize intertrial variability. For pharmacological experiments 10 µM gabazine and 5 µM 1-(2,4-Dichlorophenyl)-5-(4-iodophenyl)-4-methyl-N-1-piperidinyl-1H-pyrazole-3-carboxamide (AM251) were applied and were purchased from (Sigma-Aldrich). Membrane properties of human neurons did not show significant changes for up to 20 h after slicing, but recordings included in the analysis were arbitrarily terminated 15 h after slice preparation. Data were analyzed with Fitmaster (HEKA) and Origin 7.5 (OriginLab) Data are given as mean±standard deviation (S.D.). The Mann-Whitney U-test was used to compare datasets; differences were considered significant if p<0.05. Data collection and analysis were not performed blind to the conditions of the experiments.
Firing classification analysis
First, a set of n=200 electrophysiological features had been calculated for each cell identified based on light microscopic investigation of the axonal arbour. Then a wrapper feature selection method using Support Vector Machine (SVM) was used on the cells (RCs: n=55; non-RCs: n=52) to find the best feature set which separated the group of RCs from the group of other cells. The best feature set consisted of 2 features, the maximal standard deviation of interspike-intervals (ISI SD) and the amplitude of sag in response to hyperpolarization (-100 pA). Sweeps were discarded with less than 5 spikes for the calculation of ISI SD. The sag value was calculated as the ratio of the voltages at the onset of the hyperpolarizing step and during steady state.
Measurement of impedance profile
The impedance profile was determined by sinusoidal current injections using a standard exponential chirp pattern (0.2-200 Hz, 10 s duration) generated with Patchmaster (HEKA). Measurements (7-10 traces per cell) were made at resting membrane potential and the peak to peak amplitude of the command current waveform was tuned between 40-100 pA to test subthreshold voltage responses. The impedance profile (Z) was determined for each trace by calculating the fast Fourier transform (FFT) of the voltage response and dividing the FFT component of the corresponding command current, then the impedance profiles were normalized to the value at 200 Hz. After anatomical identification of the recorded cells, the dataset was pooled according to three defined cell types then the averaged impedance plotted against input frequency. For statistical comparison of the impedance profiles, four parameters were considered: impedance at lowest frequency (Z0.2Hz); resonance magnitude (Q, the impedance magnitude at the resonance peak i.e. maximal impedance value divided by the impedance magnitude at the lowest input frequency of 0.2 Hz); and the frequency at maximum impedance (fmax).
Two-photon calcium imaging
Structural labelling of RCs was based on 40 µM Alexa Fluor 594 (Molecular Probes). We also applied 100 µM of Oregon Green 488 BAPTA-1 (Molecular Probes), in order to measure intracellular Ca2+ dynamics of pyramidal cell dendrites in the intracellular solution (see above). Imaging with multiphoton excitation was performed using a modified Zeiss LSM7 MP (Oberkochen, Germany) two-photon laser scanning system and a FemtoRose 100 TUN (R&D Ultrafast Lasers, Hungary) titanium–sapphire laser with Finesse4 pumping laser (Laser Quantum, UK) providing 100 fs pulses at 80 MHz at a wavelength of 820 nm. Fluorescence images were acquired through a x40 water-immersion objective (0.8 NA; Olympus, Japan).
Single cell reverse transcription and digital PCR
At the end of electrophysiological recordings, the intracellular content was aspirated into the recording pipettes by application of a gentle negative pressure while maintaining the tight seal. Pipettes were then delicately removed to allow outside-out patch formation, and the content of the pipettes (~1.5 μl) was expelled into a low-adsorbtion test tube (Axygen) containing 0.5 μl SingleCellProtectTM (Avidin Ltd. Szeged, Hungary) solution in order to prevent nucleic acid degradation and to be compatible with direct reverse transcription reaction. Samples were snap-frozen in liquid nitrogen and stored or immediately used for reverse transcription. Reverse transcription of individual cells was carried out in two steps. The first step was performed for 5 min at 65°C in a total reaction volume of 7.5 μl containing the cell collected in 4 μl SingleCellProtect (Avidin Ltd., Cat.No.: SCP-250), 0.45 μl TaqMan Assays (Thermo Fisher), 0.45 μl 10 mM dNTPs (Thermo Fisher, Cat.No.: 10297018, 1.5 μl 5X first-strand buffer, 0.45 μl 0.1 mol/L DTT, 0.45 μl RNase inhibitor (Thermo Fisher, Cat.No.:N8080119) and 100 U of reverse transcriptase (Superscript III, Thermo Fisher, Cat.No.: 18080055). The second step of the reaction was carried out at 55°C for 1 hour and then the reaction was stopped by heating at 75°C for 15 min. The reverse transcription reaction mix was stored at -20°C until PCR amplification.
