Abstract
Bacteria are known to use RNA, either as mRNAs encoding proteins or as non-coding small RNAs (sRNAs), to regulate numerous biological processes. However, a few sRNAs have two functions: they act as base pairing RNAs and encode a small protein with additional regulatory functions. Thus, these so called “dual-function” sRNAs can serve as both a riboregulator and an mRNA. In some cases, these two functions can act independently within the same pathway while in other cases, the base-pairing function and protein function act in different pathways. Here, we discuss the five known dual-function sRNAs: SgrS from enteric species, RNAIII and Psm-mec from Staphylococcus aureus, Pel RNA from Streptococcus pyogenes, and SR1 from Bacillus subtilis, and review their mechanisms of action and roles in regulating diverse biological processes. We also discuss the prospect of finding additional dual-function sRNAs and future challenges in studying the overlap and competition between the functions.
INTRODUCTION
Bacteria have evolved elaborate responses to sense, protect against, and help recovery from stressful fluctuations in environmental conditions. In the past decade, small regulatory RNAs (sRNAs) have emerged as important players in the post-transcriptional regulation of various stress responses. Advances in deep sequencing have led to the identification of hundreds of these sRNAs which range from 50–350 nucleotides in length thereby greatly increasing the numbers of known sRNAs (1). Usually, these sRNA regulators are thought to be non-coding and are generally presumed to act by modulating the stability and translation of mRNAs through short base-pairing interactions or by binding to and modulating the activities of RNA binding proteins.
The majority of the base pairing sRNAs prevent translation of their mRNA targets, at times leading to degradation of the sRNA-mRNA complex (2). For this category of sRNAs, base-pairing interactions occlude the ribosome binding site (RBS) of the mRNA and thus inhibit translation initiation (3, 4). There is also a small subset of sRNAs that stabilize transcripts that are otherwise prone to degradation by cellular RNases (5, 6). In addition, sRNAs that base-pair within the 5’ untranslated region (UTR) of the mRNA can open up an inhibitory secondary structure that normally prevents translation of the mRNA (7). Irrespective of the mechanism of regulation, many sRNAs require the Sm-like RNA chaperone protein Hfq. Without Hfq, sRNAs tend to be unstable and unable to regulate their respective mRNA targets (8). The sRNAs also generally require Hfq to promote sRNA-mRNA binding and to remodel RNA secondary structures. Thus far, Hfq-dependent sRNAs and their mRNA targets have been characterized through genome-wide and bioinformatic searches and co-immunoprecipitation experiments with Hfq (9–11).
The majority of sRNAs do not contain an open reading frame (ORF) and are thus thought to be non-coding and function exclusively through sRNA-mRNA or sRNA-protein interactions. However, a few sRNAs contain short ORFs and in a small number of cases, translation of these ORFs has been shown. Of these, there are fewer still that have an experimentally demonstrated function (12, 13). These sRNAs encoding small proteins form a special class of sRNAs refered to as “dual-function sRNAs”. The small proteins tended to be overlooked due to challenges related to their annotation and biochemical detection. However, recent work suggests that the prevalence of dual-function sRNAs may be greater than currently appreciated. Computational analyses of the genomes of fourteen phylogenetically diverse bacteria predicted many sRNAs encoding proteins between 10–50 amino acids. These analyses took into account sequence features and comparative genomics to quantify the sRNA ORFs under natural selection to maintain protein-coding function (14). A second major problem with respect to studies of small proteins are barriers to finding functions of this class of proteins, particularly given that tags commonly used to characterize larger proteins can have adverse effects on proteins, which may be smaller than the tag itself.
The characterized dual-function sRNAs that act by sRNA-mRNA interactions and encode small proteins will be the focus of this chapter. Other classes of multi-functional RNAs will also be considered briefly. One class of dual-function RNAs is comprised of known mRNAs that contain sRNAs derived from the 3’ UTRs of these mRNAs either by processing or by transcription from an independent promoter located within the 3’ end of the ORF (15). Another variety of dual-function sRNAs carry out regulation of mRNA targets via base-pairing but can also bind proteins and regulate the activities of these proteins (16). There is also a growing list of examples of tRNA fragments derived from processing of pre-tRNA transcripts by RNases that have been shown to base pair with sRNAs or other RNA targets to carry out a variety of regulatory effects (17).
BASE-PAIRING sRNAs THAT ENCODE CHARACTERIZED SMALL PROTEINS
Despite the challenges in identifying and characterizing the function of small proteins encoded by known sRNAs, to date, five sRNAs have been defined as dual-function RNAs with characterized base pairing and protein-coding functions. These include SgrS from enteric species (18, 19), RNAIII and Psm-mec from Staphylococcus aureus (20), Pel RNA from Streptococcus pyogenes (21), and SR1 from Bacillus subtilis (22). All of these and other potential dual-function RNAs are involved in myriad physiological responses including quorum sensing, virulence regulation, and metabolic regulation. Below we provide an overview of the individual dual-function sRNAs discussing in detail what is known about their base-pairing function, function of the encoded protein as well as the interplay between the two different functions thereby providing insight into how a dual-function sRNA coordinates the two different activities to carry out its role in the cell.
Enterobacterial SgrS
SgrS has been well characterized in E. coli and Salmonella and is currently the only known dual-function sRNA in gram-negative bacteria (Figure 1). Orthologs of SgrS are also found in numerous γ-proteobacteria such as Shigella sp., Klebsiella pneumoniae, Erwinia amylovora, Citrobacter koseri and Yersinia pestis (23). The 227-nucleotide SgrS sRNA was first discovered in a computational screen for sRNAs in E. coli (24) and later found to play an important role in mediating the cellular response to glucose-phosphate stress (25). Bacteria use specific phosphoenolpyruvate-phosphotransferase systems (PTS) to take up sugars, such as glucose and mannose, and then phosphorylate them (see chapter by Göpel, Durica-Mitic and Görke). Glucose phosphate stress occurs when sugar transport and metabolism become uncoupled. This can occur in certain mutant strains (26, 27) or in wild-type strains that take up the non-metabolizable sugar analogs α-methylglucoside (αMG) and 2-deoxyglucose (2DG) (3, 25). Accumulation of sugar phosphates or their analogs and depletion of other glycolytic intermediates results in growth inhibition (28). This stress condition is sensed by the transcription factor SgrR and allows for sgrS transcription. Both SgrS and SgrR are required for growth under glucose phosphate stress (29).
Figure 1:

Sugar phosphate stress caused due to intracellular accumulation of phosphosugars triggers expression of the transcription factor SgrR. SgrR, in turn induces transcription of the 227 nucleotide sRNA SgrS, which also encodes a small 43 aa protein SgrT (blue). The other features of this sRNA include the base-pairing region (red) and the Hfq binding region (poly U tail). To relieve the sugar phosphate stress SgrS represses translation of mRNAs coding for sugar transporters (PtsG and ManXYZ) and other mRNAs involved in various metabolic pathways (Asd, AdiY, FolE and PurR) to help restore metabolic homeostasis during stress conditions. SgrS also activates translation of a phosphatase (YigL) that dephosphorylates the phosphosugars for export out of the cell. SgrT meanwhile is expressed from SgrS later and inhibits the activity of the glucose transporter PtsG, thereby both the sRNA and the encoded small protein act together in the same pathway to combat sugar phosphate stress.
