Abstract
CRISPR base editing enables the creation of targeted single-base conversions without generating double stranded breaks. However, the efficiency of current base editors is very low in many cell types. We re-engineered the sequences of BE3, BE4Gam, and xBE3 by codon optimization and incorporation of additional nuclear localization sequences. Our collection of optimized constitutive and inducible base-editing vector systems dramatically improves the efficiency by which single nucleotide variants can be created. The re-engineered base editors enable target modification in a wide range of mouse and human cell lines, and intestinal organoids. We also show that the optimized base editors mediate efficient in vivo somatic editing in the liver of adult mice.
Keywords: Base editing, BE3, CRISPR, APOBEC
Base editors are hybrid proteins that tether DNA modifying enzymes to nuclease defective Cas9 variants. This enables the direct conversion of cytosine (C) to other bases (T, A, or G) 1–4, or adenine (A) to inosine/guanine (I/G) nucleic acids 5,6, allowing the creation or repair of disease-associated single nucleotide variants (SNVs). The BE3 base editor carries a rat APOBEC cytidine deaminase at the N-terminus of Cas9n (Cas9D10A) and a uracil glycosylase inhibitor (UGI) domain at the C-terminus. This construct has been shown to drive targeted C>T transitions at nucleotide positions 3-8 of the protospacer (Figure 1a) following transfection of plasmid DNA or ribonuclear particles (RNPs) 7, 8.
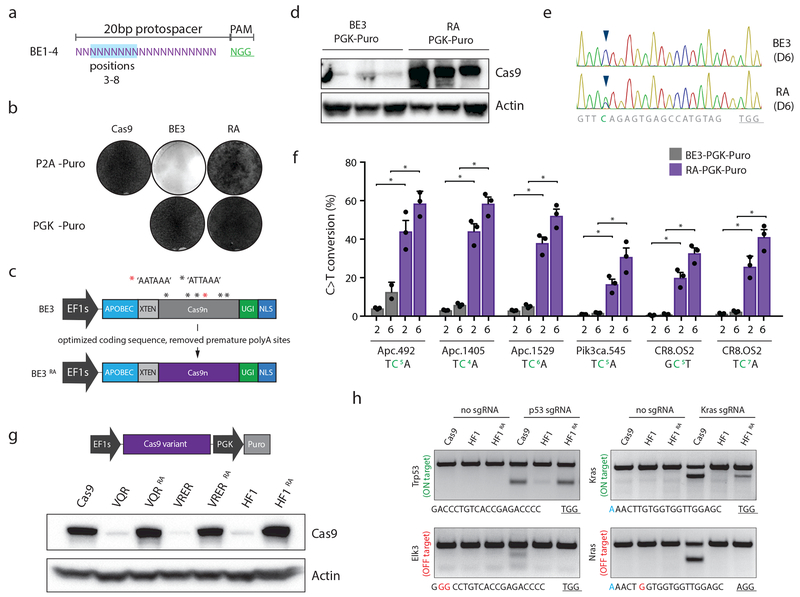
Figure 1. Optimizing the coding sequence of BE3 improves protein expression and target base editing.
a. Schematic depiction of the canonical region of target base editing. Positions 3-8 (highlighted in blue) within the protospacer are susceptible to C>T conversion by BE3. The PAM is shown in green. b. Giemsa stained NIH/3T3 cells following transduction with indicated lentiviruses, and selection in puromycin for 6 days. Representative of similar results from three independent experiments; see Supplementary Figure 1. c. Schematic representation of original BE3 (above), and codon optimized RA enzymes (below). d. Cas9 immunoblot of independently derived NIH/3T3 lines transduced with BE3 or RA (n=3). e. Sanger sequencing chromatogram showing the target region of the Apc.1405 sgRNA. Arrow highlights cytosine at position 4 that shows dramatically increased editing by RA six days following sgRNA transduction. Representative of similar results from three independent experiments; see panel f. f. Frequency of target C>T editing across 5 different sgRNA targets, 2 and 6 days following sgRNA transduction, as indicated. Graphs show mean values. Error bars represent s.d., n = 3 biologically independent samples; asterisks (*) indicate a significant difference (p<0.05) between groups, using one-way ANOVA with Sidak’s multiple comparison test. g. Western blot showing expression of original and optimized HF1 and PAM variant Cas9 proteins. Representative of similar results from three independent blots. h. T7 endonuclease assays on Trp53 and Kras target sites, and off-target sites (Elk3 and Nras, respectively) show that reassembled HF1 (HF1RA) improves on-target activity while maintaining little to no off-target cutting. Genomic target sites for each region are shown below. Of note, the slightly reduced on-target activity of HF1RA at the Kras site may be due to the G-A mismatch at position 1 of the protospacer (highlighted blue). Experiment was performed twice with similar results.
To enable base editing in difficult-to-transfect cells, we cloned a lentiviral vector in which BE3 was expressed from the EF1 short (EF1s) promoter and linked to a puromycin (puro) resistance gene via a P2A self-cleaving peptide (pLenti-BE3-P2A-Puro, BE3). Despite efficient production of viral particles and integration of the vector into target cells (Supplementary Figure 1), we could not generate puro-resistant cells (Figure 1b, Supplementary Figure 1c). To test whether this was due to low expression of the BE3-linked Puro cassette, we generated a new lentivirus whereby puro was driven by an independent (PGK) promoter (pLenti-BE3-PGK-Puro). This vector produced equivalent viral titer and target cell integration (Supplementary Figure 1), and, in contrast to BE3-P2A-Puro, enabled effective puro resistance (Figure 1b, Supplementary Figure 1c).
These data suggested that an issue in the production of BE3 protein was limiting effective base editing. During cloning of lentiviral constructs, we noted that the Cas9n DNA sequence in BE3 was not optimized for expression in mammalian cells, containing a large number of non-favored codons (Supplementary Figure 2, Supplementary Table 1) and 6 potential polyadenylation sites (AATAAA or ATTAAA) throughout the cDNA (Figure 1c); we therefore reconstructed the BE3 enzyme using an extensively optimized Cas9n sequence (Supplementary Figure 2)9. The resulting reassembled BE3 (BE3RA; hereafter RA) enabled efficient puro selection (Figure 1b, Supplementary Figure 1), markedly increased protein expression (Figure 1d), and most notably, showed up to 30-fold increase in target C>T conversion (Figure 1e,f; Supplementary Figure 3a,b). Although C>T editing increased on average 15-fold, the level of unwanted insertions and deletions (indels), or undesired (C>A or C>G) editing remained low, indicating a substantial improvement in the relative fidelity of base editing over previous versions (Supplementary Figure 3c,d). Of note, we and others10 observed similar problems with expression of high-fidelity Cas9 (HF1)11 and altered PAM specificity variants12, which share the same Cas9 cDNA as BE3. In each case, this was corrected by re-engineering the construct (Figure 1g, Supplementary Figure 4)10. The resulting increased expression of the HF1 enzyme (HF1RA) dramatically improved on-target DNA cleavage, while maintaining little or no off-target activity 13 (Figure 1h).