For digital PCR analysis, the reverse transcription reaction mixture (7.5 μl) was divided into two parts: 6 μl was used for amplification of the gene of interest and 1.5 μl cDNA was used for amplifying the housekeeping gene, GAPDH. Template cDNA was supplemented with nuclease free water to a final volume of 8 μl. 2 μl TaqMan Assays (Thermo Fisher), 10 μl OpenArray Digital PCR Master Mix (Thermo Fisher, Cat.No.: 4458095) and nuclease free water (3 μl) were mixed to obtain a total volume of 20 μl and the mixture was evenly distributed on 4 subarrays (256 nanocap-illary holes) of an OpenArray plate by using the OpenArray autoloader. Processing of the OpenArray slide, cycling in the OpenArray NT cycler and data analysis were done as previously described 70. For our dPCR protocol amplification, reactions having CT values less than 23 or greater than 33 were considered primer dimers or background signals, respectively, and excluded from the data set.
Histology and reconstruction
Following electrophysiological recordings, slices were immersed in a fixative containing 4% paraformaldehyde (for immunohistochemistry) or 4% paraformaldehyde, 15% (v/v) saturated picric acid and 1.25% glutaraldehyde (for reconstructions) in 0.1 M phosphate buffer (PB; pH=7.4) at 4°C for at least 12 h. After several washes with 0.1 M PB, slices were frozen in liquid nitrogen then thawed in 0.1 M PB, embedded in 10% gelatin and further sectioned into 60 µm slices. Sections were incubated in a solution of conjugated avidin-biotin horseradish peroxidase (ABC; 1:100; Vector Labs) in Tris-buffered saline (TBS, pH=7.4) at 4°C overnight. The enzyme reaction was revealed by 3’3-diaminobenzidine tetra-hydrochloride (0.05%) as chromogen and 0.01% H2O2 as oxidant. Sections were post fixed with 1% OsO4 in 0.1 M PB. After several washes in distilled water, sections were stained in 1% uranyl acetate, dehydrated in ascending series of ethanol. Sections were infiltrated with epoxy resin (Durcupan) overnight and embedded on glass slides. Three dimensional light microscopic reconstructions were carried out using Neurolucida system (MicroBrightField) with 100x objective. Reconstructed neurons were quantitatively analyzed with NeuroExplorer software (MicroBrightField). Axon tortuosity was measured as the average ratio of the actual axonal path and linear distance between neighboring nodes.