SgrS regulation of three different mRNAs reduces the amount of sugar phosphate accumulation in the cell by reducing synthesis of sugar transporters and enhancing dephosphorylation of the sugar phosphates. The sugar analogues αMG and 2DG are taken into the cell and then phosphorylated by PTS proteins PtsG (EIICBGlc) and ManY (EIICMan), respectively. Upon accumulation of phosphorylated αMG and 2DG in the cytoplasm, SgrS is expressed and represses translation of the two mRNA targets, ptsG (25) and manXYZ (30). SgrS base pairs with ptsG mRNA through a short region of complementarity that includes the RBS of ptsG mRNA. This base pairing interaction prevents translation of the encoded PtsG protein and allows for subsequent RNase E-dependent degradation of ptsG mRNA (25, 31, 32). SgrS again uses direct base pairing to repress the manXYZ polycistronic mRNA. Here, SgrS is able to bind manXYZ mRNA at two distinct sites; one within the manX ORF and the other in the untranslated region between manX and manY. Similar to the ptsG mRNA, SgrS binding impedes manXYZ mRNA translation and facilitates mRNA degradation by RNase E (30, 33). SgrS activates a third mRNA target, yigL, which encodes a haloacid dehalogenase-like phosphatase that dephosphorylates the phosphorylated sugars prior to their efflux. The yigL gene is in an operon with an upstream gene, pldB, and the dicistronic pldB-yigL mRNA is processed by RNase E, which results in the ‘pldB-yigL mRNA that is susceptible to further degradation. SgrS stabilizes ‘pldB-yigL mRNA by base pairing to and occluding a specific RNase E cleavage site upstream of the yigL coding region to prevent further mRNA degradation (6).
Very recently, SgrS was shown to regulate four more mRNAs: asd, adiY, folE, and purR (34). In typical sRNA-mediated repression, SgrS base pairs within the 5’ UTR of adiY and folE, which encode arginine decarboxylase gene activator and GTP cyclohydrolase I, respectively, to occlude the RBS and prevent translation initiation of these mRNAs. Similar to manXYZ, SgrS binds asd mRNA (encoding aspartate semialdehyde dehydrogenase) at two distinct sites; one that directly occludes the RBS and the other in the asd ORF. SgrS binding to both sites is required for full translational repression. Lastly, SgrS binds within the purR (encoding a repressor of purine biosynthesis) coding sequence and recruits the RNA chaperone Hfq where it can interfere with ribosome binding and directly repress purR translation. Interestingly, Hfq alone can repress purR translation while SgrS alone has a very modest effect. These enzymes and transcription factors belong to a diverse set of metabolic pathways and, while their specific role in glucose phosphate stress has not been defined, their repression by SgrS may help restore metabolic homeostasis during stress conditions.
Along with functioning as a base-pairing RNA, the SgrS sRNA encodes a 43-aa protein SgrT which also functions in the glucose-phosphate stress response (35). The E. coli sgrT coding sequence is located at the 5’ end of SgrS (nts 22 through 153) with the middle region responsible for base pairing with mRNAs (approximately nt 167 through 187) and the 3’ end responsible for binding Hfq (35). Ectopic overexpression of sgrS alleles that possess only base-pairing activity or only produced SgrT was sufficient to rescue cells from glucose phosphate stress indicating that the sRNA and small protein have redundant functions. SgrT by itself was found to have no effect on ptsG mRNA stability or translation, indicating that it must affect recovery from glucose- phosphate stress using a different mechanism (4, 18). In the years following the discovery of SgrT, the mechanism by which it allowed recovery from glucose-phosphate stress was unknown. However it has now been shown that SgrT acts to specifically inhibit the transport activity of the major glucose permease PtsG (36). The primary transporter of glucose is the glucose-PTS complex comprised of two subunits: the transmembrane EIICBGlc protein, which is a glucose permease and is encoded by ptsG, and the cytoplasmic EIIAGlc protein, which functions as an intermediate phosphotransfer protein and is encoded by crr (37). Superresolution microscopy, which showed colocalization of SgrT and PtsG in a PtsG-dependent manner, indicated SgrT binds the EIICGlc domain of PtsG to inhibit further glucose transport (36). This model was also supported by the observation of strong SgrT-mediated growth inhibition when cells were grown in glucose media requiring the IIC domain of PtsG. The inhibition of the EIICGlc domain of PtsG by SgrT possibly leads to accumulation of phosphorylated EIIAGlc, which can no longer bind and block other transport proteins (relieving inducer exclusion). Thus, together, SgrS and SgrT block further glucose-6-phosphate (G6P) accumulation, diminish intracellular G6P levels, and promote the utilization of alternative carbon sources. It is interesting to note that though the base-pairing activity of SgrS represses the synthesis of both PtsG and ManXYZ, SgrT had no effect on ManXYZ transport activity (36). Overall, these new results support the model in which the base-pairing activity of SgrS acts to inhibit new PtsG synthesis, while SgrT inhibits the activity of preexisting PtsG.
Little is known about how the base pairing function of SgrS and translation of sgrT are coordinated. The 5’ end of SgrS contains the open reading frame for SgrT and the nucleotides used for the base pairing function of SgrS are 15 nt downstream of the SgrT stop codon. Thus, a ribosome located at the sgrT stop codon would block the important base pairing nucleotides for SgrS. This block, as well as the fact that sRNA binding to mRNA targets leads to coupled SgrS-mRNA degradation, led to the hypothesis that the translation and base pairing functions of SgrS are mutually exclusive. Thus, a given SgrS molecule could serve as a substrate for sgrT translation or as an sRNA with base pairing ability, but not both simultaneously. In Salmonella, mutations that impair sgrT translation have no effect on the ability of SgrS to regulate mRNA targets via base pairing (4). However, mutations that impaired base-pairing interactions of SgrS lead to increased SgrT production. This is consistent with the model that SgrS molecules engaged in base pairing to mRNA are unavailable for translation, and thus removing base-pairing ability would increase the pool of SgrS molecules available for translation of sgrT. Lastly, it was shown that SgrT and SgrS base-pairing function act at different times during glucose-phosphate stress. SgrS RNA is produced rapidly in response to initial stress and the base-pairing function acts immediately, whereas SgrT is not detected until 30 min following the onset of stress and is required to inhibit activity of preexisting PtsG (Figure 1).
Staphylococcus aureus RNAIII
RNAIII, the first dual-function sRNA identified, is an important virulence regulator in the human pathogen S. aureus (38). Specifically, RNAIII is the effector of the staphylococcal accessory gene regulator (agr) quorum-sensing system, which has been assigned a central role in the pathogenesis of S. aureus (Figure 2) (see chapter by Westermann). The agr system decreases the expression of several cell surface proteins and increases the expression of many secreted virulence factors in the transition from late-exponential growth to stationary phase in vitro (39, 40). The agr locus comprises two operons P1 and P2 encoding transcripts RNAII and RNAIII respectively. The RNAII transcript produces four proteins AgrB, AgrD, AgrC and AgrA that make up the agr sensing mechanism. AgrD and AgrB constitute the cell density sensing cassette involved in producing the auto-inducing peptide (AIP). AgrD encodes pro-AIP, and the transmembrane protein AgrB processes pro-AIP to AIP, transporting it to the external cellular space. In late exponential phase, upon reaching critical concentration, AIP then activates the two component sensory transduction system comprised of AgrC and AgrA. AIP binds to AgrC, a kinase embedded within the membrane which in turn phosphorylates the DNA binding regulator AgrA, which is then responsible for the increased transcription of RNAII and RNAIII (41). RNAIII produced at the end of exponential phase is responsible for repressing the synthesis of early virulence genes and surface proteins (which are needed during early infection) and increasing the production of secreted factors (which are necessary for late infection) (42). Nearly all S. aureus clinical isolates from acute infections produce RNAIII, highlighting its importance as a central virulence regulator.
