Abstract
We describe dropdead1-1 (ded1), an EMS-induced recessive lesion mimic mutant of sorghum. It is characterized by the formation of spreading necrotic lesions that share many attributes with those associated with the maize lethal leaf spot1 (lls1) and Arabidopsis accelerated cell death1 (acd1) mutation. We show that as in lls1, ded1 lesions are initiated by wounding and require light for continued propagation, and that loss of chloroplast integrity is responsible for ded1 cell death. Consistent with these parallels, we demonstrate that ded1 is an ortholog of lls1 and encodes pheophorbide a oxidase (PaO) with 93% identity at the protein level. The mutant ded1 allele resulted from a stop codon-inducing single base pair change in exon 6 of the sorghum ortholog of lls1. The ded1 transcript was rapidly and transiently induced after wounding and substantially elevated in leaves containing ded1 lesions. Given that PaO is a key enzyme of the chlorophyll degradation pathway, its dysfunction would result in the accumulation of pheophorbide, a potent photosensitizer that results in the production of singlet oxygen. Consistent with this, cell death associated with ded1 lesions is most likely caused by singlet oxygen as our results exclude superoxide and H2O2 from this role. We explore the signal responsible for the propagation of lesions affecting both ded1 and lls1 lesions and find that both developmental age and ethylene increase the rate of lesion expansion in both mutants.
Introduction
A number of plant mutants undergo spontaneous cell death in the absence of any obvious stress or injury (reviewed in [1,2]). As such lesions often resemble those produced during plant interactions with pathogens, these mutants may be referred to as “disease lesions mimics” or simply “lesion mimics” [1,2]. A subset of lesion mimic mutants do encode disease signaling components resulting in constitutive hypersensitive-response [1,2]. Dominant lesion mimic mutants typically encode constitutively active alleles of genes that activate cell death. Recessive lesion mimics may encode negative regulators of cell death signaling. Recessive mutants may also encode alleles of host components (guardees) that are guarded by host immune receptors (guards) if the guardee is directly or indirectly targeted by pathogen effectors [1]. But not all lesion mimics are mutations in disease signaling components. Lesion mimics have been isolated that encode enzymes that when disrupted result in the accumulation of biochemical intermediates, including phototoxic compounds from chlorophyll metabolism. The first lesion mimic mutant was described in maize by Emerson in 1923 who described the phenotype as “blotched leaf” [3]. The observation that these maize mutants phenocopy disease symptoms was made later, by Neuffer in the 1970s [4]. Disease lesion mimics have been found in many plant species including Arabidopsis, barley and rice, where they were variously named as accelerated cell death, lesions simulating disease or spotted mutants [5–7]. More than 50 lesion mimic mutants have been described in maize and a similar number of mutants are present in Arabidopsis [1,8,9].
The lesions on lesion mimic mutants can be determinative lesions, that reach a defined maximum size, or produce propagative lesions that expand until they engulf the entire organ [1]. In the determinative class, lesions initiate often profusely but then remain restricted in size, resembling a massive number of hypersensitive cell death (HR) sites. It is presumed that lesions in the determinative class result from impairments that either lower the threshold for cell death initiation or cause the buildup of factors or metabolites to cytotoxic levels. In contrast, propagative mimics are thought to result from defects in those genes that negatively regulate cell death in plants [1]. The ubiquitous feature of lesion mimics is their association with aberrant cell death. Thus, this class of mutants should identify genes and mechanisms that control programmed cell death (PCD) [1,2].
PCD in plants plays a key role in development and the defense of plants against biotic and abiotic stresses [1,10–12]. However, our knowledge of how plants accomplish PCD remains limited. In contrast, great advances have been made in animals demonstrating the cellular machinery of a major PCD pathway, termed apoptosis, is largely conserved from worms to humans [13–19]. No such framework of genes and mechanisms has yet emerged for the control and execution of PCD in plants, although several proteases have been identified as having a role in plant PCD [20]. Triggering of PCD by defects in signaling for disease resistance results reactive oxygen species (ROS) production and constitutive expression of defense genes [21]. ROS production is a normal component of the disease resistance signaling and adaptive response to pathogens. As a result, many mutants with altered disease resistance also affect aberrant PCD or ROS production [22–24]. This inappropriate production of reaction oxygen species is a feature that is shared across all lesions mimics, whether arising from errors in metabolism or defects in defense signaling. The exploration of lesion mimic phenotypes has assisted in identifying varied mechanisms capable of inducing PCD in plants as genes underlying a number of these mutants have been cloned and characterized [9,25]. In addition to disease signaling and metabolic defects, transcriptional sensors of ROS levels have also been identified as capable of producing lesion mimic mutants. One example is the mutant LSD1, initially thought to result in cell death because of disruption of ROS associated homeostasis [6,26,27]. However, a recent study has shown that cell death associated with lsd1 is suppressed by mutations in an NLR [28].
Approximately 50% of the lesion mimic mutations result from mutations in genes not involved in disease resistance. A good proportion of these are the result of errors in metabolism, especially due to the accumulation of phototoxic intermediates. Two examples involve mutations that either block the production of chlorophyll or those that block the degradation of chlorophyll. Loss of function at acd2 in Arabidopsis blocks chlorophyll degradation and results in a lesion phenotype from the accumulation of a phototoxic catabolic intermediate that produces singlet oxygen in the light [29]. As an indication of the complexity in interpreting these mutants, overexpression of acd2 was shown to suppress cell death and disease symptom expression in an incompatible pseudomonas interaction with Arabidopsis, yet this effect of ACD2 overexpression was not dependent on chlorophyll [30]. A similar lesion phenotype is affected by mutations in lls1 in maize and acd1 in Arabidopsis. These genes encode the pheophorbide a oxidase (PaO) activity, one step upstream of acd2 in chlorophyll catabolism, and blocking these genes accumulates a phototoxic catabolic intermediate that also produces singlet oxygen in the light [31,32]. Thus, lesions from ROS production via singlet-oxygen can be powered by aberrant light capture.
These studies highlight the complexity of mechanisms regulating cell death in plants and it remains largely unaddressed how cell death associated with most lesion mimics is initiated and propagated. One exception in this regard is the afformentioned recessive mutant of maize, lls1 [33]. Initiation of lls1 lesions is triggered in response to cell damage, and propagation of these lesions by continued enlargement is fuelled by chemical conversion of photosynthetically-active light into singlet oxygen [34]. The PAO enzyme encoded by lls1 catalyzes the cleavage of the pheophorbide a macrocycle yielding the open-tetrapyrrolic backbone structure of different types of Chl catabolites found in senescent leaves and fruits [35,36]. The formation of lesions in plants lacking PaO is conserved across plants, as expected for a metabolic defect that induces ROS and cell death. Mutants in PaO in multiple plant species including the Arabidopsis accelerated cell death 1 (acd1) mutant, rice early senescence1 (eas1) mutant, VIGS of PaO in tomato, and knockdown of all the wheat PaO orthologs results in lesion formation [5,37–41].