Immunohistochemistry of biocytin-labeled cells
The recorded cells were first visualized with incubation in Cy3-conjugated streptavidin (Jackson Immunoresearch) for 2 h, diluted 1:400 in TBS. After examination by epifluorescence microscopy, the sections containing the soma of the labeled neurons were incubated in 20% normal horse serum in TBS to block nonspecific antibody-binding sites. Free-floating sections containing the soma were incubated in primary antibodies dissolved in TBS containing 0.05% NaN3 for 72 hours at room temperature. The following primary antibodies were used: rabbit-anti-pro-cholecystokinin (1:2000, polyclonal, lot TL1, gift from Andrea Varro, Liverpool University); mouse-anti-CNR1 (1:4000, IMG-CB1R-mAb001, clone IMG-3C2, lot CJ03ImmunoGenes); rabbit-anti-GABA (1:1000, A2052, polyclonal, lot 056M4834V, Sigma-Aldrich); mouse-anti-NR2F2 (1:700, Ab41859, clone H7147, lot GR15505-8, Abcam); mouse-anti-PV (1:1500, S235, clone 235, lot 10-11F, Swant); rabbit-anti-nNOS (1:1000, 160870, polyclonal, lot 189901, Cayman Chemical); rabbit-anti-NPY (1:300, T-4069, polyclonal, A00721, Peninsula Laboratories); rat-anti-somatostatin (1:50, MAB354, clone YC7, lot 2294201, Merck Millipore); rabbit-anti-calbindin (1:2000, CB-38a, polyclonal, lot 9.03, Swant); goat-anti-calretinin (1:700, CG1, polyclonal, lot 10.1, Swant); goat-anti-acetyltransferase (1:100, AB144P, polyclonal, 0608037072, Merck Millipore). After several washes in TBS, the immunoreactions were visualized with A488- or Cy5-conjugated secondary antibodies (1:500, 711-545-152 lot 119191, 715-485-150 lot 89132, 712-485-150 lot 88670, 705-485-003 lot 89133, 715-175-150 lot 105884, 705-175-147 lot 114786, Jackson Immunoresearch). The sections were mounted on slides in Vectashield (Vector Laboratories). Images were taken by confocal laser scanning microscope (LSM 880, Zeiss) using a 40x oil-immersion objective (1.4 NA). After photography, the sections were demounted, washed in 0.1 M PB, and biocytin was visualized with the avidin-biotinylated horseradish peroxidase method described above. For quantification of positive and negative immunoreactions we measured the mean fluorescence intensity of the immunostaining in the soma or in the axon terminals of the biocytin filled RCs and the fluorescence intensity of the background using the thresholding tool by ImageJ 1.48 (Jensen, 2013). Fluorescence intensity measurements were corrected with the background in every image. Finally, we classified fluorescence intensity values <2 AU as negative immunoreactions. The fluorescence intensity of cells measured positive and negative with this method were significantly different (p<0.01, MW U-test).
Electron microscopy
Axonal boutons of biocytin filled rosehip (n=6) and NGFCs (n=2) (identified based on distinctive electrophysiological properties and light microscopic investigation of the axonal arbour) were re-embedded and re-sectioned at 70 nm thickness. Digital images of serial EM sections were taken at magnifications ranging from 8,000x to 50,000x with a JEOL JEM-1400Plus electron microscope equipped with a 8 M pixel CCD camera (JEOL Ruby). Axon terminals were reconstructed in 3D and their volumes were measured using the Reconstruct software (http://synapses.clm.utexas.edu/) (n=31 boutons of RCs; n=24 boutons of NGFCs). The areas of active zones of RCs were measured at perpendicularly cut synapses, where the rigid apposition of the pre- and postsynaptic membranes was visible (n=11 active zones).
Statistics
No sample-size calculation was performed, all data were subject to statistical tests to decide whether parametric or nonparametric tests should be applied. Two-tailed test were used throughout. Single nuclei were isolated from post-mortem brains of 2 donors. This allowed us to collect nuclei from high quality specimens that met stringent quality control metrics while also confirming that transcriptomic clusters were consistent between donors and not driven by technical artifacts. No data with successful quality control were excluded from the analyses. Data from low quality nuclei samples was removed prior to downstream analysis, using a Random Forest machine learning classification approach as described58. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications10,11,14. All human specimens were controls (nonpathological) and were therefore allocated into the same experimental group. Randomization was not used. Human specimens were de-identified and assigned a unique numerical code. Researchers had access to basic information about donors (age, sex, ethnicity) as well as the unique numerical code assigned to each donor and data collection and analysis were not performed blind to the conditions of the experiments.”
Data and code availability
Custom R code and data used to generate transcriptomics related figures can be downloaded from: https://github.com/AllenInstitute/L1_rosehip. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Further information on methodology is available in Life Sciences Reporting Summary of the paper.