Figure 2:
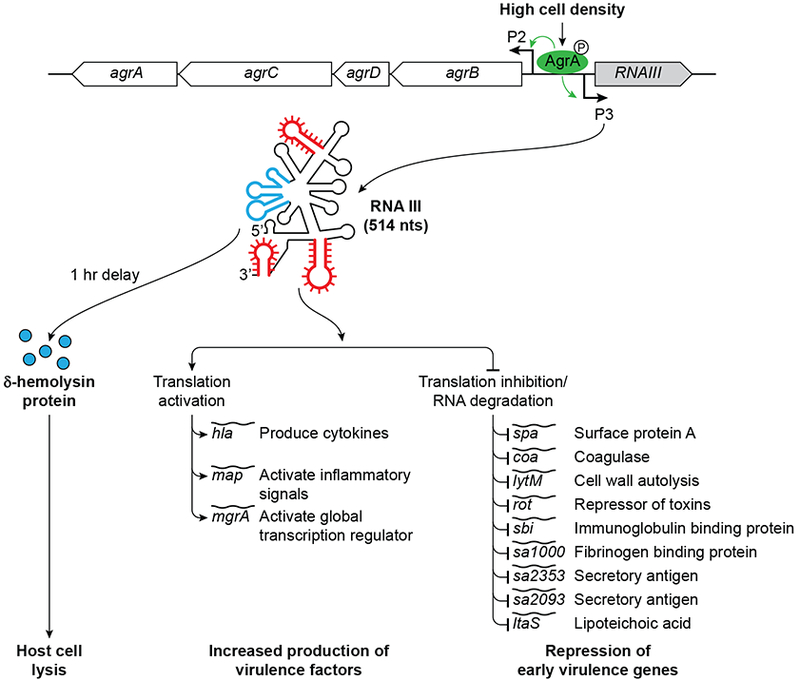
RNAIII is part of the global regulatory locus known as the accessory gene regulator (agr) locus, which encodes the components of an autoregulatory quorum-sensing system. The agr locus consists of two divergent transcripts, RNAII and RNAIII, which initiate from promoters P2 and P3, respectively. Increases in cell density lead to phosphorylation and activation of the DNA-binding response regulator AgrA. Phosphorylated AgrA in turn activates transcription from the P2 and P3 promoters; P3 activation leading to expression of RNAIII, the major effector molecule of the agr response. The secondary structure of the 514 nucleotide RNAIII consists of 14 stem loop structures with multiple base-pairing regions (red). RNAIII encodes a 26 aa delta hemolysin protein (blue, hld) but also acts as a post-transcriptional regulator of several mRNAs most of which impact virulence. The RNA activates expression of Map, alpha hemolysin and MgrA proteins by either promoting a more open secondary structure surrounding the RBS by base-pairing in the case of map and hla mRNAs or by stabilizing the RNA in the case of mgrA. RNAIII is also involved in translation inhibition and RNA degradation of various mRNAs involved in the early stages of infection.
RNAIII is 514-nt long and has a complex secondary structure composed of fourteen stem-loop (SL) motifs referred to, from 5’ to 3’, as SL1 through SL14 (43). To date, RNAIII is known to be directly involved in base pairing-dependent regulation of 12 mRNA targets, including two transcription factors. Generally, stem-loops in the 5’ UTR of RNAIII are involved in target activation while stem-loops in the 3’ region of RNAIII are involved in target repression, however there are exceptions. The 3’ end of RNAIII is more conserved among different S. aureus isolates compared to other regions of the sRNA (44). This 3’ end contains several CU-rich domains present in apical loops and unpaired regions that were found to base pair with the RBS of several target mRNAs. This base pairing blocks ribosome binding, prevents translation initiation and in some cases facilitates the subsequent degradation of the mRNA by RNase III. Using this mechanism, RNAIII represses synthesis of multiple targets, all of which are involved in the early stages of infection. These targets include: surface protein A (spa), coagulase (coa), fibrinogen-binding protein (SA1000), homologs of staphylococcal secretory antigen SsaA (sa2353 and sa2093), immunoglobulin-binding protein (sbi), lipoteichoic acid synthase (ltaS), and the major cell wall autolysin (lytM). In the case of spa repression, annealing of RNAIII to spa mRNA can inhibit the formation of the translation initiation complex but recruitment of the double-strand-specific endoribonuclease III (RNase III) by RNAIII is essential to degrade the mRNA and completely repress spa translation (45). RNAIII also represses another important target, rot mRNA, which encodes the transcription factor Rot (repressor of toxins) (46). Through its regulation of Rot, RNAIII is able to indirectly control the transcription of many secondary targets.
RNAIII activates three mRNA targets: α-hemolysin (hla), extracellular adherence Map protein (map), and the global transcription regulator MgrA (mgrA). In the absence of RNAIII, translation of hla is prevented by a stable intrinsic RNA hairpin that sequesters the RBS. Base pairing between the 5’ end of RNAIII (SL2 and SL3) and the hla 5’ UTR, releases the hairpin structure to allow ribosome binding and hla translation (47). A similar mechanism is predicted for activation of map translation after interaction with SL4 of RNAIII (48).
In contrast to hla and map, which interact with the 5’ end of RNAIII, mgrA mRNA interacts with both the 5’ end (SL2) and 3’ end (SL13) of RNAIII (49). MgrA is a global transcriptional regulator that controls more than 350 genes involved in virulence, antibiotic resistance, and biofilm formation (50–54). MgrA is transcribed from two promoters, both of which result in transcripts with unusually long 5’ UTRs. RNAIII base pairs within the 5’ UTR of only the longer transcript to stabilize the mRNA against degradation by an unknown ribonuclease and allows for increased production of MgrA. MgrA is the second master global regulator through which RNAIII can indirectly control the expression of multiple secondary targets, which significantly increases the influence of RNAIII (49).
RNAIII also contains an ORF that encodes the 26 amino acid cytotoxic peptide δ- hemolysin (hld), which targets host cell membranes and causes cell lysis (55). In contrast to other hemolytic peptides, δ-hemolysin is not active against bacteria. The ORF encoding δ-hemolysin is located towards the 5’ end of RNAIII and encompasses SL3, SL4, and SL5. Because δ-hemolysin synthesis is controlled by the levels of RNAIII, δ-hemolysin is produced in the late exponential phase. Intriguingly, the translation of hld is delayed by 1 hour after RNAIII transcription (56). The 5’ and 3’ ends of RNAIII are in close proximity suggesting that this delay may be due to intramolecular interactions of the 3’ end of RNAIII with the RBS of hld resulting in the RBS of hld being occluded in a secondary structure (43, 56). Deletion of the 3’ end of RNAIII eliminates the delay between RNAIII production and the appearance of δ-hemolysin, again suggesting the presence of translation-inhibitory structure between the 3’ end of RNAIII and the hld RBS (56). The factors mediating the conformational change in RNAIII secondary structure to allow δ-hemolysin production are not currently known.