The continued enlargement of lesions in propagative mutants, such as lls1, requires a diffusible signal. The nature of this signal in lls1 is unknown, but chloroplasts play a central role in the propagated cell-autonomous cell death in lls1 lesions [33,34]. Extensive work in Arabidopsis has pointed to the existence of multiple factors that promote or suppress cell death in response to singlet oxygen [42–45], the ROS that results from photoactivation of pheophorbide A [44,46]. Multiple studies of singlet oxygen-induced lesions have identified pathways that modulate lesion formation in response to singlet oxygen including salicylic acid and ethylene signaling [46,47]. This suggests that modulation of hormone levels, or perception, might enhance or suppress cell death in mutants that accumulate singlet-oxygen evolving metabolic intermediates. Singlet oxygen production in the chloroplast is a feature, particularly in high light, of the light harvesting process and not just an aberrant consequence of metabolic defects. Multiple chloroplast components play roles in quenching singlet oxygen, including carotenoids and tocopherols. A number of mutants have defined proteins of unknown function as modulators of singlet oxygen-induced lesions, including executer1 and executer2, and flu [42,48]. The existence of such proteins, and the modulation of singlet oxygen lesions by a wide variety of biochemical and physiological pathways, raises the possibility that singlet oxygen production could be a component of a disease response as either a mechanism to produce cytotoxic ROS during adaptive disease resistance or as a mechanism to induce cell death for nectrotropic growth of pathogens.
The focus of this paper is an EMS-induced recessive lesion mimic mutation from sorghum that we name dropdead1 (ded1). Many features of this mutation were strikingly similar to that of lls1. Here we demonstrate that the ded1 mutant results from a single base pair change in a sorghum ortholog of the maize lls1 gene. Like the lls1 mutant, ded1 lesions are mediated by a light-dependent mechanism that disrupts the integrity of chloroplasts to kill ded1 cells. Neither superoxide nor H2O2 were detectably elevated and appear not to be involved in this cell death which, given the PAO deficiency caused by ded1-1 and lls1-1 mutations, is likely mediated by singlet oxygen.
Results
Phenotypic characterization of ded1
The ded1 is a recessive mutation that originated in an EMS mutagenized M2 population of P898012 developed at Purdue University. The ded1 mutants form necrotic lesions that expand and coalesce, ultimately consuming whole leaves (Fig 1A–1D). Lesion formation is under some developmental control and initiate on mature leaves (Fig 1D). Repeated observation of successive generations of mutant families in more than five field seasons indicated that both phenotype expression and the developmental control of lesion initiation were stable and reproducible. The same phenotypes were observed in greenhouse-grown ded1-1 mutants. Lesions form on the first leaf of greenhouse-grown plants two to three weeks after emergence. Lesions initiate at or near the tip of the first leaf and progress toward the developmentally younger tissue at the base of the leaf. Over a few days, the area containing dead and dying cells increases until independently initiated lesion sectors merge with each other. This pattern of lesion formation and spread is repeated on all successive leaves and completely blights all of the foliage shortly after flowering. ded1 lesions often accumulate red pigments (presumably anthocyanins and phlobaphenes) in the center of the lesion and to a lesser extent in concentric rings, giving the lesions a characteristic “bull’s eye” appearance (Fig 1A and 1B).
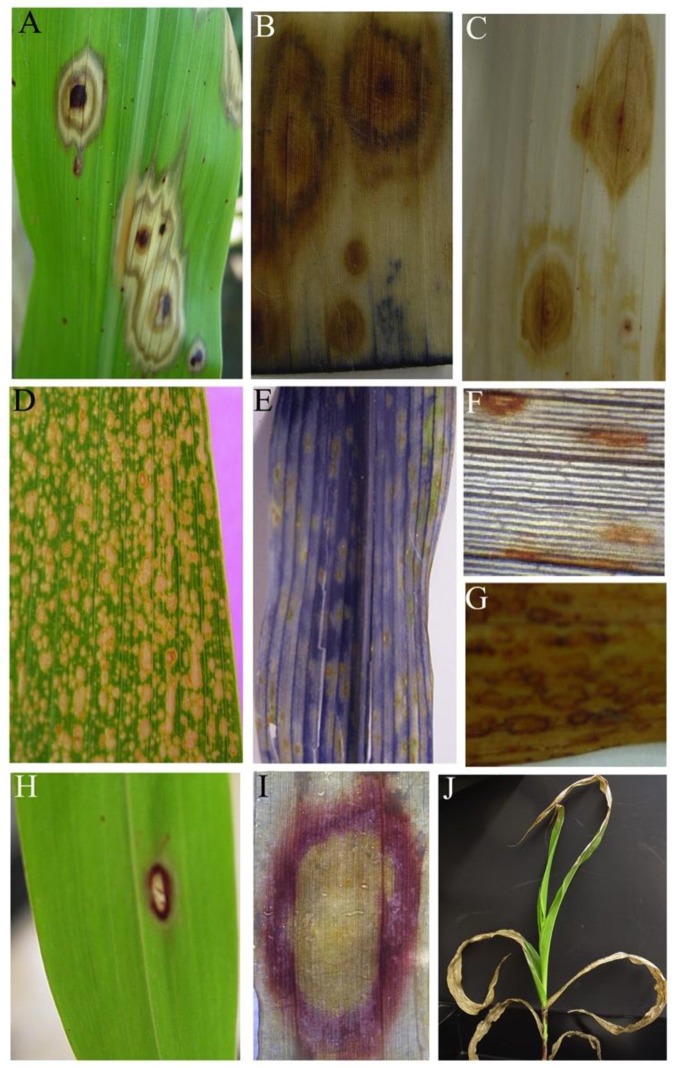
Fig 1. Phenotypic manifestation of ded1.
(A, B) Typical morphology of ded1 lesions forming spontaneously on leaves of field-grown mutants. Note the bull’s eye appearance of the lesions. (C) Typical blighting of a ded1 leaf as lesions spread and merge with each other (D) A field grown ded1 plant exhibiting developmental progression of the mutant phenotype (E) Maize lls1 lesions for comparison with ded1 lesions (F) Leaf of a ded1 plant subjected to 8 pin-prick wounds (black arrows) in two regions, one was covered with aluminum foil and one was exposed to light. Note the lack of development of lesions on the region of the leaf that had been covered with the foil (border indicated by white dashed line).
Injuring the mutant leaf tissue can trigger lesion formation in ded1-1 mutants. Mechanical penetration of leaf tissue of sufficient developmental age resulted in the rapid formation of lesions (Fig 1F). Wounding of younger leaf tissue resulted in wound sites that remained unaffected so long as the leaf was immature. However, when the leaf bearing the wound aged, competency to bear lesions developed in the injured leaf and some of the wound punctures gave rise to typical expanding ded1 lesions.
The continued expansion of ded1 lesions requires light. Covering wounded portions of mutant leaves with aluminum foil prevented the formation of ded1 lesions (Fig 1F). This phenomena was also true for preexisting lesions undergoing expansion, which could be halted by protecting leaves from light (data not shown), clearly demonstrating the importance of light in ded1 lesion ontogeny.
ded1 lesion growth is associated with structural disruption of chloroplasts
The visible phenotype of ded1 lesions, their initiation in response to physical injury, and dependence on light are similar to the maize lls1 mutant. lls1 lesions are characterized by swelling and disruption of chloroplasts [34]. To further explore the similarity of ded1 and lls1, we used transmission electron microscopy to examine the chloroplast ultrastructure during lesion formation in ded1 cells. Pin-prick wounds were made on wildtype and ded1 mutants to initiate lesions. Cells surrounding pin-prick wounds were examined and compared to uninjured tissue collected from an equivalent area of the same ded1 or wild-type leaf on the opposite side of the mid-rib. Cells were examined at 21h and 42h after wounding, and images collected from 21h tissue are shown (Figs 2 and 3). Although the lesions had progressed farther from the wound site in the 42h samples, identical ultrastructural changes were found at both 21h and 42h after wounding. Like lls1 mutants, ded1 cells from uninjured tissue were indistinguishable from wild-type cells (Figs 2a, 2c, 3a and 3c). As with lls1 cells, cells near pin-prick damaged ded1, but not wildtype or uninjured ded1 cells, exhibited dramatic changes in chloroplasts of both bundle sheath (BS) and mesophyll (M) cells (Figs 2 and 3). Chloroplasts of BS cells adjacent to the wound looked disorganized and convoluted and their thylakoid membranes appeared to have folded over upon themselves (Fig 2d). Chloroplasts in M cells adjacent to wounded ded1 cells were swollen (Fig 3d). The thylakoid membrane stacks were somewhat pulled apart in these swollen chloroplasts, but maintained a normal organization (Fig 3d). The envelope of these swollen chloroplasts appeared to be intact and no ruptured M chloroplasts were observed. Just as in lls1 mutants, the severity of chloroplast ultrastructural changes in BS and M cells decreased with the distance from the injured ded1 cells (data not shown). Taken together, these data suggest that, as in the lls1 mutation, the changes that result in cell death during propagation of lesions in ded1 plants are initiated in chloroplasts.