Nuclear localization signal (NLS) sequences at the N-terminus of Cas9 can improve the efficiency of gene targeting14. Indeed, despite the presence of a C-terminal NLS (Figure 2a), RA protein was largely excluded from the nucleus (Figure 2b). We tested two different N-terminal positions for the NLS in case the inclusion of these sequences in one location interfered with APOBEC function: 1) with a FLAG epitope tag at the N-terminus (FNLS), and 2) within the XTEN linker that bridges APOBEC and Cas9n (2X) (Figure 2a, Supplementary Figure 5a). Whereas 2X showed no obvious increase in nuclear targeting compared to RA, FNLS protein was more evenly distributed through the nucleus and cytoplasm (Figure 2b).
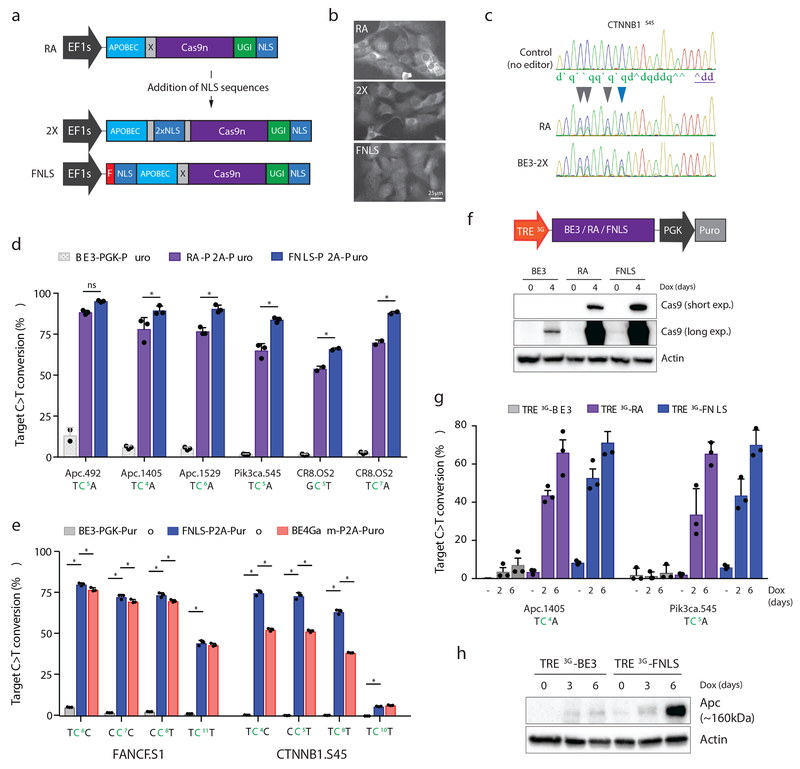
Figure 2. N-terminal NLS sequences increase the range and potency of target base editing.
a. Schematic representation of RA enzyme (above) and two new variants carrying NLS sequences within the XTEN linker (2X) or at the N-terminus (FNLS) b. Immunofluorescent staining of Cas9 in NIH/3T3 cells expressing RA, 2X, or FNLS. Experiment was repeated twice with similar results. c. Sanger sequencing chromatogram showing increased editing of the cytosine at position 10 (blue arrow) within the protospacer of a CTNNB1.S45 sgRNA. d. Frequency (%) of C>T conversion in NIH/3T3 cells transduced with RA- or FNLS-P2A-Puro lentiviral vectors 6 days following introduction of different sgRNAs, as indicated. Editing in BE3-PGK-Puro cells (from Figure 1e) is shown for comparison. e. Frequency (%) of C>T conversion in PC9 cells transduced with BE3-PGK-Puro, FNLS or BE4GamRA-P2A-Puro lentiviral vectors 6 days following introduction of different sgRNAs, as indicated. For d and e, graphs show mean values. Error bars represent s.e.m., n = 3 biologically independent samples; asterisks (*) indicate a significant difference (p<0.05) between groups, using two-way ANOVA with Tukey’s correction for multiple testing. f. Schematic representation of dox-inducible BE3 lentiviral construct and immunoblot of Cas9 in transduced and selected NIH/3T3 cells following treatment with or without dox (1μg/ml) for 4 days, as indicated. Blot was performed twice with similar results. g. Frequency (%) of C>T conversion in NIH/3T3 cells transduced with TRE3G-BE3, TRE3G-RA, or TRE3G-FNLS, and sgRNA lentiviral vectors, 0, 2 and 6 days following dox treatment. Graph shows mean values. Error bars represent s.e.m., n = 3 biologically independent experiments; asterisks (*) indicate a significant difference (p<0.05) between groups, using a two-way ANOVA with Tukey’s correction for multiple testing. h. Immunoblot showing induction of truncated (~160kDa) Apc product after target editing in NIH/3T3s expressing BE3 or FNLS. Blot was performed twice with similar results.