Staphylococcus aureus Psm-mec
The psm-mec gene is encoded on staphylococcal cassette chromosome (SCCmec), the mobile genetic element that confers antibiotic resistance to methicillin-resistant S. aureus (MRSA). The importance of the psm-mec RNA first came to light when researchers investigated why community-acquired (CA)-MRSA, which infects otherwise healthy people outside of the hospital, was more virulent and produced higher amounts of exotoxins than hospital-associated (HA)-MRSA, which infects immunocompromised patients in hospitals. They determined that the SCCmec of CA-MRSA did not contain the psm-mec locus that exists in the HA-MRSA SCCmec (20). They also found the psm-mec transcript reduced the expression of phenol-soluble modulin α (PSMα), a cytolytic toxin of S. aureus, resulting in decreased extracellular toxin production (20). However, other effects of the psm-mec locus in S. aureus on biofilm formation, cell spreading, and the expression of PSMα virulence factors are highly strain-dependent.
The psm-mec sRNA is about 143–157 nt in length with the agrA gene being the only known target encoding a positive transcription factor for the agr quorum sensing system (Figure 3) (57, 58). psm-mec RNA represses translation of the agrA transcript by directly binding within the agrA coding region (around 200 nts downstream of the start codon) which results in decreased extracellular toxin production, specifically PSMα. The exact mechanism by which psm-mec RNA base pairing causes agrA translation inhibition is still unknown. Base-pairing with Psm-mec decreases the stability of agrA mRNA in a RNase III dependent manner, however, psm-mec RNA base pairing can also decrease agrA translation independent of this decrease in agrA mRNA stability and RNase III (58). The transcription factor AgrA activates the transcription of multiple genes including rnalII, and psmα, so psm-mec RNA mediated regulation of agrA translation may indirectly control the transcription of many secondary targets. Lastly, the regulatory effects of the psm-mec RNA are highly strain-dependent for reasons that are not yet well understood (58, 59). For example, deletion of psm-mec resulted in an increase of AgrA compared to the parental strain in only 14 out of 18 isolates (58).
Figure 3:
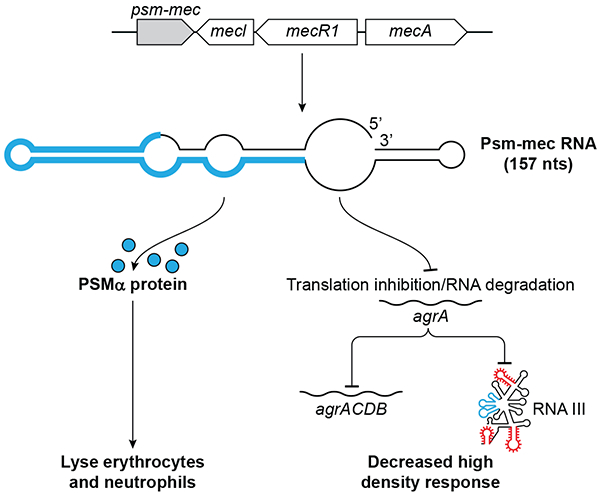
Psm-mec is located on staphylococcal cassette chromosome (SCCmec), next to the mecI/mecR1/mecA genes that confer methicillin resistance and its regulation. The 143–157 nucleotide sRNA also encodes PSM-mec a 22 aa cytolytic toxin with the ORF making most of the transcript (blue). The protein plays a role in S. aureus infection and immune evasion while the sRNA represses the translation of agrA mRNA by inhibiting translation and affecting the stability of the mRNA.
psm-mec RNA also encodes a 22-aa phenol-soluble modulin (PSM) named PSM-mec (60), with the PSM-mec ORF making up most of the transcript. This protein is expressed with an N-terminal formyl-methionine and secreted without a signal peptide (61). This small protein is a staphylococcal cytolytic toxin that plays an important role in S. aureus infection and immune evasion. The fact that PSM-mec is encoded on the SCCmec mobile genetic element makes it, to date, the only known PSM that is not encoded on the staphylococcal core genome (60). Regulation of PSM-mec production is not well understood but it has been observed that PSM-mec, like all other PSMs, is positively regulated by the Agr quorum sensing system (57, 59). In contrast to all other known PSMs, which completely lack cysteine, PSM-mec contains one cysteine residue. This cysteine residue may be important for PSM-mec secondary structure, as it is necessary for pro-inflammatory and cytolytic activities of PSM-mec (60). PSM-mec expression varies widely among MRSA isolates, which may be why the phenotypes associated with the protein such as adhesion to surfaces and biofilm formation also differ among isolates (59, 60).
Bacillus subtilis SR1
SR1, the first dual-function sRNA discovered in B. subtilis, was identified via a bioinformatics approach that searched for sRNAs in intergenic regions of the B. subtilis chromosome. The 205-nt sRNA is expressed under gluconeogenic and repressed under glycolytic conditions mainly by CcpN but also to a minor extent by CcpA, both regulators are involved in carbon catabolite repression (Figure 4) (22, 62–64).
Figure 4:
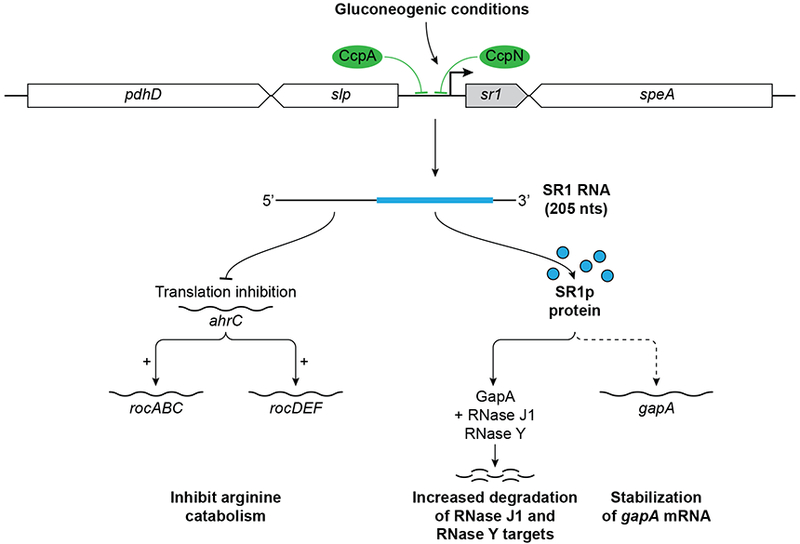
SR1 gene is encoded between pdhD and speA. Its transcription is repressed by CcpA and CcpN under glycolytic conditions. The 205 nucleotide sRNA expressed under gluconeogenic conditions and in presence of L-arginine also encodes a small 39 aa protein SR1P (blue). The ORF and the base-pairing region overlap on this sRNA. In the presence of arginine, SR1 represses translation of the ahrC mRNA, the transcriptional activator of two arginine catabolic operons rocABC and rocDEF. The small protein SR1P plays a role in gluconeogenic conditions by binding to GapA and stabilizing the gapA operon mRNA from degradation by an unknown mechanism. It also binds RNase J1 and enhances its activity. Thus, the activities of the small protein and base pairing RNA affect different pathways.