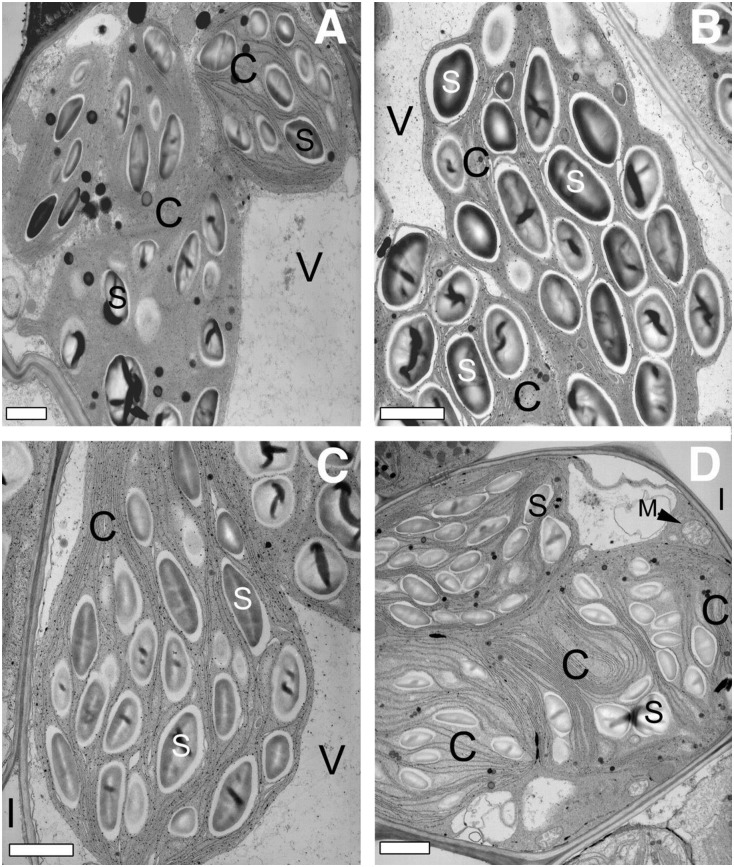
Fig 2. Transmission electron microscopy of bundle sheath cells in uninjured and injured (21 hours post wounding) wild-type and ded1 leaves.
(A) Bundle sheath cell in uninjured wild-type leaf tissue. (B) Bundle sheath cell adjoining dead cells in injured wild-type leaf tissue. (C) Bundle sheath cell in uninjured ded1 leaf tissue. (D) Bundle sheath cell adjoining dead cell in injured ded1 leaf tissue. Note the rounded appearance of the chloroplasts and folding of the thylakoid membranes. Bars = 1 μm. V = vacuole; S = starch; C = chloroplast; I = intercellular space; M = mitochondrion.
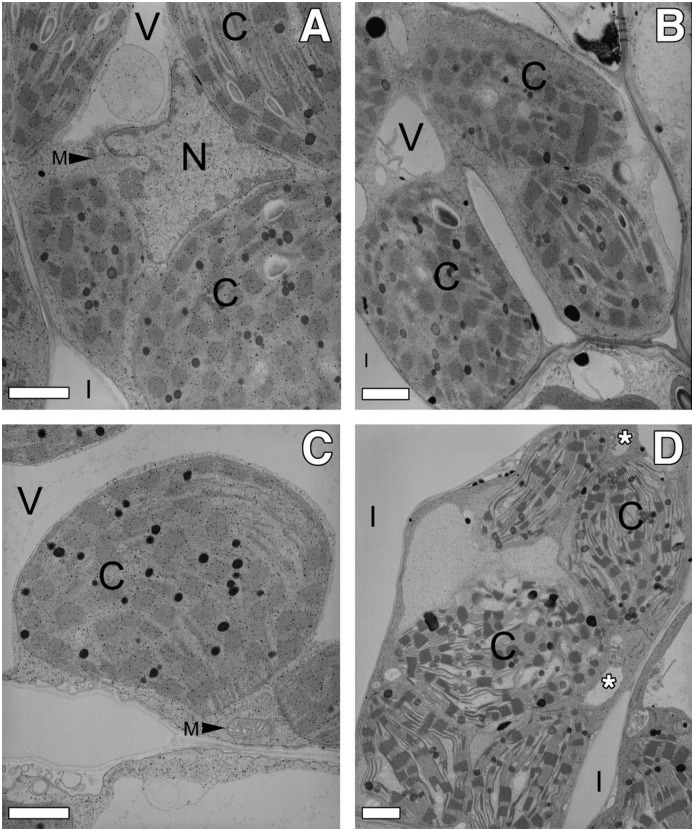
Fig 3. Transmission electron microscopy of mesophyll cells in uninjured and injured (21 hours post wounding) wild-type and ded1 leaves.
(A) Mesophyll cell in uninjured wild-type leaf tissue. (B) Mesophyll cell adjoining dead cells in injured wild-type leaf tissue. (C) Mesophyll cell in uninjured ded1 leaf tissue. (D) Mesophyll cell adjoining dead cell in injured ded1 leaf tissue. Note the swelling of the chloroplasts. Asterisks indicate location of cytoplasmic vacuoles. Bars = 1 μm. V = vacuole; C = chloroplast; I = intercellular space; M = mitochondrion; N = nucleus.
In addition to the chloroplast changes, M and BS cells of wounded ded1 plants displayed vacuolization of their cytoplasm (Figs 2d and 3d). Though their presence was clear and reproducible, the origin of these vacuoles was not clear. However, the central vacuole was conspicuously absent in all M and BS cells that exhibited severely altered chloroplast morphology, suggesting a loss of tonoplast integrity. Cytoplasmic vacuolization was never seen in the absence of chloroplast structural changes. Mitochondria, Golgi and endoplasmic reticuli (ER) in BS or M cells of injured ded1 tissue were comparatively normal, even in cells displaying dramatically altered chloroplast structure (Figs 2d and 3d). Collapsed M and BS cell corpses appeared condensed and shrunken (data not shown).
Not all of the ultrastructural changes in dying ded1 were similar to lls1. The nuclei of wounded lls1 cells displayed increased heterochromatin located at the nuclear envelope [34]. No such increase in heterochromatin was observed in the nuclei of dying ded1 M and BS cells. Thus this morphological change, which occurs in apoptotic cells [13], is not necessary for cell death associated with ded1 lesions.