In transfection-based assays, FNLS improved editing ~2-fold across multiple target positions and sgRNAs (Supplementary Figure 5b). In contrast, 2X did not alter editing within the normal target window, but significantly increased the range of editing toward cytosines at positions 10-11 in the protospacer (Figure 2c, Supplementary Figure 5b,c); the expanded range was not attributable solely to the increased length of the linker (Supplementary Figure 5c). We next generated codon-optimized 2X-P2A-Puro and FNLS-P2A-Puro lentiviral vectors and transduced mouse NIH/3T3 cells (Supplementary Figure 6). Two days following sgRNA transduction, FNLS-expressing cells showed greater than 50% C>T conversion for all sgRNAs tested (Supplementary Figure 7a), and by day six, 80-95% of all target cytosines were converted (Figure 2d). By contrast, at this timepoint, only 1/5 sgRNAs showed >80% editing with RA (Figure 2d). On average, FNLS increased editing by 35% over RA, and up to 50-fold over the original BE3 construct (Figure 2d), and produced fewer indels and non-desired (C>A and C>G) edits relative to RA (Supplementary Figure 7b,c). To confirm the re-engineered enzymes were active in multiple cell types, we transduced 3 different human cancer cell lines (PC9, H23, and DLD1) and measured editing at FANCF and CTNNB1 target sites. Although absolute editing efficiency varied, FNLS increased target C>T conversion 15 to 150-fold within the expected window (positions 3-8bp) (Figure 2e, Supplementary Figure 8a). Indels and non-desired edits were elevated in each of the cancer lines compared to 3T3s, but this was reduced by using an optimized version of the second-generation editor BE4Gam 15 (Supplementary Figures 8b and 9). The improved efficiency also increased editing at predicted off-target sites, although the overall level of off-target editing remained low (Supplementary Figure 10). As predicted from transfection experiments, the 2X construct did not alter the overall efficiency of the enzyme, but significantly extended the range of editing in both mouse and human cells (Supplementary Figure 11).
To provide a temporally controlled system for base editing, we generated (TRE3G) doxycycline (dox)-inducible constructs (Figure 2f). As expected, dox treatment drove strong induction of RA and FNLS, but limited expression of the original BE3 construct (Figure 2f). Using sgRNAs targeting Apc and Pik3ca, we observed a time-dependent generation of target missense (Pik3caE545K) and nonsense (ApcQ1405X) mutations (Figure 2g). Consistent with earlier observations, both RA and FNLS dramatically increased editing efficiency relative to the original BE3 enzyme (Figure 2g), which for Apc.1405, translated to induction of a truncated Apc protein (Figure 2h).
Together, these data demonstrate that our optimized enzymes increase the range (2X) and efficiency (FNLS) of targeted base editing. To demonstrate the utility and impact of the improved editors, we set out to engineer a series of precise and functional genetic changes in different model systems: human cancer cells, intestinal organoids, mouse embryonic stem cells, and mouse hepatocytes in vivo.
DLD1 colorectal cancer cells are sensitive to combined inhibition of Tankyrase and MEK16, 17 but WNT activating mutations in CTNNB1 are predicted to bypass this response 18. Hence, we cultured DLD1 cells carrying sgRNAs targeting codon (Serine) 45 of CTNNB1, or FANCF.S1 in Tankyrase (XAV939; 1uM) and MEK (Trametinib; 10nM) inhibitors and tracked tdTomato-positive, sgRNA-expressing cells over time (Supplementary Figure 12a). At treatment initiation, RA, 2X and FNLS-expressing cells, but not BE3, showed efficient editing (40-50%) at the FANCF control site, and CTNNB1S45F mutations at a frequency of 12-18% (Supplementary Figure 8a). In the presence of inhibitors, CTNNB1 sgRNA-transduced cells (expressing RA, 2X, or FNLS, but not the original BE3) outcompeted the non-transduced population (Figure 3a , Supplementary Figure 12b), and inhibitor-treated cells, but not DMSO-treated cells, showed enrichment of the expected S45F alteration (Figure 3b). Together, these data imply that editor-induced CTNNB1S45F mutations are functional and enable resistance to upstream WNT suppression by Tankyrase inhibitors.
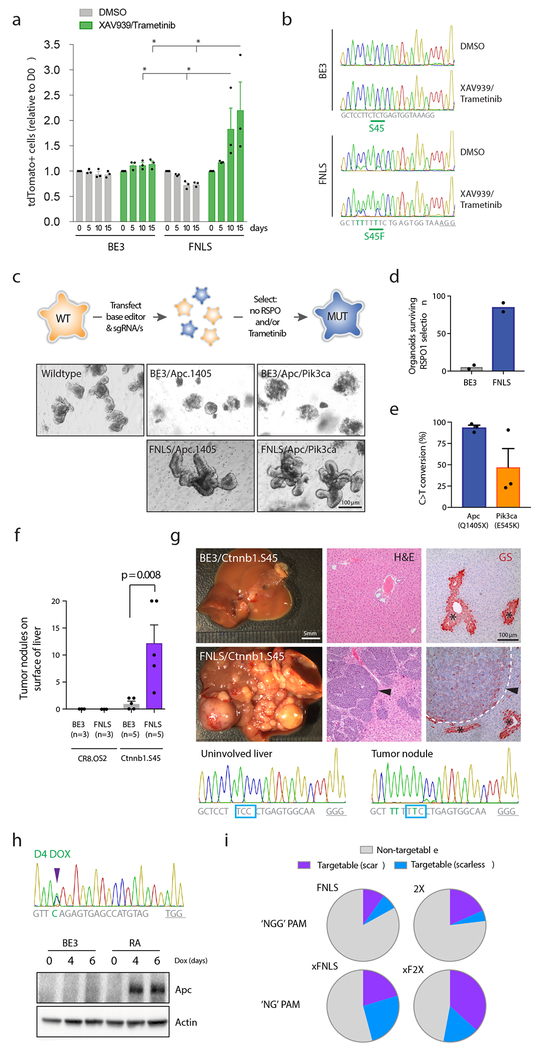
Figure 3. Optimized enzymes induce efficient base editing in a wide range of cell systems.