Supplementary Material
Acknowledgements
The authors thank the Allen Institute for Brain Science founders, Paul G. Allen and Jody Allen, for their vision, encouragement and support. This work was supported by the ERC Interimpact project (GT), the National Institute of Mental Health (USA) RFA MH 17 210 (ESL, GT), the Hungarian Academy of Sciences (GT), the National Research, Development and Innovation Office of Hungary GINOP-2.3.2-15-2016-00018, VKSZ-14-1-2015-0155 and by the National Brain Research Program, Hungary (GT). The authors thank Lena Christiansen and Fan Zhang from Illumina, Inc. for their assistance with RNA sequencing.
Footnotes
Accession codes
Author Contributions
Conceptualization, E.S.L., R.S.L., G.T.
Methodology, R.D.H., M.N., J.L.C., P.B., L.G.P., G.T.
Validation, J.L.C., S.L.D., G.M., G.T.
Formal Analysis, T.E.B., B.D.A., J.M.M., J.A.M., P.V., M.R., S.B., R.H.S., E.B., J.B., G.O., G.M.
Investigation, R.D.H., M.N., J.L.C., F.D.F., S.S., K.A.S., A.W., D.N.T., E.B., J.B., A.K.K., N.F., B.K., M.R., G.M., A.O.,G.O., G.T.
Resources, E.S.L., F.J.S., N.J.S., R.H.S., R.S.L., G.T.
Data Curation, T.E.B., B.D.A., J.M.M., J.A.M., P.V., S.B.
Writing – Original Draft, T.E.B. R.D.H., J.L.C., J.A.M., E.S.L., E.B., G.O., G.T.
Writing – Review and Editing, T.E.B., R.D.H., J.A.M., R.H.S., R.S.L., E.S.L., G.T.
Visualization, T.E.B., R.D.H., J.L.C., S.L.D., J.A.M., E.B., G.M., G.O.
Supervision, E.S.L., F.J.S., N.J.S., R.H.S., R.S.L, G.T.
Project Administration, E.S.L., S.M.S., G.T.
Funding Acquisition, E.S.L., F.J.S., R.S.L., G.T.
Competing Financial Interests
The authors declare no competing financial interest.
References
- 1.DeFelipe J, Jones EG. Cajal on the Cerebral Cortex. Oxford University Press; 1988. [Google Scholar]
- 2.Ascoli GA, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markram H, et al. Reconstruction and Simulation of Neocortical Microcircuitry. Cell. 2015;163:456–492. doi: 10.1016/j.cell.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 4.Tasic B, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science (80-. ) 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 6.Macosko E, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang X, et al. Principles of connectivity among morphologically defined cell types in adult neocortex. Science (80-. ) 2015;350:aac9462–aac9462. doi: 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyal G, et al. Unique membrane properties and enhanced signal processing in human neocortical neurons. Elife. 2016;5 doi: 10.7554/eLife.16553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B, et al. A Subtype of Inhibitory Interneuron with Intrinsic Persistent Activity in Human and Monkey Neocortex. Cell Reports. 2015;10 doi: 10.1016/j.celrep.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 10.Molnár G, et al. Complex events initiated by individual spikes in the human cerebral cortex. PLoS Biol. 2008;6:e222. doi: 10.1371/journal.pbio.0060222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molnár G, et al. Human pyramidal to interneuron synapses are mediated by multi-vesicular release and multiple docked vesicles. Elife. 2016;5 doi: 10.7554/eLife.18167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Testa-Silva G, et al. Human synapses show a wide temporal window for spike-timing-dependent plasticity. Front Synaptic Neurosci. 2010;2:12. doi: 10.3389/fnsyn.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verhoog MB, et al. Mechanisms underlying the rules for associative plasticity at adult human neocortical synapses. J Neurosci. 2013;33:17197–208. doi: 10.1523/JNEUROSCI.3158-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szegedi V, et al. Plasticity in Single Axon Glutamatergic Connection to GABAergic Interneurons Regulates Complex Events in the Human Neocortex. PLoS Biol. 2016;14:e2000237. doi: 10.1371/journal.pbio.2000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elston GN. Pyramidal cells of the frontal lobe: all the more spinous to think with. J Neurosci. 2000;20:RC95. doi: 10.1523/JNEUROSCI.20-18-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morales J, et al. Random positions of dendritic spines in human cerebral cortex. J Neurosci. 