The ded1 gene encodes the sorghum ortholog of lls1
The similarity between the ded1 and lls1 phenotypes suggested that they may result from mutations in the same gene. The sorghum ortholog of lls1, Sobic.001G504900 resides in a syntenic position and shows one-to-one orthology with the maize lls1 gene. Sobic.001G504900 was amplified and sequenced to see how it compared between the ded1 mutant and its wild-type progenitor. A PCR fragment from exon 5 through exon 7 of approximately 1.1 kb was obtained from both the ded1 mutant and progenitor DNAs. These products, designated dd57-11M and dd57-11P for mutant and progenitor amplicons, respectively, were cloned and sequenced. The ded1 mutant encodes a transversion of C to A at the 77,311,393 nucleotide position of chromosome 1 in sorghum genome (version 3.1.1), within the coding sequence of Sobic.001G504900. This substitution causes a nonsense mutation that converts the Serine encoding TCA codon at the 437 amino acid position of the Sobic.001G504900.1.p protein model to an ochre stop codon. Thus, Sobic.001G504900 in ded1-1 now encodes a truncated, and presumably non-functional, DED1 protein.
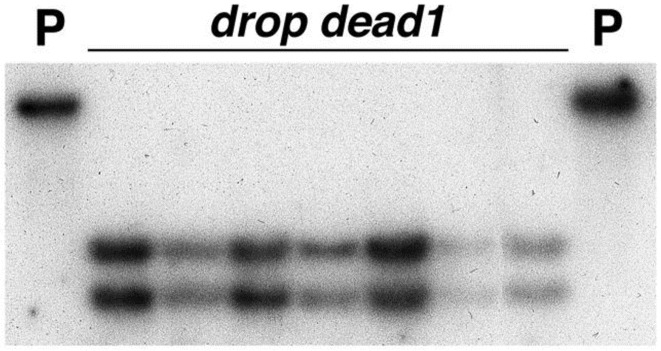
This mutation created a DraI cleavage site in the ded1-1 allele that was absent in its progenitor’s sequence. DNA from a total of 45 ded1 plants and 5 wild-type plants homozygous for the progenitor allele was isolated and digested with DraI and EcoRI restriction endonucleases. DNA blot hybridization analysis of these DNAs using dd57-11M as a hybridization probe demonstrated that the DraI/EcoRI digested DNA from wild-type plants possessed a single band of approximately 4.4-kb (Fig 4). Two bands (approximately 2.7-kb and 1.7-kb) were present in lanes containing DraI/EcoRI digested DNA isolated from ded1 plants (Fig 4). All ded1 mutants analyzed in this study were found to possess the DraI polymorphism indicating perfect co-segregation of this polymorphism and the mutant phenotype in these 45 mutants. Cosegregation was confirmed using flanking markers genotyped by Restriction Fragment Length Polymorpism (RFLP) scoring of southern blots (data not shown). These confirmed the position of DED1 and placed RFLP markers PIO000689 and PIO000603 mapped 1cM and 7cM distal to ded1 and UMC157 7cM proximal to ded1. Another marker PIO000640 was found to reside 18 cM distal to ded1.
Fig 4. DNA blot hybridization analysis demonstrating linkage of the ded1 phenotype with a restriction fragment length polymorphism generated from the single base pair change in ded1 allele.
10 μg of DNA pooled from 5 ded1 plants (labeled “dropdead1”) and 2 or 3 wild-type (labeled “P”) plants homozygous for the progenitor allele was digested with DraI and EcoRI restriction endonucleases, run on a gel, blotted and hybridized with 32P-labeled dd57-11M.
An additional F2 population was generated between ded1 and the Shanqui Red inbred line. Fifty plants from this F2 population, 45 mutant and 5 wild-type, were genotyped for these RFLP. The RFLP marker UMC 157 was found to map about 4 cM away from the ded1 locus with this population. Whether it located proximally or distally in relation to ded1 could not be established because both PIO000603 and PIO000689 segregated completely with the ded1 locus. Even PIO000640, which mapped 18 cM distal to lls1 in maize, failed to recombine with the ded1 gene in this population (data not shown). As expected, the DraI polymorphism within Sobic.001G504900 co-segregated perfectly with the mutant phenotype. These mapping data, and the presence of a nonsense mutation within Sobic.001G504900 demonstrate that ded1 is encoded by the ortholog of the maize lls1 gene.
The ded1 gene is rapidly induced by wounding in a transient fashion
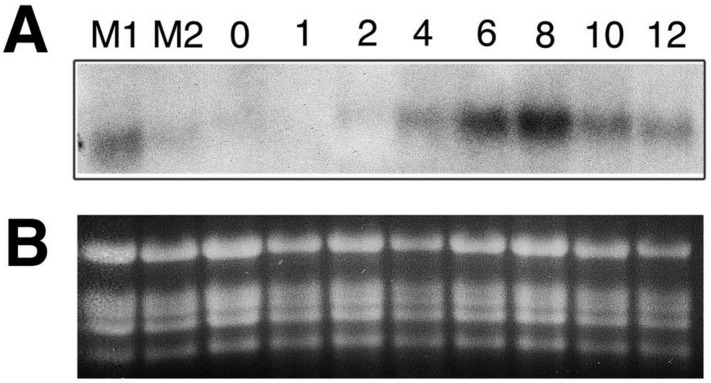
RNA gel blot analyses of DED1 transcript accumulation identified no change in transcript size, supporting the single nucleotide substitution origin of the ded1-1 allele. However, the accumulation of DED1 transcript in ded1-1 was elevated many fold, indicating a lack of nonsense-mediated decay of this allele’s gene product. The level of DED1 transcript appeared to roughly predict the number of lesions on the leaf from which the RNA was extracted (Fig 5 lanes M1 and M2). Mutant leaves that had a higher number of lesions (lane M1) had a larger increase in the level of the ded1 transcript than the mutant leaves that had a relatively fewer number of ded1 lesions (lane M2). This suggested that DED1 was induced during lesion formation and that, like the ded1 lesions, the ded1 gene expression was responsive to tissue damage resulting from spreading lesions. To investigate this possibility further, leaves of 5 week-old plants were gently rubbed with carborundum powder [49]. This maximized the number of cells experiencing the wounding stimulus. Total RNA was isolated at 1, 2, 4, 6, 8, 10 and 12h after wounding and subjected to RNA blot hybridization analysis using dd57-11 as a probe. As shown in Fig 5, a rapid and transient induction of DED1 mRNA was observed in response to wounding. The DED1 transcript started accumulating around 4h after wounding and reached the peak at 8h. This peak was followed by a sharp decline and return to the amount of the DED1 transcript detected at 4h by 12h after wounding (Fig 5).
Fig 5. RNA blot hybridization analysis of RNAs isolated from ded1 and wild-type sorghum leaves.
Wild-type leaves were wounded by gently rubbing carborundum powder down the entire length of the leaf. Total RNA was isolated from tissue collected at 1, 2, 4, 6, 8, 10 and 12h post wounding, as well as prior to wounding (0 hours). (A) 10 μg total RNA was loaded per lane, size fractionated by electrophoresis, blotted and hybridized with the 32P-labeled dd57-11M fragment. RNA in lane M2 was isolated from a leaf of a ded1 plant with a low number of lesions, whereas the RNA in Lane M1 was isolated from a ded1 plant leaf with a moderate number of lesions and contains a higher lls1 signal. (B) Ethidium bromide stained gel showing the rRNA bands was used as a loading control.