SR1 transcription is induced by L-arginine and its degradation product L-ornithine through an unknown mechanism and is involved in regulation of arginine catabolism by basepairing with its target, the ahrC mRNA (65). AhrC is the transcriptional activator of the rocABC and rocDEF arginine catabolic operons. SR1 base pairs within the coding region of ahrC mRNA potentially using 7 complementary regions that make up the 3’ half of SR1 and the central and 3’ end of ahrC mRNA (65, 66). This base-pairing interaction induces structural changes around 20 nucleotides downstream of the ahrC start codon that inhibits translation initiation by preventing binding of the ribosomal 30S subunit. Decreased AhrC consequently leads to decreased expression of the rocABC and rocDEF mRNA . Consistent with the regulation, deletion of SR1 causes increased levels of the arginine catabolism proteins RocA, RocD, and RocF (13, 65). The regulation of ahrC by SR1 was found to not require Hfq, although the Hfq is able to bind both SR1 and ahrC mRNA. Instead, Hfq is required for proper ahrC translation possibly by opening secondary structures that otherwise inhibit binding of the 30S initiation complex. This model is supported by the identification of an Hfq-binding site (5’ AAAUA) immediately upstream of the ahrC ribosome-binding site (RBS) and the observation that the ahrC mRNA is not translated in an hfq knockout strain .
In addition to its base pairing activity, SR1 encodes a 39-aa protein called SR1P. Both functions of SR1 are highly conserved; as SR1 and SR1P homologs with high structural identity have been identified in 23 species of the Bacillales order (13, 67). During the search for additional targets of SR1, gapA mRNA, encoding one of the two glyceraldehyde-3P-dehydrogenases of B. subtilis, was found to be increased in the presence of SR1. Moreover it was observed that gapA mRNA is rapidly degraded in the absence of SR1 indicating that gapA could be a potential target of SR1. However, it was also observed that SR1 did not affect transcription or translation of the gapA operon. Instead, it was shown that there is a direct interaction between SR1P and GapA and that this interaction inhibited the degradation of gapA operon mRNAs by an unknown mechanism (68). In 2016, the function of this interaction was determined. In B. subtilis, GapA plays a role in RNA degradation by binding RNase J1 and the main endonuclease RNase Y (69). SR1P binds to GapA and promotes GapA binding of RNase J1, which enhances RNase J1 activity (69). Since SR1P is only expressed under gluconeogenic conditions where the metabolic activity of GapA is not needed, SR1P modulation of the GapA ability to recruit RNases links RNA degradation to the metabolic state of the cell. Given that the SR1 RNA affects arginine catabolism and SR1P affects RNA stability, SR1 is an example of a dual-function sRNA, where the regulatory RNA and the small protein have functions in very different pathways.
Streptococcus pyogenes Pel RNA
Pel/SagA (pleiotropic effect locus/streptolysin-associated gene A) RNA was one of the first sRNAs studied in S. pyogenes. Pel is a 469-nt sRNA expressed from the first gene of the sagABCDEFGHI operon involved in the processing and export of Streptolysin S in a growth phase-dependent manner (21, 70). Deletion of pel sRNA resulted in increased transcription of various virulence factor genes such as those coding for Streptococcal inhibitor of complement (sic), NAD-glycohydrolase (nga), and M-protein (emm) and is associated with delayed maturation of the cysteine protease SpeB (21). However, the mechanism as to how Pel transcriptionally or post-transcriptionally regulates these targets remains elusive. Due to the observed regulation at the transcriptional level, Pel RNA conceivably could base pair with the mRNA of a global transcriptional regulator, however this has yet to be reported. Moreover, the effects of the Pel RNA seem to be strain-specific as pel deletion in four M1T1 S. pyogenes isolates had no effect on the transcription of the previously identified targets (71).
Pel has much similarity to RNAIII, not only with respect to its length but also because base-pairing and mRNA functions are very similar. Like RNAIII, Pel encodes a haemolytic peptide, the 53-aa Streptolysin S, a virulence factor responsible for the β- hemolytic phenotype of S. pyogenes (72). The protein is synthesized as a prepropeptide with a Gly-Gly proteolytic cleavage site that has been predicted to release a 30-aa propeptide from the 23-aa leader sequence. The remaining genes in the operon are required for efficient post-translational modification and export of Streptolysin S peptide (73).
DUAL-FUNCTION sRNAS WITH UNCHARACTERIZED PEPTIDE FUNCTIONS
Other sRNAs with established base-pairing functions could also encode small proteins. However, translation of the protein has only been observed for one: PhrS from Pseudomonas aeruginosa. As an sRNA, PhrS, whose expression requires the oxygen-responsive regulator ANR, activates translation of the quorum sensing and virulence regulator PqsR (74). PhrS binds in the 5’ UTR of an ORF upstream of the pqsR gene, whose translation is coupled to that of pqsR (74). The physiological relevance of the 37- aa protein encoded by PhrS is currently unknown. Interestingly, the ORF in phrS is more conserved than the sRNA sequence (74). Other potential dual-function sRNAs include VR-RNA from Clostridium perfringens, which controls a variety of different mRNAs resulting in activation of virulence and contains 72-aa ORF denoted hyp7, the translation of which has not been reported (75). The sRNA RivX in S. pyogenes controls transcriptional activation of the streptococcal virulence regulator Mga and also encodes a 32-aa protein (76). The Streptomyces coelicolor sRNA scr5239 inhibits translation of the dagA (encodes agarase) and metE (encodes methionine synthase) mRNAs, and contains a 33 codon ORF (77, 78). Another potential dual-function sRNA in gram-negative bacteria is the sRNA RSs0019 in Rhodobacter sphaeroides. RSs0019 is produced in response to photo-oxidative stress however, no target mRNAs have been identified and translation of the 50 codon ORF has not been observed (79–81).
OTHER TYPES OF MULTIFUNCTIONAL sRNAs/mRNAs
Although the sRNAs discussed thus far in this review function dually as both sRNA and mRNA, there are a few other sRNAs with multiple mechanisms of action. One example is the McaS sRNA of E. coli, which represses synthesis of CsgD, the master regulator of curli biogenesis, and activates synthesis of FlhD, the master regulator of flagellar biogenesis as a typical base pairing sRNA (82, 83). However, McaS also binds the global RNA-binding protein regulator CsrA, which negatively regulates pga translation. Through McaS sequestration of CsrA, pga expression increases, leading to heightened production of β−1,6 N-acetyl-D-glucosamine (PGA) and increased biofilm formation (see chapter by Romeo and Babitzke). As such, McaS is a unique sRNA with two different functions: base pairing and protein titration. Other sRNAs like GadY, Spot 42, GcvB and MicL have been shown to bind CsrA by CLIP-seq analysis. These sRNAs have primary base-pairing functions and hence binding to CsrA could indicate that, like McaS, these sRNAs might also be involved in the titration of CsrA (84).