a. Graph shows relative abundance of tdTomato-positive (sgRNA-expressing) cells in BE3 and FNLS-transduced DLD1 cells, following treatment with DMSO, or XAV939 (1μM) and Trametinib (10nM). Bars in each case represent serial passages every 5 days, starting at day 0. Graphs show mean values. Error bars represent s.e.m., n = 3 biologically independent samples; asterisks (*) indicate a significant difference (p<0.05) between groups, using a two-way ANOVA with Tukey’s correction for multiple testing. b. Chromatograms showing sequencing of the CTNNB1.S45 target site in BE3 and FNLS cells, treated with DMSO (upper) or XAV939/Trametinib (lower). Chromatogram is representative of sequencing of three independent samples with similar results. Drug treated cells show enrichment of the S45F mutation, implying it provides an advantage in XAV/Tram treated populations. c. Schematic representation of the process of editing and selection in intestinal organoids. Images show wildtype mouse small intestinal organoids following editor/sgRNA transfection and selection by RSPO1 withdrawal (6 days). Only FNLS transfected organoids show consistent outgrowth of large budding organoids in the absence of RSPO1. Images representative of three independent experiments with similar results. Transfection with tandem sgRNAs targeting Apc and Pik3ca drives the generation of compound mutant organoids that survive RSPO1 withdrawal and treatment with 25nM Trametinib (see Supplementary Figure 13). d. Number of viable organoids six days following RSPO1 withdrawal. Graphs show mean values. Error bars represent s.e.m., n = 2 biologically independent samples. e. Mean frequency of Apc.Q1405X and Pik3ca.E545K mutations in intestinal organoids following selection in RSPO1-free media, but no selection in Trametinib. Error bars represent s.e.m., n = 3 independent transfections. f. Mean number of visible tumor nodules counted of the liver of mice 4 weeks following hydrodynamic delivery of BE3 or FNLS, a mouse Ctnnb1.S45 sgRNA and Sleeping Beauty transposon-based Myc cDNA. Error bars are s.e.m., n=3-5 biologically independent animals, as indicated; statistical difference between groups calculated using a one-way ANOVA with Tukey’s correction for multiple testing. g. Representative images of tumor burden following editing of Ctnnb1 with FNLS and BE3. Right: H&E and immunohistochemical staining for Glutamine synthetase (GS; red stain) of representative sections of livers from BE3 and FNLS transfected mice. Asterisks (*) highlight pericentral hepatocytes that stain positively for GS. Arrows indicate tumors within the liver of FNLS-transfected mice. Images representative of five independent samples, with similar results. Lower: Sanger sequencing from uninvolved liver and a tumor nodule from an FNLS/Ctnnb1.S45 sgRNA-transfected mice, showing near-complete editing of the Ctnnb1 locus in tumor cells. Note: BE3 tumor nodules were too few and too small to dissect and obtain sequencing. h. Sanger sequencing chromatogram shows editing of Apc in embryonic stem cells (ESCs) following 4 days of treatment with dox (1μg/ml) and immunoblot showing induction of the expected truncated allele of Apc in RA-expressing cells, but not in BE3 cells. Blot was performed twice with similar results. i. Pie charts indicating the theoretical number of recurrent cancer-associated mutations that could be modeled using FNLS or 2X (‘NGG’ PAM) or xFNLS and xF2X (‘NG’ PAM) constructs. Purple indicates sites where only the target cytosine would be affected (scarless); blue indicates sites where creation of the desired mutation would likely be accompanied by additional C>T alterations (scar). Assumes an editing window of positions 4-8 (for FNLS, xFNLS) and 4-11 (for 2X and xF2X). See methods for details.
Truncating Apc mutations are the most common genetic events observed in human CRCs19, and drive WNT/RSPO-independent proliferation. To engineer Apc truncations, we co-transfected intestinal organoids with either BE3 or FNLS, and the Apc.1405 sgRNA (Figure 3c). FNLS-transfected cultures showed a 10-fold increase in outgrowth of RSPO1-independent organoids relative to BE3-transfected cells (Figure 3d) and carried a high frequency of targeted Apc editing (>97%)(Figure 3e) with fewer than 1% indels. Co-delivery of two tandem arrayed sgRNAs (Apc.1405 and Pik3ca.545) produced ApcQ1405X / Pik3caE545K double mutant organoids (Figure 3c,e) that could survive and expand in the presence of a MEK inhibitor (Trametinib; 25nM) (Supplementary Figure 13a,b), as has been described for HDR-generated PIK3CAE545K mutations in human organoids20.
In hepatocellular carcinoma (HCC), CTNNB1 mutations are the primary mechanism of WNT-driven tumorigenesis. To explore the potential of base editors to drive tumor formation in vivo, we introduced BE3 or FNLS, a mouse Ctnnb1.S45 sgRNA, and Myc cDNA to the livers of adult mice via hydrodynamic transfection. After 4 weeks, 3/5 BE3 transfected animals showed 1-2 small tumor nodules on the liver, whereas FNLS-transfected mice had dramatically increased disease burden with all mice (5/5) carrying multiple tumors (Figure 3f). Tumors resembled HCCs with trabecular and solid growth pattern, and showed upregulation of the WNT target, Glutamine synthetase 21 (GS; Figure 3g). Tumor nodules showed near-complete editing of the Ctnnb1 locus, creating activating S45F mutations (Figure 3g).
An alternate approach to in vivo somatic base editing is the generation of temporally regulated transgenic strains, which enables the manipulation of tissues and cell types that cannot be easily transfected in vivo and avoids the potential immunogenicity of exogenous Cas9 delivery 22, 23. To this end, we generated TRE-inducible, knock-in mouse embryonic stem cells (mESCs). We chose RA for mESC targeting, as we noted low-level ‘leaky’ editing in 3T3s cells carrying TRE3G-FNLS lentivirus (Figure 2g). TRE-RA cells showed efficient dox-dependent C>T conversion and generation of predicted mutant alleles (Figure 3h, Supplementary Figure 13c). Together, these data show that optimized RA and FNLS constructs offer a flexible and efficient platform to engineer directed somatic alterations in animals.
Base editing is arguably the most direct approach to engineer disease-associated SNVs in model systems. To estimate of the number cancer-related SNVs that could be modeled with Cas9-mediated base editing, we analyzed MSK IMPACT targeted deep sequencing of more than 22,000 tumors and defined a list of 2696 recurrent mutations (observed in at least 4 individual patients). With a conservative base editing window of positions 4-8 (FNLS) and 4-11 (2X), we estimate that ~17% of cancer-associated SNVs could be engineered with FNLS, and ~23% by exploiting the expanded range of the 2X construct. Of these, approximately 40% can be generated without any collateral editing (or “scar”) at non-target cytosines (Figure 3i). In principle, using Cas9 variants with less restrictive PAM requirements (e.g. xCas9) 24, more than 50% of all mutations could be created (Figure 3i). Toward this end, we produced optimized xFNLS and xF2X constructs which enable more efficient base editing than the published xBE3 construct (Supplementary Figure 14). Notably, the xCas9-derived base editors showed lower on-target activity for both sgRNAs and cell lines tested (Supplementary Figure 14b,c). Further work will be required to assess the contexts in which these newer variants enable efficient base editing.
Here, by optimizing protein expression and nuclear targeting, we developed a range of potent base editing and Cas9 enzymes that dramatically improve DNA editing across multiple in vitro and in vivo model systems. These tools, along with similar optimized versions for adenine (ABE) base editors25, 26, will enable the rapid generation of targeted SNVs in a variety of cell systems in vitro and in vivo and will be key to implementing base editing in genetic screens, where high efficiency is essential. Moreover, the improved protein expression of our re-engineered enzymes will substantially enhance therapeutic approaches that rely on delivery of mRNA molecules 27, whereas enhanced nuclear targeting will likely improve the delivery and/or activity of RNPs 14. In all, we expect that the toolkit described herein will make base editing a feasible and accessible option for a wide range of research and therapeutic applications.