Ethylene but neither H2O2 nor superoxides were associated with the propagation of cell death in ded1-1
The dependence of ded1 lesion formation on light and the role of the ded1 gene product in chlorophyll catabolism suggests that reactive oxygen species may be involved in the formation of lesions. To address this, we assayed the production superoxide and H2O2 in actively expanding lesions on ded1-1 leaves using NBT and DAB staining (Fig 6). These histochemical assays provide in-vivo and in-situ visualization of respective oxygen free radicals with high sensitivity [50]. The Les*-101 maize mutant was used as a positive control. Presence of O2- is indicated by the blue coloration and presence of H2O2 is indicated by reddish-brown coloration, as shown in the lesions of Les*-101 mutant maize leaf lesions (Fig 6D–6H). Cell death in expanding ded1 lesions occurs at the boundary, if ROS production causes cell death we expect a positive stain reaction only at the lesion periphery similar to the red ring of anthocyanins/phlobaphenes around ded1 lesions that are protected from light (Fig 6H and 6I). No positive reaction was observed with either NBT or DAB assays in ded1-1 leaves (Fig 6B and 6C), demonstrating a lack of superoxide and H2O2 involvement in ded1 lesion ontogeny.
Fig 6. Induction and propagation of cell death associated with ded1 lesions.
(A) Actively expanding ded1 lesions. (B,C) Lack of superoxide and H2O2 production in and around ded1 lesions. Tissue shown in b was examined for superoxide using the NBT staining assay. Lesions in c were stained with DAB to detect H2O2. (D) A maize leaf exhibiting Les*-101 lesions. (E) Positive staining of a Les*-101 leaf with NBT, indicating the presence of superoxide. (F,G) Positive staining of Les*-101 lesions with DAB, indicating the presence of H2O2. (H) A ring of red pigments that forms at the periphery of ded1 lesions following protection from light for at least 48h. (I) A close up (following clearing of chlorophyll) of the red ring circling a ded1 lesion. (J) Rapid collapse of mature leaves of ded1 plants following spray with ethephon, an ethylene releasing chemical.
The requirement for wounding raised the question of what aspect of wound signaling could mediate lesion expansion in ded1-1 mutants. Previous work demonstrated that the acd1 mutant was dramatically enhanced by the application of ethylene [5]. We utilized the commercial plant growth regulator ethephon 0.01% as an aqueous spray to increase ethylene levels in planta. Ethephon is efficiently converted from a stable form into the plant hormone ethylene by cellular metabolism. Just as was the case for acd1 mutants [5] the ded1-1 mutant was profoundly sensitive to ethephon resulting in collapse of all sprayed tissue within days of application (Fig 6J). As this had not been investigated in the lls1 mutant we tested application of ethephon to lls1 and found similar dramatic induction of lesions following 0.01% ethephon spray (data not shown). The extent of lesion enhancement resulted in complete consumption of all sprayed leaves in all plants in both the ded1-1 sorghum and lls1 maize mutants. Remarkably, ethephon application induced lesions and resulted in complete collapse of all leaves in plants as young as 3 weeks-post-sowing (Fig 6J), suggesting that ethylene was sufficient to induce chlorophyll degradation and expose the defect in ded1-1.
The ded1 gene is developmentally induced in adult leaves, during pathogen stress and during osmotic stress
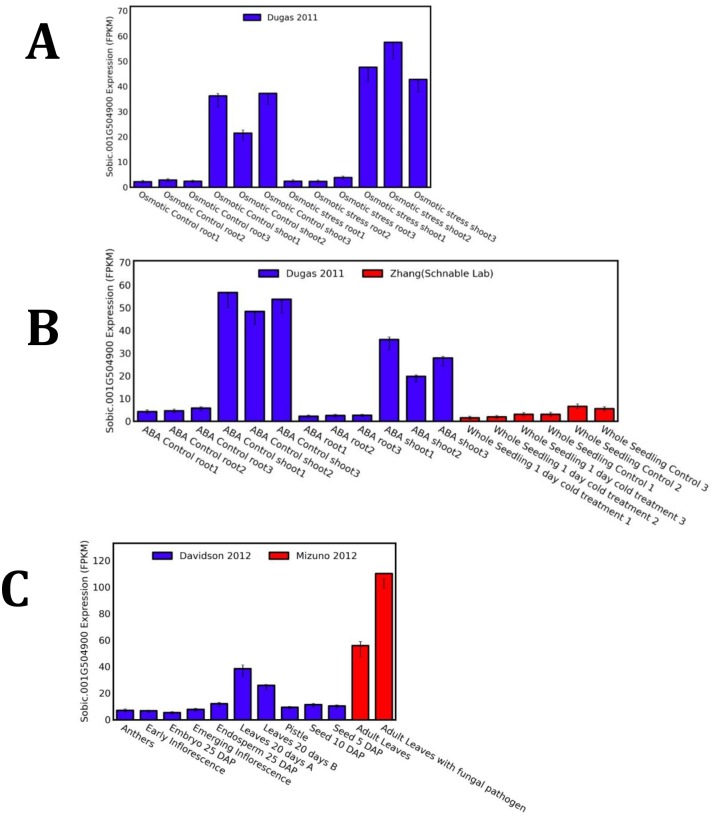
The induced expression of the Ded1 gene during wounding and ethephon treatment prompted us to survey the expression of this gene in existing RNA-seq datasets available through the qTeller database (www.qteller.com). The Ded1 transcript, normalized to facilitate comparative expression profiling, was compared across a variety of developmental stages and in response to externally applied stresses. The survey reveals that the Ded1 gene is expressed at relatively low levels in tissues other than leaves, transcript accumulation increased as leaves matured, and reached a high level in adult leaves (Fig 7A). When mature leaves were challenged by inoculation with conidia of Bipolaris sorghicola, the Ded1 transcript increased two fold (Fig 7A). The expression of the DED1 transcript increased by about 20% following osmotic stress in shoots but no detectable increase was observed in the roots (Fig 7B). One day of cold-stress had no effect on DED1 expression in whole seedlings (Fig 7C). Lastly, the application of ABA to seedling shoots and roots resulted in a ~40% reduction in DED1 transcript accumulation.
Fig 7. Transcript profiling of the DED1 gene product during development, pathogen stress, osmotic stress, and ABA application.
(A) Developmental variation in DED1 transcript accumulation (Data source—Davidson et al 2012) and in response to infection by Bipolaris sorghicola (Data source -Mizuno et al., 2012). (B) Expression of the DED1 transcript in seedling shoots and roots in response to osmotic stress (Data source Dugas et al., 2011). (C) Relative accumulation of DED1 gene product in response to ABA treatment (Data source Dugas et al., 2011) or cold stress (Data source—Zhang and Schnable Lab BioProject PRJNA343268). All RNAseq datasets are normalized as fragments per kilobase of exon per million fragments mapped (FPKM) using qTeller (http://qteller.com).
Discussion
To our knowledge, ded1 is the first lesion mimic mutation to be molecularly identified in sorghum. It is a recessive mutation of the propagative class in which lesions, once formed, expand continuously to engulf the entire leaf. It was isolated from an EMS-mutagenized M2 population by late Dr. John Axtell’s group in the Department of Agronomy at Purdue University. It was also Dr. Axtell who, after having attended a seminar on the phenotypic behavior of lls1, first predicted a relationship between these two mutants. We now establish this connection by showing that ded1-1 resulted from a single base pair change in the sorghum ortholog of the maize lls1 gene. The mutagenesis resulted in a C to A transversion, not typical of EMS mutagenesis, and it induced a premature stop codon that truncated the protein.