Another example of a multifunctional sRNA is the Gifsy-1 prophage encoded IsrK sRNA of Salmonella. Encoded on a Salmonella pathogenicity island, the IsrK RNA exists in two isoforms: the transcriptionally inactive isrK-orf45-anrP long mRNA transcript and a shorter IsrK transcript (85) (see chapter by Altuvia, Storz and Papenfort). Normally, the secondary structure in the longer isoform silences translation. However, the short IsrK isoform binds to the translationally inactive long isoform, allowing anrP translation and therefore production of the antirepressor AnrP. As a result, the lysogenic phage repressor is inactivated and expression of the antiterminator protein AntQ increased. This interference with transcription termination ultimately leads to bacterial growth arrest followed by cell death. Thus IsrK exists as two isoforms with separate functions.
Initially, it was assumed that sRNAs are encoded by independent genes, however RNA-seq approaches have revealed that there are large numbers of sRNAs that originate from either the 5’ or 3’ end of various mRNAs. The first evidence of 5’-and 3’-derived sRNAs came from cloning-based searches for sRNA (86, 87), but the first hints that these mRNA fragments could have functions came from studies of the S-adenosyl methionine (SAM) riboswitch of Listeria monocytogenes (88). This riboswitch present in the 5’ UTR of genes involved in methionine and cysteine metabolism regulates expression of the downstream genes by binding to SAM. Binding of SAM to the riboswitch results in premature termination of the transcript leading to repression of the downstream genes. However, this truncated transcript (SreA) can moonlight as an sRNA and regulate the expression of the prfA transcript in trans by base pairing with the 5’ UTR and affecting translation and stability (88). Other characterized 5’ UTR sRNAs include the 5’ UTR of irvA mRNA in Streptococcus mutans, which modulates the synthesis of a critical surface exposed lectin (gbpC) that serves as an adhesin for biofilm development. The irvA 5’ UTR base pairs with gbpC mRNA and prevents its degradation by an RNase J2-mediated pathway (89). Another example is the sRNA derived from the 5’ UTR of the IS200 transposase mRNA (tnpA), which base pairs with and represses the mRNA encoding a transcriptional regulator (invF) required for the expression of several genes encoding type III secretion system SPI1 effector proteins (90).
Currently, there are two classifications of 3’ UTR derived sRNAs: type I in which the sRNA is transcribed from a promoter embedded inside the ORF and type II in which the sRNA is derived from processing of the mRNA. For both types, the sRNAs share the terminator with the parental mRNA (15). E. coli MicL is an example of type I 3’ UTR-derived sRNA, which is transcribed from a promoter located within the coding sequence of the cutC gene and inhibits the synthesis of lipoprotein Lpp, thereby reducing envelope stress (91). Another example of this type of sRNA is DapZ of S. enterica transcribed from the promoter in the 3’ UTR of dapB mRNA and represses the translation of mRNAs of the dpp and opp operons encoding major ABC transporters (10). Two examples of 3’ UTR-derived sRNAs generated by cleavage are the 3’ UTR of S. coelicolor sodF, which represses the expression of sodN mRNA, thereby shutting off the synthesis of Ni-SOD during nickel starvation (92), and the 3’ UTR of S. enterica cpxP, which produces CpxQ sRNA, which represses multiple mRNAs encoding extracytoplasmic proteins to potentially reduce synthesis of problematic proteins to combat envelope stress (93).
It is also worth noting that, like IsrK, a number of sRNAs, particularly those derived from UTRs, have been found to be processed into multiple sizes. It is possible that the different versions act on different sets of mRNAs targets or by different mechanisms (91, 94). Hints of such nuanced regulation are coming from global analyses of sRNA-mRNA pairs where different sets of targets are observed for different forms of a sRNAs (84, 95). In general, the deep sequencing studies highlight how much remains to be learned about different cellular networks and how even the smallest fragments of RNA can have important functions in the cell. Further identification of more multifunctional RNAs and studies into uncovering their roles in the cell will help to further understand the complex regulatory networks controlling cellular physiology (96).
PERSPECTIVES
As we have discussed, there currently are only five dual-function RNAs in bacteria with reported functions. While most sRNAs identified are thought to be noncoding and this is likely in the majority of cases, there are enough examples of sRNA for which ORFs have been noted, that it is prudent to be cautious about calling a sRNA “noncoding.”
A significant challenge is documenting whether ORFs found in sRNAs are in fact translated. The challenge of identifying and detecting small proteins stems from the difficulties in their annotation and biochemical detection due to their small size (97). The short amino acid sequences make the computational identification of coding ORFs difficult due to a lack of sufficient sequence for reliable domain and homology determination, a problem compounded by the possibility of an ORF starting with a non AUG codon and the chances of start and stop codons existing in sRNA sequences by chance (14). Detection of these small proteins is limited by difficulties inherent in using standard proteomic techniques to isolate and identify proteins less than 10 kDa in size. For example, mass-spectrometry based methods to detect small proteins still suffers from false negatives due to proteolytic cleavage of the small proteins into products too small to be detected by mass spectrometry. Despite these challenges, we anticipate that the number of confirmed dual-function sRNAs will grow given the emergence of new techniques such as improved peptidomics and ribosome profiling. When more dual-function RNAs have been found and characterized, a better understanding of the properties of these RNAs likely will facilitate the development of additional tools for identifying and characterizing a putative dual-function RNA.
With the increasing discoveries of dual-function sRNAs, many questions remain. One important issue is the interplay between riboregulation and translation of the small RNA. Further understanding of this interplay undoubtedly will provide vital information regarding expression and function of the small protein. Some of the unique features that have come to light through studies of the few examples of the dual-function sRNAs include instances where the riboregulation function can influence the translation of the small protein. For example, the translation of hld is delayed by one hour after RNAIII transcription potentially because of an inhibitory structure between the 3’ end of RNAIII and the hld RBS (56). As another example, it was noted that SRP1 is not translated under the same conditions that most strongly induce sr1 transcription (65). Further studies regarding the mechanisms by which riboregulation and translation influence each other and identification of other factors involved will shed light on the interplay between the sRNA and protein function. The spatial location of the base-pairing versus the coding region of the sRNA likely is an important factor. It is also possible that other regulatory sRNAs might bind these dual-function sRNAs to influence the translation of the encoded small proteins. Moreover, little is known about the role of RNA chaperones in the interplay between the two different roles of these dual-function sRNAs. Binding of RNA chaperones to these dual-function sRNAs might influence one role versus the other by either sequestering the RNA for its role as a riboregulator or remodeling secondary structures that affect the translation of the ORF. Moreover, there might be an inherent competition between the RNA chaperones and ribosomes for binding to these sRNAs which might influence the roles of these dual-function sRNAs.
While the evolution of sRNAs is beginning to be studied (see Chapter by Dutcher and Raghavan), not much is known about the evolution of these multifunctional sRNAs. For example, did the RNAs first evolve as sRNAs or as mRNAs? SgrS was first thought to be a non-coding regulatory sRNA until the later discovery that it also encodes SgrT. In contrast, RNAIII and Pel/SagA mRNA were first found to be protein coding until the realization that the RNAs also have a regulatory function. Looking at the conservation across different species might give insight into the original function of these dual sRNAs.
Considering synthetic biology applications, dual-function RNAs are good candidates for exploitation. For example, a dual-function RNA that can inhibit both the synthesis and activity of a protein could allow for tightly regulated control of a process, and a dual-function RNA that has both base-pairing and protein-titrating activity can be used two control two different classes of genes. All in all, dual-function RNAs present an opportunity to tightly modulate gene expression using two distinct functions under specific conditions.