Online Methods
Cloning
All primers, Ultramers, and gBlocks used for cloning are listed in Supplementary Tables 2, 3, 4 and 5. pCMV-BE3-2X (CMV-2X) and pCMV-BE3-FNLS were generated by Gibson assembly, combining an XmaI-digested (2X) or NotI-digested (FNLS) pCMV-BE3 backbone with DNA Ultramers (BE3-2X NLS or T7-FLAG-NLS). Double stranded DNA from Ultramers was generated by PCR amplification using primers XTEN-NLS_F/XTEN-NLS_R, and T7-FLAG_F/T7-FLAG_R primers, respectively. pLenti-BE3-PGK-Puro (LBPP) was generated by Gibson assembly combining 4 DNA fragments: i) PCR amplified EF1s promoter (FSR-19/FSR-20), ii) PCR amplified BE3 cDNA (FSR-114/FSR-115), iii) PCR amplified PGK-Puro cassette (FSR-16/FSR-17), and iv) a BsrGI/PmeI digested pLL3-based lentiviral backbone. pLenti-BE3RA-PGK-Puro (LRPP) was generated by Gibson assembly, combining a PCR amplified BE3RA cDNA (BE3RA-PGKPuro_F/BE3RA-PGKPuro_R) and an NheI/AvrII digested BE3-PGK-Puro backbone. pLenti-FNLS-PGK-Puro (LFPP) was generated by restriction cloning a FLAG-NLS-APOBEC BamHI(blunt)/EcoRI-digested fragment, into an NheI(blunt)/EcoRI-digested pLenti-BE3RA-PGK-Puro backbone. pLenti-BE3RA-P2A-Puro (LR2P) was generated by Gibson assembly, combining 4 DNA fragments: i) a PCR amplified APOBEC-XTEN cDNA (BE3RA_APOBEC_F /BE3RA_XTEN_R), ii) PCR amplified Cas9n (BE3RA_Cas9n_F/BE3RA_Cas9n_R), iii) PCR amplified UGI (BE3RA_UGI_F/BE3RA_UGI_R), and a BamHI/NheI digested pLenti-Cas9-P2A-Puro viral backbone. Note: some wobble positions were altered within the UGI (SGGS) linker to avoid complications during Gibson assembly because of an identical region downstream of UGI. pLenti-FNLS-P2A-Puro (LF2P) was generated by restriction cloning a PCR amplified (BamHI-FLAG_F/APOBEC-RI_R) BamHI/EcoRI-digested, FLAG-NLS-APOBEC fragment into a BamHI/EcoRI digested pLenti-BE3RA-P2A-Puro backbone. pLenti-2X-P2A-Puro (LX2P) was generated by Gibson assembly, combining a PCR-amplified APOBEC-2XNLS fragment (BE3RA_APOBEC_F/BE3RA_XTEN_R) and a BamHI/XmaI digested pLenti-BE3RA-P2A-Puro backbone. pLenti-TRE3G-BE3-PGK-Puro (L3BP) was generated by Gibson assembly, combining a PCR-amplified TRE3G promoter (3G_F/3G_R), and APOBEC fragment (APOBEC_F/BE3RA_XTEN_R) with an XmaI digested pLenti-BE3-PGK-Puro backbone. pLenti-TRE3G-BE3RA-PGK-Puro (L3RP) was generated by Gibson assembly, combining a PCR-amplified TRE3G promoter (3G_F/3G_R), and APOBEC fragments (APOBEC_F/BE3RA_XTEN_R) with an XmaI digested pLenti-BE3RA-PGK-Puro backbone. pLenti-TRE3G-FNLS-PGK-Puro (L3FP) was generated by Gibson assembly, combining a PCR-amplified TRE3G promoter (3G_F/3G_R), and FNLS-APOBEC fragments (FNLS-APOBEC_F/BE3RA_XTEN_R) with an XmaI digested pLenti-BE3RA-PGK-Puro backbone. pCol1a1-TRE-BE3 (cTBE3) was generated by Gibson assembly, combining a PCR-amplified BE3 cDNA (cTRE_BE3_F/cTRE_BE3_R) with an EcoRI-digested pCol1a1-TRE backbone. pCol1a1-TRE-BE3RA (cTBE3RA) was generated by a two-step strategy. First, using Gibson assembly to introduce a PCR amplified UGI fragment (UGI_F/UGI_R) into a XhoI-digested pCol1a1-TRE-Cas9n backbone (Col1a1-TRE-Cas9n-UGI). Second, by restriction cloning a PCR-amplified, XhoI/EcoRV-digested APOBEC-XTEN-Cas9n (APOBEC_F2/APOBEC_R2) fragment into an EcoRV-digested Col1a1-TRE-Cas9n-UGI backbone. pLenti-U6-sgRNA-tdTomato-P2A-Blas (LRT2B) was generated by Gibson assembly, combining a PCR-amplified EFs-TdTomato-P2A-Blasticidin fragment (pLRT2B_EFs_F/pLRT2B_WPRE_R) with an XhoI/BsrGI-digested pLenti-U6-sgRNA-GFP (LRG) backbone. pLenti-VQR-P2A-Puro (LQ2P), pLenti-VRER-P2A-Puro (LER2P), and pLenti-HF1-P2A-Puro (LH2P) were generated by Gibson assembly, combining PCR amplified Cas9 variants (from Addgene stocks: #65771, #65773, and #72247, respectively; primers: KJ_Cas9_F/KJ_Cas9_R) with BamHI/NheI-digested pLenti-P2A-Puro backbone. pLenti-VQRRA-P2A-Puro (LQR2P), pLenti-VRERRA-P2A-Puro (LERR2P), and pLenti-HF1RA-P2A-Puro (LHR2P) were generated by Gibson assembly, combining one of two PCR amplified regions of the 3’ half of Cas9 (Cas9_RA_5F/Cas9_RA_5R or Cas9_RA_3F/Cas9_RA_3R), with gBlock fragments containing appropriate point mutations (VQR_GB, VRER_GB, or HF1_GB), and an EcoRV/NheI-digested pLenti-Cas9-P2A-Puro backbone. pLenti-xCas9RA-P2A-Puro, pLenti-xFNLS-P2A-Puro, pLenti-xF2X-P2A-Puro, and pLenti-xBE4Gam-P2A-Puro were generated by Gibson assembly of 4 PCR amplified regions (EF1s_xCas9_AF x xCas9_AR; xCas9_BF x xCas9_BR; xCas9_CF x xCas9_CR; xCas9_DF x xCas9_DR) and a BamHI/NheI digested pLenti-Cas9-P2A-Puro backbone. All constructs described above are schematized in Supplementary Figure 15.