This mutation did not change the size of the DED1 transcript but DED1 transcript accumulation increased in ded1 leaves with lesions. This revealed a feature of the ded1 gene that relates to its induction in response to cellular damage. As shown here and also for lls1 [33,34], ded1/lls1 undergoes rapid transcription but transient induction in response to cellular damage including physical injury of the tissue. This feature of the ded1 gene, along with the light- and development-dependent lesion formation is identical to the behavior of lls1 mutants in maize [33,34]. We previously reported the relationship between lls1 and the Arabidopsis acd1 mutant [32–34] and an orthologous mutant in rice was also reported [40]. The suppression of PaO by virus induced gene silencing in both tomato and wheat also resulted in lesion formation [37,41]. In all of these cases, a severe reduction of PaO function results in continuously expanding necrotic lesions. The expanding lesions in all of these systems only superficially resemble HR, and when examined, the underlying cellular and molecular phenotypes are distinct between PaO and HR induced lesions. Thus far, no evidence has been found to connect the accumulation of these phototoxic metabolites in disease signaling, though it remains formally possible that modulation of PaO contributes to an as yet undetermined plant-pathogen interaction.
The lls1 gene encodes a pheophorbide a oxygenase (PaO), a key enzyme of the chlorophyll degradation pathway activated during senescence. PaO activity has thus far been reported to be restricted to senescing tissues but our results demonstrate induction by wounding in mature yet non-senescing tissues. The lack of production of superoxide and H2O2 in and around ded1-1 lesions is consistent with ded1 also encoding a PaO. In the absence of PaO activity, the intermediate of chlorophyll degradation that is likely to accumulate is pheide a. This molecule, a potent photosensitizer, is expected to produce singlet oxygen (1O2) upon illumination, not H2O2 and superoxide.
Singlet oxygen can kill Arabidopsis cells in a genetically-controlled process in which H2O2 and superoxide play no role [51]. Singlet oxygen causes cell death in Arabidopsis, when the PaO encoded by acd1 is defective, by a process that is independent of superoxide and H2O2 [32]. Accordingly, ded1/lls1/acd1 cell death likely results from direct oxidation of membrane lipids and proteins in the chloroplast by the presence of excessive 1O2. This interpretation is supported by the fact that loss of integrity of chloroplasts is the first sign of distress in cells undergoing cell death in ded1 leaves. Indeed a close examination of the electron micrographs obtained in our study reveals that one of the first signs of chloroplast stress is the disorganization and loosening of the organization of the membranes harboring the light harvesting complexes. It is now known that at least 5 chlorophyll catabolic enzymes including PAO interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence [52].
In this context, we can now begin to understand how loss of PAO within the chlorophyll catabolic complex leads to cell death. It is known that some fluorescent (flu) mutants of Arabidopsis accumulate high levels of 1O2 when exposed to light intensities that cause oxidative damage [51]. This might be the case in lls1/ded1 mutants, in the absence of PaO. PaO is a key enzyme of chlorophyll catabolism and high levels of PAO within the larger catabolic complex are required during senescence to prevent accumulation of dangerous chlorophyll catabolites. Accumulation of the highly photoreactive catabolic intermediate, pheophorbide a, results in the light-dependent production of excessive 1O2 in chloroplasts. Anti-oxidative mechanisms within the chloroplast are apparently overwhelmed allowing damage of the chloroplast contents and thus initiating their destruction from within, and subsequently the death of the cell.
While the invocation of singlet oxygen may explain how ded1/lls1 cells die, it poses a dilemma with regard to the propagation of ded1 lesions. Singlet oxygen has such a short life (less than 100 ns) and ability to diffuse (less than 100 nm) that it is unlikely to escape ded1 chloroplasts. While this behavior of singlet oxygen is consistent with chloroplasts being the organelle mediating cell death, it will prevent this ROS from signaling cell death propagation. Furthermore, the lack of evident H2O2 or superoxide accumulation suggests these signals cannot be responsible for lesion formation in ded1. The demonstration that ethephon could trigger lesions in ded1 (Fig 6J) and lls1, similar to what was found for acd1 previously [5], indicates that ethylene promotes the propagation of PaO lesions. This likely results from increased chlorophyll catabolism in response to ethylene signal transduction. It is clear from this study and others that both physical wounding and pathogen infection (but not ABA application or osmotic/cold stress) cause increased expression of the Ded1 gene in leaves. It is widely acknowledged that ethylene, ABA, jasmonic acid (JA), and salicylic acid (SA) are positive regulators of leaf senescence and ripening. Wounding is well known to induce JA which in turn is also linked to the activation of ethylene biosynthetic genes [53]. It has been shown that JA promotes degreening via MYC2/3/4 bHLH proteins that directly bind to the Arabidopsis PAO promoter [54]. The suppression of ded1 and lls1 lesions on young leaves and activity of PaO linked to senescence also lends credence to this hypothesis. Tests in Arabidopsis seem the best way to proceed as generating double mutants of acd1 with those that are defective either in the production or perception of ethylene should be straightforward as both are readily available.
Material and methods
Plant materials
The ded1 mutant was generated by mutagenizing sorghum line P898012 with ethyl methane sulfonate (EMS) and was kindly provided by late Dr. John Axtell (Purdue University, West Lafayette, IN). Plants were grown in the field at the Agronomy Research Center at Purdue University or in pots using standard potting mix in the greenhouse at Purdue as described previously [33]. Plant phenotypes were stable across multiple years including the 1997–1999, 2002, and 2003 field seasons.
Electron microscopy
The fourth leaf of three-week-old wild-type and ded1 plants were wounded via pin pricking multiple times on one side of the mid-rib. At 21 and 42h post-wounding leaf tissue was harvested from the area surrounding, and including, the wound. Uninjured, control tissue was excised on the opposite side of the mid-rib of the injured leaf. Four wild-type and four mutant plants were examined at both time points. Tissue was fixed in 2.5% glutaraldehyde (v/v) in 100 mM sodium cacodylate buffer, pH 6.9 for 2.5h at 4°C. Tissue samples were post-fixed for 2h in 1% OsO4, 100 mM sodium cacodylate buffer, pH 6.9. The tissues were stained en block in 2% aqueous uranyl acetate for 1h, washed in deionized, distilled H2O and dehydrated through an ethanol series. The blocks were gradually infiltrated and embedded in 100% Spurr epoxy resin and polymerized at 60°C for 24h. Ultrathin sections were prepared using a diamond knife on a LKB 8800 microtome, and subsequently stained with uranyl acetate and lead citrate. The stained sections were examined on a JEOL JEM-1200EX transmission electron microscope. Images were recorded on Kodak 4489 film.
Cloning and sequencing of ded1-1 and its wild-type progenitor
Oligonucleotides complementary to exon 5 (GSP5 5' ACTTTTTCCAGTTCACAATGCC 3') and exon 7 (GSP8 5' GGTAGGCTGGGAGCGACAGTA 3') of the maize lls1 gene [33] were used to amplify an approximately 1.1-kb fragment from the sorghum ded1 mutant and its progenitor stock. These PCR products were cloned by using a TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced. The complete amplification, cloning and sequencing process was completed twice for the mutant and the progenitor alleles to enable us to distinguish true sequence polymorphisms from replication errors during amplification.