2014;34:10078–84. doi: 10.1523/JNEUROSCI.1085-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohan H, et al. Dendritic and Axonal Architecture of Individual Pyramidal Neurons across Layers of Adult Human Neocortex. Cereb Cortex. 2015;25:4839–4853. doi: 10.1093/cercor/bhv188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grindberg RV, et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacar B, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7 doi: 10.1038/ncomms11022. 11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–81. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 21.Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- 22.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–90. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olah S, et al. Output of neurogliaform cells to various neuron types in the human and rat cerebral cortex. Front Neural Circuits. 2007;1:4. doi: 10.3389/neuro.04.004.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kisvárday ZF, et al. Synapses, axonal and dendritic patterns of GABA-immunoreactive neurons in human cerebral cortex. Brain. 1990:793–812. doi: 10.1093/brain/113.3.793. [DOI] [PubMed] [Google Scholar]
- 25.Olah S, et al. Regulation of cortical microcircuits by unitary GABA-mediated volume transmission. Nature. 2009;461:1278–1281. doi: 10.1038/nature08503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varga C, Tamas G, Barzo P, Olah S, Somogyi P. Molecular and electrophysiological characterization of GABAergic interneurons expressing the transcription factor COUP-TFII in the adult human temporal cortex. Cereb Cortex. 2015;25:4430–4449. doi: 10.1093/cercor/bhv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zemankovics R, Káli S, Paulsen O, Freund TF, Hájos N. Differences in subthreshold resonance of hippocampal pyramidal cells and interneurons: the role of h-current and passive membrane characteristics. J Physiol. 2010;588:2109–2132. doi: 10.1113/jphysiol.2009.185975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katona I, Freund TF. Multiple functions of endocannabinoid signaling in the brain. Annu Rev Neurosci. 2012;35:529–58. doi: 10.1146/annurev-neuro-062111-150420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerekes BP, et al. Combined two-photon imaging, electrophysiological, and anatomical investigation of the human neocortex in vitro. Neurophotonics. 2014;1:011013. doi: 10.1117/1.NPh.1.1.011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freund TF, Buzsáki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 31.Hawrylycz MJ, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 33.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of Cortical Interneuron Subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeFelipe J. Types of neurons, synaptic connections and chemical characteristics of cells immunoreactive for calbindin-D28K, parvalbumin and calretinin in the neocortex. J Chem Neuroanat. 1997;14:1–19. doi: 10.1016/s0891-0618(97)10013-8. [DOI] [PubMed] [Google Scholar]
- 35.von Economo C. Eine neue Art Spezialzellen des Lobus cinguli and Lobus insulae. Zschr ges Neurol. Psychiatry. 1926;100:706–712. Zschr ges Neurology and Psychiatry. [Google Scholar]
- 36.Nimchinsky EA, Vogt BA, Morrison JH, Hof PR. Spindle neurons of the human anterior cingul. Ate cortex. J Comp Neurol. 1995;355:27–37. doi: 10.1002/cne.903550106. [DOI] [PubMed] [Google Scholar]
- 37.Gabbott PLA. ‘Subpial Fan Cell’ - A Class of Calretinin Neuron in Layer 1 of Adult Monkey Prefrontal Cortex. Front Neuroanat. 2016;10:28. doi: 10.3389/fnana.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somogyi P. A specific ‘axo-axonal’ interneuron in the visual cortex of the rat. Brain Res. 1977;136:345–50. doi: 10.1016/0006-8993(77)90808-3. [DOI] [PubMed] [Google Scholar]
- 40.Tamas G, Buhl EH, Somogyi P. Fast IPSPs elicited via multiple synaptic release sites by distinct types of GABAergic neuron in the cat visual cortex. J Physiol. 1997;500:715–738. doi: 10.1113/jphysiol.1997.sp022054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kisvárday ZF, Martin KAC, Whitteridge D, Somogyi P. Synaptic connections of intracellularly filled clutch cells: A type of small basket cell in the visual cortex of the cat. J Comp Neurol. 1985;241:111–137. doi: 10.1002/cne.902410202. [DOI] [PubMed] [Google Scholar]