Cell culture, transfection, and transduction
Culture:
HEK293T (ATCC CRL-3216) and DLD1 (ATCC CCL-221) cells were maintained in Dulbecco’s Modified Eagle’s Medium (Corning) supplemented with 10% (v/v) fetal bovine serum (FBS), at 37° with 5% CO2. PC9 (ATCC 32727) and NCI-H23 (ATCC: CRL-5800) were maintained in RMPI-1640 medium supplemented with 10% (v/v) fetal bovine serum, at 37° with 5% CO2. NIH/3T3 (ATCC CRL-1658) were maintained in Dulbecco’s Modified Eagle’s Medium (Corning) supplemented with 10% (v/v) bovine calf serum (CALF). Mouse KH2 embryonic stem cells were maintained on irradiated MEF feeders in M15 media containing LIF, as previously outlined 28.
Transfection:
For transfection-based editing experiments in HEK293Ts, cells were seeded on a 12-well plate at 80% confluence and co-transfected with 750ng of base editor (BE), 750ng of sgRNA expression plasmid, and 4.5μl of polyethyleminine (PEI; 1mg/ml). Cells were harvested for genomic DNA, 3 days post-transfection. For virus production, HEK293T cells were plated in a 6 well-plate and transfected 12 hours later (95% confluence) with a prepared mix in DMEM media (no supplements) containing 2.5μg of lentiviral backbone, 1.25μg of PAX2, 1.25μg of VSV-G, and 15μl of PEI (1mg/ml). 36hrs following transfection, media was replaced with target cell collection media and supernatants were harvested every 8-12hrs up to 72hrs post transfection. ESCs col1a1-targeting constructs were introduced via nucleofection in 16-well strips, using buffer P3 (Lonza Inc., V4XP-3032) in a 4D Nucleofector with X-unit attachment (Lonza Inc.). Two days following nucleofection, cells were treated with media containing 150ug/ml Hygromycin B and individual surviving clones were picked after 9-10 days of selection. Two days after clones were picked Hygromycin was removed from the media and cells were cultured in M15 thereafter. To confirm integration at the col1a1 locus we used a multiplex col1a1 PCR 28.
Transduction:
7.5 × 104 NIH/3T3, DLD1, PC9, and H23 cells were plated on 6-well plate. 24hrs following plating, cells were transduced with viral supernatants in the presence of polybrene (8μg/μl). Two days after transduction cells were selected in Puromycin (2 ug/ml) or Blasticidin S (4 μg/ml). 500k ESCs were plated in 6-well plates on gelatin and spinocculated (90 mins, 32°C, 2100 rpm) with 150 μl of concentrated lentiviral particles (using 100mg/ml polyethylene glycol, Sigma Aldrich P4338) in 1 ml of media containing polybrene (8μg/μl). After spin, media was replaced.
Fluorescent competitive proliferation assays:
DLD1 cells expressing BE3, RA, 2X, or FNLS were transduced with LRT2B-CTNNB1S45 or LRT2B-FANCFS1, selected with Blasticidin for 4 days and mixed at defined proportions with parental cells. 5 × 104 mixed cells were seeded in 96 well plates, treated with DMSO or 1μM XAV939 + 10nM Trametinib every 48h and remaining tdTomato-positive cells were tracked every 5 days by flow cytometry using a BD-Accuri C6 cytometer.
Organoid isolation, culture, transfection, and transduction
Organoid isolation was performed as previously described 29, 30. Briefly, 15 cm of the proximal small intestine was removed, flushed and washed with cold PBS. The intestine was then cut into 5 mm pieces and placed into 10 ml cold 5mM EDTA-PBS and vigorously resuspended using a 10ml pipette. The supernatant was aspirated and replaced with 10ml EDTA and placed at 4°C on a benchtop roller for 10 minutes. This was then repeated for a second time for 30 minutes. The supernatant was aspirated and then 10ml of cold PBS was added to the intestine and resuspended with a 10ml pipette. After collecting this 10ml fraction of PBS containing crypts, this was repeated and each successive fraction was collected and examined underneath the microscope for the presence of intact intestinal crypts and lack of villi. The 10ml fraction was then mixed with 10ml DMEM Basal Media (Advanced DMEM F/12 containing Pen/Strep, Glutamine, 1mM N-Acetylcysteine (Sigma Aldrich A9165-SG)) containing 10 U/ml DNAse I (Roche, 04716728001), and filtered through a 100μm filter. It was then filtered through a 70μm filter into an FBS (1ml) coated tube and spun at 1200 RPM for 3 minutes. The supernatant was aspirated and the cell pellet (purified crypts) were resuspended in basal media, mixed 1:10 with Growth Factor Reduced Matrigel (BD, 354230), and plated in multiple wells of a 48 well plate. After polymerization for 15 mins at 37C, 250μl of small intestinal organoid growth media (Basal Media containing 50 ng/mL EGF (Invitrogen PMG8043), 100ng/ml Noggin (Peprotech 250-38), and R-spondin (conditioned media) was then laid on top of the Matrigel.
Maintenance:
Media was changed on organoids every two days and they were passaged 1:4 every 5-7 days. To passage, the growth media was removed and the Matrigel was resuspended in cold PBS and transferred to a 15ml falcon tube. The organoids were mechanically disassociated using a p1000 or a p200 pipette and pipetting 50-100 times. 7 ml of cold PBS was added to the tube and pipetted 20 times to fully wash the cells. The cells were then centrifuged at 1000 RPM for 5 minutes and the supernatant was aspirated. They were then resuspended in GFR Matrigel and replated as above. For freezing, after spinning the cells were resuspended in Basal Media containing 10% FBS and 10% DMSO and stored in liquid nitrogen indefinitely.