Sorghum ded1 expression
To determine whether the expression of the ded1 gene is altered after cellular injury, leaf number 6 of five-week-old wild-type plants was wounded by gently rubbing carborundum powder along the length of the leaf. At 1, 2, 4, 6, 8, 10 and 12h post wounding, tissue was frozen and stored at -80°C until the time of RNA isolation. Control tissue was taken at 0h. Unwounded leaves were also taken from age-matched, sibling ded1 mutants, which displayed lesions. RNA isolation and RNA blot hybridization analysis with the 32P-labeled 1.1-kb sorghum ded1 PCR amplification product described above was carried out as previously described [33].
Linkage analysis of ded1 mutants
DNA was isolated from two-week-old maize or sorghum seedlings by the method of Dellaporta [55] and digested with appropriate restriction endonucleases. Following gel electrophoresis, DNA blot hybridization of the digested DNA was carried out as previously described [56]. Resulting blots were probed with the 32P-labeled 1.1-kb sorghum ded1 PCR amplification product described above, or with the inserts from the cloned RFLP probes PIO000603, PIO000689, PIO000640, PIO000044 provided by Pioneer Hi-Bred International Inc. and UMC157 acquired from the University of Missouri-Columbia’s Maize Mapping Center.
Detection of H2O2 and superoxides
The H2O2 presence in lesions was detected using the DAB uptake method as described by Thordal-Christensen et al. [50]. The leaves showing lesions were cut and placed in solution containing 1mg/ml 3,3’ diaminobenzidine (DAB-HCl) pH 3.8 and incubated at room temperature for 1 hour. The leaves were then boiled in 96% ethanol to remove chlorophyll for better visualization of coloration. The presence of H2O2 was visualized as reddish brown coloration. Superoxide was detected by nitroblue tetrazolium (NBT) staining. The NBT (1mg/ml) was dissolved in 10mM NaN3, 10mM phosphate buffer (pH 7.8). After incubation at room temperature for one hour, the leaves were cleared of chlorophyll by boiling in 96% ethanol.
Acknowledgments
All electron microscopy work was carried out at the Electron Microscopy Core Facility at the University of Missouri-Columbia. This work was supported in part by a grant from the National Science Foundation Plant Genome Research Program (1444503) to G.S.J. and B.P.D.
Data Availability
All relevant data are within the paper.
Funding Statement
The work presented was supported by grant number 1444503 from the National Science Foundation Plant Genome Research Program (https://www.nsf.gov/awardsearch/showAward?AWD_ID=1444503) to GSJ and BPD and The Agriculture Research Program at Purdue University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bruggeman Q, Raynaud C, Benhamed M, Delarue M. To die or not to die? Lessons from lesion mimic mutants. Front Plant Sci. Frontiers; 2015;6: 24 10.3389/fpls.2015.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johal GS, Hulbert SH, Briggs SP. Disease lesion mimics of maize: A model for cell death in plants. BioEssays. Wiley Subscription Services, Inc., A Wiley Company; 1995;17: 685–692. 10.1002/bies.950170805 [DOI] [Google Scholar]
- 3.Emerson RA. The inheritance of blotched leaf in maize. Cornell Univ Mem. 1923;70: 3–16. [Google Scholar]
- 4.Neuffer MG, Calvert OH. Dominant disease lesion mimics in maize. J Hered. 1975;66: 265–270. [Google Scholar]
- 5.Greenberg JT, Ausubel FM. Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. Blackwell Science Ltd; 1993;4: 327–341. 10.1046/j.1365-313X.1993.04020327.x [DOI] [PubMed] [Google Scholar]
- 6.Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. Arabidopsis mutants simulating disease resistance response. Cell. 1994;77: 565–577. 10.1016/0092-8674(94)90218-6 [DOI] [PubMed] [Google Scholar]
- 7.Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. http://www.pnas.org/content/99/11/7530.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johal GS. Disease Lesion Mimics Mutants of Maize. APSnet Featur Artic. 2007; 10.1094/APSnetFeatures-2007-0707 [DOI] [Google Scholar]
- 9.Lorrain S. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8: 263–271. 10.1016/S1360-1385(03)00108-0 [DOI] [PubMed] [Google Scholar]
- 10.Hoeberichts FA, Woltering EJ. Multiple mediators of plant programmed cell death: Interplay of conserved cell death mechanisms and plant-specific regulators. BioEssays. Wiley Subscription Services, Inc., A Wiley Company; 2003;25: 47–57. 10.1002/bies.10175 [DOI] [PubMed] [Google Scholar]
- 11.Greenberg JT. Programmed cell death: A way of life for plants. proc Natl Acad Sci USA. 1996;93: 12094–12097. Available: http://www.pnas.org/content/93/22/12094.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dangl JL. Death Don’t Have No Mercy: Cell Death Programs in Plant-Microbe Interactions. PLANT CELL ONLINE. 1996; 10.1105/tpc.8.10.1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häcker G. The morphology of apoptosis. Cell Tissue Res. 2000;301: 5–17. 10.1007/s004410000193 [DOI] [PubMed] [Google Scholar]
- 14.Zimmermann KC, Bonzon C, Green DR. The machinery of programmed cell death. Pharmacol Ther. 2001;92: 57–70. 10.1016/s0163-7258(01)00159-0 [DOI] [PubMed] [Google Scholar]
- 15.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407: 770–6. 10.1038/35037710 [DOI] [PubMed] [Google Scholar]
- 16.Thornberry NA, Lazebnik Y. Caspases: enemies within. Sci (New York, NY). 1998;281: 1312–1316. 10.1126/science.281.5381.1312 [DOI] [PubMed] [Google Scholar]
- 17.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3: E255–E263. 10.1038/ncb1101-e255 [DOI] [PubMed] [Google Scholar]
- 18.Newmeyer DD, Ferguson-Miller S. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell. 2003. pp. 481–490. 10.1016/S0092-8674(03)00116-8 [DOI] [PubMed] [Google Scholar]
- 19.Tsujimoto Y. Bcl-2 family of proteins: Life-or-death switch in mitochondria. Bioscience Reports. 2002. pp. 47–58. 10.1023/A:1016061006256 [DOI] [PubMed] [Google Scholar]
- 20.Zamyatnin AA. Plant Proteases Involved in Regulated Cell Death. Biochem Biokhimiia. 2015;80: 1701–15. 10.1134/S0006297915130064 [DOI] [PubMed] [Google Scholar]
- 21.Hu G, Richter TE, Hulbert SH, Pryor T. Disease Lesion Mimicry Caused by Mutations in the Rust Resistance Gene rp1. Plant Cell Online. 1996;8 Available: http://www.plantcell.org/content/8/8/1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell MA, R P.C. Characterization of four rice mutants with alterations in the defense response pathway. Mol Plant Path. Blackwell Science Ltd; 2005;6: 11–21. 10.1111/1468-0025.00206-i1 [DOI] [PubMed] [Google Scholar]
- 23.Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, et al. Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol. Kluwer Academic Publishers; 2007;63: 847–860. 10.1007/s11103-006-9130-y [DOI] [PubMed] [Google Scholar]
- 24.Jung Y-H, Lee J-H, Agrawal GK, Rakwal R, Kim J-A, Shim J-K, et al. The rice (Oryza sativa) Blast Lesion Mimic Mutant, blm, may confer resistance to blast pathogens by triggering multiple defense-associated signaling pathways. Plant Physiol Biochem. 2005;43: 397–406. 10.1016/j.plaphy.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 25.Buckner B, Janick-Buckner D, Gray J, Johal G. Cell-death mechanisms in maize. Trends in Plant Science. 1998. pp. 218–223. 10.1016/S1360-1385(98)01254-0 [DOI] [Google Scholar]