Clearly, studies of dual-functional sRNAs have revealed how little we understand bacterial physiology and modes of regulation. Previously thought to be junk, even the smallest RNA fragments might have important functions in the cell. Many questions remain unanswered but likely will be answered with increasing discoveries of new dual-function sRNAs and studies of the their roles and mechanism of regulation. Further breakthroughs in methodology regarding protein study and computational approaches for identification of these sRNAs and small proteins also will aid in expanding this interesting field.
Figure 5:

Pel/sagA sRNA is expressed from the pleiotropic effect locus of S.pyogenes comprising of the sagABCEDEFGHI operon. This 459 nucleotide sRNA also encodes a 53 aa protein called Streptolysin S (purple). Pel sRNA activates transcription of various mRNAs coding for different virulence factors like Sic, Nga and M protein by an unknown mechanism. The sRNA also modulates maturation of cysteine protease SpeB.
ACKNOWLEDGMENTS
M. Raina is supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. C. Bianco and A. King are supported by NIH R01 GM092830.
References
- 1.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobrovskyy M, Vanderpool CK. 2014. The small RNA SgrS: roles in metabolism and pathogenesis of enteric bacteria. Front Cell Infect Microbiol 4:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian D, Vanderpool CK. 2013. Deciphering the interplay between two independent functions of the small RNA regulator SgrS in Salmonella. J Bacteriol 195:4620–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frohlich KS, Papenfort K, Fekete A, Vogel J. 2013. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J 32:2963–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. 2013. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCullen CA, Benhammou JN, Majdalani N, Gottesman S. 2010. Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol 192:5559–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. 2002. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J 21:3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50:1111–1124. [DOI] [PubMed] [Google Scholar]
- 10.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat Rev Microbiol 9:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderpool CK, Balasubramanian D, Lloyd CR. 2011. Dual-function RNA regulators in bacteria. Biochimie 93:1943–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimpel M, Brantl S. 2017. Dual-function small regulatory RNAs in bacteria. Mol Microbiol 103:387–397. [DOI] [PubMed] [Google Scholar]
- 14.Friedman RC, Kalkhof S, Doppelt-Azeroual O, Mueller SA, Chovancova M, von Bergen M, Schwikowski B. 2017. Common and phylogenetically widespread coding for peptides by bacterial small RNAs. BMC Genomics 18:553.28732463 [Google Scholar]
- 15.Miyakoshi M, Chao Y, Vogel J. 2015. Regulatory small RNAs from the 3’ regions of bacterial mRNAs. Curr Opin Microbiol 24:132–139. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. 2013. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27:1132–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lalaouna D, Carrier MC, Masse E. 2015. Every little piece counts: the many faces of tRNA transcripts. Transcription 6:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 104:20454–20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanderpool CK, Gottesman S. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54:1076–1089. [DOI] [PubMed] [Google Scholar]
- 20.Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, Hanada Y, Han X, Kuwahara- Arai K, Hishinuma T, Baba T, Ito T, Hiramatsu K, Sekimizu K. 2011. Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog 7:e1001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. 2004. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol Microbiol 53:1515–1527. [DOI] [PubMed] [Google Scholar]
- 22.Licht A, Preis S, Brantl S. 2005. Implication of CcpN in the regulation of a novel untranslated RnA (SR1) in Bacillus subtilis. Mol Microbiol 58:189–206. [DOI] [PubMed] [Google Scholar]
- 23.Horler RS, Vanderpool CK. 2009. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res 37:5465–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15:1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanderpool CK, Gottesman S. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54:1076–1089. [DOI] [PubMed] [Google Scholar]
- 26.Kimata K, Tanaka Y, Inada T, Aiba H. 2001. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J 20:3587–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem 278:15608–15614. [DOI] [PubMed] [Google Scholar]
- 28.Richards GR, Patel MV, Lloyd CR, Vanderpool CK. 2013. Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J Bacteriol 195:4816–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanderpool CK, Gottesman S. 2007. The novel transcription factor SgrR coordinates the response to glucose-phosphate stress. J Bacteriol 189:2238–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice JB, Vanderpool CK. 2011. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res 39:3806–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamoto H, Koide Y, Morita T, Aiba H. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol 61:1013–1022. [DOI] [PubMed] [Google Scholar]
- 32.Maki K, Uno K, Morita T, Aiba H. 2008. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc Natl Acad Sci U S A 105:10332–10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice JB, Balasubramanian D, Vanderpool CK. 2012. Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proc Natl Acad Sci U S A 109:E2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bobrovskyy M, Vanderpool CK. 2016. Diverse mechanisms of post-transcriptional repression by the small RNA regulator of glucose-phosphate stress. Mol Microbiol 99:254–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 104:20454–20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lloyd CR, Park S, Fei J, Vanderpool CK. 2017. The Small Protein SgrT Controls Transport Activity of the Glucose-Specific Phosphotransferase System. J Bacteriol 199:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jahreis K, Pimentel-Schmitt EF, Bruckner R, Titgemeyer F. 2008. Ins and outs of glucose transport systems in eubacteria. FEMS Microbiol Rev 32:891 –907. [DOI] [PubMed] [Google Scholar]
- 38.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of Staphylococcal Virulence Factors Is Controlled by a Regulatory RNA Molecule. EMBO J 12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vuong C, Gotz F, Otto M. 2000. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect Immun 68:1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. [DOI] [PubMed] [Google Scholar]
- 41.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. [DOI] [PubMed] [Google Scholar]
- 42.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 12:3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. 2000. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA 6:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fechter P, Caldelari I, Lioliou E, Romby P. 2014. Novel aspects of RNA regulation in Staphylococcus aureus. FEBS Lett 588:2523–2529. [DOI] [PubMed] [Google Scholar]
- 45.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, Jacquier A, Vandenesch F, Romby P. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J 24:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21:1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morfeldt E, Taylor D, von Gabain A, Arvidson S. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 14:4569–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Mu C, Ying X, Li W, Wu N, Dong J, Gao Y, Shao N, Fan M, Yang G. 2011. RNAIII activates map expression by forming an RNA-RNA complex in Staphylococcus aureus. FEBS Lett 585:899–905. [DOI] [PubMed] [Google Scholar]