- 42.Lee AJ, et al. Canonical Organization of Layer 1 Neuron-Led Cortical Inhibitory and Disinhibitory Interneuronal Circuits. Cereb Cortex. 2015;25:2114–26. doi: 10.1093/cercor/bhu020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tremblay R, Lee S, Rudy B. GABAergic Interneurons in the Neocortex: From Cellular Properties to Circuits. Neuron. 2016;91:260–292. doi: 10.1016/j.neuron.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gjorgjieva J, Drion G, Marder E. Computational implications of biophysical diversity and multiple timescales in neurons and synapses for circuit performance. Curr Opin Neurobiol. 2016;37:44–52. doi: 10.1016/j.conb.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palmer LM, et al. The Cellular Basis of GABAB-Mediated Interhemispheric Inhibition. Science (80-. ) 2012;335 doi: 10.1126/science.1217276. [DOI] [PubMed] [Google Scholar]
- 46.Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–7. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borg I, et al. Disruption of Netrin G1 by a balanced chromosome translocation in a girl with Rett syndrome. Eur J Hum Genet. 2005;13:921–7. doi: 10.1038/sj.ejhg.5201429. [DOI] [PubMed] [Google Scholar]
- 48.Molinari F, et al. Truncating Neurotrypsin Mutation in Autosomal Recessive Nonsyndromic Mental Retardation. Science (80-. ) 2002;298:1779–1781. doi: 10.1126/science.1076521. [DOI] [PubMed] [Google Scholar]
- 49.Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. 2014;6:114–8. [PMC free article] [PubMed] [Google Scholar]
- 50.Cavanaugh SE, Pippin JJ, Barnard ND. Animal models of Alzheimer disease: historical pitfalls and a path forward. ALTEX. 2014;31:279–302. doi: 10.14573/altex.1310071. [DOI] [PubMed] [Google Scholar]
- 51.Krishnaswami SR, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–5. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 56.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–2. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aevermann B, et al. Production of a preliminary quality control pipeline for single nuclei rna-seq and its application in the analysis of cell type diversity of post-mortem human brain neocortex. Pac Symp Biocomput. 2016;22:564–575. doi: 10.1142/9789813207813_0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.R Development Core Team. R: A Language and Environment for Statistical Computing. Vol. 0. R Found Stat Comput; Vienna Austria: 2016. {ISBN} 3-900051-07-0. [Google Scholar]
- 60.Lawrence M, et al. Software for Computing and Annotating Genomic Ranges. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan J, et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat Methods. 2016;13:241–244. doi: 10.1038/nmeth.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Der Maaten LJP, Hinton GE. Visualizing high-dimensional data using t-sne. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 64.Ester M, Kriegel HP, Sander J, Xu X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining; 1996. pp. 226–231. doi: 10.1.1.71.1980. [Google Scholar]
- 65.Liu Y, Hayes DN, Nobel A, Marron JS. Statistical Significance of Clustering for High-Dimension, Low–Sample Size Data. J Am Stat Assoc. 2008;103:1281–1293. [Google Scholar]
- 66.Tasic B, et al. Shared and distinct transcriptomic cell types across neocortical areas. bioRxiv. 2017 doi: 10.1101/229542. 229542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritchie M, et al. limma powers di erential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng H, et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell. 2012;149:483–96. doi: 10.1016/j.cell.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Paxinos G, Franklin K. Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates. Academic Press; 2012. [Google Scholar]
- 70.Faragó N, et al. Digital PCR to determine the number of transcripts from single neurons after patch-clamp recording. Biotechniques. 2013;54:327–36. doi: 10.2144/000114029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom R code and data used to generate transcriptomics related figures can be downloaded from: https://github.com/AllenInstitute/L1_rosehip. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Further information on methodology is available in Life Sciences Reporting Summary of the paper.