- 26.Jabs T, Dietrich RA, Dangl JL. Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science. American Association for the Advancement of Science; 1996;273: 1853–6. 10.1126/SCIENCE.273.5283.1853 [DOI] [PubMed] [Google Scholar]
- 27.Kliebenstein DJ, Dietrich RA, Martin AC, Last RL, Dangl JL. LSD1 Regulates Salicylic Acid Induction of Copper Zinc Superoxide Dismutase in Arabidopsis thaliana. Mol Plant-Microbe Interact. The American Phytopathological Society; 1999;12: 1022–1026. 10.1094/MPMI.1999.12.11.1022 [DOI] [PubMed] [Google Scholar]
- 28.Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci. National Academy of Sciences; 2011;108: 16463–16468. 10.1073/pnas.1113726108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mach JM. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci. 2001;98: 771–776. 10.1073/pnas.98.2.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao N, Greenberg JT. Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell. 2006;18: 397–411. 10.1105/tpc.105.036251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hörtensteiner S, Kräutler B. Chlorophyll breakdown in higher plants. Biochimica et Biophysica Acta—Bioenergetics. 2011. pp. 977–988. 10.1016/j.bbabio.2010.12.007 [DOI] [PubMed] [Google Scholar]
- 32.Pruzinská A, Tanner G, Anders I, Roca M, Hörtensteiner S, Pruž Inská A, et al. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc Natl Acad Sci U S A. National Academy of Sciences; 2003;100: 15259–64. 10.1073/pnas.2036571100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray J, Close PS, Briggs SP, Johal GS. A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell. 1997;89: 25–31. 10.1016/S0092-8674(00)80179-8 [DOI] [PubMed] [Google Scholar]
- 34.Gray J, Janick-Buckner D, Buckner B, Close PS, Johal GS. Light-dependent death of maize lls1 cells is mediated by mature chloroplasts. Plant Physiol. American Society of Plant Biologists; 2002;130: 1894–1907. 10.1104/pp.008441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hörtensteiner S. Update on the biochemistry of chlorophyll breakdown. Plant Molecular Biology. 2013. pp. 505–517. 10.1007/s11103-012-9940-z [DOI] [PubMed] [Google Scholar]
- 36.Hörtensteiner S, Wüthrich KL, Matile P, Ongania KH, Kräutler B. The key step in chlorophyll breakdown in higher plants. Cleavage of pheophorbide a macrocycle by a monooxygenase. J Biol Chem. American Society for Biochemistry and Molecular Biology; 1998;273: 15335–9. 10.1074/JBC.273.25.15335 [DOI] [PubMed] [Google Scholar]
- 37.Tang C, Wang X, Duan X, Wang X, Huang L, Kang Z. Functions of the lethal leaf-spot 1 gene in wheat cell death and disease tolerance to Puccinia striiformis. J Exp Bot. 2013;64: 2955–2969. 10.1093/jxb/ert135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M, Wardzala E, Johal GS, Gray J. The wound-inducible Lls1 gene from maize is an orthologue of the Arabidopsis Acd1 gene, and the LLS1 protein is present in non-photosynthetic tissues. Plant Mol Biol. 2004;54: 175–191. 10.1023/B:PLAN.0000028789.51807.6a [DOI] [PubMed] [Google Scholar]
- 39.Tanaka R, Hirashima M, Satoh S, Tanaka A. The Arabidopsis-accelerated cell death Gene ACD1 is Involved in Oxygenation of Pheophorbide a: Inhibition of the Pheophorbide a Oxygenase Activity does not Lead to the “Stay-Green” Phenotype in Arabidopsis. Plant Cell Physiol. 2003; 10.1093/pcp/pcg172 [DOI] [PubMed] [Google Scholar]
- 40.Xie Q, Liang Y, Zhang J, Zheng H, Dong G, Qian Q, et al. Involvement of a Putative Bipartite Transit Peptide in Targeting Rice Pheophorbide a Oxygenase into Chloroplasts for Chlorophyll Degradation during Leaf Senescence. J Genet Genomics. 2016;43: 145–154. 10.1016/j.jgg.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 41.Spassieva S, Hille J. A lesion mimic phenotype in tomato obtained by isolating and silencing an Lls1 homologue. Plant Sci. 2002;162: 543–549. 10.1016/S0168-9452(01)00595-7 [DOI] [Google Scholar]
- 42.Lee KP, Kim C, Landgraf F, Apel K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci. 2007;104: 10270–10275. 10.1073/pnas.0702061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang S, Apel K, Kim C. Singlet oxygen-mediated and EXECUTER-dependent signalling and acclimation of Arabidopsis thaliana exposed to light stress. Philos Trans R Soc Lond B Biol Sci. 2014;369: 20130227 10.1098/rstb.2013.0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim C, Apel K. Singlet oxygen-mediated signaling in plants: Moving from flu to wild type reveals an increasing complexity. Photosynthesis Research. 2013. pp. 455–464. 10.1007/s11120-013-9876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meskauskiene R, Apel K. Interaction of FLU, a negative regulator of tetrapyrrole biosynthesis, with the glutamyl-tRNA reductase requires the tetratricopeptide repeat domain of FLU. FEBS Lett. 2002;532: 27–30. 10.1016/S0014-5793(02)03617-7 [DOI] [PubMed] [Google Scholar]
- 46.Kim C, Meskauskiene R, Apel K, Laloi C. No single way to understand singlet oxygen signalling in plants. EMBO Reports. 2008. pp. 435–439. 10.1038/embor.2008.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danon A, Miersch O, Felix G, Op Den Camp RGL, Apel K. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 2005;41: 68–80. 10.1111/j.1365-313X.2004.02276.x [DOI] [PubMed] [Google Scholar]
- 48.Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2001;98: 12826–31. 10.1073/pnas.221252798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agrios G. Plant Pathology 5th Edition San Diego: Acad Press; 2005; 922 10.1016/j.plantsci.2005.02.019 [DOI] [Google Scholar]
- 50.Ellis EA, Grant MB. 1 Cytochemical Localization of H 2 O 2. Plant J. 2011;196: 3–12. [DOI] [PubMed] [Google Scholar]
- 51.Gutierrez J, Gonzalez-Perez S, Garcia-Garcia F, Lorenzo O, Arellano JB. Does singlet oxygen activate cell death in Arabidopsis cell suspension cultures?: analysis of the early transcriptional defense responses to high light stress. Plant Signal Behav. 2011;6: 1937–1942. 10.4161/psb.6.12.18264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakuraba Y, Schelbert S, Park S-Y, Han S-H, Lee B-D, Andrès CB, et al. STAY-GREEN and Chlorophyll Catabolic Enzymes Interact at Light-Harvesting Complex II for Chlorophyll Detoxification during Leaf Senescence in <em>Arabidopsis</em> Plant Cell. 2012;24: 507 LP–518. Available: http://www.plantcell.org/content/24/2/507.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, et al. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell. 2017; 10.1105/tpc.17.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, et al. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015;84: 597–610. 10.1111/tpj.13030 [DOI] [PubMed] [Google Scholar]
- 55.Dellaporta S. Urea-based Plant DNA Miniprep. The Maize Handbook; 1994. p. 776 10.1007/978-1-4612-2694-9 [DOI] [Google Scholar]
- 56.Buckner B, Kelson T, Robertson D. Cloning of the y1 Locus of Maize, a Gene Involved in the Biosynthesis of Carotenoids. Plant Cell. 1990;2: 867–876. 10.1105/tpc.2.9.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.