- 49.Gupta RK, Luong TT, Lee CY. 2015. Correction for Gupta et al. , RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc Natl Acad Sci U S A 112:E7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingavale S, van Wamel W, Luong TT, Lee CY, Cheung AL. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect Immun 73:1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trotonda MP, Tamber S, Memmi G, Cheung AL. 2008. MgrA represses biofilm formation in Staphylococcus aureus. Infect Immun 76:5645–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crosby HA, Schlievert PM, Merriman JA, King JM, Salgado-Pabon W, Horswill AR. 2016. The Staphylococcus aureus Global Regulator MgrA Modulates Clumping and Virulence by Controlling Surface Protein Expression. PLoS Pathog 12:e1005604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luong TT, Newell SW, Lee CY. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J Bacteriol 185:3703–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. 2006. Transcription Profiling of the mgrA Regulon in Staphylococcus aureus. J Bacteriol 188:1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verdon J, Girardin N, Lacombe C, Berjeaud JM, Hechard Y. 2009. delta-hemolysin, an update on a membrane-interacting peptide. Peptides 30:817–823. [DOI] [PubMed] [Google Scholar]
- 56.Balaban N, Novick RP. 1995. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3’-end deletion. FEMS Microbiol Lett 133:155–161. [DOI] [PubMed] [Google Scholar]
- 57.Qin L, McCausland JW, Cheung GY, Otto M. 2016. PSM-Mec-A Virulence Determinant that Connects Transcriptional Regulation, Virulence, and Antibiotic Resistance in Staphylococci. Front Microbiol 7:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K. 2013. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog 9:e1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chatterjee SS, Chen L, Joo HS, Cheung GY, Kreiswirth BN, Otto M. 2011. Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant Staphylococcus aureus. PLoS One 6:e28781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, Chen L, Kreiswirth BN, Peschel A, Deleo FR, Otto M. 2009. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog 5:e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med 13:1510–1514. [DOI] [PubMed] [Google Scholar]
- 62.Licht A, Golbik R, Brantl S. 2008. Identification of ligands affecting the activity of the transcriptional repressor CcpN from Bacillus subtilis. J Mol Biol 380:17–30. [DOI] [PubMed] [Google Scholar]
- 63.Licht A, Brantl S. 2006. Transcriptional repressor CcpN from Bacillus subtilis compensates asymmetric contact distribution by cooperative binding. J Mol Biol 364:434–448. [DOI] [PubMed] [Google Scholar]
- 64.Licht A, Brantl S. 2009. The transcriptional repressor CcpN from Bacillus subtilis uses different repression mechanisms at different promoters. J Biol Chem 284:30032–30038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heidrich N, Chinali A, Gerth U, Brantl S. 2006. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol Microbiol 62:520–536. [DOI] [PubMed] [Google Scholar]
- 66.Heidrich N, Moll I, Brantl S. 2007. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res 35:4331–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gimpel M, Preis H, Barth E, Gramzow L, Brantl S. 2012. SR1--a small RNA with two remarkably conserved functions. Nucleic Acids Res 40:11659–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gimpel M, Heidrich N, Mader U, Krugel H, Brantl S. 2010. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol 76:990–1009. [DOI] [PubMed] [Google Scholar]
- 69.Gimpel M, Brantl S. 2016. Dual-function sRNA encoded peptide SR1P modulates moonlighting activity of B. subtilis GapA. RNA Biol 13:916–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Betschel SD, Borgia SM, Barg NL, Low DE, De Azavedo JC. 1998. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Immun 66:1671–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez N, Trevino J, Liu Z, Ho SC, Babitzke P, Sumby P. 2009. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS One 4:e7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, De Azavedo JC. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun 68:4245–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Datta V, Myskowski SM, Kwinn LA, Chiem DN, Varki N, Kansal RG, Kotb M, Nizet V. 2005. Mutational analysis of the group A streptococcal operon encoding streptolysin S and its virulence role in invasive infection. Mol Microbiol 56:681–695. [DOI] [PubMed] [Google Scholar]
- 74.Sonnleitner E, Gonzalez N, Sorger-Domenigg T, Heeb S, Richter AS, Backofen R, Williams P, Huttenhofer A, Haas D, Blasi U. 2011. The small RNA PhrS stimulates synthesis of the Pseudomonas aeruginosa quinolone signal. Mol Microbiol 80:868–885. [DOI] [PubMed] [Google Scholar]
- 75.Shimizu T, Yaguchi H, Ohtani K, Banu S, Hayashi H. 2002. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol Microbiol 43:257–265. [DOI] [PubMed] [Google Scholar]
- 76.Roberts SA, Scott JR. 2007. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol Microbiol 66:1506–1522. [DOI] [PubMed] [Google Scholar]
- 77.Vockenhuber MP, Heueis N, Suess B. 2015. Identification of metE as a second target of the sRNA scr5239 in Streptomyces coelicolor. PLoS One 10:e0120147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vockenhuber MP, Suess B. 2012. Streptomyces coelicolor sRNA scr5239 inhibits agarase expression by direct base pairing to the dagA coding region. Microbiology 158:424–435. [DOI] [PubMed] [Google Scholar]
- 79.Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. 2009. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol 74:1497–1512. [DOI] [PubMed] [Google Scholar]
- 80.Hess WR, Berghoff BA, Wilde A, Steglich C, Klug G. 2014. Riboregulators and the role of Hfq in photosynthetic bacteria. RNA Biol 11:413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muller KM, Berghoff BA, Eisenhardt BD, Remes B, Klug G. 2016. Characteristics of Pos19 - A Small Coding RNA in the Oxidative Stress Response of Rhodobacter sphaeroides. PLoS One 11:e0163425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G. 2013. Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27:1132–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Potts AH, Vakulskas CA, Pannuri A, Yakhnin H, Babitzke P, Romeo T. 2017. Global role of the bacterial post-transcriptional regulator CsrA revealed by integrated transcriptomics. Nat Commun 8:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hershko-Shalev T, Odenheimer-Bergman A, Elgrably-Weiss M, Ben-Zvi T, Govindarajan S, Seri H, Papenfort K, Vogel J, Altuvia S. 2016. Gifsy-1 Prophage IsrK with Dual Function as Small and Messenger RNA Modulates Vital Bacterial Machineries. PLoS Genet 12:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawano M, Reynolds AA, Miranda-Rios J, Storz G. 2005. Detection of 5’- and 3’-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res 33:1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31:6435–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139:770–779. [DOI] [PubMed] [Google Scholar]
- 89.Liu N, Niu G, Xie Z, Chen Z, Itzek A, Kreth J, Gillaspy A, Zeng L, Burne R, Qi F, Merritt J. 2015. The Streptococcus mutans irvA gene encodes a trans-acting riboregulatory mRNA. Mol Cell 57:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ellis MJ, Trussler RS, Charles O, Haniford DB. 2017. A transposon-derived small RNA regulates gene expression in Salmonella Typhimurium. Nucleic Acids Res 45:5470–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. 2014. MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 28:1620–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim HM, Shin JH, Cho YB, Roe JH. 2014. Inverse regulation of Fe- and Ni-containing SOD genes by a Fur family regulator Nur through small RNA processed from 3’UTR of the sodF mRNA. Nucleic Acids Res 42:2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chao Y, Vogel J. 2016. A 3’ UTR-Derived Small RNA Provides the Regulatory Noncoding Arm of the Inner Membrane Stress Response. Mol Cell 61:352–363. [DOI] [PubMed] [Google Scholar]
- 94.Hao Y, Updegrove TB, Livingston NN, Storz G. 2016. Protection against deleterious nitrogen compounds: role of sigmaS-dependent small RNAs encoded adjacent to sdiA. Nucleic Acids Res 44:6935–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melamed S, Peer A, Faigenbaum-Romm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H. 2016. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol Cell 63:884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Papenfort K, Espinosa E, Casadesus J, Vogel J. 2015. Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. Proc Natl Acad Sci U S A 112:E4772–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Storz G, Wolf YI, Ramamurthi KS. 2014. Small proteins can no longer be ignored. Annu Rev Biochem 83:753–777. [DOI] [PMC free article] [PubMed] [Google Scholar]