Transfection:
Murine small intestinal organoids were cultured in medium containing CHIR99021 (5μM) and Y-27632 (10μM) for 2 days prior to transfection. Cells suspensions were produced by dissociating organoids with TrypLE™ express (Invitrogen #12604) for 5 min at 37°C. After trypsinization, cell clusters in 300μl transfection medium were combined with 100μl DMEM/F12-Lipofectamine2000 (Invitrogen #11668)-DNA mixture (97ul-2ul-1ug) and transferred into a 48-well culture plate. The plate was centrifuged at 600g at 32°C for 60 min, followed by another 6h incubation at 37°C. The cell clusters were spun down and plated in Matrigel. For selecting organoids with Apc mutations, exogenous RSPO1 was withdrawn 2-3 days after transfection. For selection of Pik3ca alterations, organoids were cultured in medium containing Trametinib (25nM) for 1 week.
Transduction:
Organoids were prepared as described for transfection to generate small cell clusters. They were then mixed with viral supernatant and polybrene (8μg/μl) before spinoculation and incubation at 37C. Cell clusters were plated as described for transfection and, after 48 hours, organoids were selected with Puromycin (2μg/μl) for 5-7 days (including at least one passage).
Hydrodynamic delivery
All animal experiments were authorized by the regional board Karlsruhe, Germany (animal permit number G178/16) or the Institutional Animal Care and Use Committee (IACUC) at Weill Cornell Medicine (2014-0038). Eight week-old C57Bl6/N mice (Charles River, Sulzfeld, Germany) were injected with 0.9% sterile sodium-chloride solution containing 20μg pLenti-BE3-P2A-Puro or pLenti-FNLS-P2A-Puro, 10μg of respective sgRNA vector, and 5μg pT3 EF1a-myc, as well as 1µg CMV-SB13. The total injection volume corresponded to 20% of the individual mouse body weight and was injected into the lateral tail vein in 5-7 seconds. No animals were excluded from the analyses; the investigators were not blinded for the analyses.
Lentiviral titer assay
Lentiviral titers were calculated using a quantitative PCR-based kit (LV900; Applied Biological Materials Inc.), according to the manufacturer’s instructions. Briefly, 2μl of unconcentrated viral supernatant was lysed for 3mins at room temperature, and the crude lysis was used to perform qPCR amplification. The concentration of viral particles was calculated as described in the protocol (http://www.abmgood.com/High-Titer-Lentivirus-Calculation.html).
Flow Cytometry
TdTomato protein abundance was measured by calculating mean fluorescence intensity, following analysis on a BD Accuri C6 flow cytometer. Experiments described represent three independent viral transductions, each at different MOI, to account for any effect of gene dosage.
Genomic DNA isolation.
Cells were lysed in genomic lysis buffer (10 mM Tris pH 7.5, 10 mM EDTA, 0.5% SDS, 400 μg/ml Proteinase K) for at least 2hrs at 55C. Following Proteinase K heat inactivation at 95C for 15 mins, 0.5 volumes of 5M NaCl was added and centrifuged for 10mins at 15K rpm. Supernatant was mixed with 1 volume of isopropanol and DNA precipitates were washed in EtOH 70% before resuspension in 10 mM Tris pH 8.0.
Puro copy number assay
For quantification of lentiviral integrations in transduced cells we used a custom-designed Taqman copy number assay (Invitrogen) to detect the Pac (puroR) gene. Amplification was conducted on QuantStudio 6 Real-Time PCR system (Applied biosystems), using Taqman master mix reagent (Applied biosystems) and specific primers and probe (forward-5’GCGGTGTTCGCCGAGAT; reverse-5’GAGGCCTTCCATCTGTTGCT; probe (FAM) CCGGGAACCGCTCAACTC)
Protein analysis
DLD1, PC9, and 3T3 cells were scraped from a confluent well of a 6 well plate in 100ul RIPA buffer then centrifuged at 4°C at 13,000rpm to collect protein lysates. DLD1 cells were pelleted from a confluent well of a 6 well plate at 1000rpm × 4 min, resuspended in 200 ul RIPA Buffer, then centrifuged at 4°C at 13,000rpm to collect protein lysates. Organoids were collected from confluent well of a 12 well plate (~100 ul Matrigel) in 200 ul Cell Recovery Solution (Corning, #354253), incubated on ice for 20 min, then pelleted at 300 g × 5 min. The pellet was then resuspended in 20 ul RIPA buffer, and centrifuged at 4°C at 13,000rpm to collect protein lysates. ESCs were collected at the indicated time points and filtered through a 40μm cell strainer (Fisher Scientific) to remove feeders, then pelleted at 1000 rpm × 4 min and resuspended in 100 μl RIPA Buffer. Samples were centrifuged at 4°C at 13,000rpm to collect protein lysates. Antibodies used for western blot analyses were: Cas9 (BioLegend, #844301), Actin (Abcam, #ab49900), and Apc (Millipore, MABC202).
IF staining & microscopy
2 ×104 editor-expressing 3T3s were plated in a chamber slide. 24 h after cells were wash in PBS and fix in PBS-4% PFA solution 20 min at RT and incubated in permeabilization buffer (PBS-0.5% Triton X-100) for 10 min on ice. Then, cells were stained with Cas9 (BioLegend, San Diego, CA, USA, #844301) antibody at 4°C overnight. A donkey anti-mouse Alexa 594 (Thermo Fisher Scientific, Waltham, MA, USA, # A21203) was used as secondary antibody.
Immunohistochemistry
Slides containing 3μm thick liver sections were deparaffinized and rehydrated using a descending alcohol series. For antigen retrieval slides were cooked in sodium-citrate buffer (pH 6.0) in a pressure cooker for 8 minutes. Subsequently, endogenous HRP was blocked for 10 minutes in 3% H2O2. Slides were blocked with in PBS containing 5% BSA for one hour before incubation with the primary antibody (Anti-mouse Glutamine Synthetase, BD610517, BD, Heidelberg Germany) overnight (1:200 dilution in PBS/5% BSA). Slides were washed three times and staining was visualized using DAKO Real Detection System (K5003, DAKO, Hamburg, Germany) according to the manufacturers instructions.
PCR amplification for MiSeq
Target genomic regions of interest were amplified by PCR using primers pairs listed in Supplementary oligo table. PCR was performed with Herculase II Fusion DNA polymerase (Agilent Technologies, Palo Alto, CA, USA, #600675) according to the manufacturer’s instructions using 200 ng of genomic DNA as a template and under the following PCR conditions: 95°C × 2 min, 95 °C - 0:20 → 58 °C - 0:20 → 72 °C - 0:30 × 34 cycles, 72 × 3 min. PCR products were column purified (Qiagen) for analysis by Sanger sequencing or MiSeq.
Mutation detection by T7 assay
Cas9-induced mutations were detected using the T7 endonuclease I (NEB). Briefly, an approximately 500bp region surrounding the expected mutation site was PCR-amplified using Herculase II (600675, Agilent Technologies). PCR products were column purified (Qiagen) and subjected to a series of melt-anneal temperature cycles with annealing temperatures gradually lowered in each successive cycle. T7 endonuclease I was then added to selectively digest heteroduplex DNA. Digest products were visualized on a 2.5% agarose gel.
Off-target predictions
sgRNA-dependent off-target mutations were predicted from previous publication 31 or using the ‘Cas-OFFinder’ prediction tool (http://www.rgenome.net/cas-offinder) 32. Sites were prioritized as the most likely to show off-target editing if they contained the fewest mismatches and those mismatches were clustered toward the 5’end of the sgRNA.
DNA Library Preparation and MiSeq
DNA library preparations and sequencing reactions were conducted at GENEWIZ, Inc. (South Plainfield, NJ, USA). NEB NextUltra DNA Library Preparation kit was used following the manufacturer’s recommendations (Illumina, San Diego, CA, USA). Adapter-ligated DNA was indexed and enriched by limited cycle PCR. The DNA library was validated using TapeStation (Agilent Technologies, Palo Alto, CA, USA), and was quantified using Qubit 2.0 Fluorometer. The DNA library was quantified by real time PCR (Applied Biosystems, Carlsbad, CA, USA). The DNA library was loaded on an Illumina MiSeq instrument according to manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was performed using a 2×150 paired-end (PE) configuration. Image analysis and base calling was conducted by the MiSeq Control Software (MCS) on the MiSeq instrument and verified independently using a custom workflow in Geneious R11.
Identification of recurrent cancer-associated mutations
Using MSK IMPACT targeted deep sequencing of 473 cancer-relevant genes across 22,647 patient samples, we identified recurrent somatic variants present in 4 or more individual samples. This generated a list of 2,696 somatic missense, nonsense, and splice site mutations. The flanking sequences around each mutation were retrieved and queried for the presence of a relevant PAM (NGG for FNLS and 2x; NG for xFNLS and xF2X) within a specified distance downstream of the target cytosine, using the following packages (implemented in R; https://cran.r-project.org/): Bioconductor, BSgenome, Biostrings. For G>A mutations, the reverse complement strand was examined. Target cytosine (or guanines) were considered ‘editable’ if they were within positions 4-8 of the protospacer (for FNLS and xFNLS) or postitions 4-11 (for 2X and xF2X). The presence of a non-targeted cytosine in the editing window was noted and editable mutations were parsed into those in which only the target cytosine was edited (scarless) and those in which an additional cytosine is predicted to be altered (scar).
Statistics
All statistical tests used throughout the manuscript are indicated in the appropriate figure legends. In general, to compare two conditions, a two-sided Student’s t-test was used, assuming unequal variance between samples. In most cases, analyses were performed using one-way or two-way ANOVA, using Tukey’s correction for multiple comparisons. Unless otherwise stated, each replicate represents a “biologically independent experiment” which indicates either: independent cell transfections, independently transduced cell lines, or independent animals. Results of all statistical tests are available in Supplementary Table 6.
Data availability
No data sets for deposit were generated in this study. All lentiviral and expression vectors described have been deposited at Addgene for distribution.
Supplementary Material
Acknowledgements
This work was supported by a project grant from the NIH/NCI (CA195787-01), a U54 grant from the NIH/NCI (U54OD020355), project grant from the Starr Cancer Consortium (I10-0095), a Research Scholar Award from the American Cancer Society (RSG-17-202-01), and a Stand Up to Cancer Colorectal Cancer Dream Team Translational Research Grant (SU2C-AACR-DT22-17). Stand Up to Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, a scientific partner of SU2C. MPZ is supported in part by National Cancer Institute (NCI) Grant NIH T32 CA203702. EMS was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM07739 to the Weill Cornell / Rockefeller / Sloan-Kettering Tri-Institutional MD-PhD Program, and an F31 Award from the NCI/NIH under grant number 1 F31 CA224800-01. ERK is supported by an F31 NRSA predoctoral fellowship from the NCI/NIH under Award F31CA192835. FJSR was supported by the MSKCC TROT program (5T32CA160001) and is an HHMI Hanna Gray Fellow. SWL is the Geoffrey Beene Chair of Cancer Biology and an Investigator of the Howard Hughes Medical Institute. DFT is supported by the Helmholtz Association (VH-NG-1114) and by the German Research Foundation (DFG) project B05, SFB/TR 209 “Liver Cancer”. LED was supported by a K22 Career Development Award from the NCI/NIH (CA 181280-01). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Competing Financial Interests Statement
The authors declare no competing financial interests
References
- 1.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishida K et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Hess GT et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods 13, 1036–1042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Y et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods 13, 1029–1035 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox DBT et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rees HA et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun 8, 15790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim K et al. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol 35, 435–437 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Bae T, Hwang J & Kim JS Rescue of high-specificity Cas9 variants using sgRNAs with matched 5’ nucleotides. Genome Biol 18, 218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleinstiver BP et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dow LE et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 33, 390–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staahl BT et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol 35, 431–434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komor AC et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci Adv 3, eaao4774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang SM et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Schoumacher M et al. Inhibiting Tankyrases sensitizes KRAS-mutant cancer cells to MEK inhibitors via FGFR2 feedback signaling. Cancer Res 74, 3294–3305 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Mashima T et al. mTOR signaling mediates resistance to tankyrase inhibitors in Wnt-driven colorectal cancer. Oncotarget 8, 47902–47915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matano M et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nature medicine 21, 256–262 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Cadoret A et al. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene 21, 8293–8301 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Annunziato S et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev 30, 1470–1480 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D et al. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Human gene therapy 26, 432–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koblan LW et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryu SM et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol 36, 536–539 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Yin H et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol 35, 1179–1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dow LE et al. A pipeline for the generation of shRNA transgenic mice. Nature protocols (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han T et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun 8, 15945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Rourke KP et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat Biotechnol 35, 577–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae S, Park J & Kim JS Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data sets for deposit were generated in this study. All lentiviral and expression vectors described have been deposited at Addgene for distribution.